Abstract
The role that noncoding regions of the genome play in the etiology of cleft palate is not well studied. A novel method of microRNA (miR) inhibition that allows for specific miR knockdown in vivo has been developed by our laboratory. To further understand the role of miRs in palatogenesis, we used a new mouse model to inhibit specific miRs within the miR-17-92 cluster. Transgenic mice expressing inhibitory complexes for miR-17 and miR-18 manifested a clefting phenotype that was distinct from that observed in mice carrying inhibitory complexes for miR-17, miR-18, miR-19, and miR-92. An in silico candidate gene analysis and bioinformatics review led us to identify TGFBR2 as a likely target of miR-17 and miR-19 family members. Reverse transcription polymerase chain reaction (RT-PCR) experiments showed that TGFBR1 and TGFBR2 expression levels were elevated in the palates of these miR transgenic embryos at embryonic day 15.5. RT-PCR data also showed that the expression of mature miRs from the miR-17-92 cluster was significantly decreased in the transgenic embryos. Decreased expression of TGFB pathway signaling ligands was also observed. Experiments in cells showed that inhibition of miR-17 and miR-18 was sufficient to induce increases in expression of TGFB receptors, while a concomitant decrease in TGFB signaling ligands was not observed. RT-PCR of mature miR-17-92 in cells demonstrated the selectivity and specificity of inhibitory complexes. While this study builds on previous studies that have implicated miR-17-92 in the regulation of important molecular components of the TGFB signaling pathway, it is likely that interactions remain to be elucidated between miR-17-92 and as-of-yet unidentified molecules important for the control of palatogenesis. The differential regulation of palatogenesis by members of the miR-17-92 cluster indicates that several gene combinations regulate palate elevation and extension during development.
Keywords: microRNA, cleft palate, PMIS-miR-17-92, microRNA inhibitors, microRNA development, in vivo microRNAs
Introduction
An expanding body of literature suggests that a variety of genetic factors are involved in the etiology of syndromic and nonsyndromic cleft lip and palate (CL/P). While many protein-coding genes have been investigated in regard to their role in CL/P (Murray 2002; Bush and Jiang 2012; Rahimov et al. 2012), the role that noncoding regions of the genome play in the etiology of CL/P is not as well studied.
MicroRNAs (miRs) are short noncoding RNA molecules approximately 22 nucleotides long. miRs bind to complementary targets on the 3′ untranslated region (UTR) of messenger RNAs (mRNAs), attenuating mRNA translation via either mRNA strand degradation or sequestration (Bartel 2004). Through this mechanism, miRs play a broad role in the regulation of mRNA translation and have been demonstrated to play a significant part in an array of biological processes (Alvarez-Garcia and Miska 2005; Wienholds and Plasterk 2005; Iorio and Croce 2012).
Investigating the role of miRs in these processes is not always straightforward. A number of miRs have been duplicated during evolution and translocated to other regions of the genome (Maher et al. 2006; Yuan et al. 2011). This makes the complete elimination of many miR families difficult with current gene-editing strategies. Complete elimination of a duplicated miR family from the genome would require intensive gene editing and/or lengthy breeding programs to yield the models required to study global miR family elimination or knockdown, particularly in mammalian systems.
To circumvent this difficulty, we recently developed a novel method of miR inhibition, the Plasmid-Based miRNA Inhibition System (PMIS), to allow for the simultaneous knockdown of homologous miR families in vivo (Cao et al. 2016). The PMIS inhibitor complex (PMIS-IC) is composed of native, unmodified nucleic acids and can be integrated into the genome. Importantly, this enables the development of stably expressing cell and animal models that allow for the study of genome-wide miR family inhibition.
Since their initial discovery, miRs have increasingly come to the forefront of biomedical research as important noncoding regulatory elements of protein translation and as important biomarkers or therapeutic targets (Allen and Weiss 2010; Michael et al. 2010; van Rooij and Kauppinen 2014; Rupaimoole and Slack 2017). miRs come in a variety of genomic contexts, both intra- and intergenic (Ballarino et al. 2009; Godnic et al. 2013), and can be isolated or grouped into polycistronic clusters, as is the case with the widely studied miR miR-17-92 cluster (Hayashita et al. 2005; Ventura et al. 2008). miR-17-92 is a cluster of 6 highly conserved miRs from 4 families located on chromosome 13 in humans and chromosome 14 in mice (Concepcion et al. 2012). Recent studies have implicated the miR-17-92 cluster in the development of oro- and craniofacial defects (Wang et al. 2013; Cao et al. 2016).
In this study, we use PMIS-ICs that target mature miRs from the miR-17-92 cluster to analyze the effects of global miR inhibition of miR-17, miR-18, miR-19, and miR-92 family members in transgenic mice and cells. Interestingly, we have discovered that inhibition of miR-17 and miR-18 family members leads to arrest in palate formation prior to palatal shelf elevation, while inhibition of miR-17, miR-18, miR-19, and miR-92 family members leads to arrest at a later stage when palatal shelves have elevated and begun extension. We have gathered evidence to support the hypothesis that the clefting phenotype observed in PMIS mice could be at least partially attributable to aberrations in TGFB signaling. These aberrations are most likely the result of interactions between TGFBR2 and miR-17 and miR-19 family members. To our knowledge, this is the first study showing that differential inhibition of miRs in a single miR cluster can result in varied phenotypes in an animal model, and it is the first example of a group of miRs being directly linked to growth arrest in the palate. These results demonstrate the effectiveness of this novel miR inhibition strategy and shed light on possible new mechanisms of CL/P.
Materials and Methods
Animals
All animals were housed at the University of Iowa in the Office of Animal Resources and were handled in accordance with the principles and procedures of the Guide for the Care and Use of Laboratory Animals. All experimental procedures were approved per the guidelines of the University of Iowa Institutional Animal Care and Use Committee. Mice models for PMIS-miRs have been described (Cao et al. 2016). Embryos were harvested at various time points, and observation of a vaginal plug was counted as embryonic day (E) 0.5. Reverse transcription polymerase chain reaction (RT-PCR) of PMIS-ICs was performed to validate expression, and amplicons were sequenced to verify specificity. PMIS-miR constructs are available at naturemiri.com. This study conformed with ARRIVE (Animal Research: Reporting of In Vivo Experiments) guidelines for preclinical animal studies.
Whole Mount Imaging of Maxilla P0 Mice
P0 (postnatal day 0) pups were euthanized and fixed briefly in 4% paraformaldehyde. The tongue and mandible were removed to obtain an unobstructed view of the ventral maxilla. The maxilla of wild-type and cleft mutants was imaged with a standard overhead dissection microscope.
Hematoxylin and Eosin Staining of E18.5 Embryos
At day E18.5 following observation of a vaginal plug, mice were sacrificed with CO2 euthanasia. Heads were removed, skinned, and tail biopsies taken for genotyping. Heads were immediately submerged in 4% paraformaldehyde for 1 h and placed in 70% ethanol overnight. The following day, heads were dehydrated with a graded ethanol series before being cleared in xylene for 1 h. Cleared heads were placed in liquid paraffin at ~60 °C overnight. Sections between 6 and 8 μm were cut, mounted, and left to dry at 65 °C overnight. Dried slides were cleared and rehydrated before a 5- to 6-min treatment in hematoxylin. This was followed by dehydration and immersion in ethanol-based eosin solution for 45 s. Slides were then dehydrated, cleared, sealed with Cytoseal 60 (ThermoFisher), and dried overnight before imaging.
RNA Isolation and cDNA Synthesis from E15.5 Maxillary Tissue
At day E15.5 following observation of a vaginal plug, female mice were sacrificed with CO2 euthanasia. Maxillary tissues were dissected, tail biopsies taken for genotyping, and the rest of the head was discarded. Tissues were processed by flash freezing in liquid N2, homogenizing tissue, and submerging it in TRIzol Reagent (Invitrogen). RNA was isolated from homogenized tissues with the miRNeasy Mini Kit (Qiagen), and cDNA synthesis was performed with the miScript PCR System (Qiagen). All kit protocols were carried out the per manufacturer’s instructions.
RT-PCR of Murine and HEK-293 cDNAs
One microliter of 1:10-diluted cDNA product was used per 25 μL of reaction. All mRNA values were normalized to beta-actin or GAPDH, and miRNA values were normalized to RNU6B with the ΔΔct method. All mRNA primer sets were validated with melt curves and amplicon sequencing. miScript Primer Assays (Qiagen) were used for the detection of mature miRNA levels. The following qPCR primers were used (“h” and “m” denote human and mouse specific primers, respectively):
hTGFB1-For 5′- CTAATGGTGGAAACCCACAACG-3′
hTGFB1-Rev 5′- TATCGCCAGGAATTGTTGCTG-3′
mTGFB1-For 5′- CTTCAATACGTCAGACATTCGGG-3′
mTGFB1-Rev 5′- GTAACGCCAGGAATTGTTGCTA-3′
TGFB3-For 5′-AAGAAATCCATAAATTCGACATGATC-3′
TGFB3-Rev 5′-CACATTGAAGCGGAAAACCTT-3′
hTGFBR1-For 5′-ACGGCGTTACAGTGTTTCTG-3′
hTGFBR1-Rev 5′-GCACATACAAACGGCCTATCTC-3′
hTGFBR2-For 5′-GTAGCTCTGATGAGTGCAATGAC-3′
hTGFBR2-Rev 5′-CAGATATGGCAACTCCCAGTG-3′
mTGFBR1-For 5′-TCTGCATTGCACTTATGCTGA-3′
mTGFBR1-Rev 5′-AAAGGGCGATCTAGTGATGGA-3′
mTGFBR2-For 5′-GACTGTCCACTTACAAC-3′
mTGFBR2-Rev 5′-GGCAAACCGTCTCCAGAGTA-3′
Lentivirus Production and Transduction
Lentivirus production has been described (Cao et al. 2016). Briefly, a 6-cm dish of HEK 293FT cells (Invitrogen) were transfected with 2.8 μg of psPAX2, 1.9 μg of pMD2.G, and 4.5 μg of miR inhibitor or control plasmid with Fugene HD (Roche). Supernatants were collected and passed through a 0.45-μm filter 28 h after transfection. Virus was added immediately to cells after plating, and cells were cultured for 2 wk, with fresh media being supplied every 2 to 3 d. Puromycin was added for selection of stable PMIS-miR-17-18-expressing HEK-293 cells.
HEK-293 Cell Culture and Harvesting
HEK-293 cells expressing PMIS-IC constructs were cultured in Dulbecco’s Modified Eagle Medium with 10% fetal bovine serum at 37 °C. Cells collected for RT-PCR analysis were plated in 35-mm dishes and harvested approximately 48 h after plating. The media was removed and the cells washed several times with ice-cold 1× phosphate-buffered saline. Cells were then bathed in approximately 750 μL of TRIzol Reagent (Invitrogen) and warmed for 3 to 5 min at 37 °C. TRIzol-cell slurry was homogenized by pipetting before being transferred to 1.5-mL tubes. Cellular homogenate in TRIzol was then flash frozen in liquid N2 and stored at −80 °C before RNA isolation with the miRNeasy Micro Kit (Qiagen).
Micro–computed tomography Imaging and Analyses
Mouse skulls were fixed in 95% ethanol and then scanned with a Siemens Inveon Micro-CT/PET scanner. Settings of 60 kVp and 500 mA with a voxel size of 30 μm were used in the reconstruction. Reconstruction was performed with Oxirx DICOM software (Rosset et al. 2004).
Results
Newborn PMIS-miR-17-18 and PMIS-miR-17-92 Mice Have Cleft Palate
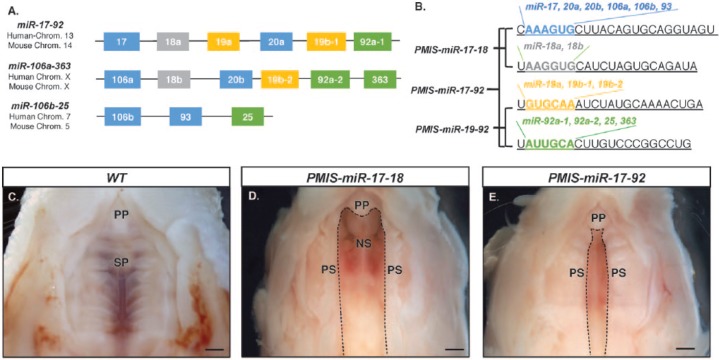
Earlier studies of miR-17-92 function indicated a possible role in craniofacial development (Wang et al. 2013; Cao et al. 2016). We sought to further probe the role of miR-17-92 in palatogenesis by creating transgenic mice with PMIS-ICs targeting the miR-17-92 cluster. One line was generated with PMIS-ICs targeting miR-17 and miR-18 family members (PMIS-miR-17-18) and another with PMIS-ICs targeting miR-19 and miR-92 family members (PMIS-miR-19-92; Fig. 1A, B). Crossing these lines yielded PMIS-miR-17-92 mice carrying PMIS-ICs for all miR-17-92 family members (Fig. 1B). Postnatal lethality was observed in several offspring from PMIS-miR-17-92 breeding pairs. Postmortem dissection of deceased mice (P0) confirmed the probable cause of death was due to complications from cleft palate. The incidence of clefting in embryos was higher, as many embryos do not survive past E16.5. Wild-type littermates (P0) with fully developed palates were used in our comparison (Fig. 1C). This examination led to the identification of 2 distinct clefting phenotypes. In PMIS-miR-17-18 mice, palatal growth arrest appeared to occur at a different stage in development, as the cleft was larger in these mice (Fig. 1D). Palatal growth arrest in PMIS-miR-17-92 mice appeared to occur after elevation of the palatal shelves, as the width of the cleft is smaller versus that of PMIS-miR-17-18 mice (Fig. 1E). The 2 observed phenotypes were consistent across all newborn mice with clefts that were examined. No evidence for clefting in PMIS-miR-19-92 mice was found.
Figure 1.
PMIS-miR-17-18 and PMIS-miR-17-92 mice are postnatal lethal with distinct clefting phenotypes. (A) The location and organization of the homologous miR-17-92 clusters. (B) The seed sequence similarities (color coded) and differences among the microRNA (miRs). PMIS-miR-17-18 were derived from miR-17-5p and miR-18a-5p. PMIS-miR-17-92 were derived from all 4 clusters (Cao et al. 2016). (C–E) Whole mount view of the ventral maxilla in P0 mice shows the different stages of palatogenesis arrest in the transgenic mice. Dashed lines outline the cleft region, if present. (C) Wild-type (WT) mouse shows complete fusion of the palatal shelves at the primary and secondary palates. (D) In the PMIS-miR-17-18 mice, the palate shelves fail to elevate, leaving large clefts in the palate. (E) In the PMIS-miR-17-92 mice, the palate shelves elevate but fail to extend to the midline and fuse. NS, nasal septum; PP, primary palate; PS, palatal shelves; SP, secondary palate.
PMIS-miR-17-18 and PMIS-miR-17-92 Palatogenesis Is Arrested at Distinct Stages
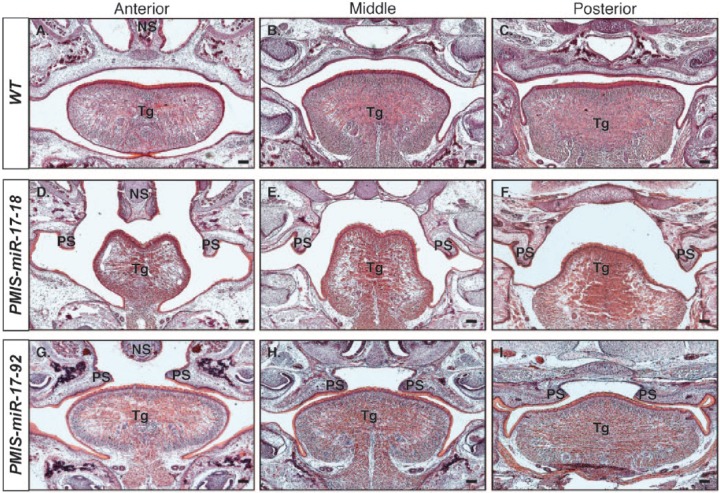
Histologic analysis of E18.5-stage embryos allowed for a more detailed view of the clefts observed in P0 mice (Fig. 2). Wild-type mice had a fully fused palate along the entire anteroposterior axis (Fig. 2A–C). Palatal shelves in PMIS-miR-17-18 mice showed no signs of elevation or extension (Fig. 2D–F). PMIS-miR-17-18 mice also had a dysmorphic tongue with stunted lateral growth. miR-17 and miR-18 appear to regulate growth of the transverse and vertical muscle group and genioglossus muscles of the tongue, which are associated with lateral growth and muscle fiber size (Sanders et al. 2013). PMIS-miR-17-92 mice had fully elevated palatal shelves that appeared to have undergone extension toward the midline, but extension was arrested before the shelves could fuse (Fig. 2G–I). Comparison of the nature of the 2 clefting phenotypes to normal palatogenesis in mice (Bush and Jiang 2012) led us to conclude that palatal growth arrest in PMIS-miR-17-18 and PMIS-miR-17-92 mice likely occurred around E13.5 and E14.5, respectively.
Figure 2.
Histologic analyses show distinct patterns of clefting at embryonic day 18.5 in PMIS-miR-17-18 and PMIS-miR-17-92 embryos. Hematoxylin and eosin staining of embryonic day 18.5 embryos shows differences in cleft palate defects observed in PMIS-miR-17-18 and PMIS-miR-17-92 embryos. (A–C) Anterior, middle, and posterior coronal sections of wild-type (WT) embryos show complete fusion of the palate. (D–F) Coronal sections of the PMIS-miR-17-18 embryos shows that arrest in palatogenesis occurs before elevation of the palatal shelves. (G–I) Coronal sections of PMIS-miR-17-92 embryos shows that arrest in palatogenesis occurs after elevation of the palate shelves but before extension and fusion at the midline. Scale bar = 100 μm. NS, nasal septum; PS, palatal shelves; Tg, tongue.
PMIS-miR-17-18, PMIS-miR-19-92, and PMIS-miR-17-92 Mice without Cleft Palate Have Craniofacial Defects
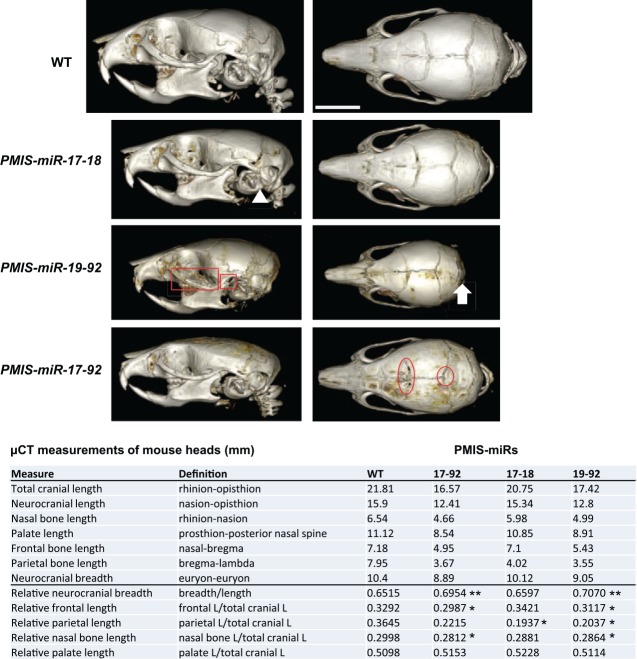
Several PMIS-miR-17-18, PMIS-miR-19-92, and PMIS-miR-17-92 mice survived past P0 to 1 to 2 mo of age. High-resolution x-ray microtomograph scans of the PMIS transgenic mice at 3 wk of age identified several craniofacial defects. We have identified suture defects, occipital bone (supraoccipital, exoccipital, and basioccipital) and interparietal defects, zygomatic arch and tympanic bulla defects, jaw length defects and apparent mandibular condyle growth defects, and microcephaly. There are differences in the cranial length and breadth in all PMIS mice as compared with wild type (Fig. 3). Quantitative measurements of total cranial length show 25% reduction in PMIS-miR-17-92 mice versus wild type and 6% reduction in the PMIS-miR-17-18 mice. Palate and frontal bone lengths are reduced 32% in PMIS-miR-17-92 and 10% in the PMIS-miR-17-18 mice when compared with wild type.
Figure 3.
MicroRNAs (miRs) within the miR-17-92 cluster differentially regulate craniofacial development. Wild-type (WT), PMIS-miR-17-18, PMIS-miR-19-92, and PMIS-miR-17-92 3-wk-old heads were analyzed by micro–computed tomography. In-depth measurements were obtained for different aspects of craniofacial growth. Quantitative measurements of total cranial length and breadth of the Plasmid-Based miRNA Inhibition System (PMIS) transgenic mice are shown as compared with WT (n = 3). The structures denoted in the figure are as follows: zygomatic arch (red rectangle), condyle (red square), tympana bulba (white triangle), occipital region (white arrow), cranial coronal suture (red oval), interparietal region (red circle). P > 0.05, n.s. *P < 0.05. **P < 0.01.
Identification of Candidate Genes Responsible for Clefting in PMIS-miR Mice
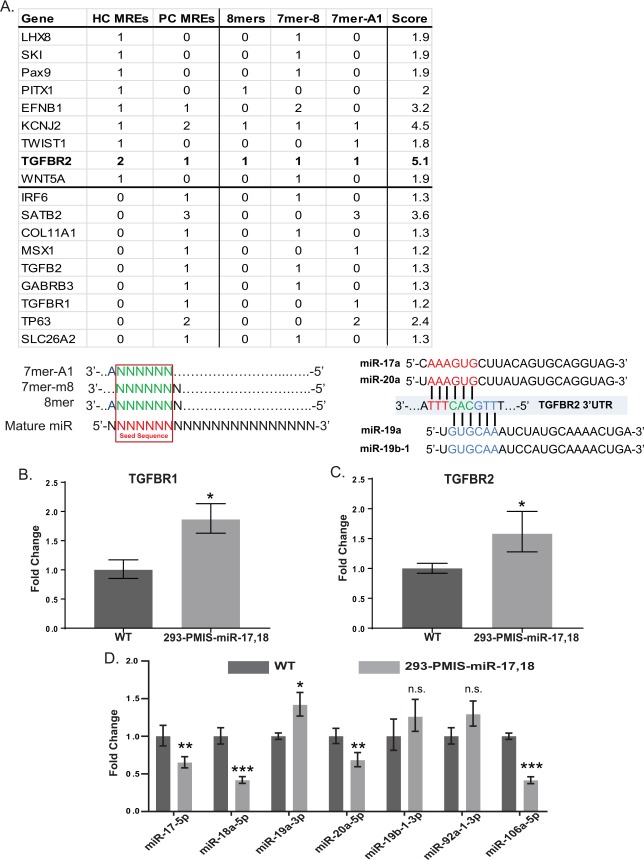
We used a tandem approach involving in silico analysis with TargetScan 7.1 (Agarwal et al. 2015) and a search of existing review literature (Murray 2002; Murray and Schutte 2004; Rahimov et al. 2012) to develop a list of candidate genes that could plausibly be affected by the inhibition of miR-17-92. We evaluated each of these candidates based on the presence of miR recognition elements (MREs) that matched canonically with predicted target sequences for miR-17-92. We used a scoring system to assign greater weights to highly conserved MREs relative to poorly conserved MREs and further refined these scores based on the type of MRE interaction that was predicted (Fig. 4A, lower panel). Scores were then assigned to each candidate gene that had at least 1 MRE identified in its 3′UTR by TargetScan. Based on these metrics, a higher score indicates a greater number of MREs, more conserved MREs, and a higher probability of endogenous interactions in cells where the gene and our miRs of interest are coexpressed. The results of this analysis are summarized in the upper panel of Figure 4A. The highest-scoring genes identified were KCNJ2, EFNB1, SATB2, and TGFBR2. TGFBR2 arose as the most viable candidate gene owing to the presence of highly conserved MREs for miR-17 and miR-19 family members, a unique feature among all candidate genes examined. Mature miR sequences for miR-17-92 and the site of the MREs for miR-17 and miR-19 family members on the TGFBR2 3′UTR are identical in humans and mice and highly conserved in vertebrates. Experiments were then performed to determine if the inhibition of miR-17-92 in PMIS mice could have an effect on the expression of genes important for TGFB signaling in the palate.
Figure 4.
Clefting candidate gene analyses for miR-17-92 family members based on miR recognition elements (MREs). (A) Candidate genes for the clefting phenotype observed in PMIS-miR-17-18 and PMIS-miR-17-92 embryos. Only candidate genes that were found to have an MRE corresponding to members of the miR-17-92 cluster are shown in the table. The top portion of the table displays genes that were found to have highly conserved MREs according to the TargetScan algorithm, while the bottom portion includes genes that have any MRE. Column 1 (Gene): Gene symbol for candidate gene. Column 2 (HC MREs): Number of highly conserved MREs. Column 3 (PC MREs): Number of poorly conserved MREs. Column 4 (8mers): Number of 8mer MREs. Column 5 (7mer-8): Number of 7mer-8 MREs. Column 6 (7mer-A1): Number of 7mer-A1 MREs. Column 7 (Score): Total point value for each gene based on columns 2–6 (HC MRE = 1, PC MRE = 0.4, 8mer = 1, 7mer-8 = 0.9, 7mer-A1 = 0.8). TGFBR2, the highest-scoring candidate gene and the subject of further analysis, is in bold. Binding schematic for the MREs considered in the analysis is depicted in the lower left panel, and specific predicted interactions between TGFBR2 and miR-17 and miR-19 family members are depicted in the lower right panel. (B) TGFB receptor expression is elevated in 293 cells that stably express PMIS (Plasmid-Based miRNA Inhibition System) inhibitor complexes specific for miR-17 and miR-18 family members. Levels of TGFBR1 are elevated in PMIS-miR-17-18 expressing 293 cells relative to vector-only controls. (C) Levels of TGFBR2 are elevated in PMIS-miR-17-18 expressing 293 cells relative to vector-only controls. (D) Levels of mature miR-17 and miR-18 family members are reduced in PMIS-miR-17-18 293 cells, while levels for miR-19 and miR-92 family members are unaffected. P > 0.05, n.s. *P < 0.05. **P < 0.01. ***P < 0.001. Values are presented as fold-change ± SE.
To validate these findings, a cellular model that stably expressed PMIS-ICs was developed with HEK-293 cells. We reasoned that since TGFBR2 has MREs for miR-17 and miR-19 family members, expression of a single PMIS-IC targeting either miR-17 or miR-19 should be sufficient to alter TGFB signaling. The TGFB receptor levels were elevated (Fig. 4B, C). However, we did not detect differences in TGFB signaling ligands, with TGFB3 being barely detectable (data not shown). We reasoned that signaling context could be more important in palatal cells undergoing rapid growth and differentiation than in a cellular monolayer, in addition to the possibility of other genetic factors driving ligand transcription that are less active in the in vitro model. The specificity of the PMIS-ICs targeting miR-17 and miR-18 was validated, with miR-17 and miR-18 mature miR family members showing reduced expression and miR-19 and miR-92 mature miR levels remaining relatively unchanged (Fig. 4D). This showed that PMIS-ICs expressed in cells were highly specific for their intended miR family targets and that inhibition of miR-17 and/or miR-18 is a likely disruptor of TGFB signaling via altered interactions with TGFBR2 mRNA.
TGFB Signaling Is Altered in PMIS-miR-17-92 Embryonic Palates
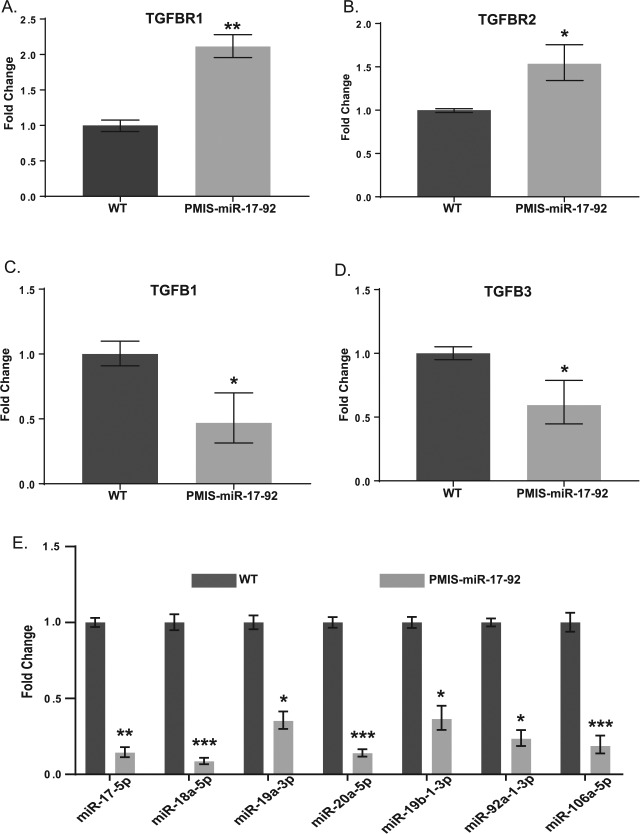
RNA was collected from the maxilla of E15.5 littermates and TGFB receptor, and ligand expression profiles in wild-type and PMIS-miR-17-92 mice were compared with RT-PCR. This showed that both TGFBR2 and TGFBR1 mRNA levels were elevated in PMIS-miR-17-92 mice (Fig. 5A, B). Reduced expression of signaling ligands TGFB1 and TGFB3 were also detected (Fig. 5C–D). Mature miRs from miR-17-92 were reduced relative to wild type, validating the effectiveness of the PMIS-ICs as in vivo inhibitors of miR activity (Fig. 5E). Levels of miR-106a, a miR-17 family member not located within the miR-17-92 cluster, were also reduced, demonstrating the effectiveness of PMIS-ICs in targeting miR-17-92 homologs elsewhere in the genome. From these data, we concluded that our strategy for inhibiting miR-17-92 was effective and that this inhibition had likely effects on TGFB signaling via altered interactions between miR-17 and miR-19 with highly conserved MREs located on TGFBR2.
Figure 5.
Inhibition of miR-17-92 family miRs results in aberrations in TGFB signaling in the maxilla of embryonic day 15.5 PMIS-miR-17-92 mice. (A) Levels of TGFBR1 are elevated in Plasmid-Based miRNA Inhibition System (PMIS) mice relative to wild type (WT). (B) Levels of TGFBR2 are elevated in PMIS mice relative to WT. (C) Levels of TGFB1 are reduced in PMIS mice relative to WT. (D) Levels of TGFB3 are reduced in PMIS mice relative to WT. (E) Levels of mature miR-17, miR-18, miR-19, and miR-92 family members are reduced in PMIS-miR-17-92 mice relative to WT. P > 0.05, n.s. *P < 0.05. **P < 0.01. ***P < 0.001. Values are presented as fold-change ± SE.
Discussion
While the groundwork for establishing a definitive link between inhibition or absence of miR-17-92 and CL/P was laid in earlier studies (Wang et al. 2013; Cao et al. 2016), in the present work we demonstrate a phenotypic difference that suggests different roles for individual miRs within miR-17-92 in the context of palatogenesis. This result further emphasizes the need to develop improved methods to study individual miRs, especially within in vivo models where effects on tissue- and organ-scale developmental processes can be fully appreciated and investigated. A challenge to miR researchers, particularly those working on redundant miRs such as those in the miR-17-92 cluster, is to develop robust model systems in which the effects of inhibiting large miR families can be thoroughly studied. With current gene-editing technologies, developing animal models where large miR families are individually knocked out of the genome is impractical. The approach taken in this study is a step toward further understanding individual miR family functions while circumventing the problems associated with generating a conventional knockout model of redundant miR families. While miRs are generally thought of as broad modulators of biological processes, this study demonstrates that they can have substantial effects on an organismal level.
Given the substantial number of miR-17-92 target genes, the possibility seems remote that the observed phenotypes are solely the result of alterations in TGFB signaling and TGFB receptor expression. The candidate gene search was limited by the effectiveness of the algorithm used to generate probable MRE matches and the precision with which 3′UTRs for our genes of interest is annotated. For example, it has been shown that Tbx1, a gene implicated in CL/P, is a direct target of the miR-17 family (Wang et al. 2010), but the 3′UTR for Tbx1 on TargetScan does not include this validated MRE due to differences in annotation. As genomic annotation increases in quality and the algorithms for identifying probable miRs and MREs progress, these issues should be less of a concern. For the time being, however, they are limitations about which investigators must remain aware. Furthermore, we have shown that the PMIS is very specific for each miR, and we have shown that the PMIS-miR-17-92 inhibitor in cells and mice has no off-target effects by functionally testing many other closely related miRs and targets (Cao et al. 2016). The specificity of the PMIS was shown to be sensitive (loss of inhibition) to 1 nucleotide change in the seed region of specific miR inhibitors.
While our data indicate that inhibition of miR-17-92 results in increased expression of TGFB receptors generally and decreased TGFB ligand expression in the palate, the role of other factors that may contribute to the distinct palatal growth arrest phenotypes in PMIS-miR-17-18 and PMIS-miR-17-92 mice remains elusive. The role of TGFBR2 in CL/P has been established in a conditional knockout model (Ito et al. 2003), as has the role of TGFB ligands (Proetzel et al. 1995; Murray and Schutte 2004). Previous studies of miR-17-92 inhibition in mouse PMCs showed that alterations in TGFBR2 and SMAD2/4 expression were possible via inhibition of miR-17 and miR-18a, respectively (Li et al. 2012). While we did not detect substantive differences in SMAD2/4 levels (data not shown), this may be attributable to the developmental time point that we chose for our analysis or the presence of nonmesenchymal tissues in our palatal tissue preparations. Determining to what extent the interplay among miR-17-92 family members plays a role in the translational repression of a variety of TGFB signaling pathway proteins could contribute further to determining the precise molecular basis for the distinct clefting phenotypes of PMIS-miR-17-18 and PMIS-miR-17-92 mice. Nonetheless, the finding that differential inhibition of miR families encoded in a single miR cluster can result in arrest of growth at different stages of palatogenesis may provide further clues into the etiologic basis of clefting.
Interestingly, it has been reported that in patients with Loeys-Dietz syndrome, a disorder linked to heterozygous loss of function mutations in TGFBR2 or TGFBR1, upregulation of proteins downstream of TGFBR2 and TGFBR1 in the signaling pathway and increased levels of pSMADs are observed (Loeys et al. 2005). While the mechanism through which this occurs is unclear, it is conceivable that canonical TGFB signaling through the TGFBR1-TGFBR2 axis is responsible for transcriptional control of regulatory factors that attenuate certain components of the intracellular TGFB signaling pathway. Somewhat counterintuitively, disrupting the signaling axis with nonfunctional TGFBR1 or TGFBR2 appears to cause increases in intracellular TGFB signaling even in the absence of extracellular signals. We speculate from this that TGFB signaling may exist in a delicate balance in the context of palatogenesis and that perturbations in signaling in either a positive or negative direction may have deleterious effects on this finely tuned developmental process.
Author Contributions
R.J. Ries, B.A. Amendt, contributed to design and analysis, drafted and critically revised the manuscript; W. Yu, N. Holton, H. Cao, contributed to design and analysis, critically revised the manuscript. All authors gave final approval and agree to be accountable for all aspects of the work.
Acknowledgments
We thank Dr. William Shalot for technical expertise with pronuclear injections. We thank the Carver Trust for supporting the high-resolution x-ray microtomograph. We thank all members of the Amendt Lab for helpful comments and suggestions.
Footnotes
This work was supported by funds from the University of Iowa Carver College of Medicine and College of Dentistry and the National Institutes of Health (grant DE13941 to B.A. Amendt).
The authors declare no potential conflicts of interest with respect to the authorship and/or publication of this article.
References
- Agarwal V, Bell GW, Nam JW, Bartel DP. 2015. Predicting effective microRNA target sites in mammalian mRNAs. Elife. 4. doi: 10.7554/eLife.05005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen KE, Weiss GJ. 2010. Resistance may not be futile: microRNA biomarkers for chemoresistance and potential therapeutics. Mol Cancer Ther. 9(12):3126–3136. [DOI] [PubMed] [Google Scholar]
- Alvarez-Garcia I, Miska EA. 2005. MicroRNA functions in animal development and human disease. Development. 132(21):4653–4662. [DOI] [PubMed] [Google Scholar]
- Ballarino M, Pagano F, Girardi E, Morlando M, Cacchiarelli D, Marchioni M, Proudfoot NJ, Bozzoni I. 2009. Coupled RNA processing and transcription of intergenic primary microRNAs. Mol Cell Biol. 29(20):5632–5638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel DP. 2004. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 116(2):281–297. [DOI] [PubMed] [Google Scholar]
- Bush JO, Jiang R. 2012. Palatogenesis: morphogenetic and molecular mechanisms of secondary palate development. Development. 139(2):231–243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao H, Yu W, Li X, Wang J, Gao S, Holton NE, Eliason S, Sharp T, Amendt BA. 2016. A new plasmid-based microRNA inhibitor system that inhibits microRNA families in transgenic mice and cells: a potential new therapeutic reagent. Gene Ther. 23(6):527–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Concepcion CP, Bonetti C, Ventura A. 2012. The miR-17-92 family of microRNA clusters in development and disease. Cancer J. 18(3):262–267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godnic I, Zorc M, Jevsinek Skok D, Calin GA, Horvat S, Dovc P, Kovac M, Kunej T. 2013. Genome-wide and species-wide in silico screening for intragenic microRNAs in human, mouse and chicken. PLoS One. 8(6):e65165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashita Y, Osada H, Tatematsu Y, Yamada H, Yanagisawa K, Tomida S, Yatabe Y, Kawahara K, Sekido Y, Takahashi T. 2005. A polycistronic microRNA cluster, mir-17-92, is overexpressed in human lung cancers and enhances cell proliferation. Cancer Res. 65(21):9628–9632. [DOI] [PubMed] [Google Scholar]
- Iorio MV, Croce CM. 2012. MicroRNA dysregulation in cancer: diagnostics, monitoring and therapeutics. A comprehensive review. EMBO Mol Med. 4(3):143–159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito Y, Yeo JY, Chytil A, Han J, Bringas P, Nakajima A, Shuler CF, Moses HL, Chai Y. 2003. Conditional inactivation of Tgfbr2 in cranial neural crest causes cleft palate and calvaria defects. Development. 130(21):5269–5280. [DOI] [PubMed] [Google Scholar]
- Li L, Shi JY, Zhu GQ, Shi B. 2012. Mir-17-92 cluster regulates cell proliferation and collagen synthesis by targeting TGFB pathway in mouse palatal mesenchymal cells. J Cell Biochem. 113(4):1235–1244. [DOI] [PubMed] [Google Scholar]
- Loeys BL, Chen J, Neptune ER, Judge DP, Podowski M, Holm T, Meyers J, Leitch CC, Katsanis N, Sharifi N, et al. 2005. A syndrome of altered cardiovascular, craniofacial, neurocognitive and skeletal development caused by mutations in TGFBR1 or TGFBR2. Nat Genet. 37(3):275–281. [DOI] [PubMed] [Google Scholar]
- Maher C, Stein L, Ware D. 2006. Evolution of arabidopsis microRNA families through duplication events. Genome Res. 16(4):510–519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michael A, Bajracharya SD, Yuen PS, Zhou H, Star RA, Illei GG, Alevizos I. 2010. Exosomes from human saliva as a source of microRNA biomarkers. Oral Dis. 16(1):34–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray JC. 2002. Gene/environment causes of cleft lip and/or palate. Clin Genet. 61(4):248–256. [DOI] [PubMed] [Google Scholar]
- Murray JC, Schutte BC. 2004. Cleft palate: players, pathways, and pursuits. J Clin Invest. 113(12):1676–1678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proetzel G, Pawlowski SA, Wiles MV, Yin M, Boivin GP, Howles PN, Ding J, Ferguson MW, Doetschman T. 1995. Transforming growth factor-[beta]3 is required for secondary palate fusion. Nat Genet. 11(4):409–414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahimov F, Jugessur A, Murray JC. 2012. Genetics of nonsyndromic orofacial clefts. Cleft Palate Craniofac J. 49(1):73–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosset A, Spadola L, Ratib O. 2004. OsiriX: an open-source software for navigating in multidimensional DICOM images. J Digit Imaging. 17(3):205–216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rupaimoole R, Slack FJ. 2017. MicroRNA therapeutics: towards a new era for the management of cancer and other diseases. Nat Rev Drug Discov. 16(3):203–222. [DOI] [PubMed] [Google Scholar]
- Sanders I, Mu L, Amirali A, Su H, Sobotka S. 2013. The human tongue slows down to speak: muscle fibers of the human tongue. Anat Rec (Hoboken). 296(10):1615–1627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Rooij E, Kauppinen S. 2014. Development of microRNA therapeutics is coming of age. EMBO Mol Med. 6(7):851–864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ventura A, Young AG, Winslow MM, Lintault L, Meissner A, Erkeland SJ, Newman J, Bronson RT, Crowley D, Stone JR, et al. 2008. Targeted deletion reveals essential and overlapping functions of the miR-17∼92 family of miRNA clusters. Cell. 132(5):875–886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Bai Y, Li H, Greene SB, Klysik E, Yu W, Schwartz RJ, Williams TJ, Martin JF. 2013. MicroRNA-17-92, a direct Ap-2α transcriptional target, modulates T-box factor activity in orofacial clefting. PLoS Genetics. 9(9):e1003785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Greene SB, Bonilla-Claudio M, Tao Y, Zhang J, Bai Y, Huang Z, Black BL, Wang F, Martin JF. 2010. Bmp signaling regulates myocardial differentiation from cardiac progenitors through a microRNA-mediated mechanism. Dev Cell. 19(6):903–912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wienholds E, Plasterk RH. 2005. MicroRNA function in animal development. FEBS Lett. 579(26):5911–5922. [DOI] [PubMed] [Google Scholar]
- Yuan Z, Sun X, Liu H, Xie J. 2011. MicroRNA genes derived from repetitive elements and expanded by segmental duplication events in mammalian genomes. PLoS One. 6(3):e17666. [DOI] [PMC free article] [PubMed] [Google Scholar]