Abstract
Tooth is made of an enamel-covered crown and a cementum-covered root. Studies on crown dentin formation have been a major focus in tooth development for several decades. Interestingly, the population prevalence for genetic short root anomaly (SRA) with no apparent defects in crown is close to 1.3%. Furthermore, people with SRA itself are predisposed to root resorption during orthodontic treatment. The discovery of the unique role of Nfic (nuclear factor I C; a transcriptional factor) in controlling root but not crown dentin formation points to a new concept: tooth crown and root have different control mechanisms. Further genetic mechanism studies have identified more key molecules (including Osterix, β-catenin, and sonic hedgehog) that play a critical role in root formation. Extensive studies have also revealed the critical role of Hertwig’s epithelial root sheath in tooth root formation. In addition, Wnt10a has recently been found to be linked to multirooted tooth furcation formation. These exciting findings not only fill the critical gaps in our understanding about tooth root formation but will aid future research regarding the identifying factors controlling tooth root size and the generation of a whole “bio-tooth” for therapeutic purposes. This review starts with human SRA and mainly focuses on recent progress on the roles of NFIC-dependent and NFIC-independent signaling pathways in tooth root formation. Finally, this review includes a list of the various Cre transgenic mouse lines used to achieve tooth root formation–related gene deletion or overexpression, as well as strengths and limitations of each line.
Keywords: dentin, odontogenesis, cell signaling, tooth regeneration, NFIC, osterix
Introduction
Tooth is composed of 2 major components: the enamel-covered crown and the cementum-covered root. There has been substantial progress in understanding crown formation and its regeneration since the molecular biology era began (Lan et al. 2014), while we know relatively little about root formation. Because there is only 1 type of odontoblast, it would be very hard to reason that odontoblasts would behave differently in crown and root. Currently, the interaction of the epithelial and mesenchymal cells is considered responsible for the initiation of root dentin formation (Nanci 2007). Based on this theory, epithelial cells from the cervical loop proliferate to form 2 layers of Hertwig’s epithelial root sheath (HERS), which then induces the adjacent dental mesenchymal cells to differentiate into odontoblasts for dentin formation (Nanci 2007; Fig. 1).
Figure 1.
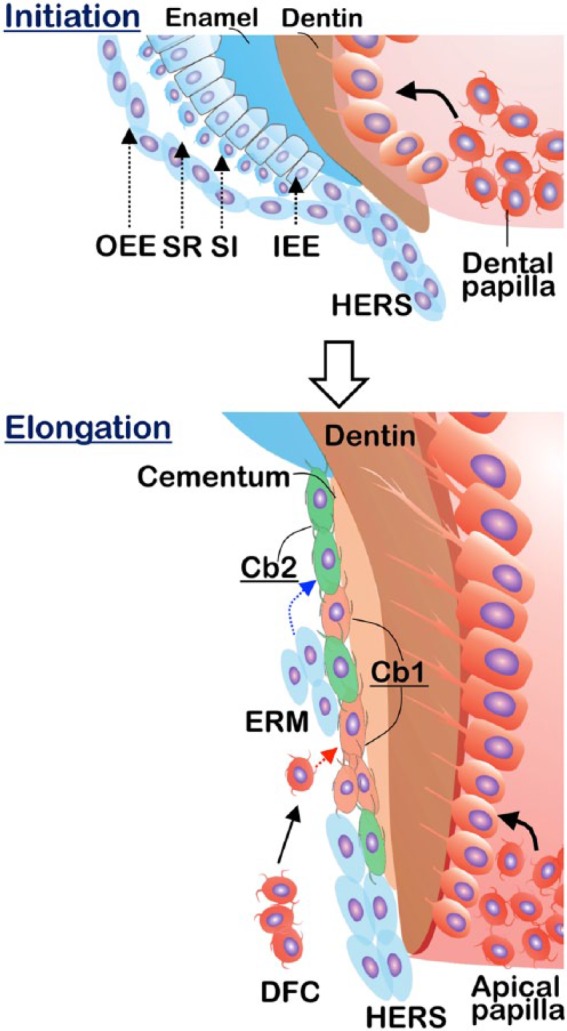
Diagram showing root morphogenesis in 2 stages: root initiation and root elongation. HERS is formed by fusion of outer enamel epithelium (OEE) and inner enamel epithelium (IEE), which marks the initiation of root formation. The odontoblasts derived from the dental papilla (apical papilla at root elongation stage) form dentin, whereas there are 2 stem cell niches giving rise to cementoblasts (Cb), including the dental follicle (dashed red arrow) and HERS (dashed blue arrow). Some HERS cells eventually become epithelial rests of Malassez (ERM). DFC, dental follicle cell; HERS, Hertwig’s epithelial root sheath; SI, stratum intermedium; SR, stellate reticulum.
Importantly, there are numerous hereditary syndromes and chromosomal anomalies that mainly affect the root structure. Technically, tooth crown regeneration does not appear to be an issue, but it is a great challenge to regrow roots for treatment of dental trauma, tooth agenesis, and periodontal diseases. In the recent decade, root research has gained substantial attention from different researchers in genetics and developmental biology. As a result, there has been impressive progress in this area, which has been extensively covered by several researchers in excellent review articles (Kumakami-Sakano et al. 2014; Luder 2015; Li et al. 2017). In the present short review, we focus on short root anomaly (SRA; the clinic relevance of the tooth root study) and roles of NFIC-dependent and NFIC-independent signaling pathways in tooth root formation.
Short Root Anomaly
The term “short root anomaly” (also called “root dwarfism”) was initially described by Lind (1972) based on radiographic measurements of the ratio of the crown versus the root in 112 children with abnormally short roots of the maxillary central incisors as compared with 100 children with normally developed roots. Currently, SRA is defined as a developmental anomaly characterized by full but short root formation with a normal crown. The diagnosis is commonly made when such a short root case appears in some family members but no other known cause is found. The etiology of SRA is largely unknown, although a familial clustering is linked to this syndrome (Lind 1972). The prevalence of SRA appears variable in different populations and ranges from 0.6% to 2.4% (Valladares Neto et al. 2013), but a Japanese research group reported a much higher population affected (Ando et al. 1976). Interestingly, all case reports of SRA are linked to permanent dentition with no such a case in primary dentition (Newman 1975).
SRA can be associated with systemic changes or syndromes (Baccetti 1998). For example, Tananuvat et al. (2014) recently identified a homozygous gene mutation in plasminogen (c.1193G>A missense mutation, leading to type I plasminogen deficiency) from a Thai girl. This affected girl displayed tapered incisor roots and thin root dentin with generalized short tooth roots and mandibular prognathism (i.e., the lower incisors and the upper incisors overlapped). The immunohistochemistry stain showed a higher level of plasminogen in the early root development versus its expression in the mouse crown odontoblast layer. Given these data, the authors proposed a specific role of plasminogen in root dentin formation. However, the plasminogen-deficient mouse model did not show teeth abnormalities but severe periodontitis, suggesting that root dentin anomaly seems not to be a causal phenotype of plasminogen deficiency and that plasminogen is not a true candidate gene for SRA (Kurtulus-Waschulewski et al. 2015).
Generally, SRA has little impact on children’s health except for a high risk of root resorption that occurs in children undergoing orthodontic treatment, which requires more careful biomechanical adaptations, periodic radiographic monitoring, clinical monitoring of tooth mobility, and permanent retention, especially for the incisors. However, orthodontic treatment is contraindicated in only extreme cases (Valladares Neto et al. 2013).
Discovery of the Specific Role of Nfic in Root Formation Points to a Different Control Mechanism in Root Formation from Crown Formation
For many years, dentin was considered 1 component in the entire tooth until the discovery of Nfic, the master gene that specifically controls tooth root rather than crown (Steele-Perkins et al. 2003; Park et al. 2007; Lee et al. 2009), which suggests a unique regulation mechanism in root formation.
NFIC belongs to the nuclear factor I family of transcription proteins, which include 3 additional members: NFIA, NFIB, and NFIX. All of these nucleus proteins bind to the consensus DNA sequence with similar affinities (Gronostajski 2000). However, each member has unique targeted tissues. For example, NFIA mainly controls brain development (das Neves et al. 1999); NFIB regulates brain and lung development (Steele-Perkins et al. 2005); and NFIX defines neural stem cell lineage (Zhou et al. 2015) and regulates neural stem cell quiescence (Martynoga et al. 2013).
NFIC is expressed in odontoblasts of crown and root, but the deletion of Nfic leads to short tooth roots with no apparent changes in molar crowns (Steele-Perkins et al. 2003). This unique root phenotype suggests a different regulation mechanism, which is distinguished from the crown. Although HERS plays an essential role in tooth root formation (detailed later), there is no gross defect in HERS in the Nfic knockout (KO) mice (Park et al. 2007). To better understand the mechanism by which the abnormal molar root occurs, Lee et al. (2009) used the Nfic KO incisor as a model to investigate the detailed changes in the cellular structure and molecular mechanisms. This group found that removing Nfic results in a great change in cell cycles, including a decrease in cell proliferation and an increase in apoptosis, which interfere with the formation of intercellular junctions and cellular polarity of the odontoblast cells. In addition, there was an increase in p-Smad2/3 and BSP expressions in the Nfic-null incisor pulp cells. In contrast, a recent report suggested that a lack of root furcation between the mesial and distal roots in the Nfic-null molar is due to an increase in cell proliferation and a reduction in cell differentiation (Kim, Bae, Yang, et al. 2015).
In fact, the role of NFIC is not restricted only in tooth root formation. An important study showed that NFIC is expressed in bone cells and that the disruption of Nfic greatly reduces osteoblast differentiation and bone formation and increases bone marrow adipocytes (Lee et al. 2014). However, overexpressing Nfic in bone marrow stem cells greatly enhances osteoblast differentiation but inhibits adipocyte differentiation, leading to more new bone formation. These results indicate that NFIC controls the cell fate between osteoblast and adipocyte differentiation. Furthermore, a lower NFIC expression in osteogenic cells is found in patients with osteoporosis.
Taken together, these findings indicate that Nfic is likely the real master gene that is required for tooth root elongation, although there has been no NFIC mutation reported in human SRA cases so far.
Osterix, One of the Key Downstream Molecules of NFIC, Plays a Critical Role in Root but Not Crown Formation
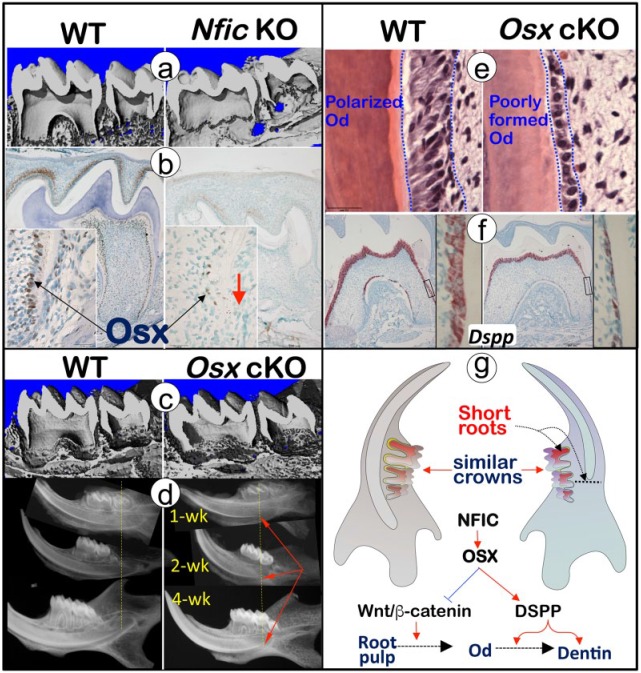
Osterix (Osx), a zinc finger–containing transcription factor, has long been known as the key factor in skeletal and cementum formation (Cao et al. 2012). A recent study demonstrated that Osx is the key downstream molecule of Nfic, based on the following evidence: 1) there is a sharp reduction in Osx expression in Nfic-null mice; 2) Osx-cKO (2.3-kb Col1α1-Cre, Osxfl/fl) mice recapture the Nfic-null root phenotypes, displaying modest changes in the tooth crown but severe defects in the molar root and incisor root analog; and 3) the in vitro overexpression of Nfic leads to a dosage-dependent increase in Osx expression that is not linked to the change in cell numbers (Fig. 2; Zhang et al. 2015). Similarly, Kim, Bae, Lee, et al. (2015), using 2 Osx-cKO mouse models (3.6-kb Col1α1-Cre, Osxfl/fl; osteocalcin-Cre, Osxfl/fl), demonstrated a crucial role of Osx in root but not crown dentin formation. Furthermore, Osx-cKO pulps displayed a lack of polarized odontoblasts but significantly more proliferative cells when compared the age-matched control, indicating an inhibitory role of Osx in cell proliferation but an enhancing role in the odontoblast differentiation (Kim, Bae, Lee, et al. 2015; Zhang et al. 2015). There is a sharp reduction of 2 critical dentin matrix proteins: DMP1 (dentin matrix protein 1; Feng et al. 2003) and DSPP (dentin sialophosphoprotein; Sreenath et al. 2003) in the cKO root rather than in the crown, supporting the notion that Osx is responsible for root-polarized odontoblast formation, partly through DMP1 and DSPP (Zhang et al. 2015). The in vitro study further confirmed that Osx overexpression increased DSPP transcription with mouse odontoblast-like cells (Chen et al. 2009; Zhang et al. 2015). Of note, there is disagreement in interpreting the Osx-cKO root phenotype. Kim, Bae, Lee, et al. (2015) claimed that the cause of the root phenotype is due to a site-specific role of Osterix in the root. This view does not address the fact that there is no apparent crown phenotype, while Osx is indeed expressed in the crown and root regions (Feng et al. 2015). It is more likely that there are other factors that compensate for Osterix function in crown but not root, which agrees with studies on Nfic, in which Nfic is expressed in the crown and root odontoblasts (Feng et al. 2015). Conventional Nfic-KO mice displayed short tooth root with limited impact on the tooth crown, indicating that NFIC and Osterix (the key downstream molecule of NFIC) are essential for root formation but their function can be compensated for by other factors in crown.
Figure 2.
Osterix, 1 of the key downstream molecules of NFIC, plays a crucial role in root but not crown formation. (a) The 2-wk-old Nfic-KO displayed short molar roots as viewed by micro–computed tomography (right panel). (b) Immunohistochemistry stains showed few positive Osx signals in the Nfic-KO odontoblasts (right panel). Red arrow indicates few positive OSX signals in the KO odontoblasts. (c) The 2-wk-old Osx-cKO molar roots (right) were short and thin according to micro–computed tomography images. (d) The Osx-cKO showed short incisor root analogue according to x-ray images. Red arrows indicate the Osx-cKO short incisor root-analogue. (e) The hematoxylin and eosin stain documented an immature odontoblast cell layer with no sign of polarization morphology in the Osx-cKO root (right panel). (f) The in situ hybridization assays displayed a great decrease in Dspp in the Osx-cKO root (right panel). (g) Osx is the key downstream molecule of NFIC, which controls root but not crown formation via its inhibitory role in cell proliferation and stimulatory function in the cell differentiation/odontoblast process formation, partially through DSPP (right panel). DSPP, dentin sialophosphoprotein; KO, knockout; Od, odontoblast; OSX, Osterix; WT, wild type. Panels A to G adapted and reproduced with permission from Zhang et al., J Bone Mineral Res, 2015, copyright John Wiley & Sons.
In summary, as the key downstream molecule of NFIC, Osx functions to promote odontoblast differentiation in root elongation but not crown formation.
Vital Roles of Wnt/β-catenin in Root Formation
It is known that β-catenin, the key downstream component of the canonical Wnt signaling pathway, is vital for bone formation (Long 2011). Interestingly, the regulation of Osx on bone cell differentiation happens via an inhibition of Wnt/β-catenin signaling (Zhang et al. 2008), suggesting that a high level of Wnt/β-catenin signaling may not be ideal for bone formation. Because bone and dentin share many similarities, several independent studies found a great impact of changes in the β-catenin levels on postnatal tooth formation. By crossing the osteocalcin-Cre that is active in all odontoblasts starting from the embryonic stage and beyond, removing β-catenin in odontoblasts completely disrupted tooth root formation with little impact on the tooth crowns (Kim et al. 2013; Zhang et al. 2013). Similarly, Han et al. (2011) reported the short root with a thin dentin layer phenotype in the overexpressed Dkk1 (a potent inhibitor of Wnt signaling) transgenic mice. Mechanism studies by Zhang et al. (2013) showed that these cKO mice displayed sharp reductions in cell proliferation by BrdU and PCNA immunostaining and a lack of expression of odontoblast markers such as Col 1, DSPP, osteocalcin, and BSP in roots with little change in crowns with the in situ hybridization assay. Thus, Zhang et al. concluded that Wnt/β-catenin signaling is functionally significant to root odontogenesis, whereas Kim et al. (2013) proposed a cell-autonomous requirement for Wnt/β-catenin signaling in the dental mesenchyme for root formation. Because neither of these groups compared its findings to the short root phenotype in the Nfic KO or tried to explain why the crown is largely unaffected in the β-catenin cKO model, it is unclear whether there is any connection between NFIC and Wnt/β-catenin signaling in root formation.
Constitutively stabilizing β-catenin studies by crossing the osteocalcin-Cre; Catnb+/lox(ex3) mice led to an acceleration in root dentin formation (including a lack of predentin and thicker dentin), although the root is short (less than half of the control root length) and disrupted (Kim et al. 2011; Bae et al. 2013). The gain of function data appears to be in agreement with the cKO data.
These studies demonstrate that Wnt/β-catenin signaling, which appears independent from NFIC, is vital for tooth root initiation, while the suitable level of Wnt/β-catenin signaling is required for proper tooth root formation.
Complex Role of TGF-β and BMP Signaling in Tooth Root Formation
Similar to Wnt/β-catenin signaling, TGF-β (transforming growth factor β) and BMP (bone morphogenetic protein) signaling is universally vital for almost all cell types during development. An in vitro study showed a complex relationship between TGF-β and NFIC in odontoblast cell reactions, in which TGF-β promotes the degradation of NFIC while NFIC downregulates the TGF-β/Smad signals (Lee et al. 2011). An in vivo study showed major defects in root formation with the impaired crown dentin formation when Tgfbr2 (TGF-β receptor 2, the key receptor for TGF-β signaling) is conditionally deleted by crossing with Sp7 (Osx)-Cre in dental mesenchyme (Wang et al. 2013). Furthermore, there is no apparent change in the expression levels of Nfic in the Tgfbr2-cKO mice, excluding the direct impact of TGF-β signaling on NFIC regulation in tooth root formation. With the same Osx-Cre line, conditional deletion of the BMP2 gene in a subset of dental pulp cells (preodontoblasts) and a subset of dental follicle cells leads to major defects in root and periodontium formation, including short roots, odontoblast dysmorphic differentiation, and the failure to form cellular cementum (Rakian et al. 2013). Interestingly, the Nfic mRNA expression is dramatically decreased in BMP2-cKO root odontoblasts, with the reduced expression of DMP1, DSPP, and Col1α1.
Overall, the TGF-β signaling is important not only for postnatal tooth root formation but also for continuous crown dentin formation without changing the NFIC signal pathway. In contrast, BMP signaling is more specific for the root formation in which NFIC is likely a key downstream molecule for root elongation.
Impact of HERS on Proper Root Formation via the Nfic Pathway
HERS is an epithelial bilayer structure formed after crown morphogenesis, which consists of the inner enamel epithelium and the outer enamel epithelium (Diekwisch 2001). The formation of HERS without a stratum intermedium or a stellate reticulum is considered to be the start of the root formation at approximately postnatal days 3 to 5 in mice (Kumakami-Sakano et al. 2014). It is currently believed that the interaction between the migrating HERS cells and the dental mesenchymal cells triggers root odontoblast formation via Smad4, the key mediator of TGF-β and BMP signaling.
The overexpression and KO studies in HERS showed that TGF-β and BMP signaling is vital for tooth root formation. A recent well-done study clearly showed that HERS cells regulate the dental mesenchyme for root formation through the Smad4-Shh-Nfic pathway (Huang et al. 2010). In this study, the authors conditionally inactivated Smad4 in the epithelial cells using K14-Cre, which led not only to abnormal enamel defects as expected but also to a severe root phenotype: a complete lack of roots in molars. This rootless phenotype is even worse than the short root phenotype in the Nfic-null mice. Further molecular studies have documented a sharp reduction in Shh in the epithelial cells and Nfic in the dental mesenchymal cells. Importantly, the ectopic application of Shh in the ex vivo culture partially restored the tooth roots and the Nfic expression in the dental mesenchyme (Huang et al. 2010). These data support the notion that HERS regulates root dentin formation partially through Smad4-Shh, which activates NFIC expression in the dental mesenchymal cells. However, future studies are required to address how Shh activates mesenchymal cells, as the direct targeting molecule of Shh is Gli1 (Briscoe and Therond 2013).
Interestingly, NFIC can downregulate the Shh signaling activity as a feedback loop through Nfic-Hhip-Shh, based on the following evidence (Liu et al. 2015): 1) Shh activity is elevated in Nfic-null mice; 2) constitutive activation of Hh signaling in dental mesenchymal progenitor cells leads to reduced proliferation and shorter roots with Gli1-CreERT2; R26SmoM2fl/fl mice, which are similar to the phenotype of Nfic-null mice; and 3) treating Nfic-null mice with an Hh inhibitor partially restores cell proliferation and root development. In addition, ChIP and RNAscope analyses suggest that NFIC binds to the promoter region of the hedgehog interacting protein (Hhip), which functions as an Hh attenuator. It is noteworthy that the inhibition of Hh signaling by injecting Hh inhibitors can also lead to short roots, with reduced apical papilla cell proliferation and impaired odontoblast differentiation (Liu et al. 2015). On the whole, these findings indicate that Shh signaling precisely regulates the root formation as Nfic and Wnt/β-catenin do.
The cell lineage tracing approach with K14-Cre; R26R mice showed that many HERS cells are detected on the surface of the root from the beginning of the root formation with a small subset of HERS cells becoming epithelial rests of Malassez (Huang et al. 2009). This study suggests that the HERS cells may directly differentiate into cementoblasts and play important roles in cementum formation. An in vitro study revealed that the HERS cells initially synthesize and secrete some enamel-related proteins and then change their morphology and produce a mineralized extracellular matrix resembling acellular cementum. This indicates that HERS cells are capable of deriving cementoblasts (Zeichner-David et al. 2003). As a consequence, the epithelial rests of Malassez, remnants of HERS in the periodontal ligament, may play a role as a stem cell niche that can give rise to new cementoblasts (Bosshardt et al. 2015).
As for the dental papilla, the apical papilla has received more attention recently, since it harbors stem cells from the apical papilla (Huang et al. 2008). Evidence is accumulating to support the hypothesis that the apical papilla appear to be the source of primary odontoblasts responsible for the root dentin formation, whereas dental pulp stem cells are likely the source of replacement odontoblasts (Huang et al. 2008; Liu et al. 2015). Cells in the apical papilla proliferate 2- to 3-fold more than those in pulp in the organ culture, with the strong potential to give rise to osteogenic/odontogenic cells (Sonoyama et al. 2008).
To sum up, 3 stem cell niches contribute to tooth root formation: the apical papilla (forming odontoblasts in root), HERS, and dental follicle (contributing to cementoblasts for cementum formation; Fig. 1).
Genes Involved in Multirooted Tooth Furcation Formation
Perhaps, one of the most intriguing challenges in root formation is to understand the mechanism by which some teeth develop a single root while others form 2 or 3 single roots. Although the formation of tooth furcation (the place where the roots fork or separate) is largely unknown at present, Hu and Simmer’s (Yang et al. 2015) laboratory recently discovered a notable taurodontism (a condition whereby the pulp chamber is vertically enlarged at the expense of the roots) with essentially no root furcation in mice lacking Wnt10a (a signaling molecule important for tooth development). Importantly, these authors found that in WNT10A mutation families, some heterozygotes exhibited molar root taurodontism. This finding may greatly stimulate more research interest in this area.
Cre Transgenic Mouse Lines Applied in Studies in Tooth Root Formation
Currently, there is no Cre transgenic mouse line that specifically targets targeting tooth root cells. However, multiple Cre transgenic lines have been used to investigate how root dentin is formed by targeting either HERS- or dental mesenchyme–derived cells (Fig. 3). At present, K14-Cre is the only transgenic line available for targeting genes expressed in HERS cells (Huang et al. 2010). The Cre lines targeting early dental mesenchyme–derived cells include Gli1-CreERT2 and Osterix-CreERT2 (mainly active in pulp cells). The cell lineage tracing (Gli1-CreERT2; R26tdTomato/+ ) and cell ablation (Gli1-CreERT2; R26DTA/+) assays revealed that the Gli1+ cells appear to be root progenitor cells and are crucial for root development, although the Gli1-derived cells are also responsible for crown dentin formation (Liu et al. 2015). Similarly, the cell lineage tracing technique showed a large number of Osx+ cells in the tooth pulp. Many of their progeny cells become root odontoblasts, as well as odontoblasts in crown, cementoblasts, alveolar bone osteoblasts, and periodontal ligament fibroblasts (Rakian et al. 2013).
Figure 3.
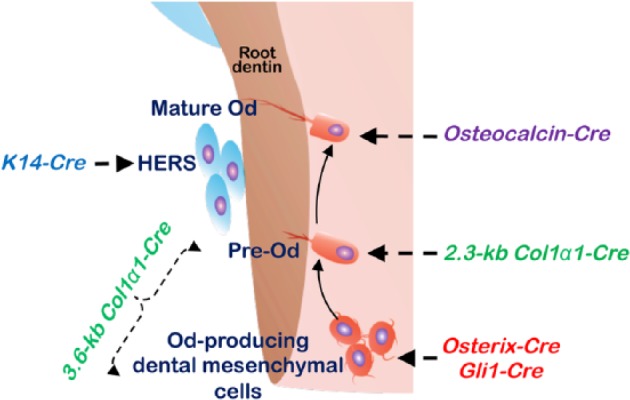
Diagram showing the Cre transgenic mouse lines applied in tooth root formation studies. HERS, Hertwig’s epithelial root sheath; Od, odontoblast.
The most common Cre lines targeting root odontoblasts are 1) 2 noninducible Cre lines—2.3-kb Col1α1-Cre (more specific to preodontoblasts) and 3.6-kb Col1α1-Cre (some pulp cells, preodontoblasts, and odontoblasts; Elefteriou and Yang 2011)—and 2) the tamoxifen-inducible 3.2-kb Col 1α1-CreERT2, which is active in preodontoblasts and odontoblasts (Maes et al. 2010). Another Cre line to target mature odontoblasts is the osteocalcin-Cre, whose activity was first observed in the developing tooth germ prior to postnatal day 0. By P4 and P6, osteocalcin-Cre was highly expressed in crown odontoblasts. At P10, the Cre was strongly expressed in the root odontoblasts (Bae et al. 2015). Additionally, some ubiquitous Cre lines, such as UBC-CreERT2, which is under the control of the human ubiquitin C promoter, have been used in root formation as well (Wang et al. 2017).
Conclusion
In the last 2 decades, there has been substantial progress in tooth root studies with the application of new techniques (including cell lineage tracing) and the gain- or loss-of-function studies of different genes (Table). These exciting findings suggest that root formation is regulated in a different and unique way when compared with crown formation. Furthermore, the new evidence demonstrates that NFIC, as the key master gene for tooth root formation, directly or indirectly interacts with Osx-Wnt/β-catenin (Fig. 2) and TGF-β-BMP-SMAD4 via an interaction between the HERS and dental mesenchyme (Fig. 4). For a potential link of the tooth root study to clinical relevance, we included the subject of the short root anomaly in this review.
Table.
Main Cre Transgenic Lines Used for the Analysis of Tooth Root Formation–Related Genes.
| Cre Transgenic Line | Examples of Gene Targeted | Root Phenotype | Reference | Original Description |
|---|---|---|---|---|
| Conventional knockout | Nfic−/− | Short roots | Steele-Perkins et al. 2003 | |
| Gli1-CreERT2 | Gli1-CreERT2; R26SmoM2fl/fl | Short roots | Liu et al. 2015 | Ahn and Joyner 2004 |
| Osterix-CreERT2 | Osx-CreERT2; PPRfl/fl | Short roots | Ono et al. 2016 | Maes et al. 2010 |
| Bmp2-cKOSp7-Cre-EGFP | Short roots | Rakian et al. 2013 | ||
| Osx-CreERT2; Tgfbr2fl/fl | Short roots | Wang et al. 2013 | ||
| 3.6-kb Col1α1-Cre | 3.6-kb Col1α1-Cre, Osxfl/fl | Short roots | Kim, Bae, Lee, et al. 2015 | Liu et al. 2004 |
| 2.3-kb Col1α1-Cre | 2.3-kb Col1α1-Cre, Osxfl/fl | Short roots | Zhang et al. 2015 | Liu et al. 2004 |
| Osteocalcin-Cre | OC-Cre; Catnb+/lox(ex3) | Short roots | Bae et al. 2013; Zhang et al. 2013 | Dacquin et al. 2002 |
| OC-Cre; Ctnnb1fl/fl | No roots | Kim et al. 2013 | ||
| OC-Cre; WlsCO/CO | Short roots | Bae et al. 2015 | ||
| K14-Cre | K14-Cre; Smad4fl/fl | No roots | Huang et al. 2010 | Dassule et al. 2000 |
| UBC-CreERT2 | UBC-CreERT2; Bmp1fl/fl, Tlll1fl/fl | Short roots | Wang et al. 2017 | Ruzankina et al. 2007 |
Figure 4.
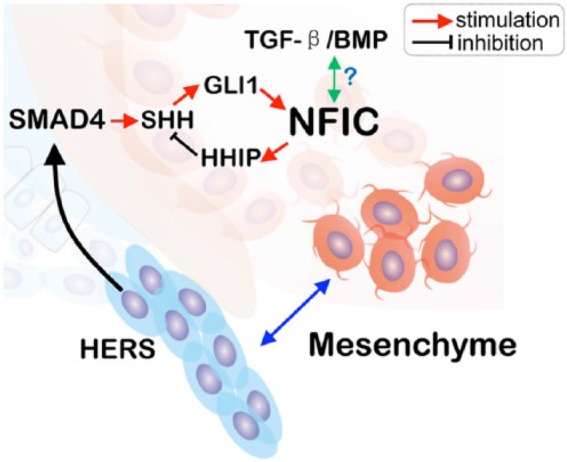
Diagram showing the current major molecular pathways regulating root formation via an interaction between Hertwig’s epithelial root sheath (HERS) and dental mesenchymal cells. BMP, bone morphogenetic protein; TGF-β, transforming growth factor β.
Future studies in this area will not only shed new light on understanding signaling pathways involved in tooth root formation but also provide a solid foundation for tooth regeneration as “the problem with tooth regeneration is the root problem” (Dr. Martha Somerman, director of National Institute of Dental and Craniofacial Research / National Institutes of Health).
Author Contributions
J. Wang, J.Q. Feng, contributed to conception, design, and data interpretation, drafted and critically revised the manuscript. Both authors gave final approval and agree to be accountable for all aspects of the work.
Footnotes
This work was partially supported from National Institutes of Health grants DE025014 and DE025659 to J.Q. Feng.
The authors declare no potential conflicts of interest with respect to the authorship and/or publication of this article.
References
- Ahn S, Joyner AL. 2004. Dynamic changes in the response of cells to positive hedgehog signaling during mouse limb patterning. Cell. 118(4):505–516. [DOI] [PubMed] [Google Scholar]
- Ando S, Aizawa K, Oshima S, Nakamura Y, Sato A, Suzuki Y, Arita I, Kaida Y, Nagayama R. 1976. Studies on the consecutive survey of succedaneous and permanent dentition in the Japanese children: part 11. On the difference between variations in the eruptive time of mandibular first premolars and their root formation. J Nihon Univ Sch Dent. 18(2):25–28. [DOI] [PubMed] [Google Scholar]
- Baccetti T. 1998. A controlled study of associated dental anomalies. Angle Orthod. 68(3):267–274. [DOI] [PubMed] [Google Scholar]
- Bae CH, Kim TH, Ko SO, Lee JC, Yang X, Cho ES. 2015. Wntless regulates dentin apposition and root elongation in the mandibular molar. J Dent Res. 94(3):439–445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bae CH, Lee JY, Kim TH, Baek JA, Lee JC, Yang X, Taketo MM, Jiang R, Cho ES. 2013. Excessive Wnt/beta-catenin signaling disturbs tooth-root formation. J Periodontal Res. 48(4):405–410. [DOI] [PubMed] [Google Scholar]
- Bosshardt DD, Stadlinger B, Terheyden H. 2015. Cell-to-cell communication—periodontal regeneration. Clin Oral Implants Res. 26(3):229–239. [DOI] [PubMed] [Google Scholar]
- Briscoe J, Therond PP. 2013. The mechanisms of hedgehog signalling and its roles in development and disease. Nat Rev Mol Cell Biol. 14(7):416–429. [DOI] [PubMed] [Google Scholar]
- Cao Z, Zhang H, Zhou X, Han X, Ren Y, Gao T, Xiao Y, de Crombrugghe B, Somerman MJ, Feng JQ. 2012. Genetic evidence for the vital function of Osterix in cementogenesis. J Bone Miner Res. 27(5):1080–1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S, Gluhak-Heinrich J, Wang YH, Wu YM, Chuang HH, Chen L, Yuan GH, Dong J, Gay I, MacDougall M. 2009. Runx2, Osx, and Dspp in tooth development. J Dent Res. 88(10):904–909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dacquin R, Starbuck M, Schinke T, Karsenty G. 2002. Mouse alpha1(i)-collagen promoter is the best known promoter to drive efficient Cre recombinase expression in osteoblast. Dev Dyn. 224(2):245–251. [DOI] [PubMed] [Google Scholar]
- das Neves L, Duchala CS, Tolentino-Silva F, Haxhiu MA, Colmenares C, Macklin WB, Campbell CE, Butz KG, Gronostajski RM. 1999. Disruption of the murine nuclear factor I-A gene (Nfia) results in perinatal lethality, hydrocephalus, and agenesis of the corpus callosum. Proc Natl Acad Sci U S A. 96(21):11946–11951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dassule HR, Lewis P, Bei M, Maas R, McMahon AP. 2000. Sonic hedgehog regulates growth and morphogenesis of the tooth. Development. 127(22):4775–4785. [DOI] [PubMed] [Google Scholar]
- Diekwisch TG. 2001. The developmental biology of cementum. Int J Dev Biol. 45(5–6):695–706. [PubMed] [Google Scholar]
- Elefteriou F, Yang X. 2011. Genetic mouse models for bone studies—strengths and limitations. Bone. 49(6):1242–1254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng JQ, Huang H, Lu Y, Ye L, Xie Y, Tsutsui TW, Kunieda T, Castranio T, Scott G, Bonewald LB, et al. 2003. The dentin matrix protein 1 (Dmp1) is specifically expressed in mineralized, but not soft, tissues during development. J Dent Res. 82(10):776–780. [DOI] [PubMed] [Google Scholar]
- Feng JQ, Zhang H, Qin C. 2015. Letter to the editor, “Osterix regulates tooth root formation in a site-specific manner.” J Dent Res. 94(9):1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gronostajski RM. 2000. Roles of the NFI/CTF gene family in transcription and development. Gene. 249(1–2):31–45. [DOI] [PubMed] [Google Scholar]
- Han XL, Liu M, Voisey A, Ren YS, Kurimoto P, Gao T, Tefera L, Dechow P, Ke HZ, Feng JQ. 2011. Post-natal effect of overexpressed DKK1 on mandibular molar formation. J Dent Res. 90(11):1312–1317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang GT, Sonoyama W, Liu Y, Liu H, Wang S, Shi S. 2008. The hidden treasure in apical papilla: the potential role in pulp/dentin regeneration and bioroot engineering. J Endod. 34(6):645–651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang X, Bringas P, Jr, Slavkin HC, Chai Y. 2009. Fate of HERS during tooth root development. Dev Biol. 334(1):22–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang X, Xu X, Bringas P, Jr, Hung YP, Chai Y. 2010. Smad4-Shh-Nfic signaling cascade-mediated epithelial-mesenchymal interaction is crucial in regulating tooth root development. J Bone Miner Res. 25(5):1167–1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim TH, Bae CH, Lee JC, Kim JE, Yang X, de Crombrugghe B, Cho ES. 2015. Osterix regulates tooth root formation in a site-specific manner. J Dent Res. 94(3):430–438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim TH, Bae CH, Lee JC, Ko SO, Yang X, Jiang R, Cho ES. 2013. Beta-catenin is required in odontoblasts for tooth root formation. J Dent Res. 92(3):215–221. [DOI] [PubMed] [Google Scholar]
- Kim TH, Bae CH, Yang S, Park JC, Cho ES. 2015. Nfic regulates tooth root patterning and growth. Anat Cell Biol. 48(3):188–194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim TH, Lee JY, Baek JA, Lee JC, Yang X, Taketo MM, Jiang R, Cho ES. 2011. Constitutive stabilization of ß-catenin in the dental mesenchyme leads to excessive dentin and cementum formation. Biochem Biophys Res Commun. 412(4):549–555. [DOI] [PubMed] [Google Scholar]
- Kumakami-Sakano M, Otsu K, Fujiwara N, Harada H. 2014. Regulatory mechanisms of Hertwig’s epithelial root sheath formation and anomaly correlated with root length. Exp Cell Res. 325(2):78–82. [DOI] [PubMed] [Google Scholar]
- Kurtulus-Waschulewski I, Wahl G, Dittrich K, Schuster V. 2015. Letter regarding the article: “Root dentin anomaly and a PLG mutation” by Tananuvat et al. Eur J Med Genet. 58(3):199–200. [DOI] [PubMed] [Google Scholar]
- Lan Y, Jia S, Jiang R. 2014. Molecular patterning of the mammalian dentition. Semin Cell Dev Biol. 25–26:61–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee DS, Choung HW, Kim HJ, Gronostajski RM, Yang YI, Ryoo HM, Lee ZH, Kim HH, Cho ES, Park JC. 2014. Nfi-C regulates osteoblast differentiation via control of Osterix expression. Stem Cells. 32(9):2467–2479. [DOI] [PubMed] [Google Scholar]
- Lee DS, Park JT, Kim HM, Ko JS, Son HH, Gronostajski RM, Cho MI, Choung PH, Park JC. 2009. Nuclear factor I-C is essential for odontogenic cell proliferation and odontoblast differentiation during tooth root development. J Biol Chem. 284(25):17293–17303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee DS, Yoon WJ, Cho ES, Kim HJ, Gronostajski RM, Cho MI, Park JC. 2011. Crosstalk between nuclear factor I-C and transforming growth factor-beta1 signaling regulates odontoblast differentiation and homeostasis. PLoS One. 6(12):e29160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Parada C, Chai Y. 2017. Cellular and molecular mechanisms of tooth root development. Development. 144(3):374–384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lind V. 1972. Short root anomaly. Scand J Dent Res. 80(2):85–93. [DOI] [PubMed] [Google Scholar]
- Liu F, Woitge HW, Braut A, Kronenberg MS, Lichtler AC, Mina M, Kream BE. 2004. Expression and activity of osteoblast-targeted Cre recombinase transgenes in murine skeletal tissues. Int J Dev Biol. 48(7):645–653. [DOI] [PubMed] [Google Scholar]
- Liu Y, Feng J, Li J, Zhao H, Ho TV, Chai Y. 2015. An Nfic-hedgehog signaling cascade regulates tooth root development. Development. 142(19):3374–3382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long F. 2011. Building strong bones: molecular regulation of the osteoblast lineage. Nat Rev Mol Cell Biol. 13(1):27–38. [DOI] [PubMed] [Google Scholar]
- Luder HU. 2015. Malformations of the tooth root in humans. Front Physiol. 6:307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maes C, Kobayashi T, Selig MK, Torrekens S, Roth SI, Mackem S, Carmeliet G, Kronenberg HM. 2010. Osteoblast precursors, but not mature osteoblasts, move into developing and fractured bones along with invading blood vessels. Dev Cell. 19(2):329–344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martynoga B, Mateo JL, Zhou B, Andersen J, Achimastou A, Urban N, van den Berg D, Georgopoulou D, Hadjur S, Wittbrodt J, et al. 2013. Epigenomic enhancer annotation reveals a key role for NFIX in neural stem cell quiescence. Genes Dev. 27(16):1769–1786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagano T, Yamaguchi H, Mikimoto K, Oida S, Kobayashi K, Shirakawa S, Gomi K. 2012. Gene expression during the formation of furcation in porcine tooth germ. J Hard Tissue Biol. 21(4):385–390. [Google Scholar]
- Nanci A. 2007. Ten cate’s oral histology—pageburst on vitalsource: development, structure, and function. Amsterdam (Netherlands): Elsevier Health Sciences. [Google Scholar]
- Newman WG. 1975. Possible etiologic factors in external root resorption. Am J Orthod. 67(5):522–539. [DOI] [PubMed] [Google Scholar]
- Ono W, Sakagami N, Nishimori S, Ono N, Kronenberg HM. 2016. Parathyroid hormone receptor signalling in osterix-expressing mesenchymal progenitors is essential for tooth root formation. Nat Commun. 7:11277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park JC, Herr Y, Kim HJ, Gronostajski RM, Cho MI. 2007. Nfic gene disruption inhibits differentiation of odontoblasts responsible for root formation and results in formation of short and abnormal roots in mice. J Periodontol. 78(9):1795–1802. [DOI] [PubMed] [Google Scholar]
- Rakian A, Yang WC, Gluhak-Heinrich J, Cui Y, Harris MA, Villarreal D, Feng JQ, MacDougall M, Harris SE. 2013. Bone morphogenetic protein-2 gene controls tooth root development in coordination with formation of the periodontium. Int J Oral Sci. 5(2):75–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruzankina Y, Pinzon-Guzman C, Asare A, Ong T, Pontano L, Cotsarelis G, Zediak VP, Velez M, Bhandoola A, Brown EJ. 2007. Deletion of the developmentally essential gene ATR in adult mice leads to age-related phenotypes and stem cell loss. Cell Stem Cell. 1(1):113–126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonoyama W, Liu Y, Yamaza T, Tuan RS, Wang S, Shi S, Huang GTJ. 2008. Characterization of apical papilla and its residing stem cells from human immature permanent teeth—a pilot study. J Endod. 34(2):166–171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sreenath T, Thyagarajan T, Hall B, Longenecker G, D’Souza R, Hong S, Wright JT, MacDougall M, Sauk J, Kulkarni AB. 2003. Dentin sialophosphoprotein knockout mouse teeth display widened predentin zone and develop defective dentin mineralization similar to human dentinogenesis imperfecta type III. J Biol Chem. 278(27):24874–24880. [DOI] [PubMed] [Google Scholar]
- Steele-Perkins G, Butz KG, Lyons GE, Zeichner-David M, Kim HJ, Cho MI, Gronostajski RM. 2003. Essential role for NFI-C/CTF transcription-replication factor in tooth root development. Mol Cell Biol. 23(3):1075–1084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steele-Perkins G, Plachez C, Butz KG, Yang G, Bachurski CJ, Kinsman SL, Litwack ED, Richards LJ, Gronostajski RM. 2005. The transcription factor gene Nfib is essential for both lung maturation and brain development. Mol Cell Biol. 25(2):685–698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tananuvat N, Charoenkwan P, Ohazama A, Ketuda Cairns JR, Kaewgahya M, Kantaputra PN. 2014. Root dentin anomaly and a PLG mutation. Eur J Med Genet. 57(11):630–635. [DOI] [PubMed] [Google Scholar]
- Valladares Neto J, Rino Neto J, de Paiva JB. 2013. Orthodontic movement of teeth with short root anomaly: should it be avoided, faced or ignored? Dental Press J Orthod. 18(6):72–85. [DOI] [PubMed] [Google Scholar]
- Wang J, Muir AM, Ren Y, Massoudi D, Greenspan DS, Feng JQ. 2017. Essential roles of bone morphogenetic protein-1 and mammalian tolloid-like 1 in postnatal root dentin formation. J Endod. 43(1):109–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Cox MK, Coricor G, MacDougall M, Serra R. 2013. Inactivation of Tgfbr2 in Osterix-Cre expressing dental mesenchyme disrupts molar root formation. Dev Biol. 382(1):27–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright JT, Hong SP, Simmons D, Daly B, Uebelhart D, Luder HU. 2008. Dlx3 c.561_562delct mutation causes attenuated phenotype of tricho-dento-osseous syndrome. Am J Med Genet A. 146(3):343–349. [DOI] [PubMed] [Google Scholar]
- Yang J, Wang SK, Choi M, Reid BM, Hu Y, Lee YL, Herzog CR, Kim-Berman H, Lee M, Benke PJ, et al. 2015. Taurodontism, variations in tooth number, and misshapened crowns in Wnt10a null mice and human kindreds. Mol Genet Genomic Med. 3(1):40–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeichner-David M, Oishi K, Su Z, Zakartchenko V, Chen LS, Arzate H, Bringas P., Jr. 2003. Role of Hertwig’s epithelial root sheath cells in tooth root development. Dev Dyn. 228(4):651–663. [DOI] [PubMed] [Google Scholar]
- Zhang C, Cho K, Huang Y, Lyons JP, Zhou X, Sinha K, McCrea PD, de Crombrugghe B. 2008. Inhibition of Wnt signaling by the osteoblast-specific transcription factor Osterix. Proc Natl Acad Sci U S A. 105(19):6936–6941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Jiang Y, Qin C, Liu Y, Ho SP, Feng JQ. 2015. Essential role of Osterix for tooth root but not crown dentin formation. J Bone Miner Res. 30(4):742–746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang R, Yang G, Wu X, Xie J, Yang X, Li T. 2013. Disruption of Wnt/beta-catenin signaling in odontoblasts and cementoblasts arrests tooth root development in postnatal mouse teeth. Int J Biol Sci. 9(3):228–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou B, Osinski JM, Mateo JL, Martynoga B, Sim FJ, Campbell CE, Guillemot F, Piper M, Gronostajski RM. 2015. Loss of Nfix transcription factor biases postnatal neural stem/progenitor cells toward oligodendrogenesis. Stem Cells Dev. 24(18):2114–2126. [DOI] [PubMed] [Google Scholar]