Abstract
The evolutionarily conserved Hippo signaling pathway is a vital regulator of organ size that fine-tunes cell proliferation, apoptosis, and differentiation. A number of important studies have revealed critical roles of Hippo signaling and its effectors Yap (Yes-associated protein) and Taz (transcriptional coactivator with PDZ binding motif) in tissue development, homeostasis, and regeneration, as well as in tumorigenesis. In addition, recent studies have shown evidence of crosstalk between the Hippo pathway and other key signaling pathways, such as Wnt signaling, that not only regulates developmental processes but also contributes to disease pathogenesis. In this review, we summarize the major discoveries in the field of Hippo signaling and what has been learned about its regulation and crosstalk with other signaling pathways, with a particular focus on recent findings involving the Hippo-Yap pathway in craniofacial and tooth development. New and exciting studies of the Hippo pathway are anticipated that will significantly improve our understanding of the molecular mechanisms of human craniofacial and tooth development and disease and will ultimately lead to the development of new therapies.
Keywords: Yap, Taz, tooth, cranial neural crest, molecular signaling, orofacial morphogenesis
Introduction
Overview of the Hippo Pathway
The Hippo pathway was first discovered in Drosophila melanogaster by using genetic screening experiments to identify growth suppressors. Since then, the Hippo pathway has emerged as an important field of research that continues to draw interest. Genetic and biochemical studies have established key roles of the Hippo pathway in regulating morphogenesis, homeostasis, disease pathogenesis, and tissue and organ regeneration. Hippo signaling affects major cellular events, such as cell fate decisions, proliferation, and cell death, largely by negatively regulating its downstream effectors Yap (Yes-associated protein) and Taz (transcriptional coactivator with PDZ binding motif; also known as Wwtr1, for WW domain–containing transcription regulator protein 1), which promote cell proliferation and survival. In most contexts, Hippo signaling negatively regulates growth by inhibiting cell proliferation and promoting cell apoptosis (Edgar 2006; Varelas 2014).
The Hippo pathway comprises evolutionarily and functionally conserved kinases (Table 1). The core kinase cascade of the Hippo pathway was first discovered in Drosophila through the identification of tumor suppressors, including Ste20-like kinase Hippo (Hpo; orthologous to Mst1 and Mst2 in mammals), WW domain–containing adaptor protein Salvador (Sav; orthologous to Sav1 in mammals and WW45 in humans), nuclear Dbf2-related family protein kinase Warts (Wts; orthologous to Lats1/2 kinases in mammals), and adaptor protein Mob as tumor suppressor (Mats; orthologous to MOB kinase activator 1A and 1B [MOB1A and MOB1B] in mammals). Recent studies also showed that the tumor suppressor Merlin (also named NF2)—a protein that is encoded by the neurofibromatosis type 2 gene and acts as a Yap inhibitor—functions as an upstream activator of Lats1/2 kinases (Hamaratoglu et al. 2006; Zhang et al. 2010; Yin et al. 2013). The upstream kinase components of the Hippo signaling pathway all converge and signal through the downstream transcriptional coactivator Yorkie (Yki). Drosophila Yki is orthologous to 2 proteins in mammals—the transcriptional coactivators Yap and Taz—which have both functional redundancy and divergent functional roles (Varelas 2014). The Drosophila Hpo-Yki pathway is homologous to the Mst-Yap/Taz pathway in mammals and functions through a phosphorylation-dependent pathway. Briefly, as shown in Figure 1, the Mst1/2 (Hpo) kinase and Sav1 (Sav) complex phosphorylates Lats1/2 (Wts) kinases. In turn, Lats1/2 (Wts) kinases further phosphorylate the downstream effectors Yap and Taz and promote the nuclear exclusion and, in some contexts, the degradation of Yap and Taz. When Hippo signaling is active, Yap and Taz interact with 14-3-3 protein, which leads to their degradation in the cytoplasm. However, when repression by Hippo signaling is absent, Yap and Taz shuttle into the nucleus, where they function as transcriptional coactivators. Yap and Taz do not directly bind DNA but instead form a complex with DNA-binding transcription factors, such as TEA domain transcription factors (TEAD), to regulate the expression of downstream genes.
Table 1.
Core Components of the Hippo-Yap Pathway in Drosophila and Mammals.
| Drosophila | Human | Mouse |
|---|---|---|
| Ste20-like kinase Hippo (Hpo) | MST1/serine/threonine kinase 4 (STK4), MST2/serine/threonine kinase 3 (STK3) | Mst1/Stk4, Mst2/Stk3 |
| WW domain–containing adaptor protein Salvador (Sav) | Salvador family WW domain–containing protein 1 (SAV1/WW45) | Sav1 |
| Nuclear Dbf2-related (NDR) family protein kinase Warts (Wts) | Large tumor suppressor kinase 1, 2 (LATS1, 2) | Lats1,2 |
| Adaptor protein Mob as tumor suppressor (Mats) | MOB kinase activator 1A and 1B (MOB1A, 1B) | Mob1a,b |
| Yorkie (Yki) | Yes-associated protein 1 (YAP1), transcriptional co-activator with PDZ binding motif (TAZ) / WW domain–containing transcription regulator protein 1 (WWTR1) | Yap1, Taz/Wwtr1 |
| Scalloped (Sd) | TEA domain transcription factors (TEAD1-4) | Tead |
Figure 1.
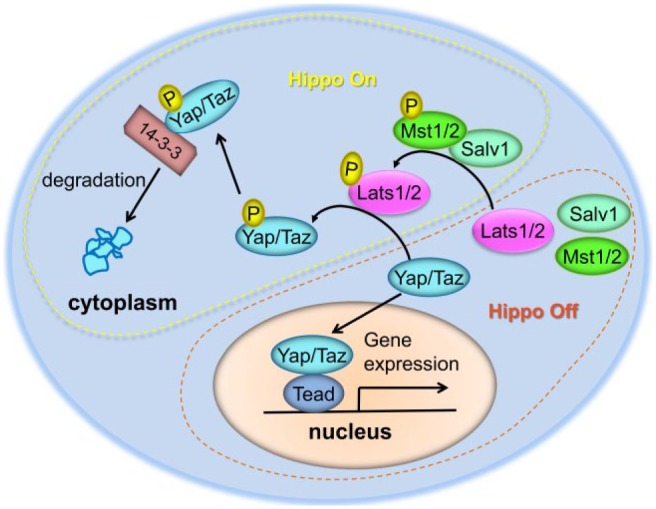
A brief summary of the intracellular Hippo pathway in mammals. The core Hippo pathway in mammals is composed of evolutionarily conserved kinases, including the Mst1/2 (Hpo in Drosophila), Sav1 (Sav in Drosophila), and Lats1/2 (Wts in Drosophila) kinases. When Hippo signaling is active, this kinase cascade is sequentially phosphorylated as shown, eventually resulting in the phosphorylation of Yap and Taz to promote their interaction with 14-3-3 protein and their degradation in the cytoplasm. Without repression by Hippo signaling, Yap and Taz can shuttle into the nucleus and bind to transcription factors (e.g., Tead) to regulate the transcription of target genes involved in different physiologic processes, such as cell proliferation, differentiation, and migration.
Hippo Pathway in Development, Regeneration, and Disease
Genetic studies in Drosophila have shown that the loss of function of any of the core Hippo kinases results in increased cell proliferation and decreased cell death, which leads to typical tissue overgrowth phenotypes (Justice et al. 1995; Xu et al. 1995; Kango-Singh et al. 2002; Tapon et al. 2002; Harvey et al. 2003; Jia et al. 2003; Pantalacci et al. 2003; Udan et al. 2003; Wu et al. 2003; Lai et al. 2005). The knockout of Hippo pathway gene Nf2, Mst1/2, or Lats2 in the mouse germline leads to embryonic lethality (McClatchey et al. 1997; McPherson et al. 2004; Yabuta et al. 2007; Zhou et al. 2009). Most Lats1 knockout mice die by postnatal day 1, but some survive postnatally with growth retardation (St John et al. 1999). Although Taz knockout mice survive to adulthood with kidney and lung defects (Hossain et al. 2007; Makita et al. 2008; Tian et al. 2010), Yap and Taz double knockout mouse embryos die before the morula stage (Nishioka et al. 2009), and the deletion of Yap alone results in embryonic lethality at embryonic day 8.5 (E8.5; Morin-Kensicki et al. 2006). To overcome the limitations of early lethality, different transgenic mouse lines and conditional knockout alleles have been established that have allowed for major breakthroughs in our understanding of the various functions of Hippo signaling in the development, homeostasis, and regeneration of different organs and tissues, as well as in disease pathogenesis and tumorigenesis (Barry and Camargo 2013; Piccolo et al. 2014; Zhou et al. 2015; Moya and Halder 2016). The phenotypes of mouse models used for studying the Hippo-Yap pathway are summarized in Table 2.
Table 2.
Phenotypes of Mouse Models Used for Studying the Hippo-Yap Pathway.
| Mouse Model: Gene | Phenotypes | Reference |
|---|---|---|
| Germline knockout | ||
| Nf2 | Failure of gastrulation initiation; most mutants are embryonic lethal between E6.5 and E7.0 with defective extraembryonic structures. | McClatchey et al. 1997 |
| Mst1 | Viable and fertile with immunologic defects. | Zhou et al. 2009 |
| Mst2 | Viable and fertile with no obvious developmental or immunologic defects. | Zhou et al. 2009 |
| Mst1/2 | Early embryonic lethality between E8.5 and E9.5. | Zhou et al. 2009 |
| Lats1 | Most mutants (60/101) died at postnatal day 1. Postnatal survivors had growth retardation, defective mammary gland development, and infertility. | St John et al. 1999 |
| Lats2 | Early embryonic lethality on or before E12.5 with defective proliferation and nervous system defects. | McPherson et al. 2004; Yabuta et al. 2007 |
| Yap | Developmental arrest and embryonic lethality at E8.5 with multiple defects, including defective yolk sac vasculogenesis, shortened body axis, and failure of chorioallantoic fusion. | Morin-Kensicki et al. 2006 |
| Taz | Development of renal cysts are detected as early as E15.5. Adulthood survivors have kidney and lung defects, suggesting Taz as a human polycystic kidney disease candidate gene. | Hossain et al. 2007; Makita et al. 2008; Tian et al. 2010 |
| Yap/Taz | Embryonic lethal before the morula stage (16 to 32 cells) | Nishioka et al. 2009 |
| Conditional knockout | ||
| Sav1 | Cardiac-specific Nkx 2.5 Cre driver. Died postnatally with heart enlargement. | Heallen et al. 2011 |
| Sav1 | Cardiac-specific Myh6-CreERT2 driver. Extended the cardiac regenerative window after apex resection and increased renewal of adult cardiomyocytes after myocardial infarction. | Heallen et al. 2013 |
| Mst1/2 | Cardiac-specific Nkx 2.5 Cre driver. E11.5 mutant hearts exhibit myocardial expansion. | Heallen et al. 2011 |
| Lats2 | Cardiac-specific Nkx 2.5 Cre driver. Myocardial expansion in E11.5 mutant hearts. | Heallen et al. 2011 |
| Lats1/2 | Cardiac-specific Myh6-CreERT2 driver. Increased cardiomyocyte proliferation and improved heart morphology and function after heart injury. | Heallen et al. 2013 |
| Yap | Cardiac-specific Nkx 2.5 Cre driver. Mutants are embryonic lethal by E10.5 with thin myocardium due to decreased proliferation. | Xin et al. 2011 |
| Yap | Cardiac-specific Tnnt2-Cre driver. Embryonic lethal by E16.5 with hypoplastic ventricles and reduced proliferation. | von Gise et al. 2012 |
| Yap | Cardiac-specific α-MHC-Cre driver. Mutants died by 11 wk with cardiomyopathy, increased apoptosis, and fibrosis. Defective neonatal heart regeneration after apex resection and worsened injury after chronic myocardial infarction. | Del Re et al. 2013; Xin et al. 2013 |
We apologize to researchers whose work is not cited here due to space constraints and reference limitations.
E, embryonic day.
Although Hippo signaling is fundamental for tissue homeostasis and regeneration, its function is like a double-edged sword; its dysregulation may lead to severe organ dysfunction or cancer progression. For example, in mice, Hippo signaling restrains heart size during embryonic cardiac development through the inhibition of Wnt signaling (Heallen et al. 2011). Newborn mouse heart can regenerate in response to injury up until postnatal day 7, but this capacity is lost in the adult mouse heart (Porrello et al. 2011; Porrello and Olson 2014). However, the cardiac-specific repression of Hippo signaling in adult mice extends the cardiac regenerative window (Heallen et al. 2013; Morikawa et al. 2015; Tao et al. 2016). This recovery of the heart’s ability to regenerate upon injury involves the regulation of cardiomyocyte proliferation, cytoskeletal remodeling, protrusion formation, and antioxidant responses (Heallen et al. 2013; Morikawa et al. 2015; Tao et al. 2016). Studies have also shown that Yap stimulates heart growth and is critical for cardiac regeneration (Xin et al. 2011; von Gise et al. 2012; Del Re et al. 2013; Xin et al. 2013; Lin et al. 2014). Yap deletion in the embryonic mouse heart causes embryonic lethality with reduced myocardial proliferation, and its deletion in the fetal mouse heart impedes the regenerative ability of the neonatal heart. Furthermore, human YAP activation in the adult mouse heart promotes cardiomyocyte proliferation without deleterious effects on cardiac function (Xin et al. 2011; von Gise et al. 2012; Del Re et al. 2013; Xin et al. 2013; Lin et al. 2014). Recently, Yap was shown to cooperate with the transcription factor Pitx2 in regulating the expression of genes critical for maintaining redox balance to promote mouse heart regeneration (Tao et al. 2016). Studies of Hippo pathway function in development, regeneration, and disease in different organs are not discussed here, but recent reviews on those topics are available elsewhere (Morgan et al. 2013; Johnson and Halder 2014; Lin and Pu 2014; Yu et al. 2015; Zhou et al. 2015; Moya and Halder 2016; Xiao et al. 2016).
Lats kinases have putative functions independent of the Hippo pathway or unrelated to Yap and Taz, implying that Lats kinases may have broader roles in regulating cell cycle machinery and mitotic fidelity and in maintaining genomic and protein stability. Genetic and biochemical studies in Drosophila have indicated that Warts, through a cell cycle–dependent phosphorylation event, functions as a negative regulator of the cell cycle and specifically interacts with Cdc2/cyclin to regulate checkpoints during mitosis (Tao et al. 1999). In addition, Lats proteins are localized at centrosomes, which are responsible for spindle pole formation during mitosis and contribute to the organization of the mitotic spindle (Iida et al. 2004; Visser and Yang 2010). Lats2 knockout cells exhibit centrosomal defects, such as centrosome amplification and fragmentation, cytokinesis defects, and genomic instability (McPherson et al. 2004; Yabuta et al. 2007). Lats proteins can also interact with and regulate various actin and microtubule cytoskeletal proteins, such as Zyxin, LIMK1, and Ajuba, indicating roles for Lats proteins in regulating cytokinesis and cell migration (Hirota et al. 2000; Yang et al. 2004; Abe et al. 2006). In addition, LATS2 was identified as a key component of retinoblastoma-induced senescence, with no connection to YAP or TAZ as effectors (Tschop et al. 2011).
To date, few germline or somatic mutations in genes encoding Hippo pathway components have been discovered by using targeted and whole-genome sequencing. However, mutations in genes encoding NF2, LATS2, and SAV1 have been frequently identified in neurofibromatosis characterized by mesothelioma and malignant peripheral nerve sheath tumors (Harvey et al. 2013; Johnson and Halder 2014). In addition, the R331W missense mutation in YAP has recently been identified as a germline risk allele for lung adenocarcinoma (Chen et al. 2015). Notably, heterozygous nonsense mutations of YAP1 have been identified in familial studies of variable phenotypes, such as orofacial clefting and intellectual disability; however, the mechanisms that cause these phenotypic alterations remain unclear (Williamson et al. 2014).
Regulation and Signaling Crosstalk of the Hippo Pathway
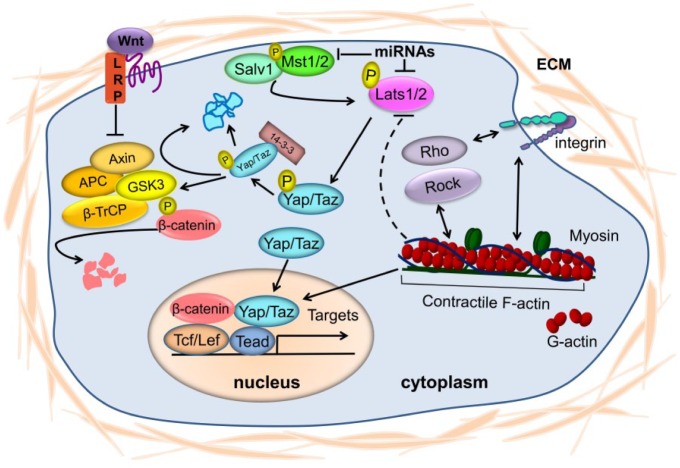
The Hippo pathway is regulated by a broad range of physiologic regulators, including cell polarity, the cytoskeleton, cell-cell contact, mechanical and hormonal signals, and genetic factors. The Hippo pathway functions as a central signal mediator and crosstalks with other major pathways, such as Wnt/β-catenin, TGF-β, Notch, and G protein–coupled receptor (GPCR) signals. In what follows, we provide a simplified yet fundamental background of the complex regulation and signal crosstalk of the Hippo pathway, summarized in Figure 2. More detailed reviews on this topic are available elsewhere (Varelas and Wrana 2012; Barry and Camargo 2013; Bernascone and Martin-Belmonte 2013; Morgan et al. 2013; Piccolo et al. 2014; Meng et al. 2016).
Figure 2.
A model for the regulation and signaling crosstalk of the Hippo pathway. As shown in the art, the Hippo pathway is regulated by mechanical signals, microRNAs, and crosstalk with Wnt, bone morphogenic protein, and TGF-β pathways. In addition, many other signals, such as G protein–coupled receptor and Notch pathway signals, can regulate or crosstalk with the Hippo pathway. ECM, extracellular matrix.
Crosstalk between Wnt/β-catenin and Hippo Pathways
Recent studies have shown extensive overlap between the Wnt/β-catenin and Hippo pathways (Varelas and Wrana 2012; Barry and Camargo 2013; Bernascone and Martin-Belmonte 2013; Morgan et al. 2013; Piccolo et al. 2014; Meng et al. 2016). Wnt has key roles in cell proliferation and differentiation, tissue development and regeneration, and tumorigenesis. In the presence of Wnt ligands, the nuclear transducer of the canonical Wnt pathway, β-catenin, accumulates in the nucleus and forms complexes with Tcf/Lef to regulate downstream gene transcription. In many contexts, the Tcf/Lef /β-catenin complex interacts with the Yap/Taz/Tead complex to activate gene expression programs (Varelas and Wrana 2012; Barry and Camargo 2013; Piccolo et al. 2014). For example, during mouse cardiac development, we found that Yap forms a complex with β-catenin in the nucleus, and they each function through their respective DNA-binding partners Tead and Tcf/Lef to activate downstream genes (Heallen et al. 2011). In addition, the deletion of Wnt/β-catenin in a Hippo-deficient heart rescues its phenotypes, such as cardiomegaly and trabecular expansion (Heallen et al. 2011).
In the absence of Wnt signaling, β-catenin is degraded in the cytoplasm by a destruction complex that consists of factors including central scaffold protein Axin, glycogen synthase kinase-3 (GSK3), adenomatous polyposis coli (APC), protein phosphatase 2A (PP2A), and casein kinase 1α (CK1α). This destruction complex degrades β-catenin through the phosphorylation of β-catenin by GSK3 and the subsequent ubiquitination of β-catenin by ubiquitin ligase β-TrCP, leading to digestion by the proteasome. Independent from their roles as Hippo signaling effectors, YAP and TAZ are sequestrated within the β-catenin destruction complex and bind Axin to recruit β-TrCP to the complex, which is critical for β-catenin degradation (Azzolin et al. 2014). The β-catenin destruction complex degrades TAZ to keep it at low levels, which occurs though the bridging of TAZ to β-TrCP by phosphorylated β-catenin (Azzolin et al. 2012). In addition, Lats2 inhibits Wnt signaling by disrupting the interaction between β-catenin and BCL9, which occurs independent of its kinase function (Li et al. 2013).
Crosstalk between the Hippo Pathway and Other Pathways
Evidence for crosstalk between the Hippo pathway and bone morphogenic protein (BMP) and TGF-β signaling has been shown by experiments revealing that YAP and TAZ bind directly to the linker region of both BMP and TGF-β Smads (Ferrigno et al. 2002; Varelas et al. 2008; Wrighton et al. 2008; Alarcón et al. 2009; Varelas et al. 2010). During neural differentiation of mouse embryonic stem cells, YAP is required both for BMP suppression and supporting Smad1-dependent transcription (Alarcón et al. 2009). TAZ and YAP have a noncanonical cytoplasmic function to retain SMAD2/3 in the cytoplasm, thereby suppressing TGF-β signaling (Ferrigno et al. 2002; Varelas et al. 2010). In addition, the binding of YAP to the inhibitory Smad7 protein enhances its inhibitory activity against TGF-β signaling, thereby reduces TGF-β receptor activity (Ferrigno et al. 2002).
Recently, Hippo and Notch pathways have been shown to converge in the development of human hepatocellular carcinoma, the control of liver cell fate, and the regulation of cranial neural crest (CNC)–derived smooth muscle differentiation, primarily through Yap and Tead’s direct regulation of Notch ligand Jagged1 and receptor Notch2 or their coregulation of downstream factors such as Cdx2 (Tschaharganeh et al. 2013; Rayon et al. 2014; Yimlamai et al. 2014; Manderfield et al. 2015).
In addition, GPCRs have been reported to function upstream of the Hippo pathway. Several GPCRs signal through YAP/TAZ; indeed, YAP/TAZ depletion was shown to inhibit cell proliferation and migration triggered by GPCR ligands (Mo et al. 2012; Yu et al. 2012).
Mechanical Signaling and the Hippo Pathway
The Hippo pathway is regulated by cell adhesion and mechanical signals from the extracellular matrix (ECM). At high cell density, cell adhesion produces a growth inhibitory signal; in turn, Hippo signaling is activated and YAP is inhibited, although YAP overexpression is sufficient for overcoming this cell-contact inhibition (Zhao et al. 2007). The ECM is a key element of the architectural signals that inform cell decisions; however, how cells determine whether to respond to the ECM and in what manner is poorly understood. Recent studies have shown that Yap and Taz function in the nuclear transduction of mechanical and cytoskeletal signals in response to changes in ECM stiffness (Dupont et al. 2011; Fernandez et al. 2011; Sansores-Garcia et al. 2011; Wada et al. 2011; Aragona et al. 2013; Calvo et al. 2013), shedding light on how intrinsic events affect the interpretation of extrinsic signals. The function of Yap/Taz in nuclear transduction in some cases may be independent of Hippo signaling and requires tension of actomyosin and Rho GTPase activity. Mechanical signals dominate as regulators of Yap/Taz over Hippo signaling, as evidenced by the finding that LATS1/2 inactivation does not increase YAP/TAZ nuclear localization in cells cultured with reduced mechanical stress (Dupont et al. 2011; Aragona et al. 2013). Instead, under low mechanical stress, filamentous actin capping and/or severing proteins such as Cofilin, CapZ, and Gelsolin limit the activity of YAP/TAZ in cells (Aragona et al. 2013). In addition, in cancer-associated fibroblasts, YAP is activated by ECM stiffening, which in turn enhances actomyosin contractility by upregulating myosin regulatory light chain (MRLC, also known as MYL9) and promotes the protumorigenic properties of cancer-associated fibroblasts (Calvo et al. 2013).
miRNA-Mediated Silencing of the Hippo Pathway
Evidence for the epigenetic regulation of the Hippo pathway has recently emerged. MicroRNAs (miRNAs)—the small, noncoding RNAs that repress gene expression by decaying the targeted mRNA or by inhibiting its translation into protein—have been identified that may have important roles in regulating the Hippo pathway. MiR-372 and miR-373, identified in a screen for oncogenic miRNAs in testicular germ cell tumor, neutralize p53-mediated cyclin-dependent kinase inhibition, possibly through the direct suppression of Lats2 expression (Voorhoeve et al. 2006).
Genetic studies in an experimental myocardial infarction model in mice have shown that the transient reexpression of the miRNA cluster miR-302-367 via mimics increased cardiomyocyte proliferation, improved cardiac function, and reduced scar formation after heart injury. Importantly, these functions are most likely due to the repression of major Hippo signaling component genes, including Mob1b, Mst1, and Lats2 (Tian et al. 2015).
Hippo Signaling in Craniofacial and Dental Development and Disease
Understanding the complex genetic architecture of craniofacial morphogenesis remains a challenge. During the last 2 decades, research in humans and animal models (e.g., chickens and mice) has advanced our understanding of the genetic basis of craniofacial development, particularly with respect to the roles of major signaling pathways, such as Bmp, sonic hedgehog (Shh), Wnt, Notch, and TGF-β pathways. Although the Hippo pathway has been shown to have pivotal roles in many different organs and tissues, as mentioned, its functions in craniofacial and tooth development and disease are largely unknown.
Hippo Signaling in Cranial Neural Crest Cells
CNC cells are a migratory cell population that originates in the dorsal neural tube and diversifies into multiple cell types, including smooth muscle cells, cartilage, bone, neurons, and glia. CNC cells are important for craniofacial development; the craniofacial skeleton, including the calvaria, mandible, and most midfacial bones, develops through the ossification of CNC-derived progenitor cells (Jiang et al. 2002; Chai and Maxson 2006).
We recently investigated the function of the Hippo pathway in craniofacial development. To specifically and effectively inactivate Yap and Taz in CNC cells, we used Wnt1Cre and Wnt1cre2SOR drivers; the Wnt1cre driver has been shown to express ectopic Wnt1 in the midbrain, whereas the Wnt1cre2SOR driver has similar recombination characteristics but does not induce ectopic Wnt activity (Wang et al. 2016). All Yap homozygous knockout compound mutants showed an embryonic lethal phenotype by E10.5, with severe craniofacial defects, such as disrupted frontonasal and mandibular structures, hemorrhage, failed neural tube closure, and neural tube vessel regression (Wang et al. 2016). In contrast to Yap homozygous compound mutants, Taz homozygous and Yap heterozygous compound mutants showed no obvious defects before E10.5 but, at later stages, developed hydrocephalus, which is characterized by excessive cerebrospinal fluid accumulation and abnormal widening of the brain space (Wang et al. 2016). In mice, Yap/Taz deficiency significantly reduced proliferation and increased cell apoptosis in the CNC-derived craniofacial structures (Wang et al. 2016). In vitro studies with a stable, multipotent mesenchymal CNC cell line named O9-1 cells also indicated that proliferation was significantly reduced in Yap/Taz knockdown cells but was significantly increased in Lats1/Lats2 knockdown cells (Wang et al. 2016). In vivo and in vitro studies indicated that Yap/Taz inactivation resulted in a deficiency of CNC cells that differentiated into smooth muscle cells (Manderfield et al. 2015; Wang et al. 2016). In CNC cells, Yap and Taz function dependently and independently of Tead to regulate the expression of downstream genes, including Notch target genes such as Jagged1 and transcription factors such as Foxc1, a gene implicated in human ocular and cerebellar malformations and in mouse vascular and hydrocephalus malformations (Manderfield et al. 2015; Wang et al. 2016).
Hippo Signaling in Tooth Development
Molecular signals involved in tooth development are also commonly involved in orofacial development, as evidenced by the frequent concurrence of orofacial defects and tooth agenesis in humans and mice. A tooth is a typical ectodermal appendage structure that develops from the oral epithelium and neural crest–derived mesenchyme. Tooth development comprises multiple stages involving different events, such as cell fate determination, proliferation, migration, and differentiation, as well as epithelial mesenchymal interactions (Jernvall and Thesleff 2000; Li and Li 2016). Tooth development initiates from the oral epithelium, which thickens into a multilayered structure called a placode. At E12.5 in mice, the placode invaginates into the underlying mesenchyme and undergoes morphologic changes to form 2 epithelial protrusions at the distal end of the tooth germ (Li et al. 2016). During the bud and cap stages, a key event is the formation of an enamel knot (EK) by E13.5, which regresses by E16. The EK, a structure considered as the tooth signaling center, regulates tooth morphogenesis and cusp growth. During tooth morphogenesis in mice, only 1 EK forms in incisors, whereas 2 EKs form in molars (Li et al. 2016).
In a study of mouse incisor development, the control of Yap expression was time dependent during tooth development and correlated with the proliferation rate (Li et al. 2011). Yap was expressed early on in most basal cells of the incisor dental epithelium, whereas at later stages, Yap was expressed mostly in the transit-amplifying cells, with high expression in the proliferating areas from the bud to eruption stages and low expression in slow-growing areas, such as the apical bud, stratum intermedium, and stellate reticulum (Li et al. 2011; Li and Li 2016). According to another study’s findings, Yap was expressed in the dental epithelial and mesenchymal tissues of mice, and YAP overexpression in dental epithelium resulted in defects of tooth morphogenesis and EK patterning (Liu et al. 2014). The overexpression of constitutively active YAP, driven by the human keratin 14 (K14) promoter, in ectoderm-derived epithelial tissues also resulted in death shortly after birth, most likely because of dehydration caused by skin defects. The teeth of mice overexpressing YAP had greatly widened dental lamina at E14.5, and tooth development either arrested at the cap stage or exhibited mislocated EKs and widened dental lamina at E16.5 (Liu et al. 2014). Mice with Yap loss of function in the dental epithelium were recently shown to have a small tooth germ caused by reduced epithelial cell proliferation (Liu et al. 2015). In E14.5 tooth germ from mice overexpressing YAP and from Yap conditional knockout mice, Hoxa1 and Hoxc13 were identified as downstream targets of Yap in oral and dental epithelial tissues, the regulation of which is probably mediated by TEAD transcription factors (Liu et al. 2015).
Interestingly, nuclear YAP was shown to be present in EKs at low levels, and its localization is regulated by cell polarity mediated through αE-catenin (Li et al. 2016). In contrast to the surrounding actively proliferating epithelium that supports continuous tissue growth, cells in the EK are postmitotic and are required for maintaining epithelial invagination and mesenchymal condensation during tooth morphogenesis. αE-catenin, encoded by Ctnna1, is the most abundant in epithelial tissues and is required specifically for the morphogenesis of ectodermal appendages (Li et al. 2016). The conditional knockout of Ctnna1 in tooth germ epithelium by using K14-Cre resulted in the accumulation of nuclear YAP in the EK region and led to failed EK formation. Importantly, the deletion of Yap/Taz restored EK formation in Ctnna1 mutants. The function of YAP was shown to be more prominent than that of TAZ, given that Ctnna1 mutants with Yap knockout and Taz heterozygosity had restored tooth germs that continued to invaginate at E16.5, whereas Ctnna1 mutants with Taz knockout and Yap heterozygosity did not (Li et al. 2016).
Journey of Hippo Signaling Research
The initial discovery of Hippo kinases in Drosophila has led to an intensive boost in research efforts focused on revealing the mechanism and function of the Hippo pathway. Although interest in the field of Hippo research has rapidly escalated, it remains a young field when compared with those of other signaling pathways, such as Wnt and Bmp pathways, especially in craniofacial and tooth studies. Nonetheless, molecular, biochemical, and genetic studies in Drosophila and vertebrates have shown that the Hippo pathway has diverse and central roles in many physiologic contexts and that it regulates development and disease processes involving cell proliferation, fate decisions, differentiation, and survival. Precise control of the Hippo pathway is essential for normal craniofacial and tooth development, but the mechanisms are still largely unknown.
Recently, exciting work utilizing mouse genetics has revealed pivotal functions of the Hippo pathway in the regeneration of several mouse organs, including the heart, liver, and intestine, suggesting that the Hippo pathway is a promising therapeutic target for regenerative medicine. The specific expression of YAP in adult murine myocardium by using recombinant adeno-associated virus subtype 9 (AAV9) significantly improved cardiac function and survival rate after myocardial infarction (Lin et al. 2014). Importantly, AAV9 has been shown to be safe in humans, producing only a minimal immune response, and is thus a feasible therapeutic agent for clinical use. Investigating potential applications of the Hippo pathway in craniofacial and tooth regenerative medicine is an important area of study.
In addition, a challenging area of research involves the regulation and crosstalk of the Hippo pathway with other pathways during craniofacial and tooth morphogenesis. It is clear that normal craniofacial and tooth morphogenesis and development require signaling pathways to be organized into coherent networks, and the disorganization or interruption of these networks results in developmental abnormalities and disease. To date, many promising findings in other fields have indicated that the Hippo pathway interacts with other essential signaling pathways, but the regulation and crosstalk of the Hippo pathway in orofacial and tooth development remain largely unexplored. We believe that this area of research is at just the beginning of an arduously long journey but one with innumerable exciting discoveries and surprises ahead.
Author Contributions
J. Wang, contributed to conception and design, drafted the manuscript; J.F. Martin, contributed to conception and design, critically revised the manuscript. Both authors gave final approval and agree to be accountable for all aspects of the work.
Acknowledgments
We appreciate the editorial assistance provided by Nicole Stancel, PhD, ELS, of the Texas Heart Institute. We apologize to researchers whose work is not cited here because of space constraints.
Footnotes
We thank the following funding sources: the American Heart Association’s National Center Scientist Development Grant (14SDG19840000 to J. Wang), the 2014 Lawrence Research Award from the Rolanette and Berdon Lawrence Bone Disease Program of Texas (to J. Wang), the National Institutes of Health (DE026561 to J. Wang; DE 023177, HL 127717, HL 130804, and HL 118761 to J.F. Martin), and the Vivian L. Smith Foundation (to J.F. Martin). J.F. Martin was supported by the LeDucq Foundation’s Transatlantic Networks of Excellence in Cardiovascular Research (14CVD01: “Defining the Genomic Topology of Atrial Fibrillation”).
The authors declare no potential conflicts of interest with respect to the authorship and/or publication of this article.
References
- Abe Y, Ohsugi M, Haraguchi K, Fujimoto J, Yamamoto T. 2006. Lats2-ajuba complex regulates gamma-tubulin recruitment to centrosomes and spindle organization during mitosis. FEBS Lett. 580(3):782–788. [DOI] [PubMed] [Google Scholar]
- Alarcón C, Zaromytidou AI, Xi Q, Gao S, Yu J, Fujisawa S, Barlas A, Miller AN, Manova-Todorova K, Macias MJ, et al. 2009. Nuclear CDKs drive Smad transcriptional activation and turnover in BMP and TGF-beta pathways. Cell. 139(4):757–769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aragona M, Panciera T, Manfrin A, Giulitti S, Michielin F, Elvassore N, Dupont S, Piccolo S. 2013. A mechanical checkpoint controls multicellular growth through Yap/Taz regulation by actin-processing factors. Cell. 154(5):1047–1059. [DOI] [PubMed] [Google Scholar]
- Azzolin L, Panciera T, Soligo S, Enzo E, Bicciato S, Dupont S, Bresolin S, Frasson C, Basso G, Guzzardo V, et al. 2014. Yap/Taz incorporation in the β-catenin destruction complex orchestrates the Wnt response. Cell. 158(1):157–170. [DOI] [PubMed] [Google Scholar]
- Azzolin L, Zanconato F, Bresolin S, Forcato M, Basso G, Bicciato S, Cordenonsi M, Piccolo S. 2012. Role of Taz as mediator of Wnt signaling. Cell. 151(7):1443–1456. [DOI] [PubMed] [Google Scholar]
- Barry ER, Camargo FD. 2013. The Hippo superhighway: signaling crossroads converging on the Hippo/Yap pathway in stem cells and development. Curr Opin Cell Biol. 25(2):247–253. [DOI] [PubMed] [Google Scholar]
- Bernascone I, Martin-Belmonte F. 2013. Crossroads of Wnt and Hippo in epithelial tissues. Trends Cell Biol. 23(8):380–389. [DOI] [PubMed] [Google Scholar]
- Calvo F, Ege N, Grande-Garcia A, Hooper S, Jenkins RP, Chaudhry SI, Harrington K, Williamson P, Moeendarbary E, Charras G, et al. 2013. Mechanotransduction and Yap-dependent matrix remodelling is required for the generation and maintenance of cancer-associated fibroblasts. Nat Cell Biol. 15(6):637–646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chai Y, Maxson RE., Jr. 2006. Recent advances in craniofacial morphogenesis. Dev Dyn. 235(9):2353–2375. [DOI] [PubMed] [Google Scholar]
- Chen HY, Yu SL, Ho BC, Su KY, Hsu YC, Chang CS, Li YC, Yang SY, Hsu PY, Ho H, et al. 2015. R331w missense mutation of oncogene Yap1 is a germline risk allele for lung adenocarcinoma with medical actionability. J Clin Oncol. 33(20):2303–2310. [DOI] [PubMed] [Google Scholar]
- Del Re DP, Yang Y, Nakano N, Cho J, Zhai P, Yamamoto T, Zhang N, Yabuta N, Nojima H, Pan D, et al. 2013. Yes-associated protein isoform 1 (Yap1) promotes cardiomyocyte survival and growth to protect against myocardial ischemic injury. J Biol Chem. 288(6):3977–3988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dupont S, Morsut L, Aragona M, Enzo E, Giulitti S, Cordenonsi M, Zanconato F, Le Digabel J, Forcato M, Bicciato S, et al. 2011. Role of Yap/Taz in mechanotransduction. Nature. 474(7350):179–183. [DOI] [PubMed] [Google Scholar]
- Edgar BA. 2006. From cell structure to transcription: Hippo forges a new path. Cell. 124(2):267–273. [DOI] [PubMed] [Google Scholar]
- Fernandez BG, Gaspar P, Bras-Pereira C, Jezowska B, Rebelo SR, Janody F. 2011. Actin-capping protein and the Hippo pathway regulate F-actin and tissue growth in Drosophila. Development. 138(11):2337–2346. [DOI] [PubMed] [Google Scholar]
- Ferrigno O, Lallemand F, Verrecchia F, L’Hoste S, Camonis J, Atfi A, Mauviel A. 2002. Yes-associated protein (Yap65) interacts with Smad7 and potentiates its inhibitory activity against TGF-beta/Smad signaling. Oncogene. 21(32):4879–4884. [DOI] [PubMed] [Google Scholar]
- Hamaratoglu F, Willecke M, Kango-Singh M, Nolo R, Hyun E, Tao C, Jafar-Nejad H, Halder G. 2006. The tumour-suppressor genes NF2/Merlin and Expanded act through Hippo signalling to regulate cell proliferation and apoptosis. Nat Cell Biol. 8(1):27–36. [DOI] [PubMed] [Google Scholar]
- Harvey KF, Pfleger CM, Hariharan IK. 2003. The Drosophila Mst ortholog, Hippo, restricts growth and cell proliferation and promotes apoptosis. Cell. 114(4):457–467. [DOI] [PubMed] [Google Scholar]
- Harvey KF, Zhang X, Thomas DM. 2013. The Hippo pathway and human cancer. Nat Rev Cancer. 13(4):246–257. [DOI] [PubMed] [Google Scholar]
- Heallen T, Morikawa Y, Leach J, Tao G, Willerson JT, Johnson RL, Martin JF. 2013. Hippo signaling impedes adult heart regeneration. Development. 140(23):4683–4690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heallen T, Zhang M, Wang J, Bonilla-Claudio M, Klysik E, Johnson RL, Martin JF. 2011. Hippo pathway inhibits Wnt signaling to restrain cardiomyocyte proliferation and heart size. Science. 332(6028):458–461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirota T, Morisaki T, Nishiyama Y, Marumoto T, Tada K, Hara T, Masuko N, Inagaki M, Hatakeyama K, Saya H. 2000. Zyxin, a regulator of actin filament assembly, targets the mitotic apparatus by interacting with h-warts/LATS1 tumor suppressor. J Cell Biol. 149(5):1073–1086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hossain Z, Ali SM, Ko HL, Xu J, Ng CP, Guo K, Qi Z, Ponniah S, Hong W, Hunziker W. 2007. Glomerulocystic kidney disease in mice with a targeted inactivation of wwtr1. Proc Natl Acad Sci U S A. 104(5):1631–1636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iida S, Hirota T, Morisaki T, Marumoto T, Hara T, Kuninaka S, Honda S, Kosai K, Kawasuji M, Pallas DC, et al. 2004. Tumor suppressor WARTS ensures genomic integrity by regulating both mitotic progression and G1 tetraploidy checkpoint function. Oncogene. 23(31):5266–5274. [DOI] [PubMed] [Google Scholar]
- Jernvall J, Thesleff I. 2000. Reiterative signaling and patterning during mammalian tooth morphogenesis. Mech Dev. 92(1):19–29. [DOI] [PubMed] [Google Scholar]
- Jia J, Zhang W, Wang B, Trinko R, Jiang J. 2003. The Drosophila Ste20 family kinase dMST functions as a tumor suppressor by restricting cell proliferation and promoting apoptosis. Genes Dev. 17(20):2514–2519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang X, Iseki S, Maxson RE, Sucov HM, Morriss-Kay GM. 2002. Tissue origins and interactions in the mammalian skull vault. Dev Biol. 241(1):106–116. [DOI] [PubMed] [Google Scholar]
- Johnson R, Halder G. 2014. The two faces of Hippo: targeting the Hippo pathway for regenerative medicine and cancer treatment. Nat Rev Drug Discov. 13(1):63–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Justice RW, Zilian O, Woods DF, Noll M, Bryant PJ. 1995. The Drosophila tumor suppressor gene warts encodes a homolog of human myotonic dystrophy kinase and is required for the control of cell shape and proliferation. Genes Dev. 9(5):534–546. [DOI] [PubMed] [Google Scholar]
- Kango-Singh M, Nolo R, Tao C, Verstreken P, Hiesinger PR, Bellen HJ, Halder G. 2002. Shar-pei mediates cell proliferation arrest during imaginal disc growth in Drosophila. Development. 129(24):5719–5730. [DOI] [PubMed] [Google Scholar]
- Lai ZC, Wei X, Shimizu T, Ramos E, Rohrbaugh M, Nikolaidis N, Ho LL, Li Y. 2005. Control of cell proliferation and apoptosis by mob as tumor suppressor, mats. Cell. 120(5):675–685. [DOI] [PubMed] [Google Scholar]
- Li CH, Li CZ. 2016. The role of Hippo signaling in tooth development. J Formos Med Assoc. 115(5):295–297. [DOI] [PubMed] [Google Scholar]
- Li CY, Hu J, Lu H, Lan J, Du W, Galicia N, Klein OD. 2016. αE-catenin inhibits Yap/Taz activity to regulate signalling centre formation during tooth development. Nat Commun. 7:12133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Chen X, Ding X, Cheng Y, Zhao B, Lai ZC, Al Hezaimi K, Hakem R, Guan KL, Wang CY. 2013. LATS2 suppresses oncogenic Wnt signaling by disrupting β-catenin/BCL9 interaction. Cell Rep. 5(6):1650–1663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Kwon HJ, Harada H, Ohshima H, Cho SW, Jung HS. 2011. Expression patterns of ABCG2, Bmi-1, Oct-3/4, and Yap in the developing mouse incisor. Gene Expr Patterns. 11(3–4):163–170. [DOI] [PubMed] [Google Scholar]
- Lin Z, Pu WT. 2014. Harnessing Hippo in the heart: Hippo/Yap signaling and applications to heart regeneration and rejuvenation. Stem Cell Res. 13(3 Pt B):571–581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Z, von Gise A, Zhou P, Gu F, Ma Q, Jiang J, Yau AL, Buck JN, Gouin KA, van Gorp PR, et al. 2014. Cardiac-specific Yap activation improves cardiac function and survival in an experimental murine MI model. Circ Res. 115(3):354–363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu M, Zhao S, Lin Q, Wang XP. 2015. Yap regulates the expression of Hoxa1 and Hoxc13 in mouse and human oral and skin epithelial tissues. Mol Cell Biol. 35(8):1449–1461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu M, Zhao S, Wang XP. 2014. Yap overexpression affects tooth morphogenesis and enamel knot patterning. J Dent Res. 93(5):469–474. [DOI] [PubMed] [Google Scholar]
- Makita R, Uchijima Y, Nishiyama K, Amano T, Chen Q, Takeuchi T, Mitani A, Nagase T, Yatomi Y, Aburatani H, et al. 2008. Multiple renal cysts, urinary concentration defects, and pulmonary emphysematous changes in mice lacking Taz. Am J Physiol Renal Physiol. 294(3):F542–F553. [DOI] [PubMed] [Google Scholar]
- Manderfield LJ, Aghajanian H, Engleka KA, Lim LY, Liu F, Jain R, Li L, Olson EN, Epstein JA. 2015. Hippo signaling is required for notch-dependent smooth muscle differentiation of neural crest. Development. 142(17):2962–2971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClatchey AI, Saotome I, Ramesh V, Gusella JF, Jacks T. 1997. The Nf2 tumor suppressor gene product is essential for extraembryonic development immediately prior to gastrulation. Genes Dev. 11(10):1253–1265. [DOI] [PubMed] [Google Scholar]
- McPherson JP, Tamblyn L, Elia A, Migon E, Shehabeldin A, Matysiak-Zablocki E, Lemmers B, Salmena L, Hakem A, Fish J, et al. 2004. Lats2/Kpm is required for embryonic development, proliferation control and genomic integrity. EMBO J. 23(18):3677–3688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng Z, Moroishi T, Guan KL. 2016. Mechanisms of Hippo pathway regulation. Genes Dev. 30(1):1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mo JS, Yu FX, Gong R, Brown JH, Guan KL. 2012. Regulation of the Hippo-Yap pathway by protease-activated receptors (PARs). Genes Dev. 26(19):2138–2143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan JT, Murphy CJ, Russell P. 2013. What do mechanotransduction, Hippo, Wnt, and TGFβ have in common? Yap and Taz as key orchestrating molecules in ocular health and disease. Exp Eye Res. 115:1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morikawa Y, Zhang M, Heallen T, Leach J, Tao G, Xiao Y, Bai Y, Li W, Willerson JT, Martin JF. 2015. Actin cytoskeletal remodeling with protrusion formation is essential for heart regeneration in Hippo-deficient mice. Sci Signal. 8(375):ra41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morin-Kensicki EM, Boone BN, Howell M, Stonebraker JR, Teed J, Alb JG, Magnuson TR, O’Neal W, Milgram SL. 2006. Defects in yolk sac vasculogenesis, chorioallantoic fusion, and embryonic axis elongation in mice with targeted disruption of Yap65. Mol Cell Biol. 26(1):77–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moya IM, Halder G. 2016. The Hippo pathway in cellular reprogramming and regeneration of different organs. Curr Opin Cell Biol. 43:62–68. [DOI] [PubMed] [Google Scholar]
- Nishioka N, Inoue K, Adachi K, Kiyonari H, Ota M, Ralston A, Yabuta N, Hirahara S, Stephenson RO, Ogonuki N, et al. 2009. The Hippo signaling pathway components lats and Yap pattern tead4 activity to distinguish mouse trophectoderm from inner cell mass. Dev Cell. 16(3):398–410. [DOI] [PubMed] [Google Scholar]
- Pantalacci S, Tapon N, Leopold P. 2003. The salvador partner Hippo promotes apoptosis and cell-cycle exit in Drosophila. Nat Cell Biol. 5(10):921–927. [DOI] [PubMed] [Google Scholar]
- Piccolo S, Dupont S, Cordenonsi M. 2014. The biology of Yap/Taz: Hippo signaling and beyond. Physiol Rev. 94(4):1287–1312. [DOI] [PubMed] [Google Scholar]
- Porrello ER, Mahmoud AI, Simpson E, Hill JA, Richardson JA, Olson EN, Sadek HA. 2011. Transient regenerative potential of the neonatal mouse heart. Science. 331(6020):1078–1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porrello ER, Olson EN. 2014. A neonatal blueprint for cardiac regeneration. Stem Cell Res. 13(3 Pt B):556–570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rayon T, Menchero S, Nieto A, Xenopoulos P, Crespo M, Cockburn K, Canon S, Sasaki H, Hadjantonakis AK, de la Pompa JL, et al. 2014. Notch and Hippo converge on Cdx2 to specify the trophectoderm lineage in the mouse blastocyst. Dev Cell. 30(4):410–422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sansores-Garcia L, Bossuyt W, Wada K, Yonemura S, Tao C, Sasaki H, Halder G. 2011. Modulating F-actin organization induces organ growth by affecting the Hippo pathway. EMBO J. 30(12):2325–2335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- St John MA, Tao W, Fei X, Fukumoto R, Carcangiu ML, Brownstein DG, Parlow AF, McGrath J, Xu T. 1999. Mice deficient of Lats1 develop soft-tissue sarcomas, ovarian tumours and pituitary dysfunction. Nat Genet. 21(2):182–186. [DOI] [PubMed] [Google Scholar]
- Tao G, Kahr PC, Morikawa Y, Zhang M, Rahmani M, Heallen TR, Li L, Sun Z, Olson EN, Amendt BA, et al. 2016. Pitx2 promotes heart repair by activating the antioxidant response after cardiac injury. Nature. 534(7605):119–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao W, Zhang S, Turenchalk GS, Stewart RA, St John MA, Chen W, Xu T. 1999. Human homologue of the Drosophila melanogaster lats tumour suppressor modulates CDC2 activity. Nat Genet. 21(2):177–181. [DOI] [PubMed] [Google Scholar]
- Tapon N, Harvey KF, Bell DW, Wahrer DC, Schiripo TA, Haber D, Hariharan IK. 2002. Salvador promotes both cell cycle exit and apoptosis in Drosophila and is mutated in human cancer cell lines. Cell. 110(4):467–478. [DOI] [PubMed] [Google Scholar]
- Tian W, Yu J, Tomchick DR, Pan D, Luo X. 2010. Structural and functional analysis of the Yap-binding domain of human TEAD2. Proc Natl Acad Sci U S A. 107(16):7293–7298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian Y, Liu Y, Wang T, Zhou N, Kong J, Chen L, Snitow M, Morley M, Li D, Petrenko N, et al. 2015. A microRNA-Hippo pathway that promotes cardiomyocyte proliferation and cardiac regeneration in mice. Sci Transl Med. 7(279):279ra38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tschaharganeh DF, Chen X, Latzko P, Malz M, Gaida MM, Felix K, Ladu S, Singer S, Pinna F, Gretz N, et al. 2013. Yes-associated protein up-regulates Jagged-1 and activates the Notch pathway in human hepatocellular carcinoma. Gastroenterology. 144(7):1530–1542. e12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tschop K, Conery AR, Litovchick L, Decaprio JA, Settleman J, Harlow E, Dyson N. 2011. A kinase shRNA screen links LATS2 and the pRB tumor suppressor. Genes Dev. 25(8):814–830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Udan RS, Kango-Singh M, Nolo R, Tao C, Halder G. 2003. Hippo promotes proliferation arrest and apoptosis in the Salvador/Warts pathway. Nat Cell Biol. 5(10):914–920. [DOI] [PubMed] [Google Scholar]
- Varelas X. 2014. The Hippo pathway effectors Taz and Yap in development, homeostasis and disease. Development. 141(8):1614–1626. [DOI] [PubMed] [Google Scholar]
- Varelas X, Sakuma R, Samavarchi-Tehrani P, Peerani R, Rao BM, Dembowy J, Yaffe MB, Zandstra PW, Wrana JL. 2008. Taz controls Smad nucleocytoplasmic shuttling and regulates human embryonic stem-cell self-renewal. Nat Cell Biol. 10(7):837–848. [DOI] [PubMed] [Google Scholar]
- Varelas X, Samavarchi-Tehrani P, Narimatsu M, Weiss A, Cockburn K, Larsen BG, Rossant J, Wrana JL. 2010. The Crumbs complex couples cell density sensing to Hippo-dependent control of the TGF-β-Smad pathway. Dev Cell. 19(6):831–844. [DOI] [PubMed] [Google Scholar]
- Varelas X, Wrana JL. 2012. Coordinating developmental signaling: novel roles for the Hippo pathway. Trends Cell Biol. 22(2):88–96. [DOI] [PubMed] [Google Scholar]
- Visser S, Yang X. 2010. Lats tumor suppressor: a new governor of cellular homeostasis. Cell Cycle. 9(19):3892–3903. [DOI] [PubMed] [Google Scholar]
- von Gise A, Lin Z, Schlegelmilch K, Honor LB, Pan GM, Buck JN, Ma Q, Ishiwata T, Zhou B, Camargo FD, et al. 2012. Yap1, the nuclear target of Hippo signaling, stimulates heart growth through cardiomyocyte proliferation but not hypertrophy. Proc Natl Acad Sci U S A. 109(7):2394–2399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voorhoeve PM, le Sage C, Schrier M, Gillis AJ, Stoop H, Nagel R, Liu YP, van Duijse J, Drost J, Griekspoor A, et al. 2006. A genetic screen implicates mirna-372 and mirna-373 as oncogenes in testicular germ cell tumors. Cell. 124(6):1169–1181. [DOI] [PubMed] [Google Scholar]
- Wada K, Itoga K, Okano T, Yonemura S, Sasaki H. 2011. Hippo pathway regulation by cell morphology and stress fibers. Development. 138(18):3907–3914. [DOI] [PubMed] [Google Scholar]
- Wang J, Xiao Y, Hsu CW, Martinez-Traverso IM, Zhang M, Bai Y, Ishii M, Maxson RE, Olson EN, Dickinson ME, et al. 2016. Yap and Taz play a crucial role in neural crest-derived craniofacial development. Development. 143(3):504–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williamson KA, Rainger J, Floyd JA, Ansari M, Meynert A, Aldridge KV, Rainger JK, Anderson CA, Moore AT, Hurles ME, et al. 2014. Heterozygous loss-of-function mutations in Yap1 cause both isolated and syndromic optic fissure closure defects. Am J Hum Genet. 94(2):295–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wrighton KH, Dai F, Feng XH. 2008. A new kid on the TGFbeta block: Taz controls Smad nucleocytoplasmic shuttling. Dev Cell. 15(1):8–10. [DOI] [PubMed] [Google Scholar]
- Wu S, Huang J, Dong J, Pan D. 2003. Hippo encodes a Ste-20 family protein kinase that restricts cell proliferation and promotes apoptosis in conjunction with salvador and warts. Cell. 114(4):445–456. [DOI] [PubMed] [Google Scholar]
- Xiao Y, Leach J, Wang J, Martin JF. 2016. Hippo/Yap signaling in cardiac development and regeneration. Curr Treat Options Cardiovasc Med. 18(6):38. [DOI] [PubMed] [Google Scholar]
- Xin M, Kim Y, Sutherland LB, Murakami M, Qi X, McAnally J, Porrello ER, Mahmoud AI, Tan W, Shelton JM, et al. 2013. Hippo pathway effector Yap promotes cardiac regeneration. Proc Natl Acad Sci U S A. 110(34):13839–13844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xin M, Kim Y, Sutherland LB, Qi X, McAnally J, Schwartz RJ, Richardson JA, Bassel-Duby R, Olson EN. 2011. Regulation of insulin-like growth factor signaling by Yap governs cardiomyocyte proliferation and embryonic heart size. Sci Signal. 4(196):ra70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu T, Wang W, Zhang S, Stewart RA, Yu W. 1995. Identifying tumor suppressors in genetic mosaics: the Drosophila lats gene encodes a putative protein kinase. Development. 121(4):1053–1063. [DOI] [PubMed] [Google Scholar]
- Yabuta N, Okada N, Ito A, Hosomi T, Nishihara S, Sasayama Y, Fujimori A, Okuzaki D, Zhao H, Ikawa M, et al. 2007. Lats2 is an essential mitotic regulator required for the coordination of cell division. J Biol Chem. 282(26):19259–19271. [DOI] [PubMed] [Google Scholar]
- Yang X, Yu K, Hao Y, Li DM, Stewart R, Insogna KL, Xu T. 2004. LATS1 tumour suppressor affects cytokinesis by inhibiting LIMK1. Nat Cell Biol. 6(7):609–617. [DOI] [PubMed] [Google Scholar]
- Yimlamai D, Christodoulou C, Galli GG, Yanger K, Pepe-Mooney B, Gurung B, Shrestha K, Cahan P, Stanger BZ, Camargo FD. 2014. Hippo pathway activity influences liver cell fate. Cell. 157(6):1324–1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin F, Yu J, Zheng Y, Chen Q, Zhang N, Pan D. 2013. Spatial organization of Hippo signaling at the plasma membrane mediated by the tumor suppressor Merlin/NF2. Cell. 154(6):1342–1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu FX, Zhao B, Guan KL. 2015. Hippo pathway in organ size control, tissue homeostasis, and cancer. Cell. 163(4):811–828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu FX, Zhao B, Panupinthu N, Jewell JL, Lian I, Wang LH, Zhao J, Yuan H, Tumaneng K, Li H, et al. 2012. Regulation of the Hippo-Yap pathway by G-protein-coupled receptor signaling. Cell. 150(4):780–791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang N, Bai H, David KK, Dong J, Zheng Y, Cai J, Giovannini M, Liu P, Anders RA, Pan D. 2010. The Merlin/NF2 tumor suppressor functions through the Yap oncoprotein to regulate tissue homeostasis in mammals. Dev Cell. 19(1):27–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao B, Wei X, Li W, Udan RS, Yang Q, Kim J, Xie J, Ikenoue T, Yu J, Li L, et al. 2007. Inactivation of Yap oncoprotein by the Hippo pathway is involved in cell contact inhibition and tissue growth control. Genes Dev. 21(21):2747–2761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou D, Conrad C, Xia F, Park JS, Payer B, Yin Y, Lauwers GY, Thasler W, Lee JT, Avruch J, et al. 2009. Mst1 and Mst2 maintain hepatocyte quiescence and suppress hepatocellular carcinoma development through inactivation of the Yap1 oncogene. Cancer Cell. 16(5):425–438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Q, Li L, Zhao B, Guan KL. 2015. The Hippo pathway in heart development, regeneration, and diseases. Circ Res. 116(8):1431–1447. [DOI] [PMC free article] [PubMed] [Google Scholar]