Abstract
The Eda pathway (Eda, Edar, Edaradd) plays an important role in tooth development, determining tooth number, crown shape, and enamel formation. Here we show that the Eda pathway also plays a key role in root development. Edar (the receptor) is expressed in Hertwig’s epithelial root sheath (HERS) during root development, with mutant mice showing a high incidence of taurodontism: large pulp chambers lacking or showing delayed bifurcation or trifurcation of the roots. The mouse upper second molars in the Eda pathway mutants show the highest incidence of taurodontism, this enhanced susceptibility being matched in human patients with mutations in EDA-A1. These taurodont teeth form due to defects in the direction of extension of the HERS from the crown, associated with a more extensive area of proliferation of the neighboring root mesenchyme. In those teeth where the angle at which the HERS extends from the crown is very wide and therefore more vertical, the mutant HERSs fail to reach toward the center of the tooth in the normal furcation region, and taurodont teeth are created. The phenotype is variable, however, with milder changes in angle and proliferation leading to normal or delayed furcation. This is the first analysis of the role of Eda in the root, showing a direct role for this pathway during postnatal mouse development, and it suggests that changes in proliferation and angle of HERS may underlie taurodontism in a range of syndromes.
Keywords: tooth, ectodermal dysplasia, epithelium, periodontium, furcation defects, ectodysplasins
Introduction
The root of a tooth is essential for anchoring the tooth in place. Defects in root size and shape can lead to problems with attachment and can affect the rest of the periodontium; therefore, an understanding of root development is essential. While crown development has been intensely studied, far less research has focused on root development. From mouse knockout studies, a number of signaling pathways and transcription factors have been indicated as being essential for normal root development, such as Shh, Wnt, Bmp, Tgfb, and Nfic (Li et al. 2017). These pathways are involved in complex signaling between the epithelium of the root, known as Hertwig’s epithelial root sheath (HERS), and the overlying preodontoblast neural crest–derived mesenchyme. Loss of Smad4 (acting downstream of Tgfb signaling) in the HERS leads to a failure in all root development (Huang et al. 2010), while loss of Smad4 in the mesenchyme alone leads to shorter roots (Gao et al. 2009). Smad4 appears to act upstream of Shh, as altering Sonic hedgehog signaling can alleviate some of the defects in the Smad4 knockout (Huang et al. 2010), with Shh switching on Nfic in the root mesenchyme and acting in a negative feedback loop (Huang et al. 2010; Liu et al. 2015).
HERS is formed as an extension of the cervical loops at the late bell stage of development. The sheath is formed of 2 layers, at the junction between the outer and inner dental epithelium. In the mouse, this process starts at postnatal day 4 (P4), with extension of this double-layered structure (Lungova et al. 2011). The HERS then extends downward to create the roots, stimulating the surrounding dental papilla mesenchyme to differentiate as odontoblasts and secrete the dentin of the root. In single-rooted teeth, the HERS extends downward in a sheet; however, to generate multiple roots, the HERS must change direction and extend horizontally to create furcations. The creation of these epithelial diaphragms has been proposed to be controlled by proliferation of the adjacent mesenchyme, with higher proliferation pushing the HERS to extend more vertically, while lower proliferation allows the HERS to bend inward to divide the roots (Sohn et al. 2014). In keeping with this finding, altered proliferation in the presumptive bifurcation regions have been identified in mouse mutants with root defects (Kim et al. 2015).
The Eda pathway has not been investigated in any detail during root development. The pathway consists of the ligand Ectodysplasin (Eda), the receptor (Edar), a tumor necrosis factor family member, and intracellular adapter protein (Edaradd). Eda is cleaved to form a soluble ligand that can then bind to Edar; it recruits Edaradd and ultimately stimulates signaling through NF-κB (Courtney et al. 2005). During early development, Eda is expressed in the forming dental placodes, with smaller placodes forming in Eda mutants (known as Tabby mutants; Pispa et al. 1999). Intriguingly, these mice can have supernumerary teeth formed from revitalization of diastema tooth buds, so hypodontia and hyperdontia can both be features in the mouse (Charles, Pantalacci, Tafforeau, et al. 2009; Prochazka et al. 2010). At the cap stage, the Eda pathway plays a crucial role in molar crown formation, with pathway mutants having molars with a reduced number of flattened cusps (Grüneberg 1966; Pispa et al. 1999; Tucker et al. 2000). This has been shown to be due to defects in the primary enamel knot at the cap stage of development (Pispa et al. 1999; Tucker et al. 2000; Ohazama et al. 2004). At the cap stage, Eda is expressed in the dental epithelium close to the oral epithelium, while Edar and Edaradd are expressed in the enamel knot (Tucker et al. 2000; Headon et al. 2001; Laurikkala et al. 2001).
Defects in these 3 components of the Eda pathway in patients leads to hypohidrotic ectodermal dysplasia (HED), which is characterized by defects in many ectodermally derived organs: skin, hair, sebaceous glands, sweat glands, and teeth (Kere et al. 1996; Monreal et al. 1999; Headon et al. 2001). HED is more common in males, as Eda is located on the X chromosome, so hemizygous males display the full phenotype. Heterozygous females with mutations in Eda have much milder symptoms with increased incidence of tooth agenesis (Lexner et al. 2007). Similar to the mouse mutants, patients display hypodontia (multiple missing teeth) and smaller teeth and have reduced numbers of cusps, producing peg-shaped teeth (Crawford et al. 1991). In addition, patients with X-linked HED (XL-HED) have root defects, including taurodontism, suggesting that the Eda pathway has an important role during root development (Lexner et al. 2007). Taurodont teeth have roots that fail to bifurcate or that bifurcate very late during root formation, with the result that the pulp chamber is very large at the expense of the roots. In the Eda mutant mouse (Tabby), variations in root pattern have been observed, with high variation in number of roots and possible cusp fusions, which are not always correlated with the size of the tooth crown (Grüneberg 1966; Charles, Pantalacci, Peterkova, et al. 2009).
A direct role for the Eda pathway in root development was suggested by the fact that Edar was identified, in a screen comparing molars and incisors in the rat, as a possible root-determining gene, with other important root genes, such as Nfic (Xing et al. 2007). We therefore decided to follow these leads and investigate root development in Eda and Edar mutant mice and compare our findings with patients having mutations in EDA-A1 (XL-HED).
Methods
Patient Scans
Dental panoramic tomography of 20 anonymized patients with confirmed EDA-A1 mutations were obtained from the hypodontia clinic at Guy’s Hospital London. Data from 15 patients (aged 6 to 16 y) were ultimately used, with the rest excluded because the roots of the permanent molars had not developed yet. Data were anonymized with only age and sex provided. The project was registered with the research and development department at Guy’s and St Thomas’s Hospital Trust.
Mice
Eda and Edar mutants were mated as previously described at The Roslin Institute (Wells et al. 2010) and in the Czech republic (Peterkova et al. 2005). Pups and adult mice were culled with schedule 1 methods as approved by the Home Office, UK. This project conforms with the ARRIVE (Animal Research: Reporting of In Vivo Experiments) guidelines.
MicroCT
Mice were scanned with a GE Locus SP micro–computed tomography (microCT) scanner and a Scanco microCT 50 scanner. Classification of teeth based on extent of taurodontism was determined with 2-dimensional planes and 3-dimensional reconstructions of individual teeth, generated with Microview software.
Histology, In Situ Hybridization, and Immunofluorescence
Wax sections were used for analysis of gene expression, protein expression, and histology (see Appendix Methods).
Results
Edar Is Expressed in HERS
Although the Eda pathway has been followed during tooth development in a number of prenatal stages, postnatal expression in the root has not been investigated. Our first step, therefore, was to assess where Eda signaling (if any) was occurring in the postnatal tooth. As Eda is a soluble ligand, we concentrated on the expression of the receptor Edar, which has a very restricted expression pattern in the enamel knot of the tooth at the cap stage (Fig. 1A, C). Edar expression, mRNA and protein, was found to be restricted to the bilayered HERS during root development, overlapping with expression of the epithelial marker E-cadherin (Fig. 1B, D, E). The restriction of expression to the epithelium agrees with expression of Edar in other ectodermal organs and suggests a direct role for this pathway in root formation. Expression levels were similar in all molar teeth.
Figure 1.
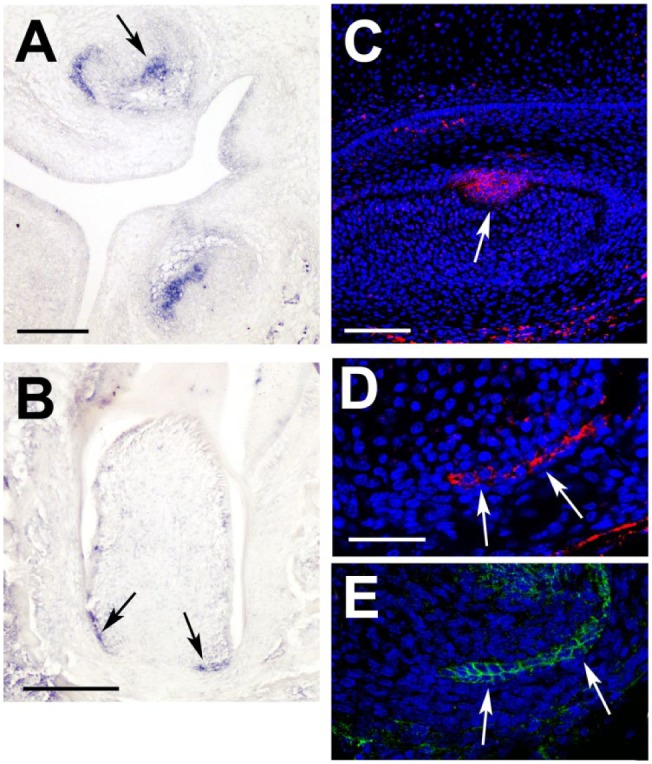
Edar is expressed in Hertwig’s epithelial root sheath (HERS) during root development. (A, B) In situ hybridization for Edar, with positive stain in blue. (A) Embryonic day 15.5 late cap stage tooth. Expression of Edar is observed in the dental epithelium, in the enamel knot (arrow), and spreading out across the inner dental epithelium, particularly on the lingual side of the tooth. (B) At postnatal day 9, Edar is expressed in the HERS (arrows) at the apical end of the tooth. (C, D) Immunofluorescence for Edar. Signal in red; nuclei labeled by DAPI (blue). (C) High-power view of the primary enamel knot (arrow) at embryonic day 15.0. (D) High power of HERS (arrows) at postnatal day 9, at the apical end of the developing tooth. (E) Serial section to panel D showing immunofluorescence for E-cadherin, a marker of epithelium overlapping with Edar in the HERS (arrows). Scale in A, B: 200 µm. C: 100 µm. D: 50 µm (same scale in E).
Taurodontism and Delayed Bifurcation of Roots in Eda Pathway Mutants
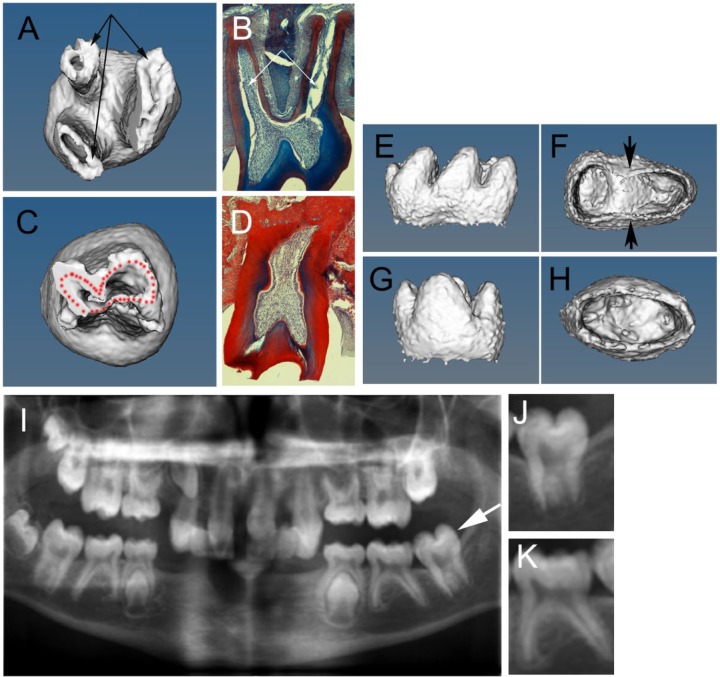
Having shown that Edar is expressed in the roots, we investigated the root defect in Eda (Tabby) and Edar (downless) mutants with microCT to produce 3-dimensional images of the molar teeth. In the mouse, the first and second lower molars (M1 and M2) have 2 roots, while the upper M1 and M2 have 3 roots, similar to the situation observed in humans. As previously reported, Eda mutants had diverse root defects, including a reduced number of roots and, in a few rare cases, an increased number of roots. Eda and Edar mutants (≥2 wk) both displayed a high incidence of taurodontism, with single roots and large pulp chambers, or delay in bifurcation, with additional variation in the length of the final root (Fig. 2A–D and data not shown). In the literature, several classifications for the degree of taurodontism in humans have been described, all with problems associated with how to accurately measure the extent of the delay in bifurcation (Jafarzadeh et al. 2008). Here we used a simple system whereby we divided taurodont teeth into those with no bifurcation (known clinically as hypertaurodont, single, or pyramidal depending on the shape of the root) and delayed bifurcation (known clinically as meso- or hypotaurodontism depending on the position of the bifurcation; n = 17 Edar-/- mice analyzed, with a total of 34 M1s and 34 M2s). From our analysis, the incidence of taurodontism was found to be higher in the upper jaw when compared with the lower jaw, with the second molars (M2) being more susceptible than the first molars (M1; Table). In total, 24.2% of upper M2s showed delayed bifurcation, with another 53% showing no bifurcation. This was in comparison with the lower M2s, which showed only a 6% incidence of delayed bifurcation and a 3% incidence of single-rooted teeth. Very few of the Edar-/- lower M1s showed a taurodont phenotype; however, the Eda mutants showed a higher incidence of taurodontism in the lower jaw (data not shown), agreeing with previous studies that demonstrated subtle differences between these 2 mutants (Charles, Pantalacci, Tafforeau, et al. 2009). At earlier stages (P9), when the roots are still growing, microCT of Eda mutants revealed early defects in the formation of the roots, with some mutant teeth having an oval apical end when compared with the figure-of-8 pattern in wild-type (WT) lower molars (Fig. 2E–H).
Figure 2.
Eda mouse mutants and patients with EDA-A1 mutations have a taurodont phenotype. (A, C, E–H) Micro–computed tomography 3-dimensional reconstructions of Eda mutant molar mice teeth. (B, D) Trichrome-stained histology sections through adult erupted second molar. (A–D) Six-month-old adult teeth. (E–H) Postnatal day 9 teeth. (A, B, E, F) Wild-type teeth. (C, D, G, H) Eda mutant molars. (A, B) Wild-type upper molar with 3 roots (arrows); 2 roots are visible in panel B. (C, D) Taurodont upper molar in Eda mutant with a single root and large pulp cavity. (E, F) Wild-type lower molar forming 2 roots with a figure-of-8 shape on the apical view (F). Arrows point to forming furcations. (G, H) Eda lower molar with an oval shape on the apical view (H) showing no signs of forming a furcation. (I) Digitalized dental panoramic tomography x-ray of a 10-y-old with X-linked hypohidrotic ectodermal dysplasia caused by a mutation in EDA-A1 with variable expression of the taurodont phenotype in addition to loss of multiple teeth. (J) High power of molar tooth highlighted by arrow in panel I displaying a taurodont phenotype. (K) Neighboring tooth with bifurcated root.
Table.
Incidence of Root Defects in Patients with Mutations in EDA-A1 and in Edar Mutant Mice.
| Upper Molars |
Lower Molars |
|||||
|---|---|---|---|---|---|---|
| M1 | M2 | Total | M1 | M2 | Total | |
| Mice | ||||||
| Delay bifurcation | 4.55 | 24.2 | 14.4 | 1.5 | 6.1 | 3.8 |
| Hypertaurodont and single root | 9.1 | 53 | 31.1 | 0 | 3 | 1.5 |
| Patients | ||||||
| Delay bifurcation | 44.8 | 37.5 | 43 | 0 | 18.2 | 4.5 |
| Hypertaurodont, single root, pyramidal | 51.7 | 62.5 | 54.1 | 16.7 | 36.7 | 20.5 |
Values are presented in percentages.
Conserved Susceptibility of Upper Molars in Human Patients
Having established that the incidence of taurodontism in Eda pathway mutants was very dependent on tooth type and position, we decided to investigate whether similar susceptibilities were also observed in human patients with mutations in EDA-A1. As previously described, patients with XL-HED (and, therefore, EDA-A1 mutations) have been shown to have a high incidence of taurodontism (Crawford et al. 1991; Lexner et al. 2007). No difference in incidence of taurodontism was referred to between upper and lower teeth. Digitalized dental panoramic tomography scans of 15 patients with EDA-A1 mutations were collected from Guy’s Hospital (Fig. 2I, J, K). Use of such scans has been shown to be a reliable method of detecting taurodontism in patients (Tulensalo et al. 1989). The molars of upper and lower jaws were examined, and the prevalence of taurodontism was calculated. As with the mice, we divided the teeth into those that showed hypertaurodontism, single, or pyramidal roots and those that showed delayed bifurcation (meso- and hypotaurodont). Overall, the patients with EDA-A1 mutations had a high incidence of taurodontism (Table). Similar to the mouse, the upper molars had a higher incidence of hypertaurodont, single, and pyramidal roots (54%) and delayed bifurcation (43%) as compared with the lower molars: 20.5% and 4.5%, respectively. Second molars also had a higher incidence of root defects than first molars in the lower jaw, although in the upper jaw, the incidence of defects was similar in M1 and M2. In general, therefore, the pattern of incidence was conserved across our mouse mutants and human patients.
HERS Morphology Is Disrupted in Eda Pathway Mutant Mice
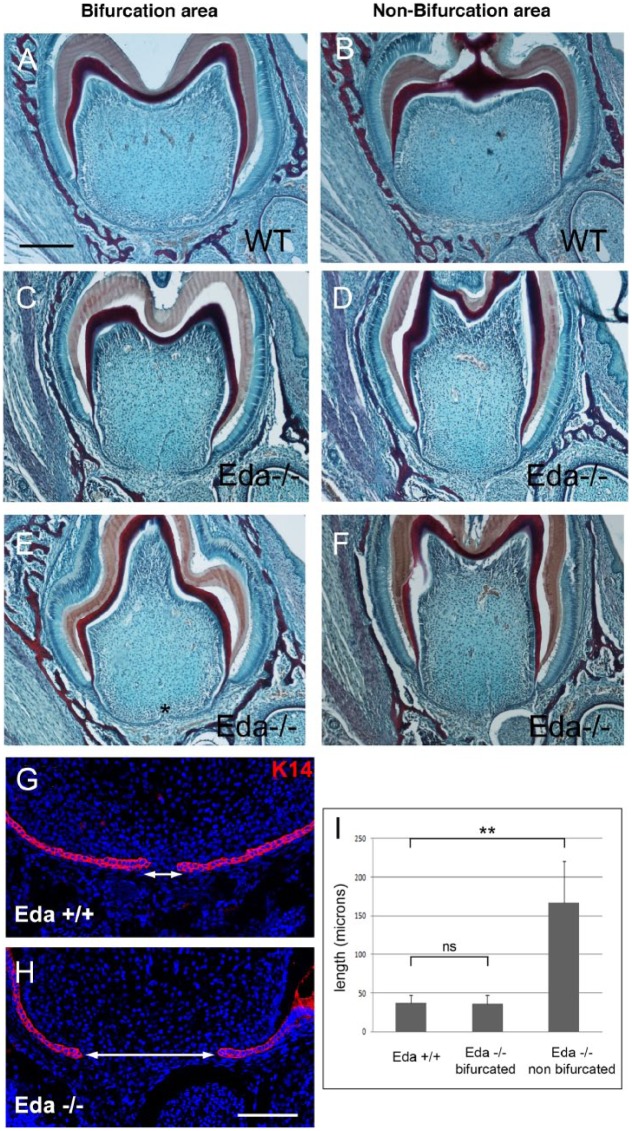
Having established that root defects are a common feature of Eda pathway mutants and XL-HED patients, we aimed to investigate the underlying mechanism behind the defect. For this, Eda mutant mice were chosen due to their slightly higher incidence of taurodontism when compared with Edar mutants, with analysis focused on the lower first molar, to allow a direct comparison of root development in mutant teeth with taurodont and bifurcated phenotypes. The simpler root pattern of the lower molars allowed for a clear selection of bifurcating and nonbifurcating regions in frontal sections, not possible in the trifurcating upper molars. As the phenotype was already evident by microCT at P9 (Fig. 2E–H), we turned to P7, when little root dentin has been laid down. In sections, it was clear that all the mutant teeth were narrower than the WTs (Fig. 3A, B vs. C–F), agreeing with the smaller overall size of Eda teeth (Charles, Pantalacci, Peterkova, et al. 2009). In WTs, the buccal and lingual sides of the HERS in the center of the tooth almost contacted each other at this stage (Fig. 3A), while they remained far apart and were shorter on either side of the furcation region (Fig. 3B; n = 3 of 3). In comparison, the Eda mutants had a highly variable phenotype, with some displaying shorter HERSs and a large distance between the 2 sides of the HERS throughout the tooth (Fig. 3C, D, I; n = 5 of 8). In contrast, in some mutant molars, the 2 sides of the HERS almost made contact in the middle, in a manner similar to WTs (n = 3 of 8; Fig. 3E, F, I). The morphology of the HERS was investigated with Keratin 14, highlighting a morphologically normal-looking bilayered structure in all the mutants (Fig. 3G, H).
Figure 3.
Hertwig’s epithelial root sheath (HERS) extension defects in Eda mutant mice at postnatal day 7. (A–F) Trichrome-stained frontal sections in the center of the tooth (bifurcation region; A, C, E) and outside the normal furcation region (B, D, F) in the lower first molar. (A, B) Wild type (WT). (C–F) Eda mutants. *Point where the 2 sides of the HERS have almost met in the mutant. (G, H) Keratin 14 expression in the bifurcation region in WT (G) and mutant (H) HERSs. Arrows indicate gap between the lingual and buccal sides of the HERS. (I) Graph showing Eda mutants can be divided into 2 groups dependent on the distance between the 2 sides of the HERS at the center of the tooth. Scale in A–F: 200 μm. G, H: 100 μm. *P < 0.05. **P < 0.01. Values are presented as mean ± SD. ns, not significant.
Altered Direction of the HERS and Proliferation in Eda Mutants
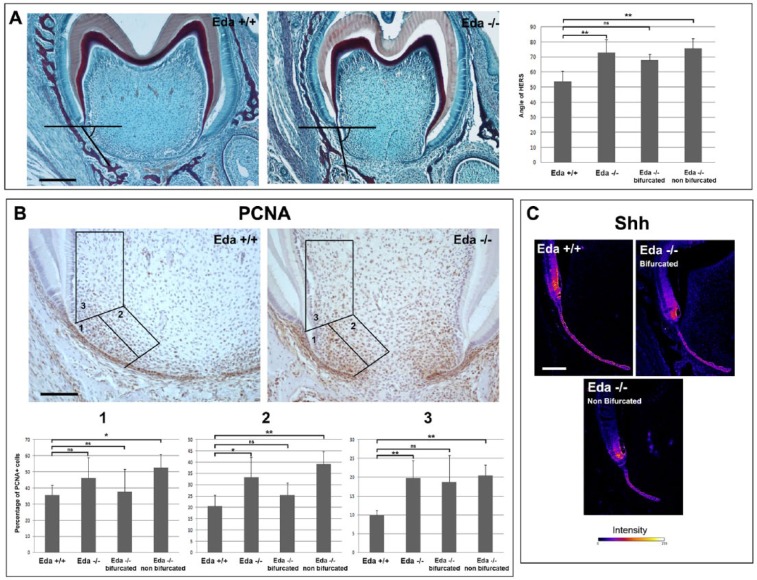
The mutant teeth appeared more elongated with alterations in the direction that the HERS extended from the crown (Fig. 3). We therefore analyzed the angle of extension in the bifurcation region (Fig. 4A) and found that the mutants had significantly wider angles, with the HERS extending more vertically (Fig. 4A). Fitting with the variable incidence of taurodontism, those mutant teeth where the buccal and lingual sides of the HERS had managed to almost reach the midline at the bifurcation region had smaller angles that were not significantly different from the WTs (Fig. 4A). In contrast, the Eda mutants with distantly spaced HERSs had much larger and therefore more vertical HERSs (Fig. 4A). In Nfic mutants, root furcation defects have been linked to changes in proliferation of the mesenchyme adjacent to the HERS (Kim et al. 2015), and differential proliferation has been suggested to influence the direction of HERS in WT roots (Sohn et al. 2014). We therefore investigated proliferation in our Eda mutants versus WTs at P7 (n = 8 Eda mutant teeth, 3 WT teeth), counting in 3 regions associated with the HERS in the center of the tooth (Fig. 4B). In all 3 mesenchymal zones counted, a larger percentage of proliferating cells were observed in those mutants that failed to form a bifurcating region (Fig. 4B). In contrast, the mutants that had managed to make a bifurcation had close to WT levels of proliferation (Fig. 4B). This suggests that the difference in proliferation may drive the HERS phenotype. The epithelial HERSs themselves had high levels of proliferation, matching that of the WT, although they appeared shorter in the center of the tooth.
Figure 4.
Proliferation and angle defects in Eda mutant mice. (A) Angle defects in the lower first molar in Eda mutant mice at postnatal day 7. Histology figures showing how angle of extension was measured with a sample wild-type (WT) and Eda mutant showing the center (normally bifurcating) part of the tooth. Graph showing statistically significant changes in angle only for those mutants with defects in formation of a bifurcation when compared with WT. Scale bar in A: 200 μm. (B) Proliferation measured by proliferating cell nuclear antigen (PCNA) in Eda mutant mice at postnatal day 7. Three boxes (1, 2, 3) were used to measure the numbers of proliferating cells to be compared with all cells in the area. Sections showing proliferation (brown) in an example WT and Eda mutant. Graphs showing the difference in percentage proliferating cells in the 3 regions shown in the images. Only mutants with a defect in bifurcation showed a significant difference in proliferation versus WTs. Scale in B: 100 μm. (C) Shh immunofluorescence. Confocal images with “fire luts” where blue is lowest intensity and white the highest. The expression of Shh in the Hertwig’s epithelial root sheath itself appears similar between WT and Eda mutant molars. Scale in C: 100 μm. *P < 0.05. **P < 0.01. Values are presented as mean ± SD. ns, not significant.
As the Edar pathway is active in the HERS (Fig. 1), this change in mesenchymal proliferation suggests that the HERSs signal back to the mesenchyme to control proliferation levels and, thereby, the direction of root development. We therefore analyzed expression of Shh, a key signaling factor expressed in the HERS that is known to signal to the mesenchyme (Khan et al. 2007). Shh has also been shown to act downstream of the Eda pathway in the skin and salivary glands (Pummila et al. 2007; Wells et al. 2010; Wells et al. 2011). Shh was expressed in the HERS and in the preameloblasts in the WT, and a very similar pattern was observed in the Eda mutant teeth at P7, although levels in the preameloblasts were potentially reduced (Fig. 4C). There was no clear difference in those teeth with a taurodont or normal bifurcation however, suggesting that this reduction does not drive the phenotype.
Discussion
Here we show for the first time that a component of the Eda pathway, Edar, is expressed during root development with a very specific expression in the developing HERS. Loss of Eda or Edar leads to a taurodont phenotype, with the upper second molars being the most sensitive to loss of this pathway. Fitting with the variable phenotype in adults, during root development, Eda mutant molars could be divided into those with normal proliferation, HERS angle, and formation of a bifurcation and those where proliferation was increased in the mesenchyme and the HERS extended in a more vertical direction. Changes in proliferation therefore appear to drive the phenotype, agreeing with the hypothesis that uniform high proliferation throughout the root mesenchyme leads to a lack of furcation (Kim et al. 2015). It would be interesting therefore to assess whether uniform proliferation underlies taurodontism in other syndromes and mutant mice.
As Edar was expressed in the HERS, this suggests another signaling factor produced by the HERS was altered in the absence of active Eda signaling. One candidate linked to the Eda pathway in other organs was Shh; however, no clear change in Shh in the HERS was observed, although there might be subtle differences in levels of expression. Another candidate could be a member of the Wnt signaling pathway, as Axin2, a target of Wnt signaling, is highly expressed in the mesenchyme adjacent to the HERS (Lohi et al. 2010) and loss and activation of the Wnt pathway lead to root defects (Bae et al. 2013; Kim et al. 2013). A taurodont phenotype is observed in patients and mice with mutations in Wnt10a, and loss of Wnt10a leads to defects in cusp pattern and hypodontia, mirroring many aspects of the Eda pathway mutant phenotype (Yang et al. 2015). In keeping with this, Wnt10a has been predicted to be a direct target of NF-κB signaling (Krappmann et al. 2004).
Our comparison of the ratio of taurodontism between human and mutant mice revealed a close similarity in the distribution of taurodontism. In both samples, the prevalence of taurodontism was higher in the upper teeth versus the lower teeth. This result is consistent with the findings that many tooth anomalies, such as taurodontism, hypodontia, and dens invagination, are more common in the upper jaws than the lower jaws (MacDonald-Jankowski and Li 1993; Shokri et al. 2014). These differences in susceptibility may be influenced by the larger number of roots that form in the upper molars in mice and humans as compared with lower molars, involving a more complex pattern of folding of the epithelium. These more complex folding patterns may be more vulnerable to changes in signaling molecules and subtle changes in proliferation. Taurodontism in general, rather than in HED patients only, is also more prevalent in second molars versus first molars (MacDonald-Jankowski and Li 1993). As the number of roots is the same in the first and second molars, the pattern of furcation cannot explain this difference in prevalence; however, the time difference between the development of first and second molars may influence the incidence of the root defect, with later-developing teeth being more susceptible.
In conclusion, we have shown that the Eda pathway has a direct role in root development, influencing proliferation and the angle of HERS and, therefore, the ability to form furcations. This pathway can thus be added to the other signaling pathways (Shh, Wnt, Tgfb, BMP) whose role in root development is just starting to be elucidated (Li et al. 2017).
Author Contributions
J.M. Fons Romero, R. Lav, contributed to data acquisition, analysis, and interpretation, critically revised the manuscript; H. Star, contributed to data acquisition, analysis, and interpretation, drafted the manuscript; S. Watkins, M. Harrison, M. Hovorakova, D. Headon, contributed to data acquisition and interpretation, critically revised the manuscript; A.S. Tucker, contributed to conception, design, and data interpretation, drafted the manuscript.
Supplementary Material
Acknowledgments
Thanks to Chris Healy for help with microCT, funded by a multiuser equipment grant from the Wellcome Trust (202852/Z/16/Z). Thanks to Max Smith for angle measurements.
Footnotes
A supplemental appendix to this article is available online.
H. Star was supported by the Ministry of Higher Education and Scientific Research, Kurdistan, Iraq, and by the Dental Institute of King’s College London. R. Lav and J.M. Fons Romero were funded by the Wellcome Trust (102889/Z/13/Z). M. Hovorakova was funded by the grant agency of the Czech Republic (14-37368G). D. Headon is supported by the Biotechnology and Biological Sciences Research Council’s Institute Strategic Programme (grant BB/P013732/1 to the Roslin Institute).
The authors declare no potential conflicts of interest with respect to the authorship and/or publication of this article.
References
- Bae CH, Lee JY, Kim TH, Baek JA, Lee JC, Yang X, Taketo MM, Jiang R, Cho ES. 2013. Excessive Wnt/beta-catenin signaling disturbs tooth-root formation. J Periodontal Res. 48(4):405–410. [DOI] [PubMed] [Google Scholar]
- Charles C, Pantalacci S, Peterkova R, Tafforeau P, Laudet V, Viriot L. 2009. Effect of eda loss of function on upper jugal tooth morphology. Anat Rec (Hoboken). 292(2):299–308. [DOI] [PubMed] [Google Scholar]
- Charles C, Pantalacci S, Tafforeau P, Headon D, Laudet V, Viriot L. 2009. Distinct impacts of Eda and Edar loss of function on the mouse dentition. PLoS One. 4(4):e4985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Courtney JM, Blackburn J, Sharpe PT. 2005. The Ectodysplasin and NFkappaB signalling pathways in odontogenesis. Arch Oral Biol. 50(2):159–163. [DOI] [PubMed] [Google Scholar]
- Crawford PJ, Aldred MJ, Clarke A. 1991. Clinical and radiographic dental findings in X linked hypohidrotic ectodermal dysplasia. J Med Genet. 28(3):181–185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Y, Yang G, Weng T, Du J, Wang X, Zhou J, Wang S, Yang X. 2009. Disruption of Smad4 in odontoblasts causes multiple keratocystic odontogenic tumors and tooth malformation in mice. Mol Cell Biol. 29(21):5941–5951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grüneberg H. 1966. The molars of the tabby mouse, and a test of the “single-active X-chromosome” hypothesis. J Embryol Exp Morphol. 15(2):223–244. [PubMed] [Google Scholar]
- Headon DJ, Emmal SA, Ferguson BM, Tucker AS, Justice MJ, Sharpe PT, Zonana J, Overbeek PA. 2001. Gene defect in ectodermal dysplasia implicates a death domain adapter in development. Nature. 414(6866):913–916. [DOI] [PubMed] [Google Scholar]
- Huang X, Xu X, Bringas P, Jr, Hung YP, Chai Y. 2010. Smad4-Shh-Nfic signaling cascade-mediated epithelial-mesenchymal interaction is crucial in regulating tooth root development. J Bone Miner Res. 25(5):1167–1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jafarzadeh H, Azarpazhooh A, Mayhall JT. 2008. Taurodontism: a review of the condition and endodontic treatment challenges. Int Endod J. 41(5):375–388. [DOI] [PubMed] [Google Scholar]
- Kere J, Srivastava AK, Montonen O, Zonana J, Thomas N, Ferguson B, Munoz F, Moran D, Clarke A, Baybayan P, et al. 1996. X-linked anhidrotic (hypohidrotic) ectodermal dysplasia is caused by mutation in a novel transmembrane protein. Nat Genet. 13(4):409–416. [DOI] [PubMed] [Google Scholar]
- Khan M, Seppala M, Zoupa M, Cobourne MT. 2007. Hedgehog pathway gene expression during early development of the molar tooth root in the mouse. Gene Expr Patterns. 7(3):239–243. [DOI] [PubMed] [Google Scholar]
- Kim TH, Bae CH, Lee JC, Ko SO, Yang X, Jiang R, Cho ES. 2013. β-catenin is required in odontoblasts for tooth root formation. J Dent Res. 92(3):215–221. [DOI] [PubMed] [Google Scholar]
- Kim TH, Bae CH, Yang S, Park JC, Cho ES. 2015. Nfic regulates tooth root patterning and growth. Anat Cell Biol. 48(3):188–194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krappmann D, Wegener E, Sunami Y, Esen M, Thiel A, Mordmuller B, Scheidereit C. 2004. The IkappaB kinase complex and NF-kappaB act as master regulators of lipopolysaccharide-induced gene expression and control subordinate activation of AP-1. Mol Cell Biol. 24(14):6488–6500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laurikkala J, Mikkola M, Mustonen T, Aberg T, Koppinen P, Pispa J, Nieminen P, Galceran J, Grosschedl R, Thesleff I. 2001. TNF signaling via the ligand-receptor pair ectodysplasin and edar controls the function of epithelial signaling centers and is regulated by Wnt and activin during tooth organogenesis. Dev Biol. 229(2):443–455. [DOI] [PubMed] [Google Scholar]
- Lexner MO, Bardow A, Hertz JM, Nielsen LA, Kreiborg S. 2007. Anomalies of tooth formation in hypohidrotic ectodermal dysplasia. Int J Paediatr Dent. 17(1):10–18. [DOI] [PubMed] [Google Scholar]
- Li J, Parada C, Chai Y. 2017. Cellular and molecular mechanisms of tooth root development. Development. 144(3):374–384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Feng J, Li J, Zhao H, Ho TV, Chai Y. 2015. An Nfic-hedgehog signaling cascade regulates tooth root development. Development. 142(19):3374–3382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohi M, Tucker AS, Sharpe PT. 2010. Expression of Axin2 indicates a role for canonical Wnt signaling in development of the crown and root during pre- and postnatal tooth development. Dev Dyn. 239(1):160–167. [DOI] [PubMed] [Google Scholar]
- Lungova V, Radlanski RJ, Tucker AS, Renz H, Misek I, Matalova E. 2011. Tooth-bone morphogenesis during postnatal stages of mouse first molar development. J Anat. 218(6):699–716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacDonald-Jankowski DS, Li TT. 1993. Taurodontism in a young adult Chinese population. Dentomaxillofac Radiol. 22(3):140–144. [DOI] [PubMed] [Google Scholar]
- Monreal AW, Ferguson BM, Headon DJ, Street SL, Overbeek PA, Zonana J. 1999. Mutations in the human homologue of mouse dl cause autosomal recessive and dominant hypohidrotic ectodermal dysplasia. Nat Genet. 22(4):366–369. [DOI] [PubMed] [Google Scholar]
- Ohazama A, Courtney JM, Tucker AS, Naito A, Tanaka S, Inoue J, Sharpe PT. 2004. Traf6 is essential for murine tooth cusp morphogenesis. Dev Dyn. 229(1):131–135. [DOI] [PubMed] [Google Scholar]
- Peterkova R, Lesot H, Viriot L, Peterka M. 2005. The supernumerary check tooth in tabby/EDA mice—a reminiscence of the premolar in mouse ancestors. Arch Oral Biol. 50(2):219–225. [DOI] [PubMed] [Google Scholar]
- Pispa J, Jung HS, Jernvall J, Kettunen P, Mustonen T, Tabata MJ, Kere J, Thesleff I. 1999. Cusp patterning defect in Tabby mouse teeth and its partial rescue by FGF. Dev Biol. 216(2):521-534. [DOI] [PubMed] [Google Scholar]
- Prochazka J, Pantalacci S, Churava S, Rothova M, Lambert A, Lesot H, Klein O, Peterka M, Laudet V, Peterkova R. 2010. Patterning by heritage in mouse molar row development. Proc Natl Acad Sci U S A. 107(35):15497–15502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pummila M, Fliniaux I, Jaatinen R, James MJ, Laurikkala J, Schneider P, Thesleff I, Mikkola ML. 2007. Ectodysplasin has a dual role in ectodermal organogenesis: inhibition of Bmp activity and induction of Shh expression. Development. 134(1):117–125. [DOI] [PubMed] [Google Scholar]
- Shokri A, Poorolajal J, Khajeh S, Faramarzi F, Kahnamoui HM. 2014. Prevalence of dental anomalies among 7- to 35-year-old people in Hamadan, Iran in 2012-2013 as observed using panoramic radiographs. Imaging Sci Dent. 44(1):7–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sohn WJ, Choi MA, Yamamoto H, Lee S, Lee Y, Jung JK, Jin MU, An CH, Jung HS, Suh JY, et al. 2014. Contribution of mesenchymal proliferation in tooth root morphogenesis. J Dent Res. 93(1):78–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tucker AS, Headon DJ, Schneider P, Ferguson BM, Overbeek P, Tschopp J, Sharpe PT. 2000. Edar/Eda interactions regulate enamel knot formation in tooth morphogenesis. Development. 127(21):4691–4700. [DOI] [PubMed] [Google Scholar]
- Tulensalo T, Ranta R, Kataja M. 1989. Reliability in estimating taurodontism of permanent molars from orthopantomograms. Community Dent Oral Epidemiol. 17(5):258–262. [DOI] [PubMed] [Google Scholar]
- Wells KL, Mou C, Headon DJ, Tucker AS. 2010. Recombinant EDA or Sonic Hedgehog rescue the branching defect in Ectodysplasin A pathway mutant salivary glands in vitro. Dev Dyn. 239(10):2674–2684. [DOI] [PubMed] [Google Scholar]
- Wells KL, Mou C, Headon DJ, Tucker AS. 2011. Defects and rescue of the minor salivary glands in Eda pathway mutants. Dev Biol. 349(2):137–146. [DOI] [PubMed] [Google Scholar]
- Xing X, Deng Z, Yang F, Watanabe S, Wen L, Jin Y. 2007. Determination of genes involved in the early process of molar root development initiation in rat by modified subtractive hybridization. Biochem Biophys Res Commun. 363(4):994–1000. [DOI] [PubMed] [Google Scholar]
- Yang J, Wang SK, Choi M, Reid BM, Hu Y, Lee YL, Herzog CR, Kim-Berman H, Lee M, Benke PJ, et al. 2015. Taurodontism, variations in tooth number, and misshapened crowns in Wnt10a null mice and human kindreds. Mol Genet Genomic Med. 3(1):40–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.