Abstract
Nonsyndromic cleft lip and/or palate (NSCL/P) is a prevalent birth defect of complex etiology. Previous studies identified mutations in ARHGAP29 associated with an increased risk for NSCL/P. To investigate the effects of ARHGAP29 in vivo, we generated a novel murine allele by inserting a point mutation identified in a patient with NSCL/P. This single-nucleotide variation of ARHGAP29 translates to an early nonsense mutation (K326X), presumably resulting in loss-of-function (LoF). Embryos from Arhgap29K326X/+ intercrosses were harvested at various time points. No homozygous Arhgap29K326X animals were found in the 45 analyzed litters, assessed as early as embryonic day 8.5 (e8.5). Coronal sectioning of e13.5 and e14.5 heads revealed that 59% of Arhgap29K326X/+ mice (n = 37) exhibited improper epithelial contact between developing oral structures, while none were observed in wild types (n = 10). In addition, Arhgap29K326X/+ embryos exhibited a significantly higher percentage of maxillary epithelium in contact with mandibular epithelium. Immunofluorescent analyses of the periderm and oral adhesions revealed the presence of Arhgap29 in periderm cells. These cells were p63 negative, keratin 17 positive, and keratin 6 positive and present at sites of adhesion, although occasionally disorganized. Oral adhesions did not appear to impair palatogenesis, as all analyzed Arhgap29K326X/+ embryos showed confluent palatal mesenchyme and epithelium at e18.5 (n = 16), and no mice were found with a cleft at birth. Collectively, our data demonstrate that ARHGAP29 is required for embryonic survival and that heterozygosity for LoF variants of Arhgap29 increases the incidence and length of oral adhesions at a critical time point during orofacial development. In conclusion, we validate the LoF nature of the human K326X mutation in vivo and reveal a previously unknown effect of Arhgap29 in murine craniofacial development.
Keywords: cleft palate, cleft lip, embryonic development, congenital abnormalities, genome-wide association study, mice
Introduction
Cleft lip and/or palate (CL/P) occurs in 1 in 1,000 live births worldwide, making it one of the most common birth defects (Beaty et al. 2016). CL/P significantly contributes to morbidity and affects social interaction and psychological well-being throughout childhood (Dixon et al. 2011). A third of CL/P cases are syndromic, while the remaining CL/P cases present with no other apparent malformations and are referred to nonsyndromic (NSCL/P; Leslie and Marazita 2013). The etiology of orofacial clefting is complex, drawing from both genetic and environmental factors. A major challenge of NSCL/P research is elucidating the contribution of genetic variants and how they interact to lead to craniofacial anomalies (Dixon et al. 2011; Leslie and Marazita 2013).
Recent advancements in CL/P research have identified numerous genetic loci associated with increased risk for NSCL/P (Beaty et al. 2016). One of the first genome-wide association studies for NSCL/P discovered 4 significant loci: IRF6, 8q24, MAFB, and ABCA4 (Beaty et al. 2010). These loci were confirmed in multiple follow-up studies in different populations (Yuan et al. 2011; Fontoura et al. 2012; Lennon et al. 2012; Beaty et al. 2013; Butali et al. 2014; Letra et al. 2014; Gowans et al. 2016; Leslie et al. 2016). A function for IRF6 and the 8q24 region in orofacial clefting has been previously described (Kondo et al. 2002; Uslu et al. 2014), but the roles of the remaining 2 loci in the genes ABCA4 and MAFB are less understood.
ABCA4 was ruled out as the etiologic gene for NSCL/P at the 1p22 locus because of its lack of expression in appropriate tissue and absence of significant genetic association with cases (Beaty et al. 2010). However, ARHGAP29 (encoding Rho GTPase activating protein 29), a nearby gene, is expressed in the palatal shelves and oral epithelium during craniofacial development. Furthermore, sequencing of ARHGAP29 in individuals with NSCL/P revealed rare coding sequence variants compared with controls (Leslie et al. 2012). Therefore, it was concluded that ARHGAP29 is the etiologic gene at this locus (Leslie et al. 2012). Subsequent studies identified functional noncoding variants in this region, affecting binding of CL/P-associated transcription factors (Liu et al. 2017). Further sequencing of ARHGAP29 yielded rare loss-of-function (LoF) variants in cases and some unaffected relatives, suggesting that 1 copy of ARHGAP29 LoF confers a moderate risk for NSCL/P (Savastano et al. 2017).
ARHGAP29 is a RhoGTPase activating protein involved in the regulation of RhoA. It is expressed in several tissues, including heart, skeletal muscle, and placenta (Saras et al. 1997). It was recently identified as a critical binding partner of Ras-interacting protein 1 during vascular tubulogenesis and a mediator of Rap1 regulation of Rho in endothelial barrier function (Barry et al. 2016). ARHGAP29 also plays a role in cancer, as its level in both circulating tumor cells and renal cancer cells is positively correlated with metastatic potential, possibly via regulation of actin dynamics (Miyazaki et al. 2017; Qiao et al. 2017). We recently demonstrated that ARHGAP29 functions in a RhoA-dependent cell migration pathway involving IRF6, linking it to a large network of genes implicated in craniofacial development (Leslie et al. 2012; Biggs et al. 2014). Irf6-deficient mice show a decrease in ARHGAP29 protein expression, suggesting that it is an effector of IRF6 (Biggs et al. 2014). While there is strong evidence of association with NSCL/P, the effect of ARHGAP29 LoF during craniofacial development has not been previously described.
In this study, we used knock-in technology in the mouse to investigate, in vivo, the function of the human mutant allele K326X in ARHGAP29. Our data support a role for ARHGAP29 in embryonic survival and murine craniofacial development.
Materials and Methods
Creation of the Arhgap29 Mutant Allele
A 13.7-Kb genomic fragment containing exons 4-12 was isolated from a C57BL/6J BAC library and cloned into a pgk-DTA targeting vector (Fig. 1A). A single A to T point mutation was engineered at base 1184 in exon 11 of the mouse Arhgap29 gene, resulting in a K to X coding change. This is at amino acid position 325 in the mouse, which is identical in surrounding sequence to 326 in human. A FRT-neo-FRT cassette was placed 3′ to exon 11 for selection in ES cells. This construct was targeted to C57BL/6 ES cells (JM8), and 3 confirmed clones were injected into B6(Cg)-Tyrc-2J/J (JAX Stock No. 0058) host blastocysts. Germline transmission was confirmed by short-range polymerase chain reaction (PCR) from the neo cassette and sequencing of the engineered point mutation. The colony was maintained on a C57BL/6NJ genetic background.
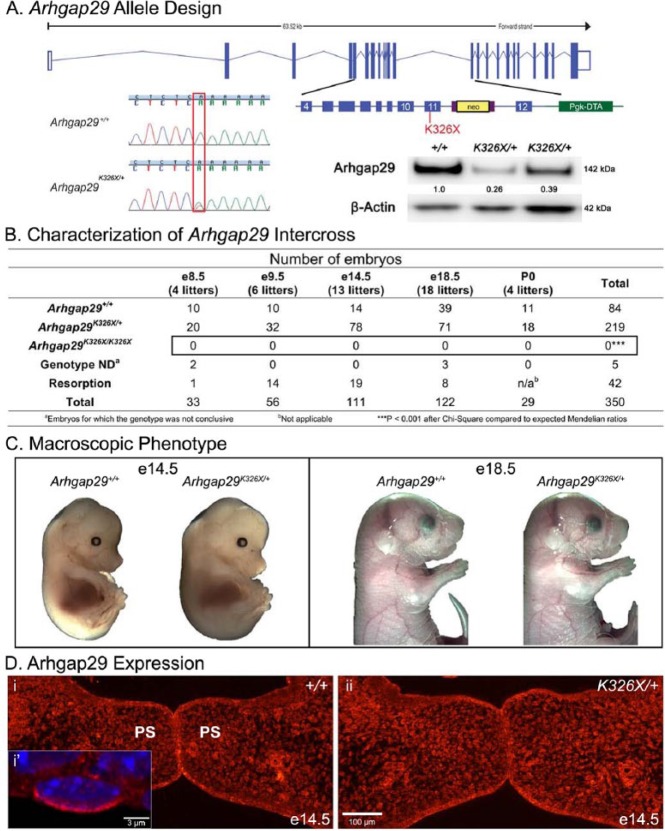
Figure 1.
Characterization of the Arhgap29K326X allele. (A) A single A to T point mutation at position 1184 in exon 11 identified in a human nonsyndromic cleft lip and/or palate patient was engineered into the murine Arhgap29, resulting in protein translation to a premature stop codon K326X. (A, inset) Western blot experiments on P4 palatal mucosa protein extracts for Arhgap29 and beta-actin. Quantification of the signal is indicated by numbers between the bands, which represent ratios of wild-type levels after normalization for actin. Note the decrease in Arhgap29 protein levels in Arhgap29K326X/+ compared with wild type. (B) Resulting Arhgap29K326X/+ mice were intercrossed and their embryos harvested at various time points. No Arhgap29K326X/K326X were found at any time point even as early as embryonic day (e) 8.5. (C) The macroscopic phenotypes of wild-type and Arhgap29K326X/+ embryos have no noticeable difference. (D) Immunofluorescent staining for Arhgap29 (red) in the palatal shelves of wild-type (i) and Arhgap29K326X/+ (ii) e14.5 embryos. (D, i′) Arhgap29 expression in the periderm (p63-negative cell on apical surface of oral epithelium).
Generation and Processing of Embryos
This procedure was approved by the Jackson Laboratory’s Animal Care and Use Committee under the National Institutes of Health guidelines for the humane care and use of laboratory research animals. It conformed with ARRIVE (Animal Research: Reporting of In Vivo Experiments) guidelines for animal studies.
Animals were mated overnight and inspected the following morning for the presence of a vaginal plug, constituting embryonic day (e) 0.5. Pregnant females were euthanized via CO2. Litters were dissected at e8.5, e9.5, e14.5, and e18.5; fixed with 4% paraformaldehyde; and embedded in paraffin for histology. Serial sections of 7 µm were collected on slides and then stained with hematoxylin and eosin.
Tissue was submitted to The Jackson Laboratory’s Transgenic Genotyping Service for genotyping with ARMS PCR, with primer sequences 5′-CTGTTTATCCAAAATTGCCTGCGATA TA-3′ (F outer), 5′-GTAGCTCGAAACAGAGACAGCTCAC A-3′ (F inner), 5′CAAACCAACACTTGAACTTAGCTGCTC T-3′ (R outer), 5′-CTGCATGCACAACAATTTTGCTTTAT A-3′ (R inner). Genotypes were verified with Sanger sequencing.
Measurement of Oral Adhesions
Oral adhesions were assessed on coronal sections and defined as regions of the oral cavity with contact between opposing epithelia. They were measured spatially in the x/y and z directions. Oral adhesions in the x/y directions were measured as previously described (Kousa et al. 2017). A representative section was selected on which the length of the maxillary epithelium was measured and compared with the length of the maxillary epithelium in contact with opposing structures. The depth of the adhesion (z direction) was calculated by counting the number of successive sections in which epithelia were in contact and multiplying by section thickness.
Western Blot Analysis
Palatal mucosa and skin were collected from 4-d-old neonates. Proteins were extracted with RIPA buffer as previously described (Biggs et al. 2014), and 20 µg of protein was separated using a Tris-acetate sodium dodecyl sulfate gel. Following transfer, membranes were incubated with antirabbit Arhgap29 antibody (Novus Biologicals) or antimouse beta-actin antibody (clone AC15, Sigma-Aldrich). Signal was detected and quantified using the Amersham Imager 600 (GE).
Fluorescent Immunostaining
Immunostaining was performed as previously described (Biggs et al. 2014). Antibodies used were keratin 17 (McGowan and Coulombe 1998), keratin 6 (Covance), E-cadherin (Santa Cruz), Arhgap29 (Novus Biologicals), p63 (clone 4A4, Santa Cruz), Irf6 (Sigma-Aldrich), goat antirabbit Alexa Fluor 568 (Invitrogen), and goat antimouse Alexa Fluor 488 (Thermo Fisher). Mountant was ProLong Diamond containing DAPI (Life Technologies). Slides were visualized with Nikon Eclipse E600 (epifluorescence) and Zeiss LSM 700 (confocal) microscopes.
Results
Arhgap29K326X/K326X Results in Embryonic Lethality
Previous sequencing of ARHGAP29 coding exons in 972 individuals with NSCL/P identified 16 nonsynonymous mutations, including 2 novel rare variants (p.S21Yfs*20 and p.Lys326X; Leslie et al. 2012). We focused on K326X because this truncation appears early in translation (exon 11 of 25), at amino acids conserved in more than 10 species, and within a Rap2 interaction domain (Leslie et al. 2012). We generated the single base change resulting in Arhgap29K326X (Fig. 1A). Heterozygotes expressed less than 50% of wild-type levels of Arhgap29 in both 4-d postnatal (P4) palatal epithelium (Fig. 1A, inset) and back skin (data not shown), confirming LoF.
To determine whether the Arhgap29K326X allele affects embryonic development in mice, embryos from intercrosses were harvested at e8.5, e9.5, e14.5, and e18.5 (Fig. 1B). No homozygous K326X mutants were identified, indicating that the mutation causes early embryonic lethality. Similar findings were observed following intercrosses of an Arhgap29 null allele (Arhgap29em1J, data not shown). Together, these results show that the homozygous K326X and em1J mutations result in early embryonic lethality. At the macroscopic level, the phenotype of the Arhgap29K326X/+ and Arhgap29em1J/+ embryos appeared identical to wild types (Fig. 1C and data not shown).
Arhgap29 Is Expressed in the Craniofacial Region and Periderm
We immunostained for Arhgap29 on coronal sections to confirm expression and determine whether Arhgap29K326X affected protein localization (Fig. 1D). Our results demonstrated the presence of Arhgap29 in the craniofacial region, as previously described (Leslie et al. 2012). Arhgap29 was strongly expressed in oral, lingual, and palatal epithelial cells and moderately in mesenchymal tissue (Fig. 1D). It was predominantly localized at the cytoplasmic membrane, although nuclear speckles were also observed, as previously described in cultured keratinocyte (Biggs et al. 2014). Arhgap29 was expressed in the periderm (p63-negative cells lining the oral epithelium), with the most prominent signal occurring on the apical side (Fig. 1D, panel i’).
Arhgap29K326X/+ Embryos Exhibit Oral Adhesions
Although the functional Arhgap29 (protein) and Arhgap29 (gene) are essential for life, we wanted to determine if the K326X allele affected craniofacial development. Our analysis was limited to K326X heterozygotes because the embryonic lethality in homozygotes precluded the formation of craniofacial structures. We examined embryonic heads at e13.5, e14.5, and e18.5 by serial coronal sectioning of entire litters (Fig. 2). Although embryos were harvested according to time-mating protocol, phenotypic variations were observed among the litters, as judged by standardized craniofacial development criteria (palatal shelves vertical alongside the tongue for e13.5; palatal shelves elevated and opposing epithelia in contact for e14.5). Because these variations may influence our analysis, we applied the following filtering criteria to each litter for further analysis: (1) the litter had to contain at least 1 wild-type for internal reference and (2) for e14.5, the phenotype of at least 1 wild-type embryo needed to demonstrate elevated palatal shelves making contact. This resulted in the analysis of 3 e13.5 litters and 5 e14.5 litters (Table).
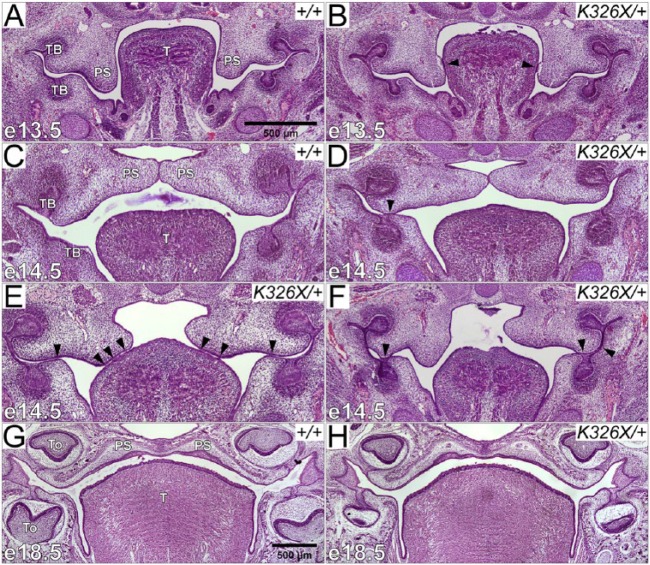
Figure 2.
Arhgap29K326X/+ embryos exhibit temporal oral adhesions. (A–H) Coronal sections of wild-type (A, C, G) and Arhgap29K326X/+ (B, D–F, H) murine embryonic heads at e13.5 (A, B), e14.5 (C–F), and e18.5 (G–H). (A) In e13.5 wild types, palatal shelves were oriented vertically alongside the tongue. (B) In Arhgap29K326X/+, a portion of oral epithelium was adherent to opposing structures, often resulting in detachment of the epithelial layer upon sectioning (arrowheads). (C) In e14.5 wild types, palatal shelves were elevated and made contact at the midline. (D–F) Contacts between opposing oral epithelia (oral adhesions) at e14.5 in Arhgap29K326X/+ are at various locations indicated by arrowheads. (G, H) e18.5 wild-type (G) and Arhgap29K326X/+ (H) exhibited no craniofacial abnormalities. PS, palatal shelf; TB, tooth bud; T, tongue; To, tooth.
Table.
Craniofacial Phenotype of Arhgap29K326X Embryos at e13.5 and e14.5.
| Palatal Shelves Elevated | Palatal Shelves in Contact | Commissure Contacta | Palatal Shelf to Tongue Adhesion | Tooth Bud Adhesionb | Any Oral Adhesionc | Potentially Deleterious Adhesiond | ||
|---|---|---|---|---|---|---|---|---|
| Arhgap29+/+ | e13.5 | 0/3 0% |
0/3 0% |
2/3 67% |
0/3 0% |
0/3 0% |
0/3 0% |
0/3 0% |
| e14.5 | 7/7 100% |
5/7 71% |
3/7 43% |
0/7 0% |
0/7 0% |
0/7 0% |
0/7 0% |
|
| Arhgap29K326X/+ | e13.5 | 0/12 0% |
0/12 0% |
10/12 83% |
4/12 33% |
7/12 58% |
9/12*
75%* |
7/12 58% |
| e14.5 | 18/25 72% |
13/25 52% |
18/25 72% |
3/25 12% |
5/25 20% |
13/25*52%* | 5/25 20% |
The maxillary and mandibular oral epithelia in the lateral commissure (lateral to the tooth bud) are in contact.
Any instance of adhesion involving the maxilla or mandibular tooth bud with opposing epithelium.
Contact between a maxillary structure (including palatal shelves) and an opposing structure. This includes contact in the commissure only if it extends medially beyond the halfway point between the maxillary and mandibular tooth buds.
The adhesion involves a not-yet elevated palatal shelf and mandibular or lingual epithelium.
P < 0.05 when compared with wild type of the same time point, Fisher exact test.
Histological observations revealed that intraoral epithelial contacts were present only in Arhgap29K326X/+ animals (75% of e13.5 and 52% of e14.5; Table, Fig. 2A, B, D-F, arrowheads). At e14.5, palatal shelves were elevated in 100% of wild types compared with only 72% of heterozygotes (Table, Fig. 2C-F), which shows a trend toward underdevelopment in Arhgap29K326X/+ animals. Adhesions were found in the midposterior palate, as defined in coronal sections by the simultaneous presence of the oral cavity, posterior nasal sinus, and eyes (Fig. 2A-F).
We characterized the location of oral adhesions within the oral cavity (Table). Thirty-two percent of heterozygotes had adhesions between the maxilla and the mandibular tooth bud (Fig. 2B, D). The palatal shelf was also adherent to the lingual epithelium in 19% of heterozygotes (Fig. 2B, E). In 59% of e13.5 and 20% of e14.5 Arhgap29K326X/+, the adhesion involved the palatal shelf before it had completely elevated (Fig. 2B, E). No oral adhesions were observed in wild-type animals.
At e14.5, Arhgap29em1J/+ embryos exhibited a similar delayed phenotype to the K326X allele, yet the incidence of oral adhesions was lower (data not shown). Of 14 embryos examined, 1 of 7 Arhgap29em1J/+ had an oral adhesion between the depressed palatal shelf and the mandibular epithelium.
Well-formed palates were observed at e18.5 in Arhgap29K326X/+ and wild-type embryos (Table, Fig. 2G, H), and no neonatal mice were reported with a cleft palate (Arhgap29K326X/+n = 22). These data suggest that the vast majority of oral adhesions seen at e13.5 and e14.5 resolve and do not impair proper palatogenesis.
Arhgap29K326X/+ Embryos Have a Significantly Higher Percentage of Oral Epithelial Contact
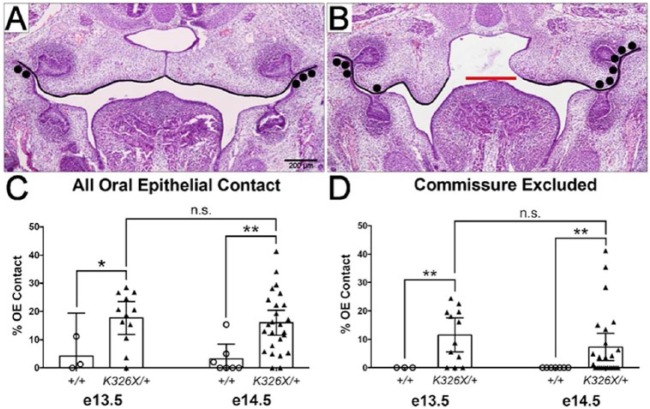
To further quantify the oral adhesions, we measured the length of maxillary epithelium in contact with an opposing structure, then divided this length by the total length of maxillary epithelium to obtain the percentage of oral epithelium in oppositional contact (Fig. 3). When all oral epithelial contacts were considered (Fig. 3A, B), Arhgap29K326X/+ had a significantly higher percentage of epithelial contact compared with wild-type embryos (Fig. 3C). As contact only in the commissure (black dots, Fig. 3A) was occasionally observed in wild-type embryos, we repeated our analysis including only adhesions beyond halfway between the tooth buds (Fig. 3D). Our results showed that 12% and 7% of the maxillary epithelium was in contact with opposing structures in e13.5 and e14.5 Arhgap29K326X/+, respectively, compared with none in wild-type embryos (Fig. 3D, P < 0.01). At e14.5, the mean depth of the contact in the z direction was 323 µm (median = 322 ± 135 µm, n = 17), which corresponds to approximately 25% of the total length of the palate.
Figure 3.
Increased length of oral epithelial contact in Arhgap29K326X/+ embryos. (A, B) Representative coronal sections used for the quantification. The length of the maxillary epithelium was measured (black lines, A, B). In embryos in which the palatal shelves were not elevated, the maxillary measurement stopped at a uniform point defined by the superior surface of the tongue (red line, B). The length of the maxillary epithelium in points of contact was also measured (black dots, A, B). (C) Percentage of oral epithelium (OE) in contact. (D) Percentage of OE in contact, excluding mild contact only in the commissures (black dots, A). *P < 0.05, **P < 0.01 following unpaired t test (with Welch’s correction for commissure excluded).
Periderm Cells Are Present at Sites of Oral Adhesions
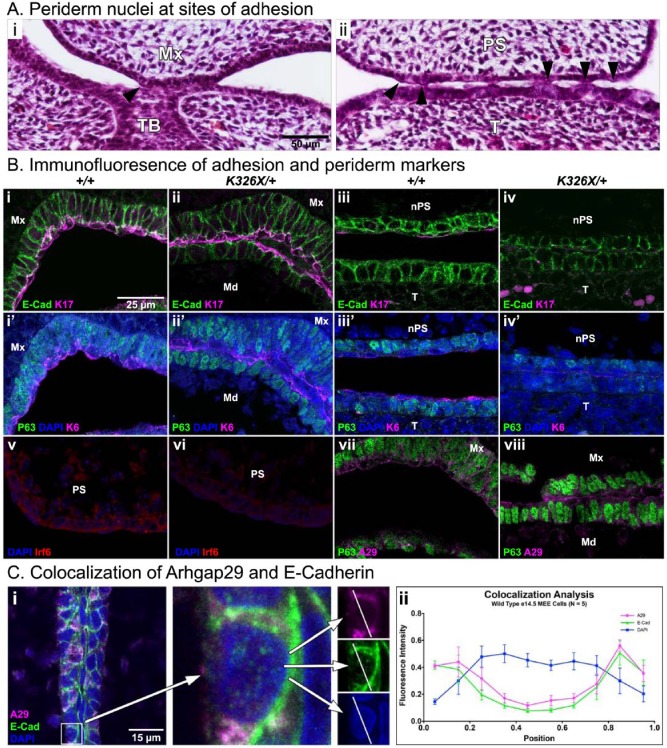
Oral adhesions have been previously attributed to altered periderm function (Richardson et al. 2009; Xiong et al. 2009; Peyrard-Janvid et al. 2014). In our histological sections, flat periderm nuclei were sometimes visible at sites of adhesion (Fig. 4A, arrowheads). Periderm cells were previously characterized by expression of keratin 17 and keratin 6 with an absence of p63 expression. We confirmed these observations in e13.5 cells along the apical surface of oral epithelia (Fig. 4B). Interestingly, there were some locations where these 2 keratins were hardly detectable, particularly at the medial sides of depressed palatal shelves and the tongue (Fig. 4B, iii, iii′, iv, iv′). We also detected Irf6 in e13.5 palatal shelves as previously described (Knight et al. 2006), and this expression was not altered in Arhgap29K326X/+ animals (Fig. 4B, v, vi). Arhgap29 was present in the oral epithelium and the periderm (Fig. 4B, vii-viii), with strong signal in periderm at the ends of adhesive spans (viii). Collectively, these data suggest that periderm cells are present in Arhgap29K326X/+ mice and maintained at the site of adhesion. However, the periderm layer often appears disorganized and occasionally discontinuous.
Figure 4.
Characterization of the oral periderm at sites of adhesion. (A, B) Histological view of e14.5 Arhgap29K326X/+ oral adhesions. Arrowheads indicate periderm nuclei present at points of contact between the maxilla and mandibular tooth bud (A, i) and the palatal shelf and tongue (A, ii). (B) Immunofluorescent staining for adhesion and periderm markers in paired littermates (genotypes designated in columns). Primary antibodies are E-cadherin (i-iv, green), keratin 17 (i-iv, magenta), p63 (i′-iv′, v-viii, green), keratin 6 (i′-ii′, magenta), Irf6 (v-vi, red), and Arhgap29 (vii-viii, magenta). i′-iv′ are sections adjacent to i-iv. Note the presence of keratin 17–positive, keratin 6–positive, and p63-negative cells (periderm) at the site of mandibular (Md) and maxillary (Mx) epithelial contact in i, i′, ii, and ii′. At an adhesion involving the nasal side of the palatal shelf (nPS) and the tongue (T; iii-iv, iii′-iv′, location the same as Fig. 2B, arrowheads), E-cadherin is present, while keratin 17 and keratin 6 expression is barely detectable. (v-vi) Irf6 is present in oral epithelium and periderm with bright speckles in nuclei, as previously described (Richardson et al. 2009). (vii-viii) Arhgap29 is expressed in the oral epithelium and the periderm, with strongest signal occurring in periderm cells at the ends of adherent spans (viii). (C) Colocalization analysis for E-cadherin and Arhgap29 at the medial edge epithelium (MEE). On a single z-plane, lines were drawn across the center of an epithelial cell (i). The fluorescence intensity along these lines was compared for each channel. Resulting values for Arhgap29 and E-cadherin were significantly correlated (P < 0.0001, Pearson correlation). Raw values were normalized and binned into discrete lengths for graphical representation (ii). Mx, maxilla; Md, mandible; TB, tooth bud; PS, palatal shelf; nPS, nasal side of palatal shelf; T, tongue.
Ectopic E-cadherin expression has previously been associated with oral adhesions (Richardson et al. 2009). To determine a potential interaction of Arhgap29 and E-cadherin, we double stained for these proteins and performed a colocalization estimate with fluorescence intensity (Fig. 4C). The signal was strongest at the plasma membrane with significant correlation in raw intensity values (P < 0.0001, Pearson correlation), suggesting the possibility that Arhgap29 can affect cellular adhesions.
Discussion
In our current study, a previously identified mutation in a patient with NSCL/P was targeted to the orthologous gene in the mouse genome, providing unique insight into the functional significance of this genetic variant in orofacial development. Using this strategy, we demonstrate a function for K326X of ARHGAP29 in craniofacial development.
Our results show that Arhgap29 is required for embryonic survival, as no Arhgap29K326X/K326X or Arhgap29em1J/em1J embryos were found at any time point. Recent studies implicating Arhgap29 in endothelial barrier function demonstratedthat Sox2-driven deletion of Arhgap29 led to embryonic death around e8.5 to e10 because of the lack of chorioallantoic fusion (Barry et al. 2016). Although we did not formally identify the cause of embryonic death, we hypothesize that the lethality of our mutants is due to the same defect. Heterozygous Arhgap29K326X/+ and Arhgap29em1J/+ embryos, however, are viable without noticeable problems, although Arhgap29K326X/+ were difficult to breed (Palmer, Rhea, Murray, and Dunnwald, personal communication, June 30, 2017). We previously hypothesized that K326X was a null allele based on the location of the K326X mutation in the gene and the nature of the genetic variant (stop codon; Leslie et al. 2012). Our current findings confirm our hypothesis that K326X leads to a nonfunctional gene product. These data are in accord with recent human sequencing studies, in which no predicted ARHGAP29 LoF variants have been identified as homozygous alleles (Leslie et al. 2012; Gowans et al. 2016; Savastano et al. 2017). Collectively, our results demonstrate that 1 functional copy of Arhgap29 is required for survival.
Arhgap29K326X showed functional relevance to craniofacial development by leading to oral adhesions during embryogenesis. Twenty percent of e14.5 Arhgap29K326X/+ embryos exhibited adhesions of the palatal shelf epithelium to another oral structure prior to complete elevation. These adhesions, even if mild, may increase the risk of a cleft palate if they restrict the movement of the palatal shelves at this critical developmental time point. Recently, a growing body of evidence has pointed to the critical role of these intraoral adhesions in the etiology of CL/P in the mouse. In particular, we previously found that similar oral adhesions occur in 11% of Irf6nl/+ e14.5 embryos, and full fusion of the oral cavity is the hallmark of all Irf6nl/nl embryos (Ingraham et al. 2006). We also detected intraoral adhesions in animals heterozygous for or lacking Grainyhead like 3 (Grhl3) (Peyrard-Janvid et al. 2014). Both IRF6 and GRHL3 mutations cause Van der Woude syndrome (Kondo et al. 2002; Peyrard-Janvid et al. 2014). Other alleles leading to oral adhesions include Notch, P63, and Jagged (Casey et al. 2006; Richardson et al. 2009; Xiong et al. 2009; Thomasonet al. 2010). Interestingly, these genes are all part of the IRF6 gene regulatory network that regulates not only proper palatogenesis but also epidermal differentiation and migration. We can now add ARHGAP29 to this network, not only based on human genetic variation but also based on the adhesion phenotype in the mouse.
The presence of oral adhesions further implicates the periderm as an important factor in craniofacial development. Studies with Grhl3, Irf6, and p63 mutant murine models demonstrate ectopic expression of E-cadherin on the outermost surface of 2 adjacent epithelial cells, often accompanied by the absence of keratin 6 or keratin 17 at the adhesion site, indicative of a lack of peridermal cells (Richardson et al. 2009; Peyrard-Janvid et al. 2014; Richardson et al. 2014). Immunostaining revealed a similar E-cadherin phenotype in our murine model. However, periderm cells were usually detected along the site of adhesion, although disorganized and with occasional discontinuity in staining. This finding suggests that the periderm is still present at the site of adhesions, implying a partial functional loss potentially because of altered differentiation of these cells. This raises further questions about the mechanism by which the 2 opposing epithelial layers adhere to one another and the function of the periderm in this process.
The generation of animal models harboring genetic variants identified in humans with cleft lip and palate are the holy grail of developmental biologists, as they allow the study, in vivo, of the function of these mutations. In the context of cleft lip and palate, this approach was used successfully with small organisms such as frogs and fish (Peyrard-Janvid et al. 2014; Liu et al. 2016). However, studies with mice were restricted to the introduction of mutations identified in syndromic forms of clefting (example: R84C mutation in IRF6 commonly found in popliteal pterygium syndrome; Richardson et al. 2006). Our allele introduces a variant identified in NSCL/P. As such, our findings expand the tools to directly study the function of common genetic variants associated with orofacial clefting. Because the K326X mutation was not completely penetrant in the family (Leslie et al. 2012), it most likely required interactions with other genetic and/or environmental predisposing covariates. Therefore, this murine model is most important for identifying these other covariates as well as expanding the role of ARHGAP29 on other biological processes associated with orofacial clefting, such as wound healing.
Author Contributions
B.J. Paul and M. Dunnwald contributed to conception, design, data acquisition, analysis, and interpretation and drafted and critically revised the manuscript; K. Palmer and J.C. Sharp contributed to data acquisition and analysis and critically revised the manuscript; C.H. Pratt and S.A. Murray contributed to conception and design and critically revised the manuscript. All authors gave final approval and agree to be accountable for all aspects of the work.
Acknowledgments
We gratefully acknowledge the support of the Central Microscopy Research Facility of the University of Iowa.
Footnotes
This research was supported by funding from National Institutes of Health (NIH)/National Institute of Arthritis and Musculoskeletal and Skin Diseases (NIAMS) to M.D. (AR067739). Additional partial financial support was provided by a grant from the NIH R37DE08559 and by the FaceBase consortium (grants DE020052 and DE020 057). B.J.P. was a recipient of a fellowship from the Iowa Center for Research by Undergraduates.
The authors declare no potential conflicts of interest with respect to the authorship and/or publication of this article.
References
- Barry DM, Koo Y, Norden PR, Wylie LA, Xu K, Wichaidit C, Azizoglu DB, Zheng Y, Cobb MH, Davis GE, et al. 2016. Rasip1-mediated Rho GTPase signaling regulates blood vessel tubulogenesis via nonmuscle myosin II. Circ Res. 119(7):810–826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beaty TH, Marazita ML, Leslie EJ. 2016. Genetic factors influencing risk to orofacial clefts: today’s challenges and tomorrow’s opportunities. F1000Res. 5:2800. eCollection 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beaty TH, Murray JC, Marazita ML, Munger RG, Ruczinski I, Hetmanski JB, Liang KY, Wu T, Murray T, Fallin MD, et al. 2010. A genome-wide association study of cleft lip with and without cleft palate identifies risk variants near MAFB and ABCA4. Nat Genet. 42(6):525–529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beaty TH, Taub MA, Scott AF, Murray JC, Marazita ML, Schwender H, Parker MM, Hetmanski JB, Balakrishnan P, Mansilla MA, et al. 2013. Confirming genes influencing risk to cleft lip with/without cleft palate in a case-parent trio study. Hum Genet. 132(7):771–781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biggs LC, Naridze RL, DeMali KA, Lusche DF, Kuhl S, Soll DR, Schutte BC, Dunnwald M. 2014. Interferon regulatory factor 6 regulates keratinocyte migration. J Cell Sci. 127(Pt 13):2840–2848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butali A, Mossey P, Adeyemo W, Eshete M, Gaines L, Braimah R, Aregbesola B, Rigdon J, Emeka C, Olutayo J, et al. 2014. Rare functional variants in genome-wide association identified candidate genes for nonsyndromic clefts in the African population. Am J Med Genet A. 164A(10):2567–2571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casey LM, Lan Y, Cho ES, Maltby KM, Gridley T, Jiang R. 2006. Jag2-Notch1 signaling regulates oral epithelial differentiation and palate development. Dev Dyn. 235(7):1830–1844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon MJ, Marazita ML, Beaty TH, Murray JC. 2011. Cleft lip and palate: understanding genetic and environmental influences. Nat Rev Genet. 12(3):167–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fontoura C, Silva RM, Granjeiro JM, Letra A. 2012. Further evidence of association of the ABCA4 gene with cleft lip/palate. Eur J Oral Sci. 120(6):553–557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gowans LJ, Adeyemo WL, Eshete M, Mossey PA, Busch T, Aregbesola B, Donkor P, Arthur FK, Bello SA, Martinez A, et al. 2016. Association studies and direct DNA sequencing implicate genetic susceptibility loci in the etiology of nonsyndromic orofacial clefts in Sub-Saharan African populations. J Dent Res. 95(11):1245–1256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingraham CR, Kinoshita A, Kondo S, Yang B, Sajan S, Trout KJ, Malik MI, Dunnwald M, Goudy SL, Lovett M, et al. 2006. Abnormal skin, limb and craniofacial morphogenesis in mice deficient for interferon regulatory factor 6 (Irf6). Nat Genet. 38(11):1335–1340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knight AS, Schutte BC, Jiang R, Dixon MJ. 2006. Developmental expression analysis of the mouse and chick orthologues of IRF6: the gene mutated in Van der Woude syndrome. Dev Dyn. 235(5):1441–1447. [DOI] [PubMed] [Google Scholar]
- Kondo S, Schutte BC, Richardson RJ, Bjork BC, Knight AS, Watanabe Y, Howard E, de Lima RL, Daack-Hirsch S, Sander A, et al. 2002. Mutations in IRF6 cause Van der Woude and popliteal pterygium syndromes. Nat Genet. 32(2):285–289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kousa YA, Roushangar R, Patel N, Walter A, Marangoni P, Krumlauf R, Klein O, Schutte BC. 2017. Irf6 and receptor tyrosine kinase signaling interact in periderm development. J Dent Res [epub ahead of print 12 July 2017] in press. doi: 10.1177/0022034517719870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lennon CJ, Birkeland AC, Nunez JA, Su GH, Lanzano P, Guzman E, Celis K, Eisig SB, Hoffman D, Rendon MT, et al. 2012. Association of candidate genes with nonsyndromic clefts in Honduran and Colombian populations. Laryngoscope. 122(9):2082–2087. [DOI] [PubMed] [Google Scholar]
- Leslie EJ, Carlson JC, Shaffer JR, Feingold E, Wehby G, Laurie CA, Jain D, Laurie CC, Doheny KF, McHenry T, et al. 2016. A multi-ethnic genome-wide association study identifies novel loci for non-syndromic cleft lip with or without cleft palate on 2p24.2, 17q23 and 19q13. Hum Mol Genet. 25(13):2862–2872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leslie EJ, Mansilla MA, Biggs LC, Schuette K, Bullard S, Cooper M, Dunnwald M, Lidral AC, Marazita ML, Beaty TH, et al. 2012. Expression and mutation analyses implicate ARHGAP29 as the etiologic gene for the cleft lip with or without cleft palate locus identified by genome-wide association on chromosome 1p22. Birth Defects Res A Clin Mol Teratol. 94(11):934–942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leslie EJ, Marazita ML. 2013. Genetics of cleft lip and cleft palate. Am J Med Genet C Semin Med Genet. 163C(4):246–258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letra A, Maili L, Mulliken JB, Buchanan E, Blanton SH, Hecht JT. 2014. Further evidence suggesting a role for variation in ARHGAP29 variants in nonsyndromic cleft lip/palate. Birth Defects Res A Clin Mol Teratol. 100(9):679–685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Leslie EJ, Carlson JC, Beaty TH, Marazita ML, Lidral AC, Cornell RA. 2017. Identification of common non-coding variants at 1p22 that are functional for non-syndromic orofacial clefting. Nat Commun. 8:14759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Leslie EJ, Jia Z, Smith T, Eshete M, Butali A, Dunnwald M, Murray J, Cornell RA. 2016. Irf6 directly regulates Klf17 in zebrafish periderm and Klf4 in murine oral epithelium, and dominant-negative KLF4 variants are present in patients with cleft lip and palate. Hum Mol Genet. 25(4):766–776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGowan KM, Coulombe PA. 1998. Onset of keratin 17 expression coincides with the definition of major epithelial lineages during skin development. J Cell Biol. 143(2):469–486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyazaki J, Ito K, Fujita T, Matsuzaki Y, Asano T, Hayakawa M, Asano T, Kawakami Y. 2017. Progression of human renal cell carcinoma via inhibition of RhoA-ROCK axis by PARG1. Transl Oncol. 10(2):142–152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peyrard-Janvid M, Leslie EJ, Kousa YA, Smith TL, Dunnwald M, Magnusson M, Lentz BA, Unneberg P, Fransson I, Koillinen HK, et al. 2014. Dominant mutations in GRHL3 cause Van der Woude Syndrome and disrupt oral periderm development. Am J Hum Genet. 94(1):23–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiao Y, Chen J, Lim YB, Finch-Edmondson ML, Seshachalam VP, Qin L, Jiang T, Low BC, Singh H, Lim CT, et al. 2017. YAP regulates actin dynamics through ARHGAP29 and promotes metastasis. Cell Rep. 19(8):1495–1502. [DOI] [PubMed] [Google Scholar]
- Richardson RJ, Dixon J, Jiang R, Dixon MJ. 2009. Integration of IRF6 and Jagged2 signalling is essential for controlling palatal adhesion and fusion competence. Hum Mol Genet. 18(14):2632–2642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson RJ, Dixon J, Malhotra S, Hardman MJ, Knowles L, Boot-Handford RP, Shore P, Whitmarsh A, Dixon MJ. 2006. Irf6 is a key determinant of the keratinocyte proliferation-differentiation switch. Nat Genet. 38(11):1329–1334. [DOI] [PubMed] [Google Scholar]
- Richardson RJ, Hammond NL, Coulombe PA, Saloranta C, Nousiainen HO, Salonen R, Berry A, Hanley N, Headon D, Karikoski R, et al. 2014. Periderm prevents pathological epithelial adhesions during embryogenesis. J Clin Invest. 124(9):3891–3900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saras J, Franzen P, Aspenstrom P, Hellman U, Gonez LJ, Heldin CH. 1997. A novel GTPase-activating protein for Rho interacts with a PDZ domain of the protein-tyrosine phosphatase PTPL1. J Biol Chem. 272(39):24333–24338. [DOI] [PubMed] [Google Scholar]
- Savastano CP, Brito LA, Faria AC, Seto-Salvia N, Peskett E, Musso CM, Alvizi L, Ezquina SA, James C, GOSgene, et al. 2017. Impact of rare variants in ARHGAP29 to the etiology of oral clefts: role of loss-of-function vs missense variants. Clin Genet. 91(5):683–689. [DOI] [PubMed] [Google Scholar]
- Thomason HA, Zhou H, Kouwenhoven EN, Dotto GP, Restivo G, Nguyen BC, Little H, Dixon MJ, van Bokhoven H, Dixon J. 2010. Cooperation between the transcription factors p63 and IRF6 is essential to prevent cleft palate in mice. J Clin Invest. 120(5):1561–1569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uslu VV, Petretich M, Ruf S, Langenfeld K, Fonseca NA, Marioni JC, Spitz F. 2014. Long-range enhancers regulating Myc expression are required for normal facial morphogenesis. Nat Genet. 46(7):753–758. [DOI] [PubMed] [Google Scholar]
- Xiong W, He F, Morikawa Y, Yu X, Zhang Z, Lan Y, Jiang R, Cserjesi P, Chen Y. 2009. Hand2 is required in the epithelium for palatogenesis in mice. Dev Biol. 330(1):131–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan Q, Blanton SH, Hecht JT. 2011. Association of ABCA4 and MAFB with non-syndromic cleft lip with or without cleft palate. Am J Med Genet A. 155A(6):1469–1471. [DOI] [PMC free article] [PubMed] [Google Scholar]