Abstract
Secondary palatogenesis occurs when the bilateral palatal shelves (PS), arising from maxillary prominences, fuse at the midline, forming the hard and soft palate. This embryonic phenomenon involves a complex array of morphogenetic events that require coordinated proliferation, apoptosis, migration, and adhesion in the PS epithelia and underlying mesenchyme. When the delicate process of craniofacial morphogenesis is disrupted, the result is orofacial clefting, including cleft lip and cleft palate (CL/P). Through human genetic and animal studies, there are now hundreds of known genetic alternations associated with orofacial clefts; so, it is not surprising that CL/P is among the most common of all birth defects. In recent years, in vitro cell-based assays, ex vivo palate cultures, and genetically engineered animal models have advanced our understanding of the developmental and cell biological pathways that contribute to palate closure. This is particularly true for the areas of PS patterning and growth as well as medial epithelial seam dissolution during palatal fusion. Here, we focus on epithelial cell-cell adhesion, a critical but understudied process in secondary palatogenesis, and provide a review of the available tools and mouse models to better understand this phenomenon.
Keywords: cleft palate, cell adhesion, epithelium, cadherins, nectins, afadin
Introduction
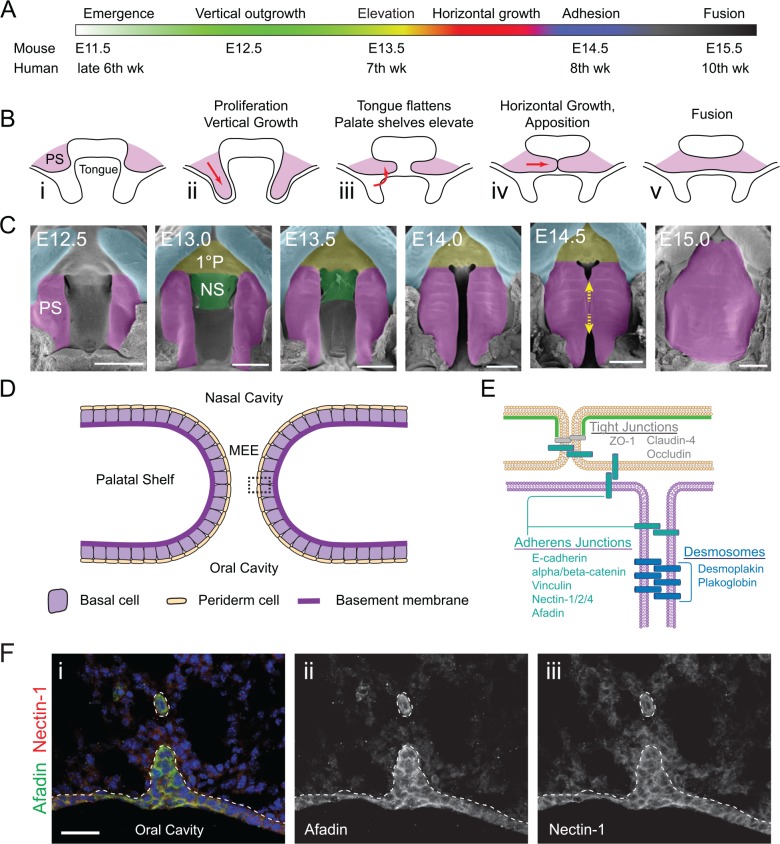
Secondary palate formation is a complex, multi-step process that involves palatal shelf (PS) 1) emergence from the maxillary prominences, 2) vertical outgrowth toward the floor of the mouth, 3) elevation above the dorsal surface of the tongue, 4) horizontal growth, 5) adhesion of the approximating medial edge epithelia (MEE) to form the medial epithelial seam (MES), and 6) fusion by dissolution of the MES (Figure 1A–C). In mice, the secondary palate forms over the course of ~3-4 days, with adhesion and fusion occurring between E14 and E15 (Walker 1956; Greene and Pratt 1976; Bush and Jiang 2012). In humans, this process occurs comparatively earlier during embryogenesis but takes longer to complete, with secondary palatogenesis taking place between the 7th and 12th weeks, and hard palate preceding soft palate closure (Danescu et al. 2015). Failure at any stage can result in a persistent gap along the midline of the roof of the oral cavity, known as cleft palate (CP).
Figure 1.
Secondary palatogenesis. (A) Timeline of morphogenetic processes that occur during palate growth and closure in mice and humans. Human data is based on the timing of hard palate closure, with soft palate fusion occurring later. (B) Schematics, in the coronal plane, of the position of the secondary palatal shelves (PS, purple) relative to the tongue during representative stages of palatogenesis. PS initiate outgrowth from the maxillary prominence at ~E11.5 to E12 (i), depending on the mouse strain (Walker 1956), and initially grow downward (ii) before elevating above the tongue at ~E13.5 to E14.0 (iii). Horizontal growth follows until opposing medial edge epithelia (MEE) meet at the midline (iv). PS fusion occurs between E14.5 and E15.5, and proceeds anteriorly and posteriorly over the course of ~6 h (Walker 1956) (v). (C) Scanning electron microscopy images of the roof of the mouth at indicated ages. (D) Cartoon depicting a coronal view of approximating palatal shelves (~E14.0). Adhesive-competent basal cells (purple) are separated from the mesenchyme by a basement membrane (dark purple). Nonadhesive periderm cells (tan) prevent the formation of intra-oral adhesions. The periderm is lost before the formation of the medial epithelial seam (MES). (E) Molecular view of inset from (D) (rotated 90°) demonstrating the distribution of cell–cell adhesions within the MEE. Proteins that have been localized to the MEE via immunohistological staining are listed. More detail regarding evidence for the proteins listed, including references, can be found in Appendix Table 1. The nonadhesive apical surface of the periderm cells is demarcated in green. (F) Afadin immunofluorescence (green, Sigma-Aldrich) in the oral side of the early MES colocalizes with nectin-1 (red, MBL D146-3). (i) Multicolor image. Panels (ii) and (iii) are isolated, greyscale images of the Afadin and nectin-1 staining, respectively. Dashed line demarcates basement membrane. Scale bars: 500 µm (C), 30 µm (F); pseudocolors: purple (PS), light blue (lip), green (NS, nasal septum), yellow (1°P, primary palate); yellow arrows in (C) indicate direction of palatal fusion. Images in (C) adapted from Facebase; timeline in (A) inspired by Bush and Jiang (2012).
A recent meta-analysis revealed 234 genes linked to CL/P in humans and 249 in mice, of which 54 are shared (Kousa et al. 2017). While many nonsyndromic and syndromic cases of orofacial clefting (OFC) are due in part to decreased mandibular growth, an unknown proportion of clefts are caused by defective mechanisms intrinsic to the palate. Since the molecular-genetic control of PS emergence and elevation, as well as MES dissolution, have been extensively reviewed elsewhere (see Cao et al. in this issue; as well as Bush and Jiang [2012]; Lan et al. [2015]; Lane and Kaartinen [2014]), here we focus on mechanisms regulating the initial steps of MEE fusion, highlighting the mouse as a model system.
Forty years ago, Greene and Pratt (1976) remarked that “adhesion between apposing epithelial surfaces appears to involve epithelial cell surface macromolecules.” More recently, the critical role of adhesion in palate closure was demonstrated by the identification of human CL/P mutations in the adherens junction (AJ) proteins Nectin-1 (NECTIN1) and E-cadherin (CDH1). In animal models, adhesion proteins including nectins, desmosomal cadherins, and other AJ proteins have been localized to the MEE (Figure 1D-F; Appendix Table 1), but comparatively little is known about their function in this epithelia. Appendix Table 2 compiles published mouse models of known cell adhesion genes and whether their role in palatogenesis has been investigated. A major hurdle to dissecting the role of adhesion proteins in palate closure is extensive functional redundancy among related family members and, in many cases (e.g., the nectins), a lack of conditional alleles.
Genes Expressed in Palatal Epithelium Underlie CL/P Disorders
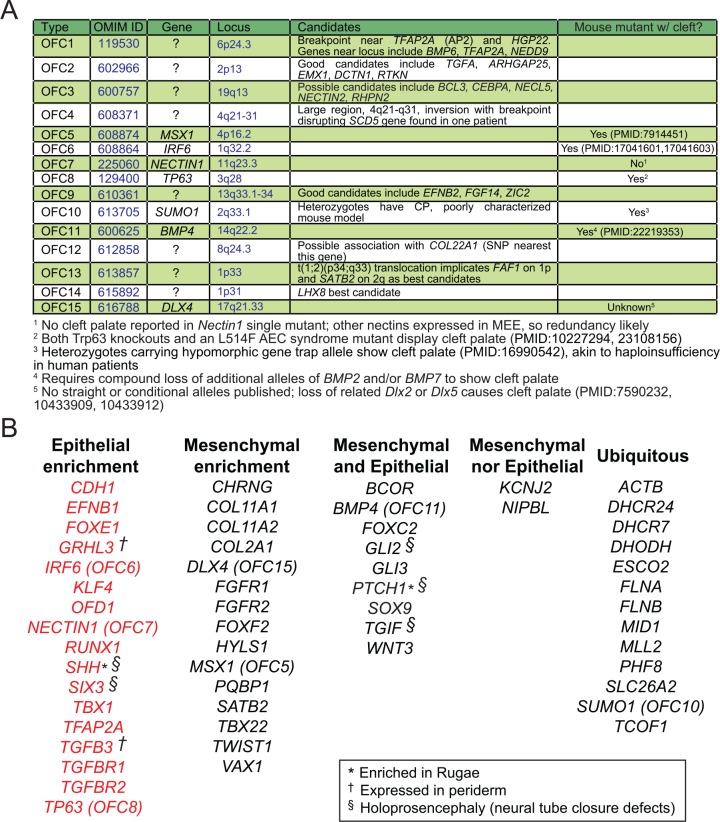
Defects in mesenchymal growth and signaling are responsible for many nonsyndromic CL/P disorders, including the Online Mendelian Inheritance in Man (www.omim.org) OFC loci listed in Figure 2A (e.g., MSX1 and DLX4). However, 3 of 7 known OMIM OFC genes are expressed in palatal epithelium, including the intensively studied transcription factors, IRF6 and TP63 (reviewed by Schutte et al. [1993]; Rinne et al. [2007]; Vanbokhoven et al. [2011]), and the cell adhesion molecule NECTIN1. To understand the relative proportions of CP genes expressed in mesenchyme v. epithelium, we examined a dataset of ~50 human CL/P loci (Dixon et al. 2011; Ma et al. 2015). We searched the primary literature, the Human Protein Atlas (http://www.proteinatlas.org/), and the Gene eXpression Database (GXD, http://www.informatics.jax.org/expression.shtml) using “Theiler stage (TS) 21: palatal shelf epithelium” (EMAPS:1736321) and “TS21: palatal shelf mesenchyme” (EMAPS: 1736421) as filters. Interestingly, most of these mRNAs/proteins are expressed in palatal epithelium, with about one-third epithelially enriched (Figure 2B). This list of genes includes transcription factors (GRHL3, KLF4, RUNX1, TBX1, IRF6, TP63), signaling molecules (SHH, TGFB3), and numerous adhesion proteins (CDH1, EFNB1/ephrin-B1, NECTIN1).
Figure 2.
Palatal gene expression and human orofacial clefting syndromes. (A) Table of nonsyndromic orofacial clefting (OFC) disorders listed in OMIM. The presence of cleft palate in corresponding mouse mutants is also noted. (B) Genes mapped to cleft palate phenotypes in humans, tabulated based on their pattern of expression. Sources: MGI Gene eXpression Database (GXD), (Dixon et al. 2011; Finger et al. 2017).
Nectins and Afadin
Nectins are a family of 4 Ig-family, transmembrane, cell-adhesion molecules that bind the obligate cytoplasmic adapter protein afadin (AFDN), which signals to the cytoskeleton by interacting with α-catenin (AJs), ZO-1/Jam-A (tight junctions, TJs), and F-actin (Mandai et al. 2015). Nectins homodimerize in cis before forming tetramers in trans (Miyahara et al. 2000). Cis-interactions are dependent on the first Ig-like loop, whereas trans-interactions involve both the first and second Ig-like loops (Yasumi et al. 2003). Heterotypic trans-interactions are favored, with nectin-1:nectin-3 showing the highest affinity, followed by nectin-2:nectin-3, and homotypic interactions showing the weakest affinity (Togashi et al. 2011). Heterotypic interactions mediate cell sorting via the expression of discrete nectins on neighboring cells (Togashi et al. 2011). Because multiple nectins are expressed on MEE/MES cells (Yoshida et al. 2012), it is tempting to speculate that similar sorting behavior may participate in MES dissolution during palatal fusion.
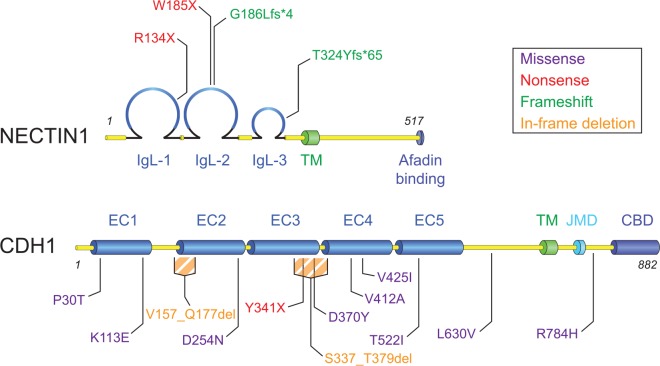
CL/P-ectodermal dysplasia syndrome, or CLPED1/OFC7 (OMIM:225060; also, Zlotogora-Ogur/Margarita Island syndrome), is characterized by numerous ectodermal phenotypes. Positional mapping and sequence analysis identified a novel, homozygous nonsense mutation (W185X) in NECTIN1 (Suzuki et al. 2000). Further familial studies characterized additional variants (T324Yfs*65, G186Lfs*4, and R134X), plus others associated with nonsyndromic CL/P (Sozen et al. 2001; Avila et al. 2006; Aslar and Tastan 2014; Yoshida et al. 2015) (Figure 3). Despite ample evidence suggesting Nectin-mediated adhesion contributes to palate fusion, molecular redundancy and embryonic lethality have made modeling these human phenotypes difficult. Multiple nectins are enriched in MEE and colocalize with afadin (Yoshida et al. 2012). However, germline knockouts of Nectin1 are viable and lack CP, even when one copy of Nectin3 is deleted (Inagaki et al. 2005). On the other hand, deletion of both Nectin1 and Nectin3 is early embryonic lethal (Yoshida et al. 2010), precluding any analysis on palate formation. Although Nectins-1, -2 and -4 are present in the MEE, no double mutant of any combination of these alleles has been generated; thus, functional redundancy could account for the lack of phenotype in single mutants. Additionally, if translated, nonsense and frameshift human NECTIN1 mutations could function as dominant-negatives. For example, when NECTIN1 is truncated after the first Ig-like loop (Figure 3), the free extracellular domain could interact with other nectins and act as a soluble inhibitor, as previously demonstrated with Fc fusions of Nectin3/Nectin1 extracellular domains (Kawakatsu et al. 2002).
Figure 3.
Mutations in NECTIN1 and CDH1 associated with cleft lip/palate (CL/P). Domain structures and sites of mutations found in patients with CL/P for NECTIN-1 (top, previously known as PVRL1) and E-cadherin/CDH1 (bottom). Mutation types are color-coded and indicated in the legend. CBD, catenin-binding domain; EC, extracellular cadherin domain; IgL, immuoglobulin-like loop; JMD, juxtamembrane domain; TM, transmembrane domain.
While mouse models have failed to recapitulate CP associated with a NECTIN1 mutation, it remains important to investigate redundancy and characterize human disease mutations. The introduction of conditional alleles, compound mouse mutants, and new genetic models for unstudied nectins (e.g., Nectin-4) represent important avenues for future research. Given the known and suspected roles of various nectins in palate closure in humans, but the lack of observed CP in individual nectin mutants, we hypothesize that loss of the obligate downstream effector of nectin signaling, afadin, might reveal whether nectins are essential for secondary palate closure. We and others (Yoshida et al. 2012) have observed co-localization of Nectin-1 and afadin in the MEE during palatal fusion (Figure 1F). Although cleft palate was not reported in an epithelial conditional knockout of afadin (Afdn) (Yoshida et al. 2014), there could be at least 2 explanations: 1) mosaic expression and function of the Cre transgene, or 2) a requirement for afadin in the periderm. It is worth noting that the Krt14-Cre transgene used in these studies (Huelsken et al. 2001; Andl et al. 2004) is only active in the basal layer and not the periderm, and it appears to be one of the weaker Krt14-Cre lines available (discussed in greater detail later). These observations suggest the need for alternative approaches to probe palate formation.
Other nectins have suspected or confirmed associations with CL/P and related disorders. NECTIN2 and the nectin-like NECL5 are separated by ~180 Mb on chromosome 19q13, a region associated with nonsyndromic CL/P (OFC3, OMIM:600757, Figure 2B). Although rare variants in both NECL5 and NECTIN2 have been linked to CL/P, the functional significance remains unclear (Warrington et al. 2006). Additionally, mutations in NECTIN4 cause ectodermal dysplasia-syndactyly syndrome (EDSS) (Brancati et al. 2010). Patient-derived keratinocytes bearing NECTIN4 mutations have impaired AJ assembly and maintenance, suggesting these alterations have functional significance in cell–cell adhesion (Fortugno et al. 2014). Interestingly, a recent study demonstrated that Nectin-4 is expressed in the MEE in a pattern similar to Nectin-1 (Richardson et al. 2017). The presence of Nectin-4 in the MEE is intriguing, as Nectin-4 and Nectin-1 interact with similar efficiency as Nectin-1 and Nectin-3, but Nectin-3 appears to be absent from the MEE (Reymond et al. 2001; Yoshida et al. 2012).
Cadherins and Catenins
Cadherins are a large family of transmembrane, Ca2+-dependent, cell–cell adhesion molecules that form the structural foundation of AJs. Mammalian classical cadherins consist of 5 N-terminal extracellular cadherin repeats, a transmembrane region (TM), an intracellular juxtamembrane domain (JMD) and a C-terminal catenin-binding domain (CBD) (Figure 3). Cell–cell adhesion is mediated by trans-dimerization between cadherin repeat homodimers. β-catenin binds directly to the CBD and recruits α-catenin, forming the minimal cadherin–catenin adhesion system. α-catenin forms numerous tertiary interactions between the AJ, the actin cytoskeleton, and other cell–cell adhesions such as nectins, TJs, and desmosomes.
E-cadherin (CDH1), the most ubiquitously expressed of the cadherins, is implicated in numerous pathologies, including hereditary diffuse gastric cancer (HDGC). Interestingly, Frebourg et al. (2006) identified multiple pedigrees of HDGC with CDH1 mutations linked to CL/P, as have other groups (Vogelaar et al. 2013; Brito et al. 2015). Several mutations are either missense or in-frame deletions, many occurring within the extracellular cadherin repeats (Figure 3). Characterization of these alleles shows functional consequences in vitro, supporting a role for cadherin-mediated adhesion in palatogenesis (Vogelaar et al. 2013).
Transmission electron microscopy and immunofluorescence studies have detected the proteins and structures that comprise AJs in the MEE/MES (Tudela et al. 2002; Kitase and Shuler 2013). However, genetic models have yet to unequivocally demonstrate a functional role for cadherins in palate closure. Germline Cdh1-null mice, like many cell adhesion mutants, are embryonic lethal (Larue et al. 1994). Whereas numerous studies have genetically ablated Cdh1 via tissue-specific Cre drivers, a specific role in palatogenesis has not been directly investigated. Two studies knocked out Cdh1 in the epidermis but the Krt14-Cre alleles used (Vasioukhin et al. 1999; Hafner et al. 2004) may not have deleted Cdh1 in the MEE early enough; there is also evidence for compensatory P-cadherin (Cdh3) upregulation upon Cdh1 loss (Tinkle et al. 2004; Tunggal et al. 2005). Since P-cadherin is also expressed in palatal epithelia (according to the MGI GXD (Finger et al. 2017) and our unpublished data), it could act redundantly with E-cadherin in palatogenesis. While epidermal-specific E/P-cadherin double mutants have been generated (Tinkle et al. 2008), palate closure was not investigated.
Interestingly, PS failed to elevate upon mesenchymal deletion of Ctnnb1 (β-catenin) with Osr2-IRES-Cre (Chen et al. 2009), whereas PS elevated and approximated but failed to fuse upon epithelial deletion with Krt14-Cre (He et al. 2011). In addition to its role in AJs, β-catenin has a well-characterized role in Wnt signaling (reviewed by Niehrs [2012]). However, these Ctnnb1-null phenotypes differ significantly from other Wnt mutants, such as Gsk3β knockouts, where PS fails to elevate (He et al. 2010a), or a stabilized Ctnnb1 mutant, which showed impaired horizontal outgrowth (He et al. 2011). These data suggest a complex mechanism, where β-catenin may play important Wnt-dependent roles in mesenchyme proliferation during PS elevation/horizontal outgrowth, in addition to cell-adhesive roles during MEE fusion.
Other Cell Adhesion Molecules
TJs are water-impermeable junctions consisting of claudin, occludin, and zonula-occludens (ZO) family members (reviewed by Zihni et al. [2016]). Several TJ proteins, including claudin-4, occludin, and ZO-1, localize to the MEE, where data suggest they form functional TJs between periderm cells (Yoshida et al. 2012). Germline null models for many TJ components are lethal due to defects in gastrulation or epidermal barrier function (Appendix Table 2). Although no data exist to support a link between human CL/P and TJ genes, it is important to explore the role TJs play in periderm formation, maintenance, and MEE fusion.
Desmosomes are cell–cell adhesions consisting of a desmoglein/desmocollin transmembrane molecules bound to a member of the intracellular keratin-binding plakophilin family. Desmogleins and desmocollins are part of the larger classical cadherin family, forming calcium-dependent dimers in trans. Desmosomal cadherins cluster at much higher density than AJ cadherins, forming hyper-adhesive junctions that ensure structural integrity of exposed epithelia. Because of their electron-dense structure, desmosomes have been readily detected in MEE and between cells forming nascent contacts in the MES by TEM (Sun et al. 1998; Mogass et al. 2000). Immunohistochemical experiments have similarly detected plakoglobin and plakophilin-1 in approximating MEE (Mogass et al. 2000; Ke et al. 2015). It is tempting to speculate that these molecules play important roles during palatal fusion by establishing strong cell–cell adhesions between approximating PS. Furthermore, the desmosomal and AJ component, plakoglobin (γ-catenin), shares functional similarities to β-catenin, activating the LEF/TCF transcription pathway (Miravet et al. 2002). However, little is known about which desmosomal components are actually expressed in the MEE/MES, and their functional role has not been assessed in mouse models.
The Eph receptor family of tyrosine kinases and their ephrin ligands have long been studied for their role in repulsive axonal guidance and boundary formation; however, through “reverse signaling”—where ephrin serves as receptor and Eph as ligand—they can also function in adhesion (reviewed in Kania and Klein [2016]). Mutations in the gene encoding ephrin-B1 (EFNB1) cause craniofrontalnasal syndrome (CFNS, OMIM:304110) which includes CL/P (Twigg et al. 2004; Wieland et al. 2004). Efnb1−/− mice present with CP (Bush and Soriano 2010), as do double mutants in the cognate receptors EphB2;EphB3 (Risley et al. 2009). Several studies have shown that the reverse-signaling function of ephrin-B1 is critical for MEE adhesion. Ectopic ephrin-B signaling forces palate fusion in chicken, which have a naturally occurring secondary palatal cleft (San Miguel et al. 2011). Additionally, EphB reverse signaling in murine palate culture can rescue the CP defect caused by transforming growth factor (TGF)-β3 inhibition (Serrano et al. 2015). EphA receptors and ligands are also being investigated but considerable redundancy has complicated genetic analyses (Agrawal et al. 2014).
Connecting the Dots between Transcription Factors and Cell Adhesion
Although direct evidence for an association between human CP phenotypes and cell adhesion molecules is currently restricted to NECTIN1, CDH1, and EFNB1, there is evidence to suggest that cell adhesions are affected in other OFC syndromes, including those linked to mutant p63 (Ferone et al. 2015). p63 (TP63) is a p53 homologue best known as a stratified epithelial-defining transcription factor (Mills et al. 1999; Yang et al. 1999). Variants in TP63 underlie multiple human CL/P syndromes, including ankyloblepharon-ectodermal dysplasia-cleft lip/palate (AEC; OMIM:106260) and ectrodactyly, ectodermal dysplasia, cleft lip/palate syndrome 3 (EEC3; OMIM:604292). Murine p63 (Trp63) germline mutants present with CP, and Trp63+/−;Irf6+/− double haploinsufficiency mutants also exhibit a CP phenotype (Yang et al. 1999; Thomason et al. 2010).
There is mounting evidence that p63 plays a complex role in palate closure, whereby its early expression in PS promotes specification of the suprabasal periderm layer, and its later downregulation in the MEE promotes reorganization of the cell–cell adhesions that facilitate periderm migration and MES dissolution (Richardson et al. 2017). The presence of the periderm is necessary to prevent aberrant intra-oral adhesions, which can prevent PS elevation and cause CP, and p63 plays a role in this process (Richardson et al. 2014; Hu et al. 2015; Richardson et al. 2017). Interestingly, ΔNp63α, though highly expressed in the PS epithelium, is downregulated during MES formation (Thomason et al. 2010), suggesting that p63 loss promotes periderm dissolution. In support of this “dual role” of p63 in palatogenesis, the Dixon lab recently demonstrated that haploinsufficiency for Trp63 can rescue CP observed in Tgfb3 mutants (where p63 expression is ectopically maintained in the MES), and that overexpression of ΔNp63α can induce CP by preventing MES dissolution (Richardson et al. 2017). This study also provides evidence that the periderm is not shed during the transition from MEE to MES but, rather, migrates out of the MES toward the nasal and oral surfaces to form “epithelial triangles,” supporting the idea that the periderm participates in the formation of nascent adhesions between the MEE.
Interestingly, NECTIN-1 is a direct target of p63, and p63 deletion results in a near-complete loss of Nectin-1 in both the epidermis and the MEE (Mollo et al. 2015). Human keratinocytes harboring the AEC L514F mutation exhibit reduced Nectin-1 expression (Mollo et al. 2015), suggesting the CP phenotype may be mediated, at least in part, by Nectin-1 loss in the MEE. Heterozygous knock-in mice expressing the p63L514F variant display fully penetrant CP, attributed to defective FGF signaling; and skin fragility, attributed to reduced expression of desmosomal proteins (Ferone et al. 2012; Ferone et al. 2013). However, whereas Irf6 and Fgfr2/3 have gathered much attention as p63 targets, emerging evidence suggests cell–cell adhesion molecules, including NECTIN1, DSC1, DSG3, DSP, and CDH3, may also be relevant (Shimomura et al. 2008; Ferone et al. 2013; Ferone et al. 2015). Perhaps the strongest evidence to date comes from in vivo transcriptional profiling and ΔNp63 CHIP-seq analyses comparing wild-type and Trp63 loss- and gain-of-function mutants, which have revealed striking alterations in the expression of AJ, desmosomal, and TJ genes (Richardson et al. 2017). Of particular interest, E14 Trp63-/- mutant PS shows reduced mRNA expression of Cdh3/P-cadherin, Nectin-1, and the desmosomal components Pkp1, Pkp3, Dsc3, Dsg2, and Dsg3. Moreover, although Nectin-4 mRNA levels are not altered in Trp63−/− mutants, Nectin-4 protein becomes mislocalized away from the basal-periderm junction to lateral basal–basal cell junctions. These data provide compelling evidence that p63 plays a critical role in regulating MEE cell–cell adhesions (Richardson et al. 2017).
Ex Vivo Palate Cultures
Before the advent of genetically engineered mice, the most widely used system to perturb palatogenesis was ex vivo palate cultures. Given the inaccessibility of the palate region, it is difficult to perform intravital imaging. However, exciting recent live-imaging studies of ex vivo cultures have granted new insights into the processes that regulate palatal fusion, including cell extrusion (Kim et al. 2015). Since the initial development of ex vivo palate culture by Moriarty et al. (1963), the technique has been adapted to include numerous model organisms that present different contexts for studying palatogenesis. Most birds and reptiles have incomplete secondary palatal fusion (Ferguson 1988), whereas chick palates can complete fusion when cultured with recombinant TGFβ3, reinforcing the importance of TGFβ in regulating palate closure (Sun et al. 1998).
One notable advantage of palatal cultures is the ability to “mix and match” PS from different organisms or genetic backgrounds. For example, it was demonstrated that LacZ+ cells from the Rosa26-lacZ mouse exhibited preferential migration in the nasal and posterior directions into the opposing LacZ− shelf during MES fusion (Jin and Ding 2006). More recently, murine PS cultures have incorporated the ever-expanding, molecular-genetic toolkit, including fluorescent reporters, genetic manipulation, and live-imaging that allow for a unique opportunity to rapidly study palatal closure (Jin and Ding 2006; Ke et al. 2015; Zhang et al. 2016; Narhi 2017).
Animal Models to Investigate Epithelial Cell–Cell Adhesion in Palate Closure
Differences in craniofacial development and structure between fish, birds, reptiles, and mammals make the mouse the most tractable genetic organism to study secondary palate fusion, as it relates to humans. Both germline and conditional mouse knockouts have been used to recapitulate human CP disorders with remarkable fidelity (see Figure 2B), and the latter has been particularly important in deciphering mesenchymal vs. epithelial contributions to CL/P. Whereas a vast arsenal of tissue-specific Cre driver lines has been used to query mesenchymal gene function in PS morphogenesis, comparatively few epithelial drivers exist, limited largely to Shh-Cre (Ahn et al. 2010), Tgfb3-Cre (Yang and Kaartinen 2007), Pitx2-Cre (Xiong et al. 2009) and Krt14-Cre.
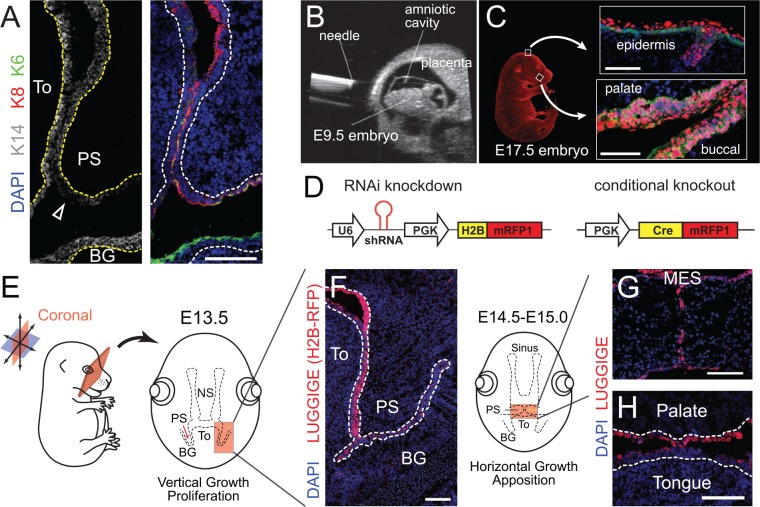
Cytokeratin-14 (Krt14 or K14) is expressed throughout the basal layer of stratified epithelia, including the epidermis, cornea, and oral epithelia. Interestingly, however, its expression at the protein level appears to be notably lower in the PS epithelium than in neighboring buccogingiva or tongue epithelium (Figure 4A). Complicating matters, there are no less than 7 distinct published Krt14-Cre alleles, of which 3 have been used to study palatogenesis (Vasioukhin et al. 1999; Dassule et al. 2000; Andl et al. 2004). Although the Millar lab allele seems to be most widely utilized in the field (Andl et al. 2004), it should be noted that there are 3 separate founder lines (designated 40, 43, and 52), which each have different levels of Cre activity. For example, in the context of lethality caused by Bmpr1a deletion, Krt14-Cre43 shows the strongest phenotype and Krt14-Cre40 shows the weakest, with Krt14-Cre52 intermediate. A summary of the various Krt14-Cre lines used to study palate closure is shown in Appendix Table 3.
Figure 4.
LUGGIGE in oral epithelia. (A) Expression of keratins in E13.5 pre-elevation palatal shelves (PS). K14 (in gray, Origene BP5009) is reduced in PS (indicated by arrowhead) as compared with nearby tongue and buccogingiva, whereas the oral keratin K6A (green, Biolegend Poly19057) and periderm marker K8 (red, Developmental Studies Hybridoma Bank TROMA-I) are present. (B, C) LUGGIGE. (B) Ultrasound image of lentiviral injection into the amniotic fluid surrounding E9.5 embryos. (C) LUGGIGE-transduced E17.5 embryo showing epithelial-specific expression of the nuclear histone H2B-mRFP1 reporter in epidermis and oral tissues. (D) Examples of lentiviral constructs harboring an shRNA and H2B-RFP1 reporter (left) for knocking down target genes, or Cre-RFP (right) for generating conditional knockouts. (E) Schematic of the palate region viewed by coronal section. (F–H) LUGGIGE can achieve high transduction in palatal cells at both early (F) and late (G, H) stages of palatogenesis. Scale bars: 100 µm. BG, buccogingiva; MES, medial epithelial seam; NS, Nasal septum; PS, palatal shelf; To, tongue.
While many groups have verified Krt14-Cre activity during embryonic palate development through reporter mice (Dassule et al. 2000; Jin and Ding 2006; Hosokawa et al. 2009; He et al. 2010b), it must be noted that the timing of reporter expression cannot be used as a surrogate for the timing of protein loss, which is affected by factors such as protein and mRNA stability. For example, the McMahon Krt14-Cre line (Dassule et al. 2000; Jax stock #018964) induced recombination of a GFP reporter for imaging purposes but was less effective at deleting Myh9 for functional analyses (Kim et al. 2015). Additionally, studies of Krt14-Cre-mediated loss of Shh highlight the significant differences that exist between these transgenic lines. In 3 studies using the McMahon line, 2 groups failed to report CP (Dassule et al. 2000; Economou et al. 2012) and one reported CP with 85% penetrance (Rice et al. 2004). Yet another study utilizing the Millar Krt14-Cre43 line reported CP with 70% penetrance (Lan and Jiang 2009). Another example is β-catenin (Ctnnb1), where CP was not reported using the Birchmeier Krt14-Cre line (Huelsken et al. 2001; Andl et al. 2004), whereas a later study using a Millar Krt14-Cre line observed CP with high penetrance (He et al. 2011). While it is possible that, in some cases, a CP phenotype may have been present but was simply overlooked, variable penetrance can be attributed to other factors, including differences in the timing of initiation of Krt14-Cre transgene expression, mosaicism, and strain differences. Furthermore, although the utility of these lines in studying palatogenesis is evident, caution is advised in the choice of which line to use, and in the application for which it is intended.
Other Approaches to Study Epithelial Contributions to Secondary Palate Closure
Viral vectors provide a powerful and versatile means to query and modulate gene function in a more rapid and high-throughput fashion than can be accomplished by traditional transgenic approaches. One elegant example of how this can be applied to study palatogenesis was recently demonstrated by Wu and colleagues (2013). Using intra-amniotic delivery of an adenovirus encoding TGFβ3 between E12.5 and E16.5, the authors showed that restoration of TGFβ3 expression specifically in the periderm could rescue the CP defect observed in TGFβ3-null mice, providing evidence that TGFβ3 is required in the periderm for it to be removed from the MEE surface prior to fusion. More recently, Ke and colleagues (2015) used lentiviral and adenoviral transduction in palatal cultures to show that IRF6 acts downstream of TGFβ3 to promote epithelial-mesenchymal transition during palate fusion.
If performed before periderm formation (E9.5), virus delivered into the amniotic space can transduce single-layered surface epithelia, leading to expression in basal cells and their progeny, including differentiated suprabasal cells and periderm (Beronja et al. 2010). Using this approach (Figure 4B–C), ultrasound-guided intra-amniotic delivery of lentiviruses harboring shRNAs can transduce the epidermis at >90% efficiency, allowing for elucidation of the genetic pathways that regulate epidermal stratification (Beronja et al. 2010; Williams et al. 2011; Williams et al. 2014). We recently showed that this technique, which we term LUGGIGE (Lentiviral Ultrasound-Guided Gene Inactivation and Gene Expression), can efficiently transduce oral epithelia at all stages of palatogenesis (Figure 4D–G), and that oral epithelial stratification is dependent on oriented cell divisions (Byrd et al. 2016). Importantly, lentiviruses can be engineered for a variety of applications (Figure 4C), including over/mis-expression, expression of disease variants, reporter gene expression, shRNA knockdown, and expression of constitutive or inducible Cre recombinase, which can be used to excise floxed alleles in a spatially and temporally controlled manner (Williams et al. 2011; Williams et al. 2014).
LUGGIGE also represents an alternative method for probing gene function in the periderm during palate fusion. Lane and Kaartinen (2014) speculated that part of the reason why Krt14-Cre conditional KOs of TGFβ family members cause CP with lower penetrance than germline mutants is due at least in part to inactivity of Krt14-Cre in the periderm. Thus, although it is likely that LUGGIGE can induce gene expression/loss earlier and more uniformly in the palatal epithelium than Krt14-Cre alleles—as has been demonstrated in the epidermis (Beronja et al. 2010)—it must also be considered that the ability to transduce the periderm may be important in the context of palate closure.
Conclusions
Adhesion between epithelial cells of apposing PS is an obligate first step of palatal fusion. Human genetic studies reveal that AJ components of both the nectin and cadherin families are essential for proper palate closure, and it is likely that other cell–cell adhesions, including desmosomes, play important roles as well. A challenge going forward will be to develop better tools, including combinations of gene knockouts and more faithful disease models mimicking the mutations found in humans. Armed with an array of powerful culture systems, imaging techniques, and animal models, researchers are poised in the next decade to make significant advances in closing the gap between suspected and known CL/P genes, and deciphering the cellular and molecular mechanisms by which they operate.
Author Contributions
K.J. Lough, K.M. Byrd, contributed to conception, design, data acquisition, analysis, and interpretation, drafted and critically revised the manuscript; D.C. Spitzer, contributed to conception, design, data acquisition, analysis, and interpretation, critically revised the manuscript; S.E. Williams, contributed to conception, design, data analysis, and interpretation, drafted and critically revised the manuscript. All authors gave final approval and agree to be accountable for all aspects of the work.
Acknowledgments
We are grateful to members of the Williams lab for helpful discussions and critical reading of the text.
Footnotes
A supplemental appendix to this article is available online.
SEM images in Figure 1 were modified from images obtained from FaceBase (www.facebase.org). The FaceBase consortium (U01DE020076) and the FaceBase Coordinating Hub (1U01DE024449-01) are funded by the National Institute of Dental and Craniofacial Research. This work was supported by a Ruth L. Kirschstein Predoctoral Individual National Research Service Award (F31 DE026956-01 to K.J.L.), Mentored Clinical Scientist Research Career Development Award (K08 DE026537-01 to K.M.B.), Extramural Loan Repayment Program for Pediatric Research (LRP-PR to K.M.B.), Center for Gastrointestinal Biology and Disease Pilot Feasibility Award (parent P30 DK034987 to S.E.W.) and a Sidney Kimmel Foundation Kimmel Scholar Award (SKF-15-065 to S.E.W.).
The authors declare no potential conflicts of interest with respect to the authorship and/or publication of this article.
References
- Agrawal P, Wang M, Kim S, Lewis AE, Bush JO. 2014. Embryonic expression of epha receptor genes in mice supports their candidacy for involvement in cleft lip and palate. Dev Dyn. 243(11):1470–1476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahn Y, Sanderson BW, Klein OD, Krumlauf R. 2010. Inhibition of wnt signaling by wise (sostdc1) and negative feedback from shh controls tooth number and patterning. Development. 137(19):3221–3231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andl T, Ahn K, Kairo A, Chu EY, Wine-Lee L, Reddy ST, Croft NJ, Cebra-Thomas JA, Metzger D, Chambon P, et al. 2004. Epithelial bmpr1a regulates differentiation and proliferation in postnatal hair follicles and is essential for tooth development. Development. 131(10):2257–2268. [DOI] [PubMed] [Google Scholar]
- Aslar D, Tastan H. 2014. Novel insertion mutation in the pvrl1 gene in turkish patients with non-syndromic cleft lip with/without cleft palate. Arch Oral Biol. 59(3):237–240. [DOI] [PubMed] [Google Scholar]
- Avila JR, Jezewski PA, Vieira AR, Orioli IM, Castilla EE, Christensen K, Daack-Hirsch S, Romitti PA, Murray JC. 2006. Pvrl1 variants contribute to non-syndromic cleft lip and palate in multiple populations. Am J Med Genet A. 140(23):2562–2570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beronja S, Livshits G, Williams S, Fuchs E. 2010. Rapid functional dissection of genetic networks via tissue-specific transduction and rnai in mouse embryos. Nat Med. 16(7):821–827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brancati F, Fortugno P, Bottillo I, Lopez M, Josselin E, Boudghene-Stambouli O, Agolini E, Bernardini L, Bellacchio E, Iannicelli M, et al. 2010. Mutations in pvrl4, encoding cell adhesion molecule nectin-4, cause ectodermal dysplasia-syndactyly syndrome. Am J Hum Genet. 87(2):265–273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brito LA, Yamamoto GL, Melo S, Malcher C, Ferreira SG, Figueiredo J, Alvizi L, Kobayashi GS, Naslavsky MS, Alonso N, et al. 2015. Rare variants in the epithelial cadherin gene underlying the genetic etiology of nonsyndromic cleft lip with or without cleft palate. Hum Mutat. 36(11):1029–1033. [DOI] [PubMed] [Google Scholar]
- Bush JO, Jiang R. 2012. Palatogenesis: Morphogenetic and molecular mechanisms of secondary palate development. Development. 139(2):231–243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bush JO, Soriano P. 2010. Ephrin-b1 forward signaling regulates craniofacial morphogenesis by controlling cell proliferation across eph-ephrin boundaries. Genes Dev. 24(18):2068–2080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byrd KM, Lough KJ, Patel JH, Descovich CP, Curtis TA, Williams SE. 2016. Lgn plays distinct roles in oral epithelial stratification, filiform papilla morphogenesis and hair follicle development. Development. 143(15):2803–2817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Lan Y, Baek JA, Gao Y, Jiang R. 2009. Wnt/beta-catenin signaling plays an essential role in activation of odontogenic mesenchyme during early tooth development. Dev Biol. 334(1):174–185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danescu A, Mattson M, Dool C, Diewert VM, Richman JM. 2015. Analysis of human soft palate morphogenesis supports regional regulation of palatal fusion. J Anat. 227(4):474–486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dassule HR, Lewis P, Bei M, Maas R, McMahon AP. 2000. Sonic hedgehog regulates growth and morphogenesis of the tooth. Development. 127(22):4775–4785. [DOI] [PubMed] [Google Scholar]
- Dixon MJ, Marazita ML, Beaty TH, Murray JC. 2011. Cleft lip and palate: Understanding genetic and environmental influences. Nat Rev Genet. 12(3):167–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Economou AD, Ohazama A, Porntaveetus T, Sharpe PT, Kondo S, Basson MA, Gritli-Linde A, Cobourne MT, Green JB. 2012. Periodic stripe formation by a turing mechanism operating at growth zones in the mammalian palate. Nat Genet. 44(3):348–351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferguson MW. 1988. Palate development. Development. 103 Suppl:41–60. [DOI] [PubMed] [Google Scholar]
- Ferone G, Thomason HA, Antonini D, De Rosa L, Hu B, Gemei M, Zhou H, Ambrosio R, Rice DP, Acampora D, et al. 2012. Mutant p63 causes defective expansion of ectodermal progenitor cells and impaired fgf signalling in aec syndrome. EMBO Mol Med. 4(3):192–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferone G, Mollo MR, Thomason HA, Antonini D, Zhou H, Ambrosio R, De Rosa L, Salvatore D, Getsios S, van Bokhoven H, et al. 2013. P63 control of desmosome gene expression and adhesion is compromised in aec syndrome. Hum Mol Genet. 22(3):531–543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferone G, Mollo MR, Missero C. 2015. Epidermal cell junctions and their regulation by p63 in health and disease. Cell Tissue Res. 360(3):513–528. [DOI] [PubMed] [Google Scholar]
- Finger JH, Smith CM, Hayamizu TF, McCright IJ, Xu J, Law M, Shaw DR, Baldarelli RM, Beal JS, Blodgett O, et al. 2017. The mouse gene expression database (gxd): 2017 update. Nucleic Acids Res. 45(D1):D730–D736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fortugno P, Josselin E, Tsiakas K, Agolini E, Cestra G, Teson M, Santer R, Castiglia D, Novelli G, Dallapiccola B, et al. 2014. Nectin-4 mutations causing ectodermal dysplasia with syndactyly perturb the rac1 pathway and the kinetics of adherens junction formation. J Invest Dermatol. 134(8):2146–2153. [DOI] [PubMed] [Google Scholar]
- Frebourg T, Oliveira C, Hochain P, Karam R, Manouvrier S, Graziadio C, Vekemans M, Hartmann A, Baert-Desurmont S, Alexandre C, et al. 2006. Cleft lip/palate and cdh1/e-cadherin mutations in families with hereditary diffuse gastric cancer. J Med Genet. 43(2):138–142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greene RM, Pratt RM. 1976. Developmental aspects of secondary palate formation. J Embryol Exp Morphol. 36(2):225–245. [PubMed] [Google Scholar]
- He F, Popkie AP, Xiong W, Li L, Wang Y, Phiel CJ, Chen Y. 2010. a. Gsk3beta is required in the epithelium for palatal elevation in mice. Dev Dyn. 239(12):3235–3246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He F, Xiong W, Wang Y, Li L, Liu C, Yamagami T, Taketo MM, Zhou C, Chen Y. 2011. Epithelial wnt/beta-catenin signaling regulates palatal shelf fusion through regulation of tgfbeta3 expression. Dev Biol. 350(2):511–519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He F, Xiong W, Wang Y, Matsui M, Yu X, Chai Y, Klingensmith J, Chen Y. 2010. b. Modulation of bmp signaling by noggin is required for the maintenance of palatal epithelial integrity during palatogenesis. Dev Biol. 347(1):109–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosokawa R, Deng X, Takamori K, Xu X, Urata M, Bringas P, Jr, Chai Y. 2009. Epithelial-specific requirement of fgfr2 signaling during tooth and palate development. J Exp Zool B Mol Dev Evol. 312B(4):343–350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu L, Liu J, Ozturk F, Gurumurthy C, Romano RA, Sinha S, Nawshad A. 2015. TGFβ3 regulates periderm removal through ΔNp63 in the developing palate. J Cell Physiol. 230(6)1212–1225. [DOI] [PubMed] [Google Scholar]
- Huelsken J, Vogel R, Erdmann B, Cotsarelis G, Birchmeier W. 2001. β-catenin controls hair follicle morphogenesis and stem cell differentiation in the skin. Cell. 105(4):533–545. [DOI] [PubMed] [Google Scholar]
- Inagaki M, Irie K, Ishizaki H, Tanaka-Okamoto M, Morimoto K, Inoue E, Ohtsuka T, Miyoshi J, Takai Y. 2005. Roles of cell-adhesion molecules nectin 1 and nectin 3 in ciliary body development. Development. 132(7):1525–1537. [DOI] [PubMed] [Google Scholar]
- Jin JZ, Ding J. 2006. Analysis of cell migration, transdifferentiation and apoptosis during mouse secondary palate fusion. Development. 133(17):3341–3347. [DOI] [PubMed] [Google Scholar]
- Kania A, Klein R. 2016. Mechanisms of ephrin-eph signalling in development, physiology and disease. Nat Rev Mol Cell Biol. 17(4):240–256. [DOI] [PubMed] [Google Scholar]
- Kawakatsu T, Shimizu K, Honda T, Fukuhara T, Hoshino T, Takai Y. 2002. Trans-interactions of nectins induce formation of filopodia and lamellipodia through the respective activation of cdc42 and rac small g proteins. J Biol Chem. 277(52):50749–50755. [DOI] [PubMed] [Google Scholar]
- Ke CY, Xiao WL, Chen CM, Lo LJ, Wong FH. 2015. Irf6 is the mediator of tgfbeta3 during regulation of the epithelial mesenchymal transition and palatal fusion. Sci Rep. 5:12791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S, Lewis AE, Singh V, Ma X, Adelstein R, Bush JO. 2015. Convergence and extrusion are required for normal fusion of the mammalian secondary palate. PLoS Biol. 13(4):e1002122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitase Y, Shuler CF. 2013. Microtubule disassembly prevents palatal fusion and alters regulation of the e-cadherin/catenin complex. Int J Dev Biol. 57(1):55–60. [DOI] [PubMed] [Google Scholar]
- Kousa YA, Mansour TA, Seada H, Matoo S, Schutte BC. 2017. Shared molecular networks in orofacial and neural tube development. Birth Defects Res. 109(2):169–179. [DOI] [PubMed] [Google Scholar]
- Lan Y, Jiang R. 2009. Sonic hedgehog signaling regulates reciprocal epithelial-mesenchymal interactions controlling palatal outgrowth. Development. 136(8):1387–1396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lane J, Kaartinen V. 2014. Signaling networks in palate development. Wiley Interdiscip Rev Syst Biol Med. 6(3):271–278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larue L, Ohsugi M, Hirchenhain J, Kemler R. 1994. E-cadherin null mutant embryos fail to form a trophectoderm epithelium. Proc Natl Acad Sci U S A. 91(17):8263–8267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma L, Shi B, Zheng Q. 2015. Targeted mutations of genes reveal important roles in palatal development in mice. Ann Plast Surg. 74(2):263–268. [DOI] [PubMed] [Google Scholar]
- Mandai K, Rikitake Y, Mori M, Takai Y. 2015. Nectins and nectin-like molecules in development and disease. Curr Topics Dev Biol. 112:197–231. [DOI] [PubMed] [Google Scholar]
- Miravet S, Piedra J, Miró F, Itarte E, García de, Herreros A, Duñach M. 2002. The transcriptional factor tcf-4 contains different binding sites for beta-catenin and plakoglobin. J Biol Chem. 277(3):1884–1891. [DOI] [PubMed] [Google Scholar]
- Miyahara M, Nakanishi H, Takahashi K, Satoh-Horikawa K, Tachibana K, Takai Y. 2000. Interaction of nectin with afadin is necessary for its clustering at cell-cell contact sites but not for its cis dimerization or trans interaction. J Biol Chem. 275(1):613–618. [DOI] [PubMed] [Google Scholar]
- Mogass M, Bringas P, Jr, Shuler CF. 2000. Characterization of desmosomal component expression during palatogenesis. Int J Dev Biol. 44(3):317–322. [PubMed] [Google Scholar]
- Mollo MR, Antonini D, Mitchell K, Fortugno P, Costanzo A, Dixon J, Brancati F, Missero C. 2015. P63-dependent and independent mechanisms of nectin-1 and nectin-4 regulation in the epidermis. Exp Dermatol. 24(2):114–119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moriarty TM, Weinstein S, Gibson RD. 1963. The development in vitro and in vivo of fusion of the palatal processes of rat embryos. J Embryol Exp Morphol. 11:605–619. [PubMed] [Google Scholar]
- Narhi K. 2017. Embryonic explant culture: Studying effects of regulatory molecules on gene expression in craniofacial tissues. Methods Mol Biol. 1537:367–380. [DOI] [PubMed] [Google Scholar]
- Niehrs C. 2012. The complex world of wnt receptor signalling. Nat Rev Mol Cell Biol. 13(12):767–779. [DOI] [PubMed] [Google Scholar]
- Reymond N, Fabre S, Lecocq E, Adelaide J, Dubreuil P, Lopez M. 2001. Nectin4/prr4, a new afadin-associated member of the nectin family that trans-interacts with nectin1/prr1 through v domain interaction. J Biol Chem. 276(46):43205–43215. [DOI] [PubMed] [Google Scholar]
- Rice R, Spencer-Dene B, Connor EC, Gritli-Linde A, McMahon AP, Dickson C, Thesleff I, Rice DP. 2004. Disruption of fgf10/fgfr2b-coordinated epithelial-mesenchymal interactions causes cleft palate. J Clin Invest. 113(12):1692–1700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson RJ, Hammond NL, Coulombe PA, Saloranta C, Nousiainen HO, Salonen R, Berry A, Hanley N, Headon D, Karikoski R, et al. 2014. Periderm prevents pathological epithelial adhesions during embryogenesis. J Clin Invest. 124(9):3891–3900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson R, Mitchell K, Hammond NL, Mollo MR, Kouwenhoven EN, Wyatt ND, Donaldson IJ, Zeef L, Burgis T, Blance R, van Heeringen SJ, et al. 2017. p63 exerts spatio-temporal control of palatal epithelial cell fate to prevent cleft palate. PLoS Genet. 13(6):e1006828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Risley M, Garrod D, Henkemeyer M, McLean W. 2009. Ephb2 and ephb3 forward signalling are required for palate development. Mech Dev. 126(3–4):230–239. [DOI] [PubMed] [Google Scholar]
- San Miguel S, Serrano MJ, Sachar A, Henkemeyer M, Svoboda KK, Benson MD. 2011. Ephrin reverse signaling controls palate fusion via a pi3 kinase-dependent mechanism. Dev Dyn. 240(2):357–364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serrano MJ, Liu J, Svoboda KK, Nawshad A, Benson MD. 2015. Ephrin reverse signaling mediates palatal fusion and epithelial-to-mesenchymal transition independently of Tgfβ3. J Cell Physiol. 230(12):2961–2972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimomura Y, Wajid M, Shapiro L, Christiano AM. 2008. P-cadherin is a p63 target gene with a crucial role in the developing human limb bud and hair follicle. Development. 135(4):743–753. [DOI] [PubMed] [Google Scholar]
- Sözen MA, Suzuki K, Tolarova MM, Bustos T, Fernández Iglesias JE, Spritz RA. 2001. Mutation of pvrl1 is associated with sporadic, non-syndromic cleft lip/palate in northern venezuela. Nat Genet. 29(2):141–142. [DOI] [PubMed] [Google Scholar]
- Sun D, Vanderburg CR, Odierna GS, Hay ED. 1998. Tgfbeta3 promotes transformation of chicken palate medial edge epithelium to mesenchyme in vitro. Development. 125(1):95–105. [DOI] [PubMed] [Google Scholar]
- Suzuki K, Hu D, Bustos T, Zlotogora J, Richieri-Costa A, Helms JA, Spritz RA. 2000. Mutations of pvrl1, encoding a cell-cell adhesion molecule/herpesvirus receptor, in cleft lip/palate-ectodermal dysplasia. Nat Genet. 25(4):427–430. [DOI] [PubMed] [Google Scholar]
- Thomason HA, Zhou H, Kouwenhoven EN, Dotto GP, Restivo G, Nguyen BC, Little H, Dixon MJ, van Bokhoven H, Dixon J. 2010. Cooperation between the transcription factors p63 and irf6 is essential to prevent cleft palate in mice. J Clin Invest. 120(5):1561–1569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tinkle CL, Lechler T, Pasolli HA, Fuchs E. 2004. Conditional targeting of e-cadherin in skin: Insights into hyperproliferative and degenerative responses. Proc Natl Acad Sci U S A. 101(2):552–557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tinkle CL, Pasolli HA, Stokes N, Fuchs E. 2008. New insights into cadherin function in epidermal sheet formation and maintenance of tissue integrity. Proc Natl Acad Sci U S A. 105(40):15405–15410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Togashi H, Kominami K, Waseda M, Komura H, Miyoshi J, Takeichi M, Takai Y. 2011. Nectins establish a checkerboard-like cellular pattern in the auditory epithelium. Science. 333(6046):1144–1147. [DOI] [PubMed] [Google Scholar]
- Tudela C, Formoso MA, Martínez T, Pérez R, Aparicio M, Maestro C, Del Río A, Martínez E, Ferguson M, Martínez-Alvarez C. 2002. Tgf-beta3 is required for the adhesion and intercalation of medial edge epithelial cells during palate fusion. Int J Dev Biol. 46(3):333–336. [PubMed] [Google Scholar]
- Tunggal JA, Helfrich I, Schmitz A, Schwarz H, Gunzel D, Fromm M, Kemler R, Krieg T, Niessen CM. 2005. E-cadherin is essential for in vivo epidermal barrier function by regulating tight junctions. EMBO J. 24(6):1146–1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Twigg SR, Kan R, Babbs C, Bochukova EG, Robertson SP, Wall SA, Morriss-Kay GM, Wilkie AO. 2004. Mutations of ephrin-B1 (EFNB1), a martker of tissue boundary formation, cause craniofrontonasal syndrome. Proc Natl Acad Sci U S A. 101(23):8652–8657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanbokhoven H, Melino G, Candi E, Declercq W. 2011. P63, a story of mice and men. J Invest Dermatol. 131(6):1196–1207. [DOI] [PubMed] [Google Scholar]
- Vasioukhin V, Degenstein L, Wise B, Fuchs E. 1999. The magical touch: Genome targeting in epidermal stem cells induced by tamoxifen application to mouse skin. Proc Natl Acad Sci U S A. 96(15):8551–8556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogelaar IP, Figueiredo J, van Rooij IA, Simoes-Correia J, van der Post RS, Melo S, Seruca R, Carels CE, Ligtenberg MJ, Hoogerbrugge N. 2013. Identification of germline mutations in the cancer predisposing gene cdh1 in patients with orofacial clefts. Hum Mol Genet. 22(5):919–926. [DOI] [PubMed] [Google Scholar]
- Walker BE, Fraser FC. 1956. Closure of the secondary palate in three strains of mice. J Embryol Exp Morphol. 4:176-89. URL accessed on 7/25/2017 at: http://garfield.library.upenn.edu/classics1988/A1988P409000001.pdf [Google Scholar]
- Warrington A, Vieira AR, Christensen K, Orioli IM, Castilla EE, Romitti PA, Murray JC. 2006. Genetic evidence for the role of loci at 19q13 in cleft lip and palate. J Med Genet. 43(6):e26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wieland I, Jakubiczka S, Muschke P, Cohen M, Thiele H, Gerlach KL, Adams RH, Wieacker P. 2004. Mutations of the ephrin-B1 gene cause craniofrontonasal syndrome. Am J Hum Genet. 74(6):1209–1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams SE, Beronja S, Pasolli HA, Fuchs E. 2011. Asymmetric cell divisions promote notch-dependent epidermal differentiation. Nature. 470(7334): 353–358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams SE, Ratliff LA, Postiglione MP, Knoblich JA, Fuchs E. 2014. Par3-mInsc and Gαi3 cooperate to promote oriented epidermal cell divisions through LGN. Nat Cell Biol. 16(8):758–769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C, Endo M, Yang BH, Radecki MA, Davis PF, Zoltick PW, Spivak RM, Flake AW, Kirschner RE, Nah HD. 2013. Intra-amniotic transient transduction of the periderm with a viral vector encoding tgfbeta3 prevents cleft palate in tgfbeta3(-/-) mouse embryos. Mol Ther. 21(1):8–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong W, He F, Morikawa Y, Yu X, Zhang Z, Lan Y, Jiang R, Cserjesi P, Chen Y. 2009. Hand2 is required in the epithelium for palatogenesis in mice. Dev Biol. 330(1):131–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang A, Schweitzer R, Sun D, Kaghad M, Walker N, Bronson RT, Tabin C, Sharpe A, Caput D, Crum C, et al. 1999. P63 is essential for regenerative proliferation in limb, craniofacial and epithelial development. Nature. 398(6729):714–718. [DOI] [PubMed] [Google Scholar]
- Yang LT, Kaartinen V. 2007. Tgfb1 expressed in the tgfb3 locus partially rescues the cleft palate phenotype of tgfb3 null mutants. Dev Biol. 312(1):384–395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasumi M, Shimizu K, Honda T, Takeuchi M, Takai Y. 2003. Role of each immunoglobulin-like loop of nectin for its cell-cell adhesion activity. Biochem Biophys Res Commun. 302(1):61–66. [DOI] [PubMed] [Google Scholar]
- Yoshida K, Hayashi R, Fujita H, Kubota M, Kondo M, Shimomura Y, Niizeki H. 2015. Novel homozygous mutation, c.400c>t (p.Arg134*), in the pvrl1 gene underlies cleft lip/palate-ectodermal dysplasia syndrome in an asian patient. J Dermatol. 42(7):715–719. [DOI] [PubMed] [Google Scholar]
- Yoshida M, Shimono Y, Togashi H, Matsuzaki K, Miyoshi J, Mizoguchi A, Komori T, Takai Y. 2012. Periderm cells covering palatal shelves have tight junctions and their desquamation reduces the polarity of palatal shelf epithelial cells in palatogenesis. Genes Cells. 17(6):455–472. [DOI] [PubMed] [Google Scholar]
- Yoshida T, Iwata T, Takai Y, Birchmeier W, Yamato M, Okano T. 2014. Afadin requirement for cytokine expressions in keratinocytes during chemically induced inflammation in mice. Genes Cells. 19(11):842–852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida T, Miyoshi J, Takai Y, Thesleff I. 2010. Cooperation of nectin-1 and nectin-3 is required for normal ameloblast function and crown shape development in mouse teeth. Dev Dyn. 239(10):2558–2569. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Dong S, Wang W, Wang J, Wang M, Chen M, Hou J, Huang H. 2016. Activation of notch1 inhibits medial edge epithelium apoptosis in all-trans retinoic acid-induced cleft palate in mice. Biochem Biophys Res Commun. 477(3):322–328. [DOI] [PubMed] [Google Scholar]
- Zihni C, Mills C, Matter K, Balda MS. 2016. Tight junctions: From simple barriers to multifunctional molecular gates. Nat Rev Mol Cell Biol. 17(9):564–580. [DOI] [PubMed] [Google Scholar]