Abstract
Background
Since its commercial release in 2011 cell-free DNA screening has been rapidly adopted as a routine prenatal genetic test. However, little is known about its performance in actual clinical practice.
Objective
To investigate factors associated with the accuracy of abnormal autosomal cell- free DNA results.
Study Design
Retrospective cohort study of 121 patients with abnormal cell-free DNA results from a referral maternal-fetal medicine practice from March 2013 to July 2015. Patients were included if cell-free DNA results for trisomy 21, trisomy 18, trisomy 13, or microdeletions (if reported by the laboratory) were positive or non-reportable. The primary outcome was confirmed aneuploidy or microarray abnormality on either prenatal or postnatal karyotype or microarray. Secondary outcomes were identifiable associations with in vitro fertilization, twins, ultrasound findings, testing platform, and testing laboratory. Kruskal-Wallis or Fisher’s exact tests were used as appropriate.
Results
121 patients had abnormal cell-free DNA results for for trisomy 21, trisomy 18, trisomy 13, and/or microdeletions. 105 patients had abnormal cell-free DNA results for for trisomy 21, trisomy 18, trisomy 13. Of these, 92 (87.6%) were positive and 13 (12.4%) were non-reportable. The results of the 92 positive cell-free DNA were for trisomy 21 (48, 52.2%), trisomy 18 (22, 23.9%), trisomy 13 (17, 18.5%), triploidy (2, 2.2%), and positive for >1 parameter (3, 3.3%). Overall, the positive predictive value of cell-free DNA was 73.5% (61/83, 95% confidence interval (CI) 63% to 82%) for all trisomies (by chromosome: trisomy 21, 83.0% (39/47, CI 69% to 92%, trisomy 18, 65.0% (13/20, CI 41% to 84%), and trisomy 13, 43.8% (7/16, CI 21% to 70%). Abnormal cell-free DNA results were associated with positive serum screening (by group: trisomy 21 17/48, 70.8%; trisomy 18 7/22, 77.8%; trisomy 13 3/17, 37.5%; non-reportable 2/13, 16.7%; p=0.004), and abnormal first trimester ultrasound (trisomy 21 25/45, 55.6%; trisomy 18 13/20, 65%; trisomy 13 6/14, 42.9%; non-reportable 1/13, 7.7%; p=0.003). There was no association between false positive rates and testing platform, but there was a difference between the four laboratories (p=0.018). Twenty-six patients had positive (n=9) or non-reportable (n=17) microdeletion results. Seven of nine screens positive for microdeletions underwent confirmatory testing; all were false positives.
Conclusions
The positive predictive value of 73.5% for cell-free DNA screening for autosomal aneuploidy is lower than reported. The positive predictive value for microdeletion testing was 0%. Diagnostic testing is needed to confirm abnormal cell-free DNA results for aneuploidy and microdeletions.
Keywords: aneuploidy, cfDNA, genetic screening, microdeletion, non-invasive prenatal testing, NIPT, prenatal diagnosis, positive predictive value, trisomy 21, trisomy 18, trisomy 13
Introduction
Introduced in 2011 to clinical practice, cell free fetal DNA (cfDNA) has changed the landscape of prenatal care, despite recent guidelines recommending that its use be limited to screening for the most common aneuploidies (T21, T18, T13) in singleton gestations [1]. Numerous case controlled studies emerged reporting high sensitivities and specificities for the most common aneuploidies with low false positive rates. The highest sensitivity (>99%) and lowest false positive rates (1%) are reported for trisomy 21 (T21) [2], with slightly lower sensitivities for trisomy 18 (T18, 97–99%) and 13 (T13, 79–92%) [3–5]. While detection rates are impressive, it is notable that specific detection rates vary depending on the laboratory used. Moreover, this data is published from studies evaluating high-risk populations, and at the time of the study, validation was limited for the general population, although recent data are encouraging [1, 6–7]. In addition, few reports have examined outcomes for microdeletion testing [8, 9], which is clinically reported despite the lack of validation or endorsement by professional societies [1].
CfDNA has become widely adopted into routine prenatal care [10], and studies have reviewed its impact on patient decision-making [11]. However, most studies have focused on large, laboratory-funded validation trials. In this study, we aimed to review all abnormal cfDNA results from a single high-volume referral Maternal-Fetal Medicine practice to evaluate the performance of cfDNA in current clinical practice. Given the frequency of our observed false positive results, we hypothesized that the test performance of cfDNA in current clinical practice is not as high as previously published.
Materials and Methods
This is a retrospective cohort study of all patients with abnormal cfDNA results from a referral Maternal-Fetal Medicine practice in Los Angeles, CA, from March 2013 to July 2015. The patient population is derived from local referrals for prenatal ultrasound screening (1st and 2nd trimester), as well as consultations for high-risk pregnancies. In total, the referring physician groups manage approximately 3000 deliveries per year, of which over 95% of patients are insured. The median maternal age of the referral base is 37.5, and approximately 50% are advanced maternal age (AMA). The ethnic breakdown of the referral population is: 60.0% Caucasian, 17.4% Asian, 11.6% Hispanic, 6.2% African-American, and 4.9% other. The majority of patients have their prenatal genetic screening tests ordered by their primary obstetric provider prior to being seen at this center. Approval for this study was granted by the Institutional Review Board at the University of California, Los Angeles.
Patients were included if cfDNA screening results for chromosomes 21, 18, 13 or microdeletions (when reported) were “abnormal’, defined as positive or (non-reportable) NR. Because the recommendation for clinical follow-up is the same as for a positive result [1, 12], tests reported as “suspected positive” or “borderline” (which was the terminology used by one of the companies in early reports, indicating higher aneuploidy risk, but not meeting cutoffs for a positive result) were considered positive for the purpose of this study. A NR result for any chromosome or individual microdeletion was categorized as NR, even if all other parameters reported (any chromosome or microdeletion) were negative. A microdeletion screen was considered abnormal and included if at least one of the reported individual microdeletions had a positive or NR result. In this study, a NR microdeletion result refers to the result stating “risk unchanged, unable to further refine risk” on the study report.
Cases were categorized into the following groups for subgroup analysis: 1) T21 positive only; 2) T18 positive only; 3) T13 positive only; 4) triploidy; 5) >1 if positive for one of T21, T18, or T13, and in addition, either positive or NR for a second independent parameter (T21, T18, T13, microdeletion, or sex chromosome); 6) NR for T21, 18, or 13; 7) negative cfDNA results but discordant with confirmatory testing; 8) only microdeletion positive; 9) only microdeletion unreportable. Screens positive for isolated sex chromosome aneuploidies were not included in this study. However, if the cfDNA test was positive or NR for an autosomal aneuploidy or microdeletion, an abnormal result for the sex chromosomes was considered as a second abnormality in the analysis. Each individual patient was counted as one case, even if the cfDNA test was repeated with a new clinical sample. For the analysis of factors associated with true versus false positive results, all positive cases for each chromosome (21, 18, or 13) with confirmed outcomes were analyzed independently, as well as in composite. For this portion of the analysis investigating factors associated with test accuracy, cases of triploidy were excluded, as these cases are rare and results may be confounded by atypical factors, such as maternal malignancy [13].
The following demographic information was abstracted from the medical record: maternal age, IVF, cell free DNA laboratory, and multiple gestation. In addition, the results of these additional screening tests were recorded if obtained: serum screening (first trimester serum screening, integrated screening, or second trimester quad screening), nuchal translucency (NT), and first and/or second trimester ultrasound findings. Positive serum screening was determined by the California Prenatal Screening Program’s age-based cutoffs for first or second trimester screening. In California, first trimester screening combines serum analyte testing with first trimester US. NT measurements were performed by sonographers and/or physicians certified by the Nuchal Translucency Quality Review Program.
The primary outcome was concordance of cfDNA results with confirmatory diagnostic testing (by chorionic villus sampling or amniocentesis) or postnatal genetic evaluation. Standard karyotype analysis was performed by a single cytogenetics laboratory. Microrray analysis was recommended in cases of microdeletion-positive screens, and was performed if the patient granted consent. Neonatal outcomes were reported by the patient’s primary obstetric provider in follow-up. Results for T21, T18, and T13 were considered independently. Secondary outcomes were associations with maternal age as a continuous variable, advanced maternal age (≥35, as a dichotomous variable), IVF, twins, US findings, testing platform, and laboratory. Pregnancy outcomes were also obtained (termination of pregnancy, spontaneous loss, normal delivery, or lost-to-followup). Positive predictive value (PPV) calculations were made based on the total number of patients with confirmatory testing with karyotype or microarray testing; as expected, some patients were either lost to follow-up or declined confirmatory testing, and so were not included in the final denominators.
Kruskal-Wallis (continuous variables) or Fisher’s exact test (categorical variables) was used as appropriate to calculate p values. Stata 14 (College Station, TX) was used for all statistical analysis.
Results
During the study period, 121 patients had abnormal cfDNA testing. Abnormal cfDNA results for T21, T18, or T13 were found in 105 patients. Of these, 92 (87.6%) were positive and 13 (12.4%) were NR. An additional 16 patients had abnormal results for microdeletions only. The sequence of patient management and outcomes is shown in Figure 1.
Figure 1. Patient Flow.
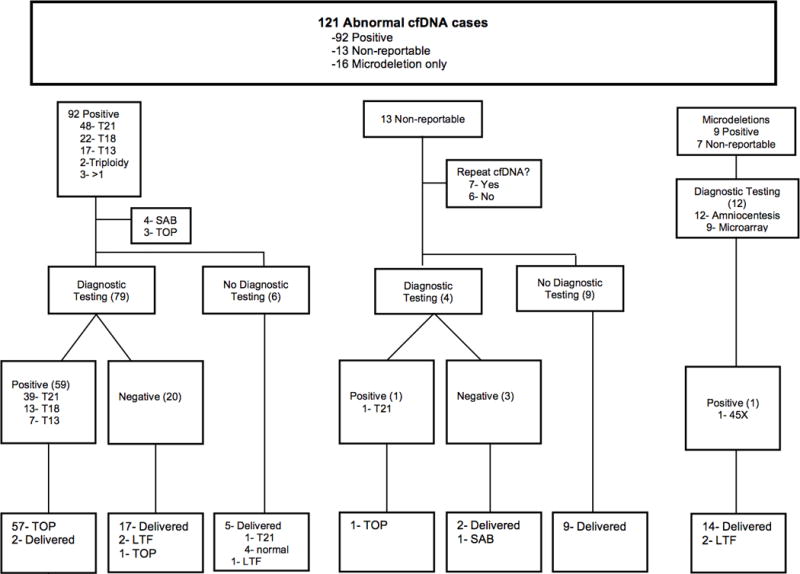
Sequence of patient management and outcomes.
Abbreviations: cfDNA, cell-free fetal DNA; T21, trisomy 21; T18, trisomy 18; T13, trisomy 13; SAB, spontaneous abortion; TOP, termination of pregnancy; LTF, lost-to-follow-up.
In the 105 patients with abnormal results for chromosomes 21, 18, or 13 on cfDNA screening (Table 1), as expected the most common positive result was T21 (48/92, 52%), followed by T18 (22/92, 24%), and T13 (17/92, 19%). There were two cases of triploidy, and three cases in which >1 parameters were abnormal. Patients with a positive results for T21, T18, T13, or triploidy were more likely to be older (p=0.007) and AMA (≥35 years old, p=0.017). When comparing between cfDNA result groups, there were significant differences noted in the distribution of test results over the four commercially-available laboratories used (p=0.006). Although fewer tests were performed by single nucleotide polymorphism (SNP) sequencing technology, SNP sequencing was more likely to give >1 positive or NR results than the other platforms (p<0.001).
Table 1.
Characteristics of patients with positive and non-reportable cfDNA results for chromosomes 21, 18, 13
| All | T21 | T18 | T13 | Triploidy | >1* | NR | p | |
|---|---|---|---|---|---|---|---|---|
| n (%) | 105 | 48 (45.7) | 22 (21.0) | 17 (16.2) | 2 (1.9) | 3 (2.9) | 13 (12.4) | |
| Median age in years (range) | 38 (27–48) | 38 (29–47) | 39 (27–48) | 38 (30–45) | 42 (40–44) | 34 (31–37) | 35 (29–41) | 0.007 |
| AMA | 87 (82.9) | 42 (87.5) | 20 (90.9) | 15 (88.2) | 2 (100) | 1 (33.3) | 7 (53.9) | 0.017 |
| IVF | 17/103 (16.5) | 5/47 (10.6) | 3/21 (14.3) | 4(23.5) | 2 (100) | 0 | 3 (23.1) | 0.066 |
| Twins | 10 (9.5) | 4(8.3) | 1 (4.6) | 3 (17.7) | 1 (50.0) | 0 | 1 (7.7) | 0.307 |
| Laboratory (n) | 0.006 | |||||||
| 1 | 37 (35.2) | 17 (35.4) | 10 (45.5) | 7 (41.2) | 0 | 0 | 3 (23.1) | |
| 2 | 38 (36.2) | 22 (45.8) | 9 (40.9) | 5 (29.4) | 0 | 1 (33.3) | 1 (7.7) | |
| 3 | 28 (26.7) | 8 (16.7) | 2 (9.1) | 5 (29.4) | 2 (100) | 2 (66.7) | 9 (69.2) | |
| 4 | 2 (1.9) | 1 (2.1) | 1 (4.6) | 0 | 0 | 0 | 0 | |
| Platformˆ | <0.001 | |||||||
| MPS | 75 (71.4) | 39 (81.3) | 19 (86.4) | 12 (70.6) | 0 | 1 (33.3) | 4 (30.8) | |
| SNP | 28 (26.7) | 8 (16.7) | 2 (9.1) | 5 (29.4) | 2 (100) | 2 (66.7) | 9 (69.3) | |
| TS | 2 (1.9) | 1 (2.1) | 1 (4.6) | 0 | 0 | 0 | 0 |
Values are n (%) unless otherwise indicated. If values were missing for certain parameters, the denominator is indicated. P-values were calculated by Kruskal-Wallis or Fisher’s exact test. Statistically significant p-values (defined as p<0.05) are indicated in bold.
Abbreviations: cfDNA, cell-free fetal DNA; T21, trisomy 21; T18, trisomy 18; T13, trisomy 13; NR, non-reportable; AMA, advanced maternal age; IVF, in vitro fertilization; MPS, massive parallel sequencing; SNP, single nucleotide polymorphism; TS, targeted sequencing.
>1 parameter was abnormal (ie: trisomy AND microdeletion, trisomy AND sex chromosome, trisomy AND NR, etc.)
MPS is used by Laboratories 1 and 2; SNP sequencing/microarray by Laboratory 3; TS by Laboratory 4.
Screening and pregnancy outcomes stratified by cfDNA results are shown in Table 2. Overall, 56/105 subjects (53.3%) underwent a form of traditional serum screening in addition to cfDNA testing. A reported positive cfDNA result for T21, T18, or T13 was associated more frequently with both a positive serum screen result (p=0.004) and 1st trimester US findings (p=0.003), when compared to patients with NR results. Positive T18 was more likely to have an NT≥3.5mm (p=0.019) compared to the other results. Abnormal findings on 1st trimester US and 2nd trimester US (if performed, n=98 and 43, respectively) are listed in Table 3. Cases with NR results were more likely to undergo repeat cfDNA screening (p<0.001), which were repeated from 1–2 times (described below).
Table 2.
Outcomes for positive and non-reportable cfDNA results for chromosomes 21, 18, 13
| All | T21 | T18 | T13 | Triploidy | >1* | NR | p | |
|---|---|---|---|---|---|---|---|---|
| n | 105 | 48 | 22 | 17 | 2 | 3 | 13 | |
| Serum Screening | 56 | 24 | 9 | 8 | 2 | 1 | 12 | <0.001 |
| FTS | 39/56 (69.6) | 23 (95.8) | 8 (88.9) | 4 (50.0) | 1 (50.0) | 1 (100) | 3 (25.0) | |
| IS | 11/56 (19.6) | 0 | 1 (111) | 2 (25.0) | 1 (50.0) | 0 | 7 (58.3) | |
| QS | 6/56 (10.7) | 1 (4.2) | 0 | 2 (25.0) | 0 | 0 | 2 (16.7) | |
| Serum Screen Positive‡ | 29/56 (51.8) | 17 (70.8) | 7 (77.8) | 3 (37.5) | 0 | 0 | 2 (16.7) | 0.004 |
| Abnormal US findings | ||||||||
| 1st TM US | 45/97 (46.4) | 25/45 (55.6) | 13/20 (65.0) | 6/14 (42.9) | 0/2 | 0/3 | 1/13 (7.7) | 0.003 |
| NT ≥3.5mm | 28/91 (30.8) | 13/41 (31.7) | 11/18 (61.1) | 3/14(21.4) | 0/2 | 0/3 | 1/13 (7.7) | 0.019 |
| 2nd TM US | 7/42 (16.7) | 3/13 (23.1) | 1/8 (12.5) | 2/10 (20) | 0/1 | 0/1 | 1/9 (11.1) | 0.963 |
| Repeat cfDNA | 11/105 (10.5) | 2 (4.2) | 0 | 0 | 2 (100) | 0/3 | 7 (53.9) | <0.001 |
| Diagnostic Testing | 84/103 (81.6) | 43/47 (91.5) | 19/22 (86.4) | 15/17 (88.2) | 0/2 | 3/3 (100) | 4/13 (30.8) | <0.001 |
| CVS | 70/103 (68.0) | 38/47 (80.9) | 15/22 (68.2) | 12/17 (70.6) | NA | 3/3 | 2/12 (16.7) | |
| Amniocentesis | 14/103 (13.6) | 5/47 (10.6) | 4/22 (18.2) | 3/17 (17.7) | NA | 0 | 2/12 (16.7) | |
| Abnormal Genetics | ||||||||
| Karyotype | 60/84 (71.4) | 37/43 (86.1) | 12/19 (63.2) | 8/15 (53.3) | NA | 2/3 (66.7) | 1/4 (25.0) | 0.010 |
| Microarray | 2/6 (33.3) | 0/1 | 1/3 | 1/1 | NA | NA | 0/1 | >0.999 |
| Pregnancy Outcome | <0.001 | |||||||
| TOP | 62/105 (59.1) | 37/48 (77.1) | 14/22 (63.6) | 8/17 (47.1) | 0/2 | 2/3 (66.7) | 1/13 (7.7) | |
| Spontaneous loss | 5/105 (4.8) | 2/48 (4.2) | 1/22 (4.6) | 1/17 (5.9) | 0/2 | 0/3 | 1/13 (7.7) | |
| Delivered | 35/105 (33.3) | 9/48 (18.8) | 4/22 (18.2) | 8/17 (47.1) | 2/2 | 1/3 (33.3) | 11/13 (84.6) | |
| Lost to follow-up | 3/105 (2.9) | 0/48 | 3/22 (13.6) | 0/17 | 0/2 | 0/3 | 0/13 | |
| No confirmed outcome | 7 | 3 | 3 | 1 | 0 | 0 | 0 | |
| Confirmed aneuploidy§ | 62/98 (63.3) | 38/45 (84.4) | 12/19 (63.2) | 9/16 (56.3) | 0/2 | 2/3 (66.7) | 1/13 (7.7) | <0.001 |
Values are n (%). If values were missing for certain parameters, the denominator is indicated. P-values were calculated by Kruskal-Wallis or Fisher’s exact test. Statistically significant p-values (defined as p<0.05) are indicated in bold.
Abbreviations: cfDNA, cell-free fetal DNA; T21, trisomy 21; T18, trisomy 18; T13, trisomy 13; NR, non-reportable; FTS, first trimester screening; IS, integrated screening; QS, quad screening; TM, trimester; US, ultrasound; NT, nuchal translucency; CVS, chorionic villus sampling; TOP, termination of pregnancy.
>1 parameter was abnormal (ie: trisomy AND microdeletion, trisomy AND sex chromosome, trisomy AND NR, etc.)
As determined by the California state prenatal screening program
Confirmed by karyotype and/or postnatal evaluation.
Table 3.
List of Abnormal Findings on First and Second Trimester Ultrasound*
| First trimester US findings (n=98) |
|---|
| NT≥3.5 (n=28) |
| Absent nasal bone (n=27) |
| Body edema (n=8) |
| Cardiac anomalies (n=6) |
| Cystic hygroma (n=7) |
| Omphalocele (n=5) |
| Limb abnormalities (n=4) |
| Fetal demise (n=3) h |
| Holoprosencephaly (n=2) |
| Abnormal skull shape (n=2) |
| Cleft lip (n=1) |
|
|
| Second trimester US findings (n=43) |
|
|
| Cardiac anomalies (n=6) |
| Limb abnormalities (n=3) |
| Enlarged nuchal fold (n=2) |
| Echogenic bowel (n=2) |
| Echogenic intracardiac focus (n=3) |
| Pyelectasis (n=2) |
| Holoprosencephaly (n=1) |
| Dandy-Walker malformation (n=1) |
Abbreviations: US, ultrasound; NT, nuchal translucency
Patients may have had more than one anomaly.
Patients with positive cfDNA results were more likely to undergo diagnostic testing compared with NR results (80/92, 87.0% vs. 4/13, 30.8%, p<0.001, Table 2). There were 98 patients that had genetic pre- or post-natal confirmation of outcomes, of which 64 (65.3%) had confirmed aneuploidy. 61/83 (73.5%) of patients with positive cfDNA for aneuploidy were true positives confirmed by karyotype (n=59) or postnatal genetic evaluation of the newborn (n=2). For T21, T18, and T13, aneuploidy was confirmed in 38/45 (84.4%), 12/19 (63.2%), and 9/16 (56.3%), respectively. In the two cases of triploidy, repeat cfDNA testing was performed and normal for each; no further testing was pursued in the context of normal US findings. One patient with cfDNA positive for T13 and serum screen positive for T21 underwent a CVS that revealed mosaic trisomy of chromosome two followed by amniocentesis that showed normal 46XX.
There were five cases of spontaneous pregnancy loss in the cohort of patients with positive or NR cfDNA (4.7%, Table 2 and Figure). Sixty percent (64/107) of patients chose to terminate their pregnancies, of which 60/64 (93.8%) had confirmed aneuploidy on diagnostic testing. One patient had a normal karyotype but an omphalocele on US. Two patients terminated based on positive cfDNA and associated US findings (one with absent nasal bone and cystic hygroma, the second with abnormal cardiac axis and micrognathia) without confirmatory testing. One patient (35 years old) terminated based on an isolated positive cfDNA result, with normal 1st trimester US.
Of the 13 NR results, seven patients underwent repeat cfDNA testing with new clinical samples (Table 2). Fetal fractions were available for 6/13 patients, and ranged from 2.7% to 12.4%. Of note, just 3/6 of these had fetal fraction < 4%. Four of these were then negative on the second repeat test. Three patients each submitted two additional samples, with a second consecutive NR result before obtaining negative results on the third sample. One of these patients had their last repeat performed by a different laboratory finally giving a negative result. Four of the 13 cases underwent confirmatory genetic testing (one of whom also had repeat cfDNA testing that was negative). Of these, 1/4 (25.0%) of NR results had T21 confirmed on karyotype and chose to terminate the pregnancy. Three patients had neither repeat cfDNA nor confirmatory testing performed. One patient had a spontaneous loss at 16 weeks; the others had normal ultrasounds and subsequently normal deliveries.
The accuracy of a positive cfDNA in predicting T21, 18, or T13 is shown in Table 4. For the composite of trisomies (T21, T18, T13 combined), the positive predictive value (PPV) was 74% (61/83, 95% confidence interval (CI) 63% to 82%). For individual chromosomes, the PPV was 83.0% (39/47, CI 69% to 92%), 65% (13/20, CI 41% to 84%), and 44% (7/16, CI 21% to 70%) for T21, T18, and T13, respectively. The PPV of cfDNA for T21 was associated with maternal age (p=0.005).
Table 4.
Factors associated with accuracy of cfDNA positive for T21, 18, 13*
| Composite T21, T18, T13 n=83 |
T21 n=47 |
T18 n=20 |
T13 n=16 |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
| ||||||||||||
| TP | FP | p | TP | FP | p | TP | FP | p | TP | FP | p | |
| n (%) | 61 (73.5) | 22 (26.5) | 39 (83.0) | 8 (17.0) | 13 (65.0) | 7 (35.0) | 7 (43.8) | 9 (56.3) | ||||
| Median age (range) | 39 (30–48) | 36 (29–40) | 0.001 | 39 (30–47) | 35.5 (29–38) | 0.005 | 39 (30–48) | 39 (35–40) | 0.6021 | 39 (35–45) | 37 (30–40) | 0.135 |
| AMA | 56/61 (91.8) | 17/22 (77.3) | 0.120 | 36/39 (92.3) | 5/8 (62.5) | 0.053 | 11/13 (84.6) | 7/7 (100) | 0.521 | 7/7 | 7 (77.8) | 0.475 |
| IVF | 10/60 (16.7) | 2/22 (9.1) | 0.499 | 5/38 (13.2) | 0/8 | 0.569 | 2/13 (15.4) | 1/7 (14.3) | >0.999 | 3 (42.9) | 1 (11.1) | 0.262 |
| Twins | 7/61 (11.5) | 1/22 (4.5) | 0.675 | 4/39 (10.3) | 0/8 | >0.999 | 1/13 (7.7) | 0/7 | >0.999 | 1 (14.3) | 2 (22.2) | >0.999 |
| Laboratory (n) | 0.018 | 0.275 | 0.311 | 0.563 | ||||||||
| 1 (32) | 28 (45.9) | 4 (18.2) | 16 (41.0) | 1 (12.5) | 7 (53.9) | 2 (28.6) | 3 (42.3) | 3 (33.3) | ||||
| 2 (34) | 19 (31.2) | 15 (68.2) | 14 (35.9) | 6 (75.0) | 5 (30.8) | 5 (71.4) | 1 (14.3) | 4 (44.4) | ||||
| 3 (16) | 13 (21.3) | 3 (13.6) | 8 (20.5) | 1 (12.5) | 2 (15.4) | 0 | 3 (42.9) | 2 (22.2) | ||||
| 4 (1) | 1 (16) | 0 | 1 (2.6) | 0 | 0 | 0 | 0 | 0 | ||||
| Platformˆ | 0.661 | >0.999 | 0.521 | 0.596 | ||||||||
| MPS (66) | 47 (77.1) | 19 (86.4) | 30 (76.9) | 7 (87.5) | 11 (84.6) | 7/7 (100) | 4 (57.1) | 7 (77.8) | ||||
| SNP (16) | 13 (21.3) | 3 (13.6) | 8 (20.5) | 1 (12.5) | 2 (15.4) | 0 | 3 (42.9) | 2 (22.2) | ||||
| TS (1) | 1 (16) | 0 | 1 (2.6) | 0 | 0 | 0 | 0 | 0 | ||||
| Serum Screen Positive† | 21/25 (84.0) | 5/16 (31.3) | 0.001 | 16 (94.1) | 1 (14.3) | <0.001 | 3/4 (75.0) | 3/5 (60.0) | >0.999 | 2/4 (50) | 1/4 (25.0) | >0.999 |
| US findings | ||||||||||||
| 1st TM US | 39/57 (68.4) | 1/19 (5.3) | <0.001 | 23/36 (63.9) | 0/8 | 0.001 | 11/13 (84.6) | 1/6 (16.7) | 0.010 | 4/7 (57.1) | 1/6 (16.7) | 0.266 |
| NT >3.5mm | 25/54 (46.3) | 1/19 (5.3) | 0.002 | 13/34 (38.2) | 0/8 | 0.043 | 9/12 (75) | 1/5 (20.0) | 0.101 | 2/7 (28.6) | 1/7 (14.3) | >0.999 |
| 2nd TM US | 5/12 (41.7) | 1/19 (5.3) | 0.022 | 3/6 (50.0) | 0/8 | 0.055 | 0/2 (0) | 1/5 (20.0) | >0.999 | 2/2 (100) | 0/8 (0) | 0.022 |
| Pregnancy Outcome | <0.001 | <0.001 | <0.001 | 0.041 | ||||||||
| TOP | 58/61 (95.1) | 1/22 (4.5) | 37 (94.9) | 0/8 | 13/13 (100) | 1/7 (14.3) | 6/7 (85.7) | 2/9 (22.2) | ||||
| Delivered | 3/61 (4.9) | 19/22 (86.4) | 2 (5.1) | 8 | 0 | 4/7 (57.1) | 1/7 (14.3) | 7/9 (77.8) | ||||
| Lost to follow-up | 0 | 2/24 (9.1) | 0 | 0 | 0 | 2/7 (28.6) | 00 | |||||
Values are n (%) unless otherwise noted. P-values were calculated by Kruskal-Wallis or Fisher’s exact test. Statistically significant p-values (defined as p<0.05) are indicated in bold.
Abbreviations: cfDNA, cell-free fetal DNA; T21, trisomy 21; T18, trisomy 18; T13, trisomy 13; TP, true positive; FP, false positive; AMA, advanced maternal age; IVF, in vitro fertilization; MPS, massive parallel sequencing; SNP, single nucleotide polymorphism; TS, targeted sequencing; TM, trimester; US, ultrasound; NT, nuchal translucency; TOP, termination of pregnancy.
Confirmed by karyotype and/or postnatal evaluation.
As determined by the California state prenatal screening program
MPS is used by Laboratories 1 and 2; SNP sequencing/microarray by Laboratory 3; TS by Laboratory 4.
There was no association between a false positive result for the composite of T21, T18, and T13 and testing platform (p=0.661, Table 4), but there was a significant difference in rates of false positive results between the laboratories (p=0.018). There was no difference in maternal age when stratified by laboratory (data not shown). When stratified by individual aneuploidy, we were unable to detect a significant difference between either laboratory or platform for any of the three trisomies, likely due to insufficient power in the subgroup analysis. As expected, true positive cfDNA results were associated with positive serum screening (p<0.001), abnormal 1st trimester US findings (p<0.001), NT ≥3.5 (p=0.002), and abnormal 2nd trimester ultrasound (p=0.022). Overall, 58/61 (95.1%) of true positive cases ended in termination.
In all, there were 63 cases of aneuploidy in our cohort. The rates of concordance between cfDNA results and karyotypes for T21, T18, and T13 were 97.5% (39/40), 86.7% (13/15), and 87.5% (7/8), respectively. The cases that were not detected by cfDNA screening are summarized in Table 5. We identified three cases of false negative cfDNA testing, although we did not have access to complete outcomes for screen-negative cases and thus cannot determine false negative rates. Cases 1 and 2 initially had negative cfDNA results for T21, T18, and T13 that were found to be discordant with subsequent karyotyping. For both of these patients, abnormal ultrasound findings prompted diagnostic testing. T13 was identified in Case 1. Case 2 was found to have mosaic ring chromosome 18, a rare disorder with partial deletions of chromosome 18. Case 3 had a positive T13 cfDNA result, with positive T18 on karyotyping, and so was a false positive for T13, and a false negative for T18. Case 4 had a NR cfDNA test, and an abnormal 1st trimester US. Karyotype confirmed T21. The cfDNA test was not repeated.
Table 5.
Aneuploidy cases missed by cfDNA
| Case | Age (y) | cfDNA Result | Serum Screening | NT (mm) | US findings | Karyotype | Outcome |
|---|---|---|---|---|---|---|---|
| 1 | 40 | Negative | None | NA | Holoprosencephaly, single umbilical artery | 47XX, +13 | TOP |
| 2 | 30 | Negative | None | 3.6 | Cystic hygroma | 46XX, mosaic chr 18 ring | TOP |
| 3 | 38 | +Trisomy 13 | None | 6.4 | Ventriculomegaly, holoprosencephaly, polydactyly | 47XY, +18 | TOP |
| 4 | 41 | Non-reportable | FTS (1/5 risk trisomy 21) | 5.9 | Absent nasal bone | 47XX, +21 | TOP |
Abbreviations: cfDNA, cell-free fetal DNA; NT, nuchal translucency; US, ultrasound; bottom, termination of pregnancy; FTS, first-trimester screen
A total of 43 cases had microdeletion results reported, all ordered by primary obstetricians prior to referral for consultation and evaluation. Microdeletion testing is only offered by 2 of the 4 companies included in this study. Of the microdeletion results in this cohort, 26 had abnormal results for at least one microdeletion (nine positive and 17 NR) (Table 6). DiGeorge syndrome (chromosome 22q11.2 deletion) was the most commonly reported positive result (4/9), while Angelman’s syndrome was the most common single microdeletion that was NR. Of the 17 patients with NR results, eight were NR for all of the reported microdeletions. Compared to a NR result, a positive microdeletion result was associated with negative cfDNA result for T21, T18, or T13 (p=0.012). Non-reportable microdeletions were more likely to have an inconclusive sex chromosome result on cfDNA screening as well (p=0.025). Fifteen cases had only an abnormal microdeletion result, with normal cfDNA results for chromosomes 21, 18, 13, and the sex chromosomes. There were no correlations found between positive cfDNA microdeletion results and serum screening results, abnormal ultrasound findings, or abnormal outcome. Of the 17 patients who underwent diagnostic testing, only ten had microarray sent despite thorough genetic counseling. Of the nine microdeletion screen-positive results, seven were confirmed to be false positive by microarray. Of note, two of the patients with positive microdeletion results (both for DiGeorge syndrome) who underwent diagnostic testing declined microarray testing despite thorough genetic counseling, with 45X and 46XY returning as karyotype results.
Table 6.
Characteristics of patients with positive or non-reportable cfDNA results for microdeletions.
| All | Microdeletion result
|
|||
|---|---|---|---|---|
| Positive | Non-reportable | p | ||
| n (%) | 26 | 9 (34.6) | 17 (65.4) | |
| Median age (range) | 35 (26–43) | 33 (27–41) | 35 (26–43) | 0.484 |
| AMA | 14 (53.9) | 4 (44.4) | 10 (58.8) | 0.683 |
| IVF | 5 (20.8) | 0 | 5 (31.3) | 0.130 |
| Twins | 1 (3.9) | 0 | 1 (5.9) | >0.999 |
| MD result | 0.004 | |||
| DiGeorge Syndrome | 5 (19.2) | 4 (44.4) | 1 (5.9) | |
| Angelman’s | 9 (34.6) | 2 (22.2) | 7 (41.2) | |
| Cri du chat | 4 (15.4) | 3 (33.3) | 1 (5.9) | |
| All affected | 8 (30.8) | 0 | 8 (100) | |
| T21/18/13 result | 0.012 | |||
| Positive | 3 (11.5) | 0 | 3 (17.7) | |
| Non-reportable | 7 (26.9) | 0 | 7 (41.2) | |
| Negative | 16 (61.5) | 9 (100) | 7 (41.2) | |
| Sex chromosome result | 0.025 | |||
| Normal | 16 (61.5) | 8 (88.9) | 8 (47.1) | |
| Monosomy X | 2 (7.7) | 1 (111) | 1 (5.9) | |
| Inconclusive | 8 (30.8) | 0 | 8 (47.1) | |
|
| ||||
| Serum Screen Positive* | 14 | 0/4 | 1/10 (10.0) | >0.999 |
| Abnormal US findings | ||||
| 1st TM US | 1/24 (4.2) | 0/8 | 1/16 (6.3) | >0.999 |
| NT ≥3.5mm | 0/24 | NA | NA | |
| 2nd TM US | 3/20 (15.0) | 1/8 (12.5) | 2/12 (16.7) | >0.999 |
| Repeat cfDNA | 7/26 (26.9) | 0/9 | 7/17 (41.2) | 0.058 |
| Diagnostic Testing | 17/25 | 9/9 | 8/16 | 0.005 |
| CVS | 12/25 | 8/9 | 4/16 | |
| Amniocentesis | 5/25 | 1/9 | 4/16 | |
| Abnormal Genetics | ||||
| Karyotype | 3/17 (17.7) | 1/9 (11.1) | 2/8 (25.0) | 0.576 |
| Microarray | 0/10 | 0/7 | 0/3 | |
| Pregnancy Outcome | 0.213 | |||
| TOP | 1/26 (3.9) | 0/9 | 1/17 (5.9) | |
| Spontaneous loss | 1/26 (3.9) | 0/9 | 1/17 (5.9) | |
| Delivered | 22/26 (84.6) | 7/9 (77.8) | 15/17 (88.2) | |
| Lost to follow-up | 2/26 (7.7) | 2/9 (22.2) | 0/17 | |
| No confirmed outcome | 2/26 | 2/9 | 0/17 | |
| Confirmed microdeletion† | 0/24 | 0/9 | 0/17 | |
| Confirmed aneuploidy† | 3/25 (12.0) | 1/8 (12.5) | 2/17 (11.8) | >0.999 |
Values are n (%). If values were missing for certain parameters, the denominator is indicated. P-values were calculated by Kruskal-Wallis or Fisher’s exact tests as appropriate. Statistically significant p-values (defined as p<0.05) are indicated in bold.
Abbreviations: cfDNA, cell-free fetal DNA; AMA, advanced maternal age; IVF, in vitro fertilization; T21, trisomy 21; T18, trisomy 18; T13, trisomy 13; TM, trimester; US, ultrasound; NT, nuchal translucency; CVS, chorionic villus sampling; TOP, termination of pregnancy.
As determined by the California state prenatal screening program
Confirmed by karyotype/microarray and/or postnatal evaluation.
Comment
We conducted a retrospective cohort study of all positive (including suspected) and NR results for cfDNA screening for T21, T18, T13, and microdeletions in a single-center perinatal referral setting. We found that the overall PPV for cfDNA testing was 83%, 65%, and 44% for T21, T18, and T13, respectively. The seven (of nine) cases of positive microdeletion screens that underwent confirmatory testing were false positives. There was a significant difference in composite test performance by laboratory.
In our cohort, the PPV for T21, T18, and T13 were lower than expected (previously described to be 91%, 77%, and 44%, respectively) [13–15]. Although we included 3 cases of suspected T21 in our positive group (which were all false positives), even with the exclusion of these cases, the PPV of 88% remains lower than expected. This data supports current recommendations that caution against making clinical decisions on cfDNA results alone without diagnostic testing. Furthermore, Norton, et al [6] recently reported that approximately 3% of NR cfDNA results were aneuploidy. Of the 13 NR cfDNA cases in our cohort, most decided to undergo repeat cfDNA testing, whereas only four elected to undergo diagnostic testing, of which one was indeed found to have aneuploidy. Depending on the likelihood of other reasons for a NR results (such as early gestational age or low fetal fraction), it may be reasonable to automatically offer karyotype after a NR result. This also highlights the importance of the individual laboratories’ varying internal cutoffs for declaring a test positive, negative, or NR. For example, in our cohort, Laboratory 2 had the lowest rate of NR results, but a higher proportion of false positive cases. Thus a more conservative definition of a “positive” test will protect test accuracy, at the risk of lower test performance. At the present time, not all laboratories report the fetal fraction. We advocate for standard reporting of fetal fraction, as this information is important for the interpretation of NR results.
In our cohort, there were 10 cases of abnormal results in twin gestations. The most recent guidelines from the American Congress of Obstetrics and Gynecology and the Society for Maternal Fetal Medicine do not recommend cfDNA for aneuploidy screening in multiple gestations due to limited evidence demonstrating its efficacy [1]. However, a recent study by our group examining cfDNA in 602 twin pregnancies from a single laboratory using MPS revealed that test performance was comparable to singletons [16]. Our small sample size here clearly limits our ability to examine outcomes for cfDNA in twin pregnancies, but further studies should provide more evidence as to test performance in this special population.
Wang et al. examined concordance between cytogenetics and 109 positive cfDNA results from four U.S. laboratories, and similarly found lower PPV than the published literature [17]. Our study builds upon their study by examining factors associated with discordance, as well as including our experience with microdeletion results. Interestingly, we did not detect a significant difference in the rate of false positive cases between the different technological platforms used for cfDNA testing, but there was a significant difference among the laboratories. Although we are eager to information on which laboratory is “best”, a true comparison of the different companies is beyond the scope of this study. Nonetheless, our results suggest that choice of laboratory does matter, and future large studies are clearly needed to evaluate comparative test performance, considering they are laboratory-developed and an independent validation of advertised test performance has not been performed.
The accuracy of a positive cfDNA result was associated with positive serum screening results. Although cfDNA as a screening modality for T21, T18, and T13 was introduced with the goal of replacing standard serum screening, our data demonstrates that over half of patients referred to our practice still undergo both tests, although it is unclear whether this was patient- or provider-driven. New 2016 guidelines now recommend no further screening if one form of testing has been done [1]. There were five cases of aneuploidy that were detected by cfDNA, but missed by first trimester serum screening; however, this study was not designed to compare the performance of the two screening strategies.
Our results also demonstrate the influence of prenatal genetic testing on pregnancy outcomes. In this cohort from Los Angeles area, 60% of patients chose to terminate the pregnancy. Of note, one patient chose to terminate based on positive cfDNA results alone, despite reassuring ultrasound evaluations and extensive counseling to await confirmatory testing. This represents a difficult, but not uncommon, scenario in which the clinician’s and genetic counselor’s advice is perceived to be at odds with the advertised low screen positive rate and high sensitivity of cfDNA testing. We continue to advocate for increased education on the nature and limitations of this screening test [18].
To date, few studies have looked at outcomes from microdeletion testing. Although performance data is lacking and routine screening is not currently recommended by national societies, patients receive results for an increasing number of microdeletion syndromes. At present, microdeletion testing may increase the emotional, physical, and financial burden of “abnormal testing” without clear and defined benefit to screening. Indeed, in our cohort, 80% (12/16) of patients with isolated abnormal microdeletion results underwent diagnostic testing, all of which returned with normal microarray results, yielding a PPV of 0%. Larger studies are needed to evaluate the performance and utility of this testing and are only beginning to be reported [9].
A significant strength of this study is its generalizability to common clinical practice in a high risk, insured population. The majority of patients were referred to our practice with cfDNA or serum screening tests that were already ordered by the patients’ primary obstetric provider. In clinical practice, the choice for which laboratory to use is largely driven by financial considerations (such as patient insurance coverage and out-of-pocket cost), as all companies currently report similar performance statistics. Furthermore, this study compares the results of different commercial tests and the platforms currently available in the United States. Inherent differences between testing platforms and individual laboratory cutoffs require investigation, as this information may provide clinical guidance as to which test may perform better in specific patient populations.
The primary limitation of this study is the retrospective design, which limits the scope of our study. For example, we do not have data on the total number of cfDNA tests performed during this time period, and so lack the total denominator required to evaluate test specificity. Furthermore, we do not have body mass index (BMI) values for the patients, which is associated with NR results [1]. Finally, although this is a large cohort for a single high-risk referral center, our sample size was too small for comprehensive analysis of the individual aneuploidies. For example, a post-hoc power analysis for T21 in our sample size of 48 positive tests, using a predicted PPV of 88% for a 38-year-old woman provided by The Perinatal Quality Foundation (perinatalquality.org), revealed that we would be able to detect a difference of 8.5% in expected vs actual PPV, with an alpha of 0.05 and a beta of 0.8. Here, we found only a 5% difference in expected vs actual PPV.
In summary, our study provides evidence that in current clinical practice, the PPV of cfDNA testing is lower than reported, even in our high-risk referral population. As cfDNA screening is increasingly offered to low-risk populations where the prevalence of aneuploidies is lower, the PPV will be even lower than what we report here. This information is important for counseling patients that will undergo this testing, and reinforces diagnostic testing as essential follow-up for positive screening results. It also points out the need of an independent registry of all cfDNA cases to understand test performance in real clinical practice, which includes detection rate, false positive rate, PPV, and NPV, not just PPV. We hope that such an endeavor can be realized through cooperation between competing commercial laboratories, in the interest of improving patient care.
Condensation.
The real-world positive predictive value of cell-free DNA screening for aneuploidy and microdeletions in clinical practice may be lower than advertised.
Acknowledgments
Financial support: SGV was supported by the National Institutes of Health/National Institute of Child Health and Human Development Reproductive Scientist Development Program (K12 HD000849).
Footnotes
Disclosures: LDP and NSS are on the speaker bureau for Illumina and LDP served on their medical advisory board. LDP is Treasurer of the Perinatal Quality Foundation. The remaining authors report no conflict of interest.
Presented in part at the Society for Maternal-Fetal Medicine 35th Annual Meeting, Atlanta, GA, February 1–6, 2016.
References
- 1.American College of Obstetricians and Gynecologists. Practice Bulletin No. 163: Screening for Fetal Aneuploidy. Obstet Gynecol. 2016 May; doi: 10.1097/AOG.0000000000001406. 2016. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 2.Bianchi DW, Platt LD, Goldberg JD, et al. Genome-wide fetal aneuploidy detection by maternal plasma DNA sequencing. Obstet Gynecol. 2012;119:890–901. doi: 10.1097/AOG.0b013e31824fb482. [DOI] [PubMed] [Google Scholar]
- 3.Norton ME, Brar H, Weiss J, et al. Non-Invasive Chromosomal Evaluation (NICE) Study: results of a multicenter prospective cohort study for detection of fetal trisomy 21 and trisomy 18. Am J Obstet Gynecol. 2012;207:137.e1–8. doi: 10.1016/j.ajog.2012.05.021. [DOI] [PubMed] [Google Scholar]
- 4.Palomaki GE, Deciu C, Kloza EM, et al. DNA sequencing of maternal plasma reliably identifies trisomy 18 and trisomy 13 as well as Down syndrome: an international collaborative study. Genet Med. 2012;14:296–305. doi: 10.1038/gim.2011.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nicolaides KH, Syngelaki A, Gil M, et al. Validation of targeted sequencing of single-nucleotide polymorphisms for non-invasive prenatal detection of aneuploidy of chromosomes 13, 18, 21, X and Y. Prenat Diagn. 2013;33:575–9. doi: 10.1002/pd.4103. [DOI] [PubMed] [Google Scholar]
- 6.Norton ME, Jacobsson B, Swamy GK, et al. Cell-free DNA analysis for noninvasive examination of trisomy. N Engl J Med. 2015 Apr 23;372(17):1589–97. doi: 10.1056/NEJMoa1407349. [DOI] [PubMed] [Google Scholar]
- 7.Zhang H1, Gao Y, Jiang F, et al. Non-invasive prenatal testing for trisomies 21, 18 and 13: clinical experience from 146,958 pregnancies. Ultrasound Obstet Gynecol. 2015 May;45(5):530–8. doi: 10.1002/uog.14792. [DOI] [PubMed] [Google Scholar]
- 8.Wapner RJ, Babiarz JE, Levy B, et al. Expanding the scope of noninvasive prenatal testing: detection of fetal microdeletion syndromes. Am J Obstet Gynecol. 2015;212:332.e1–9. doi: 10.1016/j.ajog.2014.11.041. [DOI] [PubMed] [Google Scholar]
- 9.Lefkowitz RB, Tynan JA, Liu T, et al. Clinical validation of noninvasive prenatal test for genome-wide detection of fetal copy number variants. Am J Obstet Gynecol. 2016 doi: 10.1016/j.ajog.2016.02.030. pii: S0002-9378(16)00318-5 [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 10.Platt LD, Janicki MB, Prosen T, et al. Impact of noninvasive prenatal testing in regionally dispersed medical centers in the United States. Am J Obstet Gynecol. 2014;211:368.e1–7. doi: 10.1016/j.ajog.2014.03.065. [DOI] [PubMed] [Google Scholar]
- 11.Dobson L, Reiff E, Little S, et al. Patient Choice and Clinical Outcomes Following Positive Noninvasive Prenatal Screening for Aneuploidy with Cell-free DNA (cfDNA) Prenat Diagn. 2016 Mar 3; doi: 10.1002/pd.4805. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 12.Society for Maternal-Fetal Medicine Publications Committee. Prenatal aneuploidy screening using cell-free DNA. Number 36. Am J Obstet Gynecol. 2015;212(6):711–6. doi: 10.1016/j.ajog.2015.03.043. [DOI] [PubMed] [Google Scholar]
- 13.Bianchi DW, Chudova D, Sehnert AJ, et al. Noninvasive prenatal testing and incidental detection of occult maternal malignancies. JAMA. 2015;314:162–9. doi: 10.1001/jama.2015.7120. [DOI] [PubMed] [Google Scholar]
- 14.Bianchi DW, Parker RL, Wentworth J, et al. DNA sequencing versus standard prenatal aneuploidy screening. N Engl J Med. 2014;370:799–808. doi: 10.1056/NEJMoa1311037. [DOI] [PubMed] [Google Scholar]
- 15.Gil MM, Quezada MS, Revello R, Akolekar R, Nicolaides KH. Analysis of cell-free DNA in maternal blood in screening for fetal aneuploidies: updated meta-analysis. Ultrasound Obstet Gynecol. 2015:249–66. doi: 10.1002/uog.14791. [DOI] [PubMed] [Google Scholar]
- 16.Fosler L, Winters P, Jones KW, et al. Aneuploidy screening using noninvasive prenatal testing in twin pregnancies. Ultrasound Obstet Gynecol. 2016 May 19; doi: 10.1002/uog.15964. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang JC, Sahoo T, Schonberg S, et al. Discordant noninvasive prenatal testing and cytogenetic results: a study of 109 consecutive cases. Genet Med. 2015;17(3):234–6. doi: 10.1038/gim.2014.92. [DOI] [PubMed] [Google Scholar]
- 18.Han CS, Platt LD. Noninvasive prenatal testing: need for informed enthusiasm. Am J Obstet Gynecol. 2014;211(6):577–80. doi: 10.1016/j.ajog.2014.09.012. [DOI] [PubMed] [Google Scholar]


