Abstract
Robust predictive models are essential to manage the risk of radiation-induced carcinogenesis. Chronic exposure to cosmic rays in the context of the complex deep space environment may place astronauts at high cancer risk. To estimate this risk, it is critical to understand how radiation-induced cellular stress impacts cell fate decisions and how this in turn alters the risk of carcinogenesis. Exposure to the heavy ion component of cosmic rays triggers a multitude of cellular changes, depending on the rate of exposure, the type of damage incurred and individual susceptibility. Heterogeneity in dose, dose rate, radiation quality, energy and particle flux contribute to the complexity of risk assessment. To unravel the impact of each of these factors, it is critical to identify sensitive biomarkers that can serve as inputs for robust modeling of individual risk of cancer or other long-term health consequences of exposure. Limitations in sensitivity of biomarkers to dose and dose rate, and the complexity of longitudinal monitoring, are some of the factors that increase uncertainties in the output from risk prediction models. Here, we critically evaluate candidate early and late biomarkers of radiation exposure and discuss their usefulness in predicting cell fate decisions. Some of the biomarkers we have reviewed include complex clustered DNA damage, persistent DNA repair foci, reactive oxygen species, chromosome aberrations and inflammation. Other biomarkers discussed, often assayed for at longer points post exposure, include mutations, chromosome aberrations, reactive oxygen species and telomere length changes. We discuss the relationship of biomarkers to different potential cell fates, including proliferation, apoptosis, senescence, and loss of stemness, which can propagate genomic instability and alter tissue composition and the underlying mRNA signatures that contribute to cell fate decisions. Our goal is to highlight factors that are important in choosing biomarkers and to evaluate the potential for biomarkers to inform models of post exposure cancer risk. Because cellular stress response pathways to space radiation and environmental carcinogens share common nodes, biomarker-driven risk models may be broadly applicable for estimating risks for other carcinogens.
Keywords: Biomarkers, Modeling, Cancer risk, HZE, Space radiation
1. Introduction
1.1. Criteria for modeling cancer risk from space radiation exposure
Technological revolution, medical advances and life style changes have increased human exposure to radiation in recent years. The noted increase in cancer incidence with age has in part been attributed to these exposures. The National Cancer Institute (NCI) (de Gonzalez et al., 2012), National Institute of Occupational Health and Safety (NIOSH) (Kocher et al., 2008), Environmental Protection Agency (EPA) (Pawel DJ et al., 2012; Pawel DJ, 2013), and NASA (Cucinotta et al., 2012) have developed cancer risk models for diverse applications that have significant overlap. All of these models primarily rely on epidemiological data from the Life Span Study (LSS) of Japanese atomic bomb survivors to estimate cancer risk for high dose rate low-LET (linear energy transfer) exposures. Using a dose and dose rate effectiveness factor (DDREF), these results are being extrapolated to predict effectiveness of lower doses and dose rates of various radiation qualities. To estimate DDREF, BEIR VII performed a Bayesian analysis using LSS data, cancer incidence and life shortening in animal models and chromosome aberrations (CA) in human somatic cells with estimates for low-LET radiation that rely primarily on epidemiology and cancer incidence data in animal models (NCRP, 2012). The former three models use a modified version of BEIR VII results. Modeling radiation quality and dose rate effects are especially critical for NASA due to the wide spectrum of ions of various charges and energies in space that are rarely seen on earth (Cucinotta et al., 2012). To meet this challenge, the current NASA model (Cucinotta et al., 2012) uses a combination of BEIR VII and UNSCEAR 2006 (United Nations Scientific Committee on the Effects of Atomic Radiation, 2008). In addition to epidemiology data, this model incorporates tumor induction data as well as astronaut and mouse translocation data for DDREF. Radiation quality estimates for high-LET radiation are made based largely on experimental data. As in vivo cancer incidence studies in humans are technically challenging, in addition to incorporating data from animal models of carcinogenesis, surrogate biomarkers of cancer risk are being widely used to measure effects directly in human cells to shed light on the underlying biological mechanisms. Some of these endpoints include cell transformation, CAs and DNA damage response and mutation assessments (Kocher et al., 2008; Kocher et al., 2005). In the current model, radiation quality factors are being calculated based on tumor incidence, survival, CAs and mutations. A recent improvement to NASA’s model includes the use of quality factors using data from cancer incidence in mouse models (Cucinotta, 2015). Given the degree of uncertainty in estimating risk, and the long latency of cancer development, it is projected that incorporating data from early biomarkers with the potential to predict long term biological effects will provide an effective strategy for early cancer risk prediction.
1.2. Characteristics of a good biomarker for modeling risk from GCR
In space, cells are impacted by charged particles from protons to uranium with energies of particular importance to human exposures, ranging from ~tens of GeV/n to 100 GeV/n. It has been projected that for an astronaut traveling to Mars, every cell nucleus in an astronaut’s body would be hit by a proton or secondary electron (e.g., electrons of the target atoms ionized by the HZE ion) every few days and by an HZE ion about once a month (Cucinotta et al., 1998). To extrapolate risk from GCR exposure, it is critical that biomarkers used to predict risk are sensitive to different doses, dose-rates and radiation qualities in the cosmic ray spectrum. This is especially true as estimation of risk at low doses and dose-rates (~0.1 mSv min−1) has a degree of uncertainty due to paucity of human epidemiological studies at these exposure levels. It is well known that biomarkers can be temporally classified. Biomarkers of exposure such as initial radiation-induced DNA damage foci and CAs are good predictors of the radiation dose received. Biomarkers that are assessable before, during and after radiation dose can measure individual differences in susceptibility and predict inherent risk of radiation-induced health effects. Persistent biomarkers are measures of the late effects of radiation exposure and can estimate how radiation exposure can influence cell fate choice. As cancer is a long term effect, a biomarker panel for cancer risk prediction should allow temporal classification, where exposure and susceptibility effects can be linked with various cell fate decisions and cumulatively modeled to predict cancer risk. Given the complexity of the space radiation spectrum, a thorough evaluation of the multitude of confounding factors that influence cancer risk, requires that predictive biomarkers be quantifiable, easy to measure, have low-cost detection platforms, and have the ability to be detected across various tissue types. Ideally the biomarker should have limited variability within the normal population and have detection assays that are sensitive, specific, reproducible and lend themselves to high-throughput screening techniques. Validated early biomarkers with robust predictive capability are not only useful to predict the immediate cellular and physiological consequences of exposure, but are in fact the “Holy Grail” for early cancer prediction. Here we review the relevance and predictive ability of several biomarkers in modeling the short-term and long-term biological consequences of space radiation exposure.
1.3. Impact of high-LET radiation damage on cell fate decisions
The space radiation environment is truly heterogeneous and is comprised of a complex mixture of ions from galactic cosmic radiation (GCR), trapped particles in the van Allen radiation belts and other secondary particles generated in both the spacecraft and Earth’s atmosphere (Badhwar, 1997). Despite their relatively low fluence in space, compared to other types of radiation, the consequences of high charge and energy (HZE) particle exposures have important implications. At typical energies, these particles have high-LET values, defined as LET > 10 keV/µm. These evoke qualitatively different, more complex cellular damage than low-LET radiation at the same dose (Nikjoo et al., 2001; Friedland et al., 2011).
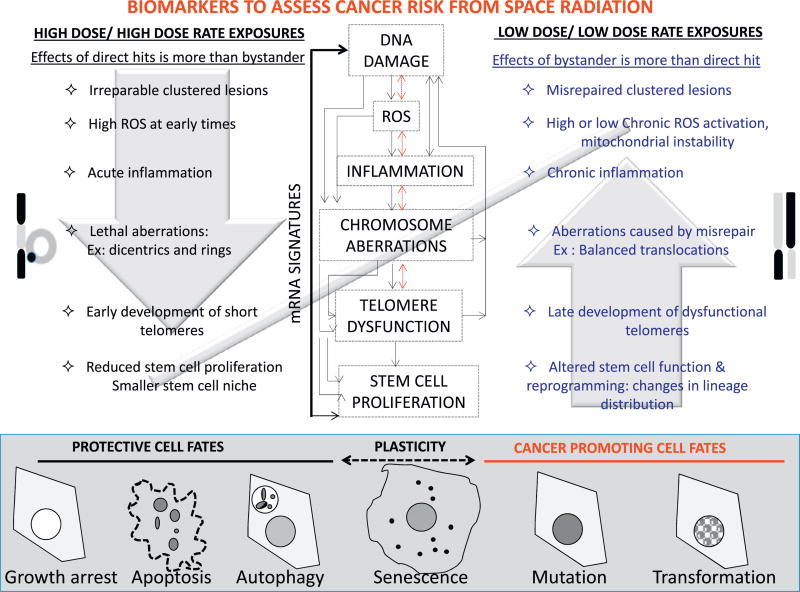
While complex damage is uncommon for endogenous reactive oxygen species or low-LET radiation, it has been associated with the increased relative biological effectiveness (RBE) of densely ionizing radiation (Durante and Cucinotta, 2008). Clustered DNA damage (CDD) seems to create short fragments that challenge the repair machinery, and target cells for growth arrest, cell death or cellular senescence if unrepaired. Misrejoining of unrepairable ends of DNA with other radiation-induced double-strand breaks leads to large DNA deletions and CAs that can significantly increase the mutation load (Cucinotta and Durante, 2006). At the cellular level, both DNA lesions and reactive oxygen species (ROS) can cause cellular inflammation, and potentiate an inflammatory state across generations. Cells have built-in regulatory mechanisms that serve as intrinsic barriers to carcinogenesis. This includes activation of p16 or p53 that prevent the proliferation of cells with damaged DNA or telomeres, and targets cells with shortened or aberrant telomeres for cell death or senescence. The damage induced by high-LET radiation could potentially overcome these protective barriers and allow the uncontrolled proliferation of damaged cells. These events can potentiate genomic instability in the progeny and in theory initiate tumor formation. Understanding the impact of radiation quality, dose and dose rate on the cellular mechanisms that balance the scales between homeostasis and cancer initiation, is essential to both predict cancer risk and prevent the carcinogenic effects of space radiation exposure.
2. DNA lesions as biomarkers of high-LET radiation exposure
2.1. Types of DNA lesions
Ionizing radiation (IR) produces more than 70 chemically distinct DNA lesions, including base and sugar modifications, abasic sites, single- and double-strand DNA breaks, and DNA-DNA and DNA-protein crosslinks (Ravanat et al., 2012; Nikitaki et al., 2015). These lesions can arise either from direct ionization of DNA (direct effect) or as a result of chemical reactions of the DNA with hydroxyl radicals and other reactive oxygen species that are produced by radiolysis of nearby water molecules (indirect effect). Although a somewhat greater fraction of high-LET damage is attributable to the direct effect (Hirayama et al., 2009), the overall spectrum of chemical products is similar for low- and high-LET radiation and does not provide a basis for discriminating between the two.
However, low- and high-LET radiation damage differs in the spatial distribution of DNA lesions. When high-LET radiation tracks pass through cells, they deposit energy inhomogeneously to produce CDD, also known as local multiply damaged sites. Densely ionizing, high-LET radiation produces much more complex clusters (Ward, 1981) than low-LET radiation, reviewed in Hada and Georgakilas (2008). Multi-scale modeling suggests that the frequency of clusters containing two or more double-strand breaks (DSBs) increases steeply with LET, and the frequency of lesions containing only a single DSB declines (Friedland et al., 2011). Locally multiply damaged sites (LMDS) can also be detected and characterized experimentally. Early work used an approach based on in situ digestion of cellular DNA with endonucleases that incise the DNA backbone at sites of base oxidation (Sutherland et al., 2000). When oxidized bases occur in close proximity to each other on opposite strands, or in close proximity to a single-strand break, enzymatic incision will create a frank DSB that can be detected by electrophoretic methods. This assay readily detects CDD produced by low-LET radiation, but saturates once a single DSB is present, and so is unable to detect the additional complexity associated with high-LET radiation exposure (Hada and Sutherland, 2006). Recent work using a different approach, involving electron microscopic imaging of chromatin-bound DSB repair proteins (Lorat et al., 2015) has shown that high-LET radiation exposure can create astonishingly CDD, especially in heterochromatin, where more than 60 broken DNA ends have been seen within a small area. These highly complex DSB clusters were completely absent in low-LET irradiated cells.
One of the consequences of highly complex, CDD is the generation of small DNA fragments with lengths corresponding to the distance between adjacent DSBs. Modeling predicts that exposure of human cells to 1 Gy 56Fe ions will produce 20–50 DNA fragments of < 1000 bp, depending on the energy. This is about 30 times the yield predicted for 1 Gy of γ-rays (Campa et al., 2009). Although experimental data are limited, an early study showed that high-LET irradiated cells release about twice as many ~40 bp DNA fragments as compared to low-LET irradiated cells (Wang et al., 2008). In light of the recent data showing the existence of very complex clusters following high-LET radiation, these could well be underestimates.
2.2. DNA lesion detection sensitivity, response to dose and dose rate
Each of the different types of DNA lesions discussed in the preceding section could potentially serve as a biomarker of radiation exposure (reviewed in Ravanat et al., 2012). Oxidized DNA bases and other chemical products of radiation damage are excised by cellular DNA repair mechanisms and can be measured directly in accessible body fluids. Alternatively, they can be detected, prior to repair, in chromosomal DNA extracted from cells obtained from oral mucosa or blood. Levels of oxidized bases and other radiation products increase linearly with dose. For DNA irradiated in the native cellular environment, the number of oxidized bases is about 0.2 per 106 bp per Gy (Mohsin Ali et al., 2004).
The sensitivity of methods based on direct analysis of radiation products is limited mainly by the relatively high background levels, which can be attributed to reaction of DNA with endogenous ROS. Endogenous ROS are formed as byproducts of normal metabolism, and although levels vary between individuals, they tend to increase with stress, metabolic rate, and chronological age. Estimates of the steady state burden of oxidized bases vary widely, but levels may be assumed to be on the same order as the increment attributable to 1 Gy of radiation exposure, and are perhaps much higher in some instances (Mohsin Ali et al., 2004).
Another difficulty with direct measurements of radiation products is that they turn over rapidly as the result of cellular DNA repair processes. For this reason, they are not useful for estimating cumulative damage arising from the chronic, low dose rate exposures that are of concern for human space travelers. Finally, most of the available methods for chemical determination of radiation products require that they be removed from their original context through enzymatic digestion of DNA. Critical information about their original spatial distribution in DNA is therefore lost.
Many of the same limitations apply to analysis of DNA single-strand breaks (SSBs), which, like oxidized bases, are among the most abundant radiation-induced DNA lesions. SSBs can be measured by in situ single-cell electrophoresis under denaturing (alkaline) conditions (referred to as the “Comet assay” based on the comet-like tails that emerge from permeabilized cells). The comet assay has a practical sensitivity of about 0.2 Gy, which again is limited by the background levels of SSBs attributable to endogenous ROS. Similar to oxidized bases, SSBs are repaired very rapidly, and again are not useful for detection of cumulative damage arising from exposures at very low dose rates. Analysis of SSBs also does not differentiate between high- and low-LET radiation-induced DNA damage.
DSBs are produced much less efficiently than SSBs (0.005 per 106 bp per Gy for low-LET radiation), but are a much more specific marker of radiation damage, because endogenous ROS do not generate DSBs to an appreciable extent (except in the specialized context of DNA replication, where collapsed replication forks produce a special category of “single-ended” DSBs). The sensitivity of physical methods for DSB detection is limited by the small absolute numbers of DSBs produced at low doses. In situ single cell electrophoresis under native (neutral pH) conditions has a practical sensitivity of 2 Gy. An alternative method, pulsed field gel electrophoresis, is slightly less sensitive, although it is highly quantitative. It should be noted that electrophoretic assays quantify the number of DSB-containing local multiply damaged sites, rather than DSBs directly, and thus tend to underreport true values for high-LET radiation. Due to the limited sensitivity of physical methods of DSB detection, they have largely been supplanted by cytological methods based on counting of repair foci, which are regions of modified chromatin associated with unrepaired DSBs. Each unrepaired DSB leads initially to the formation of one repair focus, and the method is thus highly sensitive. Our best estimates of the burden of endogenous DSBs comes from counting repair foci, which are present at a level of 0.05 per cell in non-cycling fibroblasts (Rothkamm and Lobrich, 2003) and 0.1 per cell in primary human lymphocytes (Lobrich et al., 2005). For lymphocytes, inter-individual variability is on the order of 2-fold. Based on these estimates of the endogenous burden, the dose of radiation required to double the number of endogenous DSBs is the order of 0.001 Gy, well within the range of interest for human exposures. Although repair foci are produced by both low- and high-LET radiation, high-LET foci are more persistent, which provides some basis for discriminating between high- and low-LET radiation exposures. The dynamics of repair foci and potential confounding factors with these assays are discussed in more detail in the next section.
As is evident from the preceding discussion, the repertoire of available biomarkers of high-LET DNA damage is currently not ideal, as all of the available approaches suffer from some short-comings: limited sensitivity, susceptibility to confounding factors, or an inability to discriminate between high- and low-LET damage. An ideal biomarker for high-LET radiation exposure would be one that is sensitive to the complexity of LMDS. Although such markers have yet to be fully developed and validated, direct detection of repair proteins bound to DNA ends in situ, is one approach that appears promising. Whereas repair foci arise as a secondary response to radiation and are relatively large (hundreds of nanometers), binding of repair proteins is a primary response that occurs within seconds, and the resulting complexes are small (tens of nanometers or less). The highest available resolution is obtained by electron microscopic imaging, although this may be difficult to implement at scale for biomarker analysis (Lorat et al., 2015). Recent reports indicate, however, that binding of proteins involved in non-homologous end joining (NHEJ) repair (e.g., Ku protein) can be detected at single molecule sensitivity by optical fluorescence microscopy, using a combination of optimized extraction methods (which remove the background of unbound proteins) and super-resolution imaging (Britton et al., 2013; Reid et al., 2015). Although these methods have yet to be applied for detection of high-LET damage, they could, in principle, be used to estimate the complexity of high-LET radiation-induced LMDS based on the number of DNA ends within a restricted volume. Similar optical fluorescence methods can be applied for detection of single-strand DNA binding proteins, which are markers of homologous recombination (HR) and alternative end-joining repair (Yajima et al., 2013). Although super-resolution imaging technology may be too complex for flight measurements, it could be applied for post mission analysis of archived specimens.
A more speculative approach for quantification of complex local multiply damaged sites is based on release of characteristic small DNA fragments. A better understanding of their lifetime and the metabolic fate of these fragments, together with improved analytical methods for detection, will be required before this approach can be adopted for widespread use.
2.3. Non-targeted DNA lesions
Any discussion of high-LET radiation-induced DNA damage should include mention of DNA lesions arising as a result of non-targeted, or bystander effect. There are many examples where contact, co-culture, or exposure to conditioned media cause non-irradiated cells to take on characteristics of directly irradiated cells, including activation of the DNA damage response, oxidative stress, and induction of mutations and genome instability (reviewed in Morgan and Sowa, 2015 and Blyth and Sykes, 2011). The LET dependence of bystander effects is not well established and may be model-dependent. It has been shown that fibroblasts exposed to high-LET (56Fe) particles generate media-transmissible signals that lead to decreased clonogenic survival, an increased frequency of micronuclei and γ-H2AX foci-positive cells in non-irradiated fibroblasts (Yang et al., 2007a, 2007b). Separately, it has been shown in high-LET irradiated tumor studies that bystander cells increase the use of error-prone DSB repair pathways, similar to an effect seen in directly irradiated cells (Li et al., 2015). The nature of the DNA lesions in bystander cells, and in particular whether there is any evidence of complex LMDS similar to those in directly irradiated cells, remains to be fully investigated.
2.4. Relationship between radiation-induced DNA lesions and cell fate decisions
A lower dose of high-LET radiation, relative to low-LET reference radiation, is sufficient to initiate given biological endpoints, for example, a specified level of cell killing or cancer risk. This ratio of doses defines the “relative biological effectiveness” (RBE) of a given radiation type. For high-LET charged particles, RBE’s are typically in the range of 2 to 5, depending on the experimental model. RBE increases with LET and reaches a maximum value at approximately 100 keV/µm for many endpoints (reviewed in Blakely 2012).
The RBE value for high-LET radiation is thought to be determined, in large part, by the complexity of LMDS and the rate of repair of DSBs present within those sites. There is considerable evidence that DSBs induced by high-LET radiation are repaired more slowly than those induced by low-LET radiation (Asaithamby et al., 2008; Reynolds et al., 2012), and to some extent by different pathways. It may be the case that DSBs embedded in local multiply damaged sites are simply poor substrates for DNA repair enzymes. However, there is also evidence that short single- and double-stranded DNA fragments, produced from local multiply damaged sites, bind to and inhibit proteins in the NHEJ pathway, increasing the reliance on slower, alternative repair mechanisms (Pang et al., 2011; Yuan et al., 2015). Consistent with impaired NHEJ repair as an underlying factor, RBE values for high-LET radiation decrease to unity in cells that are genetically deficient for XRCC5 or LIG4, two important NHEJ genes (Wang et al., 2010; Okayasu et al., 2006). Also consistent with this idea, increased expression of APE1, an enzyme that increases the yield of short DNA fragments by processing clusters of oxidized bases into frank DSBs, selectively increases the RBE for high-LET radiation (Wang et al., 2014).
Modeling studies also support a strong relationship between DSB complexity and RBE. The relation between the LET dependence for calculated yields of DSBs of various complexity and cell inactivation was first studied with the simulation code PARTRAC for proton and α-particle irradiation (Friedland et al., 2005). RBE values for various ion types for cell inactivation of human fibroblast cells reaches a maximum of about 4 with LETs between 100 and 200 keV/µm, whereas those for DSB+ (+ indicating greater complexity of DSB) reach a maximum of about 2 at an LET of about 70 keV/µm and DSB++ culminate with RBE values above 10 at LET values of about 200 keV/µm (Tsuruoka et al., 2005). A weighted sum with cell killing probabilities of 8%, 2% and 0.5% for the categories of DSB++, DSB+ and DSB, respectively, has been found in overall agreement with the LET dependence of measured RBE and cell inactivation cross sections (Friedland et al., 2011).
3. DNA damage response foci
The seminal discovery that the histone variant H2AX was specifically modified (phosphorylated) at sites of DSBs (Rogakou et al., 1998) gave rise to a quantitative surrogate marker for DSB measurement (Nelms et al., 1998). As γ-H2AX is proposed to recruit DNA repair proteins to DSB sites (Paull et al., 2000), γ-H2AX kinetics is considered a robust marker for assessment of repair and resolution of damage. They have extensively been used to quantify DSBs in radiotherapy patients (Sak and Stuschke, 2010) and used as a biomarker for aging and disease (reviewed in Mah et al., 2010). Quantification of γ-H2AX by immunofluorescence and flow cytometry is unambiguous and has proven to be reliable in detecting even minor DNA repair defects (Wang et al., 2005) in both G0/G1 and G2 phases (Lobrich et al., 2010) of the cell cycle.
While flow cytometry has the advantage of being quick and high throughput, and can seamlessly differentiate cell cycle stages, it primarily provides just total intensity/cell. In contrast, immunofluorescent microscopy allows users to quantify damage by identifying visible bright foci after exposure to ionizing radiation. These radiation induced foci (RIF) require validated computational code for automatic quantification as manual counting has led to statistical uncertainty (Rothkamm and Lobrich, 2003), subjective bias and lack of reproducibility. There are many other DNA repair proteins forming RIF (e.g. 53BP1, MDC1, MRE11, ATM, DNA-PKcs, etc.). Early algorithms for RIF quantification were based on threshold approaches after initial enhancement of the spot (foci) enhancement by image processing (Costes et al., 2006; Bocker and Iliakis, 2006), but such approaches fail to detect all foci when large intensity variations were observed. Pattern recognition algorithms have resolved this issue opening the door to high throughput 3D foci detection with reproducible data across different research laboratories (Neumaier et al., 2012; Runge et al., 2012). Other factors affecting reproducibility are the type of microscope used, baseline foci levels, cell type, passage number and oxygen levels in culture.
The conundrum lies in whether the increase and persistence of RIF higher than baseline levels is an indicator of genomic instability. Impaired DNA repair is thought to be a major factor contributing to tumor development (Lengauer et al., 1998; Thompson and Schild, 2002). Indeed, higher amounts of basal DNA damage, as evidenced by an increase in the number of basal γ-H2AX foci is often observed in different types of tumors (Warters et al., 2005; Bonner et al., 2008). A dramatic increase in the frequency of complex CAs in radio-resistant tumor cells was observed after carbon ion irradiation (Hofman-Huther et al., 2004). Subsequent cytogenetic studies revealed exposure to HZE radiation can cause a much higher level of complex chromosomal rearrangements (Loucas and Cornforth, 2001; Durante et al., 2002). Based on these observations, it is tempting to speculate that the increase in RIF number and their persistence after high-LET radiation exposure is a marker of underlying genomic instability due to the induction of complex CAs.
3.1. Relationship between RIF and dose, dose rate, radiation quality, and time
The dependence of RIF on dose, dose rate and LET remains an open debate. As far as the dose dependence, the number of RIF appears to be proportional to dose for low-LET doses < 1 Gy (Moroni et al., 2013) with numbers reaching a maximum between 10–20 RIF/Gy a few hours post IR. However, for doses > 1 Gy, some laboratories show in human breast epithelial cells that the number of 53BP1 RIF saturates with increasing doses (Neumaier et al., 2012; Georgescu et al., 2015) while RIF appear faster (Neumaier et al., 2012). Dose saturation is not as pronounced if one reports the total amount of 53BP1 recruited within all RIF in the nucleus (Georgescu et al., 2015). In this latter work, live cell imaging of RIF suggests saturation observed after high doses of low-LET is the result of coalescence of nearby DSB into single RIF. In contrast, peripheral blood mononuclear cells (PBMCs) extracted in patients who underwent computed tomography (CT) or radiotherapy show a good linear relationship between dose and γ-H2AX numbers (Redon et al., 2009), a result confirmed with macaque PBMCs (Redon et al., 2010). A recent study on dose-response relationships after whole blood irradiation in patients receiving molecular radiotherapy (MRT) with I-131 and Lu-177 showed a linear relationship between the number of γ-H2AX and 53BP1 foci per nucleus and the absorbed dose to the blood for both radionuclides studied (Eberlein et al., 2015).
For high-LET radiation, there are two possible geometrical configurations for exposing cells. If one exposes the plate perpendicular to the beam, then the number of RIF does not match the expected number of DSB but matches instead the number of track traversals, with one large and bright RIF per particle traversal, indicating these RIF are made of multiple DSBs (Costes et al., 2006). In contrast, when cells are exposed parallel to the beam, tracks of several foci can be visualized along the linear trajectory of particle traversal (Jakob et al., 2003; Aten et al., 2004). Again, it is interesting to note as LET increases, the number of RIF along the tracks reaches a maximum of ~1 RIF/µm for LET’s larger than ~150 keV/µm in non-malignant human breast cells (Vadhavkar et al., 2014) or in cancer cell lines stained for γ-H2AX or XRCC1 (Jakob et al., 2009). Thus, as energy deposition along the track increases with LET, the number of foci remains the same but each focus has more DSBs (Costes et al., 2010). On the other hand, total intensity of RIF labeled by γ-H2AX along the tracks shows a linear dependence with LET as high as 400 keV/µm (Vadhavkar et al., 2014).
The time required for a cell to repair the induced damage and/or the kinetics of RIF resolution is dependent on radiation quality. Remarkably, RIF are induced within seconds to minutes following IR, and typically reach a maximum at 0.5 to 1 h post IR and diminish as DNA repair proceeds (Rogakou et al., 1998; Costes et al., 2006; Anderson et al., 2001; Rothkamm et al., 2003; Leatherbarrow et al., 2006; Costes et al., 2007; Markova et al., 2007). It is well known that the CDD induced by HZE particle radiation takes longer to repair (Neumaier et al., 2012; Saha et al., 2013; Saha et al., 2014; Whalen et al., 2008).
Several labs have shown evidence of a non-targeted effect at low and acute doses (Burdak-Rothkamm et al., 2007; Pereira et al., 2014). Two potential cytokines, transforming growth factor beta (TGF-beta) and nitric oxide (NO), have been proposed to be instrumental in forming foci in bystander cells (Dickey et al., 2009). It is interesting to note that RIF can be induced inside the nucleus of a cell by simply irradiating the cytoplasm of cells with active mitochondrial function (Tartier et al., 2007). Very low doses of either low or high-LET radiation (1 cGy) can elicit the same level of 53BP1 foci in both irradiated and neighboring cells. However, when pre-irradiating cells with 1 GeV protons up to three hours before, cells become unresponsive to the non-targeted response (Yang et al., 2011a). This same group showed that for doses below 1 cGy and for doses as low as 0.5 mGy, a constant 53BP1 foci offset is observed and it is the same in both irradiated and non-irradiated bystander cells, suggesting that foci observed at such dose levels are primarily induced by ROS and reflect the systemic activation of wound signaling (Yang et al., 2011b). Given the fact that low dose rates involve constant exposure to extremely low doses of radiation, non-targeted effects are therefore a major confounding factor for interpreting low dose-rate RIF.
3.2. Pattern of RIF and relationship to cell fate decisions
DNA repair foci, in combination with other repair kinases (ATR, ATM) and the downstream checkpoint proteins (Chk1, Chk2) and effectors (p53) can affect cellular proliferation, replication and cell cycle progression until the repair process is complete (Sancar et al., 2004; Houtgraaf et al., 2006). Alternatively, cells with unrepaired DNA damage may also permanently exit the cell cycle and become senescent (Jaiswal and Lindqvist, 2015). It appears that sustained p53 activation results in cell cycle exit, while oscillating pulses of p53 ensures cells resume proliferation after DNA damage (Purvis et al., 2012). Because of their complex nature, CDD are often refractory to repair (Asaithamby and Chen, 2011). Persistence of γ-H2AX and 53BP1 foci at the DSB sites after HZE radiation indicates the irreparability of the CDD. If cells continue to proliferate with damaged DNA, it can cause unwanted mutation in the DNA leading to genomic instability, one of the hall marks of carcinogenesis (Jackson and Bartek, 2009). To prevent this, cells with unrepaired DNA damage often undergo cell death (Roos and Kaina, 2006) by necrosis, apoptosis or autophagic cell death (Edinger and Thompson, 2004). Phosphorylation of p53 by E2F1 is a central theme in deciding whether a cell with unrepaired damaged DNA will undergo cell death (Wu and Levine, 1994; Qin et al., 1994).
DDR checkpoint signaling is not always stringent as evidenced by early G2/M progression in cells with CDD. This results in these cells entering mitosis with unrepaired CDD and eventually accumulating carcinogenic mutations (Camacho et al., 2010; Lobrich and Jeggo, 2007). Results obtained from various in vivo experiments confirm that the CDD has potent biological effects, such as mutagenicity and carcinogenicity (Shikazono et al., 2009; Singleton et al., 2002). The studies done by multicolor banding in situ hybridization (mBAND) have shown that high-LET IR produces a large number of chromosomal inversions involving complex rearrangements of both inter- and intra-chromosome exchanges (Hada et al., 2007). Another study found that high-LET irradiated immortalized human bronchial (BEP2D) and breast (MCF-10F) cells undergo malignant transformation through a series of successive steps and finally become tumorigenic in nude mice (Hall and Hei, 2003). Overall, from these observations it is clear that CDD is difficult to repair and proper checkpoint signaling and cell fate decisions are of prime importance to prevent genomic instability and onset of carcinogenesis.
3.3. Ability of RIF to predict cancer risk from high-LET exposures
Persistent RIF post IR are thought to be good predictors of radiation sensitivity (Dikomey et al., 1998). Interestingly, low dose rate exposures seem to be better at identifying individuals with DNA repair deficiencies. For example, a patient with one mutated allele of ATM showed persistent foci after 24 h of chronic exposure to γ-rays, whereas the same cells exposed to an acute dose of 1 Gy showed no difference in RIF levels as compared to normal controls (Kato et al., 2006a).
To address the question of biomarker specificity for high-LET radiation-induced cancer risk, one should focus on DNA damage specific to this exposure. One potential idea is to focus on complex DSBs. These DSBs are specific to high-LET radiation and speculated to be repaired inefficiently by NHEJ, but competently in G2 by HR. A role for HR is demonstrated by clonogenic survival assays that show deficient HR leads to a hypersensitivity to high-LET radiation (Asaithamby et al., 2011). However, NHEJ repair has still been shown to play a more important role as compared to HR in defining radiosensitivity after exposure to high-LET radiation (Takahashi et al., 2014). But perhaps higher or persistent levels of Rad51 RIF may provide a biomarker for high-LET exposures. The clustering of nearby DSBs into single RIF after high doses of low-LET (Neumaier et al., 2012; Georgescu et al., 2015) was described in the previous section and since HZE particles deposit extremely high doses along very narrow tracks (< 1 µm diameter), a “DSB clustering phenotype” will always occur along HZE tracks independent of the dose. In turn, DSB proximity leads to increased mis-rejoining and mutation rates (Costes et al., 2001). Thus, higher levels or more persistent “DSB clustering” is a potential and promising easy way to identify individuals at an increased cancer risk from high-LET radiation.
4. ROS
Exposure to low- and high-LET radiation induces ROS and is associated with temporal and mechanistically distinct responses (Li et al., 2014; Sridharan et al., 2015). At low levels, ROS function in signal transduction by modifying reactive cysteine residues modulating the activity of phosphatases, kinases, ion channels and transcription factors (Reczek and Chandel, 2015). ROS activity and specificity is controlled by specific homeostatic pathways, which exert effects via localizing generation, controlling target proximity and neutralization by antioxidants, and detoxifying pathways (for examples see (D’Autreaux and Toledano, 2007)). Oxidative stress results from an imbalance between formation and neutralization of intermediaries, resulting in damage to proteins, lipids and DNA. ROS produced in response to radiation mediate multiple acute and chronic biological outcomes, including modulating cell proliferation and death, cell fate and differentiation, inflammation and disease.
4.1. Measurement of cellular ROS and oxidative stress
The approach most widely employed to measure ROS in vitro is by direct incubation of metabolically active cells with redox sensitive probes. Caveats of this approach are poor specificity and modification of the redox status of the cell (Kalyanaraman et al., 2012). Novel probes of higher specificity have been developed based on boronate chemistry and on redox sensitive proteins to overcome some of the limitations. They can be targeted to different subcellular compartments, used to monitor ROS production in vivo and have sufficient sensitivity to detect changes in basal metabolic rate (Salin et al., 2015). Indirect methods are based on monitoring effects on target molecules and on antioxidant systems as surrogates for ROS activity, including the accumulation of adducts in proteins such as carbonylation, lipid peroxidation (TBARS, MDA and 4-HNE) and oxidized DNA bases as single molecules and/or combined measurements (for a comprehensive review relevant to the use of oxidative stress as biomarkers see Halliwell and Whiteman, 2004). Many of these markers are amenable to measurements in blood and urine and have been employed in numerous epidemiological studies demonstrating that oxidative stress biomarkers increase in association with multiple diseases, including cancer (reviewed in Frijhoff et al., 2015).
Although oxidative stress biomarkers in blood do reflect redox changes following a wide range of treatments on specific tissues in animals (reviewed in Margaritelis et al., 2015), there is a poor understanding of the biological processes reflected by these reporters in the plasma and urine (Il’yasova et al., 2012). Such biomarkers are highly sensitive to environmental and lifestyle factors, and have a significant inter- and intra-individual variability (Kato et al., 2006b). While several studies show association of these types of markers with cancer, more studies are needed to validate them against known inducers of oxidative stress in humans and in prospective studies to be valuable as risk predictors (Loft et al., 2012, 2013).
4.2. Effect of dose, dose rate and radiation quality on ROS level
ROS are generated at multiple time points following exposure to radiation, and are associated with distinct responses. ROS resulting from the interaction of photons with water are generated in a spatial and density distribution uniquely distinctive between low- and high-LET radiation (Goodhead, 1988), are very short-lived and inflict mostly damage to macromolecules. ROS also are critical signaling intermediaries in the biological responses elicited by radiation. Responses to an acute exposure (within the first 24 h) include bystander effects and cellular stress responses associated with the activation and function of DNA damage response pathways (Yang et al., 2007; Klammer et al., 2015). Elevated ROS have been shown (in vitro and in vivo) to persist for weeks (Werner et al., 2014; Tseng et al., 2014; Baulch et al., 2015), months and years following exposure to high-LET radiation in bone marrow (Chang et al., 2015), intestinal cells (Datta et al., 2012) and the CNS (Limoli et al., 2007). The effect of radiation quality on these secondary increases in ROS associated with the cellular stress response are still poorly understood, as some studies have found a greater effectiveness of high- over low-LET radiation in fibroblasts (Buonanno et al., 2011; Laurent et al., 2013; Dettmering et al., 2015), while not in other cell types (Werner et al., 2014; Baulch et al., 2015).
The effect of dose on ROS levels depends on the amount of time elapsed following exposure. ROS increase within the first 24 h post exposure are proportional to dose in neuronal stem cells (Tseng et al., 2014; Giedzinski et al., 2005). However, the most prevalent observations are that the detected ROS level increases over a narrow dose range, is not directly proportional to the dose and levels off with increasing dose (Werner et al., 2014; Baulch et al., 2015; Limoli et al., 2007; Dettmering et al., 2015). This observation suggests a threshold-type mechanism consistent with the activation of a metabolic process or enzyme, rather than the result of accumulated damage. The effect of the dose rate on ROS production has been examined in very few systems, but evidence suggests qualitatively similar ROS responses at low- and high-dose rates. For example, human neural stem cells exposed to 250 MeV protons fail to show any difference in induced levels of ROS when exposed to either 20–25 c/h or 25–50 cGy/min (Tseng et al., 2013).
4.3. Evidence for ROS in non-targeted effects
ROS are one of many candidate molecules proposed to mediate the bystander effect (reviewed in Li et al., 2014). ROS induced in the context of the bystander response appear to be independent of LET (Yang et al., 2007; He et al., 2014). While non-targeted radiation effects represent an active area of research, a clear understanding of the precise role of ROS is lacking because of the potential participation of ROS in multiple components of these non-targeted effects. ROS could be involved in the elaboration of the signal by the producing (targeted) cell, or function as the intercellular signal or message, or function in the receiving mechanism transducing the signal and the mediator of a biological response in the unirradiated, receiving cell (for some examples see Klammer et al., 2015). So, although it is clear that ROS play a role in effects seen in bystander cells, it is difficult to know exactly what role they play and how this role may vary with dose, dose-rate or radiation quality. Therefore this is an area of active investigation that requires further study.
4.4. Extrapolation of current ROS experimental results to GCR exposures
ROS are a potential biomarker for GCR exposure as ROS increase is pervasively detected in multiple cell systems and tissues, in response to protons and heavy ions varying in mass, energy, fluence or dose (Baulch et al., 2015; Tseng et al., 2013; Alan Mitteer et al., 2015). While changes in LET/energy imparted by a particle appear to be less important, a critical factor may be the dose needed to achieve a threshold response following a single exposure (Baulch et al., 2015). Even though significant increases in overall protein carbonylation were detected in extracts of fibroblasts exposed to doses as low as 0.2 cGy (Gonon et al., 2013), an important unknown is whether the radiation environment in deep space will be sufficient to elicit ROS production in tissues in vivo (Held, 2009; Kronenberg and Cucinotta, 2012). Nonetheless, the sensitivity of ROS as a biomarker is supported by increased oxidative stress markers detected in erythrocytes (Rizzo et al., 2012), skin (Mao et al., 2014), and in the retinas of mice (Mao et al., 2013) after their return from space flight. An additional advantage for ROS as a biomarker is the persistence of the responses providing a wide window of opportunity for detection. However, to be useful as a biomarker to predict exposure effects, the mechanisms involved in ROS generation need to be elucidated.
4.5. Cell fate decisions and ROS patterns of expression
Antioxidants mitigate various short- and long-term effects of high-LET radiation exposure, even when administered post irradiation, suggesting that ROS generated in response to radiation and persisting over time are consequential for these outcomes (reviewed in Kennedy, 2014). Early ROS production modulates outcomes resulting from DNA damage load and complexity (Hada and Sutherland, 2006; Mitteer et al., 2015). ROS also regulate DNA damage responses and DNA repair by influencing the function of multiple DNA repair proteins, such as interfering with Ku binding to DNA, Ape1 and increasing ATM activity (reviewed in Sridharan et al., 2015).
ROS are both reporters and effectors of late tissue injury. At 24 weeks post exposure to whole body proton irradiation (1 Gy dose), hematopoietic stem cells (HSC) display increased ROS levels and are reduced in numbers and impaired in function (Chang et al., 2015). HSCs tightly regulate ROS levels, as low levels of ROS are required for proliferation, while higher levels promote differentiation and exhaustion of the undifferentiated pools (reviewed in Zhou et al., 2014). Similarly, in the central nervous system, unregulated high levels of ROS are also detrimental, as exhibited by cognitive dysfunction due to high ROS levels being reduced via overexpression of catalase (Parihar et al., 2015). A major difficulty in the interpretation of the biological consequences of redox changes in the absence of further understanding of the mechanisms involved is that the contribution of ROS is implicated based on the absence of an effect when antioxidants are used. However, experiments using antioxidants must be carefully interpreted as they may prevent adaptive responses (Kennedy, 2014; Zhou et al., 2014) in addition to having deleterious or confounding effects.
4.6. Ability of ROS measurements to predict cancer risk
Theoretically, ROS could be a biomarker suitable for defining cancer risk for multiple reasons. First, oxidative stress is a feature of most stages of cancer regardless of tissue of origin (Hussain et al., 2003), thus expected to reflect multiple tumor types, at various stages. Accordingly, markers for oxidative stress are elevated in the blood of cancer patients. Second, HZE-induced ROS increase during post irradiation events could influence cancer development processes such as cell initiation, inactivation and tissue repopulation by promoting cell death, senescence, and produce growth factors and cytokines that mediate bystander signaling (Sawant et al., 2001). Increased ROS levels correlate with cell transformation in the post irradiation period (Redpath and Gutierrez, 2001). Stem cells, which are a cell type relevant for cancer initiation, exhibit robust ROS responses and are susceptible to ROS-mediated effects on their function. Lastly, initial studies suggest that ROS play a role in heavy ion-induced carcinogenesis. HZE-induced tumor incidence and burden can be reduced by antioxidant supplementation of the diet in animals (Kennedy, 2014). ROS are critical components of the non-targeted effects, which have been proposed to play a significant role in low-dose HZE-induced carcinogenesis (Cucinotta and Chappell, 2010; Weil et al., 2014). ROS might also be indirectly associated with carcinogenesis by promoting inflammation, which is causally associated with all the steps of carcinogenesis including initiation, promotion, and progression.
5. Radiation-induced immune/inflammatory changes
Immune/ inflammatory changes post exposure to IR are very pertinent to the ensuing cellular response. The predominant proteins induced are key transcription factors NF-κB and STATs that regulate pro-inflammatory cytokines such as TNF-α, IL-1, IL-6, GM-CSF, MCP-1 (Di Maggio et al., 2015). Immune cells secrete both pro (IL-6, IL-1 and TNF-α) and anti-inflammatory cytokines (IL-10, 12) in response to IR (Formenti and Demaria, 2009; Sergei et al., 2010). The cytokine levels obtained following exposures will tend to vary dependent on the assay used to measure them. For instance, IL-6, when measured in epithelial cells 24 h post exposure to 1.2 Gy of X-rays, shows a radiation-dependent increase via ELISA, whereas RNA transcripts increase much earlier (by 1 h), peaking at 2 h and declining to baseline by 8–24 h (Beetz et al., 1997). Also, the cytokine RNA profiles vary depending on whether they are derived from in vitro or in vivo measurements. For example, in vivo exposure (20 Gy) does not induce IFN-γ/ TNF-α, but in vitro exposure does. IL-1 and IL-6 are known to increase following in vivo exposures, but in vitro increases are not noted, likely due to a higher background in these experiments. Thus, such studies may provide more consistent findings and insights into the mechanisms affected if both protein levels and the function of macrophages/ primitive erythroid cells (PECs) are obtained (Chiang et al., 2007).
Recently a protein complex called the inflammasome was found in several immune cells post IR. It was caused by activation of caspase-1, triggered by cell death and could provide a novel acute and chronic immune response marker (Lorat et al., 2015). This is the first evidence of a clear protein complex whose downstream signaling causes cell death, suggesting a possible biomarker that is more robust compared to mere changes in hematopoietic numbers or levels of cytokines, that are not specifically altered by IR alone (Pernot et al., 2012). In addition, similar to these in vitro studies, in vivo evidence from the atomic bomb survivors (Life Span Study) and Chernobyl survivors, points to a similar common profile of impaired cellular immunity, increased humoral response and higher inflammation (Annex D, United Nations Scientific Committee on the Effects of Atomic Radiation, 2006) following radiation exposures.
5.1. Immune/inflammation baseline levels, normal variation and radiation-induced susceptibility
Baseline levels of proteins and cytokines can vary between different populations or individuals, depending on a number of factors including geographical location and health status. For example, IL-6 was elevated in persons having an increased risk of myocardial infarction (Ridker et al., 2000). A great degree of variability was observed in the various subsets of murine immune cells following radiation exposure (Gridley et al., 2002). B cells revealed the most sensitivity, followed by T cells, NK cells and then by the more radioresistant macrophages, DC and granulocytes. The sensitivity of immune cells to radiation is further influenced by age, sex and genetic background as exhibited by studies in mice and human cells (Lindsay et al., 2007).
5.2. Impact of dose, dose-rate and radiation quality on immune/inflammatory changes
Dose, radiation quality and time, all impact immune response to IR. High doses of low-LET IR suppress splenic monocytes. In human peripheral blood lymphocytes (PBL), X-rays induced dose-dependent increases in TNFSS4 and CCL2 mRNA, two inflammation associated genes (Wang et al., 2014). In coronary artery endothelial cells, mir146, a key miRNA regulator in both the innate and adaptive immune response, was up-regulated by both single and fractionated high doses of radiation. Several chemokines showed a direct correlation with miR146 levels. In addition, other miRNAs showed inverse correlations with immune response genes emphasizing the role of miRNAs in inflammation (Palayoor et al., 2014). Low doses suppressed apoptosis and increased the number of splenocytes, reflecting a radiation-induced redistribution between different compartments of the hematopoietic system (Bogdándi et al., 2010). However, single low doses of low-LET radiation (0.5–1 Gy) also hampered adhesion, induced apoptosis, and reduced iNOS. A lower oxidative burst in macrophages exhibiting a biphasic (non-linear) response was also shown to modulate inflammation (Rödel et al., 2012).
Lymphocytes, one of the hematopoietic types of cells showing the most sensitivity to radiation, were depleted with exposure to medium from cells exposed to high doses (> 20 0–40 0 mGy) of radiation, although the degree of depletion was dependent on the species. Depletion of PBL was observed over the first few days post radiation exposure and corresponded with an increase in CAs. This depletion has served as an established measurement of IR induced immune response (Williams et al., 2010), and is thought to be caused by cell death, altered cell trafficking, inhibition of normal cell differentiation and/or failure of correct homing of hematopoietic progenitors. However, at lower doses (< 0.2 Gy/d) and low dose rates (< 0.1 Gy/h) there is a latent immuno-stimulatory effect involving both acquired and innate immunity (United Nations Scientific Committee on the Effects of Atomic Radiation, 2006), although the degree of this effect was dependent on the strain of mice used. Another subset of cells, the hematopoietic progenitors, show significantly reduced homing capacity following fractionated radiation, as opposed to a single dose (Singh et al., 2009; Storb et al., 1994), implying that more damage may occur in the host stromal environment with fractionated doses. Post exposure, immune cells ability to return to homeostasis takes months and they can exhibit an increase in adaptive immunity that enhances radioresistance upon subsequent exposures (Seed, 1996). Total body irradiation (TBI) can result in a loss of blood cells, resulting in the acute radiation syndrome often seen at high doses, irrespective of the type of radiation or dose rate (Sanzari et al., 2013). However, a protracted low-dose/ low-dose-rate (LDR) exposure of γ-rays to the skin showed a reduced immuno-reactivity compared to an acute gamma-ray or proton exposure, and was perhaps due to an adaptive-type of response (Mao et al., 2013).
Autoimmunity was evident in mice following a fractionated lymphoid exposure (2.5 Gy of γ-radiation/ day, 5 times/ week equal to a total dose of 42.5 Gy). This autoimmunity could be prevented by inoculation of CD4+ but not CD8+ T cells (Sakaguchi et al., 1994). In contrast, low dose γ-radiation (single acute 0.5 Gy exposure) has been used to attenuate arthritis, lupus and multiple sclerosis, and is thought to work by decreasing IL-6, and up-regulating Treg cells. However, this same treatment can exacerbate asthma, atopic dermatitis and Hashimoto’s Thyroiditis by decreasing IFNγ and up-regulating CD4+ T cells. These studies in total suggest that high doses of IR may cause immunosuppression, but that following low doses results can differ depending on the cell type.
How dose rate may affect cellular immune responses is unclear. Some studies report normal ratios of T cells (CD3−/CD4+) with chronic low dose rates (1.2 m /h) (Ina and Sakai, 2005). A γ-ray study revealed that irrespective of dose rate there was activation of a pro-inflammatory response with some significantly altered cytokines including notch ligand Jag 2, which has a critical role in development of lymphocytes and NK cells. This response likely reflects the anti-tumor immune response of the thymus. However, at high dose rates, immunosuppression was typically observed (Shina et al., 2011). Therefore, in total, these studies reveal that changes in dose rate may result in different and variable effects.
Cytokine expression post radiation peaks approximately 4–24 h post IR, with pro-inflammatory cytokines, such as TNF-α, IFN-γ from monocytes/ macrophages and ILs from T/ B cells (Kusunoki and Hayashi, 2008), aiding in immune recovery at the expense of disturbed homeostasis. This increase in inflammatory cells and cytokines results in a chronic increase in ROS (Ryan, 2012), altering cell differentiation and cell fate. There is some evidence that high doses of IR induce a rapid decrease in all types of immune cells in vivo, followed by regeneration by day 7. However, low doses show no evidence of regeneration by day 7, thus additional studies with longer time points post exposure are needed to reach definitive conclusions regarding how time and dose influence immune responses (Bogdándi et al., 2010).
Radiation quality has a significant effect on the immune response. For example, in vitro studies show little change with low doses (0.05–0.5 Gy) of low-LET IR in terms of cytokines levels, surface antigen expression or maturation of DC (Jahns et al., 2011). Although at a dose of 1 Gy or greater some non-significant induction (IL1β and TNFα, MCP1) or suppression (IL-10, IL-8) of certain cytokines was seen. However, following HZE exposure more significant changes were observed, including the expression of ICAM-1 resulting in vascular dysfunction (Cherry et al., 2012), the alteration of tissue architecture, and lymphoid cell distribution in spleen and thymus tissues following carbon ions exposure (Erofeeva et al., 2000; Grigorenko et al., 1998). 56Fe ion exposure resulted in persistent increases in circulating B-cells after total body irradiation, although these changes were not observed consistently (Gridley et al., 2002; Pecaut and Gridley, 2011). A recent review (Girdhani et al., 2013) summarizes the effects of protons in repressing neovascularization and immune responses, which then can affect cancer progression and metastasis. Protons, depending on dose, can inhibit angiogenesis, invasion and inflammatory cytokines’ release (Girdhani et al., 2012). Thus, radiation quality can play a unique role in modifying or exacerbating the immune response.
5.3. Ability of immune/inflammatory experimental results to be extrapolated to GCR type exposures
The mixed field, chronic, low-dose exposures in space pose a unique challenge to measure immune/ inflammatory changes. Gridley et al. (2002) showed that lymphoid cells and tissues are markedly affected by high-LET radiation at relatively low doses, with some changes that persist long after exposure, and that different consequences may be induced by different densities of ionizing particles. Thus simultaneous exposure to multiple radiation qualities at different energies could lead to a broader spectrum of immune dysfunction than currently anticipated (Gridley et al., 2002). The immune profile of Apollo astronauts showed fluctuations in PBL, but all were within normal limits and cells were immunocompetent (Kimzey et al., 1975). A shift in Th2 was also seen which might cause an increased risk for autoimmune disorders. However, the genetic bias for cytokine profiles of different mouse strains is known and this information could be used in selection of astronaut populations that may exhibit a more radioresistant genotype and be better suited for the long space travel. In surveying the literature, immune changes measured with low- and high-LET radiation have overall shown a huge variability in results based on alterations in dose and radiation quality studies, thus extrapolating immune changes observed in these in vitro and in vivo rodent studies to GCR exposures for astronauts in space will likely be challenging.
5.4. Immune/inflammatory changes relationship to cell fate decisions
Immune/ inflammatory changes affect many other biological systems. For example, low doses of radiation causes a decrease in cytokine secretion and antigen uptake in differentiated bone marrow dendritic cells in vivo. This pattern implies a shift towards immune tolerance rather than immune activation. Low doses of radiation appear to modulate the immune and hematopoietic system to prevent autoimmunity (Chun et al., 2013). Immune responses and senescence appear tightly linked in that IR induced senescent cells are likely a source of inflammatory mediators that appear to enhance tumor progression (Davalos et al., 2010). Low dose protracted exposure (5 cGy/every 12 h to a total dose of 10 Gy) to X-rays induced senescence in human mammary fibroblasts by fostering an oncogenic environment (Chuang et al., 2005). Factors released from senescent cells induce proinflammatory cytokines that likely include signaling NK cells to clear senescent debris, and resulting in a chronic inflammatory response (Freund et al., 2010). A recent study demonstrates that IR impairs T cell activation by altering metabolic reprogramming (Li et al., 2015). Together these studies imply that a chronic inflammatory phenotype seems to drive tumor cell progression.
5.5. Ability of inflammation/immune changes to predict cancer risk from high-LET exposures
The immune response following radiation is a double edged sword: immune surveillance is able to get rid of preclinical cancers, however due to chronic inflammation and inappropriate hormonal response tumor promotion/ progression is stimulated (Prestwich et al., 2008). A limited amount of research has been performed on the effects of the immune response following radiation, and most studies have been done using relatively higher doses of protons and γ-rays (1–3 Gy), and even fewer studies using heavy ions (Kennedy, 2014; Gridley et al., 2002). However, these studies suggest that immune / inflammatory changes can serve as a biomarker for cancer following high-LET exposure due to the major effects observed in lymphoid cells at very low doses (Gridley et al., 2002). IL-8 secreted from normal prostate epithelial cells was strongly correlated with cancer aggressiveness (Neveu et al., 2013). A more detailed study of what these immune changes mean in relation to cancer outcome is needed. As there have been few studies addressing the need for low dose (< 0.5 Gy or lower)/ low dose rate for both low/ high-LET radiation induced immune response (Blakely and Chang, 2007), we reiterate that for immune response alterations to be used as a clear biomarker, its role in mechanisms affecting cell fate decisions, such as metabolism changes which shift effects to promote tumor progression needs to be defined. In irradiated mice, radiation-induced genomic instability can result from specific cell interactions and macrophages via cytokines and ROS/ RNS expression (Barcellos-Hoff et al., 2015). These results caused the authors to conclude that cellular interactions augment radiogenic cancer risk and that the central player is the innate immune system.
5.6. Evidence for non-targeted immune/inflammatory responses
Both the targeted (TE) and non-targeted effects (NTE) of IR result in inflammatory or immunosuppressive effects (Prestwich et al., 2008). Due to inter-dependence of TE and NTE, NTE creates a milieu where cancer progresses (Barcellos-Hoff et al., 2014). In spleens of head-only 56Fe exposed mice there is an increase in B cells following a 5 Gy dose. This effect was likely due to the radiation injury causing cellular proliferation of activated B-cells, and antibody production against the released brain specific self- antigens (Pecaut and Gridley, 2011). NTE were also recently observed following relatively low doses of hypo-fractionated protons (1 Gy) or HZE ions (0.5 Gy) following thoracic spinal column exposures, and found to increase a microglial marker in the rat brain and NFκB in heart tissue (Suresh Kumar et al., 2015). Thus, a survey of the literature in this area allows us to conclude that NTE is observed as a consequence of an altered immune response, and in response to both single and hypo-fractionated doses of both low and high-LET radiation.
6. Chromosome aberrations (CA)
Aberrations in chromosome structure and number are a major consequence of misrepair of DSBs and chromosome mis-seggregation caused by ionizing radiation. This is especially significant as chromosome instability is not just an accepted hallmark of cancer but also a primary driver of tumor initiation and tumor growth. Both numerical alterations in chromosomes and several types of structural aberrations have been noted in tumors. The types of aberrations include those resulting from intra chromosomal exchanges (terminal deletions, interstitial deletions, centric rings, acentric rings and pericentric inversions) and two types that result from exchanges between chromosomes (dicentrics and reciprocal translocations). These alterations can be distinguished via cytogenetic analysis of Giemsa stained chromosomes (Mitelman et al., 2005). Giemsa (G) banding, the current gold standard and the first technique used to assess aberrations, produces a characteristic-staining pattern that precisely differentiates each chromosome based on its heterochromatic regions. Copy number changes greater than 6Mb and 9Mb can be discriminated with ~400–500 bands respectively. However, this technique is labor intensive and smaller aberrations can be detected only if they alter the density and/or pattern of bands in the region of interest (Geiersbach et al., 2014). Newer techniques such as FISH (fluorescence in situ hybridization) and m-FISH (multiplex-FISH) have leveraged the use of multi-colored probes to distinguish individual chromosomes; thus increasing the ease of detection. Nevertheless neither of these techniques allow the simultaneous high-resolution assessment of all types of chromosome rearrangements (Bailey and Cornforth, 2007), as detection resolution is in the Mb range (Cornforth et al., 2002). To overcome this limitation, recent models such as BDSTRACKS (biological damage by stochastic tracks) (Ponomarev et al., 2014) that can simulate smaller translocations on a theoretical level have been used to complement experimental results. This improvement in detection sensitivity using theoretical modeling is especially useful to assess radiation quality effects of high-LET radiation.
6.1. Baseline levels and individual variation in CAs
A number of studies have reported intra- and inter-individual variations of baseline frequencies of CAs in various human cell lines. Individual factors such as age, gender, life style factors, (smoking, alcohol drinking, dietary habits, vitamin intake, and physical activity), occupational exposure and geographical location seem to account for a significant proportion of this variability. Combined analysis of the impact of each of these variables, as in the study by Bolognesi et al. (1997), emphasizes the need to take into account the potential confounding effect of each variable in the design of bio-monitoring studies based on CAs. As differences in CA levels can be sensitively detected with various modifying factors, CA may be a very useful biomarker to accurately assess the additional genotoxic risk contributed by space radiation exposure (Hagmar et al., 1998).
6.2. Impact of dose, dose rate, radiation quality and time on CAs
Most studies have noted that dose dependence of CAs can be linear or linear-quadratic, depending on the ion and the energy. The number of CAs has been shown to increase with an increase in LET. Assessing the dose-rate dependence of CAs is more challenging as immediate repair post DNA damage has an influence on the persistence of accumulated CAs over time. For low-LET radiation, the yield of CAs progressively diminishes at lower doses and dose rates (Cornforth et al., 2002) and results in a dose response curve that exhibits a pronounced upward curvature at high dose rates and close to linearity at low dose rates (LDR). For low doses of high-LET radiation, the relationship between CAs and exposure is still uncertain, as only a small proportion of cells are directly hit. Estimates of risk from low doses at LDR exposures are often extrapolated using data from Japanese atomic bomb survivors using either a linear or linear-quadratic model. Radiation risk estimates for space exposures are extrapolated from epidemiological data obtained on Earth for cohorts exposed predominantly to acute doses of gamma rays, making results highly problematic and error-prone. The uncertainty can be reduced, if risk estimates are compared directly to space radiation-induced biological alterations, i.e. by detecting biomarkers such as CAs in astronauts. CDD from high-LET radiation have a significant impact on the complexity of the aberrations observed. High-LET radiation is known to result in a higher yield of complex exchanges, or exchanges involving greater than two breaks in two or more chromosomes, and to greatly impact cellular cytotoxicity. In comparison to low-LET, wherein the DNA damage at LDR is a small additional component close to the background level of DNA damage, heavy ions cause a great yield of complex CAs in the population of directly hit cells. At lower doses and dose rates, the overall fraction of the cells with one direct hit is much smaller and is of primary importance for human space missions, as this type of exposure more closely mimics the average cumulative fluence for a mission to Mars. Statistical models have extrapolated rules for dose and dose-rate dependence for different repair mechanisms, as reviewed in Sachs et al. (1997), with results based on the number of independent hits per cell. These rules differentiate between “1-track action” and “2-track action”. The dose and dose-rate dependencies are more complicated for 2-track action than for 1-track as the former includes cumulative effect from two independent radiation hits. As the number of tracks is linearly proportional to dose, 2-track action usually produces an approximately quadratic yield with dose.
When dose is given over a protracted time, 2-track action usually produces a smaller effect as repair can take place before the second hit. However, the yield of 1-track action is linearly proportional to dose and is independent of dose-rate. mFISH and spectral karyotyping techniques have led to an explosive increase in cytogenetic data, which, together with computer-assisted modeling, has provided new insights into the formation of radiation-induced CAs (Ponomarev et al., 2014). In mammalian cells, mechanistic evidence on dose-response/dose-rate relationships suggests complex aberrations occur by a breakage-and-reunion mechanism during NHEJ in G0/G1. The persistence of CAs is also an important factor for human space flight as most differentiated cells like neurons, CAs persist until the death of the organism or until the cell undergoes programmed cell death or mitotic death. However, in lymphocytes, although CAs might not last for the lifetime of a person, they do persist for the duration of a typical space flight. Assessing risk from the gradual accumulation of CAs in an astronaut during deep space missions is a major challenge as current experimental data uses primarily acute exposures or at best fractionated exposures for both for cells and animals.
6.3. Evidence for non-targeted CAs
Several studies provide evidence of a NTE for induction of chromosomal aberrations in bystander cells with high-LET exposures. For example, a non-linear dose-response for CAs has been noted in both lymphocytes and fibroblasts at very low doses of high-LET radiation exposure (Hada et al., 2014a, 2014b). It has been suggested that these non-targeted effects could be caused by aberrant cell signaling, involving nitric oxide and TGF-β, or by doses from delta-rays that do not directly traverse the cell nuclei. These results invoke a new paradigm to explain the shape of these dose-response relationships that postulates that the linear dose-response relationship observed for the induction of chromosome aberrations following exposure to high-LET radiation is a combination of bystander and direct effects (Geard et al., 2002). Similar results have been noted with alpha particle exposures. It has been demonstrated that an alpha-particle transversal produces chromosome damage in both the directly “hit” cell and in the “bystander cells” that receive very little energy deposition. This bystander effect results in the low dose induction of chromosome aberrations (Ponnaiya et al., 2007). As the dose increases, the frequency of bystander chromosome aberrations remains constant and the frequency of directly induced aberrations continues to increase. The combination of both bystander and direct effects is thought to contribute to the linear increase in chromosome aberrations even at very low doses. Understanding the contribution of NTEs to both dose and the nature and complexity of the resulting CAs will be essential to define its significance in defining cancer risk.
6.4. Extrapolation of CAs experimental results to GCR type exposures
Thus far, the majority of studies assessing radiation-induced CAs have used human cells and animal models and single ion or mixed beam exposures that mimic exposure to a specific set of ions at selected doses and dose rates. Since the material of the spacecraft, shielding and the human body itself can change the particle energy and create secondary fragments of the original GCR particle, more ground-based studies are required to understand how the energy and type of particles influence the yields of various types of CA. Recent expansion of the beam capabilities at NSRL, Brookhaven National Laboratory to simulate GCR exposure will aid in mimicking exposures closer to those found in space. One major caveat in extrapolating rodent and murine results to humans is that mice have a different spectrum of cancers and are more prone to developing leukemia than humans who develop more solid tumors. Combining rodent results with studies in lymphocytes from astronauts might provide a better estimate of risk from long-term missions. NASA assesses cytogenetic damage in lymphocytes of astronauts and uses these data to validate and develop risk-assessment models for characterizing excess health risk from space-radiation exposure. Chromosomal aberrations in peripheral blood lymphocytes (PBLs) are a unique biomarker that can provide simultaneous information on dose, dose equivalent and risk, and have been extensively measured in astronauts during the past 10 years. Recent results show a fast time-dependent decay of chromosomal aberrations in blood lymphocytes after space flight and a lack of correlation between translocations and cumulative dose in astronauts involved in multiple missions. This lack of correlation with cumulative dose may in part be attributed to individual variability in the kinetics of repair post cosmic radiation exposure (Durante, 2005).
6.5. Relationship between CAs and cell fate decisions
The presence of CAs in a cell can have a profound impact on cell fate. Basically there are a few cell fates that are possible for a cell containing CAs. If the CA does not disrupt any vital genes important for normal cell function then there will likely be no phenotype change. However, if lethal aberrations such as dicentrics are induced, a cell can undergo apoptosis or cell death. In the context of carcinogeneisis, this would likely be a protective mechanism that prevents the proliferation of damaged cells. Other cellular processes, such as cell cycle arrest and mitotic death can result from similar types of aberrations. Thus, all of these options result in a similar outcome whereby proliferation of genetically damaged cells is prevented (Jaiswal and Lindqvist, 2015). Another cell fate choice that has the highest impact on cancer risk is when a cell containing a CA does not result in lethality, but instead continues to proliferate. This non-lethal genomic damage can propagate genomic instability with every cell division and thus potentiate carcinogenesis.
6.6. CAs ability to predict cancer risk following high-LET exposures
There is evidence for recurring translocations that provide transforming events for the development of leukemia and lymphoma. However, although cytogenetic aberrations are commonly seen in solid tumors, they typically do not contribute to tumor development and have different dose- and dose-rate dependencies (Albertson et al., 2003). A number of studies have used CA’s yields as a biomarker of cancer risk (Bonassi et al., 2008). However, CA’s have been noted in various tumors independent of radiation exposure. It is clear from numerous studies that many tumors contain high levels and heterogeneity in CA that is often associated with a higher tumor grade and poorer prognosis. However, it is not clear whether CA’s initiate tumorigenesis or are simply a reflection of the chromosomal instability within the tumor (Giam and Rancati, 2015). Although predicting radiation-induced cancers in different population cohorts based on microdosimetry is difficult, the current risk projected from high doses incorporates effects of dose-rate, and is in part based on the linear-quadratic model and yields of CA’s (Fry et al., 1997). We expect that an increased knowledge of RBEs for radiation-induced CAs will help mission planners develop better risk reduction strategies (Panganiban et al., 2013).
7. Mutations
Mutations are essential features of cancer (Hanahan and Weinberg, 2011) and ionizing radiation was the first environmental mutagen identified (Muller, 1927). Mutations can serve as bioindicators of radiation effects as a function of dose, dose rate and radiation quality. Studies make use of specific genetic loci with screening strategies that allow for the detection of mutational events through biochemical assays. Often, the mutations identified using these strategies are not directly linked to tumor formation. However, information gathered in these studies can guide our understanding of the types of mutational events likely to occur at loci directly linked to the formation of radiogenic tumors.
The loss of function for tumor suppressor genes requires mutation of two alleles to enable tumor formation. Model systems allow quantification of mutations at heterozygous autosomal loci. In these models, one copy of a target gene, such as thymidine kinase (TK1, or Tk) or adenine phosphor-ribosyltransferase (APRT, or Aprt), has been inactivated and a mutagenic event that results in the loss of function of the second copy of the gene can be selected using a chemical that kills normal cells. Mutations can also occur in dominant oncogenes. In this case, mutation of a single allele can lead to tumorigenesis (Land et al., 1983).
Ionizing radiation produces mutations by various mechanisms, including single base-pair substitutions (Grosovsky et al., 1988), small insertions and deletions, loss of the entire gene, multi-locus deletions, recombination-mediated events leading to loss of heterozygosity (LOH), and chromosome loss (a form of aneuploidy) (reviewed in (Turker et al., 2009; Wiese et al., 2001). Studies using single copy genes, such as the X-linked hypoxanthine phosphoribosyltransferase (HPRT, or Hprt) gene, detect a subset of the radiation-induced events seen at an autosomal locus (Liber et al., 1989). Mutation at the CD59 locus in human × hamster hybrid cells has been used to demonstrate that ionizing radiation often produces chromosomal scale events, including whole chromosome loss, although such events are often lethal in normal human cells (Kraemer et al., 2001).
7.1. Ability to detect mutations at low doses and dose-rates
In early work, hybrid male mice were irradiated and mated with un-irradiated females that harbored seven recessive loci. Mutations were assessed in these seven marker loci in the offspring. The dose-response for mutant frequency for these dispersed genomic regions was linear, regardless of the dose-rate. Low dose-rate exposures (0.8 cGy min−1 or less) of low-LET radiation delivered to stem cell spermatogonia induced only ~1/3 as many mutations in the offspring as those delivered as single, high dose-rate exposure (Russell et al., 1958; Russell and Kelly, 1982). The reduction in mutant frequency at lower dose-rates was later associated with a decline in “large lesion” mutations (Russell and Hunsicker, 2012). Since these data reflect radiation effects on the irradiated male germ line, it is more challenging to extrapolate these findings to somatic cells in adults, where most human cancers develop.
Other systems have examined mutations generated via specific molecular mechanisms. The pUN mouse monitors deletion formation. The dose-response for pUN mutations following acute X-ray exposure was linear down to 1 cGy. (Schiestl et al., 1994). Radiation-induced autosomal mutations were also studied in Dlb-1 heterozygous mice. Mutations were measured in intestinal stem cells and their progeny, cells thought to be important in carcinogenesis. Fewer γ–ray-induced mutations occurred when the dose rate was lowered to 0.01 Gy min−1 (Winton et al., 1989).
Not all studies confirm a sparing effect for low dose-rate exposure to low-LET radiations. The frequencies of HPRT mutations in a human lymphoblast cell line following protracted X-ray exposure could not be distinguished from mutant frequencies measured after acute exposure, indicating no dose-rate effect in this cell type (Grosovsky and Little, 1985). Other studies using the same cell line have found an inverse dose rate effect for autosomal TK1 mutation (Amundson and Chen, 1996; Brenner et al., 1996).
7.2. Comparison of in vitro and in vivo mutation results and considerations of cross-species extrapolation
The Aprt heterozygous mouse model allows the translation of mutation studies from in vitro to in vivo. Mutations were characterized in kidney cells irradiated in vitro, or in cells retrieved from kidneys of irradiated mice. All of the types of mutational events that occurred in kidney cells exposed in vitro also occurred in vivo (Ponomareva et al., 2002). The chromosomal milieu of Aprt in the mouse is similar to that for TK1 in human cells. This aids in cross-species extrapolations of mutational risk, although one class of radiation-induced mutations (whole chromosome loss) that can be observed in the Aprt mouse model is not detected in the human cells.
7.3. Tissue specific effects of radiation mutagenesis – low-LET
Radiation mutagenesis has been examined in the LacZ mouse, in which many copies of the bacterial LacZ gene were introduced in the germline, and are present in all tissues of the progeny (Boerrigter et al., 1995). No cell division is needed in the tissue to detect mutants. X-ray-induced mutations were studied in lung, spleen and liver (Gossen et al., 1995). Proton-induced mutations were studied in brain and spleen (Chang et al., 2005). Mutant frequencies differed for each tissue as a function of dose and time, demonstrating tissue specificity for one form of low-LET radiation encountered in space flight (Chang et al., 2005). While the LacZ mouse model cannot detect chromosomal scale changes, the Aprt heterozygous mouse model can measure very large, tissue specific mutational events (Ponomareva et al., 2002). Both the strain background and the developmental age of the animal are important when examining radiogenic Aprt mutations (see e.g. Liang et al., 2007).
7.4. Effects of LET and dose rate on mutant frequency
The effects of ionization density on mutant frequency have been well characterized. An LET dependence was seen for the induction of Hprt mutants in Chinese hamster V79 cells (Thacker et al., 1979) and for the induction of HPRT mutants in human fibroblasts (Cox and Masson, 1979; Chen et al., 1994). The maximum effectiveness for the induction of HPRT mutants was in the LET range from 90–200 keV/µm.
The frequencies of HPRT mutants and autosomal TK1 mutants were measured in TK6 human lymphoid cells as a function of LET. More mutants were detected for the TK1 locus, regardless of the LET (Kronenberg et al., 1995). The greatest yield of TK1 mutants was achieved at 61 keV/µm, after which it declined sharply as LET increased. In contrast, the yield of HPRT mutants reached a maximum at 95 keV/µm, but remained relatively flat as LET increased. Recent studies using a different human lymphoblast cell line confirmed the maximum in the LET response for TK1 mutations (Liber et al., 2014). In vitro and in vivo studies on the induction of autosomal Aprt mutants in mouse kidney epithelium also demonstrate a strong effect of LET. Densely ionizing Fe ions were substantially more mutagenic than sparsely ionizing protons, whether the results are considered in terms of dose or in terms of particle fluence (Kronenberg et al., 2009; Kronenberg et al., 2013).
The effects of dose-rate on high-LET mutagenesis are less well studied. Protracted exposure of human lymphoblasts to fast neutrons at very low dose-rates resulted in an increased mutant frequency as the dose-rate was lowered (Kronenberg, 1991). The effect of dose-fractionation on the induction of Aprt mutants was studied in proton-exposed mouse kidney epithelial cells. Mutant frequencies were lower when the dose was fractionated but there also was less cytotoxicity, leading to only a very slight reduction in the mutational burden in the tissue compared with the results for an acute exposure (Kronenberg et al., 2013).
7.5. Non-targeted effects on mutation frequency
Non-targeted effects contribute to charged particle mutagenesis, particularly at low fluence. A charged particle microbeam enabled the targeted delivery of 20 alpha particles to 20% of the AL cells in the population. No mutations should arise in the directly irradiated AL cells, since this high alpha-particle dose is lethal to the targeted cells. Yet the frequency of CD59-deficient mutants was increased in the bulk culture, suggesting that the non-irradiated cells were the source of the increased mutant fraction (Zhou et al., 2000). Additional microbeam studies indicated that bystander mutagenesis was a major component of the overall mutational response at low fluence (Zhou et al., 2001).
7.6. Molecular approaches – results following low-LET or high-LET exposures
It has been well documented that ionizing radiation can create small mutations (Grosovsky et al., 1988; Liber et al., 1986). When the mutational system is less restrictive, ionizing radiation often produces large genomic changes. The TK1 locus in human cells, or the Aprt locus in mice, each are able to detect large deletion mutations and complex mutation (Ponomareva et al., 2002; Yandell et al., 1990). High-LET radiation produces large mutations at very low particle fluences that include chromosomal rearrangements and/or chromosome loss (Wiese et al., 2001; Kraemer et al., 2001; Hryciw et al., 2015). In contrast, mutations that arise from the non-targeted effects of irradiation resemble spontaneous mutants and not the majority of the directly induced mutants seen at short times following exposure (Chang and Little, 1992).
7.7. Relationship between mutations and cancer
Second step autosomal mutations play important roles in heterozygous cancer models that have assessed charged particle-induced tumorigenesis, including Apc mutation leading to intestinal tumors (Trani et al., 2014; Datta et al., 2013), Pten mutation leading to gliomas (McEllin et al., 2010), and Sfpi1 (PU.1) in acute myeloid leukemia (AML) (Genik et al., 2014). Studies of radiogenic AML in mice have examined PU.1 mutations on chromosome two. Deletion of one allele of PU.1 was proposed as a candidate bioindicator for radiogenic AML (Peng et al., 2009). Point mutation of the second PU.1 allele is not radiation related, and mutation of other genes located near PU.1 may be particularly important in radiogenic AML in different mouse strains. A study on the induction of radiation-induced glioblastoma suggests that radiogenic loss of the tumor suppressor Ink4b can elevate tumorigenesis in sensitized mouse models, and that amplification of the Met oncogene is also a common feature of radiation-induced tumors (Camacho et al., 2015).
7.8. New technologies to identify mutations and links to carcinogenic mechanisms
New approaches to the analysis of mutations are becoming available with the advent of dense SNP arrays, next-generation DNA and RNA sequencing, ChIP sequencing, and advanced molecular cytogenetic approaches. Associating radiation-induced changes with particular point mutations, loss of heterozygosity, or other events that may be causal to tumor formation will be challenging, given the wide variety of molecular events that lead to radiation-induced mutations and the large number of driver and passenger mutations found in human tumors (Bozic et al., 2010; Vogelstein et al., 2013).
7.9. Mammalian mutation assays with relevance to space flight conditions
Some mutation assays are particularly sensitive for the detection of mutations induced by low to moderate doses of charged particle radiation (Wiese et al., 2001; Kronenberg et al., 1995; Liber et al., 2014). A low fluence of heavy ions, including exposures as low as an average of one particle traversal per cell nucleus, can produce “radiation signature” mutations including deletions and complex mutations that often reflect chromosomal rearrangements (Hryciw et al., 2015). Furthermore, mutation experiments have been performed with mammalian cells on the ISS under specialized conditions (Yatagai et al., 2011). Together, these results indicate the utility of mutation assays to evaluate at least some of the essential features of the carcinogenic process at space-relevant doses.
8. Telomere dysfunction and carcinogenesis
Telomeres, the protective caps at the end of the chromosome, play an important role in maintaining genome stability. Telomere length is determined by the number of repetitive TTAGGG sequences at the ends of the chromosome and varies between species. The binding and interaction of ssDNA and dsDNA binding proteins to this backbone allows the formation of a uniquely folded tertiary structure. This structure prevents both chromosomal end fusions and the initiation of DNA damage responses that can propagate chromosomal instability (Blackburn et al., 2000). Both the minimum length of the telomere and maintenance of the nucleoprotein structure impact its function in safeguarding chromosome ends. Maintaining this length is inherently challenging as telomeres lose ~50–100 bps with each cell division, due to the end replication problem of DNA polymerase. This problem is overcome by telomerase (TERT) that synthesizes and elongates telomere repeats to maintain length. Although stem or progenitor cells still contain telomerase, the limited or lack of telomerase activity in normal differentiated somatic cells typically results in telomere shortening and governs the replicative life span of each cell (Broccoli et al., 1995; Hiyama et al., 1995). Studies have shown that in addition to restricting stem cell function, regeneration, and organ maintenance (Henriques and Ferreira, 2012), the systemic effects of telomere dysfunction include an increased cancer risk (Ju et al., 2006). Unlimited proliferation caused by the reactivation of telomerase is a hallmark of cancer and allows for the continued maintenance of cells with short telomeres. Genome wide association studies have revealed the TERT-CLPTM1L locus as a susceptibility factor for multiple malignancies. The association of TERT polymorphisms (rs2853691, rs2736100 and rs451360), deregulation and expression changes in TERT and other proteins that bind telomere ends with the progression and prognosis of cancers, all point to the key role of telomere regulation in maintaining cellular homeostasis.
8.1. Caveats specific to measuring telomere length
The gold standard for measuring telomere length is Terminal Restriction Fragment (TRF) Southern blot. DNA restriction digests are run on a pulse-field gel and an approximate average telomere length within the population is determined using a probe specific to the telomere sequence. As repeats lack restriction sites of interest, addition of sub-telomeric regions impacts the accuracy of measurement, especially at the single cell level. qPCR can be used to quantify the abundance of telomeric DNA in a sample. Q-FISH or telomere FISH is an alternate method that uses fluorescently-tagged peptide nucleic acid (PNA) probes to hybridize to telomere repeats in chromosome spreads. Although this labor-intensive method requires a fair number of cells, the ability to detail individual telomere lengths at a resolution of ~200 bps, and also estimate the amount of telomeric DNA in interstitial sites is a proven advantage. A modification of this technique to support high-throughput analysis using flow cytometry is called FLOW-FISH. This method requires smaller amounts of DNA and has the ability to define average telomere length in the entire cell population. Recently a novel assay, ADDIT (addition of de novo initiated telomeres), was developed to enable the assessment of new repeats at a single telomere and has revealed the importance of proteins such as ATM in telomere maintenance (Lee et al., 2015).
8.2. Relationship of telomere length and telomerase activity to dose, radiation quality and time
A number of studies have reported an increase in telomerase activity with radiation exposure (Hande et al., 1998; Leteurtre et al., 1997). This induction appears to be cell type specific (Finnon et al., 2000; Sishc et al., 2015) and exhibits dose-dependence and temporal kinetics. Increase in activity has been noted as early as 0.5 h and up to 24 h following exposure of fibroblasts and lymphoblasts to low-LET radiation. Interestingly, studies in TK6 and WTK1 lymphoblasts reveal a dose-dependent increase in telomere length fourteen days post radiation exposure, suggesting different mechanisms in play regulating telomere dysfunction at these late times. Of note, the induction of telomerase activity at low doses of radiation exposure is independent of wild-type TP53.
Very few studies have compared the effect of low and high-LET radiation on telomere length. High-LET exposures typically appear to increase telomere length at both early and late time points (Berardinelli et al., 2014). In contrast, low-LET radiation appears to either cause an initial shortening or no change in length, depending on cell type and subsequent lengthening at long times, similar to high-LET radiation (Neuhof et al., 2001; Ojima et al., 2004). Reactivation of telomerase post high-LET exposure has been postulated to be the cause behind early telomere elongation. Studies indicate that telomere length increase can occur in the absence of telomerase activation, even at doses up to 10 Gy of X-rays or protons suggesting a telomerase-independent mechanism for elongation (Berardinelli et al., 2010). Both short and long-term studies on PBLs from cancer patients undergoing radiotherapy (15–74 Gy) reveal that although telomere length distribution remains unchanged, there is a significant dose-dependent decrease in the number of short telomeres (< 4.4 kb) (Maeda et al., 2013). It has been speculated that following radiation cells with shorter telomeres could be selectively lost, or radiation specifically targets only shorter telomeres for elongation.
Some studies have shown that the immediate effects of radiation on telomere length are dependent on cell type, dose and LET. While length changes were absent post low doses (0.1–1 Gy) in AG1522 fibroblasts with both low or high-LET radiation (Nieri et al., 2013), another study showed telomere elongation specific to high-LET proton exposure (4 Gy) in human HFF2 fibroblasts (Berardinelli et al., 2013, 2011). As telomeres make up less than 1% of the total genomic DNA (Fumagalli et al., 2012), it is unlikely that the effect of radiation on telomeres of the entire population is caused by direct ionization during the initial exposure. Given this, and the presence of telomere length changes in unirradiated bystander cells and the progeny of radiated cells, it is plausible that telomere dysfunction post–radiation is caused by errors in telomere maintenance. The sensitivity of telomere length to initial and persistent ROS (Henle et al., 1999) supports the existence of a non-targeted effect on telomere length regulation.
Maintenance of telomere length appears to be strictly regulated by various proteins. One such example is ATM kinase, which is activated by radiation exposure and essential for normal length elongation (Lee et al., 2015), but dispensable for lengthening of short telomeres (Feldser et al., 2006). Studies using immortal human cells suggest that both ATM and ATR are required to localize telomerase to telomeres (Tong et al., 2015). The fine regulation of telomere length by dose, radiation quality and persistence of effects with time makes this an attractive biomarker to interrogate long-term carcinogenic effects of radiation.
8.3. Consequences of telomere changes on cell fate
Changes in telomere structure, length and function are all thought to contribute to cell fate decisions associated with degenerative events from aging to cancer. Dysfunctional or damaged telomeres can influence the choice of major cell fate decisions. First, the vulnerability of exposed ends associated with telomere attrition can promote chromosome end fusions (de Lange, 1998; Kim Sh et al., 2002) and potentiate genomic instability through subsequent cycles of breakage and fusion (McClintock, 1941; Artandi et al., 2000). This initiates the cellular choice towards carcinogenesis. To prevent the potentiation of genomic instability, cells have developed built-in tumor suppressor mechanisms. So as an alternate choice, when telomere lengths reach a critical threshold, cells either die or respond by initiating replicative senescence, a barrier that limits proliferation of cells with potentially aberrant genomes. Apoptosis is also increased in cells with shorter telomeres (Ilyenko et al., 2011). It is speculated that telomeres themselves could potentially acquire a senescence signaling state in a progressive and stochastic manner, as they shorten (Blackburn, 2000). Given that each cell has 92 telomeres, the nature of the initiating telomere signal that influences senescence instead of tumor initiation and vice versa is highly debated.
Recent studies have shown that reactivation of telomerase can trigger cellular reprogramming and enrichment of putative stem cell populations post radiation exposure (Sishc et al., 2015). The contribution of the mean telomere length, the length of the shortest telomere, and the degree of telomerase activation that drives these cell fate decisions is unclear. Studies have suggested that as little as five dysfunctional/damaged telomeres are sufficient to trigger the signal for senescence (Kaul et al., 2012) and in yeast a single short telomere is sufficient to trigger senescence (Xu et al., 2013; Abdallah et al., 2009). The degree of telomere dysfunction, rate of loss of telomere repeats, length of the shortest telomere are all factors that likely play a critical role in choosing either to maintain homeostasis by inducing cell death and senescence or in altering the steady state by reprogramming or immortalizing the cell to promote carcinogenesis. As telomere dysfunction can alter the fate of a cell in several directions, length changes might not directly point to carcinogenesis risk post high-LET exposure. However, indirectly assessing the different cell fates triggered as a consequence of telomere length changes could provide a cumulative measure of how the nature of the exposure can tilt the balance towards cancer development.
8.4. Efficacy of using telomere length assessments to extrapolate consequences of GCR exposure
Sensitivity of telomeres to constituents of low dose, low dose-rate GCR exposure and our ability to relate telomere phenotypes to long term consequences are two key factors that determine the robustness of telomeres as a biomarker. The heterogeneous nature of the GCR spectrum poses a huge challenge to design and interpret experiments using simulated GCR exposures. Thus far, current studies have primarily been carried out using mono energetic beams of single HZE ions e.g. low- (1 keV/µm) and high-LET protons (28.5 keV/µm), carbon ions (50 keV/µm) (Berardinelli et al., 2010, 2015), or Fe ions (1 GeV/n) (Sishc et al., 2015) at doses ranging from 1–8 Gy, in comparison to X-rays. The temporal decision to lengthen or shorten telomeres or reactivate telomerase depends on various factors. While telomere lengthening caused by X-rays is delayed and noted only 2 weeks post exposure (Ojima et al., 2004), the impact of protons (28.5 keV/µm) can be observed within 24 h (Sgura et al., 2006) and shows significance mainly at higher doses (Berardinelli et al., 2010). This lengthening effect is also observed in vivo in exposed Chernobyl workers who exhibit longer telomeres years post initial exposure (Reste et al., 2014). It is speculated that both recombination events during chromosome healing, newly synthesized telomere repeats due to telomerase reactivation and alternative telomerase independent lengthening using the ALT pathway (Berardinelli et al., 2010) are all potential underlying mechanisms of this lengthening process. In contrast, studies have also shown that in certain cell types telomeres are shortened 24 h after exposure to high doses of gamma radiation (18–40 Gy) (Abdallah et al., 2009). These results suggest that the nature of length change induced, the underlying mechanisms and the time required to bring forth this change could be unique to each radiation quality and cell type. These data should be carefully integrated to interpret telomere related phenotypes from simulated mixed field GCR exposures to better project risk from actual missions.
Given the heterogeneity in telomere length within each cell, it is essential to also examine the proportional distribution of telomere lengths within a cell. As cells with short telomeres can die, it would be critical to avoid misinterpretation if the study population were skewed by assessment of just the surviving population. It is widely accepted that metabolic pathways and telomere signals interact reciprocally in inducing senescence. Mitochondrial dysfunction has been known to affect telomere dependent senescence and conversely telomere signals may induce mitochondrial failure (Passos et al., 2007; Sahin et al., 2011). Thus, input from mitochondrial dysfunction would be indispensable to extrapolating the consequences of GCR exposure. The large number factors that play into interpretation of the consequences of telomere length changes, makes it a challenging yet potentially useful biomarker for risk projection. Further studies using relevant chronic low doses and dose rates of exposure will be required to assess the impact of these parameters on telomere length and its biological consequences.
9. Stem cell changes
The well-established notion of the existence of tissue stem cells, capable of both self-renewal and of replenishing the cells of various tissue types, is exciting for its potential extrapolation to the cancer setting and the modulation of radiation-induced cancer risk. It has been proposed that a small population of normal stem derived cells may exist, that is capable of regenerating and perpetuating the heterogeneous tumor (Reya et al., 2001). This has been bolstered by findings in myeloma that there are intrinsic variations in proliferation among cancer cells (Park et al., 1971) and, in the case of solid tumors, to also generate tumors in animal models (Hamburger and Salmon, 1977). If such proves to be the case generally, tracking stem and cancer stem cell (CSC) kinetics might vastly improve our appreciation of cancer risk. This is because properties of the putative cancer stem cell, the entity that perpetuates the cancer, will likely be quite different from those of the ‘typical’ cancer cell from which we have, heretofore, inferred the properties of the cancer as a whole.
Accordingly, it has proven useful to look at normal stem cell biology for clues to tracking the origin and behavior of CSC cells, and for methods to assay this critical population and perhaps its immediate forerunner cells. Given one of the clearest displays of the stem cell hierarchy is in the hematopoietic system, it is not surprising that acute myeloid leukemia (AML) also displays a population hierarchy. The cell surface markers CD34+ /CD38− (also marking normal hematopoietic stem cells) identify a subpopulation of cancer cells in AML – the cancer stem cells – that can be grafted into SCID mice, where they home to the bone marrow and proliferate to produce cell morphologies and a dissemination pattern characteristic of those seen in the AML patients (Lapidot et al., 1994).
In solid tumors, evidence exists for the role of CSCs in tumors of the brain and breast. Oncogene expression, or loss of tumor suppressor genes in stem cells, has specifically been argued to be a necessary event for malignant transformation (Barker et al., 2009; Perez-Caro et al., 2009). In glioma, a CD133+ subpopulation demonstrated a capacity for self-renewal, proliferation and differentiation, and was shown to give rise to the phenotypic equivalents of tumor cells from the patient (Singh et al., 2003). In breast cancer, a minor population composed of CD44+/CD24 (−/low) lineage-cells could be passaged continuously, and each time produce tumors with a heterogeneous mix of these and cells with other markers, similar to actual breast cancers (Al-Hajj et al., 2003). Glimpsing what may be the ultimate in CSC selection – marking for migratory potential reminiscent of embryonic stem cells – cells with a CD44+/CD24− phenotype were found to exhibit enhanced invasive properties, indicative of an enrichment for a metastatic sub-fraction within this phenotype (Sheridan et al., 2006). Sheridan et al. speculate that the CD44+/CD24− phenotype may define the expression of the group of genes involved in invasion.
9.1. Defining changes cancer stem cell populations
While defining cancer stem cell fractions based on cell surface antigen markers has been productive, it has largely traded the more formidable task of tracking markers for the disease with the simpler one of tracking associated immunogenic (CD) biomarkers using antibodies. In AML, for instance, the actual genetic patterns one witnesses are mutations; common ones being in nucleophos-min 1 (NPM1) and Fms-like tyrosine kinase 3 (FLT3) (Foran, 2010). So, although immunologic markers have been called ‘prospective’ identifiers of stem cell status, it is important to consider that the cells are being identified based on the enrichment of these markers in various functional assays in vitro or in vivo (e.g. proliferation, migration, invasion) that are themselves surrogates for actual stem cell status. These markers, therefore offer limited promise of predictiveness beyond what the surrogate assays themselves provide. Another problem stemming from the correlation, rather than the strict identification, of the immunologic marker with stem cell status is that it may not be definitive for the stem cell state. CD133, for example, although it has been strongly linked to CSC status, its expression is not exclusively linked to the CSC phenotype (Miranda-Lorenzo et al., 2014). In such cases, alternative detections are sometimes possible, but the problem of specificity is compounded by the low frequency of CSCs in the cancer cell population. The task faced for improving marker detection at the level of CSC creation would thus appear to be to more directly connect intrinsic cancer stem cell properties to assayable endpoints that may still be efficiently screened for across populations in bulk.
Prospects for this goal may be limited, however, by CSC status being a rare stochastically-generated state upon a cell that may for the most part be limited to a rare candidate population. Not surprisingly, stem cell changes giving rise to cancer have yet to be defined in any generalizable way. On the other hand, it has been reported that the induction of a cancer-permissive stroma by the tumor is a prerequisite to cancer progression after the fact, without which a nascent cancer is maintained at a microscopic size (Mueller and Fusenig, 2002; Beacham and Cukierman, 2005). This being the case, detection of changes to stromal permissiveness in connection with cancer stem cell detection may constitute a leap in the field by providing a de facto means of cancer detection without having to detect the actual conversion event.
Understanding cancer stem cell growth, like its normal stem cell counterpart, in response to radiation must be undertaken in the context of the population hierarchy, since the dynamics of expansion of the component hierarchical subpopulations is highly interdependent. Further, given the evident time dependence of both the structure and recovery of this hierarchy after any alteration to its equilibrium, one would expect an exquisite dependence of expansion of CSCs on the nature, strength, and duration of the perturbing influence. This is indeed the case with radiation, where strong dependencies on dose, dose rate, radiation quality and the passage of time have been observed.
9.2. Sensitivity of stem cells to radiation
Again, normal stem cell biology furnishes some guidance, albeit imperfect. Addressing radiosensitivity, it is currently thought that adult stem cells tend to be differentially sensitive compared to their more committed progenitors. While hematopoietic stem cells have been found to be generally more resistant than their more committed progenitors and mammary stem cells are more resistant to X-ray-induced apoptosis than progenitor cells from mammospheres. By contrast, stem cells toward the base of the small intestinal crypt are more sensitive to irradiation than small intestine progenitors located higher in the crypt (Liu et al., 2014). Similarly, cancer stem cells differ in their sensitivity in comparison to the remaining tumor cell population, but in this case, with a uniform tendency toward greater radioresistance. This has been attributed to a combination of effects, including improved free radical scavenging, low proteasome activity, and activation of the DNA damage response (Pajonk et al., 2010). All these properties, highly specific to the stem cell compartment, may offer potentially more reliable functionally-based marker alternatives to immunologic biomarkers.
9.3. Relationship between stem cell changes and cancer risk
Surprisingly, since it is now known almost all adults possess latent cancers (Black and Welch, 1993), how the intricacies of stem cell dynamics play into cancer risk from radiation exposure may depend as much on the perturbation of the dynamics of preexisting CSCs and their descendants in latent cancers as it does any de novo creation of cancer cells from normal stem cells. Adding further complexity, the CSC hierarchy, like its normal counterpart, may indirectly influence the CSC compartment through feedback contact effects and spatial constraints, thereby significantly modifying cancer progression toward clinical realization (Enderling et al., 2009; Hillen et al., 2013). It is therefore not enough to know the direct effects on their numbers to assess either radiation response on the one hand, or progression to clinical cancer on the other. Indeed, we observed that radiation, through the “non-targeted” effect of perturbation of associated non-CSC subpopulation dynamics, can have a profound indirect effect on the overall CSC composition of a pre-clinical cancer in a manner that depends exquisitely on the size of a dose, how it is delivered, and the time of subsequent observation (Enderling et al., 2009; Hillen et al., 2013; Gao et al., 2014). Given the greater sensitivity of the non-CSC population, this indirect mechanism is made all the more important. Studying a putative CSC population of CD133+ glioma cells following three fractions of 2 Gy doses of photon radiation, we found the resultant CSC fraction was higher than could be explained based on simulations of CSC symmetric and asymmetric division kinetics, to the extent of favoring a strong reprogramming component, i.e., an actual conversion from a non-CSC to a CSC. The results find strong support in other studies where equilibrium between putative CSC and non-CSC states have been observed (Gupta et al., 2011; Roesch et al., 2010; Sehl et al., 2015). Gao et al. (2014) and Gupta et al. (2011) also employed a mathematical modeling framework to reach that conclusion, in that case a stochastic Markov chain approach, and argued that the results are consistent with a finding that an interconversion of state from non-CSC to CSC is taking place. Using data from cell line and mouse xenograft experiments, Wicha and colleagues (Sehl et al., 2015) examined tumor growth dynamics in response to alterations in the rate of symmetric self-renewal of breast CSCs, and confirmed that small changes in CSC behavior can give rise to the Gompertzian growth pattern observed in breast tumors. In summary, the effect of radiation on the CSC population must be understood in terms of its effect on the hierarchy as a whole, since the hierarchy acts to dynamically define the numbers of, and occasionally might even serve as a source of, CSCs.
9.4. Extrapolating experimental assessments of stem cell changes to GCR exposures in space flight
Extrapolating stem cell changes observed in experimental systems to GCR type exposures should be productive, insofar as perturbations to the hierarchy as a whole can impact stem cell cycling and the resulting likelihood of carcinogenic conversion (or expansion of an existing latent CSC population in the case of pre-existing disease). This non-targeted effect, beyond direct stem-cell ‘hits’, expands the effective target range, which may facilitate bridging interpretations at moderate doses down to the low-dose limit. In this connection, a controversial study by Tomasetti and Vogelstein (Tomasetti and Vogelstein, 2015) would suggest an increase in stem cell cycling, for whatever reason, would increase the rate of carcinogenic conversion of a stem cell. On the other hand, that study was a statistical one based on a comparison of frequencies of cancers arising de novo in tissues with differing stem cell cycling rates, and so would not take into account the effect of concurrent radiation-induced alterations to the stem cell hierarchy and more broadly, the stem cell niche (Li and Neaves, 2006), as would be expected in the space radiation setting. Thus, it may not be so much a question of the ability to extrapolate findings to the GCR setting, as it is we rely on the proper hierarchy- and niche-respecting experimental platforms to make such extrapolations.
10. mRNA expression
Proteins play an integral role in the majority of molecular functions in a living organism. Thus understanding level and activity of relevant proteins is key to dissecting the cellular response to ionizing radiation. The abundance of messenger RNA (mRNA) has been used as an approximate measure for the levels of proteins, as mRNA transcripts can be assayed and measured in a relatively inexpensive manner (Guo et al., 2008). This measurement may be impacted by a number of factors such as DNA sequence variation among individuals (Duan et al., 2013), intra/inter-population differences (Hasegawa et al., 2015; Storey et al., 2007), variability in baseline cell/tissue expression (Petryszak et al., 2014) and experimental artifacts (Cheng et al., 2012). Currently, a higher fold change for significance is recommended to account for these variations and make robust comparisons (Cheng et al., 2012).
10.1. Gene expression changes as a sensitive biomarker to differentiate dose, dose rate and temporal response
Gene expression profiling has been widely utilized to understand how the number of mRNA transcripts is impacted by radiation quality, dose, dose rate, and time. Studies have extensively investigated dose-responsiveness of specific human genes, especially those relevant to bio-dosimetry. Although a subset of genes showed a dose response for the first few hours post exposure, this pattern however disappeared with time. For instance, Amundson et al. have shown that when using human PBLs exposed to gamma irradiation, the expression of CDKN1A, DDB2 and XPC genes exhibited a linear dose-response relationship up to 48 h post irradiation, which declined thereafter (Amundson et al., 2000). A similar result was observed in another study involving human PBL exposed to gamma irradiation (Kang et al., 2003). Furthermore, it has been observed that the slope of the dose response curve decreases with an increase in dose (Paul and Amundson, 2008). Ionizing radiation also induces different numbers of mRNA transcripts depending on radiation attributes. For example, it has been shown that the number of radiation-induced genes increased with increasing dose and time after exposure in a study that assayed mRNA expression at various doses and times post γ-rays (Knops et al., 2012). In addition, the number of significantly up-regulated genes was greater than down-regulated genes at all doses and time points after exposure (Albrecht et al., 2012). A comprehensive review of the “time effect” of ionizing radiation on mRNA expression can be found in a review paper by Roy et al. (2009).
Using an ex-vivo blood irradiation model and doses of 0.56 Gy, 2.23 Gy and 4.45 Gy at two different dose rates (1.03 Gy/min and 3.1 mGy/min), it was reported that the number of genes differentially expressed at 1.03 Gy/min was higher than those expressed at 3.1 mGy/min for the same dose of X-rays (Ghandhi et al., 2015). These results were confirmed by a related study that showed that the magnitude of mRNA transcripts was lower for low-dose rate exposure (Paul et al., 2015). Notably, high-LET particles have been shown to perturb more genes in comparison to low-LET radiation (Chauhan et al., 2014; Turtoi et al., 2010). Furthermore, the number and magnitude of mRNA transcripts increased with an increase in the energy of HZE particles (Ding et al., 2005). A more comprehensive study involving a wide spectrum of qualities and gene sets is essential to validate these results. To accomplish this, our group has used literature mining to identify possible candidate genes for this proposed study (Table A, Appendix). We predict that this list of 150 genes that respond to IR exposure will provide a starting point to build a mRNA biomarker signature to assess radiation quality and dose rate effects for high-LET exposure.
10.2. mRNA expression at low and acute doses
Very few studies have detailed mRNA expression at low and acute doses of radiation. Some studies have shown that a transcriptional response (similar to what was observed in cells directly hit by alpha-particle radiation) was observed in non-targeted cells. However, cellular response to bystander irradiation was delayed by 30 min. compared to those that were directly hit by radiation (Ghandhiet al., 2010). Ermakov et al. (2009) identified the regulation of caspase-3, DNA-binding receptors and Toll-like receptor 9 (TLR9) in bystander cells following low-doses of X-rays. A systematic investigation of the gene expression patterns in a human mesenchymal stem cell line exposed to a range of doses (0.01, 0.05, 0.2 or 1 Gy) of Cs at various time points (1, 4, 12 or 48 h) suggests a highly non linear relationship between radiation dose and transcriptional response (Jin et al., 2008). Out of subset (16%) of genes that were altered in response to 0.1 Gy and 2 Gy of IR, a higher proportion was strongly influenced by exposure time (Jin et al., 2008). More detailed experiments are still required to understand bystander effects following high-LET exposure.
10.3. mRNA’s ability to define molecular pathways important in radiation-induced cancer
Molecular pathway activation was investigated for several IR experiments using breast, colorectal and lung cancer tissues. DNA microarray gene expression data for breast, colorectal and non-small cell lung cancer radiation experiments were obtained from the publicly available Gene Expression Omnibus (GEO). Table B (Appendix) lists the gene expression datasets used. Quality control checks involved generation of residual plots, and normalization via the AffyPLM package from Bioconductor. As an additional quality control step, Normalized Unscaled Standard Errors (NUSE) plots were used (Bolstad, 2005). Upon completion of the quality control measures, single-channel array normalization (SCAN) was used, which normalized each sample individually by modeling and removing probe and array-specific background noise using only data from within each array (Piccolo et al., 2012). Batch effects were minimized using ComBat (Johnson et al., 2006). Pathway information comprised of common gene symbols was obtained from Wiki Pathways (Pico et al., 2008). Table C (Appendix) lists the top 20 molecular pathways activated by IR exposure in different tissue types. Activated pathways include p53, cell cycle, DNA excision repair, DNA replication (purine metabolism, pyrimidine metabolism), all of which are hallmarks of radiation exposure and cancer. For IR, many of the remaining pathways reflect MAPK, ERK, p39, WNT signaling (not shown), along with biochemical processes related to molecular degradation for IR and/or metabolism and cellular processes during cellular proliferation in cancer. In our analysis of gene expression datasets, not surprisingly, the epidermal growth factor receptor (EGFR) pathway was highlighted as one of the significant pathways activated by radiation exposure. IR-induced EGFR activation has been shown to promote cell proliferation after single and repeated exposures, a response that is blocked by selective inhibition of EGFR tyrosine phosphorylation. We observed that EGFR was up-regulated in cancer as well. Activation of the EGFR signal transduction pathway has been shown to enhance cellular processes involved in tumor growth and progression, angiogenesis, invasion, and metastasis. Moreover, increased expression of EGF has been observed in a wide variety or tumors, including non-small cell lung cancer and squamous cell carcinoma of the head and neck (Lee et al., 2008). Similarly, detailing which pathways are activated by radiation in different tissue types will be useful in creating customized mRNA biomarker expression panels specific to each tissue type.
10.4. Extrapolation of experimental transcription results to GCR exposures in space
Transcriptional studies using heavy ions have revealed increased pathway activity for cellular growth and proliferation, cell cycle changes, cell death, DNA replication, repair, and recombination (Ding et al., 2013, 2015; Roy et al., 2008). However, these have largely been phenomenological observations related to the hallmarks of cancer and the cell lines used, rather than clear patterns elucidating sequential and temporal changes leading to tumor development. The question of whether or not current or future data from experimental studies of high-LET exposures can support reliable extrapolation to astronaut risk from exposure to chronic, low dose mixtures of ions and spallation fragments remains to be addressed. Current studies on transcription typically employ RNASeq analysis, which provides intronic and exonic information, as well as results for variants (SNPs) and isoforms from alternative splicing. This level of granularity is considerably more informative than the existing historical results spawned from the use of 3′ biased probe-based bead and oligonucleotide microarray data. Going forward, as stability of the replication fork is a major threat posed by high-LET-induced clustered lesions, there would be significant merit in transcriptional studies targeting prevention of fork collapse, including SMARCAL1 and AH2, which stabilize stalled forks during replication stress (Bansbach et al., 2009). Areas that are in need of further investigation include the effect of high-LET ion exposure on lesion bypass, sister chromatid template switching and HR for repair at late time points (Chang and Cimprich, 2009). In addition, little is known surrounding high-LET effects on mono-to-poly PCNA ubiquitination, dual ATR-checkpoint and PCNA-DDR activation and 3′–5′ clamp specificity for single stalled forks, role of translesion synthesis (TLS) involving SPARTAN and PAF15, the role of Y family polymerases, and their recruitment and signaling in TLS. Thus, overall further transcriptional studies of high-LET exposures are necessary to identify how different DNA repair pathways are regulated and coordinated as a whole, how cell cycle phase and chromatin context influence these pathways, and how DNA is repaired during active DNA replication. The current gaps in knowledge will require considerable resources for identifying molecular targets for therapy and radioprotection in the context of DDR.
10.5. Relationship between cell fate decisions and mRNA expression patterns
In reviewing the public gene expression datasets that are currently available, there are very few studies involving low doses of high-LET ions or mixtures of ions whose data would be amenable for addressing the questions raised in previous sections. This is mostly due to the lack of large-scale, multi-organ, multi-charge/energy/dose transcriptional in vivo animal studies, which would be needed for assessing risks. Studies have shown that each cell type has its own unique characteristic spectrum of gene expression alterations elicited by low doses that is distinct from high doses, and other unirradiated and radiated human tissues. Comparing expression values of mRNA extracted from skin biopsies following in vivo radiation exposure, suggests that exposure to low doses of radiation cause a significant, transient up-regulation of zinc finger proteins, keratins, BMP receptor, BAG and cyclins while other proteins such as TNF, interleukins, heat shocks proteins and S100 are down regulated (Davalos et al., 2010). However, the actual impact of these alterations on cell fate is unclear. A DNA microarray study used to examine the gene expression profiles in human lymphocytes from four healthy volunteers after 0.1 Gy low dose radiation or 0.25 or 0.5 Gy high dose radiation generated a 30 gene consensus signature of IR response genes (Fachin et al., 2007). Of these DNA repair and stress response, cell growth and cell differentiation, general metabolism and transcription regulation were among the main biological pathways that were primarily affected by low dose and only modestly by high dose (Fachin et al., 2007). Interestingly, subset (CD4+)-specific repression of certain genes and associated down regulation of pathways involved in oxidative phosphorylation and protein biosynthesis, highlights the fact that pathway choice varies even amongst subsets of the same cell type (Gruel et al., 2008). Another study showed that a low dose (0.05 Gy) triggers the activation of the immune response (cytokine and chemokine signaling pathways, immune-related and inflammation processes), high doses preferentially activate genes in the p53 signaling pathway, DDR and apoptosis (El-Saghire et al., 2013), all part of the characteristic radiation stress response. These data suggest that low dose and high dose exposures trigger distinct molecular pathways and biological processes in human blood cells. It has also been speculated that different pathways/processes and resultant cell fate choices differ in their sensitivity to dose rate of radiation exposure (Amundson et al., 2003). It is thought that the lack of significant gene expression changes below 5 cGy is indicative of the existence of a threshold for induction of low dose-responsive genes (Riecke et al., 2012). Studies have also suggested that alterations in transcription with low doses including bystander and adaptive responses are often related to protective responses that are sometimes referred to as the hormeisis effect. Although this pro-survival response appears better than the cell kill and apoptosis noted with high doses, it potentially increases the mutational load in surviving cells and could contribute more to carcinogenesis risk. Further studies using large-scale animal models are needed to test whether mRNA expression patterns at low doses may relate to various cell fate decisions and have a significant impact on cellular transformation. The well defined and reproducible transcriptional alterations in human lymphocytes triggered by doses as low as 2 cGy (Knops et al., 2012) make this a sensitive experimental system to comprehensively study radiation quality and dose rate effects on cell fate.
As discussed earlier, mutation is one of the molecular characteristics that can be used as a bioindicator in the study of radiation effects. The impact of mutation on mRNA expression, protein activity, cellular proliferation and survival has long been noted (Hanahan and Weinberg, 2011). Various groups have studied the association of somatic mutations and mRNA expression using the multidimensional genomic and transcriptomic data available from The Cancer Genome Atlas (TCGA) project (Ding et al., 2015; Bashashati et al., 2012). For instance, a recent study by Jia and Zhao (2014) using the TCGA data from multiple cancer types has shown that, among the various features of mutation, mutation type (missense and nonsense mutations) have greater impact on mRNA expression. A study involving the Japanese atomic bomb survivors diagnosed with a myelodysplastic syndrome (MDS), a disease known to be associated with mutation of AML1/RUNX1 gene, has indicated that missense mutations have a greater negative effect on the trans-activation of AML1 gene compared to truncated-type mutation (Harada et al., 2003). Mutation can also have a ripple effect on the expression of proteins that are physically or functionally interacting. In this regard, it was noted that reduced levels of NBN and RAD50 proteins were observed in patients with known mutations in the MRE11 gene (Uziel et al., 2003). This relationship between mutation and mRNA expression confirms the intertwined nature of various biomarkers with one another and suggests multiple biomarkers may provide a more ideal readout for modeling radiation-induced cancer.
11. Current efforts in biomarker modeling
A number of these biomarkers detailed in this review have been employed in modeling efforts. DNA damage has been modeled using Monte Carlo track structure codes that can accurately predict the passage of a charged particle in water as a surrogate of biological tissue. DNA damage modeling indicates that decreasing the energy of ions causes a shift toward a greater complexity of clustered DNA damage. This is validated by a marked reduction in the proportion of simple SSBs and DSBs accompanied by a substantial increase in complex SSBs and DSBs (Nikjoo et al., 2001). DNA repair foci modeling using codes such as RITRACKS to calculate the geometric pattern of repair foci in nuclei has shown that foci number and position are strongly dependent on the radiation type (Plante et al., 2013). Modifications of this code incorporating Green’s function of the diffusion equation and simulating total DNA charge enabled the effects of low energy (below the ionization threshold) electrons ability to induce SSBs and DSBs (Boudaiffa et al., 2000, Panajotovic et al., 2006; Plante and Cucinotta, 2015) and to analyze the significance of charge migration in determining lesion type and complexity respectively (Belov et al., 2015). However, how early effects such as DNA damage, repair foci and ROS relate to cellular and tissue effects at longer times post exposure are currently unclear.
The induction of ROS by mitochondria and its cellular effects have been included in several models thus far. A kinetic model was developed to predict superoxide production rates by complexes in the electron transport chain and accounts for the effects of substrates, inhibitors, changes in pH and membrane potential gradients (Markevich and Hoek, 2015). Another model incorporated antioxidant systems to account for the fact that most ROS experimental data used to inform models is collected employing techniques that detect H2O2 diffusing from mitochondria as a surrogate measurement for superoxide production (Gauthier et al., 2013). Fewer models have linked ROS production in mitochondria to other cellular processes (Jafri and Kumar, 2014). The role of ROS in apoptosis has been included in models regulating mitochondrial membrane permeabilization (Jacob et al., 2016) and in transition pore opening (Bagci et al., 2006). ROS functioning as a determinant of cell fate has also been incorporated in a multistage model of tumorigenesis, however, the robustness of this model has not been validated with experimental evidence based on predictions (Chaudhary et al., 2011). Similarly, a number of radiation biology groups (Barcellos-Hoff et al., 2014; Mantovani et al., 2008) have shown that provoking innate and acquired immune response leads to an acute and chronic inflammatory cascade, the latter being a hallmark of cancer. A good example of such integration can be found in a two pathway model proposed by Mantovani et al. (2008). Behesti et al. recently showed that inflammatory response is also a function of age (Beheshti et al., 2015). It has been difficult to extrapolate uncertainties in immune risks due to the fact that only a small number of tumor cells usually escape immune surveillance mechanisms, whereas the majority of the cells are successfully eliminated. Hence, Shah et al. (2012) predicted that a practical threshold below which the immune system eliminates cancer cells entirely is unlikely. More recently, in a review analyzing the effect of sex and gender on adaptation by the immune system in space, Kennedy et al. (2014) concluded that it has been very difficult to obtain immunologic and infectious data using a small number of animals and extrapolate results to humans undergoing spaceflight. Thus a focus of these studies should be on the astronaut population with emphasis on long term, well-studied infectious risks. Therefore, a clear non-variable model system(s) is first needed to then use to extrapolate data into risk projection models for the role of inflammation/ immune systems in cancer progression. As mechanisms associated with deregulation of immune response to IR are still not clearly understood, better animal/ cell culture models that delineate mechanisms of deregulation are needed (Sridharan et al., 2015).
A number of different mathematical modeling approaches have been devised to understand telomere length changes. Initially probabilistic (Arino et al., 1995; Olofsson and Kimmel, 1999) and simple deterministic models were developed that only took into account the gradual telomeric loss occurring during each cell division. These models were improved upon by introducing the concept of “abrupt telomere shortening” or a sudden change in length that could be caused by DNA damage, and more common when telomere length was shorter, as with increasing age (Rubelj and Vondracek, 1999). Subsequent models have added environment related components that influence telomere length, such as ROS produced by mutant mitochondria. Another model developed by Blagoev et al considered the possibility that telomerase may have a more prominent role at short telomeres (Blagoev, 2009). More recently a model describing telomere dynamics following treatment with a G4-stabilizing drug, RHPS4 was developed. In this work they first defined how telomere length would change over a few generations and then incorporated changes that would occur over a longer period of time (Hirt et al., 2014). Using these modeling approaches the authors were able to determine that higher concentrations of telomerase can cause continued telomere lengthening, which is consistent with findings in the literature.
A major limitation in the use of all of these biomarkers in detecting cancer initiation appear to be the inability to identify initiated cells or events closer to the hypothetical transition to the putative forerunner transformed cell or the cancer stem cell. Paucity of knowledge of what the critical biomarker cellular changes are is obviously a major limitation. However, one prospect for identification in solid tumors comes from a study of the cells of the small intestine, where the cancer’s origin in stem cells at the base of the crypt is thought to be resolved due to the fact the descendant cells migrate out of the crypt and slough off into the lumen. To this end, it was found Lgr5 (the product of a gene encoding for a component of the Wnt receptor complex) identifies putative stem cells at the crypt base (Barker et al., 2007), so that any ectopic occurrence of Lgr5+ cells might therefore be a potential marker of a forerunner of the CSC population that manifests as cancer. A potential roadblock to identifying prospective stem cells using the Lgr5+ marker is the plasticity and variability in expression of this marker by CSCs, a phenomenon also noted with conversions of CSCs to non-CSCs in breast cancer (Kobayashi et al., 2012). Surprisingly, the aberrant expression of a gene encoding mesenchymal bone morphogenetic protein antagonist, GREM1, has been found to initiate tumorigenesis in the colon from cells outside the stem cell niche. It operates by causing continued expression of stem cell properties in Lgr5- progenitor cells that have left the niche, resisting the normally differentiating influence of the morphogen gradient as the cells move higher along the vertical epithelial axis (Davis et al., 2015). This underscores the importance of the tissue microenvironment in determining cell fate; a parameter that also needs to factor into studies assessing stem cells.
An intermediate approach to dealing with this expanded biomarker problem, that may add less difficulty in implementation as it strives to include increasingly more dynamical considerations, is to use simpler biological assays in conjunction with more sophisticated dynamical mathematical modeling; in particular, mathematical modeling that focuses on stem cell subpopulations and generation and expansion of CSCs as limited by non-CSCs and the tissue environment. Motivating the endeavor, in what may be the most provocative study of cancer risk to date, a 2015 study Tomasetti and Vogelstein, using statistical methods, found an 80% correlation of cancer risk with the number of stem cell divisions for two-thirds of the cancers in relevant tissues. If the association between DNA alterations in proliferating stem cells and cancer risk is further validated, we envision three areas that require further investigation. Targeted studies on (1) the radiation induced stem cell proliferation (assuming de novo carcinogenesis), (2) the resulting CSC population (assuming an alternative latency-shortening paradigm), and (3) mathematical modeling of CSC interaction with its normal microenvironment (Pajonk et al., 2010; Black and Welch, 1993; Enderling et al., 2009; Gao et al., 2014) may offer a high-value correlate to cancer incidence risk, not with-standing problems pinpointing the ‘causal’ population.
Altogether, a wide array of technologies involving high-throughput assays for transcription, translation, mutation, DNA repair, and biological sequence analysis for mutation profiling will need to be leveraged before realistic projections can be made for cancer risk from GCR exposures in space. The histology and morphology of the most radiogenic and radioresistant cancers and the tissue types should also be taken into consideration in the design of transcriptional regulatory-based experiments. Advancements in the area of individual genetic susceptibility to radiation-induced cancer can also rapidly accelerate our understanding of the role of novel and known polymorphisms in cancer and radiosensitivity. Investigations are also warranted for identification of time-dependent appearance of pathogenic variants in vivo after irradiation and the role of these variants in cancer. Advances in nextgen sequencing technology will also aid in accelerating these studies.
12. Conclusions
Currently, the dominant uncertainty in space radiation risk assessment is associated with radiation quality followed by dose rate effects (Cucinotta et al., 2012). While studies like the ongoing million worker (Bouville et al., 2015) will shrink uncertainties for low-LET radiation including those from dose rate effects, the overall achievable limit on uncertainty reduction for high-LET radiation based on scaling from low-LET radiation studies and adding in empirical radiation quality and dose rate modifiers is below NASA’s current target. To overcome this barrier, NASA has developed an aggressive plan (Roadmap NHRP) to leverage a mechanistic understanding of radiation effects to develop models with better predictive capability. Validating and modeling early surrogate biomarkers of cancer risk that are temporally separated would be an integral component of this plan and will shed light on the fundamental biological processes that can trigger cancer initiation. Each of the end points reviewed here has been independently modeled for exposure effects to various environmental carcinogens. In the same way, we predict that models using these biomarkers can be used to predict cancer risk from radiation exposure based on mechanistic data (Fig. 1). Although independent models are available for each end point, the main limitations in modeling radiobiological effects are the multi-scale dimension that spans several orders of magnitude in time and space, and our ability to temporally integrate multiple endpoints to assess long-term effects. In addition, expanding the scale of time and structural complexity of simulation is exorbitantly expensive in terms of computer simulation time. The time scale, depth of details required and availability of computational resources usually guide the choice of one model over another. Most models are also based to some extent on experimental data, which might be scarce, conflicting or incorrect in some cases. When data is unavailable, empirically assigning values to parameters used in models greatly influence the accuracy of the results. We are proposing that iteratively developed models, incorporating the impact of each biomarker response (late to early) one by one will reveal the significance of the complex interdependence between different pathways in cancer development. However, relating mechanistic simulation models to i n vivo data is a huge challenge. An upcoming goal is to use knowledge based modeling of various mechanistic biomarkers of radiation effects to determine and improve quality factors and better predict cancer risk.
Fig. 1. Schematic representation of known mechanistic links between biomarkers that define cell fates, which promote or protect from cancer risk.
We have detailed the feedback network between various biomarkers of cellular stress response at early and late times post radiation exposure. For simplicity biomarkers have been defined as “early” and “late”, although some early biomarkers are also observed at later times. Early biomarkers evaluated include persistent damage, DNA repair foci, alterations in ROS and inflammation. Late biomarkers of genomic instability include mutations, chromosome aberrations and telomere length changes. The phenotype of each of these biomarkers differ post exposure to HD/HDR and LD/LDR radiation. As majority of the current data is based on HD/HDR exposures, biological effects of LD/LDR exposures that mimic GCR are speculative, need to be further validated using GCR simulator at NSRL, and hence are highlighted in blue. Summary of current literature suggests that majority of acute high-dose low-LET exposures cause complex clustered lesions that are irreparable. In contrast, although cells hit with low doses of high-LET radiation also get complex clustered damages due to intense energy deposition along the track, bystander effects appear to play a prominent role in the ensuing biological response, pathways activated and the eventual fate of the cell. Most of the irreparable clustered lesions initiate a cascade of biological processes that are so damaging that the inherent protective mechanisms prevent carcinogenesis by targeting these cells for growth arrest, apoptosis, autophagy or senescence. Cells with fewer numbers of complex clustered lesions and those with non-lethal lesions caused by high-LET radiation may, however, continue to proliferate following misrepair of the lesions. Chromosome aberrations or mutations caused by these lesions perpetuate further DNA damage, chronic ROS, chronic inflammation and telomere dysfunction, resulting in more genomic instability and in some cases cellular transformation. Cell fate choice and distribution may differ between HD/HDR and LD/LDR exposures, and is an essential area that needs further investigation. The nodes of intersection between endpoints that govern the choice between pro- and anti- survival cell fates must be factored in along with temporal mRNA/protein expression profiles, in order to obtain a clear biomarker signature for modeling and early prediction of cancer risk. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
Acknowledgments
We acknowledge that there is a large body of work that has been completed in this area and due to space limitations we could not discuss it in its entirety. Thus, we apologize in advance for important works that we were unable to cite in the current review. The authors wish to thank laboratory members and staff who have assisted in the cited work, including Tatiana Kovyrshina for QC and normalization of the array data. Special thanks to the scientists and staff at NASA Space Radiation Laboratory and Medical at Brookhaven National Laboratory, including Drs. Adam Rusek, Chiara LaTessa, and Peter Guida. We thank Dr. Stan Curtis for helpful discussions. Lastly, we would like to acknowledge the visionary efforts of Dr. Francis Cucinotta who initiated the VSBM groups for risk modeling, and has widely contributed and supported numerous research efforts in space radiation biology.
Funding
This work was supported by National Aeronautics and Space Administration NNX13AD57G, and NNX15AE06G (to A. Asaithamby); National Aeronautics and Space Administration NNJ09HC64I and the Low Dose Scientific Focus Area, United States Department of Energy DE-AC02-05CH11231 (to S.V. Costes); National Aeronautics and Space Administration NNX15AD63G (to W. Dynan), NASA Specialized Center of Research (NSCOR) NNX11AC30G (to Emory University – W. Dynan, P. Doetsch, and E. Werner); National Aeronautics and Space Administration Grants NNX13AJ01G, NNX11AK26G, and NSCOR NNJ06HA28G (to L. Hlatky); National Aeronautics and Space Administration NAS9-0.2078 (to Y. Kidane, I. Plante, and A. Ponomarev); National Aeronautics and Space Administration NNJ12HB88I (to A. Kronenberg); National Aeronautics and Space Administration NNX11AO89G and NNX13AD74G (to M. Naidu); National Aeronautics and Space Administration NNX12AO52A (to Tatiana Kovyrshina, M.S. who performed QC, normalization, and batch-effect removal of expression arrays used for pathway activation for analysis by L. Peterson). National Aeronautics and Space Administration NNJ12HD071 and NNJ13HA96I (to J. Pluth); Dose Scientific Focus Area, US Department of Energy DE-AI02-10ER64969 – J. Saha (to F. Cucinotta); Low Dose Scientific Focus Area, US Department of Energy DE-AC0205CH11231 (to A.Snijders).
Appendix
Table A.
Potential candidate genes for comprehensive study of radiation quality and dose-rate effects.
| Rank | Gene symbol | Rank | Gene symbol | Rank | Gene symbol | Rank | Gene symbol | Rank | Gene symbol |
|---|---|---|---|---|---|---|---|---|---|
| 1 | ATM | 31 | FANCD2 | 61 | ABL1 | 91 | CDT1 | 121 | ING1 |
| 2 | TP53 | 32 | XRCC4 | 62 | POLD1 | 92 | SETMAR | 122 | NFKB1 |
| 3 | ATR | 33 | RPA1 | 63 | BAX | 93 | CDK1 | 123 | IER3 |
| 4 | H2AFX | 34 | DCLRE1C | 64 | RAD17 | 94 | MCL1 | 124 | UVRAG |
| 5 | NBN | 35 | UIMC1 | 65 | JUN | 95 | ERCC1 | 125 | CUL4B |
| 6 | PRKDC | 36 | MDM2 | 66 | UBC | 96 | PLK3 | 126 | RAD1 |
| 7 | DDB2 | 37 | XRCC3 | 67 | MAPK9 | 97 | RECQL4 | 127 | PAXIP1 |
| 8 | CHEK1 | 38 | RPA2 | 68 | LIG4 | 98 | CASP3 | 128 | LIG3 |
| 9 | CHEK2 | 39 | ERCC2 | 69 | MMP1 | 99 | RNF168 | 129 | RAD23B |
| 10 | BRCA1 | 40 | RAD9A | 70 | XRCC2 | 100 | SMUG1 | 130 | RPA3 |
| 11 | PCNA | 41 | GADD45A | 71 | BLM | 101 | TLK1 | 131 | TERT |
| 12 | DDB1 | 42 | MAPK14 | 72 | NTHL1 | 102 | CLSPN | 132 | MAPK1 |
| 13 | MRE11A | 43 | PPM1D | 73 | KAT5 | 103 | HIPK2 | 133 | DCLRE1A |
| 14 | POLH | 44 | CDC25A | 74 | BRCA2 | 104 | CELF2 | 134 | GTF2H4 |
| 15 | RAD51 | 45 | APEX1 | 75 | POLK | 105 | POLE | 135 | CCNB1 |
| 16 | TP53BP1 | 46 | ERCC8 | 76 | ATRIP | 106 | RAD54L | 136 | DCLRE1B |
| 17 | XPA | 47 | PARP1 | 77 | NHEJ1 | 107 | UVSSA | 137 | VRK1 |
| 18 | MDC1 | 48 | RAD52 | 78 | BRE | 108 | WRN | 138 | REV3L |
| 19 | CDKN1A | 49 | RNF8 | 79 | TRIM29 | 109 | MSH4 | 139 | C11ORF31 |
| 20 | XRCC1 | 50 | DTL | 80 | FAM175A | 110 | RFWD2 | 140 | BRIP1 |
| 21 | XPC | 51 | RAD51B | 81 | KIN | 111 | ATMIN | 141 | RPS6KA5 |
| 22 | MAPK8 | 52 | RBX1 | 82 | UBE2B | 112 | WEE1 | 142 | GADD45B |
| 23 | RAD50 | 53 | CDC25C | 83 | PNKP | 113 | UBB | 143 | HIST1H1A |
| 24 | ERCC6 | 54 | UBE2D3 | 84 | ELAVL1 | 114 | BCL2L1 | 144 | UBE2V2 |
| 25 | MC1R | 55 | OGG1 | 85 | POLI | 115 | ATF2 | 145 | CUL1 |
| 26 | RAD18 | 56 | BARD1 | 86 | UBE2A | 116 | CDC25B | 146 | SMC1A |
| 27 | XRCC6 | 57 | REV1 | 87 | APLF | 117 | MAPKAPK2 | 147 | EXO1 |
| 28 | XRCC5 | 58 | TOPBP1 | 88 | RBBP8 | 118 | PLK1 | 148 | SPRR2B |
| 29 | CUL4A | 59 | ERCC4 | 89 | AKT1 | 119 | TP53I3 | 149 | SPRR2E |
| 30 | ERCC5 | 60 | ERCC3 | 90 | BRCC3 | 120 | RRM2B | 150 | SFR1 |
Table B.
Gene expression datasets used for molecular pathway activation studies.
| Cancer/radiation | GEO dataset | Subtype (# arrays) | Ref. |
|---|---|---|---|
| Breast cancer | GSE1456 (120 arrays) | Basal (25), ERBB2 (15), | Pawitan et al. (2005) |
| Luminal A (39), | |||
| Luminal B (23), | |||
| Normal (18) | |||
| Colorectal cancer | GSE21510, | Metastatic recurrence Stage: 2b (8), | GSE21510: Tsukamoto et al. (2011) |
| GSE27854, | Stage: 3b (32), | GSE27854: Kikuchi et al. (2013) | |
| GSE18105 | Stage: 2b (25), | ||
| (253 arrays) | Metastasis Stage: 4 (42) | GSE18105: Matsuyama et al. (2010) | |
| Stage: 2a (37), | |||
| Stage: 1 (27), | |||
| Metastatic recurrence Stage: 3b (17), | |||
| Stage: 3c (7), | |||
| Stage: 3a (7), | |||
| Metastatic recurrence Stage: 3c (5), | |||
| Metastatic recurrence Stage: 2a 5), normal (41) | |||
| Lung cancer | GSE37745, | Squamous cell (92), | GSE37745: Botling, et al. (2013) |
| GSE28582, | Adenocarcinoma (151), | GSE28582: Micke et al. (2011) | |
| GSE19804 | Large cell (46) | ||
| (349 arrays) | Normal (60) | GSE19804: Lu et al. (2010) | |
| Protons (4.5 MeV) | GSE16935 (6 arrays) | 4 h 2.5 Gy (3) vs. 4 h 0 Gy (3) | Mezentsev et al. (2011) |
| Gamma (Cs-137) | GSE1977 (30 arrays) | IR-5 Gy (15) vs. mock 0 Gy (15) | Rieger et al. (2004) |
| Iron, Gamma | GSE16518 (12 arrays) | Iron 1.67 Gy 1 GeV (3) vs. iron control (3) Gamma 5 Gy-137Cs (3) vs. gamma control (3) | Meador et al. (2011) |
Table C.
Top 20 pathways activated by IR exposure, in breast, colorectal, and non-small cell lung cancer. Activation results based on 358 pathways, with top 50 pathways having over-represented genes (red, positive rank 1 to 50) and bottom 50 pathways having under-represented genes (green, negative rank −50 to −1), when compared with expected genes based on total background gene list. Rank order determined from Z-score test based on hypergeometric distribution. Pathways listed have greatest mean rank.
References
- Abdallah P, Luciano P, Runge KW, Lisby M, Geli V, Gilson E, et al. A two-step model for senescence triggered by a single critically short telomere. Nat. Cell. Biol. 2009;11(8):988–993. doi: 10.1038/ncb1911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Hajj M, Wicha MS, Benito-Hernandez A, Morrison SJ, Clarke MF. Prospective identification of tumorigenic breast cancer cells. Proc. Natl. Acad. Sci. USA. 2003;100(7):3983–3988. doi: 10.1073/pnas.0530291100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albertson DG, Collins C, McCormick F, Gray JW. Chromosome aberrations in solid tumors. Nat. Genet. 2003;34(4):369–376. doi: 10.1038/ng1215. [DOI] [PubMed] [Google Scholar]
- Albrecht H, Durbin-Johnson B, Yunis R, Kalanetra Karen M, Wu S, Chen R, et al. Transcriptional response of ex vivo human skin to ionizing radiation: comparison between low- and high-dose effects. Radiat. Res. 2012;177(1):69–83. doi: 10.1667/rr2524.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amundson SA, Chen DJ. Inverse dose-rate effect for mutation induction by gamma-rays in human lymphoblasts. Int. J. Radiat. Biol. 1996;69(5):555–563. doi: 10.1080/095530096145562. [DOI] [PubMed] [Google Scholar]
- Amundson SA, Do KT, Shahab S, Bittner M, Meltzer P, Trent J, et al. Identification of potential mRNA biomarkers in peripheral blood lymphocytes for human exposure to ionizing radiation. Radiat Res. 2000;154(3):342–346. doi: 10.1667/0033-7587(2000)154[0342:iopmbi]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- Amundson SA, Lee RA, Koch-Paiz CA, Bittner ML, Meltzer P, Trent JM, et al. Differential responses of stress genes to low dose-rate gamma irradiation. Mol. Cancer Res. 2003;1(6):445–452. [PubMed] [Google Scholar]
- Anderson L, Henderson C, Adachi Y. Phosphorylation and rapid relocalization of 53BP1 to nuclear foci upon DNA damage. Mol. Cell. Biol. 2001;21(5):1719–1729. doi: 10.1128/MCB.21.5.1719-1729.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arino O, Kimmel M, Webb GF. Mathematical modeling of the loss of telomere sequences. J. Theor. Biol. 1995;177(1):45–57. doi: 10.1006/jtbi.1995.0223. [DOI] [PubMed] [Google Scholar]
- Artandi SE, Chang S, Lee SL, Alson S, Gottlieb GJ, Chin L, et al. Telomere dysfunction promotes non-reciprocal translocations and epithelial cancers in mice. Nature. 2000;406(6796):641–645. doi: 10.1038/35020592. [DOI] [PubMed] [Google Scholar]
- Asaithamby A, Chen DJ. Mechanism of cluster DNA damage repair in response to high-atomic number and energy particles radiation. Mutat. Res. 2011;711(1–2):87–99. doi: 10.1016/j.mrfmmm.2010.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asaithamby A, Uematsu N, Chatterjee A, Story MD, Burma S, Chen DJ. Repair of HZE-particle-induced DNA double-strand breaks in normal human fibroblasts. Radiat. Res. 2008;169(4):437–446. doi: 10.1667/RR1165.1. [DOI] [PubMed] [Google Scholar]
- Asaithamby A, Hu B, Chen DJ. Unrepaired clustered DNA lesions induce chromosome breakage in human cells. Proc. Natl. Acad. Sci. USA. 2011;108(20):8293–8298. doi: 10.1073/pnas.1016045108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aten JA, Stap J, Krawczyk PM, van Oven CH, Hoebe RA, Essers J, et al. Dynamics of DNA double-strand breaks revealed by clustering of damaged chromosome domains. Science. 2004;303(5654):92–95. doi: 10.1126/science.1088845. [DOI] [PubMed] [Google Scholar]
- Badhwar GD. The radiation environment in low-Earth orbit. Radiat. Res. 1997;148(5 Suppl):S3–10. [PubMed] [Google Scholar]
- Bagci EZ, Vodovotz Y, Billiar TR, Ermentrout GB, Bahar I. Bistability in apoptosis: roles of bax, bcl-2, and mitochondrial permeability transition pores. Biophys. J. 2006;90(5):1546–1559. doi: 10.1529/biophysj.105.068122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey S, Cornforth M. Telomeres and DNA double-strand breaks: ever the twain shall meet? Cell. Mol. Life Sci. 2007;64:2956–2964. doi: 10.1007/s00018-007-7242-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bansbach CE, Betous R, Lovejoy CA, Glick GG, Cortez D. The annealing helicase SMARCAL1 maintains genome integrity at stalled replication forks. Genes Dev. 2009;23(20):2405–2414. doi: 10.1101/gad.1839909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barcellos-Hoff MH, Adams C, Balmain A, Costes S, Demaria S, Illa-Bochaca I, et al. Systems biology perspectives on the carcinogenic potential of radiation. J. Radiat. Res. 2014;55:1145–1154. [Google Scholar]
- Barcellos-Hoff MH, Blakely EA, Burma S, Fornace AJ, Gerson S, Hlatky LH, et al. Concepts and challenges in cancer risk prediction for the space radiation environment. Life Sci. Space Res. 2015;6:92–103. doi: 10.1016/j.lssr.2015.07.006. [DOI] [PubMed] [Google Scholar]
- Barker N, van Es JH, Kuipers J, Kujala P, van den Born M, Cozijnsen M, et al. Identification of stem cells in small intestine and colon by marker gene Lgr5. Nature. 2007;449(7165):1003–1007. doi: 10.1038/nature06196. [DOI] [PubMed] [Google Scholar]
- Barker N, Ridgway RA, van Es JH, van de Wetering M, Begthel H, van den Born M, et al. Crypt stem cells as the cells-of-origin of intestinal cancer. Nature. 2009;457(7229):608–611. doi: 10.1038/nature07602. [DOI] [PubMed] [Google Scholar]
- Bashashati A, Haffari G, Ding J, Ha G, Lui K, Rosner J, et al. DriverNet: uncovering the impact of somatic driver mutations on transcriptional networks in cancer. Genome Biol. 2012;13(12) doi: 10.1186/gb-2012-13-12-r124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baulch JE, Craver BM, Tran KK, Yu L, Chmielewski N, Allen BD, et al. Persistent oxidative stress in human neural stem cells exposed to low fluences of charged particles. Redox. Biol. 2015;5:24–32. doi: 10.1016/j.redox.2015.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beacham DA, Cukierman E. Stromagenesis: the changing face of fibroblastic microenvironments during tumor progression. Semin. Cancer Biol. 2005;15(5):329–341. doi: 10.1016/j.semcancer.2005.05.003. [DOI] [PubMed] [Google Scholar]
- Beetz A, Messer G, Oppel T, Van Beuningen D, Peter RU, Kind P. Induction of Interleukin 6 by ionizing radiation in a human epithelial cell line: control by corticosteroids. Int. J. Radiat. Biol. 1997;72(1):33–43. doi: 10.1080/095530097143518. [DOI] [PubMed] [Google Scholar]
- Beheshti A, Wage J, McDonald JT, Lamont C, Peluso M, Hahnfeldt P, et al. Tumor-host signaling interaction reveals a systemic, age-dependent splenic immune influence on tumor development. Oncotarget. 2015;6(34):35419–35432. doi: 10.18632/oncotarget.6214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belov OV, Boyda DL, Plante I, Shirmovsky SE. Simulation of the charge migration in DNA under irradiation with heavy ions. Biomed. Mater. Eng. 2015;26(Suppl 1):S1937–S1944. doi: 10.3233/BME-151496. [DOI] [PubMed] [Google Scholar]
- Berardinelli F, Antoccia A, Cherubini R, De Nadal V, Gerardi S, Cirrone GA, et al. Transient activation of the ALT pathway in human primary fibroblasts exposed to high-LET radiation. Radiat. Res. 2010;174(5):539–549. doi: 10.1667/RR2127.1. [DOI] [PubMed] [Google Scholar]
- Berardinelli F, Antoccia A, Cherubini R, De Nadal V, Gerardi S, Tanzarella C, et al. Telomere alterations and genomic instability in long-term cultures of normal human fibroblasts irradiated with X rays and protons. Radiat. Prot. Dosim. 2011;143(2–4):274–278. doi: 10.1093/rpd/ncq486. [DOI] [PubMed] [Google Scholar]
- Berardinelli F, Antoccia A, Buonsante R, Gerardi S, Cherubini R, De Nadal V, et al. The role of telomere length modulation in delayed chromosome instability induced by ionizing radiation in human primary fibroblasts. Environ. Mol. Mutagen. 2013;54(3):172–179. doi: 10.1002/em.21761. [DOI] [PubMed] [Google Scholar]
- Berardinelli F, Sgura A, Di Masi A, Leone S, Cirrone GA, Romano F, et al. Radiation-induced telomere length variations in normal and in Nijmegen Break-age Syndrome cells. Int. J. Radiat. Biol. 2014;90(1):45–52. doi: 10.3109/09553002.2014.859400. [DOI] [PubMed] [Google Scholar]
- Berardinelli F, De Vitis M, Nieri D, Cherubini R, De Nadal V, Gerardi S, et al. mBAND and mFISH analysis of chromosomal aberrations and breakpoint distribution in chromosome 1 of AG01522 human fibroblasts that were exposed to radiation of different qualities. Mutat. Res. Genet. Toxicol. Environ. Mutagen. 2015;793:55–63. doi: 10.1016/j.mrgentox.2015.05.012. [DOI] [PubMed] [Google Scholar]
- Black WC, Welch HG. Advances in diagnostic imaging and overestimations of disease prevalence and the benefits of therapy. New Engl. J. Med. 1993;328(17):1237–1243. doi: 10.1056/NEJM199304293281706. [DOI] [PubMed] [Google Scholar]
- Blackburn EH, Chan S, Chang J, Fulton TB, Krauskopf A, McEachern M, et al. Molecular manifestations and molecular determinants of telomere capping. Proceedings of Cold Spring Harbor Symposium on Quantitative Biology. 2000;65:253–263. doi: 10.1101/sqb.2000.65.253. [DOI] [PubMed] [Google Scholar]
- Blackburn EH. Telomere states and cell fates. Nature. 2000;408(6808):53–56. doi: 10.1038/35040500. [DOI] [PubMed] [Google Scholar]
- Blagoev KB. Cell proliferation in the presence of telomerase. PLoS One. 2009;4(2):e4622. doi: 10.1371/journal.pone.0004622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blakely EA, Chang PY. A review of ground-based heavy-ion radiobiology relevant to space radiation risk assessment. Part II: cardiovascular and immunological effects. Adv. Space Res. 2007;40(4):461–469. [Google Scholar]
- Blakely EA. Dr. Lauriston S. Taylor lecture on radiation protection and measurements: what makes particle radiation so effective? Health Phys. 2012;103(5):508–528. doi: 10.1097/HP.0b013e31826a5b85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blyth BJ, Sykes PJ. Radiation-induced bystander effects: what are they, and how relevant are they to human radiation exposures? Radiat. Res. 2011;176(2):139–157. doi: 10.1667/rr2548.1. [DOI] [PubMed] [Google Scholar]
- Bocker W, Iliakis G. Computational Methods for analysis of foci: validation for radiation-induced gamma-H2AX foci in human cells. Radiat. Res. 2006;165(1):113–124. doi: 10.1667/rr3486.1. [DOI] [PubMed] [Google Scholar]
- Boerrigter ME, Dolle ME, Martus HJ, Gossen JA, Vijg J. Plasmid-based transgenic mouse model for studying in vivo mutations. Nature. 1995;377(6550):657–659. doi: 10.1038/377657a0. [DOI] [PubMed] [Google Scholar]
- Bogdándi EN, Balogh A, Felgyinszki N, Szatmári T, Persa E, Hildebrandt G, et al. Effect of low-dose radiation on the immune system of mice after total–body irradiation. Radiat. Res. 2010;174(4):480–489. doi: 10.1667/RR2160.1. [DOI] [PubMed] [Google Scholar]
- Bolognesi C, Abbondandolo A, Barale R, Casalone R, Dalprà L, De Ferrari M, et al. Age-related increase of baseline frequencies of sister chromatid exchanges, chromosome aberrations, and micronuclei in human lymphocytes. Cancer Epidemiol. Biomark. Prev. 1997;6(4):249–256. [PubMed] [Google Scholar]
- Bolstad B. Affy PLM: Model Based QC Assessment of Affymetrix GeneChips 2005 [Google Scholar]
- Bonassi S, Norppa H, Ceppi M, Strömberg U, Vermeulen R, Znaor A, et al. Chromosomal aberration frequency in lymphocytes predicts the risk of cancer: results from a pooled cohort study of 22,358 subjects in 11 countries. Carcinogenesis. 2008;29:1178–1183. doi: 10.1093/carcin/bgn075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonner WM, Redon CE, Dickey JS, Nakamura AJ, Sedelnikova OA, Solier S, et al. GammaH2AX and cancer. Nat. Rev. Cancer. 2008;8(12):957–967. doi: 10.1038/nrc2523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Botling J, Edlund K, Lohr M, Hellwig B, et al. Biomarker discovery in non-small cell lung cancer: integrating gene expression profiling, meta-analysis, and tissue microarray validation. Clin. Cancer Res. 2013;19(1):194–201. doi: 10.1158/1078-0432.CCR-12-1139. [DOI] [PubMed] [Google Scholar]
- Boudaiffa B, Cloutier P, Hunting D, Huels MA, Sanche L, et al. Resonant formation of DNA strand breaks by low-energy (3 to 20 eV) electrons. Science. 2000;287(5458):1658–1660. doi: 10.1126/science.287.5458.1658. [DOI] [PubMed] [Google Scholar]
- Bouville A, Toohey REJD, Boice, Beck HL, Dauer LT, Eckerman KF, et al. Dose reconstruction for the million worker study: status and guidelines. Health Phys. 2015;108(2):206–220. doi: 10.1097/HP.0000000000000231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bozic I, Antal T, Ohtsuki H, Carter H, Kim D, Chen S, et al. Accumulation of driver and passenger mutations during tumor progression. Proc. Natl. Acad. Sci. USA. 2010;107(43):18545–18550. doi: 10.1073/pnas.1010978107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner DJ, Hahnfeldt P, Amundson SA, Sachs RK. Interpretation of inverse dose-rate effects for mutagenesis by sparsely ionizing radiation. Int. J. Radiat. Biol. 1996;70(4):447–458. doi: 10.1080/095530096144923. [DOI] [PubMed] [Google Scholar]
- Britton S, Coates J, Jackson SP. A new method for high-resolution imaging of Ku foci to decipher mechanisms of DNA double-strand break repair. J. Cell. Biol. 2013;202(3):579–595. doi: 10.1083/jcb.201303073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broccoli D, Young JW, de Lange T. Telomerase activity in normal and malignant hematopoietic cells. Proc. Natl. Acad. Sci. USA. 1995;92(20):9082–9086. doi: 10.1073/pnas.92.20.9082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buonanno M, de Toledo SM, Pain D, Azzam EI. Long-term consequences of radiation-induced bystander effects depend on radiation quality and dose and correlate with oxidative stress. Radiat. Res. 2011;175(4):405–415. doi: 10.1667/RR2461.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burdak-Rothkamm S, Short SC, Folkard M, Rothkamm K, Prise KM. ATR-dependent radiation-induced gamma H2AX foci in bystander primary human astrocytes and glioma cells. Oncogene. 2007;26(7):993–1002. doi: 10.1038/sj.onc.1209863. [DOI] [PubMed] [Google Scholar]
- Camacho CV, Mukherjee B, McEllin B, Ding LH, Hu B, Habib AA, et al. Loss of p15/Ink4b accompanies tumorigenesis triggered by complex DNA double-strand breaks. Carcinogenesis. 2010;31(10):1889–1896. doi: 10.1093/carcin/bgq153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camacho CV, Todorova PK, Hardebeck MC, Tomimatsu N, Gil del Alcazar CR, Ilcheva M, et al. DNA double-strand breaks cooperate with loss of Ink4 and Arf tumor suppressors to generate glioblastomas with frequent Met amplification. Oncogene. 2015;34(8):1064–1072. doi: 10.1038/onc.2014.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campa A, Alloni D, Antonelli F, Ballarini F, Belli M, Dini V, et al. DNA fragmentation induced in human fibroblasts by 56Fe ions: experimental data and Monte Carlo simulations. Radiat. Res. 2009;171(4):438–445. doi: 10.1667/RR1442.1. [DOI] [PubMed] [Google Scholar]
- Chang DJ, Cimprich KA. DNA damage tolerance: when it’s OK to make mistakes. Nat. Chem. Biol. 2009;5(2):82–90. doi: 10.1038/nchembio.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang WP, Little JB. Persistently elevated frequency of spontaneous mutations in progeny of CHO clones surviving X-irradiation: association with delayed reproductive death phenotype. Mutat. Res. 1992;270(2):191–199. doi: 10.1016/0027-5107(92)90130-t. [DOI] [PubMed] [Google Scholar]
- Chang PY, Bakke J, Orduna J, Lin S, Doppalaudi R. Proton-induced genetic damage in lacZ transgenic mice. Radiat. Res. 2005;164(4 Pt 2):481–486. doi: 10.1667/rr3322.1. [DOI] [PubMed] [Google Scholar]
- Chang J, Feng W, Wang Y, Luo Y, Allen AR, Koturbash I, et al. Whole–body proton irradiation causes long-term damage to hematopoietic stem cells in mice. Radiat. Res. 2015;183(2):240–248. doi: 10.1667/RR13887.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaudhary SU, Shin SY, Won JK, Cho KH. Multiscale modeling of tumorigenesis induced by mitochondrial incapacitation in cell death. IEEE Trans. Biomed. Eng. 2011;58(10):3028–3032. doi: 10.1109/TBME.2011.2159713. [DOI] [PubMed] [Google Scholar]
- Chauhan V, Howland M, Wilkins R. Identification of gene-based responses in human blood cells exposed to alpha particle radiation. BMC Med. Genom. 2014;7:43. doi: 10.1186/1755-8794-7-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen DJ, Tsuboi K, Nguyen T, Yang TC. Charged-particle mutagenesis II. Mutagenic effects of high energy charged particles in normal human fibroblasts. Adv. Space Res. 1994;14(10):347–354. doi: 10.1016/0273-1177(94)90487-1. [DOI] [PubMed] [Google Scholar]
- Cheng W-C, Shu W-Y, Li C-Y, Tsai M-L, Chang C-W, Chen C-R, et al. Intra- and inter-individual variance of gene expression in clinical studies. PLoS One. 2012;7(6):e38650. doi: 10.1371/journal.pone.0038650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherry JD, Liu B, Frost JL, Lemere CA, Williams JP, Olschowka JA, et al. Galactic cosmic radiation leads to cognitive impairment and increased Aβ plaque accumulation in a mouse model of Alzheimer’s disease. PLoS One. 2012;7(12):e53275. doi: 10.1371/journal.pone.0053275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiang C-S, Chen F-H, Hong J-H, Jiang P-S, Huang H-L, Wang C-C, et al. Functional phenotype of macrophages depends on assay procedures. Int. Immunol. 2007;20(2):215–222. doi: 10.1093/intimm/dxm137. [DOI] [PubMed] [Google Scholar]
- Chuang EYY, Little JB, Yuan ZM. Cellular mechanisms for low-dose ionizing radiation–induced perturbation of the breast tissue microenvironment. Cancer Res. 2005;65(15):6734–6744. doi: 10.1158/0008-5472.CAN-05-0703. [DOI] [PubMed] [Google Scholar]
- Chun SH, Park G-Y, Han YK, Kim SD, Kim JS, Lee CG, et al. Effect of low dose radiation on differentiation of bone marrow cells into dendritic cells. Dose Response. 2013;11(3):374–384. doi: 10.2203/dose-response.12-041.Lee. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornforth M, Bailey S, Goodwin E. Dose responses for chromosome aberrations produced in noncycling primary human fibroblasts by alpha particles, and by gamma rays delivered at sublimiting low dose rates. Radiat. Res. 2002;158:43–53. doi: 10.1667/0033-7587(2002)158[0043:drfcap]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- Costes S, Sachs R, Hlatky L, Vannais D, Waldren C, Fouladi B. Large–mutation spectra induced at hemizygous loci by low-LET radiation: evidence for intrachromosomal proximity effects. Radiat. Res. 2001;156(5 Pt 1):545–557. doi: 10.1667/0033-7587(2001)156[0545:lmsiah]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- Costes SV, Boissiere A, Ravani S, Romano R, Parvin B, Barcellos-Hoff MH. Imaging features that discriminate between foci induced by high- and low-LET radiation in human fibroblasts. Radiat. Res. 2006;165(5):505–515. doi: 10.1667/RR3538.1. [DOI] [PubMed] [Google Scholar]
- Costes SV, Ponomarev A, Chen JL, Nguyen D, Cucinotta FA, Barcellos-Hoff MH. Image-based modeling reveals dynamic redistribution of DNA damage into nuclear sub-domains. PLoS Comput. Biol. 2007;3(8):e155. doi: 10.1371/journal.pcbi.0030155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costes SV, Chiolo I, Pluth JM, Barcellos-Hoff MH, Jakob B. Spatiotemporal characterization of ionizing radiation induced DNA damage foci and their relation to chromatin organization. Mutat. Res. 2010;704(1–3):78–87. doi: 10.1016/j.mrrev.2009.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox R, Masson WK. Mutation and inactivation of cultured mammalian cells exposed to beams of accelerated heavy ions. III. Human diploid fibroblasts. Int. J. Radiat. Biol. Relat. Stud. Phys. Chem. Med. 1979;36(2):149–160. doi: 10.1080/09553007914550901. [DOI] [PubMed] [Google Scholar]
- Cucinotta FA, Chappell LJ. Non-targeted effects and the dose response for heavy ion tumor induction. Mutat. Res. 2010;687(1–2):49–53. doi: 10.1016/j.mrfmmm.2010.01.012. [DOI] [PubMed] [Google Scholar]
- Cucinotta FA, Durante M. Cancer risk from exposure to galactic cosmic rays: implications for space exploration by human beings. Lancet Oncol. 2006;7(5):431–435. doi: 10.1016/S1470-2045(06)70695-7. [DOI] [PubMed] [Google Scholar]
- Cucinotta FA, Nikjoo H, Goodhead DT. The effects of delta rays on the number of particle-track traversals per cell in laboratory and space exposures. Radiat. Res. 1998;150(1):115–119. [PubMed] [Google Scholar]
- Cucinotta FA, Kim MY, Chappell LJ. Space Radiation Cancer Risk Projections and Uncertanities. 2012 NASA TP 2013-217375. [Google Scholar]
- Cucinotta FA. A new approach to reduce uncertainties in space radiation cancer risk predictions. PLoS One. 2015;10(3):e0120717. doi: 10.1371/journal.pone.0120717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Autreaux B, Toledano MB. ROS as signalling molecules: mechanisms that generate specificity in ROS homeostasis. Nat. Rev. Mol. Cell. Biol. 2007;8(10):813–824. doi: 10.1038/nrm2256. [DOI] [PubMed] [Google Scholar]
- Datta K, Suman S, Kallakury BV, Fornace AJ., Jr Exposure to heavy ion radiation induces persistent oxidative stress in mouse intestine. PLoS One. 2012;7(8):e42224. doi: 10.1371/journal.pone.0042224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datta K, Suman S, Kallakury BV, Fornace AJ., Jr Heavy ion radiation exposure triggered higher intestinal tumor frequency and greater beta-catenin activation than gamma radiation in APC(Min/+) mice. PLoS One. 2013;8(3):e59295. doi: 10.1371/journal.pone.0059295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davalos AR, Coppe J-P, Campisi J, Desprez P-Y. Senescent cells as a source of inflammatory factors for tumor progression. Cancer Metastasis Rev. 2010;29(2):273–283. doi: 10.1007/s10555-010-9220-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis H, Irshad S, Bansal M, Rafferty H, Boitsova T, Bardella C, et al. Aberrant epithelial GREM1 expression initiates colonic tumorigenesis from cells out-side the stem cell niche. Nat. Med. 2015;21(1):62–70. doi: 10.1038/nm.3750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Gonzalez ABAA, Veiga LHS, Rajaraman P, Thomas BA, Hoffman FO, et al. RadRAT: a radiation risk assessment tool for lifetime cancer risk projection. J. Radiol. Protoc. 2012;32:205–222. doi: 10.1088/0952-4746/32/3/205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Lange T. Telomeres and senescence: ending the debate. Science. 1998;279(5349):334–335. doi: 10.1126/science.279.5349.334. [DOI] [PubMed] [Google Scholar]
- Dettmering T, Zahnreich S, Colindres-Rojas M, Durante M, Taucher-Scholz G, Fournier C. Increased effectiveness of carbon ions in the production of reactive oxygen species in normal human fibroblasts. J. Radiat. Res. 2015;56(1):67–76. doi: 10.1093/jrr/rru083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Maggio FA, Minafra L, Forte GI, Cammarata FP, Lio D, Messa C, et al. Portrait of inflammatory response to ionizing radiation treatment. J. Inflamm. 2015;12(14):1–11. doi: 10.1186/s12950-015-0058-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickey JS, Baird BJ, Redon CE, Sokolov MV, Sedelnikova OA, Bonner WM. Intercellular communication of cellular stress monitored by gamma-H2AX induction. Carcinogenesis. 2009;30(10):1686–1695. doi: 10.1093/carcin/bgp192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dikomey E, Dahm-Daphi J, Brammer I, Martensen R, Kaina B. Correlation between cellular radiosensitivity and non-repaired double-strand breaks studied in nine mammalian cell lines. Int. J. Radiat. Biol. 1998;73(3):269–278. doi: 10.1080/095530098142365. [DOI] [PubMed] [Google Scholar]
- Ding L-H, Shingyoji M, Chen F, Chatterjee A, Kasai K-E, Chen David J. Gene expression changes in normal human skin fibroblasts induced by HZE-particle radiation. Radiat. Res. 2005;164(4 Pt 2):523–526. doi: 10.1667/rr3350.1. [DOI] [PubMed] [Google Scholar]
- Ding LH, Park S, Peyton M, Girard L, Xie Y, Minna JD, et al. Distinct transcriptome profiles identified in normal human bronchial epithelial cells after exposure to gamma-rays and different elemental particles of high Z and energy. BMC Genom. 2013;14:372. doi: 10.1186/1471-2164-14-372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding LH, Park S, Xie Y, Girard L, Minna JD, Story MD. Elucidation of changes in molecular signalling leading to increased cellular transformation in oncogenically progressed human bronchial epithelial cells exposed to radiations of increasing LET. Mutagenesis. 2015;30(5):685–694. doi: 10.1093/mutage/gev028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan J, Shi J, Ge X, Dölken L, Moy W, He D, et al. Genome-wide survey of interindividual differences of RNA stability in human lymphoblastoid cell lines. Sci. Rep. 2013;3:1318. doi: 10.1038/srep01318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durante M, Cucinotta FA. Heavy ion carcinogenesis and human space exploration. Nat. Rev. Cancer. 2008;8(6):465–472. doi: 10.1038/nrc2391. [DOI] [PubMed] [Google Scholar]
- Durante M, George K, Wu H, Cucinotta FA. Karyotypes of human lymphocytes exposed to high-energy iron ions. Radiat. Res. 2002;158(5):581–590. doi: 10.1667/0033-7587(2002)158[0581:kohlet]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- Durante M. Biomarkers of space radiation risk. Radiat. Res. 2005;164(4 Pt 2):467–473. doi: 10.1667/rr3359.1. [DOI] [PubMed] [Google Scholar]
- Eberlein U, Peper M, Fernandez M, Lassmann M, Scherthan H. Calibration of the gamma-H2AX DNA double strand break focus assay for internal radiation exposure of blood lymphocytes. PLoS one. 2015;10(4):e0123174. doi: 10.1371/journal.pone.0123174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edinger AL, Thompson CB. Death by design: apoptosis, necrosis and autophagy. Curr. Opin. Cell Biol. 2004;16(6):663–669. doi: 10.1016/j.ceb.2004.09.011. [DOI] [PubMed] [Google Scholar]
- El-Saghire H, Thierens H, Monsieurs P, Michaux A, Vandevoorde C, Baatout S. Gene set enrichment analysis highlights different gene expression profiles in whole blood samples X-irradiated with low and high doses. Int. J. Radiat. Biol. 2013;89(8):628–638. doi: 10.3109/09553002.2013.782448. [DOI] [PubMed] [Google Scholar]
- Enderling H, Anderson AR, Chaplain MA, Beheshti A, Hlatky L, Hahnfeldt P. Paradoxical dependencies of tumor dormancy and progression on basic cell kinetics. Cancer Res. 2009;69(22):8814–8821. doi: 10.1158/0008-5472.CAN-09-2115. [DOI] [PubMed] [Google Scholar]
- Ermakov AV, Konkova MS, Kostyuk SV, Egolina NA, Efremova LV, Veiko NN. Oxidative stress as a significant factor for development of an adaptive response in irradiated and nonirradiated human lymphocytes after inducing the bystander effect by low-dose X-radiation. Mutat. Res. 2009;669(1–2):155–161. doi: 10.1016/j.mrfmmm.2009.06.005. [DOI] [PubMed] [Google Scholar]
- Erofeeva LM, Sapin MR, Grigorenko DE. The thymus of mice at different periods after irradiation with fast carbon ions. Morfologiia. 2000;117(1):42–476. [PubMed] [Google Scholar]
- Fachin AL, Mello SS, Sandrin-Garcia P, Junta CM, Donadi EA, Passos GA, et al. Gene expression profiles in human lymphocytes irradiated in vitro with low doses of gamma rays. Radiat. Res. 2007;168(6):650–665. doi: 10.1667/RR0487.1. [DOI] [PubMed] [Google Scholar]
- Feldser D, Strong MA, Greider CW. Ataxia telangiectasia mutated (Atm) is not required for telomerase-mediated elongation of short telomeres. Proc. Natl. Acad. Sci. USA. 2006;103(7):2249–2251. doi: 10.1073/pnas.0511143103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finnon P, Silver AR, Bouffler SD. Upregulation of telomerase activity by X-irradiation in mouse leukaemia cells is independent of Tert, Terc, Tnks and Myc transcription. Carcinogenesis. 2000;21(4):573–578. doi: 10.1093/carcin/21.4.573. [DOI] [PubMed] [Google Scholar]
- Foran JM. New prognostic markers in acute myeloid leukemia: perspective from the clinic. Hematology Am society/the Education Program of the American Society of Hematology, 2010. American Society of Hematology Education Program. 2010:47–55. doi: 10.1182/asheducation-2010.1.47. [DOI] [PubMed] [Google Scholar]
- Formenti SC, Demaria S. Systemic effects of local radiotherapy. Lancet Oncol. 2009;10:718–726. doi: 10.1016/S1470-2045(09)70082-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freund A, Orjalo AV, Desperez P-Y, Campisi J. Inflammatory networks during cellular senescence: causes and consequences. Trends Mol. Med. 2010;16(5):238–246. doi: 10.1016/j.molmed.2010.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedland W, Dingfelder M, Jacob P, Paretzke HG. Calculated DNA double-strand break and fragmentation yields after irradiation with He ions. Radiat. Phys. Chem. 2005;72:279–286. [Google Scholar]
- Friedland W, Dingfelder M, Kundrat P, Jacob P. Track structures, DNA targets and radiation effects in the biophysical Monte Carlo simulation code PARTRAC. Mutat. Res. 2011;711(1–2):28–40. doi: 10.1016/j.mrfmmm.2011.01.003. [DOI] [PubMed] [Google Scholar]
- Frijhoff J, Winyard PG, Zarkovic N, Davies SS, Stocker R, Cheng D, et al. Clinical relevance of biomarkers of oxidative stress. Antioxid. Redox Signal. 2015;23(14):1144–1170. doi: 10.1089/ars.2015.6317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fry R, Grosovsky A, Hanawalt P, Jostes R, Little J, Morgan W, et al. The impact of biology on risk assessment–workshop of the national research council’s board on radiation effects research. Radiat. Res. 1997;150:695–705. [PubMed] [Google Scholar]
- Fumagalli M, Rossiello F, Clerici M, Barozzi S, Cittaro D, Kaplunov JM, et al. Telomeric DNA damage is irreparable and causes persistent DNA-damage-response activation. Nat. Cell Biol. 2012;14(4):355–365. doi: 10.1038/ncb2466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao X, McDonald JT, Naidu M, Hahnfeldt P, Hlatky L. A proposed quantitative index for assessing the potential contribution of reprogramming to cancer stem cell kinetics. Stem Cells Int. 2014;2014:249309. doi: 10.1155/2014/249309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gauthier LD, Greenstein JL, Cortassa S, O’Rourke B, Winslow RL. A computational model of reactive oxygen species and redox balance in cardiac mitochondria. Biophys. J. 2013;105(4):1045–1056. doi: 10.1016/j.bpj.2013.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geard C, Jenkins-Baker G, Marino S, Ponnaiya B. Novel approaches with track segment Alpha particles and cell co-cultures in studies of bystander effects. Radiat. Prot. Dosim. 2002;99(1–4):233–236. doi: 10.1093/oxfordjournals.rpd.a006771. [DOI] [PubMed] [Google Scholar]
- Geiersbach KB, Gardiner AE, Wilson A, Shetty S, Bruyère H, Zabawski J, et al. Subjectivity in chromosome band–level estimation: a multicenter study. Genet. Med. 2014;16:170–175. doi: 10.1038/gim.2013.95. [DOI] [PubMed] [Google Scholar]
- Genik PC, Vyazunova I, Steffen LS, Bacher JW, Bielefeldt-Ohmann H, McKercher S, et al. Leukemogenesis in heterozygous PU.1 knockout mice. Radiat. Res. 2014;182(3):310–315. doi: 10.1667/RR13738.1. [DOI] [PubMed] [Google Scholar]
- Georgescu W, Osseiran A, Rojec M, Liu Y, Bombrun M, Tang J, et al. Characterizing the DNA damage response by cell tracking algorithms and cell features classification using high-content time-lapse analysis. PLoS One. 2015;10(6):e0129438. doi: 10.1371/journal.pone.0129438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghandhi SA, Ming L, Ivanov VN, Hei TK, Amundson SA. Regulation of early signaling and gene expression in the alpha-particle and bystander response of IMR-90 human fibroblasts. BMC Med. Genom. 2010;3:31. doi: 10.1186/1755-8794-3-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghandhi SA, Smilenov LB, Elliston CD, Chowdhury M, Amundson SA. Radiation dose-rate effects on gene expression for human biodosimetry. BMC Med. Genom. 2015;8:22. doi: 10.1186/s12920-015-0097-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giam M, Rancati G. Aneuploidy and chromosomal instability in cancer: a jackpot to chaos. Cell Div. 2015;10:3. doi: 10.1186/s13008-015-0009-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giedzinski E, Rola R, Fike JR, Limoli CL. Efficient production of reactive oxygen species in neural precursor cells after exposure to 250 MeV protons. Radiat. Res. 2005;164(4 Pt 2):540–544. doi: 10.1667/rr3369.1. [DOI] [PubMed] [Google Scholar]
- Girdhani S, Lamont C, Hahnfeldt P, Abdollahi ALH. Proton irradiation suppresses angiogenic genes and impairs cell invasion and tumor growth. Radiat. Res. 2012;178(1):33–45. doi: 10.1667/rr2724.1. [DOI] [PubMed] [Google Scholar]
- Girdhani S, Sachs RK, Hlatky L. Biological effects of proton radiation: what we know and don’t know. Radiat. Res. 2013;179(3):257–272. doi: 10.1667/RR2839.1. [DOI] [PubMed] [Google Scholar]
- Gonon G, Groetz JE, de Toledo SM, Howell RW, Fromm M, Azzam EI. Nontargeted stressful effects in normal human fibroblast cultures exposed to low fluences of high charge, high energy (HZE) particles: kinetics of biologic responses and significance of secondary radiations. Radiat. Res. 2013;179(4):444–457. doi: 10.1667/RR3017.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodhead DT. Spatial and temporal distribution of energy. Health Phys. 1988;55(2):231–240. doi: 10.1097/00004032-198808000-00015. [DOI] [PubMed] [Google Scholar]
- Gossen JA, Martus HJ, Wei JY, Vijg J. Spontaneous and X-ray-induced deletion mutations in a LacZ plasmid-based transgenic mouse model. Mutat. Res. 1995;331(1):89–97. doi: 10.1016/0027-5107(95)00055-n. [DOI] [PubMed] [Google Scholar]
- Gridley DS, Pecaut MJ, Nelson GA. Total-body irradiation with high-let particles: acute and chronic effects on the immune system. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2002;282(3):R677–R88. doi: 10.1152/ajpregu.00435.2001. [DOI] [PubMed] [Google Scholar]
- Grigorenko DE, Sapin MR, Erofeeva ML. Splenic lymphoid tissue of mice after irradiation with fast carbon ions. Morfologiia. 1998;114(5) [PubMed] [Google Scholar]
- Grosovsky AJ, Little JB. Evidence for linear response for the induction of mutations in human cells by x-ray exposures below 10 rads. Proc. Natl. Acad. Sci. USA. 1985;82(7):2092–2095. doi: 10.1073/pnas.82.7.2092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grosovsky AJ, de Boer JG, de Jong PJ, Drobetsky EA, Glickman BW. Base substitutions, frameshifts, and small deletions constitute ionizing radiation-induced point mutations in mammalian cells. Proc. Natl. Acad. Sci. USA. 1988;85(1):185–188. doi: 10.1073/pnas.85.1.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruel G, Voisin P, Vaurijoux A, Roch-Lefevre S, Gregoire E, Maltere P, et al. Broad modulation of gene expression in CD4 + lymphocyte subpopulations in response to low doses of ionizing radiation. Radiat. Res. 2008;170(3):335–344. doi: 10.1667/RR1147.1. [DOI] [PubMed] [Google Scholar]
- Guo Y, Xiao P, Lei S, Deng F, Xiao Gary G, Liu Y, et al. How is mRNA expression predictive for protein expression? A correlation study on human circulating monocytes. Acta Biochim. Biophys. Sin. 2008;40(5):426–436. doi: 10.1111/j.1745-7270.2008.00418.x. [DOI] [PubMed] [Google Scholar]
- Gupta PB, Fillmore CM, Jiang G, Shapira SD, Tao K, Kuperwasser C, et al. Stochastic state transitions give rise to phenotypic equilibrium in populations of cancer cells. Cell. 2011;146(4):633–644. doi: 10.1016/j.cell.2011.07.026. [DOI] [PubMed] [Google Scholar]
- Hada M, Georgakilas AG. Formation of clustered DNA damage after high-LET irradiation: a review. J. Radiat. Res. 2008;49(3):203–210. doi: 10.1269/jrr.07123. [DOI] [PubMed] [Google Scholar]
- Hada M, Sutherland BM. Spectrum of complex DNA damages depends on the incident radiation. Radiat. Res. 2006;165(2):223–230. doi: 10.1667/rr3498.1. [DOI] [PubMed] [Google Scholar]
- Hada M, Cucinotta FA, Gonda SR, Wu H. mBAND analysis of chromosomal aberrations in human epithelial cells exposed to low- and high-LET radiation. Radiat. Res. 2007;168(1):98–105. doi: 10.1667/RR0759.1. [DOI] [PubMed] [Google Scholar]
- Hada M, George K, Chappell L, Cucinotta FA. Chromosomal aberrations in human lymphocytes and fibroblasts after exposure to very low doses of high--LET radiation. J. Radiat. Res. 2014;55:i50–i1. [Google Scholar]
- Hada M, Chappell L, Wang M, George K, Cucinotta F. Induction of chromosomal aberrations at fluences of less than one HZE particle per cell nucleus. Radiat. Res. 2014;182:368–379. doi: 10.1667/RR13721.1. [DOI] [PubMed] [Google Scholar]
- Hagmar L, Bonassi S, Strömberg U, Brøgger A, Knudsen L, Norppa H, et al. Chromosomal aberrations in lymphocytes predict human cancer: a report from the European Study Group on Cytogenetic Biomarkers and Health (ESCH) Cancer Res. 1998;58(18):4117–4121. [PubMed] [Google Scholar]
- Hall EJ, Hei TK. Genomic instability and bystander effects induced by high--LET radiation. Oncogene. 2003;22(45):7034–7042. doi: 10.1038/sj.onc.1206900. [DOI] [PubMed] [Google Scholar]
- Halliwell B, Whiteman M. Measuring reactive species and oxidative damage in vivo and in cell culture: how should you do it and what do the results mean? Br. J. Pharmacol. 2004;142(2):231–255. doi: 10.1038/sj.bjp.0705776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamburger AW, Salmon SE. Primary bioassay of human tumor stem cells. Science. 1977;197(4302):461–463. doi: 10.1126/science.560061. [DOI] [PubMed] [Google Scholar]
- Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144(5):646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- Hande MP, Lansdorp PM, Natarajan AT. Induction of telomerase activity by in vivo X-irradiation of mouse splenocytes and its possible role in chromosome healing. Mutat. Res. 1998;404(1–2):205–214. doi: 10.1016/s0027-5107(98)00115-8. [DOI] [PubMed] [Google Scholar]
- Harada H, Harada Y, Tanaka H, Kimura A, Inaba T. Implications of somatic mutations in the AML1 gene in radiation-associated and therapy-related myelodysplastic syndrome/acute myeloid leukemia. Blood. 2003;101(2):673–680. doi: 10.1182/blood-2002-04-1010. [DOI] [PubMed] [Google Scholar]
- Hasegawa Y, Taylor D, Ovchinnikov DA, Wolvetang EJ, de Torrenté L, Mar Jessica C. Variability of gene expression identifies transcriptional regulators of early human embryonic development. PLoS Genet. 2015;11(8):e1005428. doi: 10.1371/journal.pgen.1005428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He M, Dong C, Konishi T, Tu W, Liu W, Shiomi N, et al. Differential effects of p53 on bystander phenotypes induced by gamma ray and high-LET heavy ion radiation. Life Sci. Space Res. 2014;1:53–59. doi: 10.1016/j.lssr.2014.02.003. [DOI] [PubMed] [Google Scholar]
- Held KD. Effects of low fluences of radiations found in space on cellular systems. Int. J. Radiat. Biol. 2009;85(5):379–390. doi: 10.1080/09553000902838558. [DOI] [PubMed] [Google Scholar]
- Henle ES, Han Z, Tang N, Rai P, Luo Y, Linn S. Sequence-specific DNA cleavage by Fe2 +-mediated fenton reactions has possible biological implications. J. Biol. Chem. 1999;274(2):962–971. doi: 10.1074/jbc.274.2.962. [DOI] [PubMed] [Google Scholar]
- Henriques CM, Ferreira MG. Consequences of telomere shortening during lifespan. Curr. Opin. Cell Biol. 2012;24(6):804–808. doi: 10.1016/j.ceb.2012.09.007. [DOI] [PubMed] [Google Scholar]
- Hillen T, Enderling H, Hahnfeldt P. The tumor growth paradox and immune system-mediated selection for cancer stem cells. Bull. Math. Biol. 2013;75(1):161–184. doi: 10.1007/s11538-012-9798-x. [DOI] [PubMed] [Google Scholar]
- Hirayama R, Ito A, Tomita M, Tsukada T, Yatagai F, Noguchi M, et al. Contributions of direct and indirect actions in cell killing by high-LET radiations. Radiat. Res. 2009;171(2):212–218. doi: 10.1667/RR1490.1. [DOI] [PubMed] [Google Scholar]
- Hirt BV, Wattis JA, Preston SP. Modelling the regulation of telomere length: the effects of telomerase and G-quadruplex stabilising drugs. J. Math. Biol. 2014;68(6):1521–1552. doi: 10.1007/s00285-013-0678-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiyama K, Hirai Y, Kyoizumi S, Akiyama M, Hiyama E, Piatyszek MA, et al. Activation of telomerase in human lymphocytes and hematopoietic progenitor cells. J. Immunol. 1995;155(8):3711–3715. [PubMed] [Google Scholar]
- Hofman-Huther H, Scholz M, Rave-Frank M, Virsik-Kopp P. Induction of reproductive cell death and chromosome aberrations in radioresistant tumour cells by carbon ions. Int. J. Radiat. Biol. 2004;80(6):423–435. doi: 10.1080/09553000410001702319. [DOI] [PubMed] [Google Scholar]
- Houtgraaf JH, Versmissen J, van der Giessen WJ. A concise review of DNA damage checkpoints and repair in mammalian cells. Cardiovasc. Revascularization Med. 2006;7(3):165–172. doi: 10.1016/j.carrev.2006.02.002. [DOI] [PubMed] [Google Scholar]
- Hryciw G, Grygoryev D, Lasarev M, Ohlrich A, Dan C, Madhira R, et al. Accelerated (48)Ti ions induce autosomal mutations in mouse kidney epithelium at low dose and fluence. Radiat. Res. 2015;184(4):367–377. doi: 10.1667/RR14130.1. [DOI] [PubMed] [Google Scholar]
- Hussain SP, Hofseth LJ, Harris CC. Radical causes of cancer. Nat. Rev. Cancer. 2003;3(4):276–285. doi: 10.1038/nrc1046. [DOI] [PubMed] [Google Scholar]
- Il’yasova D, Scarbrough P, Spasojevic I. Urinary biomarkers of oxidative status. Clin. Chim. Acta. 2012;413(19–20):1446–1453. doi: 10.1016/j.cca.2012.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ilyenko I, Lyaskivska O, Bazyka D. Analysis of relative telomere length and apoptosis in humans exposed to ionising radiation. Exp. Oncol. 2011;33(4):235–238. [PubMed] [Google Scholar]
- Ina Y, Sakai K. Activation of immunological network by chronic low–dose-rate irradiation in wild-type mouse strains: analysis of immune cell populations and surface molecules. Int. J. Radiat. Biol. 2005;81(10):721–729. doi: 10.1080/09553000500519808. [DOI] [PubMed] [Google Scholar]
- Jackson SP, Bartek J. The DNA-damage response in human biology and disease. Nature. 2009;461(7267):1071–1078. doi: 10.1038/nature08467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacob SF, Wurstle ML, Delgado ME, Rehm M. An analysis of the truncated Bid- and ROS-dependent spatial propagation of mitochondrial permeabilization waves during apoptosis. J. Biol. Chem. 2016;291(9):4603–4613. doi: 10.1074/jbc.M115.689109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jafri MS, Kumar R. Modeling mitochondrial function and its role in disease. Prog. Mol. Biol. Transl. Sci. 2014;123:103–125. doi: 10.1016/B978-0-12-397897-4.00001-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jahns J, Anderegg U, Saalbach A, Rosin B, Patties I, Glasow A, et al. Influence of low dose irradiation on differentiation, maturation and T-cell activation of human dendritic cells. Mutat. Res. 2011;709– 710:32–39. doi: 10.1016/j.mrfmmm.2011.02.007. [DOI] [PubMed] [Google Scholar]
- Jaiswal H, Lindqvist A. Bystander communication and cell cycle decisions after DNA damage. Front. Genet. 2015;6:63. doi: 10.3389/fgene.2015.00063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakob B, Scholz M, Taucher-Scholz G. Biological imaging of heavy charged– particle tracks. Radiat. Res. 2003;159(5):676–684. doi: 10.1667/0033-7587(2003)159[0676:biohct]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- Jakob B, Splinter J, Taucher-Scholz G. Positional stability of damaged chromatin domains along radiation tracks in mammalian cells. Radiat. Res. 2009;171(4):405–418. doi: 10.1667/RR1520.1. [DOI] [PubMed] [Google Scholar]
- Jia P, Zhao Z. VarWalker:Personalized Mutation Network Analysis of Putative Cancer Genes from Next-Generation Sequencing Data. Plos Comput Biol. 2014;10(2):1–13. doi: 10.1371/journal.pcbi.1003460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin YW, Na YJ, Lee YJ, An S, Lee JE, Jung M, et al. Comprehensive analysis of time- and dose-dependent patterns of gene expression in a human mesenchymal stem cell line exposed to low-dose ionizing radiation. Oncol. Rep. 2008;19(1):135–144. [PubMed] [Google Scholar]
- Johnson WE, Li C, Rabinovic A, Johnson WE, Li C, Rabinovic A. Adjusting batch effects in microarray expression data using empirical bayes methods. Biostatistics. 2006;8(1):118–127. doi: 10.1093/biostatistics/kxj037. Print Biostatistics. 2006;8(1): 118-27. [DOI] [PubMed] [Google Scholar]
- Ju YJ, Park JE, Juhn KM, Jeong J, Yun M, Park MJ, et al. Chromosomal end fusion resulting from telomere erosion increases susceptibility to radiation via multinucleation: effect of p53. Int. J. Oncol. 2006;29(4):753–763. [PubMed] [Google Scholar]
- Kalyanaraman B, Darley-Usmar V, Davies KJ, Dennery PA, Forman HJ, Grisham MB, et al. Measuring reactive oxygen and nitrogen species with fluorescent probes: challenges and limitations. Free Radic. Biol. Med. 2012;52(1):1–6. doi: 10.1016/j.freeradbiomed.2011.09.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang C-M, Park K-P, Song J-E, Jeoung D-I, Cho C-K, Kim T-H, et al. Possible biomarkers for ionizing radiation exposure in human peripheral blood lymphocytes. Radiat. Res. 2003;159(3):312–319. doi: 10.1667/0033-7587(2003)159[0312:pbfire]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- Kato TA, Nagasawa H, Weil MM, Little JB, Bedford JS. Levels of gamma-H2AX Foci after low-dose-rate irradiation reveal a DNA DSB rejoining defect in cells from human ATM heterozygotes in two at families and in another apparently normal individual. Radiat. Res. 2006;166(3):443–453. doi: 10.1667/RR3604.1. [DOI] [PubMed] [Google Scholar]
- Kato I, Ren J, Heilbrun LK, Djuric Z. Intra- and inter-individual variability in measurements of biomarkers for oxidative damage in vivo: nutrition and breast health study. Biomarkers. 2006;11(2):143–152. doi: 10.1080/13547500600565693. [DOI] [PubMed] [Google Scholar]
- Kaul Z, Cesare AJ, Huschtscha LI, Neumann AA, Reddel RR. Five dys-functional telomeres predict onset of senescence in human cells. EMBO Rep. 2012;13(1):52–59. doi: 10.1038/embor.2011.227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennedy AR, Crucian B, Huff JL, Klein SL, Morens D, Murasko D, et al. Effects of sex and gender on adaptation to space: immune system. J. Women’s Health. 2014;23(11):956–958. doi: 10.1089/jwh.2014.4913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennedy AR. Biological effects of space radiation and development of effective countermeasures. Life Sci. Space Res. 2014;1:10–43. doi: 10.1016/j.lssr.2014.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kikuchi A, Ishikawa T, Mogushi K, Ishiguro M, et al. Identification of NUCKS1 as a colorectal cancer prognostic marker through integrated expression and copy number analysis. Int. J. Cancer. 2013;132(10):2295–2302. doi: 10.1002/ijc.27911. [DOI] [PubMed] [Google Scholar]
- Kim Sh SH, Kaminker P, Campisi J. Telomeres, aging and cancer: in search of a happy ending. Oncogene. 2002;21(4):503–511. doi: 10.1038/sj.onc.1205077. [DOI] [PubMed] [Google Scholar]
- Kimzey SL, Fischer CL, Johnson PC, Ritzman SE, Magel CE. Hematology and immunology studies. Biomedical Results of Apollo NASA SP-368. Apollo NASA. 1975;368:213–222. [Google Scholar]
- Klammer H, Mladenov E, Li F, Iliakis G. Bystander effects as manifestation of intercellular communication of DNA damage and of the cellular oxidative status. Cancer Lett. 2015;356(1):58–71. doi: 10.1016/j.canlet.2013.12.017. [DOI] [PubMed] [Google Scholar]
- Knops K, Boldt S, Wolkenhauer O, Kriehuber R. Gene expression in low- and high-dose-irradiated human peripheral blood lymphocytes: possible applications for biodosimetry. Radiat. Res. 2012;178(4):304–312. doi: 10.1667/rr2913.1. [DOI] [PubMed] [Google Scholar]
- Kobayashi S, Yamada-Okabe H, Suzuki M, Natori O, Kato A, Matsubara K, et al. LGR5-positive colon cancer stem cells interconvert with drug-resistant LGR5-negative cells and are capable of tumor reconstitution. Stem Cells. 2012;30(12):2631–2644. doi: 10.1002/stem.1257. [DOI] [PubMed] [Google Scholar]
- Kocher DC, Apostoaei AI, Hoffman FO. Radiation effectiveness factors for use in calculating probability of causation of radiogenic cancers. Health Phys. 2005;89(1):3–32. doi: 10.1097/01.hp.0000154172.48895.45. [DOI] [PubMed] [Google Scholar]
- Kocher DCAA, Henshaw RW, Hoffman FO, Schubauer-Berigan MK, Stancescu DO, et al. Interactive radioepidemiological program (IREP): a web based tool for estimating probability of causation/assigned share of radiogenic cancers. Health Phys. 2008;95:119–147. doi: 10.1097/01.HP.0000291191.49583.f7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraemer SM, Vannais DB, Kronenberg A, Ueno A, Waldren CA. Gamma-ray mutagenesis studies in a new human-hamster hybrid, A(L)CD59(+/−), which has two human chromosomes 11 but is hemizygous for the CD59 gene. Radiat. Res. 2001;156(1):10–19. doi: 10.1667/0033-7587(2001)156[0010:grmsia]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- Kronenberg A, Cucinotta FA. Space radiation protection issues. Health Phys. 2012;103(5):556–567. doi: 10.1097/HP.0b013e3182690caf. [DOI] [PubMed] [Google Scholar]
- Kronenberg A, Gauny S, Criddle K, Vannais D, Ueno A, Kraemer S, et al. Heavy ion mutagenesis: linear energy transfer effects and genetic linkage. Radiat. Environ. Biophys. 1995;34(2):73–78. doi: 10.1007/BF01275209. [DOI] [PubMed] [Google Scholar]
- Kronenberg A, Gauny S, Kwoh E, Connolly L, Dan C, Lasarev M, et al. Comparative analysis of cell killing and autosomal mutation in mouse kidney epithelium exposed to 1 GeV/nucleon iron ions in vitro or in situ. Radiat. Res. 2009;172(5):550–557. doi: 10.1667/RR1804.1. [DOI] [PubMed] [Google Scholar]
- Kronenberg A, Gauny S, Kwoh E, Grossi G, Dan C, Grygoryev D, et al. Comparative analysis of cell killing and autosomal mutation in mouse kidney epithelium exposed to 1 GeV protons in vitro or in vivo. Radiat. Res. 2013;179(5):511–520. doi: 10.1667/RR3182.1. [DOI] [PubMed] [Google Scholar]
- Kronenberg A. Perspectives on fast-neutron mutagenesis of human lymphoblastoid cells. Radiat. Res. 1991;128(1 Suppl):S87–S93. [PubMed] [Google Scholar]
- Kusunoki Y, Hayashi T. Long-lasting alterations of the immune system by ionizing radiation exposure: implications for disease development among atomic bond survivors. Int. J. Radiat. Biol. 2008;84(1):1–14. doi: 10.1080/09553000701616106. [DOI] [PubMed] [Google Scholar]
- Land H, Parada LF, Weinberg RA. Cellular oncogenes and multistep carcinogenesis. Science. 1983;222(4625):771–778. doi: 10.1126/science.6356358. [DOI] [PubMed] [Google Scholar]
- Lapidot T, Sirard C, Vormoor J, Murdoch B, Hoang T, Caceres-Cortes J, et al. A cell initiating human acute myeloid leukaemia after transplantation into SCID mice. Nature. 1994;367(6464):645–648. doi: 10.1038/367645a0. [DOI] [PubMed] [Google Scholar]
- Laurent C, Leduc A, Pottier I, Prevost V, Sichel F, Lefaix JL. Dramatic increase in oxidative stress in carbon-irradiated normal human skin fibroblasts. PLoS One. 2013;8(12):e85158. doi: 10.1371/journal.pone.0085158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leatherbarrow EL, Harper JV, Cucinotta FA, O’Neill P. Induction and quantification of gamma-H2AX foci following low and high-LET-irradiation. Int. J. Radiat. Biol. 2006;82(2):111–118. doi: 10.1080/09553000600599783. [DOI] [PubMed] [Google Scholar]
- Lee HC, An S, Lee H, Woo SH, Jin HO, Seo SK, Choe TB, Yoo DH, Lee SJ, Hong YJ, Park MJ, Rhee CH, Park IC, Hong SI. Activation of epidermal growth factor receptor and its downstream signaling pathway by nitric oxide in response to ionizing radiation. Mol. Cancer Res. 2008;6(6):996–1002. doi: 10.1158/1541-7786.MCR-08-0113. [DOI] [PubMed] [Google Scholar]
- Lee SS, Bohrson C, Pike AM, Wheelan SJ, Greider CW. ATM Kinase is required for telomere elongation in mouse and human cells. Cell Rep. 2015;13(8):1623–1632. doi: 10.1016/j.celrep.2015.10.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lengauer C, Kinzler KW, Vogelstein B. Genetic instabilities in human cancers. Nature. 1998;396(6712):643–649. doi: 10.1038/25292. [DOI] [PubMed] [Google Scholar]
- Leteurtre F, Li X, Gluckman E, Carosella ED. Telomerase activity during the cell cycle and in gamma-irradiated hematopoietic cells. Leukemia. 1997;11(10):1681–1689. doi: 10.1038/sj.leu.2400784. [DOI] [PubMed] [Google Scholar]
- Li L, Neaves WB. Normal stem cells and cancer stem cells: the niche matters. Cancer Res. 2006;66(9):4553–4557. doi: 10.1158/0008-5472.CAN-05-3986. [DOI] [PubMed] [Google Scholar]
- Li M, Gonon G, Buonanno M, Autsavapromporn N, de Toledo SM, Pain D, et al. Health risks of space exploration: targeted and nontargeted oxidative injury by high-charge and high-energy particles. Antioxid. Redox Signal. 2014;20(9):1501–1523. doi: 10.1089/ars.2013.5649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Doho G, Zheng X, Jella KK, Li S, Wang Y, et al. Co-culturing with high-charge and energy particle irradiated cells increases mutagenic joining of enzymatically induced DNA double-strand breaks in nonirradiated cells. Radiat. Res. 2015;184(3):249–258. doi: 10.1667/RR14092.1. [DOI] [PubMed] [Google Scholar]
- Li H-H, Wang Y-W, Chen R, Zhou B, Ashwell JD, Fornace AL., Jr Ionizing radiation impairs T cell activation by affecting metabolic reprogramming. Int. J. Biol. Sci. 2015;11(7):726–736. doi: 10.7150/ijbs.12009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang L, Deng L, Mendonca MS, Chen Y, Zheng B, Stambrook PJ, et al. X-rays induce distinct patterns of somatic mutation in fetal versus adult hematopoietic cells. DNA Repair. 2007;6(9):1380–1385. doi: 10.1016/j.dnarep.2007.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liber HL, Leong PM, Terry VH, Little JB. X-rays mutate human lymphoblast cells at genetic loci that should respond only to point mutagens. Mutat. Res. 1986;163(1):91–97. doi: 10.1016/0027-5107(86)90062-x. [DOI] [PubMed] [Google Scholar]
- Liber HL, Yandell DW, Little JB. A comparison of mutation induction at the tk and hprt loci in human lymphoblastoid cells; quantitative differences are due to an additional class of mutations at the autosomal tk locus. Mutat. Res. 1989;216(1):9–17. doi: 10.1016/0165-1161(89)90018-6. [DOI] [PubMed] [Google Scholar]
- Liber HL, Idate R, Warner C, Bailey SM. Radiation quality and mutagenesis in human lymphoblastoid cells. Radiat. Res. 2014;182(4):390–395. doi: 10.1667/RR13817.1. [DOI] [PubMed] [Google Scholar]
- Limoli CL, Giedzinski E, Baure J, Rola R, Fike JR. Redox changes induced in hippocampal precursor cells by heavy ion irradiation. Radiat. Environ. Biophys. 2007;46(2):167–172. doi: 10.1007/s00411-006-0077-9. [DOI] [PubMed] [Google Scholar]
- Lindsay KJ, Coates PJ, Lorimore SA, Wright EG. The genetic basis of tissue responses to ionizing radiation. Br. J. Radiol. 2007;80(Special Issue 1):S2–S6. doi: 10.1259/bjr/60507340. [DOI] [PubMed] [Google Scholar]
- Liu JC, Lerou PH, Lahav G. Stem cells: balancing resistance and sensitivity to DNA damage. Trends Cell Biol. 2014;24(5):268–274. doi: 10.1016/j.tcb.2014.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lobrich M, Jeggo PA. The impact of a negligent G2/M checkpoint on genomic instability and cancer induction. Nat. Rev. Cancer. 2007;7(11):861–869. doi: 10.1038/nrc2248. [DOI] [PubMed] [Google Scholar]
- Lobrich M, Rief N, Kuhne M, Heckmann M, Fleckenstein J, Rube C, et al. In vivo formation and repair of DNA double-strand breaks after computed tomography examinations. Proc. Natl. Acad. Sci. USA. 2005;102(25):8984–8989. doi: 10.1073/pnas.0501895102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lobrich M, Shibata A, Beucher A, Fisher A, Ensminger M, Goodarzi AA, et al. gamma H2AX foci analysis for monitoring DNA double-strand break repair: strengths, limitations and optimization. Cell Cycle. 2010;9(4):662–669. doi: 10.4161/cc.9.4.10764. [DOI] [PubMed] [Google Scholar]
- Loft S, Svoboda P, Kawai K, Kasai H, Sorensen M, Tjonneland A, et al. Association between 8-oxo-7,8-dihydroguanine excretion and risk of lung cancer in a prospective study. Free Radic. Biol. Med. 2012;52(1):167–172. doi: 10.1016/j.freeradbiomed.2011.10.439. [DOI] [PubMed] [Google Scholar]
- Loft S, Olsen A, Moller P, Poulsen HE, Tjonneland A. Association between 8-oxo-7,8-dihydro-2′-deoxyguanosine excretion and risk of postmenopausal breast cancer: nested case-control study. Cancer Epidemiol. Biomark. Prev. 2013;22(7):1289–1296. doi: 10.1158/1055-9965.EPI-13-0229. [DOI] [PubMed] [Google Scholar]
- Lorat Y, Brunner CU, Schanz S, Jakob B, Taucher-Scholz G, Rube CE. Nanoscale analysis of clustered DNA damage after high-LET irradiation by quantitative electron microscopy–the heavy burden to repair. DNA Repair. 2015;28:93–106. doi: 10.1016/j.dnarep.2015.01.007. [DOI] [PubMed] [Google Scholar]
- Loucas BD, Cornforth MN. Complex chromosome exchanges induced by gamma rays in human lymphocytes: an mFISH study. Radiat. Res. 2001;155(5):660–671. doi: 10.1667/0033-7587(2001)155[0660:cceibg]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- Lu TP, Lee JM, Hsu CP, et al. Identification of a novel biomarker, SEMA5A, for non-small cell lung carcinoma in nonsmoking women. Cancer Epid. Biomarkers Prev. 2010;19(10):2590–2597. doi: 10.1158/1055-9965.EPI-10-0332. [DOI] [PubMed] [Google Scholar]
- Maeda T, Nakamura K, Atsumi K, Hirakawa M, Ueda Y, Makino N. Radiation-associated changes in the length of telomeres in peripheral leukocytes from inpatients with cancer. Int. J. Radiat. Biol. 2013;89(2):106–109. doi: 10.3109/09553002.2013.734945. [DOI] [PubMed] [Google Scholar]
- Mah LJ, El-Osta A, Karagiannis TC. Gamma H2AX as a molecular marker of aging and disease. Epigenetics. 2010;5(2):129–136. doi: 10.4161/epi.5.2.11080. [DOI] [PubMed] [Google Scholar]
- Mantovani AP, Sica A, Balkwill F. Cancer-related inflammation. Nature. 2008;454(7203):436–444. doi: 10.1038/nature07205. [DOI] [PubMed] [Google Scholar]
- Mao XW, Pecaut MJ, Stodieck LS, Ferguson VL, Bateman TA, Bouxsein M, et al. Spaceflight environment induces mitochondrial oxidative damage in ocular tissue. Radiat. Res. 2013;180(4):340–350. doi: 10.1667/RR3309.1. [DOI] [PubMed] [Google Scholar]
- Mao Xw, Pecaut MJ, Cao JD, Moldovan M, Gridley DS. Low-dose radiation modifies skin response to acute gamma-rays and protons. In vivo. 2013;27(6):695–700. [PubMed] [Google Scholar]
- Mao XW, Pecaut MJ, Stodieck LS, Ferguson VL, Bateman TA, Bouxsein ML, et al. Biological and metabolic response in STS-135 space-flown mouse skin. Free Radic. Res. 2014;48(8):890–897. doi: 10.3109/10715762.2014.920086. [DOI] [PubMed] [Google Scholar]
- Margaritelis NV, Veskoukis AS, Paschalis V, Vrabas IS, Dipla K, Zafeiridis A, et al. Blood reflects tissue oxidative stress: a systematic review. Biomarkers. 2015;20(2):97–108. doi: 10.3109/1354750X.2014.1002807. [DOI] [PubMed] [Google Scholar]
- Markevich NI, Hoek JB. Computational modeling analysis of mitochondrial superoxide production under varying substrate conditions and upon inhibition of different segments of the electron transport chain. Biochim. Biophys. Acta. 2015;1847(6–7):656–679. doi: 10.1016/j.bbabio.2015.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markova E, Schultz N, Belyaev IY. Kinetics and dose-response of residual 53BP1/gamma-H2AX foci: co-localization, relationship with DSB repair and clonogenic survival. Int. J. Radiat. Biol. 2007;83(5):319–329. doi: 10.1080/09553000601170469. [DOI] [PubMed] [Google Scholar]
- Matsuyama T, Ishikawa T, Mogushi K, Yoshida T, et al. MUC12 mRNA expression is an independent marker of prognosis in stage II and stage III colorectal cancer. Int. J. Cancer. 2010;127(10):2292–2299. doi: 10.1002/ijc.25256. [DOI] [PubMed] [Google Scholar]
- McClintock B. the stability of broken ends of chromosomes in zea mays. Genetics. 1941;26(2):234–282. doi: 10.1093/genetics/26.2.234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McEllin B, Camacho CV, Mukherjee B, Hahm B, Tomimatsu N, Bachoo RM, et al. PTEN loss compromises homologous recombination repair in astrocytes: implications for glioblastoma therapy with temozolomide or poly(ADP-ribose) polymerase inhibitors. Cancer Res. 2010;70(13):5457–5464. doi: 10.1158/0008-5472.CAN-09-4295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meador JA, Ghandhi SA, Amundson SA. p53-independent downregulation of histone gene expression in human cell lines by high- and low-let radiation. Radiat. Res. 2011;175(6):689–699. doi: 10.1667/rr2539.1. [DOI] [PubMed] [Google Scholar]
- Miranda-Lorenzo I, Dorado J, Lonardo E, Alcala S, Serrano AG, Clausell-Tormos J, et al. Intracellular autofluorescence: a biomarker for epithelial cancer stem cells. Nat. Methods. 2014;11(11):1161–1169. doi: 10.1038/nmeth.3112. [DOI] [PubMed] [Google Scholar]
- Mezentsev A, Amundson SA. Global gene expression responses to low- or high-dose radiation in a human three-dimensional tissue model. Radiat Res. 2011;175(6):677–688. doi: 10.1667/RR2483.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Micke P, Edlund K, Holmberg L, Kultima HG, et al. Gene copy number aberrations are associated with survival in histologic subgroups of non-small cell lung cancer. J. Thoracic Oncology. 2011;6(11):1833–1844. doi: 10.1097/JTO.0b013e3182295917. [DOI] [PubMed] [Google Scholar]
- Mitelman F, Mertens F, Johansson B. Prevalence estimates of recurrent balanced cytogenetic aberrations and gene fusions in unselected patients with neoplastic disorders. Genes Chromosome Cancer. 2005;43(4):350–366. doi: 10.1002/gcc.20212. [DOI] [PubMed] [Google Scholar]
- Mitteer AR, Wang Y, Shah J, Gordon S, Fager M, Butter PP, et al. Proton beam radiation induces DNA damage and cell apoptosis in glioma stem cells through reactive oxygen species. Sci. Rep. 2015;5:13961. doi: 10.1038/srep13961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohsin Ali M, Kurisu S, Yoshioka Y, Terato H, Ohyama Y, Kubo K, et al. Detection of endonuclease III- and 8-oxoguanine glycosylase-sensitive base modifications in gamma-irradiated DNA and cells by the aldehyde reactive probe (ARP) assay. J. Radiat. Res. 2004;45(2):229–237. doi: 10.1269/jrr.45.229. [DOI] [PubMed] [Google Scholar]
- Morgan WF, Sowa MB. Non-targeted effects induced by ionizing radiation: mechanisms and potential impact on radiation induced health effects. Cancer Lett. 2015;356(1):17–21. doi: 10.1016/j.canlet.2013.09.009. [DOI] [PubMed] [Google Scholar]
- Moroni M, Maeda D, Whitnall MH, Bonner WM, Redon CE. Evaluation of the gamma-H2AX assay for radiation biodosimetry in a swine model. Int. J. Mol. Sci. 2013;14(7):14119–14135. doi: 10.3390/ijms140714119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller MM, Fusenig NE. Tumor-stroma interactions directing phenotype and progression of epithelial skin tumor cells. Differ.: Res. Biol. Divers. 2002;70(9–10):486–497. doi: 10.1046/j.1432-0436.2002.700903.x. [DOI] [PubMed] [Google Scholar]
- Muller HJ. Artificial transmutation of the gene. Science. 1927;66(1699):84–87. doi: 10.1126/science.66.1699.84. [DOI] [PubMed] [Google Scholar]
- NCRP. Uncertainties in the Estimation of Radiation Risks and Probability of Disease Causation. Bethesda MD: 2012. NCRP report no. 171. [Google Scholar]
- Nelms BE, Maser RS, MacKay JF, Lagally MG, Petrini JH. In situ visualization of DNA double-strand break repair in human fibroblasts. Science. 1998;280(5363):590–592. doi: 10.1126/science.280.5363.590. [DOI] [PubMed] [Google Scholar]
- Neuhof D, Ruess A, Wenz F, Weber KJ. Induction of telomerase activity by irradiation in human lymphoblasts. Radiat. Res. 2001;155(5):693–697. doi: 10.1667/0033-7587(2001)155[0693:iotabi]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- Neumaier T, Swenson J, Pham C, Polyzos A, Lo AT, Yang P, et al. Evidence for formation of DNA repair centers and dose-response nonlinearity in human cells. Proc. Natl. Acad. Sci. USA. 2012;109(2):443–448. doi: 10.1073/pnas.1117849108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neveu B, Moreel X, Deschênes-Rompré M-P, LaRue H, Ayari C, Fradet Y, et al. IL-8 secretion in primary cultures of prostate cells is associated with prostate cancer aggressiveness. Res. Rep. Urol. 2013;6:27–34. doi: 10.2147/RRU.S58643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nieri D, Berardinelli F, Sgura A, Cherubini R, De Nadal V, Gerardi S, et al. Cyogenetics effects in AG01522 human primary fibroblasts exposed to low doses of radiations with different quality. Int. J. Radiat. Biol. 2013;89(9):698–707. doi: 10.3109/09553002.2013.797126. [DOI] [PubMed] [Google Scholar]
- Nikitaki Z, Hellweg CE, Georgakilas AG, Ravanat JL. Stress-induced DNA damage biomarkers: applications and limitations. Front. Chem. 2015;3:35. doi: 10.3389/fchem.2015.00035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikjoo H, O’Neill P, Wilson WE, Goodhead DT. Computational approach for determining the spectrum of DNA damage induced by ionizing radiation. Radiat. Res. 2001;156(5 Pt 2):577–583. doi: 10.1667/0033-7587(2001)156[0577:cafdts]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- Ojima M, Hamano H, Suzuki M, Suzuki K, Kodama S, Watanabe M. Delayed induction of telomere instability in normal human fibroblast cells by ionizing radiation. J. Radiat. Res. 2004;45(1):105–110. doi: 10.1269/jrr.45.105. [DOI] [PubMed] [Google Scholar]
- Okayasu R, Okada M, Okabe A, Noguchi M, Takakura K, Takahashi S. Repair of DNA damage induced by accelerated heavy ions in mammalian cells proficient and deficient in the non-homologous end-joining pathway. Radiat Res. 2006;165(1):59–67. doi: 10.1667/rr3489.1. [DOI] [PubMed] [Google Scholar]
- Olofsson P, Kimmel M. Stochastic models of telomere shortening. Math. Biosci. 1999;158(1):75–92. doi: 10.1016/s0025-5564(98)10092-5. [DOI] [PubMed] [Google Scholar]
- Pajonk F, Vlashi E, McBride WH. Radiation resistance of cancer stem cells: the 4 R’s of radiobiology revisited. Stem Cells. 2010;28(4):639–648. doi: 10.1002/stem.318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palayoor ST, ohn-Aryankalayil J, Makinde AY, Falduto MT, Magnuson SR, Cole-man CN. Differential expression of stress and immune response pathway transcripts and miRNAs in normal human endothelial cells subjected to fractionated or single-dose radiation. Mol. Cancer Res. 2014;12(7):1002–1015. doi: 10.1158/1541-7786.MCR-13-0623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panajotovic R, Martin F, Cloutier P, Hunting D, Sanche L. Effective cross sections for production of single-strand breaks in plasmid DNA by 0.1 to 4.7 eV electrons. Radiat. Res. 2006;165(4):452–459. doi: 10.1667/rr3521.1. [DOI] [PubMed] [Google Scholar]
- Pang D, Winters TA, Jung M, Purkayastha S, Cavalli LR, Chasovkikh S, et al. Radiation-generated short DNA fragments may perturb non-homologous end-joining and induce genomic instability. J. Radiat. Res. 2011;52(3):309–319. doi: 10.1269/jrr.10147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panganiban R-AM, Snow AL, Day RM. Mechanisms of radiation toxicity in transformed and non-transformed cells. Int. J. Mol. Sci. 2013;14(8):15931–15958. doi: 10.3390/ijms140815931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parihar VK, Allen BD, Tran KK, Chmielewski NN, Craver BM, Martirosian V, et al. Targeted overexpression of mitochondrial catalase prevents radiation-induced cognitive dysfunction. Antioxid. Redox Signal. 2015;22(1):78–91. doi: 10.1089/ars.2014.5929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park CH, Bergsagel DE, McCulloch EA. Mouse myeloma tumor stem cells: a primary cell culture assay. J. Natl. Cancer Inst. 1971;46(2):411–422. [PubMed] [Google Scholar]
- Passos JF, Saretzki G, von Zglinicki T. DNA damage in telomeres and mitochondria during cellular senescence: is there a connection? Nucleic Acids Res. 2007;35(22):7505–7513. doi: 10.1093/nar/gkm893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul S, Amundson SA. Development of gene expression signatures for practical radiation biodosimetry. Int. J. Radiat. Oncol. Biol. Phys. 2008;71(4):1236–1244. doi: 10.1016/j.ijrobp.2008.03.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul S, Smilenov LB, Elliston CD, Amundson SA. Radiation dose-rate effects on gene expression in a mouse biodosimetry model. Radiat. Res. 2015;184(1):24–32. doi: 10.1667/RR14044.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paull TT, Rogakou EP, Yamazaki V, Kirchgessner CU, Gellert M, Bonner WM. A critical role for histone H2AX in recruitment of repair factors to nuclear foci after DNA damage. Curr. Biol. 2000;10(15):886–895. doi: 10.1016/s0960-9822(00)00610-2. [DOI] [PubMed] [Google Scholar]
- Pawel DJ, Puskin JS. U.S. environmental protection agency radiogenic risk models and projections for the U.S. population. Health Phys. 2012;102:646–656. [Google Scholar]
- Pawel DJ. U.S. Environmental Protection Agency Radiogenic Risk Projections: Uncertainty Analysis. Health Phys. 2013;103:26–40. doi: 10.1097/HP.0b013e31826119ed. [DOI] [PubMed] [Google Scholar]
- Pecaut MJ, Gridley DS. Impact of head-only iron ion radiation on the peripheral LPS response. In vivo. 2011;25:903–911. [PubMed] [Google Scholar]
- Pawitan Y, Bjohle J, Amler L, Borg AL, et al. Gene expression profiling spares early breast cancer patients from adjuvant therapy: derived and validated in two population-based cohorts. Breast Cancer Res. 2005;7(6):R953–R964. doi: 10.1186/bcr1325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng Y, Brown N, Finnon R, Warner CL, Liu X, Genik PC, et al. Radiation leukemogenesis in mice: loss of PU.1 on chromosome 2 in CBA and C57BL/6 mice after irradiation with 1 GeV/nucleon 56Fe ions, X rays or gamma rays. Part I. Experimental observations. Radiat. Res. 2009;171(4):474–483. doi: 10.1667/RR1547.1. [DOI] [PubMed] [Google Scholar]
- Pereira S, Malard V, Ravanat JL, Davin AH, Armengaud J, Foray N, et al. Low doses of gamma-irradiation induce an early bystander effect in zebrafish cells which is sufficient to radioprotect cells. PLoS One. 2014;9(3):e92974. doi: 10.1371/journal.pone.0092974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Caro M, Cobaleda C, Gonzalez-Herrero I, Vicente-Duenas C, Bermejo-Rodriguez C, Sanchez-Beato M, et al. Cancer induction by restriction of oncogene expression to the stem cell compartment. EMBO J. 2009;28(1):8–20. doi: 10.1038/emboj.2008.253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pernot E, Hall J, Baatout S, Benotmane M. Ionizing radiation biomarkers for potential use in epidemiological studies. Mutat. Res. 2012;751(2):258–286. doi: 10.1016/j.mrrev.2012.05.003. [DOI] [PubMed] [Google Scholar]
- Petryszak R, Burdett T, Fiorelli B, Fonseca Nuno A, Gonzalez-Porta M, Hastings E, et al. Expression Atlas update–a database of gene and transcript expression from microarray- and sequencing-based functional genomics experiments. Nucleic Acids Res. 2014;42(Database issue):D926–D932. doi: 10.1093/nar/gkt1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piccolo SR, Sun Y, Campbell JD, Lenburg ME, Bild AH, Johnson WE. A single-sample microarray normalization method to facilitate personalized-medicine workflows. Genomics. 2012;100(6):337–344. doi: 10.1016/j.ygeno.2012.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pico AR, Kelder T, Van Iersel MP, Hanspers K, Conklin BR, Evelo C. WikiPathways: pathway editing for the people. PLoS Biol. 2008;6(7):e184. doi: 10.1371/journal.pbio.0060184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plante I, Cucinotta FA. Simulation of the radiolysis of water using Green’s functions of the diffusion equation. Radiat. Prot. Dosim. 2015;166(1–4):24–28. doi: 10.1093/rpd/ncv179. [DOI] [PubMed] [Google Scholar]
- Plante I, Ponomarev AL, Cucinotta FA. Calculation of the energy deposition in nanovolumes by protons and HZE particles: geometric patterns of initial distributions of DNA repair foci. Phys. Med. Biol. 2013;58(18):6393–6405. doi: 10.1088/0031-9155/58/18/6393. [DOI] [PubMed] [Google Scholar]
- Ponnaiya B, Jenkins-Baker G, Randers-Pherson G, Geard CR. Quantifying a bystander response following microbeam irradiation using single-cell RT-PCR analyses. Exp. Hematol. 2007;35(4 Suppl 1):64–68. doi: 10.1016/j.exphem.2007.01.013. [DOI] [PubMed] [Google Scholar]
- Ponomarev AL, George K, Cucinotta FA. Generalized time-dependent model of radiation-induced chromosomal aberrations in normal and repair-deficient human cells. Radiat. Res. 2014;181(3):284–292. doi: 10.1667/RR13303.1. [DOI] [PubMed] [Google Scholar]
- Ponomareva ON, Rose JA, Lasarev M, Rasey J, Turker MS. Tissue-specific deletion and discontinuous loss of heterozygosity are signatures for the mutagenic effects of ionizing radiation in solid tissues. Cancer Res. 2002;62(5):1518–1523. [PubMed] [Google Scholar]
- Prestwich RJ, Errington E, Hatfield P, Merrick AE, Selby PJ, Melcher A. The immune system — is it relevant to cancer development, progression and treatment? Clin. Oncol. 2008;20(2):100–112. doi: 10.1016/j.clon.2007.10.011. [DOI] [PubMed] [Google Scholar]
- Purvis JE, Karhohs KW, Mock C, Batchelor E, Loewer A, Lahav G. p53 dynamics control cell fate. Science. 2012;336(6087):1440–1444. doi: 10.1126/science.1218351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin XQ, Livingston DM, Kaelin WG, Jr, Adams PD. Deregulated transcription factor E2F-1 expression leads to S-phase entry and p53-mediated apoptosis. Proc. Natl. Acad. Sci. USA. 1994;91(23):10918–10922. doi: 10.1073/pnas.91.23.10918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rödel F, Frey B, Gaipl U, Keilholz L, Fournier C, Manda K, et al. Modulation of inflammatory immune reactions by low-dose ionizing radiation: molecular mechanisms and clinical application. Curr. Med. Chem. 2012;19(12):1741–1750. doi: 10.2174/092986712800099866. [DOI] [PubMed] [Google Scholar]
- Ryan JL. Ionizing Radiation, The good, the bad, and the ugly. J. Investig. Dermatol. 2012;132:985–993. doi: 10.1038/jid.2011.411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravanat JL, Cadet J, Douki T. Oxidatively generated DNA lesions as potential biomarkers of in vivo oxidative stress. Curr. Mol. Med. 2012;12(6):655–671. doi: 10.2174/156652412800792651. [DOI] [PubMed] [Google Scholar]
- Reczek CR, Chandel NS. ROS-dependent signal transduction. Curr. Opin. Cell Biol. 2015;33:8–13. doi: 10.1016/j.ceb.2014.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redon CE, Dickey JS, Bonner WM, Sedelnikova OA. gamma-H2AX as a biomarker of DNA damage induced by ionizing radiation in human peripheral blood lymphocytes and artificial skin. Adv. Space Res. 2009;43(8):1171–1178. doi: 10.1016/j.asr.2008.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redon CE, Nakamura AJ, Gouliaeva K, Rahman A, Blakely WF, Bonner WM. The use of gamma-H2AX as a biodosimeter for total-body radiation exposure in non-human primates. PLoS One. 2010;5(11):e15544. doi: 10.1371/journal.pone.0015544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redpath JL, Gutierrez M. Kinetics of induction of reactive oxygen species during the post-irradiation expression of neoplastic transformation in vitro. Int. J. Radiat. Biol. 2001;77(11):1081–1085. doi: 10.1080/09553000110065807. [DOI] [PubMed] [Google Scholar]
- Reid DA, Keegan S, Leo-Macias A, Watanabe G, Strande NT, Chang HH, et al. Organization and dynamics of the nonhomologous end-joining machinery during DNA double-strand break repair. Proc. Natl. Acad. Sci. USA. 2015;112(20):E2575–E2584. doi: 10.1073/pnas.1420115112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rieger KE, Chu G. Portrait of transcriptional responses to ultraviolet and ionizing radiation in human cells. Nucl. Acids Res. 2004;32(16):4786–4803. doi: 10.1093/nar/gkh783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reste J, Zvigule G, Zvagule T, Kurjane N, Eglite M, Gabruseva N, et al. Telomere length in Chernobyl accident recovery workers in the late period after the disaster. J. Radiat. Res. 2014;55(6):1089–1100. doi: 10.1093/jrr/rru060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reya T, Morrison SJ, Clarke MF, Weissman IL. Stem cells, cancer, and cancer stem cells. Nature. 2001;414(6859):105–111. doi: 10.1038/35102167. [DOI] [PubMed] [Google Scholar]
- Reynolds P, Anderson JA, Harper JV, Hill MA, Botchway SW, Parker AW, et al. The dynamics of Ku70/80 and DNA-PKcs at DSBs induced by ionizing radiation is dependent on the complexity of damage. Nucleic Acids Res. 2012;40(21):10821–10831. doi: 10.1093/nar/gks879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ridker PM, Rifai N, Stampfer MJ, Hennekens CH. Plasma concentration of interleukin-6 and the risk of future myocardial infarction among apparently healthy men. Circulation. 2000;101:1767–1772. doi: 10.1161/01.cir.101.15.1767. [DOI] [PubMed] [Google Scholar]
- Riecke A, Rufa CG, Cordes M, Hartmann J, Meineke V, Abend M. Gene expression comparisons performed for biodosimetry purposes on in vitro peripheral blood cellular subsets and irradiated individuals. Radiat. Res. 2012;178(3):234–243. doi: 10.1667/rr2738.1. [DOI] [PubMed] [Google Scholar]
- Rizzo AM, Corsetto PA, Montorfano G, Milani S, Zava S, Tavella S, et al. Effects of long-term space flight on erythrocytes and oxidative stress of rodents. PLoS One. 2012;7(3):e32361. doi: 10.1371/journal.pone.0032361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roadmap NHRP. [accessed 1/26/2016]; https://humanresearchroadmap.nasa.gov/
- Roesch A, Fukunaga-Kalabis M, Schmidt EC, Zabierowski SE, Brafford PA, Vultur A, et al. A temporarily distinct subpopulation of slow-cycling melanoma cells is required for continuous tumor growth. Cell. 2010;141(4):583–594. doi: 10.1016/j.cell.2010.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogakou EP, Pilch DR, Orr AH, Ivanova VS, Bonner WM. DNA double-stranded breaks induce histone H2AX phosphorylation on serine 139. J. Biol. Chem. 1998;273(10):5858–5868. doi: 10.1074/jbc.273.10.5858. [DOI] [PubMed] [Google Scholar]
- Roos WP, Kaina B. DNA damage-induced cell death by apoptosis. Trends Mol. Med. 2006;12(9):440–450. doi: 10.1016/j.molmed.2006.07.007. [DOI] [PubMed] [Google Scholar]
- Rothkamm K, Lobrich M. Evidence for a lack of DNA double-strand break repair in human cells exposed to very low x-ray doses. Proc. Natl. Acad. Sci. USA. 2003;100(9):5057–5062. doi: 10.1073/pnas.0830918100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothkamm K, Kruger I, Thompson LH, Lobrich M. Pathways of DNA double-strand break repair during the mammalian cell cycle. Mol. Cell. Biol. 2003;23(16):5706–5715. doi: 10.1128/MCB.23.16.5706-5715.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy D, Guida P, Zhou G, Echiburu-Chau C, Calaf GM. Gene expression profiling of breast cells induced by X-rays and heavy ions. Int. J. Mol. Med. 2008;21(5):627–636. doi: 10.3892/ijmm.21.5.627. [DOI] [PubMed] [Google Scholar]
- Roy L, Gruel G, Vaurijoux A. Cell response to ionising radiation analysed by gene expression patterns. Ann. dell’Istituto Super. Sanita. 2009;45(3):272–277. [PubMed] [Google Scholar]
- Rubelj I, Vondracek Z. Stochastic mechanism of cellular aging–abrupt telomere shortening as a model for stochastic nature of cellular aging. J. Theor. Biol. 1999;197(4):425–438. doi: 10.1006/jtbi.1998.0886. [DOI] [PubMed] [Google Scholar]
- Runge R, Hiemann R, Wendisch M, Kasten-Pisula U, Storch K, Zophel K, et al. Fully automated interpretation of ionizing radiation-induced gamma H2AX foci by the novel pattern recognition system AKLIDES(R) Int. J. Radiat. Biol. 2012;88(5):439–447. doi: 10.3109/09553002.2012.658468. [DOI] [PubMed] [Google Scholar]
- Russell LB, Hunsicker PR. The effect of dose rate on the frequency of specific-locus mutations induced in mouse spermatogonia is restricted to larger lesions; a retrospective analysis of historical data. Radiat. Res. 2012;177(5):555–564. doi: 10.1667/rr2853.1. [DOI] [PubMed] [Google Scholar]
- Russell WL, Kelly EM. Mutation frequencies in male mice and the estimation of genetic hazards of radiation in men. Proc. Natl. Acad. Sci. USA. 1982;79(2):542–544. doi: 10.1073/pnas.79.2.542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell WL, Russell LB, Kelly EM. Radiation dose rate and mutation frequency. Science. 1958;128(3338):1546–1550. doi: 10.1126/science.128.3338.1546. [DOI] [PubMed] [Google Scholar]
- Sachs RK, Hahnfeld P, Brenner D. The link between low-LET dose-response relations and the underlying kinetics of damage production/repair/misrepair. Int. J. Radiat. Biol. 1997;72:351–374. doi: 10.1080/095530097143149. [DOI] [PubMed] [Google Scholar]
- Saha J, Wang M, Cucinotta FA. Investigation of switch from ATM to ATR signaling at the sites of DNA damage induced by low and high-LET radiation. DNA Repair. 2013;12(12):1143–1151. doi: 10.1016/j.dnarep.2013.10.004. [DOI] [PubMed] [Google Scholar]
- Saha J, Wilson P, Thieberger P, Lowenstein D, Wang M, Cucinotta FA. Biological characterization of low-energy ions with high-energy deposition on human cells. Radiat. Res. 2014;182(3):282–291. doi: 10.1667/RR13747.1. [DOI] [PubMed] [Google Scholar]
- Sahin E, Colla S, Liesa M, Moslehi J, Muller FL, Guo M, et al. Telomere dysfunction induces metabolic and mitochondrial compromise. Nature. 2011;470(7334):359–365. doi: 10.1038/nature09787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sak A, Stuschke M. Use of gammaH2AX and other biomarkers of double-strand breaks during radiotherapy. Semin. Radiat. Oncol. 2010;20(4):223–231. doi: 10.1016/j.semradonc.2010.05.004. [DOI] [PubMed] [Google Scholar]
- Sakaguchi N, Miyai K, Sakaguchi S. onizing radiation and autoimmunity. Induction of autoimmune disease in mice by high dose fractionated total lymphoid irradiation and its prevention by inoculating normal T cells. J. Immunol. 1994;152(5):2586–2595. [PubMed] [Google Scholar]
- Salin K, Auer SK, Rudolf AM, Anderson GJ, Cairns AG, Mullen W, et al. Individuals with higher metabolic rates have lower levels of reactive oxygen species in vivo. Biol. Lett. 2015;11(9):20150538. doi: 10.1098/rsbl.2015.0538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sancar A, Lindsey-Boltz LA, Unsal-Kacmaz K, Linn S. Molecular mechanisms of mammalian DNA repair and the DNA damage checkpoints. Annu. Rev. Biochem. 2004;73:39–85. doi: 10.1146/annurev.biochem.73.011303.073723. [DOI] [PubMed] [Google Scholar]
- Sanzari JK, Wan X, Krigsfeld G, Wroe AJ, Gridley DS, Kennedy AR. The effects of gamma and proton radiation exposure on hematopoietic cell counts in the ferret model. Gravity Space Res. 2013;1(1):79–94. [PMC free article] [PubMed] [Google Scholar]
- Sawant SG, Randers-Pehrson G, Geard CR, Brenner DJ, Hall EJ. The by-stander effect in radiation oncogenesis: I. Transformation in C3H 10T1/2 cells in vitro can be initiated in the unirradiated neighbors of irradiated cells. Radiat. Res. 2001;155(3):397–401. doi: 10.1667/0033-7587(2001)155[0397:tbeiro]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- Schiestl RH, Khogali F, Carls N. Reversion of the mouse pink-eyed unstable mutation induced by low doses of x-rays. Science. 1994;266(5190):1573–1576. doi: 10.1126/science.7985029. [DOI] [PubMed] [Google Scholar]
- Seed TM. hematopoietic tissue repair under chronic low dose irradiation. Adv. Space Res. 1996;18(1–2):65–70. doi: 10.1016/0273-1177(95)00792-d. [DOI] [PubMed] [Google Scholar]
- Sehl ME, Shimada M, Landeros A, Lange K, Wicha MS. Modeling of cancer stem cell state transitions predicts therapeutic response. PLoS One. 2015;10(9):e0135797. doi: 10.1371/journal.pone.0135797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sergei I, Grivennikov FRG, 2, Michael, Karin Immunity, inflammation and cancer. Cell. 2010;140(6):883–889. doi: 10.1016/j.cell.2010.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sgura A, Antoccia A, Berardinelli F, Cherubini R, Gerardi S, Zilio C, et al. Telomere length in mammalian cells exposed to low- and high-LET radiations. Radiat. Prot. Dosim. 2006;122(1–4):176–179. doi: 10.1093/rpd/ncl478. [DOI] [PubMed] [Google Scholar]
- Shah DJ, Sachs RK, Wilson DJ. Radiation-induced cancer: a modern view. Br. J. Radiol. 2012;85(1020):e1166–e73. doi: 10.1259/bjr/25026140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheridan C, Kishimoto H, Fuchs RK, Mehrotra S, Bhat-Nakshatri P, Turner CH, et al. CD44+/CD24− breast cancer cells exhibit enhanced invasive properties: an early step necessary for metastasis. Breast Cancer Res. 2006;8(5) doi: 10.1186/bcr1610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shikazono N, Noguchi M, Fujii K, Urushibara A, Yokoya A. The yield, processing, and biological consequences of clustered DNA damage induced by ionizing radiation. J. Radiat. Res. 2009;50(1):27–36. doi: 10.1269/jrr.08086. [DOI] [PubMed] [Google Scholar]
- Shina SC, Leeb K-M, Kanga YM, Kimb K, Limb SA, Yanga KH, et al. Differential expression of immune-associated cancer regulatory genes in low- versus high-dose-rate irradiated AKR/J mice. Genomics. 2011;97(6):358–363. doi: 10.1016/j.ygeno.2011.01.005. [DOI] [PubMed] [Google Scholar]
- Singh SK, Clarke ID, Terasaki M, Bonn VE, Hawkins C, Squire J, et al. Identification of a cancer stem cell in human brain tumors. Cancer Res. 2003;63(18):5821–5828. [PubMed] [Google Scholar]
- Singh AK, Chen J, Calado R, Sowers A, Mitchell JB, Barrett AJ. Total body irradiation with lung dose-reduction does not improve hematopoietic cell homing to bone marrow during allogeneic transplantation. Bone Marrow Transpl. 2009;45(1):25–30. doi: 10.1038/bmt.2009.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singleton BK, Griffin CS, Thacker J. Clustered DNA damage leads to complex genetic changes in irradiated human cells. Cancer Res. 2002;62(21):6263–6269. [PubMed] [Google Scholar]
- Sishc BJ, Nelson CB, McKenna MJ, Battaglia CL, Herndon A, Idate R, et al. Telomeres and telomerase in the radiation response: implications for instability, reprograming, and carcinogenesis. Front. Oncol. 2015;5:257. doi: 10.3389/fonc.2015.00257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sridharan DM, Asaithamby A, Bailey SM, Costes SV, Doetsch PW, Dynan WS, et al. Understanding cancer development processes after HZE-particle exposure: roles of ROS, DNA damage repair and inflammation. Radiat. Res. 2015;183(1):1–26. doi: 10.1667/RR13804.1. [DOI] [PubMed] [Google Scholar]
- Sridharan DM, Asaithamby A, Bailey SM, Costes S, Doetsch PD, Dynan WS, et al. Understanding cancer development processes after HZE-particle exposure: roles of ROS, DNA damage repair and inflammation. Radiat. Res. 2015;183(1–26) doi: 10.1667/RR13804.1. [DOI] [PubMed] [Google Scholar]
- Storb R, Raff RF, Appelbaum FR, Deeg J, Graham TC, Schuen Fractionated versus single-dose total body iradiation at low and high dose rates to condition canine littermates for DLA-identical marrow grafts. Blood. 1994;83(11):3384–3389. [PubMed] [Google Scholar]
- Storey JD, Madeoy J, Strout Jeanna L, Wurfel M, Ronald J, Akey Joshua M. Gene-expression variation within and among human populations. Am. J. Human Genet. 2007;80(3):502–509. doi: 10.1086/512017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suresh Kumar MA, Peluso M, Chaudhary P, Dhawan J, Beheshti A, Manickam K, et al. Fractionated radiation exposure of rat spinal cords leads to latent neuro-inflammation in brain, cognitive deficits, and alterations in Apurinic Endonuclease 1. PLoS One. 2015;10(7):e0133016. doi: 10.1371/journal.pone.0133016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutherland BM, Bennett PV, Sidorkina O, Laval J, et al. Clustered DNA damages induced in isolated DNA and in human cells by low doses of ionizing radiation. Proc. Natl. Acad. Sci. USA. 2000;97(1):103–108. doi: 10.1073/pnas.97.1.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi A, Kubo M, Ma H, Nakagawa A, Yoshida Y, Isono M, et al. Non-homologous end-joining repair plays a more important role than homologous recombination repair in defining radiosensitivity after exposure to high-LET radiation. Radiat. Res. 2014;182(3):338–344. doi: 10.1667/RR13782.1. [DOI] [PubMed] [Google Scholar]
- Tartier L, Gilchrist S, Burdak-Rothkamm S, Folkard M, Prise KM. Cytoplasmic irradiation induces mitochondrial-dependent 53BP1 protein relocalization in irradiated and bystander cells. Cancer Res. 2007;67(12):5872–5879. doi: 10.1158/0008-5472.CAN-07-0188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thacker J, Stretch A, Stephens MA. Mutation and inactivation of cultured mammalian cells exposed to beams of accelerated heavy ions. II. Chinese ham-ster V79 cells. Int J. Radiat. Biol. Relat. Stud. Phys. Chem. Med. 1979;36(2):137–148. doi: 10.1080/09553007914550891. [DOI] [PubMed] [Google Scholar]
- Thompson LH, Schild D. Recombinational DNA repair and human disease. Mutat. Res. 2002;509(1–2):49–78. doi: 10.1016/s0027-5107(02)00224-5. [DOI] [PubMed] [Google Scholar]
- Tomasetti C, Vogelstein B. Cancer etiology. Variation in cancer risk among tissues can be explained by the number of stem cell divisions. Science. 2015;347(6217):78–81. doi: 10.1126/science.1260825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong AS, Stern JL, Sfeir A, Kartawinata M, de Lange T, Zhu XD, et al. ATM and ATR signaling regulate the recruitment of human telomerase to telomeres. Cell Rep. 2015;13(8):1633–1646. doi: 10.1016/j.celrep.2015.10.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trani D, Nelson SA, Moon BH, Swedlow JJ, Williams EM, Strawn SJ, et al. High-energy particle-induced tumorigenesis throughout the gastrointestinal tract. Radiat. Res. 2014;181(2):162–171. doi: 10.1667/RR13502.1. [DOI] [PubMed] [Google Scholar]
- Tseng BP, Lan ML, Tran KK, Acharya MM, Giedzinski E, Limoli CL. Characterizing low dose and dose rate effects in rodent and human neural stem cells exposed to proton and gamma irradiation. Redox. Biol. 2013;1:153–162. doi: 10.1016/j.redox.2013.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tseng BP, Giedzinski E, Izadi A, Suarez T, Lan ML, Tran KK, et al. Functional consequences of radiation-induced oxidative stress in cultured neural stem cells and the brain exposed to charged particle irradiation. Antioxid. Redox Signal. 2014;20(9):1410–1422. doi: 10.1089/ars.2012.5134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukamoto S, Ishikawa T, Iida S, Ishiguro M, et al. Clinical significance of osteoprotegerin expression in human colorectal cancer. Clin Cancer Res. 2011;17(8):2444–2450. doi: 10.1158/1078-0432.CCR-10-2884. [DOI] [PubMed] [Google Scholar]
- Tsuruoka C, Suzuki M, Kanai T, Fujitaka K. LET and ion species dependence for cell killing in normal human skin fibroblasts. Radiat. Res. 2005;163(5):494–500. doi: 10.1667/rr3360. [DOI] [PubMed] [Google Scholar]
- Turker MS, Connolly L, Dan C, Lasarev M, Gauny S, Kwoh E, et al. Comparison of autosomal mutations in mouse kidney epithelial cells exposed to iron ions in situ or in culture. Radiat. Res. 2009;172(5):558–566. doi: 10.1667/RR1805.1. [DOI] [PubMed] [Google Scholar]
- Turtoi A, Brown I, Schläger M, Schneeweiss FHA. Gene expression profile of human lymphocytes exposed to (211)At alpha particles. Radiat. Res. 2010;174(2):125–136. doi: 10.1667/RR1659.1. [DOI] [PubMed] [Google Scholar]
- UNSCEAR Report II. 2006 [Google Scholar]
- United Nations Scientific Committee on the Effects of Atomic Radiation. Effects of Ionizing Radiation. United Nations; New York: 2008. [Google Scholar]
- Uziel T, Lerenthal Y, Moyal L, Andegeko Y, Mittelman L, Shiloh Y. Requirement of the MRN complex for ATM activation by DNA damage. EMBO J. 2003;22(20):5612–5621. doi: 10.1093/emboj/cdg541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vadhavkar N, Pham C, Georgescu W, Deschamps T, Heuskin AC, Tang J, et al. Combinatorial DNA damage pairing model based on X-ray-induced foci predicts the dose and LET dependence of cell death in human breast cells. Radiat. Res. 2014;182(3):273–281. doi: 10.1667/RR13792.1. [DOI] [PubMed] [Google Scholar]
- Vogelstein B, Papadopoulos N, Velculescu VE, Zhou SLA, Diaz, Kinzler KW. Cancer genome landscapes. Science. 2013;339(6127):1546–1558. doi: 10.1126/science.1235122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Pluth JM, Cooper PK, Cowan MJ, Chen DJ, Yannone SM. Artemis deficiency confers a DNA double-strand break repair defect and Artemis phosphorylation status is altered by DNA damage and cell cycle progression. DNA Repair. 2005;4(5):556–570. doi: 10.1016/j.dnarep.2005.02.001. [DOI] [PubMed] [Google Scholar]
- Wang H, Wang X, Zhang P, Wang Y. The Ku-dependent non-homologous end-joining but not other repair pathway is inhibited by high linear energy transfer ionizing radiation. DNA Repair. 2008;7(5):725–733. doi: 10.1016/j.dnarep.2008.01.010. [DOI] [PubMed] [Google Scholar]
- Wang H, Zhang X, Wang P, Yu X, Essers J, Chen D, et al. Characteristics of DNA-binding proteins determine the biological sensitivity to high-linear energy transfer radiation. Nucleic Acids Res. 2008;38(10):3245–3251. doi: 10.1093/nar/gkq069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Wang X, Chen G, Zhang X, Tang X, Park D, et al. Distinct roles of Ape1 protein, an enzyme involved in DNA repair, in high or low linear energy transfer ionizing radiation-induced cell killing. J. Biol. Chem. 2014;289(44):30635–30644. doi: 10.1074/jbc.M114.604959. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Wang P, Guo F, Wang X, Li J, Guo YYLÜ. X-ray-induced changes in the expression of inflammation-related genes in human peripheral blood. Int. J. Mol. Sci. 2014;15(11):19516–19534. doi: 10.3390/ijms151119516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward JF. Some biochemical consequences of the spatial distribution of ionizing radiation-produced free radicals. Radiat. Res. 1981;86(2):185–195. [PubMed] [Google Scholar]
- Warters RL, Adamson PJ, Pond CD, Leachman SA. Melanoma cells express elevated levels of phosphorylated histone H2AX foci. J. Investig. Dermatol. 2005;124(4):807–817. doi: 10.1111/j.0022-202X.2005.23674.x. [DOI] [PubMed] [Google Scholar]
- Weil MM, Ray FA, Genik PC, Yu Y, McCarthy M, Fallgren CM, et al. Effects of 28Si ions, 56Fe ions, and protons on the induction of murine acute myeloid leukemia and hepatocellular carcinoma. PLoS One. 2014;9(7):e104819. doi: 10.1371/journal.pone.0104819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werner E, Wang H, Doetsch PW. Opposite roles for p38MAPK-driven responses and reactive oxygen species in the persistence and resolution of radiation-induced genomic instability. PLoS One. 2014;9(10):e108234. doi: 10.1371/journal.pone.0108234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whalen MK, Gurai SK, Zahed-Kargaran H, Pluth JM. Specific ATM-mediated phosphorylation dependent on radiation quality. Radiat. Res. 2008;170(3):353–364. doi: 10.1667/RR1354.1. [DOI] [PubMed] [Google Scholar]
- Wiese C, Gauny SS, Liu WC, Cherbonnel-Lasserre CL, Kronenberg A. Different mechanisms of radiation-induced loss of heterozygosity in two human lymphoid cell lines from a single donor. Cancer Res. 2001;61(3):1129–1137. [PubMed] [Google Scholar]
- Williams JP, Brown SL, McBride WH. Animal Models for Medical counter-measures to Radiation exposure. Radiat. Res. 2010;173(4):557–578. doi: 10.1667/RR1880.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winton DJ, Peacock JH, Ponder BA. Effect of gamma radiation at high- and low-dose rate on a novel in vivo mutation assay in mouse intestine. Mutagenesis. 1989;4(5):404–406. doi: 10.1093/mutage/4.5.404. [DOI] [PubMed] [Google Scholar]
- Wu X, Levine AJ. p53 and E2F-1 cooperate to mediate apoptosis. Proc. Natl. Acad. Sci. USA. 1994;91(9):3602–3606. doi: 10.1073/pnas.91.9.3602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Z, Duc KD, Holcman D, Teixeira MT. The length of the shortest telomere as the major determinant of the onset of replicative senescence. Genetics. 2013;194(4):847–857. doi: 10.1534/genetics.113.152322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yajima H, Fujisawa H, Nakajima NI, Hirakawa H, Jeggo PA, Okayasu R, et al. The complexity of DNA double strand breaks is a critical factor enhancing end-resection. DNA Repair. 2013;12(11):936–946. doi: 10.1016/j.dnarep.2013.08.009. [DOI] [PubMed] [Google Scholar]
- Yandell DW, Dryja TP, Little JB. Molecular genetic analysis of recessive mutations at a heterozygous autosomal locus in human cells. Mutat. Res. 1990;229(1):89–102. doi: 10.1016/0027-5107(90)90011-r. [DOI] [PubMed] [Google Scholar]
- Yang H, Anzenberg V, Held KD. Effects of heavy ions and energetic protons on normal human fibroblasts. Radiat. Biol. Radioecol./Rossiiskaia Akad. Nauk. 2007;47(3):302–306. [PubMed] [Google Scholar]
- Yang H, Anzenberg V, Held KD. The time dependence of bystander responses induced by iron-ion radiation in normal human skin fibroblasts. Radiat. Res. 2007;168(3):292–298. doi: 10.1667/RR0864.1. [DOI] [PubMed] [Google Scholar]
- Yang H, Magpayo N, Held KD. Targeted and non-targeted effects from combinations of low doses of energetic protons and iron ions in human fibroblasts. Int. J. Radiat. Biol. 2011;87(3):311–319. doi: 10.3109/09553002.2010.537431. [DOI] [PubMed] [Google Scholar]
- Yang H, Magpayo N, Rusek A, Chiang IH, Sivertz M, Held KD. Effects of very low fluences of high-energy protons or iron ions on irradiated and by-stander cells. Radiat. Res. 2011;176(6):695–705. doi: 10.1667/rr2674.1. [DOI] [PubMed] [Google Scholar]
- Yatagai F, Honma M, Takahashi A, Omori K, Suzuki H, Shimazu T, et al. Frozen human cells can record radiation damage accumulated during space flight: mutation induction and radioadaptation. Radiat. Environ. Biophys. 2011;50(1):125–134. doi: 10.1007/s00411-010-0348-3. [DOI] [PubMed] [Google Scholar]
- Yuan Y, Britton S, Delteil C, Coates J, Jackson SP, Barboule N, et al. Single-stranded DNA oligomers stimulate error-prone alternative repair of DNA double-strand breaks through hijacking Ku protein. Nucleic Acids Res. 2015;43(21):10264–10276. doi: 10.1093/nar/gkv894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou H, Randers-Pehrson G, Waldren CA, Vannais D, Hall EJ, Hei TK. Induction of a bystander mutagenic effect of alpha particles in mammalian cells. Proc. Natl. Acad. Sci. USA. 2000;97(5):2099–2104. doi: 10.1073/pnas.030420797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou H, Suzuki M, Randers-Pehrson G, Vannais D, Chen G, Trosko JE, et al. Radiation risk to low fluences of alpha particles may be greater than we thought. Proc. Natl. Acad. Sci. USA. 2001;98(25):14410–14415. doi: 10.1073/pnas.251524798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou D, Shao L, Spitz DR. Reactive oxygen species in normal and tumor stem cells. Adv. Cancer Res. 2014;122:1–67. doi: 10.1016/B978-0-12-420117-0.00001-3. [DOI] [PMC free article] [PubMed] [Google Scholar]