SUMMARY
The EGLN (also called PHD) prolyl hydroxylase enzymes and their canonical targets, the HIFα subunits, represent the core of an ancient oxygen-monitoring machinery used by metazoans. In this review we highlight recent progress in understanding the overlapping versus specific roles of EGLN enzymes and HIF isoforms and discuss how feedback loops based on recently identified noncoding RNAs introduce additional layers of complexity to the hypoxic response. Based on novel interactions identified upstream and downstream of EGLNs, an integrated network connecting oxygen-sensing functions to metabolic and signaling pathways is gradually emerging with broad therapeutic implications.
Keywords: Hypoxia, Oxygen, EGLN, PHD, HIF1α, HIF2α, metabolism, noncoding RNA, miRNA, lncRNA, kidney cancer, HIF2 inhibitors, EGLN inhibitors
The Core of Oxygen Sensing in Metazoans
How animal cells adapt to variations in ambient oxygen concentration was a recurring question during the 20th century, as groups from diverse fields joined the quest for an elusive “sensor”. Studies of the prototypical hypoxia-responsive mRNA encoding EPO led to the identification over 20 years ago of a hypoxia-inducible activity called hypoxia-inducible factor or “HIF”. Contrary to the prevailing expectations at that time, HIF was soon detected in a wide variety of hypoxic cells and tissues and not just specialized cells dedicated to producing EPO. Subsequent studies showed that HIF is a heterodimer consisting of alpha subunit, such as the first alpha subunit to be cloned, hypoxia-inducible factor alpha (HIF1α), and a beta subunit. The alpha subunit is a basic helix–loop–helix, PAS domain–containing, DNA-binding protein that is rapidly degraded in normoxic cells (Kaelin and Ratcliffe, 2008). As oxygen tension decreases, HIFα becomes progressively more stable and binds its partner HIFβ, the oxygen- insensitive protein product of the ARNT gene. The resulting complex undergoes nuclear translocation, binds to hypoxia-response elements (HREs) and transcriptionally activates hundreds of genes involved in low oxygen adaptation (Kaelin and Ratcliffe, 2008; Semenza, 2012).
The discovery that cells lacking the pVHL tumor suppressor protein accumulate high levels of hypoxia-inducible mRNAs, and do not degrade HIF, even when oxygen is plentiful ultimately led to the recognition that pVHL is the substrate recognition module of the ubiquitin ligase that targets HIFα for proteasomal degradation under normoxic conditions. While the discovery of HIF, and its regulation by pVHL, provided key mechanistic insights into the coordinated activation of the hypoxic transcriptome, it did not immediately reveal the oxygen sensor’s identity. A major step forward was the demonstration that two conserved prolyl residues within the region of HIFα that is recognized by pVHL, called the oxygen-dependent degradation domain, are enzymatically hydroxylated in an oxygen-dependent manner (Ivan et al., 2001; Jaakkola et al., 2001; Masson et al., 2001; Yu et al., 2001). Hydroxylation of either (or both) of these prolyl residues generates a high-affinity binding site for pVHL, leading to HIFα’s polyubiquitylation and destruction in well-oxygenated cells. HIF prolyl hydroxylation is mediated by the EGLN (also called PHD) 2-oxoglutarate (2OG)–dependent dioxygenases, which require oxygen, iron, and 2-oxoglutarate to function (Bruick and McKnight, 2001; Epstein et al., 2001; Ivan et al., 2002). Importantly, the EGLNs have relatively low oxygen affinities and hence are poised to sense oxygen in a physiologically relevant concentration range (Kaelin and Ratcliffe, 2008).
The EGLNs are highly susceptible to self-inactivation as a result of autooxidiation. As a result, antioxidants such as ascorbate are usually included when measuring EGLN activity in vitro (Flashman et al., 2010; Knowles et al., 2003). Likewise, EGLN activity in cells can be modulated by reactive oxygen species and by intracellular cysteine, which protects the EGLNs from oxidative damage (see also below).
Overall, the EGLN-HIF system is remarkably well-conserved throughout evolution, being recognizable in the simplest known animal, the placozoan Trichoplax adhaerens (Loenarz et al., 2011). The extraordinary diversification of lifeforms during the past 500 million years was necessarily associated with higher variability in oxygen tension within larger and more complex animals, as well as during development, and it appears that the ancestral oxygen sensing was subjected to evolutionary pressure to evolve into a more sophisticated machinery. Thus, the genome of higher organisms such as mammals typically generates three canonical HIF prolyl hydroxylases, encoded by EGLN1, 2 and 3 genes (with the commonly used aliases PHD2, PHD1 and PHD3, respectively), three HIFα subunits (encoded by HIF1A, EPAS1/HIF2A and HIF3A) and two HIFβ partners (encoded by ARNT1 and 2). Numerous studies based on isoform-specific genetic inactivation have revealed both overlapping and specific functions, with important implications for physiology, disease and drug development (Kaelin and Ratcliffe, 2008; Semenza, 2012). The importance of this pathway is underscored by the discovery of EGLN1 and EPAS1 genetic polymorphisms in human populations living at extremely high altitudes (Bigham and Lee, 2014).
While EGLN1 is widely recognized as the central hypoxia/oxygen sensor with respect to HIF (Berra et al., 2003; Schofield and Ratcliffe, 2004), all three isoforms exhibit enzymatic properties consistent with sensing roles, having Km values for O2 above anticipated cellular and tissue oxygen levels (Ehrismann et al., 2007; Hirsila et al., 2003), and EGLN2 and EGLN3 contribute to the regulation of HIF in certain settings. For example, acutely eliminating Egln1 in the mouse liver leads to a pulsatile induction of the canonical HIF-target Epo, presumably reflecting compensation by Egln2 and Egln3, while eliminating all 3 paralogs leads to sustained, high-level, hepatic Epo production (Minamishima and Kaelin, 2010; Querbes et al., 2012). A fourth prolyl hydroxylase termed P4H-TM (alternatively known as PHD4), possessing an endoplasmic reticulum transmembrane domain, was characterized by Myllyharju, Koivunen and colleagues and shown to control HIF stability and erythropoietin production in vivo (Koivunen et al., 2007; Laitala et al., 2012). Overall, however, significantly less is known about this enzyme compared to the “original” members of the family.
It perhaps makes sense, from an evolutionary standpoint, that oxygen monitoring in more complex organisms such as mammals utilizes multiple distinct EGLNs that have different oxygen affinities and that are capable of performing specific, in addition to common, biochemical functions (Table 1). For example, physiologic oxygen tension varies dramatically between organs, from as high as 80–100 mmHg in the lung alveoli and arterial blood, to 1–2 mmHg in the renal papilla (Leichtweiss et al., 1969), too wide a window for a single enzymatic sensor. Multiple differences are discernible between mammalian EGLN isoforms. All three EGLNs hydroxylate the highly conserved Pro564 in HIF1α, but only EGLN1 and 2 can modify the more recently evolved Pro402 (Berra et al., 2003; Chowdhury et al., 2016). Additionally, the isoforms differ in their affinity for specific HIF isoforms, for example EGLN3 exhibits preference for HIF2α (Appelhoff et al., 2004). There is also growing evidence that each of the three EGLNs has specific non-HIF targets, as discussed below. A summary of EGLN isoform characteristics is provided in Table 1.
Table 1.
Summary of EGLN isoform distinguishing features
| Enzyme name |
Preferred HIF isoform |
Specific features |
|---|---|---|
| EGLN1 (PHD2) | HIF1α (both NTAD and CTAD) | Lowest O2 affinity (main sensor); Knockout embryonically lethal |
| EGLN2 (PHD1) | HIF2α (both NTAD and CTAD) | Estrogen-inducible; Transcript not induced by hypoxia; Potential oncogene |
| EGLN3 (PHD3) | HIF2α (only CTAD) | Regulator of apoptosis; Multiple non-HIF target candidates |
As noted by Schofield and colleagues (Markolovic et al., 2015), the 2001 discovery that hydroxylation can play physiologically-relevant roles in transcriptional regulation has generated considerable interest in the larger class of 2-OG dioxygenases, with its approximately 60 members. An important question is whether any of these enzymes, in addition to EGLNs, also operate as oxygen sensors under physiological conditions. While, by definition, diatomic oxygen is necessary for the function of all these enzymes, most exhibit extremely high affinity for oxygen, meaning that they theoretically can remain active until cells are virtually anoxic (Salminen et al., 2015; Sanchez-Fernandez et al., 2013). For specific members however, evidence of sensor function has been reported. As summarized by Rocha and colleagues (Shmakova et al., 2014), moderate hypoxia (1–3% O2) leads to a global increase in H3K9me2, H3K9me3 and H3K36me3 in cells, suggesting that hypoxia directly (or indirectly) inhibits certain Jumonji C domain-containing (JMJC) histone demethylases such as KDM5A, KDM4D and KDM4E. The available information, although still limited, indicates that KDM4E, similar to EGLNs, has a relatively low affinity for oxygen compared to other dioxygenases (Shmakova et al., 2014) thus strengthening its sensor credentials. Interestingly, many JMJC KDMs, like EGLN1 and EGLN3, are induced by hypoxia, (and where studied, HIF), potentially to compensate for decreased activity (Shmakova et al., 2014).
Another special case is Factor Inhibiting HIF (FIH1/HIF1AN), which is a stable component of chromatin-bound HIF. HIFα has two transactivation domains, called the N-terminal and C-terminal transactivation domains (NTAD and CTAD). Rather than acting on proline, FIH hydroxylates a specific asparagine in HIFα’s CTAD (Asn803) (Hewitson et al., 2002; Lando et al., 2002). In contrast to EGLN-mediated hydroxylation, which dramatically enhances a protein-protein interaction (HIF-pVHL), Asn803 hydroxylation disrupts the interaction between HIF and its p300/CBP coactivators (Markolovic et al., 2015), thereby crippling the CTAD. Based on its higher oxygen affinity compared to EGLNs, FIH maintains sufficient activity at intermediate levels of hypoxia that are sufficient to stabilize HIF1α, thus adding a second oxygen – mediated checkpoint in the pathway. In contrast to EGLNs, FIH is relatively promiscuous, as it can hydroxylate a broad spectrum of additional substrates, including Notch (Coleman et al., 2007), cytoskeletal ankyrin family proteins (Yang et al., 2011a) and the TRPV3 ion channel (Karttunen et al., 2015). Furthermore, in addition to the normally preferred asparaginyl, FIH can also hydroxylate histidinyl and aspartyl residues (Yang et al., 2011b). Although the roles of FIH in the broader response to hypoxia are still being unraveled, its preference of HIF1α over HIF2α has several practical implications. First, drugs that block EGLN (see also below), but not FIH, can stimulate EPO without inducing VEGF because the former is driven by HIF2 and the latter normally by the HIF1 CTAD. Second, the presence of FIH does not prevent pVHL-defective kidney cancers from coopting the HIF program because these tumors are driven largely by HIF2 rather than HIF1 (Cho and Kaelin, 2016). From the standpoint of normal physiology, the genetic inactivation of Fih in mice does not, surprisingly, generate an obvious phenotype (in stark contrast to inactivation of Egln1, Hif1a, or Vhl). Based on its ancient origin and preservation, however, one can speculate that FIH confers a fitness advantage by fine-tuning diverse cellular pathways.
HIF1 and HIF2: Related, But Far from Identical
Next generation sequencing technology has enabled an increasingly comprehensive characterization of the transcription program set in motion by hypoxia and the HIFs. Overall, some HIF targets are induced when diverse cell types are subjected to in vitro hypoxia, while others, such as the abovementioned EPO, are highly tissue-specific. Upon closer examination, the hypoxic response exhibits additional layers of complexity, including kinetic differences. Below we will examine the mechanistic basis for the known similarities and differences between HIF1 and HIF2, and the resulting implications for disease and therapy.
HIF1 has been traditionally described as the driver of metabolic responses to hypoxia, including the control of most glycolytic enzymes, such as phosphofructokinase (PFKFB3), lactate dehydrogenase A (LDHA) and pyruvate kinase (PKM). Its metabolic role has been expanded to maintenance of intracellular pH, via targets such as monocarboxylate transporter 4 (MCT4) and carbonic anhydrase 9 (CA-IX). Furthermore, HIF1 induction triggers a multi-pronged suppression of mitochondrial respiration that includes transcriptional induction of pyruvate dehydrogenase kinases (PDK1 and PDK3 isoforms) (Keith et al., 2011; Kim et al., 2006). Furthermore, as shown by Semenza’s group, HIF1 reciprocally regulates mitochondrial COX4 subunit expression by activating transcription of the gene encoding COX4-2, but also by inducing the gene encoding LONP1, a mitochondrial protease that degrades COX4-1. This switch from COX4-1 to COX4-2 decreases the activity of cytochrome oxidase complex and reduces oxygen consumption (Fukuda et al., 2007). On the other hand, HIF2 is viewed as predominantly responsible for hypoxic induction of genes linked to growth signals, including TGFA and PDGFB, the cell-cycle, such as CCND1, stem cell biology, such as OCT4, invasion, such as MMP2 and MMP13, and erythropoiesis, such as EPO.
Overall, fundamental differences between HIF1 and HIF2 exist and understanding these should be key for rational pharmacological targeting of the oxygen sensing pathway. What accounts, at the molecular level, for the differences between HIF siblings? It should be noted that the consensus sequence for a Hypoxia Response Element (HRE), (G/C/T)ACGTGC(G/C), is both short, degenerate, and unable to discriminate between HIF1 and HIF2. On the other hand, protein-protein interactions at promoters and enhancers with a diverse panel of transcription factors, co-regulators and chromatin remodelers appear to exhibit important isoform differences. In short, the ability of a given HRE to support activation by HIF1, HIF2, both, or neither is likely influenced by chromatin accessibility and neighboring transcription factors.
HIF1 and HIF2 differential interactions with two central growth-promoting drivers, MYC and mTORC1, provide key explanations for at least some of the functional contrasts between the isoforms. In brief, hypoxic induction of HIF1 prevents MYC from associating with its partner MAX and with SP1 transcription factor on chromatin, the net result being suppression of MYC-dependent transactivation. Additionally, HIF1 induces MAX interactor 1 (MXI1), which further inhibits the expression of MYC targets (Dang et al., 2008). A caveat is necessary however, as the antagonism between HIF1 and MYC is in fact more nuanced, depending on their relative abundance. When MYC family members are highly overexpressed, such as in neuroblastoma, they collaborate with HIF1 to boost glycolysis, and overcome the inhibitory effects of HIF1 highlighted above, thus allowing proliferation under decreased O2 availability (Keith et al., 2011). On the other hand, HIF2α facilitates the formation of an active MYC complex and activates a hypoxic pro-growth program. Similar contrast has been reported with respect to mTORC1 function. HIF1, but not HIF2, induces the expression of DNA-damage-inducible transcript 4 (DDIT4/ REDD1), which releases TSC2 from the sequestering effects of 14-3-3 proteins, leading to mTORC1 complex inhibition (Keith et al., 2011). Therefore, HIF1 and HIF2 can, at least in some contexts, have opposing roles on cell proliferation and growth, such as appears to be the case in pVHL-defective kidney cancers (Cho and Kaelin, 2016).
Optimization of EGLN-HIF System by Positive and Negative Feedbacks
Oxygen sensing machinery is subject to regulatory feedbacks, both positive and negative, that involve a broad spectrum of components, including proteins, metabolites and the more recently appreciated noncoding RNAs. Such mechanisms may play critical roles for the differences in timing between HIF1 and HIF2 induction during hypoxia. Thus HIF1 is generally thought to coordinate the acute response, with its protein level peaking within the first 12 hours, followed by gradual decrease. In contrast, HIF2 exhibits a more delayed induction followed by a stable plateau pointing to a role in chronic adaptive responses (Koh and Powis, 2012).
One important feedback appears to involve EGLN hydroxylases themselves. EGLN1 and 3 consistently score as HIF transcriptional targets. One could speculate that their gradual loss of function in low oxygen is compensated in part by increased abundance.
A more recently appreciated, yet insufficiently understood, set of feedbacks linked to oxygen sensing involves noncoding RNAs (Gee et al., 2014). The noncoding transcriptome comprises tens of thousands of RNAs performing biological functions without being translated into proteins (Cech and Steitz, 2014). Benefitting from technical advances in RNA sequencing, studies performed over the past decade have connected a variety of noncoding RNAs to hypoxia responses. With the important caveat that independent validation is not yet available for most of these candidates, they may add an important tissue-specific dimension to the hypoxic response. Indeed, it is generally agreed that noncoding RNAs exhibit higher tissue variability than do coding transcripts. Based on their ability to interact with an astonishingly diverse spectrum of protein and nucleic acid targets, noncoding RNAs are likely to introduce previously unsuspected regulatory feedbacks in the hypoxic response.
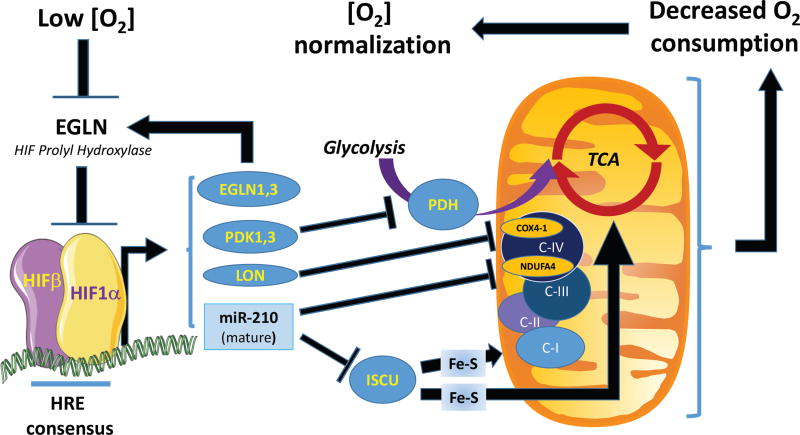
Historically, the first family of noncoding RNAs to be linked to hypoxia were microRNAs (miRNAs)(Kulshreshtha et al., 2007b). These are short oligoribonucleotides (approximately 22nt in length) known to repress the expression of target gene by promoting mRNAs degradation and/or translation blockade. While low oxygen tension globally downregulates miRNAs biogenesis (Rupaimoole et al., 2014; van den Beucken et al., 2014), a select few mature miRNAs are induced by hypoxia in multiple cell types. In particular, miR-210 has been identified as direct HIF target by multiple groups and is consistently induced by hypoxia in normal and transformed cells (Chan et al., 2009; Gee et al., 2014; Kulshreshtha et al., 2007a). As summarized in Fig. 1, arguably the most robust targets of miR-210 are ISCU, an assembly factor for Fe-S complexes required for ETC activity (Chan and Loscalzo, 2010; Favaro et al., 2010), and NDUFA4, which is an integral component of ETC complex IV (Balsa et al., 2012; Fasanaro et al., 2009). Elevated miR-210 expression in hypoxia downregulates ISCU and NDUFA4, thus reducing electron transfer complex activity. This noncoding arm of HIF may reinforce the HIF1-mediated suppression of mitochondrial respiration via the coding gene products discussed above. Collectively, these suppressive effects on mitochondrial respiration are thought to decrease oxygen consumption and thereby help restore cellular oxygenation.
Figure 1.
Model of cooperation between coding genes and miR-210 downstream of HIF1. Feedback loop leading to renormalization of local O2 tension as a result of decreased local consumption.
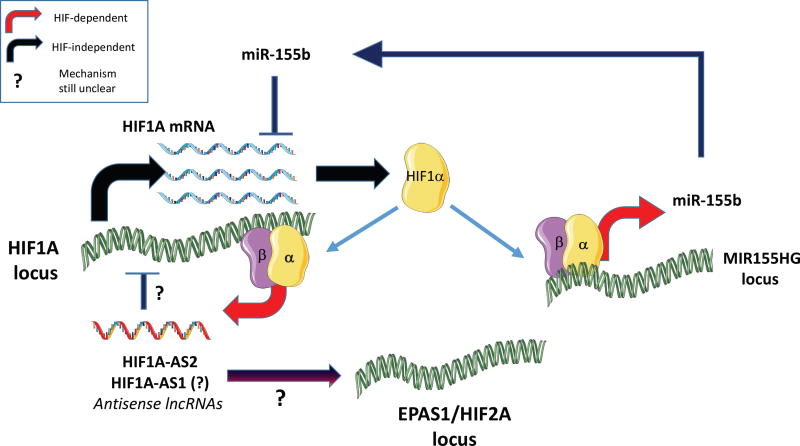
Multiple studies have provided evidence for miRNA-based feedbacks that affect the expression of HIF itself, in hypoxic or non-hypoxic contexts. For example miR-218 was shown to be downregulated in mesenchymal glioblastoma and involved in resistance to chemotherapy. Mechanistically, low miR-218 releases multiple RTK effectors from its inhibitory control, leading to activation of hypoxia-inducible factors, most notably HIF2α (Mathew et al., 2015). As shown by Cormac Taylor’s group, induction of miR-155 by hypoxia appears to be part of a complex feedback mechanism that generates an oscillatory pattern for HIF1 transcriptional activity (Bruning et al., 2011). Given the generally acknowledged subtle effects of miRNAs, especially when expressed at physiological levels, and taking into account their probable distribution between many targets, a measurable impact on HIF abundance may require cooperative action. As shown by Maria Czyzyk-Krzeszka’s group in the context of VHL-deficient clear cell renal carcinoma (Mikhaylova et al., 2012), responses involving coding and noncoding transcripts can cooperate. In particular, VHL induces miR-204, which downregulates MAP1LC3B (LC3B) and as consequence decreases macro-autophagic activity. In parallel, VHL, by suppressing HIF, also transcriptionally suppresses LC3C. This dual mechanism of autophagy blockade appears to contribute to VHL tumor-suppressor function (Mikhaylova et al., 2012). More recently, Celeste Simon and colleagues generated evidence of a HIF-independent mechanism that shifts the balance between the pro- and anti-tumorigenic effects of HIFα isoforms (Mathew et al., 2014). In a subset of clear cell renal cell carcinomas that expresses both isoforms, they have identified consistent downregulation miR-30c-2-3p and miR-30a-3p. As these miRNAs target HIF2α, but not HIF1α, a decrease in their expression selectively increases the abundance of the former, pro-oncogenic, HIFα isoform.
The more recently studied long-noncoding RNAs (lncRNAs) form a vast and heterogeneous family of transcripts larger than 200nt in length. Chronologically, one of the first genomic loci reported to generate a hypoxia-inducible lncRNAs was HIF1A gene itself (Thrash-Bingham and Tartof, 1999). The negative strand of this locus produces multiple antisense noncoding transcripts in response to hypoxia, the highest expressed being HIF1-AS2 (Mineo et al., 2016). This is in contrast to HIF1A mRNA, which is typically downregulated or unchanged in hypoxia. The antisense HIF transcript is important for the hypoxic process itself, as its knockdown blunts the induction of HIF2α and that of important HIF targets (NDRG1, VEGFA and ADM). While a direct connection to HIF itself remains elusive, the above study identified several RNA-binding proteins interacting with HIF1A-AS2, including IGF2BP2 and DHX9, which may explain its importance for the hypoxic response. Consistently, loss of HIF1A-AS2 suppresses tumor cell viability and delays xenograft growth (Mineo et al, 2016). The various feedbacks involving noncoding RNAs described above are summarized in Fig 2.
Figure 2.
Model of noncoding RNA-based feedback circuits optimizing the transition from a HIF1 to a HIF2-based hypoxic response.
EGLN-HIF Beyond Conventional Hypoxic Responses
It has become evident that EGLN enzymes respond to other inputs in addition to hypoxia. For example, mutations in fumarate hydratase (FH) and succinate dehydrogenase subunits (SDHA, SDHC, SDHD) identified in rare kidney cancers and neuroendocrine tumors disrupt the normal TCA cycle metabolite flow and cause the accumulation of fumarate and succinate, respectively, which then inhibit the EGLNs by competing with their natural co-substrate, 2-oxoglutarate. It was recently shown that acidosis and hypoxia, commonly coexisting in the tumor microenvironment, favor the accumulation of L-2-hydroxyglutarate (Intlekofer et al., 2017; Nadtochiy et al., 2016), an endogenous 2-oxoglutarate antagonist (Xu et al., 2011). Theoretically, this process could reinforce EGLN1inhibition in solid tumors. A recent study provides evidence that EGLN1 has an even wider metabolite-sensing capacity. Triple negative breast cancer cells secrete glutamate, which inhibits the xCT glutamate-cystine antiporter, leading to intracellular cysteine depletion. EGLN1 appears to be particularly sensitive to absence of cysteine, and undergoes oxidative self-inactivation, ultimately resulting in HIF1α accumulation (Briggs et al., 2016)..
EGLN substrates other than the HIFs are beginning to emerge and might provide previously unsuspected connections between oxygen sensing, nutrient sensing, and growth responses. For example, AKT1 and AKT2 kinases, which control growth and metabolism, are negatively regulated by EGLN1-mediated hydroxylation under normoxia (Guo et al., 2016). A case reminiscent of the EGLN-HIF-VHL relationship is provided by NDRG family member 3 (NDRG3). Under normoxic conditions, NDRG3 is marked for VHL-dependent ubiquitination by EGLN1-mediated prolyl hydroxylation. Interestingly, its hypoxic stabilization, which contributes to Raf-ERK pathway activation, also requires direct binding of lactate, thus establishing a novel connection between oxygen sensing, metabolic reprogramming and oncogenic signaling (Lee et al., 2015). Another example of EGLN function outside of the HIF network is provided by EGLN2. Hydoxylation of FOXO3a by EGLN2 destabilized FOXO3a by preventing its interaction with the USP9x deubiquitinase (Zheng et al., 2014). EGLN3-mediated hydroxylation of Acetyl-CoA Carboxylase (ACC2) leads to decreased fatty acid oxidation under normoxia and high nutrient availability (German et al., 2016). Pyruvate kinase M2, the proliferogenic splice form of pyruvate kinase, appears to be another major metabolic enzyme serving as hydroxylation substrate. Hydroxylation of prolines 403/408 by EGLN3 stimulates PKM2 catalytic activity and also allows PKM2, through direct binding, to promote HIF transcriptional activity (Luo et al., 2011). Furthermore, Stamler and colleagues (Xie et al., 2009) showed that EGLN3 can also hydroxylate the β2 adrenergic receptor at two proline residues (Pro-382 and -395) in normoxic conditions. This event triggers the recruitment of the pVHL ubiquitin ligase complex and subsequent proteasomal degradation of the receptor. These results unveil a surprisingly broad reach of hydroxylation-based signaling and may suggest new avenues for translational applications. An important caveat is that all the hydroxylation targets summarized above, with the exception of HIFα, await independent validation.
Pharmacological Inhibition of HIF
The knowledge that solid tumors contain hypoxic regions fueled considerable interest in the development of HIF inhibitors, with virtually all early efforts focused on HIF1. Progress was impeded, however, by the prevailing dogma that bHLH-PAS domain proteins were “undruggable”. Bruick and Gardner, however, identified a druggable hydrophobic pocket in the HIF2α PAS B domain, which led to the development of the first generation of small molecule HIF2 inhibitors (Scheuermann et al., 2009). Two of these compounds, the lead compound PT2385 and the related tool compound PT2399, selectively disrupt HIF2α’s interaction with ARNT and suppress pVHL-defective kidney cancers in preclinical models (Cho and Kaelin, 2016). Based on the promising preclinical data, PT2385 has entered clinical trials with early signs of activity. Nonetheless, de novo and acquired resistance to PT2385/2399 has already been documented in the laboratory (Cho and Kaelin, 2016). Current work is aimed at circumventing this resistance as well as identifying other tumor types where HIF2 plays a role in tumor maintenance. A cautionary tale in this regard is provided by the analysis of tumors, such as KRAS-driven lung cancers in genetically-engineered mice, where disruption of HIF2 actually accelerates tumor growth (Mazumdar et al., 2010). This again underscores the importance of context with respect to the HIF response.
Likewise, HIF1 may still hold significant value as target in solid tumors. In general, HIF1 contributes to the Warburg Effect, which is believed to provide building blocks for anabolism, and promotes survival under hypoxic conditions through cell-intrinsic changes in, for example, ATP synthesis and turnover, and cell-extrinsic changes, such as induction of angiogenesis. As discussed above, mTORC1 blockade by HIF1 may benefit tumor cells residing in the perinecrotic areas where very low or absent oxygen is accompanied by nutrient deprivation and acidity. Cell survival under such conditions includes strategies such as suppression of ATP-intensive processes (e.g. protein translation, lipid synthesis), activation of mitophagy, and exit from the cell-cycle, all processes mediated by HIF1 rather than HIF2. Indeed, the HIF2-driven growth program would be incompatible with cell survival under these circumstances. HIF1 targeting may also be valuable as part of combined interventions, for example with radiation or antiangiogenic agents (McIntyre and Harris, 2015). The recent discovery that the HIF1 PAS domains, like the HIF2 PAS domain, contain hydrophobic pockets should stimulate efforts to identify direct HIF1 antagonists (Wu et al Nature 2015). In addition, acriflavine and proflavine were identified in a phenotypic screen for HIF1 inhibitors and appear to target a HIF1α/ARNT interface (Wilkins et al., 2016). Many other compounds have been identified that can, at least indirectly, downregulate HIF1 (Xia et al., 2012). A caveat with these indirect inhibitors is that HIF1 turns over very rapidly and therefore will be one of the first proteins to disappear when cells are confronted with a toxic agent that can decrease global transcription or translation.
Pharmacological Activation of the Hypoxic Response
Many diseases of the developed world are linked to inadequate oxygen delivery, including anemia, myocardial infarction, and stroke. The realization that HIFs are negatively regulated by enzymes opened the way for the development of a new class of pharmacological agents capable of stabilizing HIF and activating the hypoxic response. The first generation of EGLN inhibitors have advanced to phase II/III clinical trials for patients with anemia linked to chronic renal failure and appears promising, based on preclinical data, for treating other types of anemia, such as anemia of chronic disease, as well as diseases linked to regional ischemia (Maxwell and Eckardt, 2016).
Acknowledgments
WGK’s work is supported by the Howard Hughes Medical Institute, MI’s work is supported by NIH R01CA155332.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Appelhoff RJ, Tian YM, Raval RR, Turley H, Harris AL, Pugh CW, Ratcliffe PJ, Gleadle JM. Differential function of the prolyl hydroxylases PHD1, PHD2, and PHD3 in the regulation of hypoxia-inducible factor. J Biol Chem. 2004;279:38458–38465. doi: 10.1074/jbc.M406026200. [DOI] [PubMed] [Google Scholar]
- Balsa E, Marco R, Perales-Clemente E, Szklarczyk R, Calvo E, Landazuri MO, Enriquez JA. NDUFA4 is a subunit of complex IV of the mammalian electron transport chain. Cell Metab. 2012;16:378–386. doi: 10.1016/j.cmet.2012.07.015. [DOI] [PubMed] [Google Scholar]
- Berra E, Benizri E, Ginouves A, Volmat V, Roux D, Pouyssegur J. HIF prolyl-hydroxylase 2 is the key oxygen sensor setting low steady-state levels of HIF-1alpha in normoxia. EMBO J. 2003;22:4082–4090. doi: 10.1093/emboj/cdg392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bigham AW, Lee FS. Human high-altitude adaptation: forward genetics meets the HIF pathway. Genes Dev. 2014;28:2189–2204. doi: 10.1101/gad.250167.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briggs KJ, Koivunen P, Cao S, Backus KM, Olenchock BA, Patel H, Zhang Q, Signoretti S, Gerfen GJ, Richardson AL, et al. Paracrine Induction of HIF by Glutamate in Breast Cancer: EglN1 Senses Cysteine. Cell. 2016;166:126–139. doi: 10.1016/j.cell.2016.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruick RK, McKnight SL. A conserved family of prolyl-4-hydroxylases that modify HIF. Science. 2001;294:1337–1340. doi: 10.1126/science.1066373. [DOI] [PubMed] [Google Scholar]
- Bruning U, Cerone L, Neufeld Z, Fitzpatrick SF, Cheong A, Scholz CC, Simpson DA, Leonard MO, Tambuwala MM, Cummins EP, et al. MicroRNA-155 promotes resolution of hypoxia-inducible factor 1alpha activity during prolonged hypoxia. Mol Cell Biol. 2011;31:4087–4096. doi: 10.1128/MCB.01276-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cech TR, Steitz JA. The noncoding RNA revolution-trashing old rules to forge new ones. Cell. 2014;157:77–94. doi: 10.1016/j.cell.2014.03.008. [DOI] [PubMed] [Google Scholar]
- Chan SY, Loscalzo J. MicroRNA-210: a unique and pleiotropic hypoxamir. Cell Cycle. 2010;9:1072–1083. doi: 10.4161/cc.9.6.11006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan SY, Zhang YY, Hemann C, Mahoney CE, Zweier JL, Loscalzo J. MicroRNA-210 controls mitochondrial metabolism during hypoxia by repressing the iron-sulfur cluster assembly proteins ISCU1/2. Cell Metab. 2009;10:273–284. doi: 10.1016/j.cmet.2009.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho H, Kaelin WG. Targeting HIF2 in Clear Cell Renal Cell Carcinoma. Cold Spring Harb Symp Quant Biol. 2016 doi: 10.1101/sqb.2016.81.030833. [DOI] [PubMed] [Google Scholar]
- Chowdhury R, Leung IK, Tian YM, Abboud MI, Ge W, Domene C, Cantrelle FX, Landrieu I, Hardy AP, Pugh CW, et al. Structural basis for oxygen degradation domain selectivity of the HIF prolyl hydroxylases. Nat Commun. 2016;7:12673. doi: 10.1038/ncomms12673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coleman ML, McDonough MA, Hewitson KS, Coles C, Mecinovic J, Edelmann M, Cook KM, Cockman ME, Lancaster DE, Kessler BM, et al. Asparaginyl hydroxylation of the Notch ankyrin repeat domain by factor inhibiting hypoxia-inducible factor. J Biol Chem. 2007;282:24027–24038. doi: 10.1074/jbc.M704102200. [DOI] [PubMed] [Google Scholar]
- Dang CV, Kim JW, Gao P, Yustein J. The interplay between MYC and HIF in cancer. Nat Rev Cancer. 2008;8:51–56. doi: 10.1038/nrc2274. [DOI] [PubMed] [Google Scholar]
- Ehrismann D, Flashman E, Genn DN, Mathioudakis N, Hewitson KS, Ratcliffe PJ, Schofield CJ. Studies on the activity of the hypoxia-inducible-factor hydroxylases using an oxygen consumption assay. Biochem J. 2007;401:227–234. doi: 10.1042/BJ20061151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Epstein AC, Gleadle JM, McNeill LA, Hewitson KS, O'Rourke J, Mole DR, Mukherji M, Metzen E, Wilson MI, Dhanda A, et al. C. elegans EGL-9 and mammalian homologs define a family of dioxygenases that regulate HIF by prolyl hydroxylation. Cell. 2001;107:43–54. doi: 10.1016/s0092-8674(01)00507-4. [DOI] [PubMed] [Google Scholar]
- Fasanaro P, Greco S, Lorenzi M, Pescatori M, Brioschi M, Kulshreshtha R, Banfi C, Stubbs A, Calin GA, Ivan M, et al. An integrated approach for experimental target identification of hypoxia-induced miR-210. J Biol Chem. 2009;284:35134–35143. doi: 10.1074/jbc.M109.052779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Favaro E, Ramachandran A, McCormick R, Gee H, Blancher C, Crosby M, Devlin C, Blick C, Buffa F, Li JL, et al. MicroRNA-210 regulates mitochondrial free radical response to hypoxia and krebs cycle in cancer cells by targeting iron sulfur cluster protein ISCU. PLoS One. 2010;5:e10345. doi: 10.1371/journal.pone.0010345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flashman E, Davies SL, Yeoh KK, Schofield CJ. Investigating the dependence of the hypoxia-inducible factor hydroxylases (factor inhibiting HIF and prolyl hydroxylase domain 2) on ascorbate and other reducing agents. Biochem J. 2010;427:135–142. doi: 10.1042/BJ20091609. [DOI] [PubMed] [Google Scholar]
- Fukuda R, Zhang H, Kim JW, Shimoda L, Dang CV, Semenza GL. HIF-1 regulates cytochrome oxidase subunits to optimize efficiency of respiration in hypoxic cells. Cell. 2007;129:111–122. doi: 10.1016/j.cell.2007.01.047. [DOI] [PubMed] [Google Scholar]
- Gee HE, Ivan C, Calin GA, Ivan M. HypoxamiRs and cancer: from biology to targeted therapy. Antioxid Redox Signal. 2014;21:1220–1238. doi: 10.1089/ars.2013.5639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- German NJ, Yoon H, Yusuf RZ, Murphy JP, Finley LW, Laurent G, Haas W, Satterstrom FK, Guarnerio J, Zaganjor E, et al. PHD3 Loss in Cancer Enables Metabolic Reliance on Fatty Acid Oxidation via Deactivation of ACC2. Mol Cell. 2016;63:1006–1020. doi: 10.1016/j.molcel.2016.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo J, Chakraborty AA, Liu P, Gan W, Zheng X, Inuzuka H, Wang B, Zhang J, Zhang L, Yuan M, et al. pVHL suppresses kinase activity of Akt in a proline-hydroxylation-dependent manner. Science. 2016;353:929–932. doi: 10.1126/science.aad5755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hewitson KS, McNeill LA, Riordan MV, Tian YM, Bullock AN, Welford RW, Elkins JM, Oldham NJ, Bhattacharya S, Gleadle JM, et al. Hypoxia-inducible factor (HIF) asparagine hydroxylase is identical to factor inhibiting HIF (FIH) and is related to the cupin structural family. J Biol Chem. 2002;277:26351–26355. doi: 10.1074/jbc.C200273200. [DOI] [PubMed] [Google Scholar]
- Hirsila M, Koivunen P, Gunzler V, Kivirikko KI, Myllyharju J. Characterization of the human prolyl 4-hydroxylases that modify the hypoxia-inducible factor. J Biol Chem. 2003;278:30772–30780. doi: 10.1074/jbc.M304982200. [DOI] [PubMed] [Google Scholar]
- Intlekofer AM, Wang B, Liu H, Shah H, Carmona-Fontaine C, Rustenburg AS, Salah S, Gunner MR, Chodera JD, Cross JR, et al. L-2-Hydroxyglutarate production arises from noncanonical enzyme function at acidic pH. Nat Chem Biol. 2017;13:494–500. doi: 10.1038/nchembio.2307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivan M, Haberberger T, Gervasi DC, Michelson KS, Gunzler V, Kondo K, Yang H, Sorokina I, Conaway RC, Conaway JW, et al. Biochemical purification and pharmacological inhibition of a mammalian prolyl hydroxylase acting on hypoxia-inducible factor. Proc Natl Acad Sci U S A. 2002;99:13459–13464. doi: 10.1073/pnas.192342099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivan M, Kondo K, Yang H, Kim W, Valiando J, Ohh M, Salic A, Asara JM, Lane WS, Kaelin WG., Jr HIFalpha targeted for VHL-mediated destruction by proline hydroxylation: implications for O2 sensing. Science. 2001;292:464–468. doi: 10.1126/science.1059817. [DOI] [PubMed] [Google Scholar]
- Jaakkola P, Mole DR, Tian YM, Wilson MI, Gielbert J, Gaskell SJ, von Kriegsheim A, Hebestreit HF, Mukherji M, Schofield CJ, et al. Targeting of HIF-alpha to the von Hippel-Lindau ubiquitylation complex by O2-regulated prolyl hydroxylation. Science. 2001;292:468–472. doi: 10.1126/science.1059796. [DOI] [PubMed] [Google Scholar]
- Kaelin WG, Jr, Ratcliffe PJ. Oxygen sensing by metazoans: the central role of the HIF hydroxylase pathway. Mol Cell. 2008;30:393–402. doi: 10.1016/j.molcel.2008.04.009. [DOI] [PubMed] [Google Scholar]
- Karttunen S, Duffield M, Scrimgeour NR, Squires L, Lim WL, Dallas ML, Scragg JL, Chicher J, Dave KA, Whitelaw ML, et al. Oxygen-dependent hydroxylation by FIH regulates the TRPV3 ion channel. J Cell Sci. 2015;128:225–231. doi: 10.1242/jcs.158451. [DOI] [PubMed] [Google Scholar]
- Keith B, Johnson RS, Simon MC. HIF1alpha and HIF2alpha: sibling rivalry in hypoxic tumour growth and progression. Nat Rev Cancer. 2011;12:9–22. doi: 10.1038/nrc3183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JW, Tchernyshyov I, Semenza GL, Dang CV. HIF-1-mediated expression of pyruvate dehydrogenase kinase: a metabolic switch required for cellular adaptation to hypoxia. Cell Metab. 2006;3:177–185. doi: 10.1016/j.cmet.2006.02.002. [DOI] [PubMed] [Google Scholar]
- Knowles HJ, Raval RR, Harris AL, Ratcliffe PJ. Effect of ascorbate on the activity of hypoxia-inducible factor in cancer cells. Cancer Res. 2003;63:1764–1768. [PubMed] [Google Scholar]
- Koh MY, Powis G. Passing the baton: the HIF switch. Trends Biochem Sci. 2012;37:364–372. doi: 10.1016/j.tibs.2012.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koivunen P, Tiainen P, Hyvarinen J, Williams KE, Sormunen R, Klaus SJ, Kivirikko KI, Myllyharju J. An endoplasmic reticulum transmembrane prolyl 4-hydroxylase is induced by hypoxia and acts on hypoxia-inducible factor alpha. J Biol Chem. 2007;282:30544–30552. doi: 10.1074/jbc.M704988200. [DOI] [PubMed] [Google Scholar]
- Kulshreshtha R, Ferracin M, Negrini M, Calin GA, Davuluri RV, Ivan M. Regulation of microRNA expression: the hypoxic component. Cell Cycle. 2007a;6:1426–1431. [PubMed] [Google Scholar]
- Kulshreshtha R, Ferracin M, Wojcik SE, Garzon R, Alder H, Agosto-Perez FJ, Davuluri R, Liu CG, Croce CM, Negrini M, et al. A microRNA signature of hypoxia. Mol Cell Biol. 2007b;27:1859–1867. doi: 10.1128/MCB.01395-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laitala A, Aro E, Walkinshaw G, Maki JM, Rossi M, Heikkila M, Savolainen ER, Arend M, Kivirikko KI, Koivunen P, et al. Transmembrane prolyl 4-hydroxylase is a fourth prolyl 4-hydroxylase regulating EPO production and erythropoiesis. Blood. 2012;120:3336–3344. doi: 10.1182/blood-2012-07-441824. [DOI] [PubMed] [Google Scholar]
- Lando D, Peet DJ, Gorman JJ, Whelan DA, Whitelaw ML, Bruick RK. FIH-1 is an asparaginyl hydroxylase enzyme that regulates the transcriptional activity of hypoxia-inducible factor. Genes Dev. 2002;16:1466–1471. doi: 10.1101/gad.991402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee DC, Sohn HA, Park ZY, Oh S, Kang YK, Lee KM, Kang M, Jang YJ, Yang SJ, Hong YK, et al. A lactate-induced response to hypoxia. Cell. 2015;161:595–609. doi: 10.1016/j.cell.2015.03.011. [DOI] [PubMed] [Google Scholar]
- Leichtweiss HP, Lubbers DW, Weiss C, Baumgartl H, Reschke W. The oxygen supply of the rat kidney: measurements of int4arenal pO2. Pflugers Arch. 1969;309:328–349. doi: 10.1007/BF00587756. [DOI] [PubMed] [Google Scholar]
- Loenarz C, Coleman ML, Boleininger A, Schierwater B, Holland PW, Ratcliffe PJ, Schofield CJ. The hypoxia-inducible transcription factor pathway regulates oxygen sensing in the simplest animal, Trichoplax adhaerens. EMBO Rep. 2011;12:63–70. doi: 10.1038/embor.2010.170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo W, Hu H, Chang R, Zhong J, Knabel M, O'Meally R, Cole RN, Pandey A, Semenza GL. Pyruvate kinase M2 is a PHD3-stimulated coactivator for hypoxia-inducible factor 1. Cell. 2011;145:732–744. doi: 10.1016/j.cell.2011.03.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markolovic S, Wilkins SE, Schofield CJ. Protein Hydroxylation Catalyzed by 2-Oxoglutarate-dependent Oxygenases. J Biol Chem. 2015;290:20712–20722. doi: 10.1074/jbc.R115.662627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masson N, Willam C, Maxwell PH, Pugh CW, Ratcliffe PJ. Independent function of two destruction domains in hypoxia-inducible factor-alpha chains activated by prolyl hydroxylation. EMBO J. 2001;20:5197–5206. doi: 10.1093/emboj/20.18.5197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathew LK, Huangyang P, Mucaj V, Lee SS, Skuli N, Eisinger-Mathason TS, Biju K, Li B, Venneti S, Lal P, et al. Feedback circuitry between miR-218 repression and RTK activation in glioblastoma. Sci Signal. 2015;8:ra42. doi: 10.1126/scisignal.2005978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathew LK, Lee SS, Skuli N, Rao S, Keith B, Nathanson KL, Lal P, Simon MC. Restricted expression of miR-30c-2-3p and miR-30a-3p in clear cell renal cell carcinomas enhances HIF2alpha activity. Cancer Discov. 2014;4:53–60. doi: 10.1158/2159-8290.CD-13-0291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxwell PH, Eckardt KU. HIF prolyl hydroxylase inhibitors for the treatment of renal anaemia and beyond. Nat Rev Nephrol. 2016;12:157–168. doi: 10.1038/nrneph.2015.193. [DOI] [PubMed] [Google Scholar]
- Mazumdar J, Hickey MM, Pant DK, Durham AC, Sweet-Cordero A, Vachani A, Jacks T, Chodosh LA, Kissil JL, Simon MC, et al. HIF-2alpha deletion promotes Kras-driven lung tumor development. Proc Natl Acad Sci U S A. 2010;107:14182–14187. doi: 10.1073/pnas.1001296107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McIntyre A, Harris AL. Metabolic and hypoxic adaptation to anti-angiogenic therapy: a target for induced essentiality. EMBO Mol Med. 2015;7:368–379. doi: 10.15252/emmm.201404271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikhaylova O, Stratton Y, Hall D, Kellner E, Ehmer B, Drew AF, Gallo CA, Plas DR, Biesiada J, Meller J, et al. VHL-regulated MiR-204 suppresses tumor growth through inhibition of LC3B-mediated autophagy in renal clear cell carcinoma. Cancer Cell. 2012;21:532–546. doi: 10.1016/j.ccr.2012.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minamishima YA, Kaelin WG., Jr Reactivation of hepatic EPO synthesis in mice after PHD loss. Science. 2010;329:407. doi: 10.1126/science.1192811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mineo M, Ricklefs F, Rooj AK, Lyons SM, Ivanov P, Ansari KI, Nakano I, Chiocca EA, Godlewski J, Bronisz A. The Long Non-coding RNA HIF1A-AS2 Facilitates the Maintenance of Mesenchymal Glioblastoma Stem-like Cells in Hypoxic Niches. Cell Rep. 2016;15:2500–2509. doi: 10.1016/j.celrep.2016.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nadtochiy SM, Schafer X, Fu D, Nehrke K, Munger J, Brookes PS. Acidic pH Is a Metabolic Switch for 2-Hydroxyglutarate Generation and Signaling. J Biol Chem. 2016;291:20188–20197. doi: 10.1074/jbc.M116.738799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Querbes W, Bogorad RL, Moslehi J, Wong J, Chan AY, Bulgakova E, Kuchimanchi S, Akinc A, Fitzgerald K, Koteliansky V, et al. Treatment of erythropoietin deficiency in mice with systemically administered siRNA. Blood. 2012;120:1916–1922. doi: 10.1182/blood-2012-04-423715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rupaimoole R, Wu SY, Pradeep S, Ivan C, Pecot CV, Gharpure KM, Nagaraja AS, Armaiz-Pena GN, McGuire M, Zand B, et al. Hypoxia-mediated downregulation of miRNA biogenesis promotes tumour progression. Nat Commun. 2014;5:5202. doi: 10.1038/ncomms6202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salminen A, Kauppinen A, Kaarniranta K. 2-Oxoglutarate-dependent dioxygenases are sensors of energy metabolism, oxygen availability, and iron homeostasis: potential role in the regulation of aging process. Cell Mol Life Sci. 2015;72:3897–3914. doi: 10.1007/s00018-015-1978-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez-Fernandez EM, Tarhonskaya H, Al-Qahtani K, Hopkinson RJ, McCullagh JS, Schofield CJ, Flashman E. Investigations on the oxygen dependence of a 2-oxoglutarate histone demethylase. Biochem J. 2013;449:491–496. doi: 10.1042/BJ20121155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheuermann TH, Tomchick DR, Machius M, Guo Y, Bruick RK, Gardner KH. Artificial ligand binding within the HIF2alpha PAS-B domain of the HIF2 transcription factor. Proc Natl Acad Sci U S A. 2009;106:450–455. doi: 10.1073/pnas.0808092106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schofield CJ, Ratcliffe PJ. Oxygen sensing by HIF hydroxylases. Nat Rev Mol Cell Biol. 2004;5:343–354. doi: 10.1038/nrm1366. [DOI] [PubMed] [Google Scholar]
- Semenza GL. Hypoxia-inducible factors in physiology and medicine. Cell. 2012;148:399–408. doi: 10.1016/j.cell.2012.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shmakova A, Batie M, Druker J, Rocha S. Chromatin and oxygen sensing in the context of JmjC histone demethylases. Biochem J. 2014;462:385–395. doi: 10.1042/BJ20140754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thrash-Bingham CA, Tartof KD. aHIF: a natural antisense transcript overexpressed in human renal cancer and during hypoxia. J Natl Cancer Inst. 1999;91:143–151. doi: 10.1093/jnci/91.2.143. [DOI] [PubMed] [Google Scholar]
- van den Beucken T, Koch E, Chu K, Rupaimoole R, Prickaerts P, Adriaens M, Voncken JW, Harris AL, Buffa FM, Haider S, et al. Hypoxia promotes stem cell phenotypes and poor prognosis through epigenetic regulation of DICER. Nat Commun. 2014;5:5203. doi: 10.1038/ncomms6203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkins SE, Abboud MI, Hancock RL, Schofield CJ. Targeting Protein-Protein Interactions in the HIF System. Chem Med Chem. 2016;11:773–786. doi: 10.1002/cmdc.201600012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia Y, Choi HK, Lee K. Recent advances in hypoxia-inducible factor (HIF)-1 inhibitors. Eur J Med Chem. 2012;49:24–40. doi: 10.1016/j.ejmech.2012.01.033. [DOI] [PubMed] [Google Scholar]
- Xie L, Xiao K, Whalen EJ, Forrester MT, Freeman RS, Fong G, Gygi SP, Lefkowitz RJ, Stamler JS. Oxygen-regulated beta(2)-adrenergic receptor hydroxylation by EGLN3 and ubiquitylation by pVHL. Sci Signal. 2009;2:ra33. doi: 10.1126/scisignal.2000444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu W, Yang H, Liu Y, Yang Y, Wang P, Kim SH, Ito S, Yang C, Wang P, Xiao MT, et al. Oncometabolite 2-hydroxyglutarate is a competitive inhibitor of alpha-ketoglutarate-dependent dioxygenases. Cancer Cell. 2011;19:17–30. doi: 10.1016/j.ccr.2010.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang M, Chowdhury R, Ge W, Hamed RB, McDonough MA, Claridge TD, Kessler BM, Cockman ME, Ratcliffe PJ, Schofield CJ. Factor-inhibiting hypoxia-inducible factor (FIH) catalyses the post-translational hydroxylation of histidinyl residues within ankyrin repeat domains. FEBS J. 2011a;278:1086–1097. doi: 10.1111/j.1742-4658.2011.08022.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang M, Ge W, Chowdhury R, Claridge TD, Kramer HB, Schmierer B, McDonough MA, Gong L, Kessler BM, Ratcliffe PJ, et al. Asparagine and aspartate hydroxylation of the cytoskeletal ankyrin family is catalyzed by factor-inhibiting hypoxia-inducible factor. J Biol Chem. 2011b;286:7648–7660. doi: 10.1074/jbc.M110.193540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu F, White SB, Zhao Q, Lee FS. HIF-1alpha binding to VHL is regulated by stimulus-sensitive proline hydroxylation. Proc Natl Acad Sci U S A. 2001;98:9630–9635. doi: 10.1073/pnas.181341498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng X, Zhai B, Koivunen P, Shin SJ, Lu G, Liu J, Geisen C, Chakraborty AA, Moslehi JJ, Smalley DM, et al. Prolyl hydroxylation by EglN2 destabilizes FOXO3a by blocking its interaction with the USP9x deubiquitinase. Genes Dev. 2014;28:1429–1444. doi: 10.1101/gad.242131.114. [DOI] [PMC free article] [PubMed] [Google Scholar]