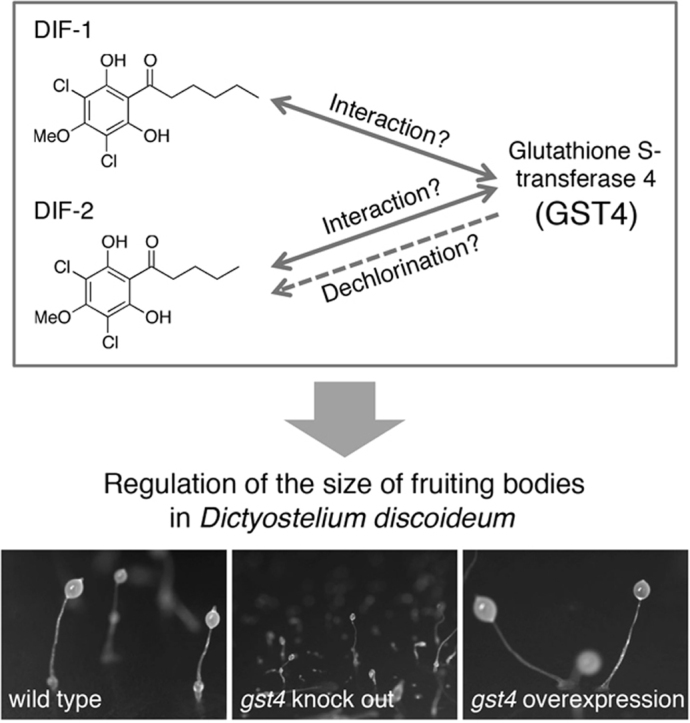
Abstract
In the development of the cellular slime mold Dictyostelium discoideum, two chlorinated compounds, the differentiation-inducing factors DIF-1 and DIF-2, play important roles in the regulation of both cell differentiation and chemotactic cell movement. However, the receptors of DIFs and the components of DIF signaling systems have not previously been elucidated. To identify the receptors for DIF-1 and DIF-2, we here performed DIF-conjugated affinity gel chromatography and liquid chromatography–tandem mass spectrometry and identified the glutathione S-transferase GST4 as a major DIF-binding protein. Knockout and overexpression mutants of gst4 (gst4– and gst4OE, respectively) formed fruiting bodies, but the fruiting bodies of gst4– cells were smaller than those of wild-type Ax2 cells, and those of gst4OE cells were larger than those of Ax2 cells. Both chemotaxis regulation and in vitro stalk cell formation by DIFs in the gst4 mutants were similar to those of Ax2 cells. These results suggest that GST4 is a DIF-binding protein that regulates the sizes of cell aggregates and fruiting bodies in D. discoideum.
Abbreviations: DIF-1, differentiation-inducing factor 1, 1-(3,5-dichloro-2,6-dihydroxy-4-methoxyphenyl)hexan-1-one; DIF-2, differentiation-inducing factor-2, 1-(3,5-dichloro-2,6-dihydroxy-4-methoxyphenyl)pentan-1-one; DIF-1-NH2, amino derivative of DIF-1, 6-amino-1-(3,5-dichloro-2,6-dihydroxy-4-methoxyphenyl)hexan-1-one; GSH, glutathione; GST, glutathione S-transferase; THPH, 1-(2,4,6-trihydroxyphenyl)hexan-1-one; LC/MS/MS, liquid chromatography–mass-mass spectrometry (liquid chromatography–tandem mass spectrometry)
Keywords: Cellular slime mold, Dictyostelium discoideum, DIF-1, DIF-2, Glutathione S-transferase
Graphical abstract
Highlights
-
•
Differentiation-inducing factors (DIFs) regulate development of D. discoideum.
-
•
Glutathione S-transferase 4 (GST4) was identified as a major DIF-binding protein.
-
•
Fruiting bodies of gst4 knockout mutant were smaller than wild-type fruiting bodies.
-
•
Fruiting bodies of gst4 overexpression mutant were larger than wild-type fruiting bodies.
-
•
GST4 and DIFs may regulate the size of fruiting bodies in D. discoideum.
1. Introduction
The cellular slime mold Dictyostelium discoideum is an excellent model organism for analyses of both chemotaxis and cell differentiation. These vegetative amoebae grow by ingesting bacteria and, on starvation, start morphogenesis; during morphogenesis, the cells gather to form a slug-shaped multicellular aggregate, in which cells differentiates into two distinct cell types, prespore and prestalk cells. Eventually, the cells form a fruiting body consisting of spores and a multicellular stalk [1], [2].
Cyclic AMP and chlorinated alkylphenones, specifically differentiation-inducing factors 1 and 2 (DIF-1 and DIF-2) (Fig. 1A), play pivotal roles in the development of D. discoideum. Extracellular cAMP is not only essential for cell differentiation, but it also acts as a chemoattractant when the cells gather to form a multicellular aggregate [1], [2]. DIF-1 and DIF-2 initially were identified as the factors that induce in vitro stalk cell differentiation in D. discoideum [3], [4], [5], [6]. DIF-1 is more active than DIF-2 in inducing stalk cell differentiation under submerged assay conditions; DIF-2 has ~40% of the specific activity of DIF-1 [6], [7]. In addition, DIF-1 has been shown to inhibit prespore differentiation [8], [9] and prestalk-to-prespore conversion [10], promote prespore-to-prestalk conversion [8], [11], [12], and induce basal disc formation [13], [14].
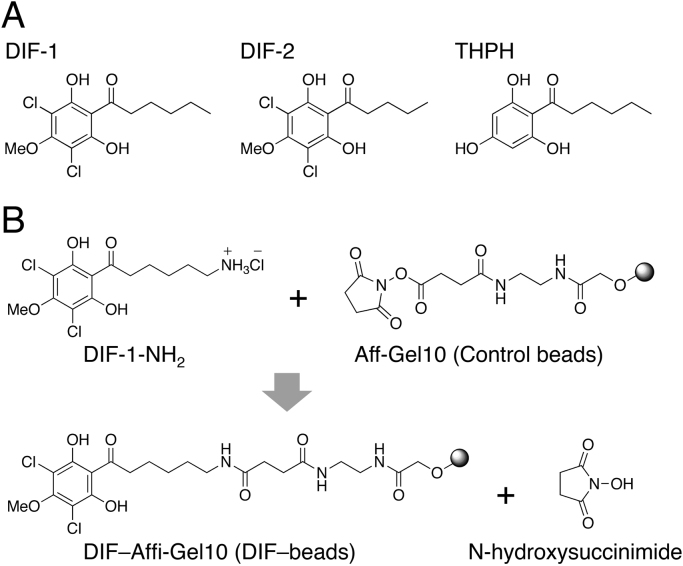
Fig. 1.
(A) Chemical structures of DIF-1, DIF-2, and THPH. DIF-1, 1-(3,5-dichloro-2,6-dihydroxy-4-methoxyphenyl)hexan-1-one; DIF-2, 1-(3,5-dichloro-2,6-dihydroxy-4-methoxyphenyl)pentan-1-one; THPH, 1-(2,4,6-trihydroxyphenyl)hexan-1-one. (B) Scheme of the coupling reaction for DIF-1-NH2 [6-amino-1-(3,5-dichloro-2,6-dihydroxy-4-methoxyphenyl)hexan-1-one hydrochloride] and Affi-Gel 10 resin. DIF–Affi-Gel 10 resin (DIF–beads) was prepared as described previously [18].
Recently, we demonstrated that DIF-1 and DIF-2 also function as negative and positive modulators, respectively, of chemotactic cell movement toward cAMP and that the mechanisms by which DIFs modulate chemotaxis differ—at least in part—from those used to induce cell differentiation [15], [16]. Despite the importance of DIF-1 and DIF-2 in D. discoideum development, the cellular signaling systems involving DIFs, including their receptors, remain to be elucidated.
To identify the receptor(s) for DIFs, we here performed affinity chromatography by using DIF-conjugated resin and recovered several DIF-binding proteins in D. discoideum, one of which was identified as glutathione S-transferase 4 (GST4), by using liquid chromatography–tandem mass spectrometry (LC/MS/MS). Knockout and overexpression mutants of gst4 (gst4– and gst4OE cells, respectively) formed fruiting bodies, but gst4– fruiting bodies were smaller than those of wild-type cells, and the fruiting bodies of gst4OE were larger than those of wild-type cells. Our results suggest that GST4 is a DIF-binding protein that regulates the sizes of cell aggregates and fruiting bodies during D. discoideum development.
2. Materials and methods
2.1. Cells and reagents
D. discoideum Ax2 cells were used in this study. Affi-Gel 10 resin was purchased from Bio-Rad (Hercules, CA, USA). DIF-1, DIF-2, and THPH (Fig. 1A) were synthesized as described previously [17], dissolved in ethanol or dimethyl sulfoxide (DMSO), and stored at −20 °C. DIF-1-NH2 (Fig. 1B) was synthesized as described previously [18].
2.2. Coupling of DIF-1-NH2 to Affi-Gel 10 resin
DIF-1-NH2 was coupled to Affi-Gel 10 resin at 4 °C to produce DIF–Affi-Gel 10 (DIF–beads) according to the manufacturer's instructions (Fig. 1B) as described previously [18]. The DIF–beads were washed well with 0.1 M phosphate-buffered saline and kept at 4 °C until use.
2.3. Preparation of cell extracts for DIF-affinity chromatography
Ax2 cells were grown axenically at 21 °C in HL-5 medium. Cells were collected by centrifugation (500×g, 3 min) and allowed to develop for 7 h on a 2% (w/v) agar plate (90 mm) at 21 °C (5×108 cells/plate). Then the cells (at loose aggregate stage) were harvested by using a salt solution (10 mM NaCl, 10 mM KCl), placed into 1.5-mL microcentrifuge tubes, and collected by centrifugation (800×g, 30 s). Cell pellets were washed with 1 mL of salt solution and then stored at –70 °C until use. The frozen cells in a microcentrifuge tube (~2.5×108 cells) were lysed with 1 mL of TBS-T (10 mM Tris-HCl pH 7.4, 150 mM NaCl, 0.05% Tween 20, and the Complete protease inhibitor cocktail [Roche Diagnostics, Mannheim, Germany]) for 10 min at 4 °C on a rotating platform. Lysates were spun (12,000×g, 10 min) twice to remove cell debris, and the resulting supernatant (cell extract) was used for affinity purification of DIF-binding proteins.
2.4. DIF-affinity chromatography, SDS-PAGE, and protein identification
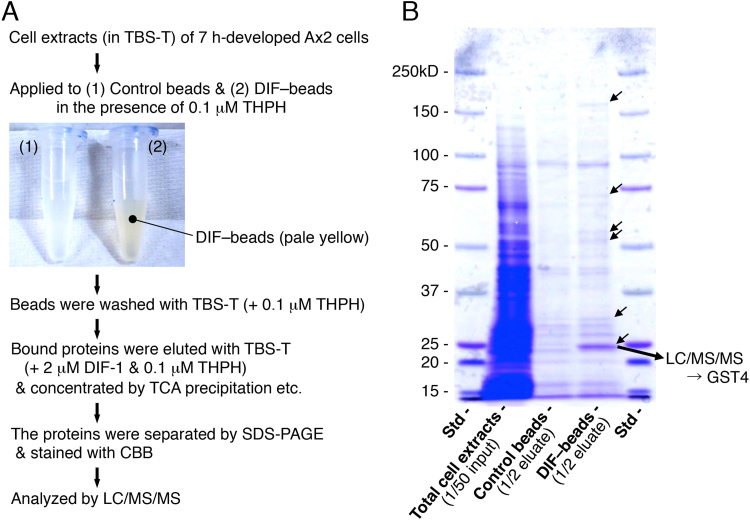
DIF-binding proteins were purified as described in Fig. 2A. Briefly, Affi-Gel 10 resin (control beads) and DIF–beads (approximately 0.6 mL of each) were placed in 1.5-mL microcentrifuge tubes, washed well with TBS-T, and then washed several times with TBS-T containing 100 nM THPH (a non-bioactive analog of DIF-1). Then, 1 μL of 0.1 mM THPH was added to 1 mL of cell extract in TBS-T (final THPH concentration, 100 nM) to block the non-specific binding of cell proteins to beads, and 0.5 mL of cell extract was applied to control beads and DIF–beads in tubes, which were rotated gently for 1 h on a microcentrifuge tube rotator (MTR-103, As One Corporation, Osaka, Japan) at 4 °C. The beads were then pelleted, the supernatants were removed, and the beads were washed 7 times with 0.7 mL of TBS-T containing 100 nM THPH. Control and DIF–beads were incubated for 1 h (with rotation at 4 °C) with 0.5 mL each of TBS-T containing 100 nM THPH and 20 μM DIF-1, to elute the bound proteins. After the beads were pelleted, 0.45 mL of each supernatant (eluate) from the control and DIF–beads was collected in each of two 1.5-mL microcentrifuge tubes.
Fig. 2.
(A) Scheme for the identification of DIF-binding proteins in D. discoideum. Ax2 cells were developed on agar plates for 7 h, lysed in 0.05% Tween 20 in Tris-buffered saline (TBS-T), and incubated with control beads or DIF–beads in the presence of 0.1 μM THPH to reduce non-specific protein binding to beads. After beads were washed with TBS-T containing THPH, the bound proteins were eluted by using TBS-T containing 2 μM DIF-1. The affinity-purified proteins were separated by SDS-PAGE and stained with Coomassie brilliant blue. The proteins that were specifically present in the DIF-beads eluate were identified by using LC/MS/MS. (B) Photo of the SDS-PAGE gel stained with Coomassie brilliant blue. Total cell extracts, eluates of control and DIF–beads, and molecular mass standards (Std) were subjected to SDS-PAGE, and the resulting gel was stained; arrows indicate a major band and minor bands unique to the DIF-beads lane. The ~25-kD protein band present in the DIF-beads lane was harvested, and the proteins in the band were analyzed by LC/MS/MS. Ultimately, the major protein in the band was identified as the glutathione S-transferase GST4.
To concentrate the eluted proteins, 4.5 μL of 2% (w/v) deoxycholate was added to each eluate; the tubes were mixed well and allowed to stand at 4 °C for 30 min. Then, 50 μL of 100% (w/v) trichloroacetic acid was added to the eluates, mixed well, and allowed to stand at 4 °C for 1 h. After centrifugation (12,000×g, 20 min, 4 °C), supernatants were discarded and, to remove remaining trichloroacetic acid, the precipitated proteins were suspended in 1 mL of cold acetone and incubated on ice for 5 min. After centrifugation (12,000×g, 20 min, 4 °C), supernatants were discarded, and the precipitated proteins were air-dried and suspended in 30 μL of SDS sample buffer [80 mM Tris–HCl pH 6.8, 2% (w/v) SDS, 0.1 M dithiothreitol, 20% (w/v) glycerol, the Complete protease inhibitor cocktail] containing 3 mM NaOH (for neutralization of any remaining trichloroacetic acid).
The sample proteins were separated by SDS–PAGE (5–20% gradient) and stained with Coomassie brilliant blue (CBB) solution (0.25% [w/v] CBB, 45% [v/v] methanol, 10% [v/v] acetic acid). The gel was washed in 45% methanol, 10% acetic acid several times and then in 10% methanol, 10% acetic acid to remove excess CBB. The protein bands specifically present in the eluate of DIF–beads (Fig. 2B) were harvested. Each protein-containing gel slice was digested with trypsin, and the gel digests were analyzed by LC/MS/MS as described previously [19], except that we used an LTQ Orbitrap XL mass spectrometer (Thermo Fisher Scientific, Waltham, MA, USA). Product ion data were searched against all biological kingdoms in the NCBI non-redundant protein sequence (nr) database by using the Mascot search engine (Matrix Science, Boston, MA, USA).
2.5. Generation of gst4-null and gst4-overexpression mutants
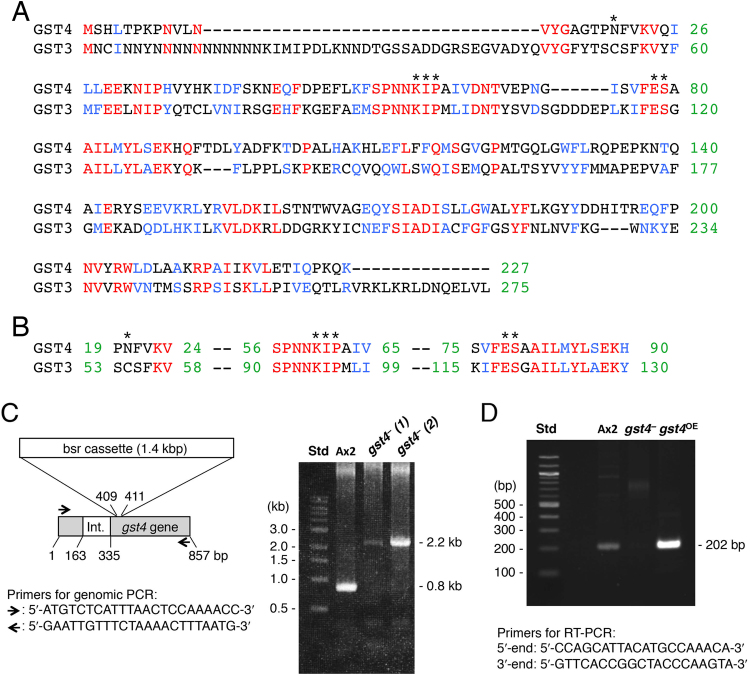
The gene-targeting construct for generating the gst4-null mutant was prepared as follows. First, the target region of gst4 was amplified from genomic DNA by using the primers described in Fig. 3C. The blasticidin S resistance gene-expression cassette (bsr) was then inserted into the gst4 coding region by using a fusion PCR technique, and the linear construct was amplified by PCR [20]. To construct the overexpression vector, the full-length gst4 gene was amplified from cDNA prepared from aggregation-stage mRNA by using an oligo-dT primer and verified by DNA sequencing. The PCR product was inserted into the cloning site of pHK12neo, as described previously [21].
Fig. 3.
(A) Deduced amino acid sequences of GST3 and GST4. GST3 and GST4 were aligned by using the ClustalW program. Identical (red) and similar (blue) amino acid residues are indicated. The identity between the two GSTs was calculated to be 29.5%, their similarity was 55.9%, and the E-value was 2e−28 in BlastP. (B) Sequence alignment around the predicted GSH binding site. Asterisks mark the key conserved residues of the predicted GSH binding site [23]. (C) Generation of the gst4− mutant. gst4– cells were generated by using the indicated gene-targeting construct. Ax2 cells and 2 clones of the gst4– mutant underwent genomic PCR amplification. The PCR products were separated on an agarose gel and visualized by ethidium bromide staining. One of the clones, gst4–(2), was used for the present study. Int., intron; Std, molecular size standard. (D) Generation of the gst4OE mutant. The gst4OE mutant was generated as described in the Materials and Methods section, and gst4 mRNA expression in Ax2 cells and the gst4 mutants was assessed by semi-quantitative RT-PCR amplification with the indicated primers.
Ax2 cells were electroporated with 10 μg of the generated DNA fragment or empty vector, and transformants were selected by using either blasticidin S (Funakoshi, Tokyo, Japan) for gene-targeting experiments or G418 (Sigma-Aldrich, Buchs, Switzerland) for the overexpression assays, as described previously [22].
Expression of gst4 mRNA in the wild-type and transformed cells was assessed by semi-quantitative RT-PCR as follows. Total RNA was prepared by using RNeasy mini kits (Qiagen, Düsseldorf, Germany) followed by cDNA synthesis by using Superscript II (Invitrogen, Carlsbad, CA, USA) with random DNA hexamers as primers. RT-PCR was performed by using the primers shown in Fig. 3D, as described previously [15].
2.6. Comparison of the phenotypes of Ax2 and gst4 mutants
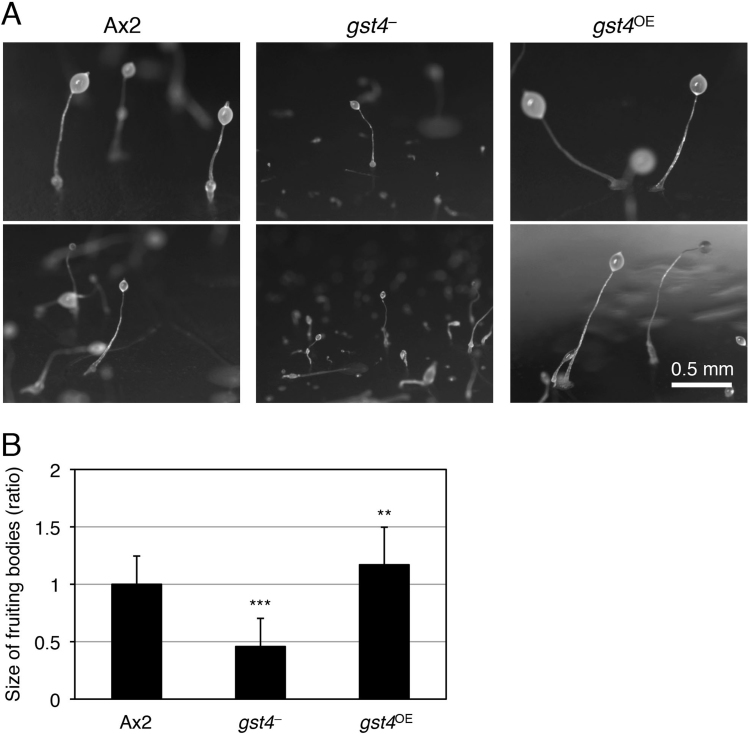
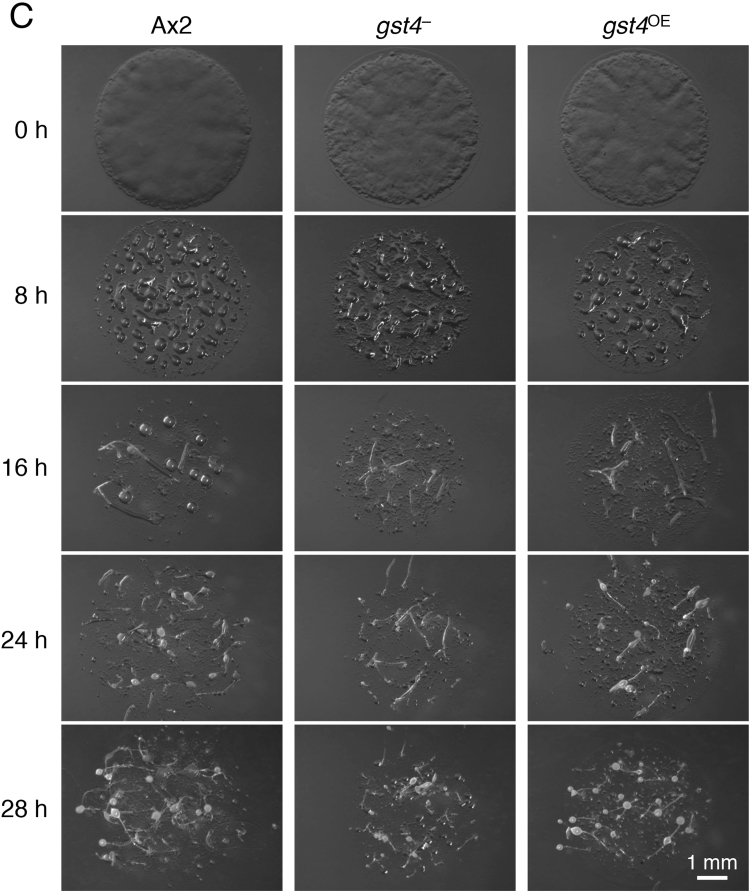
All cells were grown in HL5 medium at 21 °C. Blasticidin S (10 μg/mL) or G418 (20 μg/mL) was added to cultures of gst4– or gst4OE cells in HL5 medium, respectively. To observe fruiting body formation in the presence of bacteria (Fig. 4A), each strain was inoculated on a 5LP plate (0.5% [w/v] lactose and 0.5% [w/v] DIFCO peptone with 1.5% [w/v] agar) with Klebsiella aerogenes and incubated for 4–5 days at 21 °C. The fruiting bodies were observed through a stereomicroscope (model SZX12, Olympus, Tokyo, Japan). For the time-course observation of development (Fig. 4C and Supplemental Fig. 2), cells in the early exponential growth phase were harvested and washed 3 times with a phosphate buffer solution (10 mM Na2HPO4, 10 mM KH2PO4, pH 6.5) and then plated on 1.5% (w/v) non-nutrient agar plates at 5×106, 1×106, and 2×105 cells/cm2. Developmental processes were observed through the stereomicroscope at the indicated time points.
Fig. 4.
Developmental phenotypes of gst4 mutants. (A) Ax2, gst4–, and gst4OE cells were grown and developed in association with bacteria on agar to form fruiting bodies. Two representative photos of the fruiting bodies are shown. Bar=0.5 mm. (B) Comparison of the size of fruiting bodies. The lengths of the fruiting bodies of Ax2, gst4–, and gst4OE in the photos were measured, and the relative lengths [mean±SD (bars); n=50] are shown. **, P<0.01;***, P<0.001 versus Ax2 cells. The fruiting bodies of gst4– were smaller than those of Ax2, whereas the fruiting bodies of gst4OE were larger than those of Ax2. (C) Ax2, gst4–, and gst4OE cells were grown in an axenic medium, and starved cells were plated on agar (at the high cell density of 5×106 cells/cm2) and allowed to develop to form fruiting bodies. The cells were evaluated at the indicated time points, and representative photos are shown. Note that photos are not necessarily those of the same cell drops because continual time-course observation of a cell drop causes it to dry out and disturbs development and morphogenesis. Bar=1 mm.
2.7. Chemotaxis assay: small-population assay
Chemotaxis toward cAMP was assessed by using Ax2, gst4–, and gst4OE strains, as described previously [15].
2.8. Stalk cell induction assay
Ax2, gst4–, and gstOE cells were grown, starved, and incubated at 21 °C in 35-mm tissue culture dishes (5×106 cells/dish) in 2 mL of stalk salt solution (2 mM NaCl, 10 mM KCl, 1 mM CaCl2, 50 μg/mL penicillin, 100 μg/mL streptomycin sulfate, and 10 mM Mes-KOH [pH 6.2]) containing 5 mM cAMP. After 20 h of incubation, cells were washed 3 times with stalk salt solution and incubated for an additional 28 h in 2 mL of stalk salt solution in the presence of 0.2% (v/v) DMSO (vehicle) or DIF-1 (10 or 100 nM) or DIF-2 (10 or 100 nM). At 48 h, the stalk cell population (% stalk cells among total cells) was assessed by using phase-contrast microscopy; generally, more than 150 cells/dish were evaluated.
2.9. Statistical analysis
Statistical analysis was performed by using the unpaired Student's t-test (two-tailed). P values less than 0.05 were considered to indicate significant differences.
3. Results
3.1. Identification of DIF-binding proteins in Ax2 cells: DIF-affinity chromatography and LC/MS/MS
For DIF-affinity chromatography, we first synthesized a chemically responsive derivative of DIF-1, DIF-1-NH2, which we used to generate DIF–beads (DIF-conjugated resin) as described in Fig. 1 [18].
D. discoideum cells at the aggregation and mound stages are expected to express DIF-binding proteins (or DIF receptors), because DIF-1 and DIF-2 regulate cell differentiation and chemotaxis toward cAMP at these stages [6], [15], [16]. Therefore, to obtain DIF-binding proteins, we extracted the cellular proteins from Ax2 cells that had developed for 7 h, exposed control and DIF–beads to the protein extracts, and then applied eluted proteins to SDS-PAGE. Several bands were present specifically in the DIF-beads lane (Fig. 2B: arrows), which we then analyzed by LC/MS/MS. Several of these unique bands are still under investigation; however, we identified a major protein in the gel (~25 kD band) as a glutathione S-transferase (GST), GST4 (dictyBase gene ID, DDB_G0271892 [http://dictybase.org]). There are 21 potential GSTs in the D. discoideum genome [23]. The open reading frame of the gst4 gene consists of 684 nucleotides (Supplemental Fig. 1); GST4 comprises 227 amino-acid residues (Fig. 3A) and has a molecular mass of 26 kD. Accordingly, we anticipate that GST4 is a DIF-binding protein or a protein that is associated with a DIF-binding protein.
3.2. Phenotypes of gst4 mutants
To investigate the physiologic role(s) of GST4 in D. discoideum development, we prepared two mutant strains, gst4-null (gst4–) and gst4-overexpression (gst4OE) cells, according to the strategies described in Fig. 3C and the Materials and Methods section. We then compared the fruiting body formation of the mutants and the parental strain, Ax2, in association with bacteria (Fig. 4A). Both mutants formed fruiting bodies that were normal in shape, but the gst4– fruiting bodies were significantly smaller than those of Ax2, whereas gst4OE fruiting bodies were significantly larger than those of Ax2 (Fig. 4B). next evaluated the developmental processes of Ax2 and the mutant strains at various cell densities on agar (Fig. 4C and Supplemental Fig. 2). Again, the fruiting bodies of gst4– were smaller than those of Ax2, whereas those of gst4OE were larger than those of Ax2 (Fig. 4C). These results suggest that GST4 regulates the sizes of cell aggregates and fruiting bodies via interaction with DIFs. In addition, at low cell densities, the morphogenesis (i.e., aggregation) of gst4– cells was somewhat slower than that of Ax2 cells, whereas that of gst4OE cells was more rapid (Fig. 4C and Supplemental Fig. 2). These findings imply that GST4 is involved in chemotactic cell movement or another cellular signaling system.
3.3. Effects of DIF-1 and DIF-2 on chemotaxis and in vitro stalk cell formation in gst4 mutants
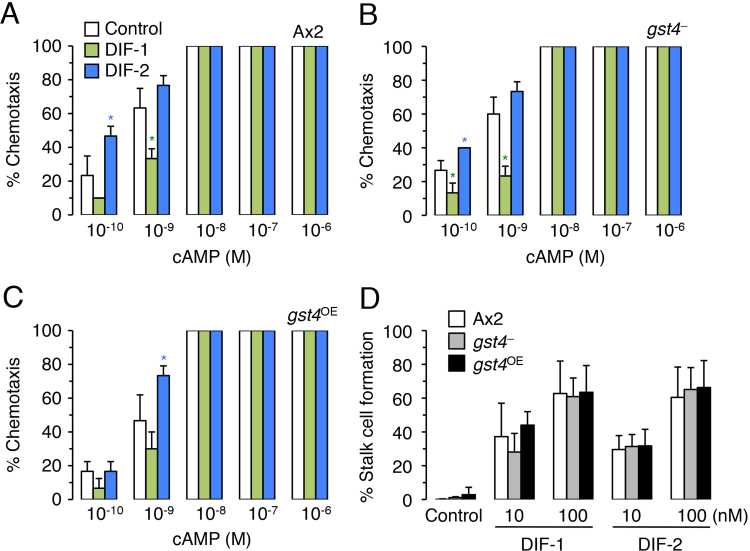
To investigate the potential role of GST4 in the DIF signaling systems, we examined the effects of DIF-1 and DIF-2 on chemotaxis and in vitro stalk cell formation in the gst4 mutants (Fig. 5). As described previously [15], [16], DIF-1 suppressed the chemotactic movement of Ax2 cells in shallow cAMP gradients, whereas DIF-2 promoted this behavior (Fig. 5A); the same pattern occurred in both gst4 mutants (Fig. 5BCE). In contrast, no significant differences in in vitro stalk cell induction in response to DIF-1 and DIF-2 were noted among Ax2 cells and the gst4 mutants (Fig. 5D). These results suggest that GST4 itself is not a DIF receptor that regulates chemotaxis or stalk cell differentiation.
Fig. 5.
Effects of DIF-1 and DIF-2 on chemotaxis and stalk cell formation in Ax2, gst4–, and gst4OE. (A–C) Ax2 (A), gst4– (B), and gst4OE (C) cells were starved for 6 h, and cell droplets were placed on PB agar containing 3 mM caffeine (control) plus 10 nM DIF-1 or DIF-2. Cells were assayed for chemotaxis toward the indicated doses of cAMP (10 cell droplets were examined for each cAMP concentration). Data are the mean±SD (bars) for triplicate samples. *, P<0.05 versus control cells. (D) Ax2, gst4–, and gst4OE cells were incubated for 20 h with cAMP (5 mM), washed free of the additive, and further incubated for 28 h with DMSO (0.2%) or DIF-1 (10 and 100 nM) or DIF-2 (10 and 100 nM). Stalk cell formation was assessed by phase-contrast microscopy, and the mean values±SD (bars) from four independent experiments are presented. No significant difference in stalk cell formation was noted between the strains.
4. Discussion
4.1. Functions of DIFs in D. discoideum
DIFs were first identified as the factors that induce prestalk and stalk cell differentiation in D. discoideum [3], [4], [5], [6]. To date, the signaling systems underlying the actions of DIFs have been only partially elucidated. The transcription factors DimA and DimB are involved in prestalk cell induction by DIF-1 [24], [25], [26], and DIF-1 has been suggested to induce prestalk–stalk cell differentiation, at least in part, by raising intracellular calcium or proton concentrations [27], [28], [29], [30], [31]. In contrast, DIFs also are known to function as modulators of chemotactic cell movement in shallow cAMP gradients, at least in part, by affecting intracellular cGMP levels [15], [16]. However, the receptors that mediate the functions of DIFs currently are unknown.
In the present study, to identify the receptors for DIF, we performed DIF-conjugated affinity gel chromatography and LC/MS/MS: we identified GST4 as a major DIF-binding protein. In addition, we found that GST4 in combination with DIF-1 or DIF-2 (or both) may regulate the size of fruiting bodies. If confirmed, this regulatory role regarding the size of fruiting bodies would be a novel function of DIFs in D. discoideum.
4.2. Role of GST4 in D. discoideum development and morphogenesis
GSTs are generally categorized into four major groups: cytosolic, microsomal, mitochondrial (also known as kappa class GSTs), and bacterial fosfomycin-resistance GSTs [32], [33], [34]. GSTs are well known as major phase II detoxification enzymes that catalyze the conjugation of glutathione (GSH) to xenobiotic and endobiotic substrates in both prokaryotic and eukaryotic cells [35], [36], [37]. In addition, GSTs have diverse cellular functions, including those in anti-oxidative stress responses and the catalysis of conjugation with endogenous ligands and of various metabolic reactions other than detoxification.
In D. discoideum, GSH is required for cell growth, transition from the growth to the developmental phase, and GSH levels can affect the culmination of cellular development and the determination of cell fate [38], [39], [40], [41]. Although the detailed roles of 21 potential cytosolic GSTs in D. discoideum remain to be elucidated, the enzyme DIF dechlorinase that directly binds to and degrades DIF-1 was recently identified to be GST3/DrcA (DIF reductive dechlorinase encoded by the gene drcA, DDB_G0293840) [23]. In addition, experiments using tritium-labeled DIF-1, recombinant GST3/DrcA, and drcA mutants showed that GST3 likely is essential and sufficient for DIF-1 dechlorination [23]; this finding indicates that GST4 is unlikely to be a DIF-1 dechlorinase.
In contrast, GST3/DrcA belongs to the Ure2p-like class of proteins [23]. Ure2p is a Saccharomyces cerevisiae transcription factor whose C-terminal region adopts a cytosolic GST fold, whereas the N-terminal region is implicated in amyloid fibril formation and heritable prion activity [34], [36], [42]. In addition, Ure2p has peroxidase activity in its native and amyloid fibrillar forms [43]. Given that GST4 also belongs to the Ure2p-like class [23] and contains 5 of the 6 key conserved amino acid residues of the GSH-binding site (Fig. 3AB), we surmise that GST4 is a DIF-binding protein. However, Cys54 of GST3/DrcA is strongly implicated as being required for DIF-1 dechlorination [23]. In contrast, GST4 has an asparagine (Asp20) in place of cysteine at the corresponding position (Fig. 3B), another feature that supports the idea that GST4 is not a DIF-1 dechlorinase.
Taken together, our findings suggest various possible roles for GST4. First, GST4 could be an as-yet-unidentified DIF-2 dechlorinase (although GST3/DrcA may also be a DIF-2 dechlorinase). Second, GST4 may be a direct or indirect regulator (such as a DIF-binding protein or a protein that associates with a DIF-binding protein, respectively) for the physiologic activities of DIF-1 or DIF-2 (or both), even though the gst4 mutants did not demonstrate any marked defects in chemotaxis or stalk cell formation under the assay conditions (Fig. 5). Regardless, DIFs in combination with GST4 may regulate the sizes of cell aggregates and fruiting bodies. Third, GST4 by itself (without DIFs) may function as a regulator of the size of cell aggregates and fruiting bodies—e.g., GST4 may affect cAMP oscillation in some way.
In conclusion, by using DIF-affinity chromatography and LC/MS/MS, we here identified GST4 as a candidate DIF-binding protein that may modulate the developmental regulation of cell aggregation in D. discoideum. Although the potential mechanisms we have suggested remain to be verified, our current results may provide new insights into DIF signaling systems and GST functions in D. discoideum.
Acknowledgements
This work was supported in part by a Grant-in-aid for Scientific Research from the Ministry of Education, Culture, Sports, Science, and Technology of Japan (no. 24590110 and 15K07964).
Footnotes
Supplementary data associated with this article can be found in the online version at http://dx.doi.org/10.1016/j.bbrep.2016.09.006.
Transparency document associated with this article can be found in the online version at http://dx.doi.org/10.1016/j.bbrep.2016.09.006.
Appendix A. Supplementary material
Supplementary material
.
Appendix B. Transparency document
Supplementary material
.
Supplementary material
.
Supplementary material
.
Supplementary material
.
References
- 1.Konijn T.M., van de Meene J.G.C., Bonner J.T., Barkley D.S. The acrasin activity of adenosine-3′,5′-cyclic phosphate. Proc. Natl. Acad. Sci. USA. 1967;58:1152–1154. doi: 10.1073/pnas.58.3.1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Darmon M., Brachet P., Pereira da Silva L.H. Chemotactic signals induce cell differentiation in Dictyostelium discoideum. Proc. Natl. Acad. Sci. USA. 1975;72:3163–3166. doi: 10.1073/pnas.72.8.3163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Town D.D., Gross J.D., Kay R.R. Cell differentiation without morphogenesis in Dictyostelium discoideum. Nature. 1976;262:717–719. doi: 10.1038/262717a0. [DOI] [PubMed] [Google Scholar]
- 4.Morris H.R., Taylor G.W., Masento M.S., Jermyn K.A., Kay R.R. Chemical structure of the morphogen differentiation inducing factor from Dictyostelium discoideum. Nature. 1987;328:811–814. doi: 10.1038/328811a0. [DOI] [PubMed] [Google Scholar]
- 5.Morris H.R., Masento M.S., Taylor G.W., Jermyn K.A., Kay R.R. Structure elucidation of two differentiation inducing factors (DIF-2 and DIF-3) from the cellular slime mould Dictyostelium discoideum. Biochem. J. 1988;249:903–906. doi: 10.1042/bj2490903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kay R.R., Flatman P., Thompson C.R.L. DIF signalling and cell fate. Semin. Cell Dev. Biol. 1999;10:577–585. doi: 10.1006/scdb.1999.0341. [DOI] [PubMed] [Google Scholar]
- 7.Masento M.S., Morris H.R., Taylor G.W., Johnson S.J., Skapski A.C., Kay R.R. Differentiation-inducing factor from the slime mould Dictyostelium discoideum and its analogues. Biochem. J. 1988;256:23–28. doi: 10.1042/bj2560023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kay R.R., Jermyn K.A. A possible morphogen controlling differentiation in Dictyostelium. Nature. 1983;303:242–244. doi: 10.1038/303242a0. [DOI] [PubMed] [Google Scholar]
- 9.Berks M., Kay R.R. Combinatorial control of cell differentiation by cAMP and DIF-1 during development of Dictyostelium discoideum. Development. 1990;110:977–984. doi: 10.1242/dev.110.3.977. [DOI] [PubMed] [Google Scholar]
- 10.Inouye K. Control of cell type proportions by a secreted factor in Dictyostelium discoideum. Development. 1989;107:605–609. doi: 10.1242/dev.107.3.605. [DOI] [PubMed] [Google Scholar]
- 11.Early A., Williams J.G. A Dictyostelium prespore-specific gene is transcriptionally repressed by DIF in vitro. Development. 1988;103:519–524. doi: 10.1242/dev.103.3.519. [DOI] [PubMed] [Google Scholar]
- 12.Kubohara Y., Okamoto K. DIF-resistant expression of a prespore-specific gene in Dictyostelium in vitro development. Exp. Cell Res. 1993;204:382–384. doi: 10.1006/excr.1993.1047. [DOI] [PubMed] [Google Scholar]
- 13.Saito T., Kato A., Kay R.R. DIF-1 induces the basal disc of the dictyostelium fruiting body. Dev. Biol. 2008;317:444–453. doi: 10.1016/j.ydbio.2008.02.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yamada Y., Nuñez-Corcuera B., Williams J.G. DIF-1 regulates Dictyostelium basal disc differentiation by inducing the nuclear accumulation of a bZIP transcription factor. Dev. Biol. 2011:77–86. doi: 10.1016/j.ydbio.2011.03.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kuwayama H., Kubohara Y. Differentiation-inducing factor-1 and -2 function also as modulators for Dictyostelium chemotaxis. PLoS One. 2009;4:e6658. doi: 10.1371/journal.pone.0006658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kuwayama H., Kikuchi H., Oshima Y., Kubohara Y. Artificial compounds differentially control Dictyostelium chemotaxis and cell differentiation. Cell Struct. Funct. 2011;36:21–26. doi: 10.1247/csf.10018. [DOI] [PubMed] [Google Scholar]
- 17.Gokan N., Kikuchi H., Nakamura K., Oshima Y., Hosaka K., Kubohara Y. Structural requirements of Dictyostelium differentiation-inducing factors for their stalk-cell-inducing activity in Dictyostelium cells and anti-proliferative activity in K562 human leukemic cells. Biochem. Pharmacol. 2005;70:676–685. doi: 10.1016/j.bcp.2005.06.002. [DOI] [PubMed] [Google Scholar]
- 18.Kubohara Y., Kikuchi H., Nakamura K., Matsuo Y., Oshima Y. Preparation of an antibody that recognizes and neutralizes Dictyostelium differentiation-inducing factor-1. Biochem. Biophys. Res. Commun. 2010;396:364–369. doi: 10.1016/j.bbrc.2010.04.098. [DOI] [PubMed] [Google Scholar]
- 19.Edmondson R.D., Vondriska T.M., Biederman K.J., Zhang J., Jones R.C., Zheng Y., Allen D.L., Xiu J.X., Cardwell E.M., Pisano M.R., Ping P. Protein kinase C epsilon signaling complexes include metabolism- and transcription/translation-related proteins: complimentary separation techniques with LC/MS/MS. Mol. Cell. Proteom. 2002;1:421–433. doi: 10.1074/mcp.m100036-mcp200. [DOI] [PubMed] [Google Scholar]
- 20.Kuwayama H., Obara S., Morio T., Katoh M., Urushihara H., Tanaka Y. PCR-mediated generation of a gene disruption construct without the use of DNA ligase and plasmid vectors. Nucleic Acid Res. 2002;30(E2) doi: 10.1093/nar/30.2.e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kuwayama H., Miyanaga Y., Urushihara H., Ueda M. A RabGAP regulates life-cycle duration via trimeric G-protein cascades in Dictyostelium discoideum. PLoS One. 2013;8:e81811. doi: 10.1371/journal.pone.0081811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kuwayama H., Nagasaki A. Desalted deep sea water increases transformation and homologous recombination efficiencies in Dictyostelium discoideum. J. Mol. Microbiol. Biotechnol. 2008;14:157–162. doi: 10.1159/000107371. [DOI] [PubMed] [Google Scholar]
- 23.Velazquez F., Peak-Chew S.Y., Fernández I.S., Neumann C.S., Kay R.R. Identification of a eukaryotic reductive dechlorinase and characterization of its mechanism of action on its natural substrate. Chem. Biol. 2011;18:1252–1260. doi: 10.1016/j.chembiol.2011.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Thompson C.R.L., Fu Q., Buhay C., Kay R.R., Shaulsky G. A bZIP/bRLZ transcription factor required for DIF signaling in Dictyostelium. Development. 2004;131:513–523. doi: 10.1242/dev.00939. [DOI] [PubMed] [Google Scholar]
- 25.Huang E., Blagg S.L., Keller T., Katoh M., Shaulsky G., Thompson C.R.L. bZIP transcription factor interactions regulate DIF responses in Dictyostelium. Development. 2006;133:449–458. doi: 10.1242/dev.02240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhukovskaya N.V., Fukuzawa M., Yamada Y., Araki T., Williams J.G. The Dictyostelium bZIP transcription factor DimB regulates prestalk-specific gene expression. Development. 2006;133:439–448. doi: 10.1242/dev.02190. [DOI] [PubMed] [Google Scholar]
- 27.Inouye K. Induction by acid load of the maturation of prestalk cells in Dictyostelium discoideum. Development. 1988;104:669–681. [Google Scholar]
- 28.Kubohara Y., Okamoto K. Cytoplasmic Ca2+ and H+ concentrations determine cell fate in Dictyostelium discoideum. FASEB J. 1994;8:869–874. doi: 10.1096/fasebj.8.11.8070636. [DOI] [PubMed] [Google Scholar]
- 29.Schaap P., Nebl T., Fisher P.R. A slow sustained increase in cytosolic Ca2+ levels mediates stalk gene induction by differentiation inducing factor in Dictyostelium. EMBO J. 1996;15:5177–5183. [PMC free article] [PubMed] [Google Scholar]
- 30.Azhar M., Kennady P.K., Pande G., Nanjundiah V. Stimulation by DIF causes an increase of intracellular Ca2+ in Dictyostelium discoideum. Exp. Cell Res. 1997;230:403–406. doi: 10.1006/excr.1996.3420. [DOI] [PubMed] [Google Scholar]
- 31.Kubohara Y., Arai A., Gokan N., Hosaka K. Pharmacological evidence that stalk cell differentiation involves increases in the intracellular Ca2+and H+ concentrations in Dictyostelium discoideum. Dev. Growth Differ. 2007;49:253–264. doi: 10.1111/j.1440-169X.2007.00920.x. [DOI] [PubMed] [Google Scholar]
- 32.Hayes J.D., Flanagan J.U., Jowsey I.R. Glutathione transferases. Annu. Rev. Pharmacol. Toxicol. 2005;45:51–88. doi: 10.1146/annurev.pharmtox.45.120403.095857. [DOI] [PubMed] [Google Scholar]
- 33.Frova C. Glutathione transferases in the genomics era: new insights and perspectives. Biomol. Eng. 2006;23:149–169. doi: 10.1016/j.bioeng.2006.05.020. [DOI] [PubMed] [Google Scholar]
- 34.Morel M., Ngadin A.A., Droux M., Jacquot J.P., Gelhaye E. The fungal glutathione S-transferase system. Evidence of new classes in the wood-degrading basidiomycete Phanerochaete chrysosporium. Cell Mol. Life Sci. 2009;66:3711–3725. doi: 10.1007/s00018-009-0104-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sheehan D., Meade G., Foley V.M., Down C.A. Structure, function and evolution of glutathione transferases: implications for classification of non-mammalian members of an ancient enzyme superfamily. Biochem. J. 2001;360:1–16. doi: 10.1042/0264-6021:3600001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Oakley A.J. Glutathione transferases: new functions. Curr. Opin. Struct. Biol. 2005;15:716–723. doi: 10.1016/j.sbi.2005.10.005. [DOI] [PubMed] [Google Scholar]
- 37.Allocati N., Federici L., Masulli M., Ilio C. Di. Glutathione transferases in bacteria. FEBS J. 2009;276:58–75. doi: 10.1111/j.1742-4658.2008.06743.x. [DOI] [PubMed] [Google Scholar]
- 38.Kim B.J., Choi C.H., Lee C.H., Jeong S.Y., Kim J.S., Kim B.Y., Yim H.S., Kang S.O. Glutathione is required for growth and prespore cell differentiation in Dictyostelium. Dev. Biol. 2005;284:387–398. doi: 10.1016/j.ydbio.2005.05.034. [DOI] [PubMed] [Google Scholar]
- 39.Kim J.S., Seo J.H., Kang S.O. Glutathione initiates the development of Dictyostelium discoideum through the regulation of YakA. Biochim. Biophys. Acta. 1843;2014:664–674. doi: 10.1016/j.bbamcr.2013.12.014. [DOI] [PubMed] [Google Scholar]
- 40.Choi C.H., Kim B.J., Jeong S.Y., Lee C.H., Kim J.S., Park S.J., Yim H.S., Kang S.O. Reduced glutathione levels affect the culmination and cell fate decision in Dictyostelium discoideum. Dev. Biol. 2006;295:523–533. doi: 10.1016/j.ydbio.2006.03.038. [DOI] [PubMed] [Google Scholar]
- 41.Choi C.H., Park S.J., Jeong S.Y., Yim H.S., Kang S.O. Methylglyoxal accumulation by glutathione depletion leads to cell cycle arrest in Dictyostelium. Mol. Microbiol. 2008;70:1293–1304. doi: 10.1111/j.1365-2958.2008.06497.x. [DOI] [PubMed] [Google Scholar]
- 42.Chan J.C., Oyler N.A., Yau W.M., Tycko R. Parallel beta-sheets and polar zippers in amyloid fibrils formed by residues 10-39 of the yeast prion protein Ure2p. Biochemistry. 2005;44:10669–10680. doi: 10.1021/bi050724t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bai M., Zhou J.M., Perrett S. The yeast prion protein Ure2 shows glutathione peroxidase activity in both native and fibrillar forms. J. Biol. Chem. 2004;279:50025–50030. doi: 10.1074/jbc.M406612200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material
Supplementary material
Supplementary material
Supplementary material
Supplementary material