Abstract
Background
Kynurenine aminotransferase 3 (KAT3) catalyzes the transamination of Kynurenine to kynurenic acid, and is identical to cysteine conjugate beta-lyase 2 (CCBL2) and glutamine transaminase L (GTL). GTL was previously purified from the rat liver and considered as a liver type glutamine transaminase. However, because of the substrate overlap and high sequence similarity of KAT3 and KAT1, it was difficult to assay the specific activity of each KAT and to study the enzyme localization in animals.
Methods
KAT3 transcript and protein levels as well as enzyme activity in the liver and kidney were analyzed by regular reverse transcription-polymerase chain reaction (RT-PCR), real time RT-PCR, biochemical activity assays combined with a specific inhibition assay, and western blotting using a purified and a highly specific antibody, respectively.
Results
This study concerns the comparative biochemical characterization and localization of KAT 3 in the mouse. The results showed that KAT3 was present in both liver and kidney of the mouse, but was much more abundant in the kidney than in the liver. The mouse KAT3 is more efficient in transamination of glutamine with indo-3-pyruvate or oxaloacetate as amino group acceptor than the mouse KAT1.
Conclusions
Mouse KAT3 is a major glutamine transaminase in the kidney although it was named a liver type transaminase.
General significance
Our data highlights KAT3 as a key enzyme for studying the nephrotoxic mechanism of some xenobiotics and the formation of chemopreventive compounds in the mouse kidney. This suggests tissue localizations of KAT3/GTL/CCBL2 in other animals may be carefully checked.
Abbreviations: CCBL, cysteine conjugate beta-lyase; GTK, glutamine transaminase K; GTL, glutamine transaminase L; KAT, kynurenine aminotransferase; KYNA, kynurenic acid; PLP, pyridoxal-5′-phosphate.
Keywords: Kynurenine aminotransferase, Kynurenic acid, Kynurenine, Cysteine conjugate beta-lyase 2, Glutamine transaminase, Aminotransferase
Highlights
-
•
Mouse kynurenine aminotransferase 3 (KAT3) was specifically inhibited by methionine.
-
•
Mouse KAT3 is more abundant in the kidney than in the liver.
-
•
Mouse KAT3 is a major glutamine transaminase in the kidney although it was named a liver transaminase.
1. Introduction
Kynurenine aminotransferase (KAT) catalyzes the transamination of kynurenine to kynurenic acid (KYNA). KYNA is the only known endogenous antagonist of the N-methyl-d-aspartate subtype of glutamate receptors [1], [2], [3], [4]. It is also the antagonist of the α7-nicotinic acetylcholine receptor [5], [6], [7], [8]. Because of the pivotal role KYNA plays, KATs have been paid a great attention for their functions in the central nervous system (CNS) [9], [10]. In addition to the roles in CNS, KYNA is also involved in the control of the cardiovascular function [11] and physiological blood pressure [12], [13]. In humans, rats and mice, four KAT enzymes, KATs 1, 2, 3 and 4 were reported [9]. Among the four mammalian KATs, KAT1 and KAT3 share similar genomic structure and high sequence identity [14] and therefore likely have overlapped biological functions. KAT1 is identical to glutamine transaminase K (GTK), a kidney type transaminase and cysteine conjugate beta-lyase 1 (CCBL1). Recently, it was demonstrated that KAT3 is not only identical to cysteine conjugate beta-lyase 2 (CCBL2), but also to glutamine transaminase L (GTL), a liver type glutamine transaminase [15]. Based on the biochemical property of mouse KAT3, it efficiently catalyzes the transamination reaction of glutamine, methionine, phenylalanine and cysteine [16].
Although the involvement of KAT1 and KAT3 in brain KYNA production has frequently been discussed [9], their specific activities, and mRNA transcripts are much higher in livers and kidneys than in brains [14], [17]. GTK is referred to as KAT1 in most cases [18], [19], [20], but the fact is that glutamine is a much better substrate of the enzyme than is kynurenine [20]. Moreover, glutamine is one of the most abundant free amino acids in the body and central to many of the pathways of intermediary metabolism [21]. The concentration of glutamine in most tissues, including the liver and kidney, is in the mM range [22], [23], whereas the concentration of kynurenine in liver has been reported to be about 0.4 μM [24]. Therefore, glutamine transamination activity of GTK and GTL may play a fundamental role in metabolism of glutamine.
Glutamine is metabolized by mechanisms of the glutaminase and glutaminase II pathways. It is suggested that the glutaminase II pathway is quantitatively important in humans [25]. Glutaminase II pathway consists of a glutamine transaminase coupled to ω-amidase. Transamination of glutamine results in formation of the corresponding α-ketoglutaramate which is hydrolyzed by ω-amidase to α-ketoglutarate and ammonia [26]. This glutaminase II pathway plays many roles in animals and humans. Glutamine transaminases function as repair enzymes that salvage a number of α-keto acids, and play roles in nitrogen and sulfur homeostasis, 1-carbon metabolism, and the formation of chemopreventive metabolites of seleno-amino acids. The glutamine transaminases, including GTK and GTL may contribute to the detoxification of halogenated alkenes and possibly other xenobiotic electrophiles, many of which are environmental contaminants [25], [27]. In addition, the “glutamine addiction” of many tumors suggests that the glutamine transaminases may have a fundamental, influential role in regulating cancer progression although the role of the glutaminase II pathway in cancer biology has been less studied. Considering the above facts and the importance of glutamine metabolism, the mouse KAT3/GTL/CCLB2 may play more important roles in kidney and liver than in the brain. In this paper, we investigated its expressions, localizations and enzyme activity in the liver and kidney, and assessed its potential roles regarding transamination activity towards glutamine and others except kynurenine.
2. Methods
2.1. Expression and purification of recombinant mouse KAT1 and KAT3
Both recombinant mouse KAT1 and KAT3 were expressed and purified as previously described [28]. Briefly, amplified KAT1 and KAT3 sequences were cloned into an Impact™-CN plasmid (New England Biolabs) for expression of a fusion protein containing a chitin-binding domain. Transformed Escherichia coli cells were used to produce the recombinant proteins. The expressed proteins were purified using affinity purification, DEAE-Sepharose, Mono-Q and gel-filtration chromatography. The purified recombinant KAT1 and KAT3 were concentrated to 10 mg mL−1 protein in 10 mM phosphate buffer (pH 7.5) containing 40 mM pyridoxal-5′-phosphate (PLP) and 10 mM b-mercaptoethanol using a Centricon YM-50 concentrator (Millipore). Protein concentration was tested by a protein assay kit from Bio-Rad (Hercules, CA) using bovine serum albumin as a standard.
2.2. Glutamine transaminase and KAT activity assay
KAT activity assay was based on previously described methods [28]. Briefly, a reaction mixture of 100 μL, containing 5 mM l-kynurenine, 2 mM glyoxylate or other α-keto acid (for co-substrate test), 40 μM PLP, and 5 μg of recombinant protein, was prepared using 100 mM potassium phosphate buffer (pH 7.5). The mixture was incubated for 15 min at 38 °C, and the reaction stopped by adding an equal volume of 0.8 M formic acid. The supernatant of the reaction mixture, obtained by centrifugation at 15,000g for 10 min, was analyzed for the product, KYNA, by high-performance liquid chromatography (HPLC) with ultraviolet detection at 330 nm.
A glutamine transaminase activity assay was developed here. A reaction mixture of 100 µl consisted of 2 µg of purified KAT enzymes, 5 mM glutamine, 2 mM phenylpyruvate, and 40 µM PLP in 100 mM boric acid buffer, pH 9.0, in 15 min at 38 °C. The reaction mixture was incubated for 15 min at 38 °C and the reaction was stopped by adding an equal volume of 0.8 M formic acid into the reaction mixture. The mixture was centrifuged for 10 min at 15,000g and supernatant (5 µl) was injected into an HPLC reverse-phase column (150×4.6 mm, Varian, Palo Alto, CA) for analysis. The formation of transamination product, phenylalanine was monitored by an on-line UV detector at a wavelength of 257 nm.
2.3. Co-substrate specificity of mouse KAT1 and KAT3
To determine the substrate specificity for α-keto acids, 16 α-keto acids were individually tested for their ability to function as an amino group acceptor for mouse KAT1 and KAT3. Each of the 16 α-keto acids were assayed at 2 mM in the presence of 5 mM kynurenine in the 100 mL reaction mixture prepared in 100 mM phosphate buffer, including 40 μM PLP. The rate of KYNA production was determined as described in the KAT activity assay.
2.4. Glutamine activity assay for mouse tissue crude proteins
Three female and three male mice were sacrificed and their livers and kidneys were immediately removed and transferred into a protein extract buffer (50 mL of 50 mM Tris–acetate buffer containing 40 mM PLP, 10 mM β-mercaptoethanol, 2 mM EDTA, and 1 mM PMSF at pH 8.0). The livers and kidneys were homogenized in a pre-cooled homogenizer, separately. The mixture was centrifuged at 20,000g, 4 °C, for 20 min. The supernatant was collected, and dialyzed overnight at 4 °C against the protein extract buffer with a 50 kDa molecular weight cutoff membrane. The dialyzed crude protein extracts were used for enzyme activity assay, inhibition assay and western blotting. The crude protein concentration was determined by a protein assay kit from Bio-Rad (Hercules, CA) using bovine serum albumin as a standard. A crude extract sample containing 20 μg protein was used in 100 μl of the same typical reaction mixture as was used in the recombinant protein activity assay. The mixture was incubated at 38 °C for 2 h.
2.5. Western blot analysis
2.5.1. Purification of anti mouse KAT3 antibody
In order to get specific anti-mouse KAT3 antibody without cross-reacting with mouse KAT1, the very similar protein to KAT3, we purified KAT3 antibodies and verified the specificity by western-blotting. Briefly, the anti mouse KAT3 rabbit polyclonal antibody was obtained from Santa Cruz (Cat#SC-67378). The antiserum was diluted into 2× volumes with ice cooled sterile saline, and passed through 0.45 μm filter. Saturated ammonium sulfate solution was added into the sample up to 50% saturation, and stored at 4 °C for 4 h. The mixture was centrifuged at 15 kg for 10 min at 4 °C, and the precipitated protein was dissolved in 1× volume of ice-cooled saline. 1/2 volume saturated ammonium sulfate was added in the solution up to 33% saturation, and kept at 4 °C for at least 4 h. After centrifugation, the precipitated protein (IgG) was dissolved in PBS, and subjected to further affinity purification. The antibody was first absorbed by the recombinant mouse KAT1 protein fused with a chitin-binding domain and fixed in a chitin resin (New England BioLabs). The resin was washed with PBS while all flowed through protein that was not bound to KAT1 protein was collected and concentrated using a Millipore protein concentrator with 10 kDa molecular weight cutoff. The specificity of the concentrated protein was tested by western blot analysis using horseradish peroxidase catalyzing color change.
2.5.2. Western blotting
Twenty μg crude liver or kidney protein sample was boiled for 5 min in Laemmli sample buffer and then separated by SDS-PAGE at 12% polyacrylamide using a Hoefer SE 260 mini-vertical gel electrophoresis apparatus at 180 V. Separated proteins in the polyacrylamide gel were transferred onto a PVDF membrane (GE Healthcare) in CAPS buffer (10 mM CAPS, pH11.0) at 400 mA using a Hoefer TE 22 transfer apparatus. The membrane was blocked with 5% BSA in 1× PBST (PBS with Tween 20) and incubated with the purified primary antibody (1:500 diluted in 1× PBST with 1% BSA) overnight at 4 °C. The membrane was washed three times for 10 min each in PBST, and incubated with goat anti-rabbit IgG (whole molecule) conjugated with HRP (1:10,000 in 1× PBST with 0.5% BSA) (Sigma-Aldrich) for 90 min. After extensive wash, the membrane was incubated in 2.0 mM 3,3′-diaminobenzidine solution prepared in PBS for 5 min. The image of immunoblot was digitalized with the Alphaimager HP system (Alpha Innotech) and saved as TIF format.
2.6. RT-PCR and qRT-PCR analyses
2.6.1. RT-PCR
The kidney or liver tissues from three females and three males were grinded to powder in liquid nitrogen. Approximately 50 mg of kidney or liver power was used to extract total RNA using a mirVana miRNA Isolation Kit (Ambion, Inc.) according to the manufacturer's instructions. Total RNA was treated with RNase free DNAase. After heat-inactivation, total RNA was reverse transcribed to the first-strand cDNA using SuperScript™ III First-Strand Synthesis System (Invitrogen) with oligo (dT) 18 primer. The mRNA transcripts of mouse KAT1 and KAT3 were analyzed using RT-PCR. A specific primer pair for KAT1 (Forward: 5′-GAGCTGGAGCTGGTGGCTGC-3′ and Reverse: 5′-GCGCTGCCGATGGTCAGTGT-3′) that amplifies a 152 bp product and specific primer pairs for KAT3 (Forward: 5′-GCTGACCTTTGCGTCAAGCACG-3′ and Reverse: 5′-GGGCCAATGCTCCAGCCGAG-3′) that amplifies a 188 bp DNA fragment were designed and used for PCR amplification. A mouse β-actin gene was used as a control (Forward primer: 5′-GCGGACTGTTACTGAGCTGCGT-3′ and Reverse primer: 5′-TGCTGTCGCCTTCACCGTTCC-3′. Product length=217 bp). The PCR reaction consisted of 2 min at 94 °C, 30 cycles (each cycle: 94 °C for 30 s, 60 °C for 30 s, 72 °C for 1 min), and a final extension at 72 °C for 10 min. All final products were analyzed by 1% agarose gel electrophoresis. The image was digitalized with the Alphaimager HP system (Alpha Innotech).
2.6.2. qRT-PCR analyses
To confirm the RT-PCR results, six different female (3) and male (3) mice were chosen for qRT-PCR experiment. The total RNA extraction and the first-strand cDNA synthesis were same as described above. The qRT-PCR amplifications were performed with ROCHE LightCycler 96 using a FastStart Essential DNA Green Master (Roche). Each sample was amplified in triplicate. The primer sequences were: Ccbl1/kat1 sense 5′-GGAGATGGACCCACTCAAGAAT-3′ and antisense 5′- GCCTGAAAGGCTGTGAACAAG-3′; Ccbl2/kat3 sense 5′-AACCCCGGCGACACCTA-3′, and antisense 5′- TGATCGTTCTCCTGGTTCCAA-3′; Gapdh sense 5′-TGCACCACCAACTGCTTAGC-3′, and antisense 5′- CAGTCTTCTGAGTGGCAGTGATG-3′, which are different from the above RT-PCR primers. The qRT-PCR amplification conditions were: denaturation at 94 °C for 2 min followed by 30–35 cycles of 94 °C for 10 s and 72 °C for 30 s. Fluorescence values were recorded at 72 °C. Melting curve analysis was performed to confirm specific amplification. Relative expression levels of target genes were calculated by the 2−ΔΔCt method [29].
2.7. Statistical analysis
The results of enzyme activities and relative transcript levels were analyzed by Student's t-test.
3. Results
3.1. Comparison of co-substrate specificities of mouse KAT1 and KAT3
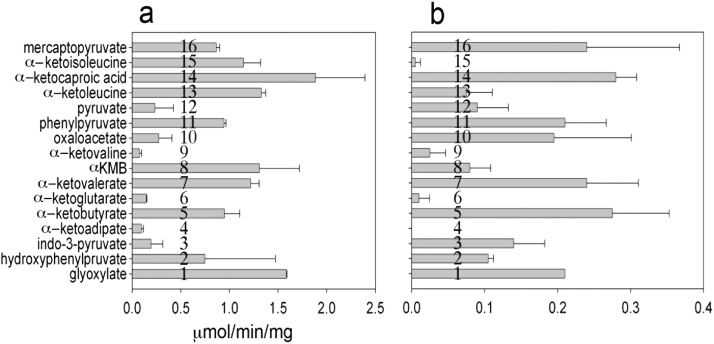
For comparison, two KATs were tested for KAT activity towards 16 different α-ketoacids. They share many α-ketoacids as co-substrates, showing relatively high activities with glyoxylate, α-Ketoisocaproic acid, phenylpyruvate, α-ketovalerate, mercaptopyruvate and αKMB. With respect to the specificities of these co-substrates, both enzymes behaved similarly. However, differences between mouse KAT1 and KAT3 became evident with α-ketoisoleucine, oxaloacetate or indo-3-pyruvate. Mouse KAT1 showed high activity with α-ketoisoleucine, and little activity with indo-3-pyruvate and oxaloacetate, while mouse KAT3 displayed high activity with indo-3-pyruvate and oxaloacetate, and little activity with α-ketoisoleucine (Fig. 1). The co-substrate profile of mouse KAT1 was essentially the same as human KAT1 except the mouse enzyme showed a high activity with α-ketoisoleucine, while the activity of human KAT1 was undetectable under the same reaction conditions [20].
Fig. 1.
Transamination activity of two KATs towards different α-ketoacids. Purified recombinant mouse KAT1 and KAT3 were incubated with each of 16 α-ketoacids at 2 mM in the presence of 5 mM kynurenine in a reaction mixture including 40 μM PLP and 100 mM phosphate, pH 7.5. The activity was quantified by the amount of KYNA produced in the reaction mixture. (A) mouse KAT1; (B) mouse KAT3.
3.2. Glutamine transaminase activity assay using phenylpyruvate as a co-substrate
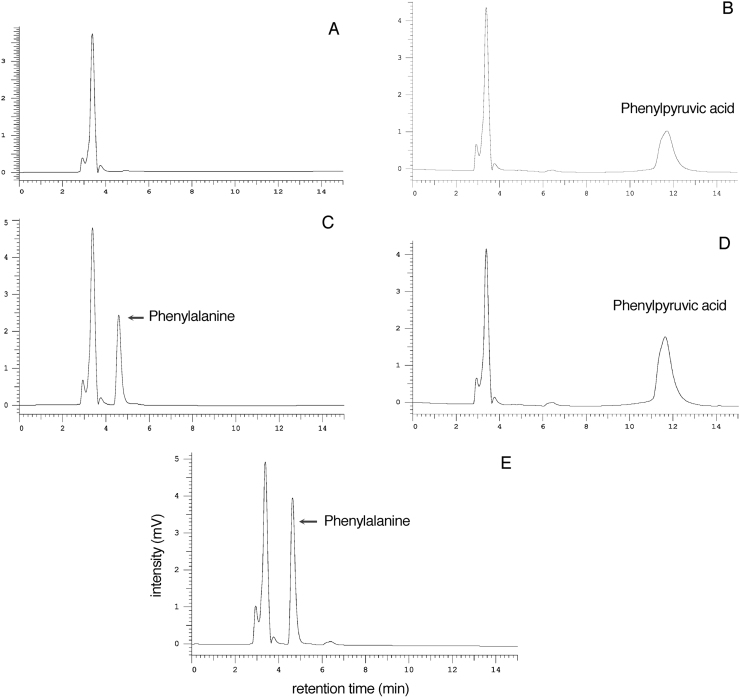
A glutamine transaminase activity assay using HPLC UV–vis detection was developed here. The enzyme reaction mixture (100 μl) includes 5 mM glutamine, 2 mM phenylpyruvate and 2 μg recombinant mouse KAT3 or KAT1, 100 mM boric acid buffer, pH 9.0. The mixture was incubated at 38 °C for 15 min. The reaction was stopped by adding an equal volume of 0.8 M formic acid. The mixture was centrifuged and the supernatant was injected into an HPLC column (150×4.6 mm, Varian, Palo Alto, CA) for analysis. The mobile phase consists of 10 mM potassium phosphate (monobasic) buffer containing 10% (v/v) acetonitrile for the analysis of phenylalanine in the reaction mixtures. The formation of transamination product, phenylalanine was monitored by an on-line UV detector at a wavelength of 257 nm. The activity assays showed both mouse KAT1 and KAT3 had glutamine transaminase activities using phenylpyruvate as a co-substrate. Fig. 2 showed chromatograms of the assay using mouse KAT3 as an example. In the figure, phenylpyruvate was consumed completely by the enzyme to produce phenylalanine. Table 1 showed enzyme kinetic parameters towards glutamine and phenylalanine.
Fig. 2.
HPLC UV–vis detection of glutamine transaminase activity. The reaction mixtures, standards and buffers were mixed with an equal volume of 0.8 M formic acid before being injected into an HPLC reverse-phase column (150×4.6 mm, Varian, Palo Alto, CA) for analysis. The mobile phase consists of 10 mM potassium phosphate (monobasic) buffer containing 10% (v/v) acetonitrile for the analysis of phenylalanine in the reaction mixtures. The formation of transamination product, phenylalanine was monitored by an in-line UV detector at a wavelength of 257 nm. A, B, C, and D illustrate chromatograms of boric acid buffer, 1 mM phenylpyruvate standard, 1 mM phenylalanine standard, and the reaction mixture without incubation, respectively. Chromatogram E illustrates the product, phenylalanine (arrowed) formed in 100 μl reaction mixture including 5 mM glutamine, 2 mM phenylpyruvate and 2 μg recombinant mouse KAT3, 100 mM boric acid buffer, pH 9.0, in 15 min at 38 °C. The reaction was stopped by adding an equal volume of 0.8 M formic acid.
Table 1.
Kinetic parameters of mKAT 1 & 3 towards glutamine and phenylpyruvate.
| Km (mM) | kcat (min−1) | kcat/Km (min−1 mM−1) | ||
|---|---|---|---|---|
| mKAT I/GTK | Glutamine | 2.3±0.4 | 410.5±23.7 | 178.5 |
| Phenylpyruvate | 0.8±0.3 | 300.2±16.3 | 375.3 | |
| mKAT3/GTL | Glutamine | 0.7±0.3 | 160.0±15.0 | 228.6 |
| Phenylpyruvate | 0.6±0.2 | 190.0±28.3 | 316.7 |
The activities were measured as described in the Method section. The Km and kcat were derived by using varying concentrations (0.2–20 mM) of individual substrate in the presence of 20 mM of glutamine or 5 mM phenylpyruvate. The parameters were calculated by fitting the Michaelis–Menten equation to the experimental data using the enzyme kinetics module. Results are means±SE.
3.3. Inhibition of glutamine transaminase activities of mouse KAT1 and KAT3
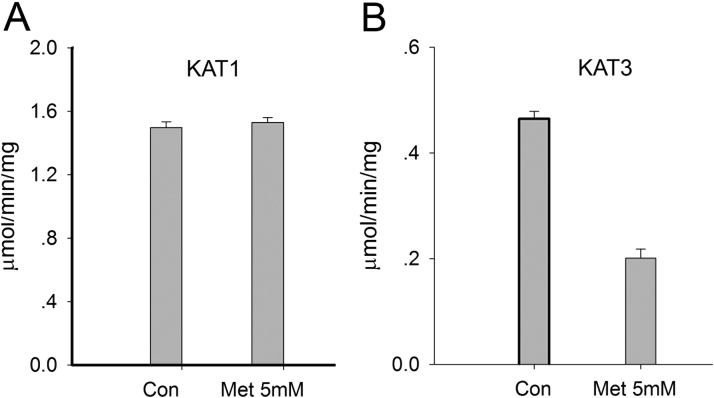
In order to find the biochemical differences of KAT1 and KAT3, inhibition of two enzymes were investigated. Based on the previous reports regarding their biochemical characteristics [16], [28], the inhibitor, methionine was tested for the inhibition of glutamine transaminations catalyzed by mouse KAT1 and KAT3. When the enzyme activity was assayed in a typical reaction mixture (5 mM glutamine, 2 mM phenylpyruvate, and 2 μg mouse KAT3 or KAT1) in the presence of 5 mM inhibitor, methionine, no noticeable inhibitory effect on mouse KAT1-catalyzed glutamine transamination was observed, however methionine greatly decreased the rate of glutamine transamination (57%) of mouse KAT3 (Fig. 3). The decrease in the rate of KAT3-catalyzed reaction in the presence of the inhibitors is apparently due to competitive inhibition of mouse KAT3.
Fig. 3.
Inhibition of glutamine transaminase activity of mouse recombinant KAT1 and KAT3 by methionine. The reaction mixture consisted of 5 mM glutamine, 2 mM phenylpyruvate, 40 μM PLP, 2 μg recombinant protein, KAT1 or KAT3 in 100 mL 100 mM boric acid buffer, pH 9.0. The mixture was incubated at 38 °C for 15 min and the reaction was stopped by adding an equal volume of 0.8 M formic acid. Measurement of phenylalanine product was performed by HPLC with UV detection at a wavelength of 257 nm. Panel B shows methionine significantly inhibited KAT3 activity.
3.4. KAT and glutamine transaminase activities of mouse kidney and liver crude proteins
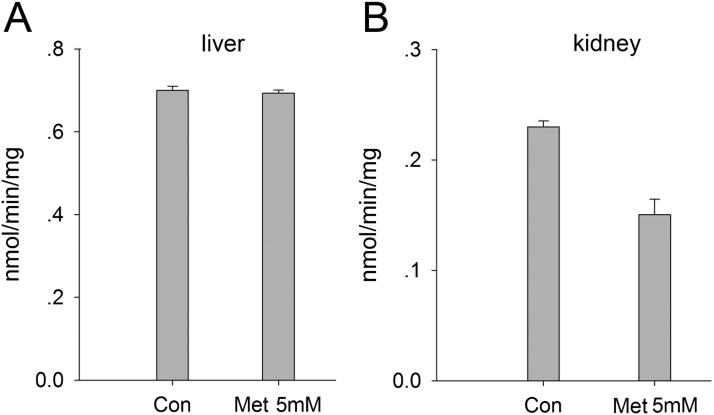
At the identical conditions as used above in the co-substrate specificity and inhibition studies of mouse KAT1 and KAT3 enzymes, the liver and kidney crude proteins showed KAT and glutamine transaminase activities (Fig. 4). In particular, the crude liver proteins showed higher KAT and glutamine transaminase activities than did the kidney crude proteins. By estimating KAT1 and KAT3 contribution to the detected enzyme activities in the liver and kidney, we did specific inhibition tests for the crude proteins. The results showed that enzyme activity of the kidney crude proteins was inhibited by methionine, and the crude proteins from mouse liver were not inhibited significantly by the inhibitor (Fig. 4). This result indicated that mouse KAT3/GTL/CCBL2 is mainly present in kidney in mouse, not in liver.
Fig. 4.
Inhibition of glutamine transaminase activity of mouse liver and kidney crude protein extracts by methionine. The reaction mixture consisted of 5 mM glutamine, 2 mM phenylpyruvate, 40 μM PLP, 20 μl crude liver protein extract or kidney protein extract in 100 mL of 100 mM boric acid buffer, pH 9.0. The mixture was incubated at 38 °C for 2 h and the reaction was stopped by adding an equal volume of 0.8 M formic acid. Measurement of phenylalanine was performed by HPLC with UV detection at a wavelength of 257 nm. Panel B shows methionine significantly inhibited KAT activity in the kidney.
3.5. Expressions of kat3 and kat1 genes in the mouse liver and kidney
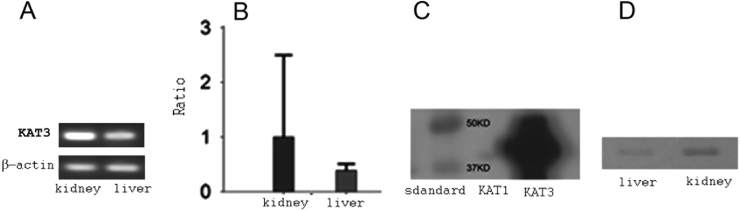
The expressions of kat1 and kat3 genes in the mouse liver and kidney were analyzed in six mice (three males and three females) by RT-PCR (Fig. 5A) and qRT-PCR (Fig. 5B). kat1 and kat3 showed different and measureable expression levels, and both genes were expressed in both liver and kidney of the mouse. However, kat1 was expressed in a higher level in the liver than in the kidney, while kat3 was expressed in a higher level in the kidney than in the liver.
Fig. 5.
mRNA and protein levels of KATs in the mouse liver and kidney. The mRNA transcripts of mouse kat1 and kat3 in the liver and kidney were analyzed using both RT-PCR and qRT-PCR. All final products in RT-PCR experiments were analyzed by 1% agarose gel electrophoresis (A). Relative expression levels of kat1 and kat3 genes were shown as percentages of gapdh gene expression in qRT-PCR tests (B). The specificity of the purified mouse KAT3 antibody was tested by Western blotting. Both recombinant mouse KAT1 and KAT3 proteins were run in a SDS-PAGE, transferred to a PVDF membrane and immuno-stained with the purified mouse KAT3 antibody. The antibody recognized the recombinant mouse KAT3 protein very well, without cross-reaction with recombinant mouse KAT1 (C). Using the purified mouse KAT3 antibody, KAT3 protein was detected both in liver and kidney, but a more intensive band was seen in the kidney (D).
3.6. Western blot analysis
In order to reveal KAT3 localization in the mouse kidney and liver, we did western-blotting using the purified anti KAT3 antibody, which was purified from the anti-serum produced in a rabbit. The antibody had no cross-reaction with mouse KAT1 by western-blotting (Fig. 5C). The results showed that KAT3 protein was found both in the mouse liver and kidney, but more abundant in the kidney (Fig. 5D).
4. Discussion
It has been reported that glutamine transaminase has at least two forms: K-form (GTK) purified from kidney, and l-form (GTL) purified from liver in rats [30], [31], [32]. GTK has been cloned and linked to KAT1 [33], [34], [35], and GTL protein has been linked to KAT3 [15]. Thus, it is considered that GTK is a main glutamine transaminase in the rat kidney tissue; and GTL is a main glutamine transaminase in the rat liver [17]. The annotation of mouse homologs of glutamine transaminases was based on the sequence identity, which may not provide information of tissue localization of the enzymes. In this study, we provided evidence that the mouse KAT3/GTL was found both in the liver and kidney of the mouse, but the mouse KAT3/GTL was more abundant in the kidney than in the liver, which contrasts to the rat homolog (rat KAT3/GTL) that is abundant in the liver. This conclusion is supported by multiple experimental results. First of all, both independent RT-PCR and qRT-PCR experiments showed that relative mRNA levels of kat3 gene were higher in the kidney than in the liver of the mouse compared with those of house keeping genes. Secondly, previous studies found that the mouse KAT3 was inhibited by equimolar concentration methionine, while human KAT1 and mouse KAT1 were not significantly inhibited by equimolar concentration methionine [9], [16], [28]. Therefore, inhibition levels of methionine could be used to distinguish natural enzyme activity of the mouse KAT1/GTK and KAT3/GTL from tissues. Glutamine transaminase activity of the mouse kidney crude protein was significantly inhibited by methionine, while the enzyme activity from the mouse liver crude protein was not significantly inhibited, suggesting that KAT3/GTL is more abundant in the mouse kidney than in the liver. Finally, western blotting using purified anti-mouse KAT3/GTL antibody further demonstrated the protein level of KAT3/GTL is also more abundant in the kidney than in the liver. Note that GTL means liver type glutamine transaminase, which is not proper for naming the mouse homologue, but GTL is retained for the mouse homologue, mouse KAT3/GTL/CCBL2, for biochemical and gene structure similarities in this report.
The tissue localization difference may raise a question as to why the mouse KAT3/GTL tissue localization is different from the rat KAT3/GTL. To answer why mouse GTL is abundant in the kidney instead of the liver, we may look over any differences in biochemical property and substrate availability between the mouse and rat enzymes. Both selenomethionine and Se-methyselenocysteine are substrates of rat KAT1/GTK, human KAT1/GTK and mouse KAT3/GTL in the presence of α-keto-γ-methiolbutyrate as amine acceptor [15], [25], [36]. Selenomethionine is a relatively good aminotransferase substrate of recombinant mouse KAT3/GTL, suggesting that it selectively metabolizes selenomethionine to α-keto-γ-methylselenobutyrate. However, it seems that KAT3/GTL is more efficient in transamination of both selenomethionine and Se-methyselenocysteine than is KAT1/GTK. Here, we reported that this enzyme was largely present in the kidney of the mouse, suggesting that dietary selenomethionine and Se-methyselenocysteine can be converted to the corresponding α-keto acids, α-keto-γ-methylselenobutyrate and β-methylselenopyruvate, respectively in the mouse kidney. Selenium in the kidney tissues of swine was greater than in the liver when it was provided as selenomethionine [37]. The greatest concentrations were achieved in the kidneys and adrenals in rats when given selenomethionine [38]. In pigs, selenium concentration was generally the highest in the kidney after administration of selenomethionine [39]. However, in the mouse, the liver absorbed the injected selenomethionine more efficiently than did any other tissues, showing the highest selenium concentrations [40]. Regarding the fact that selenomethionine is a relatively good substrate of the mouse KAT3/GTL, and possibly rat and human KAT3/GTL enzymes, the differences of tissue preference of selenomethionine between different animals (higher concentration in the kidneys of the pig and rat, higher concentration in the liver of the mouse) may explain the different tissue localization of KAT3/GTL between mice and the others (rats and pigs), although the mechanism is not yet understood. The kidney plays a major role in the interorgan metabolism of glutamine [41]. As reviewed in the introduction, glutamine transaminases play a role in acid-base homeostasis for producing urinary ammonia production in the kidney. The excretion of ammonia in the urine facilitates the elimination of strong metabolic acids, such as sulfuric acid that arises during the catabolism of methionine and cysteine [41], [42]. Thus, the mouse KAT3/GTL is a key glutamine transaminase in mice for the acid-base homeostasis in the kidney.
In addition, α-Keto-γ-methylselenobutyrate and β-methylselenopyruvate are potent inhibitors of histone deacetylases and therefore may be chemoprotective [43], [44]. Understanding the extent of in situ conversion of seleno amino acid to α-keto-γ-methylselenobutyrate and β-methylselenopyruvate by tissue specific glutamine transaminases (GTL or GTK) may be practical in interpreting treatment results of cancers in different tissues. Based on what we reported about the mouse GTL localization, a further study may be needed to confirm the tissue localization of human GTK and GTL in order to understand the treatment results of cancers using seleno amino acids.
It was suggested that the glutaminase II pathway is quantitatively important in humans [25]. However, there is no evidence showing the same phenomenon in the mouse. Here we provided indirect evidence for that the glutaminase II pathway is more important than the glutaminase pathway in metabolizing glutamine in the mouse kidney based on the enzymes biochemical parameter comparison. Km for glutamine of the rat glutaminase (a key enzyme in glutaminase pathway) is 2.6 mM [45] and Km for glutamine of the human kidney type glutaminase isozyme (GLS2) is 4 mM. The Km value (0.7 mM) for glutamine of the mouse KAT3/GTL, we reported here, was much smaller than Km values of both rat glutaminase and human GLS2 (both for glutaminase pathway), which suggests mouse KAT3/GTL will likely catalyze glutamine more than did the others. If the mouse glutaminase and human GLS2 behave the same as rat or human homologues, the reaction may favor the glutaminase II pathway instead of the glutaminase pathway. However, not having the enzyme kinetic parameters of the mouse glutaminase and GLS2, we may not make a definite conclusion for the major role the glutaminase II pathway plays in the mouse kidney.
It has been established that KAT1 and KAT3 are multifunctional aminotransferases. Kinetic analysis of the enzymes towards different amino acids showed that the enzyme is efficient in catalyzing the transamination of glutamine, phenylalanine, leucine, kynurenine, tryptophan, methionine, tyrosine, histidine, cysteine and aminobutyrate. The large spectrum of amino acid substrates of KAT1 and KAT3 supports the proposed role of the enzyme in sparing the essential amino acids methionine, histidine, phenylalanine and tyrosine [46] and provides a mechanism to maintain a continual equilibrium among the amino acids [47]. These enzymes therefore appear to play a role in the homeostatic metabolic mechanism for the preservation of amino acid balance in which glutamine, a dietary non-essential amino acid, functions to maintain the tissue levels of amino acids and to prevent loss of essential amino acid carbon chains [31], [32]. Apparently, KAT3/GTL is a major aminotransferase in playing such an important role in the mouse kidney.
Acknowledgments
The work was funded by a special cooperation funds for Science and Technology of Hainan Province (No. KJHZ2015-31) and in part by an open fund of the State Key Laboratory of Veterinary Etiological Biology, Lanzhou Veterinary Research Institute, Chinese Academy of Agricultural Sciences (Grant No. SKLVEB2014KFKT001).
Footnotes
Supplementary data associated with this article can be found in the online version at 10.1016/j.bbrep.2016.09.008.
Appendix A. Transparency document
Supplementary material
.
References
- 1.Leeson P.D., Iversen L.L. The glycine site on the NMDA receptor: structure-activity relationships and therapeutic potential. J. Med. Chem. 1994;37:4053–4067. doi: 10.1021/jm00050a001. [DOI] [PubMed] [Google Scholar]
- 2.Perkins M.N., Stone T.W. An iontophoretic investigation of the actions of convulsant kynurenines and their interaction with the endogenous excitant quinolinic acid. Brain Res. 1982;247:184–187. doi: 10.1016/0006-8993(82)91048-4. [DOI] [PubMed] [Google Scholar]
- 3.Stone T.W., Perkins M.N. Actions of excitatory amino acids and kynurenic acid in the primate hippocampus: a preliminary study. Neurosci. Lett. 1984;52:335–340. doi: 10.1016/0304-3940(84)90184-8. [DOI] [PubMed] [Google Scholar]
- 4.Birch P.J., Grossman C.J., Hayes A.G. Kynurenic acid antagonises responses to NMDA via an action at the strychnine-insensitive glycine receptor. Eur. J. Pharmcol. 1988;154:85–87. doi: 10.1016/0014-2999(88)90367-6. [DOI] [PubMed] [Google Scholar]
- 5.Pereira E.F., Hilmas C., Santos M.D., Alkondon M., Maelicke A., Albuquerque E.X. Unconventional ligands and modulators of nicotinic receptors. J. Neurobiol. 2002;53:479–500. doi: 10.1002/neu.10146. [DOI] [PubMed] [Google Scholar]
- 6.Hilmas C., Pereira E.F., Alkondon M., Rassoulpour A., Schwarcz R., Albuquerque E.X. The brain metabolite kynurenic acid inhibits alpha7 nicotinic receptor activity and increases non-alpha7 nicotinic receptor expression: physiopathological implications. J. Neurosci. 2001;21:7463–7473. doi: 10.1523/JNEUROSCI.21-19-07463.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Alkondon M., Pereira E.F., Yu P., Arruda E.Z., Almeida L.E., Guidetti P., Fawcett W.P., Sapko M.T., Randall W.R., Schwarcz R., Tagle D.A., Albuquerque E.X. Targeted deletion of the kynurenine aminotransferase ii gene reveals a critical role of endogenous kynurenic acid in the regulation of synaptic transmission via alpha7 nicotinic receptors in the hippocampus. J. Neurosci. 2004;24:4635–4648. doi: 10.1523/JNEUROSCI.5631-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Stone T.W. Kynurenic acid blocks nicotinic synaptic transmission to hippocampal interneurons in young rats. Eur. J. Neurosci. 2007;25:2656–2665. doi: 10.1111/j.1460-9568.2007.05540.x. [DOI] [PubMed] [Google Scholar]
- 9.Han Q., Cai T., Tagle D.A., Li J. Structure, expression, and function of kynurenine aminotransferases in human and rodent brains. Cell. Mol. Life Sci. 2010;67:353–368. doi: 10.1007/s00018-009-0166-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Notarangelo F.M., Pocivavsek A. Elevated kynurenine pathway metabolism during neurodevelopment: implications for brain and behavior. Neuropharmacology. 2016 doi: 10.1016/j.neuropharm.2016.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Colombari E., Sato M.A., Cravo S.L., Bergamaschi C.T., Campos R.R., Jr., Lopes O.U. Role of the medulla oblongata in hypertension. Hypertension. 2001;38:549–554. doi: 10.1161/01.hyp.38.3.549. [DOI] [PubMed] [Google Scholar]
- 12.Kwok J.B., Kapoor R., Gotoda T., Iwamoto Y., Iizuka Y., Yamada N., Isaacs K.E., Kushwaha V.V., Church W.B., Schofield P.R., Kapoor V. A missense mutation in kynurenine aminotransferase-1 in spontaneously hypertensive rats. J. Biol. Chem. 2002;277:35779–35782. doi: 10.1074/jbc.C200303200. [DOI] [PubMed] [Google Scholar]
- 13.Wang Y., Liu H., McKenzie G., Witting P.K., Stasch J.P., Hahn M., Changsirivathanathamrong D., Wu B.J., Ball H.J., Thomas S.R., Kapoor V., Celermajer D.S., Mellor A.L., Keaney J.F., Jr., Hunt N.H., Stocker R. Kynurenine is an endothelium-derived relaxing factor produced during inflammation. Nat. Med. 2010;16:279–285. doi: 10.1038/nm.2092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yu P., Li Z., Zhang L., Tagle D.A., Cai T. Characterization of kynurenine aminotransferase III, a novel member of a phylogenetically conserved KAT family. Gene. 2006;365:111–118. doi: 10.1016/j.gene.2005.09.034. [DOI] [PubMed] [Google Scholar]
- 15.Pinto J.T., Krasnikov B.F., Alcutt S., Jones M.E., Dorai T., Villar M.T., Artigues A., Li J., Cooper A.J. Kynurenine aminotransferase III and glutamine transaminase L are identical enzymes that have cysteine S-conjugate beta-lyase activity and can transaminate l-selenomethionine. J. Biol. Chem. 2014;289:30950–30961. doi: 10.1074/jbc.M114.591461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Han Q., Robinson H., Cai T., Tagle D.A., Li J. Biochemical and structural properties of mouse kynurenine aminotransferase III. Mol. Cell. Biol. 2009;29:784–793. doi: 10.1128/MCB.01272-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cooper A.J., Meister A. Isolation and properties of a new glutamine transaminase from rat kidney. J. Biol. Chem. 1974;249:2554–2561. [PubMed] [Google Scholar]
- 18.Han Q., Robinson H., Cai T., Tagle D.A., Li J. Structural insight into the inhibition of human kynurenine aminotransferase i/glutamine transaminase K. J. Med. Chem. 2009;52:2786–2793. doi: 10.1021/jm9000874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rossi F., Han Q., Li J., Li J., Rizzi M. Crystal structure of human kynurenine aminotransferase I. J. Biol. Chem. 2004;279:50214–50220. doi: 10.1074/jbc.M409291200. [DOI] [PubMed] [Google Scholar]
- 20.Han Q., Li J., Li J. pH dependence, substrate specificity and inhibition of human kynurenine aminotransferase I. Eur. J. Biochem. 2004;271:4804–4814. doi: 10.1111/j.1432-1033.2004.04446.x. [DOI] [PubMed] [Google Scholar]
- 21.Wilmore D.W. Glutamine and the gut. Gastroenterology. 1994;107:1885–1886. doi: 10.1016/0016-5085(94)90836-2. [DOI] [PubMed] [Google Scholar]
- 22.Kaiser L.G., Schuff N., Cashdollar N., Weiner M.W. Age-related glutamate and glutamine concentration changes in normal human brain: 1H MR spectroscopy study at 4 T. Neurobiol. Aging. 2005;26:665–672. doi: 10.1016/j.neurobiolaging.2004.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Newsholme P., Procopio J., Lima M.M., Pithon-Curi T.C., Curi R. Glutamine and glutamate their central role in cell metabolism and function. Cell Biochem. Funct. 2003;21:1–9. doi: 10.1002/cbf.1003. [DOI] [PubMed] [Google Scholar]
- 24.Saito K., Fujigaki S., Heyes M.P., Shibata K., Takemura M., Fujii H., Wada H., Noma A., Seishima M. Mechanism of increases in l-kynurenine and quinolinic acid in renal insufficiency. Am. J. Physiol. Ren. Physiol. 2000;279:F565–F572. doi: 10.1152/ajprenal.2000.279.3.F565. [DOI] [PubMed] [Google Scholar]
- 25.Cooper A.J.L., Dorai T., Dorai B., Krasnikov B.F., Li J., Hallen A., Pinto J.T. Role of glutamine transaminases in nitrogen, sulfur, selenium, and 1-carbon metabolism. In: Rajendram R., Preedy R.V., Patel B.V., editors. Glutamine in Clinical Nutrition. Springer; New York, NY: 2015. pp. 37–54. [Google Scholar]
- 26.Meister A., Sober H.A., Tice S.V., Fraser P.E. Transamination and associated deamidation of asparagine and glutamine. J. Biol. Chem. 1952;197:319–330. [PubMed] [Google Scholar]
- 27.Cooper A.J., Kuhara T. Alpha-Ketoglutaramate: an overlooked metabolite of glutamine and a biomarker for hepatic encephalopathy and inborn errors of the urea cycle. Metab. Brain Dis. 2014;29:991–1006. doi: 10.1007/s11011-013-9444-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Han Q., Cai T., Tagle D.A., Li J. Thermal stability, pH dependence and inhibition of four murine kynurenine aminotransferases. BMC Biochem. 2010;11:19. doi: 10.1186/1471-2091-11-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Livak K.J., Schmittgen T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 30.Cooper A.J. Purification of soluble and mitochondrial glutamine transaminase K from rat kidney. Use of a sensitive assay involving transamination between l-phenylalanine and alpha-keto-gamma-methiolbutyrate. Anal. Biochem. 1978;89:451–460. doi: 10.1016/0003-2697(78)90374-3. [DOI] [PubMed] [Google Scholar]
- 31.Cooper A.J., Meister A. Comparative studies of glutamine transaminases from rat tissues. Comp. Biochem. Physiol. B. 1981;69B:137–145. [Google Scholar]
- 32.Cooper A.J., Meister A. Glutamine transaminase L from rat liver. Methods Enzymol. 1985;113:338–343. doi: 10.1016/s0076-6879(85)13046-6. [DOI] [PubMed] [Google Scholar]
- 33.Perry S.J., Schofield M.A., MacFarlane M., Lock E.A., King L.J., Gibson G.G., Goldfarb P.S. Isolation and expression of a cDNA coding for rat kidney cytosolic cysteine conjugate beta-lyase. Mol. Pharm. 1993;43:660–665. [PubMed] [Google Scholar]
- 34.Mosca M., Cozzi L., Breton J., Speciale C., Okuno E., Schwarcz R., Benatti L. Molecular cloning of rat kynurenine aminotransferase: identity with glutamine transaminase K. FEBS Lett. 1994;353:21–24. doi: 10.1016/0014-5793(94)01003-x. [DOI] [PubMed] [Google Scholar]
- 35.Alberati-Giani D., Malherbe P., Kohler C., Lang G., Kiefer V., Lahm H.W., Cesura A.M. Cloning and characterization of a soluble kynurenine aminotransferase from rat brain: identity with kidney cysteine conjugate beta-lyase. J. Neurochem. 1995;64:1448–1455. doi: 10.1046/j.1471-4159.1995.64041448.x. [DOI] [PubMed] [Google Scholar]
- 36.Cooper A.J., Pinto J.T., Krasnikov B.F., Niatsetskaya Z.V., Han Q., Li J., Vauzour D., Spencer J.P. Substrate specificity of human glutamine transaminase K as an aminotransferase and as a cysteine S-conjugate beta-lyase. Arch. Biochem. Biophys. 2008;474:72–81. doi: 10.1016/j.abb.2008.02.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Panter K.E., Hartley W.J., James L.F., Mayland H.F., Stegelmeier B.L., Kechele P.O. Comparative toxicity of selenium from seleno-DL-methionine, sodium selenate, and Astragalus bisulcatus in pigs. Fundam. Appl. Toxicol. 1996;32:217–223. [PubMed] [Google Scholar]
- 38.Thomson C.D., Stewart R.D. Metabolic studies of (75Se)selenomethionine and (75Se)selenite in the rat. Br. J. Nutr. 1973;30:139–147. doi: 10.1079/bjn19730015. [DOI] [PubMed] [Google Scholar]
- 39.Tian J.Z., Yun M.S., Ju W.S., Long H.F., Kim J.H., Kil D.Y., Chang J.S., Cho S.B., Kim Y.Y., Han I.K. Effects of dietary selenium supplementation on growth performance, selenium retention in tissues and nutrient digestibility in growing-finishing pigs. Asian-Austral. J. Anim. Sci. 2006;19:55–60. [Google Scholar]
- 40.Suzuki Y., Hashiura Y., Sakai T., Yamamoto T., Matsukawa T., Shinohara A., Furuta N. Selenium metabolism and excretion in mice after injection of (82)Se-enriched selenomethionine. Metallomics: Integr. Biom. Sci. 2013;5:445–452. doi: 10.1039/c3mt20267d. [DOI] [PubMed] [Google Scholar]
- 41.Brosnan J.T. The 1986 Borden award lecture. The role of the kidney in amino acid metabolism and nutrition. Can. J. Physiol. Pharm. 1987;65:2355–2362. doi: 10.1139/y87-373. [DOI] [PubMed] [Google Scholar]
- 42.Brosnan J.T., Lowry M., Vinay P., Gougoux A., Halperin M.L. Renal ammonium production--une vue canadienne. Can. J. Physiol. Pharm. 1987;65:489–498. doi: 10.1139/y87-084. [DOI] [PubMed] [Google Scholar]
- 43.Nian H., Bisson W.H., Dashwood W.M., Pinto J.T., Dashwood R.H. Alpha-keto acid metabolites of organoselenium compounds inhibit histone deacetylase activity in human colon cancer cells. Carcinogenesis. 2009;30:1416–1423. doi: 10.1093/carcin/bgp147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pinto J.T., Lee J.I., Sinha R., MacEwan M.E., Cooper A.J. Chemopreventive mechanisms of alpha-keto acid metabolites of naturally occurring organoselenium compounds. Amino Acids. 2011;41:29–41. doi: 10.1007/s00726-010-0578-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Michalik M., Nelson J., Erecinska M. Glutamate production in islets of Langerhans: properties of phosphate-activated glutaminase. Metabolism. 1992;41:1319–1326. doi: 10.1016/0026-0495(92)90102-g. [DOI] [PubMed] [Google Scholar]
- 46.Cooper A.J. The role of glutamine transaminase K (GTK) in sulfur and alpha-keto acid metabolism in the brain, and in the possible bioactivation of neurotoxicants. Neurochem. Int. 2004;44:557–577. doi: 10.1016/j.neuint.2003.12.002. [DOI] [PubMed] [Google Scholar]
- 47.Han Q., Fang J., Li J. Kynurenine aminotransferase and glutamine transaminase K of Escherichia coli: identity with aspartate aminotransferase. Biochem. J. 2001;360:617–623. doi: 10.1042/0264-6021:3600617. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material