Abstract
Members of the RNaseA family are present in various tissues and secretions but their function is not well understood. Some of the RNases are proposed to participate in host defence. RNase4 and RNase5 are present in cows' milk and have antimicrobial activity. However, their presence in many tissues and secretions has not been characterised. We hypothesised that these two RNases are present in a range of tissues and secretions where they could contribute to host defence. We therefore, determined the relative abundance of RNase4 and RNase5 mRNA as well as protein levels in a range of host defence related and other tissues as well as a range of secretions in cattle, using real time PCR and western blotting. The two RNases were found to be expressed in liver, lung, pancreas, mammary gland, placenta, endometrium, small intestine, seminal vesicle, salivary gland, kidney, spleen, lymph node, skin as well as testes. Corresponding proteins were also detected in many of the above tissues, as well as in seminal fluid, mammary secretions and saliva. This study provides evidence for the presence of RNase4 and RNase5 in a range of tissues and secretions, as well as some major organs in cattle. The data are consistent with the idea that these proteins could contribute to host defence in these locations. This work contributes to growing body of data suggesting that these proteins contribute to the physiology of the organism in a more complex way than acting merely as digestive enzymes.
Abbreviations: RNase, ribonuclease; EAR, Eosinophil-associated ribonuclease; qPCR, quantitative polymerase chain reaction; PBMC, peripheral blood mononuclear cells; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; HRP, horseradish peroxidase; IgG, immunoglobulin G
Keywords: RNases, Host defence, Secretion, Mastitis
Highlights
-
•
RNase4 and RNase5 are present in several tissues and secretions in cattle.
-
•
mRNA and protein levels of the RNases correlate in various tissues analysed.
-
•
The RNases could contribute to host defence in these tissues and secretions.
1. Introduction
The mammalian ribonuclease (RNase) superfamily comprises eight canonical RNases of 14–16 kDa, for which orthologues or paralogues are present in all vertebrates, as well as additional RNases present in only some mammals. All members of this superfamily are secretory proteins that share sequence similarity with bovine pancreatic RNaseA and contain 6–8 cysteine residues that are necessary for forming their native three dimensional structures. Some, but not all RNases of this family, have RNase activity, which is associated with a conserved catalytic motif CKXXNTF [1].
RNases are present in a wide range of tissues and secretions, as well as some major organs. RNaseA is secreted by the pancreas and has been suggested to play a role in digestion [2]. RNase5 is secreted by endothelial cells and has been proposed to play a role in angiogenesis [3], [4]. RNase4 and RNase5 are also present in milk and colostrum [5], [6], [7]. An RNase, named seminal RNase, has also been reported in seminal fluid [8], [9], and RNase3 has been reported to be present in saliva [10]. RNases are also associated with immune cells. For example, RNase2 and RNase3 are present in neutrophils and eosinophils [11], [12], [13], [14], Eosinophil-Associated RNase11 (EAR11) has been detected in macrophages [15], and monocytes have been reported to secrete RNase4 [16], [17]. Paneth cells present in the intestine of mice have also been shown to secrete RNase5 [18]. RNases are also expressed in major internal organs, with RNase6 being reported to be present in kidney [19], while RNase2, RNase3 and RNase6 all have been reported to be present in liver [20]. Thus, existing knowledge of RNases expression presents an incomplete picture of their physiological role.
Several members of the RNaseA family have been reported to have host defence associated activities [21], [22], [23], [24]. Recombinant mouse RNase5 has been shown to have antimicrobial activity against some a number of microbial pathogens [18], [25], and RNase4 and RNase5 from cows' milk have been reported to have antimicrobial activity against Candida albicans [5]. Cows' milk RNase5 (angiogenin-1), angiogenin-2 and RNase4 have been shown to enhance the antimicrobial activity of lactoferrin and lactofericin against a range of Gram-negative and Gram-positive bacteria [26]. The protein sequences of RNase4 and RNase5 are similar (45% amino acid sequence identity) and possess key features of the RNase A superfamily such as a secretion leader sequence, 6–8 cysteine residues for forming the three-dimensional structure and the conserved catalytic triad. Both these RNases possess ribonuclease activity, but the activity of RNase5 is significantly less potent than that of RNase4 [27].
RNase3 has been shown to possess potent bactericidal activity against Escherichia coli and Staphylococcus aureus [28], [29], [30]. In addition, RNase7 from human skin has been demonstrated to have antimicrobial activity against Enterococcus facieum and S. aureus [31], [32]. Recently, both RNase6 and RNase7 have been shown to have antimicrobial activity against a number of uropathogenic bacteria [33]. Moreover, mRNA and protein levels of RNase6 and RNase7 were up-regulated in the infected urinary tract [33], [34]. Besides these antimicrobial activities, some RNases have also been shown to possess antiviral activity. RNase2 and RNase3 are closely related, both biochemically and functionally [35]. These RNases have been demonstrated to reduce the infectivity of respiratory syncytial virus and human immunodeficiency virus in cell culture [36], [37]. Other activities have also been reported. For example, RNase3 has been reported to have antihelminthic activity, with high toxicity demonstrated against Schistosoma mansoni, Brugia pahangi and Trichinella spiralis [38], [39], [40]. RNase3 has also been reported to have cytotoxic activity against bronchial epithelial cells [41], and bovine seminal RNase has been shown to have cytotoxic activity on proliferating lymphocytes [42]. Furthermore, some RNases have been proposed to act as chemoattractants. For example, RNase2 and EAR2 have been demonstrated to stimulate the migration of immature human dendritic cells [43]. Collectively, these observations suggest that some of the RNases may contribute to host defence. The means through which they might play this role have been proposed for some RNases [30], [44], [45], [46], [47], while for others, this remains poorly characterised.
Host defence in cattle has several unique features, and resistance to infection in this species is an economically significant trait. Many aspects of host defence in cattle are incompletely understood, including the possible involvement of the RNases. Only limited data are available on expression pattern of RNases in cattle, with RNase4 and RNase5 expression reported only in the mammary gland [5], [26]. Therefore, the purpose of this study was to investigate the abundance of mRNA transcripts of RNase4 and RNase5 in cattle in a range of tissues, in order to address whether expression occurs beyond the mammary gland, and thereby better understand their possible roles in host defence in this species. The mRNA transcript accumulation of RNase4 and RNase5 in a range of host-defence related tissues was assessed by quantitative PCR (qPCR). The protein abundance of these two RNases was also measured in a range of tissues and secretions using western blotting. The data show that RNase4 and RNase5 are most highly expressed in the liver and are present in a wider range of host-defence related tissues and secretions than has previously been reported. These findings support the concept that RNase4 and RNase5 play a role in host defence in cattle.
2. Materials and methods
2.1. Reagents
All the chemicals used in the present study were purchased from Sigma-Aldrich (St. Louis, MO), unless otherwise stated.
2.2. Tissue and secretion collection
All animal manipulations and tissue collection was performed with the approval of the Ruakura Animal Ethics committee. All tissues were collected from New Zealand Holstein-Friesian dairy cattle. Male reproductive tract tissues were collected after slaughtering a healthy, 3 year old dairy bull. Fresh tissues were snap frozen in liquid nitrogen (N2) and powdered under liquid N2 using a mortar and pestle and stored at −80 °C. For the collection of seminal fluid, the ejaculatory duct was removed from the slaughtered bull and its contents were removed by squeezing the duct from one end. All other tissues including pancreas, spleen, endometrium, kidney, salivary gland, lungs, liver, lymph node, small intestine, skin, mammary gland, brain, and placenta were collected from a healthy non-pregnant cow during peak lactation, slaughtered 12 h after the last milking. Liver and mammary gland tissues were also obtained from this cows as well as two separate cows. Colostrum, milk and serum was collected from a separate cow, 6 h after calving. Saliva was obtained from a healthy, heifer using a hollow steel bit connected to a vacuum pump as previously described [48].
2.3. Bovine PBMC preparation
Bovine peripheral blood mononuclear cells (PBMCs) were purified from fresh bovine blood using a Lymphoprep (Axis-Shield PoC AS, Oslo, Norway) solution according to a previously described method [49], with minor modifications as follows. Peripheral blood was collected into vacationers (BD, Biosciences) containing the anticoagulant acid citrate dextrose (ACD) via jugular vein puncture of a cow. A 16 mL portion of fresh blood was diluted by mixing it with 17 mL of calcium and magnesium free Hank’s Balanced Salt Solution (HBSS) (Invitrogen, New Zealand). This diluted blood was then layered carefully onto 12 mL of Lymphoprep (Axis-Shield PoC AS, Oslo, Norway) in a 50 mL tube. The sample was then centrifuged at 800×g for 40 min at 20 °C, without using the brake, in a swing-bucket rotor. The pale grey cell layer containing monocytes and lymphocytes was recovered by aspiration from the Lymphoprep–buffer interface. The cells were diluted in 40 mL of calcium and magnesium free HBSS medium and centrifuged again at 250×g for 7 min at 4 °C. The supernatant was discarded and 3 volumes of erythrocyte lysis buffer (15 mM NH4Cl, 1 mM KHCO3 and 10 mM EDTA) was added to the residual cell suspension and incubated for 5 min at room temperature to lyse the contaminating erythrocytes. The cells were then washed with phosphate buffered saline (PBS) and re-suspended in 5 mL of RPMI-1640 culture medium. The viability of PBMCs was determined by using a trypan blue exclusion assay and found to be >95%.
2.4. RNA isolation and cDNA synthesis
A 100 mg portion of each powdered bovine tissue sample, was thawed and homogenized in 3 mL of TRIzol reagent (Invitrogen, Auckland, New Zealand) and total RNA was isolated according to the manufacturer's instructions. For successful RNA isolation from pancreas, the tissue was stored in RNAlater® (Ambion, CA) and RNA was isolated according to manufacturer's instructions. The quantity of the purified RNA was determined using a Nanodrop ND-1000 Analyser (NanoDrop Technologies, Wilmington, DE, USA). This purified RNA was incubated for 1 h at 37 °C in DNase-I (New England Biolab) per 50 µL containing 10 µg RNA to digest genomic DNA. After the incubation, 1 µL of 0.5 M EDTA (pH 8.0) was added to the mixture, which was then incubated at 75 °C for 10 min to inactivate the DNase-I enzyme. The RNA was then precipitated with 3 M Sodium Acetate, pH 5.2 and dissolved in diethylpyrocarbonate- (DEPC) treated water. The RNA was then quantified by spectrophotometric analysis using a Nanodrop ND-1000 Analyser (NanoDrop Technologies, Wilmington, DE, USA). The 260/280 ratios of the purified RNA samples were between 1.9 and 2.0 indicating high purity. The integrity of the RNA was confirmed by denaturing agarose gel electrophoresis. The RNA was then reverse transcribed into cDNA using the Super script II First-Strand Synthesis System (Life technologies, NZ) according to the manufacturer's instructions. Briefly, 1 µg of DNase-I-treated total RNA from the tissue was reverse transcribed using Oligo dT (0.5 µg) as the primer and 50 units of Super script II RT in a total of 20 µL.
2.5. Messenger RNA transcript analysis of RNase4 and RNase5 by real-time quantitative PCR
The relative abundance of RNase4 and RNase5 mRNA transcripts was determined by qPCR using a Corbett Rotorgene 6000 instrument (Qiagen) with SYBR ExTaq Mix (Takara, Japan) according to a previously published method [50]. The PCR program comprised an initial denaturation for 3 min, followed by 40 cycles of 95 °C for 10 s, and 60 °C for 25 s. All cDNA mixtures were diluted 1/10, and 2 µL of this diluted cDNA was subjected to qPCR amplification in a 20 µL reaction volume using RNase4 and RNase5 specific primer pairs as previously described [51]. The expression levels of the genes were determined by a previously described method [52]. Briefly, the mRNA transcript accumulation of RNase4 and RNase5 were quantified relative to the geometric mean of three housekeepers namely beta-tubulin, glyceraldehyde 3-phosphate dehydrogenase (GAPDH), and cyclophilin-A, and normalized for different amplification efficiencies. All amplification efficiencies were between 1.6 and 1.8 (data not shown). Each sample was measured in triplicate. A no template control, an RT negative control, and a dissociation curve analysis were performed in each amplification. These analyses produced a single discrete peak for all the primer pairs, showing that the reaction product contained a single amplicon.
Analyses of mRNA expression of RNase4 and RNase5 was performed on tissues obtained from a number ofcattle. Liver and mammary gland tissues were collected from three individual cows, while all other tissues were from a single a bull (for the male-specific tissues) and a cow (or all other tissues). The reason for this collection strategy was to demonstrate that the expression of mRNA transcripts of RNase4 and RNase5 has very low variability among different biological replicates using liver and mammary tissues, before measuring their relative abundance in a wider set of tissues from a single animal.
2.6. Preparation of bovine milk RNases
RNase4 and RNase5 were prepared from bovine milk as previously described (14). Briefly, bulk milk was obtained after skimming and homogenization at a dairy factory and subjected to three successive rounds of cation exchange chromatography that were greater than 99% pure with no detectable contaminating proteins as assessed by SDS gel electrophoresis. The identity of the RNases was confirmed by matrix-assisted laser desorption ionisation time of-flight (MALDI-TOF) mass spectrometry. The RNaseA purified from bovine pancreas was purchased from Sigma Aldrich (St. Louis, MO) (Catalogue # R6513).
2.7. Western blotting
Powdered frozen tissues (100 mg), as described above from a cow and a bull, were added to 1 mL of cold Low Salt buffer (10 mM HEPES, 1.5 mM MgCl2, 10 mM KCl, 0.5 mM DTT, 100 mM sodium ortho-vanadate and 0.2 mM PMSF) containing Complete Protease Inhibitor Cocktail (Roche, Auckland, New Zealand). This tissue suspension was sonicated using a Vibracell sonicator (Sonics and Material Inc., Danbury, USA) for 3 cycles of 15 s bursts on ice with a 2 min cooling between each cycle. After the final sonication, samples were centrifuged at 500×g for 10 min at 4 °C to remove cellular debris. The total protein concentration of the resulting supernatant was determined using the Bradford protein assay [53]. Each sample was then diluted in sodium dodecyl sulphate (SDS) sample buffer (10% Glycerol, 5% β-mercaptoethanol, 2% SDS and 62.5 mM Tris, pH 6.8) to obtain a final protein concentration ranging from 5 mg/mL protein. A 105 µg portion of total protein was subjected to electrophoresis on a 12% SDS-polyacrylamide gels and then transferred to nitrocellulose membranes (Amersham Biosciences, UK) by electroblotting. As a positive control, 50 ng of purified RNase4 or RNase5 were also included in each gel. The membrane was blocked with 4% fat-free milk and then probed with a 1/5000 dilution of polyclonal rabbit IgG antibody raised against full length bovine RNase4 or RNase5 produced using a method and antibodies as previously described [5]. The membrane was then probed with horseradish peroxidase (HRP) conjugated anti-rabbit immunoglobulin G raised in goat (Sigma, St. Louis, MO) at a 1/25,000 dilution. After washing, the signals were visualized by chemiluminescence as described previously [54]. The membrane was exposed to a photographic film for up to 3 min before developing using an X-ray developer.
3. Results
3.1. RNase4 and RNase5 mRNA in bovine tissues
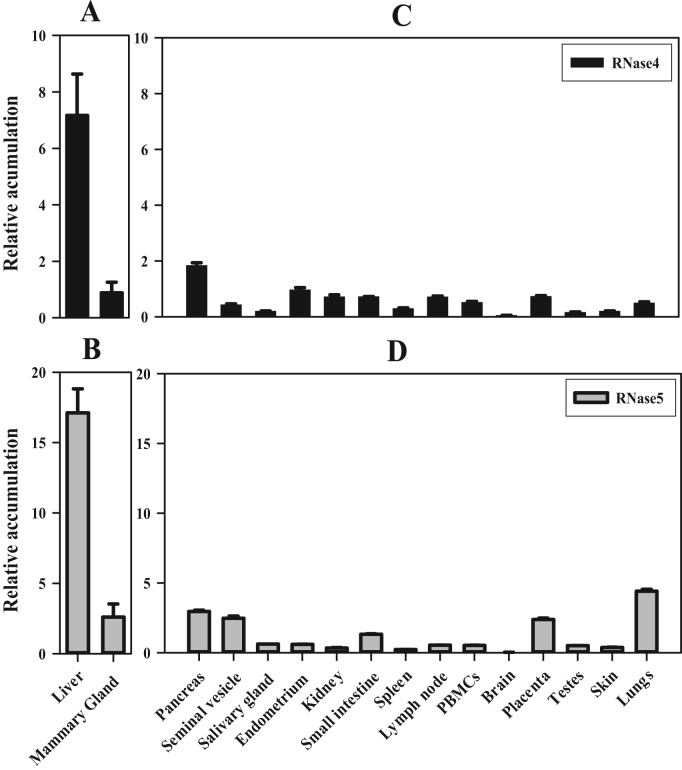
In order to provide evidence for a host defence function for RNase4 and RNase5 in tissues and secretions other than milk, their relative mRNA transcript abundance was examined in endometrium, isolated peripheral blood mononuclear cells (PBMCs), small intestine, skin, lymph node, salivary gland, pancreas, lung, seminal vesicle, testes as well as the mammary gland. These tissues were chosen because they are exposed to pathogens or are known to respond to them. Abundance of RNase4 and RNase5 was also analysed in some major organs; liver, kidney, spleen, and brain, as these tissues are known to express other members of the RNase family. Both RNase4 and RNase5 mRNA transcripts were present in all tissues analysed except brain, although the abundance differed markedly among the tissues (Fig. 1). RNase4 and RNase5 transcripts were found to be the most abundant in liver, with other tissues having a far lesser abundance (Fig. 1). Pancreas, seminal vesicle, small intestine, lung and placenta also contained substantive amounts of RNase4 and RNase5 mRNA transcripts, similar to that of mammary tissue. However, salivary gland, endometrium, skin, kidney, spleen, lymph node, PBMCs, and testes had a relatively low abundance of RNase4 and RNase5 transcripts. Analysis of liver and mammary tissue from multiple cattle showed that inter-animal biological variation was moderate (maximum SE±1.7 in liver and 0.93 in mammary gland) and that the differences between the two types of tissue were significant (p=0.01 for RNase4 and 0.001 for RNase5). Overall, these results show that liver rather than the mammary gland is the predominant site of expression of RNase4 and RNase5 mRNA transcripts in cattle. Furthermore, the results show that expression is not limited to these two tissues but also occurs in a range of exocrine and immune-associated tissues and cells. These results establish that RNase4 and 5 expression occurs more widely among tissues than has been previously reported in any species.
Fig. 1.
Tissue wide mRNA transcripts accumulation of RNase4 and RNase5 in cattle. A total of 1 µg of DNase-I-treated RNA from the indicated tissues was reverse transcribed into cDNA. The resulting cDNA preparation was diluted 1/10 and 2 µL of that cDNA was subjected to qPCR to analyse the accumulation of the mRNA transcripts of bovine RNase4 and RNase5. The accumulation of RNase4 and RNase5 mRNA transcripts is represented relative to the geometric mean of the mRNA transcript accumulation of three reference genes, i.e. beta-tubulin, GAPDH, and cyclophilin A. Data in panels A and B show the mean (±SE) from analysis of tissue from three individual cattle, while panels C and D show triplicate analyses of one individual cow.
3.2. Assessment of cross reactivity of RNase4 and RNase5 polyclonal antibodies
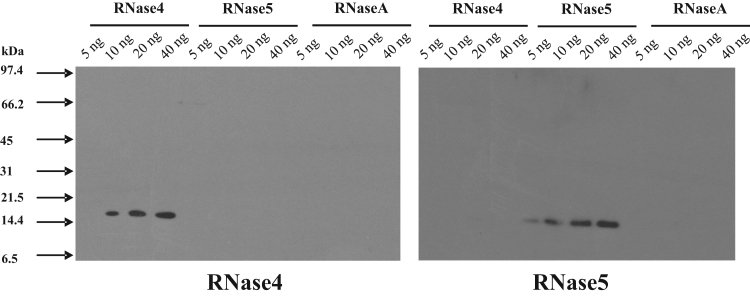
The specificity of the polyclonal antibodies raised against full length bovine RNase4 and RNase5 proteins was determined by western blotting using highly pure RNase4, RNase5, and RNaseA.
The result confirmed that RNase4 polyclonal antibodies specifically detect bovine RNase4 and do not cross-react with either bovine RNase5 with which it shares closest sequence similarity among other members of the RNase family, or a second member of the family, RNaseA (Fig. 2). Similarly, the RNase5 polyclonal antibodies detected only RNase5 and not RNase4 or RNaseA (Fig. 2).
Fig. 2.
Assessment of cross-reactivity of RNase4 and RNase5 polyclonal antibodies. Cross-reactivity of RNase4 and RNase5 polyclonal antibodies was evaluated by western blotting. Varying amounts of purified RNase4, RNase5 and bovine pancreatic RNaseA (as indicated) were subjected to SDS-PAGE and then transferred onto a nitrocellulose membrane. This was probed with affinity purified primary anti-RNase4 or anti-RNase5 antibodies and subsequently with HRP conjugated goat anti-rabbit IgG antibody. The signals were visualized by enhanced chemiluminescence followed by exposure to X-ray film for 30 s.
3.3. Abundance of RNase4 and RNase5 protein among bovine tissues and secretions
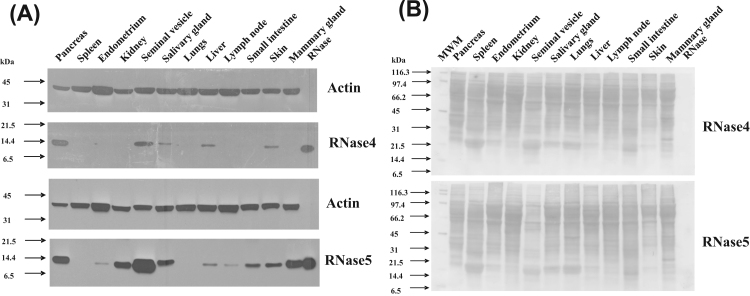
The abundance of RNase4 and RNase5 protein among a similar range of tissues as described above was evaluated by western blotting. In addition, a range of secretions and body fluids were also assessed. These analyses aimed to determine if RNase4 and RNase5 mRNA abundance are correlated with the abundance of their corresponding proteins within each of these tissues, and whether or not the RNases are present in a range of biological fluids.
The RNase4 protein was found to be most abundant in pancreas and seminal vesicle of all the tissues analysed (Fig. 3A). It was also present at lesser abundance in salivary gland, endometrium, skin as well as the kidney (Fig. 3A). RNase4 was not detected in the mammary gland, small intestine, spleen or lymph node. Overall, RNase5 was found to have a broadly similar pattern of tissue distribution to that of RNase4 (Fig. 3A). However, some differences in the pattern of expression were observed between the two RNases in some tissues. RNase5 was detected in the small intestine and lymph node, in contrast to RNase4. Also, the relative abundance of the RNase protein did not correlate with the relative abundance of its corresponding mRNA in some tissues, specifically liver, salivary gland, and mammary gland. Notably, in liver, only a weak signal was observed for the RNase4 and RNase5 proteins despite this tissue having the highest abundance of mRNA transcripts for both the RNases. Similarly, a large difference was observed between the levels of RNase4 and RNase5 mRNA and protein in mammary tissue. These results can be explained if both tissues secrete almost all of the RNase4 and 5 they produce. However, further experimentation would be required to prove this unequivocally. The blots were stained for total protein with Ponceau S to confirm equal loading (Fig. 3B). Control blots probed with secondary antibody alone produced no signal, confirming that the signal was due to the primary antibody (data not shown).
Fig. 3.
Abundance of RNase4 and RNase5 protein in the indicated tissues in cattle. (A). Abundance of RNase4 (upper two panels) and RNase5 (lower two panels) in pancreas, spleen, endometrium, kidney, salivary gland, lungs, liver, lymph node, small intestine, skin, and mammary gland tissues of a cow as evaluated by western blot. A total of 105 µg of total protein was subjected to SDS-PAGE and then transferred onto a nitrocellulose membrane. This was probed with affinity purified primary anti-RNase4 or anti-RNase5 antibodies and subsequently with HRP conjugated goat anti-rabbit IgG antibody. The signals were visualized by enhanced chemiluminescence followed by exposure to X-ray film for 3 min (B). Ponceau S stained images of the membranes is shown to indicate the protein loaded in Fig. 3A.
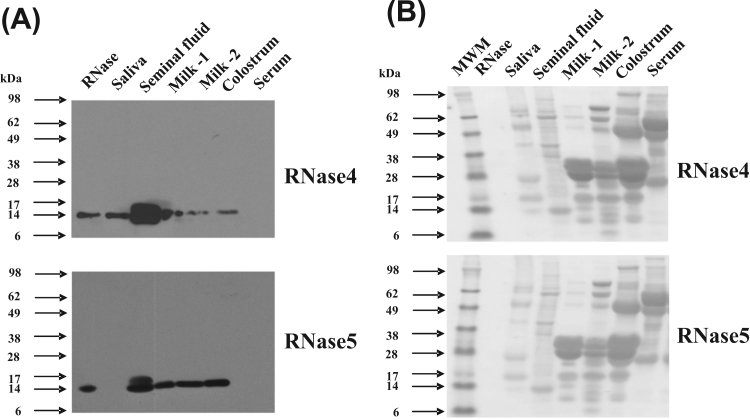
The presence of RNase4 and RNase5 was also examined in a range of additional secretions and body fluids. Surprisingly, RNase4 and RNase5 proteins were both most abundant in seminal fluid compared with the other fluids analysed (Fig. 4A), with the relative abundance in this secretion being at least twice that in milk. This suggests that RNase4 and RNase5 may be important proteins of the seminal fluid and play a role in protecting seminal vesicle and sperm from pathogens. Another band of slightly higher molecular weight was also observed in seminal fluid sample in both the western blots. This could represent a multimeric form, a complex with another protein or it could be the result of post-translational modification of the protein in this secretion. Which of these possible explanations is the case has not been further investigated. Both RNase4 and RNase5 were present in colostrum at approximately the same abundance as in milk (Fig. 4A). Only RNase4, and not RNase5, was detected in saliva. Also, neither RNase4 nor RNase5 was detected in serum. The blot was stained for total protein with Ponceau S to confirm approximately equal loading (Fig. 4B). Overall, these findings show that RNase4 and RNase5 proteins are present in a range of secretions.
Fig. 4.
Presence of RNase4 and RNase5 protein in the indicated body fluids in cattle. (A). Presence of RNase4 (upper panel) and RNase5 (lower panel) in saliva, seminal fluid, milk-1 (uninfected), milk-2 (mastitic), colostrum, and serum in cattle as evaluated by western blotting. A total of 40 μg of protein from various fluids or secretions were subjected to SDS-PAGE and then transferred to nitrocellulose membrane. This membrane was then probed with anti-RNase4 or anti-RNase5 antibodies followed by probing with HRP conjugated goat anti-rabbit antibody. The signals were visualized by enhanced chemiluminescence followed by exposure to X-ray film for 2 min (B). Ponceau S stained images of the membranes is shown to indicate the protein loaded in Fig. 4A.
4. Discussion
This study has revealed that the RNase4 and RNase5 genes are most highly expressed in the liver and that their mRNAs and corresponding proteins are present in a wider range of tissues than previously reported in studies of human, cattle, and rodents [5], [7], [18], [26], [55], [56], [57]. The dynamics of variation of the levels of RNase4 and RNase5 within populations of cattle is not addressed in this study. All of the tissues in which these RNases are present have a host defence function capability, so the findings are consistent with the idea that RNase4 and RNase5 play a role in host defence. The relative abundance of RNase4 and RNase5 mRNA transcripts among tissues appears to correlate with their corresponding proteins in most of the tissues, but not in liver, mammary tissue, and salivary gland. One possible explanation for this is the particularly high secretory capacity of these tissues leading to a high flux of RNases through the intracellular space and accumulation in the extracellular space.
The observation made in this study, that high levels of mRNA transcripts of RNase4 and RNase5 are present in bovine liver, is in accordance with the previously reported high levels of these RNases in the livers of both humans and mice [33], [55], [56], [57], [58]. Since all members of this family are secreted proteins and their genes contain a secretion leader sequence, it is likely that these RNases are secreted into the extracellular space from liver. However, neither RNase was detected in serum. It is possible that the RNases are present in serum but that their detection is limited by the dilution effect consequent on the very high concentration of major serum proteins such as albumin. Further research is required to confirm the secretion rate and fate of RNase4 and RNase5 in bovine liver.
Previous work has shown that RNase4 and RNase5 purified from cows’ milk have antimicrobial activity against the yeast, C. albicans, but not against the mastitis causing pathogen S. uberis [5]. It is possible, but not yet demonstrated, that the RNases have anti-fungal properties in vivo and at least part of their function is to kill fungal pathogens that have invaded the mammary gland, the oral cavity, the intestinal tract, or the reproductive tract, all of which have access to the external environment and are likely to be exposed to pathogens. RNase7 from human skin has been demonstrated to have antimicrobial activity against a range of cutaneous microbes [25], [32], [59], [60]. In addition, RNase3, RNase6 and RNase7 have been shown to possess antimicrobial activity [29], [30], [33], [34], [61]. RNase2, one of the major secretory proteins of human eosinophils does not possess broad-spectrum antibacterial activity [61], but it displays antiviral activity against a range of viruses [37], [62], [63]. In addition, RNase3 has been shown to have anti-viral as well as anti-parasitic activities [36], [38], [39], [40]. Together, these findings suggest that antimicrobial activity could be a common function among several members of the RNaseA superfamily.
Adding further weight to a host defence role of the RNases, some of the RNases are induced upon infection in host defence related tissues. RNase2 has been shown to be induced in response to LPS [64], the intestinal pathogenic bacterium Clostridium difficile, and the airway epithelial microbe S. aureus [24], [65]. RNase6 has been reported to be induced in response to infection in the urinary tract in humans and mice [33], [34]. RNase7 is up-regulated in skin cells during infection with various pathogenic microbes [25], [31], [32], [59]. Together, these findings suggest that these RNases may play an important role as effector proteins in a range of tissues.
Tissues that are exposed to the environment secrete a range of antimicrobial proteins [66], [67]. A number of host-defence related proteins such as cathelicidins and defensins are known to be up-regulated in response to infection in some tissues, including the small intestine [68], [69]. Since RNases are known to withstand high temperature and low pH and are reported to be resistant to some proteases [70], [71], it is conceivable that at least some of the RNase4 and RNase5 present in colostrum and milk could remain intact after ingestion and be present in an active form in the intestine where they may contribute to optimising the commensal microbial population of the newborn. Verification of this idea awaits further investigation.
Author contributions
SKG performed all the experiments, and designed the experiments, analysed data and prepared the manuscript. BH contributed to the conception of the study. TTW conceived the study, contributed in designing the experiments, coordinated the team, and guided preparation of the manuscript. All authors read and approved the final manuscript.
Acknowledgments
This work was supported by a New Zealand Ministry of Science and Innovation Research contract (C10X0707) ‘Host Defence Proteins in Milk' awarded to TTW, and Post Graduate Research Scholarship from the University of Otago and AgResearch awarded to SKG.
Footnotes
Transparency document associated with this article can be found in the online version at doi:10.1016/j.bbrep.2016.09.001.
Contributor Information
Sandeep K. Gupta, Email: sandeep.gupta@agresearch.co.nz.
Brendan J. Haigh, Email: brendan.haigh@agresearch.co.nz.
Thomas T. Wheeler, Email: tom.rita@xtra.co.nz.
Appendix A. Transparency document
Supplementary material
References
- 1.Beintema J.J., Kleineidam R.G. The ribonuclease A superfamily: general discussion. Cell Mol. Life Sci. 1998;54(8):825–832. doi: 10.1007/s000180050211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barnard E. Biological function of pancreatic ribonuclease. Nature. 1969;221:340–344. doi: 10.1038/221340a0. [DOI] [PubMed] [Google Scholar]
- 3.Fett J.W., Strydom D.J., Lobb R.R., Alderman E.M., Bethune J.L., Riordan J.F., Vallee B.L. Isolation and characterization of angiogenin, an angiogenic protein from human carcinoma cells. Biochemistry. 1985;24(20):5480–5486. doi: 10.1021/bi00341a030. [DOI] [PubMed] [Google Scholar]
- 4.Pavlov N., Frendo J.-L., Guibourdenche J., Degrelle S.A., Evain-Brion D., Badet J. Angiogenin expression during early human placental development; association with blood vessel formation. BioMed. Res. Int. 2014;2014:781632. doi: 10.1155/2014/781632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Harris P., Johannessen K.M., Smolenski G., Callaghan M., Broadhurst M.K., Kim K., Wheeler T.T. Characterisation of the anti-microbial activity of bovine milk ribonuclease4 and ribonuclease5 (angiogenin) Int. Dairy J. 2010;20(6):400–407. [Google Scholar]
- 6.Strydom D.J., Bond M.D., Vallee B.L. An angiogenic protein from bovine serum and milk-purification and primary structure of angiogenin-2. Eur. J. Biochem. 1997;247(2):535–544. doi: 10.1111/j.1432-1033.1997.00535.x. [DOI] [PubMed] [Google Scholar]
- 7.Raven l-A., Cocks B.G., Pryce J.E., Cottrell J.J., Hayes B.J. Genes of the RNASE5 pathway contain SNP associated with milk production traits in dairy cattle. Genet. Sel. Evol. 2013;45(1):1–11. doi: 10.1186/1297-9686-45-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.D’Alessio G., Donato A.D., Parente A., Piccoli R. Seminal RNase: a unique member of the ribonuclease superfamily. Trends Biochem. Sci. 1991;16(3):104–106. doi: 10.1016/0968-0004(91)90042-t. [DOI] [PubMed] [Google Scholar]
- 9.Sica F., Pica A., Merlino A., Russo Krauss I., Ercole C., Picone D. The multiple forms of bovine seminal ribonuclease: structure and stability of a C-terminal swapped dimer. FEBS Lett. 2013;587(23):3755–3762. doi: 10.1016/j.febslet.2013.10.003. [DOI] [PubMed] [Google Scholar]
- 10.Schmekel B., Ahlner J., Malmström M., Venge P. Eosinophil cationic protein (ECP) in saliva: a new marker of disease activity in bronchial asthma. Respir. Med. 2001;95(8):670–675. doi: 10.1053/rmed.2001.1123. [DOI] [PubMed] [Google Scholar]
- 11.Sur S., Glitz D.G., Kita H., Kujawa S.M., Peterson E.A., Weiler D.A., Kephart G.M., Wagner J.M., George T.J., Gleich G.J. Localization of eosinophil-derived neurotoxin and eosinophil cationic protein in neutrophilic leukocytes. J. Leukoc. Biol. 1998;63(6):715–722. doi: 10.1002/jlb.63.6.715. [DOI] [PubMed] [Google Scholar]
- 12.Durack D., Ackerman S., Loegering D., Gleich G. Purification of human eosinophil-derived neurotoxin. Proc. Natl. Acad. Sci. USA. 1981;78(8):5165–5169. doi: 10.1073/pnas.78.8.5165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Awad A., Yassine H., Barrier M., Vorng H., Marquillies P., Tsicopoulos A., Duez C. Natural killer cells induce eosinophil activation and apoptosis. PLoS ONE. 2014;9(4):e94492. doi: 10.1371/journal.pone.0094492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Karawajczyk M., Peterson C.G.B., Venge P., Garcia R.C. An extragranular compartment of blood eosinophils contains eosinophil protein X/eosinophil-derived neurotoxin (EPX/EDN) Inflammation. 2013;36(2):320–329. doi: 10.1007/s10753-012-9549-z. [DOI] [PubMed] [Google Scholar]
- 15.Cormier S., Yuan S., Crosby J., Protheroe C., Dimina D., Hines E., Lee N., Lee J. T(H)2-mediated pulmonary inflammation leads to the differential expression of ribonuclease genes by alveolar macrophages. Am. J. Respir. Cell Mol. Biol. 2002;27(6):678–687. doi: 10.1165/rcmb.4882. [DOI] [PubMed] [Google Scholar]
- 16.Egesten A., Dyer K.D., Batten D., Domachowske J.B., Rosenberg H.F. Ribonucleases and host defense: identification, localization and gene expression in adherent monocytes in vitro. BBA. 1997;1358(3):255–260. doi: 10.1016/s0167-4889(97)00081-5. [DOI] [PubMed] [Google Scholar]
- 17.Wong K.L., JJ-Y Tai, Wong W.-C., Han H., Sem X., Yeap W.-H., Kourilsky P., Wong S.-C. Gene expression profiling reveals the defining features of the classical, intermediate, and nonclassical human monocyte subsets. Blood. 2011;118(5):e16–e31. doi: 10.1182/blood-2010-12-326355. [DOI] [PubMed] [Google Scholar]
- 18.Hooper L.V., Stappenbeck T.S., Hong C.V., Gordon J.I. Angiogenins: a new class of microbicidal proteins involved in innate immunity. Nat. Immunol. 2003;4(3):269–273. doi: 10.1038/ni888. [DOI] [PubMed] [Google Scholar]
- 19.Irie M., Nitta R., Ohgi K., Niwata Y., Watanabe H., Iwama M., Beintema J., Sanda A., Takizawa Y. Primary structure of a non-secretory ribonuclease from bovine kidney. J. Biochem. 1988;104(2):289–296. doi: 10.1093/oxfordjournals.jbchem.a122460. [DOI] [PubMed] [Google Scholar]
- 20.Zhao W., Kote-Jarai Z., van Santen Y., Hofsteenge J., Beintema J.J. Ribonucleases from rat and bovine liver: purification, specificity and structural characterization. Biochem. Biophys. Res. Commun. 1998;1384(1):55–65. doi: 10.1016/s0167-4838(97)00213-6. [DOI] [PubMed] [Google Scholar]
- 21.Gupta S.K., Haigh B.J., Griffin F.J., Wheeler T.T. The mammalian secreted RNases: mechanisms of action in host defence. Innate Immun. 2013;19(1):86–97. doi: 10.1177/1753425912446955. [DOI] [PubMed] [Google Scholar]
- 22.Rosenberg H.F. Eosinophil-Derived Neurotoxin (EDN/RNase 2) and the mouse Eosinophil-Associated RNases (mEars): expanding roles in promoting host defense. Int. J. Mol. Sci. 2015;16(7):15442–15455. doi: 10.3390/ijms160715442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Boix E., Nogues M.V. Mammalian antimicrobial proteins and peptides: overview on the RNase A superfamily members involved in innate host defence. Mol. Biosyst. 2007;3(5):317–335. doi: 10.1039/b617527a. [DOI] [PubMed] [Google Scholar]
- 24.Hosoki K., Nakamura A., Kainuma K., Sugimoto M., Nagao M., Hiraguchi Y., Tanida H., Tokuda R., Wada H., Nobori T. Differential activation of eosinophils by bacteria associated with Asthma. Int. Arch. Allergy Immunol. 2013;161(suppl 2):16–22. doi: 10.1159/000350338. [DOI] [PubMed] [Google Scholar]
- 25.Abtin A., Eckhart L., Mildner M., Ghannadan M., Harder J., Schroder J.-M., Tschachler E. Degradation by stratum corneum proteases prevents endogenous RNase inhibitor from blocking antimicrobial activities of RNase 5 and RNase 7. J. Invest Dermatol. 2009;129(9):2193–2201. doi: 10.1038/jid.2009.35. [DOI] [PubMed] [Google Scholar]
- 26.Murata M., Wakabayashi H., Yamauchi K., Abe F. Identification of milk proteins enhancing the antimicrobial activity of lactoferrin and lactoferricin. J. Dairy Sci. 2013;96(8):4891–4898. doi: 10.3168/jds.2013-6612. [DOI] [PubMed] [Google Scholar]
- 27.Gupta S.K. University of Otago; Dunedin, New Zealand: 2013. The Role of Cows' Milk RNases in Host Defence, Ph.D thesis. [Google Scholar]
- 28.Ester B., Marc T., Daniel S., Maria Victoria N. The antipathogen activities of eosinophil cationic protein. Curr. Pharm. Biotechnol. 2008;9(3):141–152. doi: 10.2174/138920108784567353. [DOI] [PubMed] [Google Scholar]
- 29.Persson T., Andersson P., Bodelsson M., Laurell M., Malm J., Egesten A. Bactericidal activity of human eosinophilic granulocytes against escherichia coli. Infect. Immun. 2001;69(6):3591–3596. doi: 10.1128/IAI.69.6.3591-3596.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pulido D., Moussaoui M., Nogués M.V., Torrent M., Boix E. Towards the rational design of antimicrobial proteins. FEBS J. 2013;280(22):5841–5852. doi: 10.1111/febs.12506. [DOI] [PubMed] [Google Scholar]
- 31.Köten B., Simanski M., Gläser R., Podschun R., Schröder J.-M., Harder J. RNase 7 contributes to the cutaneous defense against Enterococcus faecium. PLoS One. 2009;4(7):e6424. doi: 10.1371/journal.pone.0006424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Simanski M., Dressel S., Glaser R., Harder J. RNase 7 protects healthy skin from Staphylococcus aureus colonization. J. Investig. Dermatol. 2010;130(12):2836–2838. doi: 10.1038/jid.2010.217. [DOI] [PubMed] [Google Scholar]
- 33.Becknell B., Eichler T., Beceiro S., Li B., Easterling R., Carpenter A.R., James C., McHugh K.M., Hains D.S., Partida-Sanchez S. Ribonucleases 6 and 7 have antimicrobial function in the human and murine urinary tract. Kidney Int. 2015;87(1):151–161. doi: 10.1038/ki.2014.268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Spencer J.D., Schwaderer A.L., Wang H., Bartz J., Kline J., Eichler T., DeSouza K.R., Sims-Lucas S., Baker P., Hains D.S. Ribonuclease 7, an antimicrobial peptide up-regulated during infection, contributes to microbial defense of the human urinary tract. Kidney Int. 2013;83(4):615–625. doi: 10.1038/ki.2012.410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gleich G.J., Loegering D.A., Bell M.P., Checkel J.L., Ackerman S.J., McKean D.J. Biochemical and functional similarities between human eosinophil-derived neurotoxin and eosinophil cationic protein: homology with ribonuclease. Proc. Natl. Acad. Sci. USA. 1986;83(10):3146–3150. doi: 10.1073/pnas.83.10.3146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Domachowske J.B., Dyer K.D., Adams A.G., Leto T.L., Rosenberg H.F. Eosinophil cationic protein/RNase 3 is another RNase A-family ribonuclease with direct antiviral activity. Nucleic Acids Res. 1998;26(14):3358. doi: 10.1093/nar/26.14.3358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rugeles M.T., Trubey C.M., Bedoya V.I., Pinto L.A., Oppenheim J.J., Rybak S.M., Shearer G.M. Ribonuclease is partly responsible for the HIV-1 inhibitory effect activated by HLA alloantigen recognition. AIDS. 2003;17(4):481–486. doi: 10.1097/00002030-200303070-00002. [DOI] [PubMed] [Google Scholar]
- 38.Ackerman S., Gleich G., Loegering D., Richardson B., Butterworth A. Comparative toxicity of purified human eosinophil granule cationic proteins for schistosomula of Schistosoma mansoni. Am. J. Trop. Med. Hyg. 1985;34(4):735–745. doi: 10.4269/ajtmh.1985.34.735. [DOI] [PubMed] [Google Scholar]
- 39.Hamann K., Barker R., Loegering D., Gleich G. Comparative toxicity of purified human eosinophil granule proteins for newborn larvae of Trichinella spiralis. J. Parasitol. 1987;73(3):523–529. [PubMed] [Google Scholar]
- 40.Hamann K., Gleich G., Checkel J., Loegering D., McCall J., Barker R. In vitro killing of microfilariae of Brugia pahangi and Brugia malayi by eosinophil granule proteins. J. Immunol. 1990;144(8):3166–3173. [PubMed] [Google Scholar]
- 41.Chang K.C., Lo C.W., Fan T., Chang M.D.T., Shu C.W., Chang C.H., Chung C.T., Fang S., Chao C.C., Tsai J.J. TNF-a mediates eosinophil cationic protein-induced apoptosis in BEAS-2 B cells. BMC Cell Biol. 2010;11(1):6. doi: 10.1186/1471-2121-11-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sinatra F., Callari D., Viola M., Longombardo M., Patania M., Litrico L., Emmanuele G., Lanteri E., D’Alessandro N., Travali S. Bovine seminal RNase induces apoptosis in normal proliferating lymphocytes. Int J. Clin. Lab. Res. 2000;30(4):191–196. doi: 10.1007/s005990070006. [DOI] [PubMed] [Google Scholar]
- 43.Yang D., Rosenberg H.F., Chen Q., Dyer K.D., Kurosaka K., Oppenheim J.J. Eosinophil-derived neurotoxin (EDN), an antimicrobial protein with chemotactic activities for dendritic cells. Blood. 2003;102(9):3396–3403. doi: 10.1182/blood-2003-01-0151. [DOI] [PubMed] [Google Scholar]
- 44.Torrent M., Badia M., Moussaoui M., Sanchez D., Nogués M.V., Boix E. Comparison of human RNase 3 and RNase 7 bactericidal action at the Gram-negative and Gram-positive bacterial cell wall. FEBS J. 2010;277(7):1713–1725. doi: 10.1111/j.1742-4658.2010.07595.x. [DOI] [PubMed] [Google Scholar]
- 45.Pulido D., Moussaoui M., Andreu D., Nogués M.V., Torrent M., Boix E. Antimicrobial action and cell agglutination by the eosinophil cationic protein are modulated by the cell wall lipopolysaccharide structure. Antimicrob. Agents Chemother. 2012;56(5):2378–2385. doi: 10.1128/AAC.06107-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Torrent M., Pulido D., Valle J., Nogués M.V., Andreu D., Boix E. Ribonucleases as a host-defence family: evidence of evolutionarily conserved antimicrobial activity at the N-terminus. Biochem. J. 2013;456(1):99–108. doi: 10.1042/BJ20130123. [DOI] [PubMed] [Google Scholar]
- 47.Torrent M., Pulido D., Nogués M.V., Boix E. Exploring new biological functions of amyloids: bacteria cell agglutination mediated by host protein aggregation. PLoS Pathog. 2012;8(11):e1003005. doi: 10.1371/journal.ppat.1003005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rajan G.H., Morris C.A., Carruthers V.R., Wilkins R.J., Wheeler T.T. The relative abundance of a salivary protein, bSP30, is correlated with susceptibility to bloat in cattle herds selected for high or low bloat susceptibility. Anim. Genet. 1996;27(6):407–414. doi: 10.1111/j.1365-2052.1996.tb00507.x. [DOI] [PubMed] [Google Scholar]
- 49.Bøyum A. Separation of blood leucocytes, granulocytes and lymphocytes. Tissue Antigens. 1974;4(3):269–274. [PubMed] [Google Scholar]
- 50.Livak K.J., Schmittgen T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods. 2001;25(4):402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 51.Wheeler T.T., Maqbool N.J., Gupta S.K. Mapping, phylogenetic and expression analysis of the RNase (RNase A) locus in cattle. J. Mol. Evol. 2012;74(5–6):237–248. doi: 10.1007/s00239-012-9502-7. [DOI] [PubMed] [Google Scholar]
- 52.Berg D.K., Smith C.S., Pearton D.J., Wells D.N., Broadhurst R., Donnison M., Pfeffer P.L. Trophectoderm lineage determination in cattle. Dev. Cell. 2011;20(2):244–255. doi: 10.1016/j.devcel.2011.01.003. [DOI] [PubMed] [Google Scholar]
- 53.Bradford M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976;72(1–2):248–254. doi: 10.1006/abio.1976.9999. [DOI] [PubMed] [Google Scholar]
- 54.Smolenski G.A., Broadhurst M.K., Stelwagen K., Haigh B.J., Wheeler T.T. Host defence related responses in bovine milk during an experimentally induced Streptococcus uberis infection. Proteome Sci. 2014;12(1):19. doi: 10.1186/1477-5956-12-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Dyer K.D., Rosenberg H.F. The mouse RNase 4 and RNase 5/ang 1 locus utilizes dual promoters for tissue-specific expression. Nucleic Acids Res. 2005;33(3):1077–1086. doi: 10.1093/nar/gki250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Dyer K.D., Rosenberg H.F., Zhang J. Isolation, characterization, and evolutionary divergence of mouse rnase 6: evidence for unusual evolution in rodents. J. Mol. Evol. 2004;59(5):657–665. doi: 10.1007/s00239-004-2657-0. [DOI] [PubMed] [Google Scholar]
- 57.Futami J., Tsushima Y., Murato Y., Tada H., Sasaki J., Seno M., Yamada H. Tissue-specific expression of pancreatic-type RNases and RNase inhibitor in humans. DNA Cell Biol. 1997;16(4):413–419. doi: 10.1089/dna.1997.16.413. [DOI] [PubMed] [Google Scholar]
- 58.Sasso M.P., Lombardi M., Confalone E., Carsana A., Palmieri M., Furia A. The differential pattern of tissue-specific expression of ruminant pancreatic type ribonucleases may help to understand the evolutionary history of their genes. Gene. 1999;227(2):205–212. doi: 10.1016/s0378-1119(98)00586-1. [DOI] [PubMed] [Google Scholar]
- 59.Harder J., Schröder J. RNase 7, a novel innate immune defense antimicrobial protein of healthy human skin. J. Biol. Chem. 2002;277(48):46779–46784. doi: 10.1074/jbc.M207587200. [DOI] [PubMed] [Google Scholar]
- 60.Zanger P., Holzer J., Schleucher R., Steffen H., Schittek B., Gabrysch S. Constitutive expression of the antimicrobial peptide rnase 7 is associated with staphylococcus aureus infection of the skin. J. Infect. Dis. 2009;200(12):1907–1915. doi: 10.1086/648408. [DOI] [PubMed] [Google Scholar]
- 61.Lehrer R.I., Szklarek D., Barton A., Ganz T., Hamann K., Gleich G. Antibacterial properties of eosinophil major basic protein and eosinophil cationic protein. J. Immunol. 1989;142(12):4428–4434. [PubMed] [Google Scholar]
- 62.Domachowske J.B., Dyer K.D., Bonville C.A., Rosenberg H.F. Recombinant human eosinophil-derived neurotoxin/rnase 2 functions as an effective antiviral agent against respiratory syncytial virus. J. Infect. Dis. 1998;177(6):1458–1464. doi: 10.1086/515322. [DOI] [PubMed] [Google Scholar]
- 63.Liu J., Li Y.-H., Xue C.-F., Ding J., Gong W.-D., Zhao Y., Huang Y. Targeted ribonuclease can inhibit replication of hepatitis B virus. World J. Gastroenterol. 2003;9(2):295–299. doi: 10.3748/wjg.v9.i2.295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yang D., Chen Q., Rosenberg H.F., Rybak S.M., Newton D.L., Wang Z.Y., Fu Q., Tchernev V.T., Wang M., Schweitzer B. Human Ribonuclease A superfamily members, eosinophil-derived neurotoxin and pancreatic ribonuclease, induce dendritic cell maturation and activation. J. Immunol. 2004;173(10):6134–6142. doi: 10.4049/jimmunol.173.10.6134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hosoki K., Nakamura A., Nagao M., Hiraguchi Y., Tokuda R., Wada H., Nobori T., Fujisawa T. Differential activation of eosinophils by ‘probiotic’ bifidobacterium bifidumbifidobacterium bifidum and ‘pathogenic’ clostridium difficileclostridium difficile. Int. Arch. Allergy Immunol. 2010;152(suppl 1):83–89. doi: 10.1159/000312131. [DOI] [PubMed] [Google Scholar]
- 66.Hancock R.E., Diamond G. The role of cationic antimicrobial peptides in innate host defences. Trends Microbiol. 2000;8(9):402–410. doi: 10.1016/s0966-842x(00)01823-0. [DOI] [PubMed] [Google Scholar]
- 67.Hiemstra P. Epithelial antimicrobial peptides and proteins: their role in host defence and inflammation. Paediatr. Respir. Rev. 2001;2(4):306–310. doi: 10.1053/prrv.2001.0165. [DOI] [PubMed] [Google Scholar]
- 68.Ganz T. Defensins: antimicrobial peptides of innate immunity. Nat. Rev. Immunol. 2003;3(9):710–720. doi: 10.1038/nri1180. [DOI] [PubMed] [Google Scholar]
- 69.Ramanathan B., Davis E.G., Ross C.R., Blecha F. Cathelicidins: microbicidal activity, mechanisms of action, and roles in innate immunity. Microbes Infect. 2002;4(3):361–372. doi: 10.1016/s1286-4579(02)01549-6. [DOI] [PubMed] [Google Scholar]
- 70.Kunitz M., McDonald M.R. Ribonuclease – crystalline – from beef pancrease. Biochem. Prep. 1953;3:9–19. [Google Scholar]
- 71.Arnold U., Rücknagel K.P., Schierhorn A., Ulbrich-Hofmann R. Thermal unfolding and proteolytic susceptibility of Ribonuclease A. Eur. J. Biochem. 1996;237(3):862–869. doi: 10.1111/j.1432-1033.1996.0862p.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material