Abstract
The molecular mechanisms underlying the phagocytosis of apoptotic cells need to be elucidated in more detail because of its role in immune and inflammatory intractable diseases. We herein developed an experimental method to investigate phagocytosis quantitatively using the fruit fly Drosophila, in which the gene network controlling engulfment reactions is evolutionally conserved from mammals. In order to accurately detect and count engulfing and un-engulfing phagocytes using whole animals, Drosophila embryos were homogenized to obtain dispersed cells including phagocytes and apoptotic cells. The use of dispersed embryonic cells enables us to measure in vivo phagocytosis levels as if we performed an in vitro phagocytosis assay in which it is possible to observe all phagocytes and apoptotic cells in whole embryos and precisely quantify the level of phagocytosis. We confirmed that this method reproduces those of previous studies that identified the genes required for the phagocytosis of apoptotic cells. This method allows the engulfment of dead cells to be analyzed, and when combined with the powerful genetics of Drosophila, will reveal the complex phagocytic reactions comprised of the migration, recognition, engulfment, and degradation of apoptotic cells by phagocytes.
Keywords: Immunology, Issue 126, phagocytosis, apoptotic cells, Drosophila, embryo, hemocyte, in vivo
Introduction
In metazoan animals, e.g. the nematode Caenorhabditis elegans, the fruit fly Drosophila melanogaster, and mice and humans, a large number of cells undergo apoptosis during development to shape their bodies and in adulthood to maintain homeostasis1,2. Apoptotic cells need to be rapidly removed because they induce inflammation in the surrounding tissues by releasing immunogenic intracellular materials if not completely removed3. In order to facilitate rapid removal, apoptotic cells present so-called eat-me signals that are recognized by the engulfment receptors of phagocytes, and are eliminated by phagocytosis3,4,5,6. Thus, phagocytosis plays a crucial role in the maintenance of host homeostasis, and hence, elucidating the molecular mechanisms underlying the phagocytosis of apoptotic cells is of importance.
The mechanisms responsible for the phagocytosis of apoptotic cells appear to be evolutionally conserved among species in the nematodes, flies, and mice7. Several phagocytosis assays are currently available to assess the engulfment of apoptotic cells in these model animals. In C. elegans, 131 somatic cells undergo programmed cell death during development, and cell corpses are phagocytosed by neighboring cells, which are non-professional phagocytes8. Thus, counting the number of remaining cell corpses in C. elegans indicates the level of phagocytosis in vivo. By searching for nematode mutants that show an increased number of dead cells, several genes required for phagocytosis have been identified and genetically characterized9,10,11,12.
Ex vivo phagocytosis assays with primary culture phagocytes, generally macrophages, are often utilized in mice. Apoptotic cells are prepared using cell lines such as Jurkat cells, and are mixed with primary phagocytes. After an incubation for several hours, the total numbers of phagocytes and engulfing phagocytes are counted in order to assess the level of phagocytosis. As a sophisticated modification of this method, Nagata's group developed an ex vivo phagocytosis assay with cells expressing a caspase-resistant ICAD (inhibitor of caspase-activated DNase), in which apoptotic cells do not undergo apoptotic DNA fragmentation, but DNA is still cleaved when cells are phagocytosed. When these cells are used as apoptotic targets in a phagocytosis assay, only the DNA of engulfed apoptotic cells is fragmented, and stained by TdT-mediated dUTP nick end labeling (TUNEL). Therefore, the level of apoptotic cell engulfment is measured by counting TUNEL signals in a mixture of phagocytes and apoptotic cells13.
In D. melanogaster, professional phagocytes named hemocytes, Drosophila macrophages, are responsible for the phagocytosis of apoptotic cells14,15. In addition to in vitro phagocytosis assays with culture cell lines, in vivo phagocytosis assays with whole Drosophila embryos are available. Drosophila embryos are a powerful tool for examining the level of apoptotic cell engulfment because many cells undergo apoptosis and are phagocytosed by hemocytes during embryonic development14,15,16. An example of an in vivo phagocytosis assay is the method developed by Franc's group. In their method, hemocytes are detected by the immunostaining of peroxidasin, a hemocyte marker, apoptotic cells are stained using the nuclear dye, 7-amino actinomycin D in whole Drosophila embryos, and the number of double positive cells is counted as a signal of phagocytosis17. Another example of a phagocytosis assay on embryos is based on the concept of Nagata's method described above; however, in vivo phagocytosis is evaluated using the embryos of dCAD (Drosophila caspase-activated DNase) mutant flies18,19. These in vivo phagocytosis assays are useful for directly observing phagocytosis in situ. However, difficulties are associated with excluding any possible bias in the step of counting phagocytosing cells because it is hard to observe all phagocytes and apoptotic cells in whole embryos due to its thickness.
In order to overcome this limitation, we developed a new phagocytosis assay in Drosophila embryos. In our method, in order to easily count phagocytosing hemocytes, whole embryos are homogenized to prepare dispersed embryonic cells. Phagocytes are detected by the immunostaining of a phagocyte marker, and apoptotic cells are detected by TUNEL with these dispersed embryonic cells. The use of dispersed embryonic cells enables us to measure in vivo phagocytosis levels as if we performed an in vitro phagocytosis assay that precisely quantifies the level of phagocytosis. All genotypes of flies may be employed in this assay if they develop to stage 16 of embryos20, the developmental stage at which apoptotic cell clearance by phagocytosis is the most abundant. This method has the advantage of assessing the level of phagocytosis quantitatively, and, thus, may contribute to the identification of new molecules involved in the phagocytosis of apoptotic cells in vivo.
Protocol
1. Preparation
- Preparation of fresh grape juice agar plates
- Add 100 mL of water to 4.4 g of agar, and heat the mixture in a microwave oven to dissolve agar.
- Add 80 mL of fresh grape juice, 5 mL of acetic acid, and 5 mL of ethanol to agar solution.
- Pour approximately 1.5 mL of the solution with a pipette onto each glass slide, and allow it to solidify.
- Preparation of 6 cm dish agarose plates
- Add 50 mL water to 0.5 g of agarose, and heat the mixture in a microwave oven to dissolve agarose.
- Pour approximately 2.5 mL of agarose solution onto a 6 cm dish with a pipette.
2. Stage 16 Embryo Collection
Collect adult flies, and add 200 females and 200 males into a 50 mL conical tube.
Put a plate of fresh grape juice agar on yeast into the 50-mL tube, and close with a sponge cap.
Incubate the flies at 16 °C in the light for 2 - 3 days. Change the plate once a day.
Move the flies to the dark at 25 °C for 1 h.
Change the old plate to a new one without yeast. Allow flies to lay eggs for 2 h.
Collect the plate and incubate at 16 °C for 26 h.
Collect embryos from the plate with a paint brush into 1 mL of PBS containing 0.2% (v/v) Triton X-100.
Wash embryos twice with 1 mL of PBS containing 0.2% (v/v) Triton X-100.
In order to remove the chorion, add 1.2 mL of sodium hypochlorite solution (2.2 - 3.4% Cl) to embryos for 3 min.
Wash embryos 4 times with 1 mL of PBS containing 0.2% (v/v) Triton X-100.
Place embryos on 6-cm dish agarose plates. Pick up stage 16 embryos under a microscope as described by Roberts20. Collect approximately 50 embryos in a 1.5-mL Treff micro test tube with a micropipette.
3. Preparation of Embryonic Cells
Wash embryos twice with 150 µL of PBS.
Homogenize embryos 30 times in a 1.5 mL micro test tube and pellet mixer in the presence of 200 µL of collagenase (0.25% (w/v)) in PBS.
Disperse embryonic cells by pipetting 10 times, and move the cell suspension to a 1.5 mL tube. Incubate cells at 37 °C for 1 min in a water bath. Repeat this step twice.
Add 800 µL of PBS to the cell suspension, and centrifuge at 1400 x g at 4 °C for 5 min.
Remove the supernatant and add 200 µL of trypsin (0.25% (w/v)) in PBS to the cells. Disperse embryonic cells by pipetting 50 times.
Filtrate the cell suspension with a 70 µm Cell Strainer.
In order to stop trypsin activity, add 40 µL heat-inactivated fetal bovine serum to the filtrated cell suspension. Add 800 µL of PBS and centrifuge at 1,400 x g at 4 °C for 5 min.
Remove the supernatant, suspend precipitated cells in 200 µL of PBS, and centrifuge at 1400 x g at 4 °C for 5 min.
Remove the supernatant, and suspend precipitated cells in 30 µL of PBS.
Mount the prepared cell suspension on aminopropyl triethoxysilane-coated glass slides, and wait for 10 - 15 min until cells attach to the glass slides.
Remove the remaining solution on the glass slides with a pipette. Mount 60 - 70 µL of PBS containing 4% (w/v) PFA (paraformaldehyde) onto cells for 10 - 15 min for fixation.
Remove the fixation solution, and place glass slides into PBS for more than 1 min.
4. Staining of Hemocytes
- Immunostaining with an anti-Croquemort antibody
- Serially soak glass slides in methanol for 10 min, in PBS containing 0.2% (v/v) Triton X-100 for 10 min, and then in PBS for 10 min.
- Mount 20 µL of 5% (v/v) whole swine serum in PBS containing 0.2% (v/v) Triton X-100 on cells for blocking. Incubate slides at room temperature for 20 min.
- Soak glass slides in PBS containing 0.2% (v/v) Triton X-100 for 10 min 5 times.
- Soak glass slides in PBS for 10 min.
- Mount 20 µL of alkaline phosphatase-labeled anti-rat IgG (0.5% (v/v)) in PBS containing 0.2% (v/v) Triton X-100 and 5% (v/v) whole swine serum on cells at room temperature for 1 h.
- Soak glass slides in PBS containing 0.2% (v/v) Triton X-100 for 10 min 5 times.
- Soak glass slides in buffer containing 100 mM Tris-HCl, pH 9.5, 100 mM NaCl, and 50 mM MgCl2 for 10 min.
- Mount phosphatase substrate solution containing 0.23 mg/mL 5-bromo-4-chloro-3-indolyl-phosphate (BCIP), 0.35 mg/mL nitro blue tetrazolium (NBT), 100 mM Tris-HCl, pH 9.5, 100 mM NaCl, and 50 mM MgCl2 on cells.
- Observe cells under a microscope for proper staining. When strong purple signals appear in the granules of hemocytes, remove the substrate solution, and soak glass slides in buffer consisting of 10 mM Tris-HCl, pH 8.5 and 1 mM EDTA. Staining for approximately 5 - 10 min is recommended.
- Immunostaining with an anti-GFP antibody (another option for the staining of hemocytes)
- Serially soak glass slides in methanol for 10 min, PBS containing 0.2% (v/v) Triton X-100 for 10 min, and PBS for 10 min.
- Mount 20 µL of 5% (v/v) whole swine serum in PBS containing 0.2% (v/v) Triton X-100 on cells for blocking. Incubate slides at room temperature for 20 min.
- Remove the solution on glass slides, and mount 20 µL of the mouse anti-GFP antibody (1% (v/v))/PBS containing 0.2% (v/v) Triton X-100 on cells. Incubate slides at room temperature overnight.
- Soak glass slides in PBS containing 0.2% (v/v) Triton X-100 for 10 min 5 times.
- Soak glass slides in PBS for 10 min.
- Mount 20 µL of alkaline phosphatase-labeled anti-mouse IgG (0.5% (v/v)) in PBS containing 0.2% (v/v) Triton X-100 and 5% (v/v) whole swine serum on cells at room temperature for 1 h.
- Soak glass slides in PBS containing 0.2% (v/v) Triton X-100 for 10 min 5 times.
- Soak glass slides in buffer consisting of 100 mM Tris-HCl, pH 9.5, 100 mM NaCl, and 50 mM MgCl2 for 10 min.
- Mount the phosphatase substrate solution containing 0.23 mg/mL BCIP, 0.35 mg/mL NBT, 100 mM Tris-HCl, pH 9.5, 100 mM NaCl, and 50 mM MgCl2 on cells.
- Observe cells under a microscope for proper staining. When purple signals appear in whole hemocytes, remove the substrate solution and soak slides in buffer consisting of 10 mM Tris-HCl, pH8.5 and 1 mM EDTA. Staining for approximately 10 - 15 min is recommended.
5. Stain Apoptotic Cells by TUNEL
Soak glass slides in PBS for 5 min.
Repeat with new PBS.
Mount Equilibration buffer (See Materials Table) on cells for 10 min.
Remove the solution from glass slides, and mount 20 µL of TdT solution containing 5 µL of terminal deoxynucleotidyl transferase and 15 µL of Reaction Buffer on cells at 37 °C for 1 h.
Remove the solution from glass slides, and soak slides in buffer consisting of 0.5 mL STOP/Wash buffer and 17 mL of water.
Soak slides in PBS for 5 min 3 times.
Mount 20 µL of Anti-Digoxigenin-Peroxidase on cells at room temperature for 30 min.
Soak glass slides in PBS for 5 min 4 times.
Soak glass slides in peroxidase substrate solution consisting of 30 mL of 50 mM Tris-HCl, pH7.5 containing a full micro spatula of 3,3'-diaminobenzidine tetrahydrichloride (DAB), and 0.002% (v/v) H2O2 for 30 s.
Repeat step 5.9 until apoptotic cells are stained brown. Soak glass slides in water to stop the peroxidase reaction.
Enclose cells with water for observation.
6. Measure the Level of Phagocytosis of Apoptotic Cells
Observe samples under a light microscope in order to assess the level of phagocytosis. Count Croquemort-positive cells or GFP-positive cells as hemocytes, TUNEL-positive cells as apoptotic cells, and double positive cells as phagocytosing hemocytes.
The phagocytic index is defined as the number of double positive cells to the total number of Croquemort-positive cells or GFP-positive cells. It is recommended that more than 300 hemocytes are observed.
Representative Results
In order to examine the phagocytosis of apoptotic cells, Drosophila embryos of developmental stage 16 were collected and prepared as dispersed cells. Hemocytes, Drosophila macrophages, were stained by immunocytochemistry for the hemocyte marker "Croquemort"17,22 using a specific antibody19,21, and apoptotic cells were stained by TUNEL in dispersed embryonic cells (Figure 1A). Croquemort, a Drosophila CD36-related receptor, is specifically expressed on all embryonic hemocytes22, and has been shown genetically to be involved in the clearance of apoptotic cells in Drosophila embryos17. Croquemort-positive cells have purple signals in the small granules of their cells. TUNEL-positive cells show a brown signal in whole corpuses. TUNEL-positive cells are smaller than other cells, and some are inside Croquemort-positive cells, which are considered to be phagocytosed dead cells. Between 2 and 10% Croquemort-positive cells and 1 - 5% TUNEL-positive cells are normally observed in all embryonic cells.
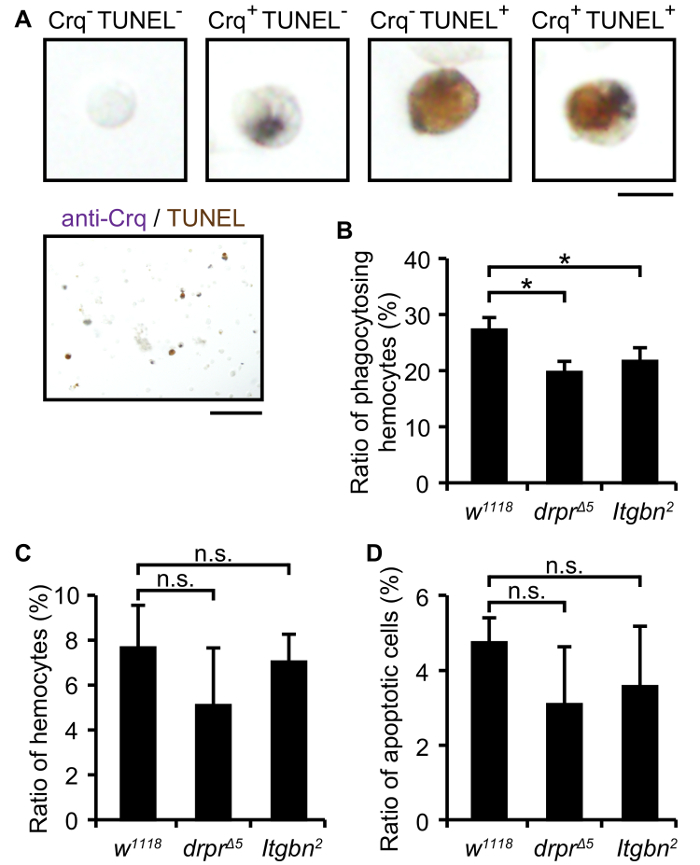
In Drosophila, there are two signaling pathways for the engulfment of apoptotic cells. Two phagocytosis receptors for the corresponding pathways, Draper and integrin αPS3βν, independently recognize dead cells19,21,23,24,25. Draper is a transmembrane protein which possesses atypical epidermal growth factor (EGF)-like sequences in the extracellular region and the two phosphorylatable motifs NPxY and YxxL in the intracellular portion26. Integrin αPS3βν is also a transmembrane receptor consisting of a heterodimer of α and β subunits25. Figure 1B shows the ratio of phagocytosing hemocytes to total hemocytes in wild-type or drpr or Itgbn single mutant embryos. By counting all Croquemort-positive cells (total hemocytes) and Croquemort- and TUNEL-double positive cells (phagocytosing hemocytes), we calculated the phagocytic index as the number of phagocytosing hemocytes to the total number of hemocytes. The phagocytic index was lower in the drpr or Itgbn mutant than in the wild type, while the numbers of hemocytes and apoptotic cells were similar (Figure 1C-D), indicating that these two genes are required for apoptotic cell engulfment, as previously reported.
When an anti-Croquemort antibody is not available or mutant lines with altered Croquemort expression are examined, we need to select another option to detect hemocytes. Embryos with a GAL4 driver controlled by a hemocyte-specific promoter (srpHemo-GAL4), and the GFP gene controlled by an upstream activation sequence (UAS) including the GAL4-binding site (UAS-EGFP), have GFP-labeled hemocytes27, which enables us to detect hemocytes using anti-GFP. Figure 2A shows an image obtained after detecting hemocytes and apoptotic cells with immunostaining using an anti-GFP antibody, and TUNEL in embryonic cells with srpHemo-GAL4UAS-EGFP. While an anti-Croquemort antibody stains small granules in cells, an anti-GFP antibody stains whole hemocytes. Similar to Figure 1A, 2 - 6% GFP-positive cells and 1 - 5% TUNEL-positive cells are observed in all embryonic cells. Some TUNEL-positive cells are detected inside GFP-positive cells, which are considered to be phagocytosed dead cells. RNAi-mediated knockdown is easily applied to assess any genes for their involvement in phagocytosis by crossing flies with the UAS-dsRNA of the candidate genes with srpHemo-GAL4UAS-EGFP. The RNAi-mediated knockdown of drpr or Itgbn reduced the phagocytic index (Figure 2B), whereas the numbers of hemocytes and apoptotic cells in embryonic cells was comparable (Figure 2C-2D), indicating again that these phagocytosis receptors are involved in the phagocytosis of apoptotic cells. Similar phagocytic indexes are obtained from both hemocyte detection methods, suggesting that both are compatible.
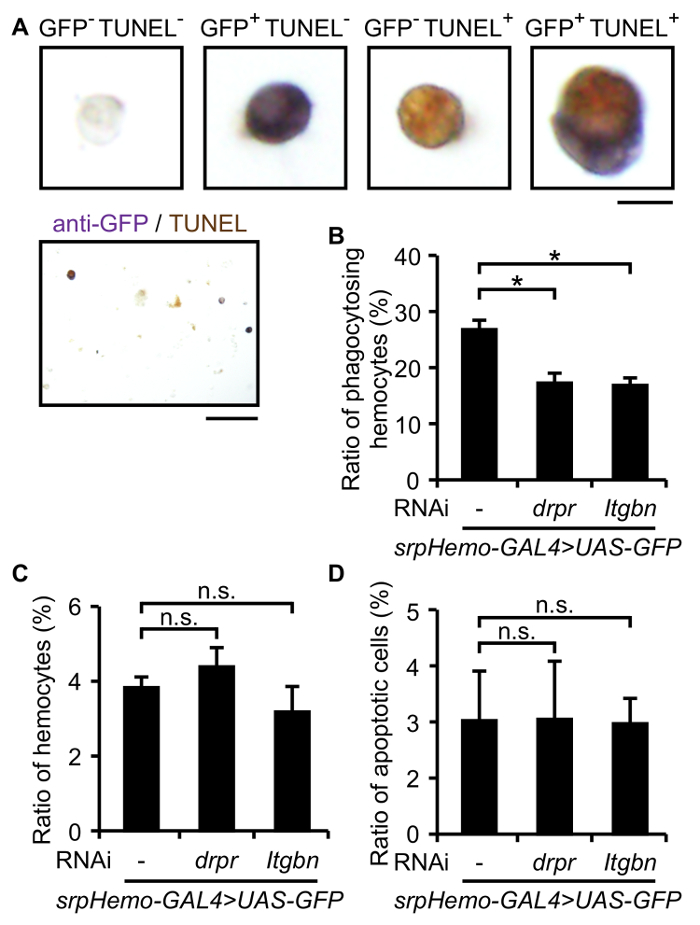
Figure 1:Staining of Embryonic Cells by an Anti-Croquemort Antibody and TUNEL. (A) Bright field images of dispersed embryonic cells from the w1118 strain by immunostaining with an anti-Croquemort antibody and TUNEL. Magnified views of representative positively or negatively stained cells (top 4 panels) and a low-power field (bottom panel) are shown. Scale bars, 5 µm (top); 50 µm (bottom). (B) The ratio of Croquemort-positive hemocytes with the TUNEL signal to all Croquemort-positive hemocytes in the drpr or Itgbn mutant was analyzed with dispersed embryonic cells. (C) The ratio of Croquemort-positive hemocytes to all cells in the drpr or Itgbn mutant was analyzed with dispersed embryonic cells. (D) The ratio of TUNEL-positive apoptotic cells to all cells in the drpr or Itgbn mutant was analyzed with dispersed embryonic cells. Genotypes; w1118(wild-type control), drprΔ5 (drpr mutant), and Itgbn2 (integrin βν mutant). Data were representative of three independent experiments and analyzed by the Student's t-test. Approximately 300 Croquemort-positive hemocytes were observed in each experiment, and values represent the mean±S.D. of three microscopic fields. *; p<0.05, n.s.; difference not significant. Please click here to view a larger version of this figure.
Figure 2: Staining of Embryonic Cells by Anti-GFP and TUNEL. (A) Bright field images of dispersed embryonic cells from the srpHemo-GAL4UAS-EGFP/+ strain by immunostaining with an anti-GFP antibody and TUNEL. Magnified views of representative positively or negatively stained cells (top 4 panels) and a low-power field (bottom panel) are shown. Scale bars = 5 µm (top); 50 µm (bottom). (B) The ratio of GFP-positive hemocytes with the TUNEL signal to all GFP-positive hemocytes in RNAi-mediated knockdown flies of drpr or Itgbn was analyzed with dispersed embryonic cells. (C) The ratio of GFP-positive hemocytes to all cells in RNAi-mediated knockdown flies of drpr or Itgbn was analyzed with dispersed embryonic cells. (D) The ratio of TUNEL-positive apoptotic cells to all cells in RNAi-mediated knockdown flies of drpr or Itgbn was analyzed with dispersed embryonic cells. Genotypes; RNAi - : y w/w; srpHemo-GAL4 UAS-EGFP/+, RNAi drpr: y w/w; srpHemo-GAL4 UAS-EGFP/+; UAS-drpr-IR/+, RNAi Itgbn: y w/w; srpHemo-GAL4 UAS-EGFP/+; UAS-Itgbn-IR/+. Data were representative of three independent experiments and analyzed by the Student's t-test. Approximately 300 GFP-positive hemocytes were observed in each experiment, and values represent the mean±S.D. of three microscopic fields. *; p <0.05, n.s.; difference not significant. Please click here to view a larger version of this figure.
Discussion
We herein described a phagocytosis assay using Drosophila embryos. By using dispersed embryonic cells to measure phagocytosis quantitatively, hemocytes, Drosophila professional phagocytes, are immunostained for the hemocyte marker Croquemort or srpHemo-driven GFP, and apoptotic cells are detected by TUNEL in this protocol. The level of phagocytosis is expressed as a phagocytic index by counting the total number of hemocytes and phagocytosing hemocytes. The use of dispersed embryonic cells enables us to observe all phagocytes and apoptotic cells in whole embryos and to precisely quantify the level of phagocytosis. In addition, our method detects hemocytes and apoptotic cells with dyes that are observable under a light microscope, not a fluorescence microscope. This facilitates the assessment of phagocytosis levels because it is possible to observe hemocytes and apoptotic cells simultaneously without changing any filter.
The following points are critical for staining embryonic cells. In the fixation step of embryonic cells, fresh PFA prepared within two weeks needs to be used. In the immunostaining of Croquemort, the incubation of cells with anti-Croquemort needs to be performed at 4 °C. In TUNEL staining, the incubation of cells with DAB needs to be performed by carefully checking under a microscope to avoid excessive staining. TUNEL visualizes fragmented DNA by detecting 3'-OH, which is abundant in apoptotic cells. However, normal cells also have 3'-OH in the 3'-terminal of DNA, but at a lower level than that in apoptotic cells. Therefore, an incubation of cells with DAB for too long will stain not only apoptotic cells, but also normal cells. The observation of cells every 15 or 30 s during DAB staining is recommended to prevent this.
In this protocol, although quantification of the level of phagocytosis is fast and precise, some information such as the site of engulfment or distribution of hemocytes is lost. We previously showed that this assay and an in situ phagocytosis assay using whole embryos produced similar results in terms of the phagocytic index21. Therefore, we recommend performing an in situ phagocytosis assay in parallel in order to accurately interpret the results obtained in some cases.
Drosophila have another professional phagocyte, glial cells, in the central nervous system. Although we do not describe how to assess the phagocytosis of apoptotic neurons by glial cells in embryos in this protocol, the same protocol is applied to quantify the level of engulfment as that reported by Kuraishi et al., EMBO.J21. Likewise, this assay suggests that TUNEL positive cells in Drosophila embryos are likely to be phagocytosed by non-professional phagocytes. Phagocytosis by those unidentified phagocytes with unknown engulfment receptors might explain the result (Figure 1D) that the total number of apoptotic cells in embryonic cells seems not to be increased in a phagocytosis gene mutant.
In conclusion, this protocol has successfully led to the discovery of genes required for the phagocytosis of apoptotic cells by professional phagocytes.24,25,28. Based on the evolutionary conservation of the molecular mechanisms underlying phagocytosis7, revealing gene functions in Drosophila embryos that involve complex phagocytosis reactions may provide meaningful insights into the phagocytic clearance of dead cells, which is related to inflammatory disorders and autoimmune diseases in mammals29.
Disclosures
The authors declare that they have no competing financial interests.
Acknowledgments
We are grateful to Kaz Nagaosa and Akiko Shiratsuchi for their advice.
References
- Danial NN, Korsmeyer SJ. Cell death: critical control points. Cell. 2004;116(2):205–219. doi: 10.1016/s0092-8674(04)00046-7. [DOI] [PubMed] [Google Scholar]
- Vaux DL, Korsmeyer SJ. Cell death in development. Cell. 1999;96(2):245–254. doi: 10.1016/s0092-8674(00)80564-4. [DOI] [PubMed] [Google Scholar]
- Savill J, Fadok V. Corpse clearance defines the meaning of cell death. Nature. 2000;407(6805):784–788. doi: 10.1038/35037722. [DOI] [PubMed] [Google Scholar]
- Lauber K, Blumenthal SG, Waibel M, Wesselborg S. Clearance of apoptotic cells: getting rid of the corpses. Mol Cell. 2004;14(3):277–287. doi: 10.1016/s1097-2765(04)00237-0. [DOI] [PubMed] [Google Scholar]
- Ravichandran KS, Lorenz U. Engulfment of apoptotic cells: signals for a good meal. Nat Rev Immunol. 2007;7(12):964–974. doi: 10.1038/nri2214. [DOI] [PubMed] [Google Scholar]
- Ravichandran KS. Beginnings of a good apoptotic meal: the find-me and eat-me signaling pathways. Immunity. 2011;35(4):445–455. doi: 10.1016/j.immuni.2011.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakanishi Y, Nagaosa K, Shiratsuchi A. Phagocytic removal of cells that have become unwanted: implications for animal development and tissue homeostasis. Dev Growth Differ. 2011;53(2):149–160. doi: 10.1111/j.1440-169X.2010.01224.x. [DOI] [PubMed] [Google Scholar]
- Reddien PW, Horvitz HR. The engulfment process of programmed cell death in caenorhabditis elegans. Annu Rev Cell Dev Biol. 2004;20:193–221. doi: 10.1146/annurev.cellbio.20.022003.114619. [DOI] [PubMed] [Google Scholar]
- Hedgecock EM, Sulston JE, Thomson JN. Mutations affecting programmed cell deaths in the nematode Caenorhabditis elegans. Science. 1983;220(4603):1277–1279. doi: 10.1126/science.6857247. [DOI] [PubMed] [Google Scholar]
- Ellis RE, Jacobson DM, Horvitz HR. Genes required for the engulfment of cell corpses during programmed cell death in Caenorhabditis elegans. Genetics. 1991;129(1):79–94. doi: 10.1093/genetics/129.1.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gumienny TL. CED-12/ELMO, a novel member of the CrkII/Dock180/Rac pathway, is required for phagocytosis and cell migration. Cell. 2001;107(1):27–41. doi: 10.1016/s0092-8674(01)00520-7. [DOI] [PubMed] [Google Scholar]
- Hsu TY, Wu YC. Engulfment of apoptotic cells in C. elegans is mediated by integrin alpha/SRC signaling. Curr Biol. 2010;20(6):477–486. doi: 10.1016/j.cub.2010.01.062. [DOI] [PubMed] [Google Scholar]
- Hanayama R. Identification of a factor that links apoptotic cells to phagocytes. Nature. 2002;417(6885):182–187. doi: 10.1038/417182a. [DOI] [PubMed] [Google Scholar]
- Tepass U, Fessler LI, Aziz A, Hartenstein V. Embryonic origin of hemocytes and their relationship to cell death in Drosophila. Development. 1994;120(7):1829–1837. doi: 10.1242/dev.120.7.1829. [DOI] [PubMed] [Google Scholar]
- Gold KS, Bruckner K. Macrophages and cellular immunity in Drosophila melanogaster. Semin Immunol. 2015;27:357–368. doi: 10.1016/j.smim.2016.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abrams JM, White K, Fessler LI, Steller H. Programmed cell death during Drosophila embryogenesis. Development. 1993;117(1):29–43. doi: 10.1242/dev.117.1.29. [DOI] [PubMed] [Google Scholar]
- Franc NC, Heitzler P, Ezekowitz RA, White K. Requirement for croquemort in phagocytosis of apoptotic cells in Drosophila. Science. 1999;284(5422):1991–1994. doi: 10.1126/science.284.5422.1991. [DOI] [PubMed] [Google Scholar]
- Mukae N, Yokoyama H, Yokokura T, Sakoyama Y, Nagata S. Activation of the innate immunity in Drosophila by endogenous chromosomal DNA that escaped apoptotic degradation. Genes Dev. 2002;16(20):2662–2671. doi: 10.1101/gad.1022802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manaka J. Draper-mediated and phosphatidylserine-independent phagocytosis of apoptotic cells by Drosophila hemocytes/macrophages. J Biol Chem. 2004;279(46):48466–48476. doi: 10.1074/jbc.M408597200. [DOI] [PubMed] [Google Scholar]
- Roberts DB. Drosophila: A Practical Approach. Oxford: IRL Press; 1998. pp. 3868–3878. [Google Scholar]
- Kuraishi T, et al. Pretaporter, a Drosophila protein serving as a ligand for Draper in the phagocytosis of apoptotic cells. Embo j. 2009;28(24):3868–3878. doi: 10.1038/emboj.2009.343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franc NC, Dimarcq JL, Lagueux M, Hoffmann J, Ezekowitz RA. Croquemort, a novel Drosophila hemocyte/macrophage receptor that recognizes apoptotic cells. Immunity. 1996;4(5):431–443. doi: 10.1016/s1074-7613(00)80410-0. [DOI] [PubMed] [Google Scholar]
- Kuraishi T. Identification of calreticulin as a marker for phagocytosis of apoptotic cells in Drosophila. Exp Cell Res. 2007;313(3):500–510. doi: 10.1016/j.yexcr.2006.10.027. [DOI] [PubMed] [Google Scholar]
- Nagaosa K. Integrin betanu-mediated phagocytosis of apoptotic cells in Drosophila embryos. J Biol Chem. 2011;286(29):25770–25777. doi: 10.1074/jbc.M110.204503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nonaka S, Nagaosa K, Mori T, Shiratsuchi A, Nakanishi Y. Integrin alphaPS3/betanu-mediated phagocytosis of apoptotic cells and bacteria in Drosophila. J Biol Chem. 2013;288(15):10374–10380. doi: 10.1074/jbc.M113.451427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita Y, Nagaosa K, Shiratsuchi A, Nakanishi Y. Role of NPxY motif in Draper-mediated apoptotic cell clearance in Drosophila. Drug Discov Ther. 2012;6(6):291–297. [PubMed] [Google Scholar]
- Bruckner K, et al. The PDGF/VEGF receptor controls blood cell survival in Drosophila. Dev Cell. 2004;7(1):73–84. doi: 10.1016/j.devcel.2004.06.007. [DOI] [PubMed] [Google Scholar]
- Nonaka S, et al. Signaling Pathway for Phagocyte Priming upon Encounter with Apoptotic Cells. J Biol Chem. 2017;292(19):8059–8072. doi: 10.1074/jbc.M116.769745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanayama R. Autoimmune disease and impaired uptake of apoptotic cells in MFG-E8-deficient mice. Science. 2004;304(5674):1147–1150. doi: 10.1126/science.1094359. [DOI] [PubMed] [Google Scholar]


