Abstract
The encoding of biological information that is accessible to future generations is generally achieved via changes to the DNA sequence. Long-lived inheritance encoded in protein conformation (rather than sequence) has long been viewed as paradigm-shifting but rare. The best characterized examples of such epigenetic elements are prions, which possess a self-assembling behavior that can drive the heritable manifestation of new phenotypes. Many archetypal prions display a striking N/Q-rich sequence bias and assemble into an amyloid fold. These unusual features have informed most screening efforts to identify new prion proteins. However, at least three known prions (including the founding prion, PrPSc) do not harbor these biochemical characteristics. We therefore developed an alternative method to probe the scope of protein-based inheritance based on a property of mass action: the transient overexpression of prion proteins increases the frequency at which they acquire a self-templating conformation. This paper describes a method for analyzing the capacity of the yeast ORFeome to elicit protein-based inheritance. Using this strategy, we previously found that >1% of yeast proteins could fuel the emergence of biological traits that were long-lived, stable, and arose more frequently than genetic mutation. This approach can be employed in high throughput across entire ORFeomes or as a targeted screening paradigm for specific genetic networks or environmental stimuli. Just as forward genetic screens define numerous developmental and signaling pathways, these techniques provide a methodology to investigate the influence of protein-based inheritance in biological processes.
Keywords: Cellular Biology, Issue 126, Prion, yeast, screen, protein folding, high throughput, epigenetic inheritance, protein-based inheritance
Introduction
Biological systems frequently experience transient fluctuations in protein abundance. Whether these have a lasting impact in shaping the phenotype of an organism or of future generations remains unclear. The best-known instances of this biology involve a rare class of proteins, prions, which drive the emergence of heritable traits without genome modification. Instead, these proteinaceous and infectious particles transmit phenotypes via self-perpetuating changes to protein conformation1,2. This type of inheritance was discovered as the cause of the unusual inheritance patterns of a devastating neurodegenerative disease. However, studies in organisms ranging from fungi to mammals3,4,5,6,7,8,9,10 have since revealed that prion-like elements can confer adaptive value. Nonetheless, prions have been viewed as a fascinating but rare biological oddity.
This prevailing wisdom is in part held because the characterization of protein-based inheritance has long been restricted by a small set of examples. Recent systematic screening efforts have widened this picture significantly by identifying several new bona fide prions11 and almost two dozen protein domains12 with the capacity to fuel prion-like conformational conversion. However, because these approaches have generally focused on strong amino acid sequence biases, the prions that have been discovered share the biochemical properties of the founding yeast prions [PSI+]13,14, [URE3]15, and [RNQ+]11,16. These include: 1) modular domains that are rich in long polymeric stretches of asparagine (N) and glutamine (Q), 2) assembly into an amyloid [PRION+] conformation17,18,19, and 3) complete reliance on disaggregase Hsp104 function for faithful propagation from mother to daughter13,20,21. Indeed, many bona fide prions, including [GAR+], [Het-s], and even the original prion (PrPSc), would be missed under such stringent criteria. Perhaps more importantly, they would be unable to capture any novel mechanisms of protein-based inheritance22. Thus, the true biological breadth of such phenomena may be far more common in nature than previously assumed.
To investigate this question, a high-throughput, proteome-wide strategy was employed. A hallmark of all prions, including PrPSc, [GAR+], and [Het-s], is that the transient overexpression of the causal proteins strongly increases the rate of prion acquisition15,23,24,25,26. We took advantage of this feature to systematically ask, across the entire yeast ORFeome, if stable protein-based, epigenetic states could be initiated by transiently inducing the overexpression of individual proteins. It is well known that protein overexpression can alter phenotypes27. However, prion proteins are unusual because their temporary overproduction produces a change in phenotype that is heritable for many hundreds of generations after the initial overexpression. We previously took advantage of this feature, as well as the unusual inheritance patterns of protein-based genetic elements, to identify dozens of proteins that are capable of heritably re-wiring phenotypic landscapes without altering the genome28. Although some identified proteins were previously known as prions, most were not, underscoring the power of this approach to uncover new forms of protein-based inheritance.
Protocol
1. Initial Overexpression
Transform the yeast cells (in this case, BY4741 MAT a haploids) previously grown in YPD liquid (10 g of yeast extract, 20 g of peptone, and 20 g of glucose per 1 L) with the desired candidate constructs from the FLEXGene ORFeome library (yeast ORFs under the control of a galactose-inducible promoter in the URA3-marked centromericplasmid backbone, pBY01129).
Use autoclaved toothpicks to pick four separate colonies from these transformations to serve as biological replicates. Grow them for 48-72 h in 150 µL of SRaffinose-URA (0.74 g of CSM-URA, 6.7 g of yeast nitrogen base without amino acids, and 20 g of raffinose per 1 L) in 96-well plates. Inoculate colonies from the first ORF in well A1 of all plates, colonies from the second ORF in A2 of all plates, etc. NOTE: All growth steps are conducted at 30 °C and at atmospheric levels of CO2 (407.05 ppm), unless stated otherwise.
Confirm that the cultures are saturated (cells visible by eye at the bottom of each well) and use a liquid handling robot to inoculate a 1:4 array of 1 µL from each 96-well plate well into 4 separate wells of a 384-well plate filled with 45 µL of SGal-URA (0.74 g of CSM-URA, 6.7 g of yeast nitrogen base without amino acids, and 20 g of galactose per 1 L) per well. NOTE: This creates a composite plate in which 4 biological replicates of each ORF are arrayed in a square pattern.
At the same time, prepare separate 384-well plates containing 45 µL of SGal-URA with the stressors of interest (e.g., manganese chloride at 20 mM) per well. Inoculate this plate in the same manner as in step 1.3. NOTE: To achieve the greatest dynamic range for phenotypic detection, a 2-fold dilution series around the LD50 concentration of a given stressor is recommended. This can be done beforehand to determine the optimum concentration at which to observe both enhanced and reduced growth.
As a final parallel control, inoculate a third 384-well plate in the same manner, with the same stressor included in medium that will not induce plasmid expression. Ensure that the plate contains 45 µL of SD-URA (0.74 g of CSM-URA, 6.7 g of yeast nitrogen base without amino acids, and 20 g of glucose per 1 L) per well and is inoculated as in steps 1.3 and 1.4.
Immediately place plates with cells on a microplate stacker and set the protocol for a 72-h continuous loop, measuring the OD600 with a microplate reader equipped with stackers at room temperature and atmospheric CO2 (407.05 ppm). Note: The frequency of the growth measurement will depend on the number of plates (ultimately determined by the number of genes and conditions the investigator wants to probe) used in the run. The more plates used, the longer it will take the plate reader to complete a loop and thus the longer time between measurements (measurement time per plate is ~45 s, depending on the instrumentation employed).
- After the growth measurements, use the same liquid handling robot to transfer 1 µL per well of the SGal-URA-induced cultures (i.e., those that experienced protein overexpression) to new 384-well plates containing 45 µL of SD-URA per well (medium that does not permit protein expression of the plasmid).
- In parallel, perform analogous inoculations of a second set of 384-well plates containing 45 µL of SD-URA per well from the cultures that were grown in SD-URA in the presence of the stressors (i.e., those from step 1.5).
- Grow the plates for 48 h at 30 °C to saturation in a humidified chamber (i.e., a sealable plastic bin with a damp paper towel inside).
Take the plates from the previous step and reinoculate 1 µL per well in 384-well plates containing 45 µL of SD-URA. Then, perform a separate reinoculation of 1 µL per well from the same source plate into a separate 384-well plate containing 45 µL of SD-URA with the stressor. NOTE: This will determine if the sensitivity/resistance phenotypes to a stressor persist after the protein overexpression has ceased.
Immediately place plates with cells on a microplate stacker and set the protocol for a 48-h continuous loop measuring the OD600 with a microplate reader at room temperature and atmospheric CO2 (407.05 ppm).
- Export the time versus OD600 measurements as an XY table using the plate reader software. Group columns of the OD600 for each biological replicate together and calculate the mean. Create an XY plot of time versus OD600 to generate growth curves.
- Compare the growth rates of cultures that had been subjected to a past overexpression (i.e., those originally induced in galactose-containing medium before reinoculation in SD-URA) versus cultures that had not (i.e., those propagated in glucose), as described previously30.
- For cultures that show significant growth differences in response to a given stressor, dependent upon ancestral protein overexpression, take 1 µL from each biological replicate, dilute it in 10 mL of water, and then plate 50 µL on plates containing 5-FOA (0.74 g of CSM-URA, 1 g of 5-FOA, 50 mg of uracil, 6.7 g of yeast nitrogen base without amino acids, 20 g of glucose, and 20 g of agar per liter). Grow at 30 °C for 3 days.
- If this results in too many or too few colonies per plate, adjust the dilution factor accordingly. Note: 5-FOA is converted into a toxic intermediate by the uracil biosynthesis pathway. This will cause a loss of the URA3-marked expression plasmid transformed in the first step.
Pick 8-32 single colonies with autoclaved toothpicks and pin them to 96-well plates containing 150 µL of SD-CSM. Grow to saturation for 48-72 h in a humidified chamber at 30 °C. Use these cultures to inoculate two new sets of 96-well plates containing 150 µL of SD-CSM, both with and without the stressors from step 1.11.
Place the plates on a microplate stacker and set the protocol to measure the OD600 on a 48-h continuous loop.
Analyze the data as in step 1.10 and confirm that the significant growth differences seen in step 1.10 are maintained after plasmid loss. NOTE: These cells harbor stable phenotypic states that might be indicative of protein-based inheritance.
Test the cells that maintained the induced phenotypes for classic hallmarks of protein-based inheritance (see below).
2. Tests for Prion-like Inheritance
- Chaperone-mediated "curing"
- Test Hsp104 chaperone dependence.
- Pick a colony of a yeast strain harboring a stable phenotypic state, streak to single colonies on a YPD plate (10 g of yeast extract, 20 g of peptone, 20 g of glucose, and 20 g of agar per 1 L) containing 3 mM GdnHCl and grow at 30 °C for 3 days. In parallel, streak out a colony from a naïve strain to single colonies on YPD+ 3 mM GdnHCl as a control in the same manner.
- Repeat 2 more times on fresh YPD plates containing 3 mM GdnHCl.
- Because GdnHCl exposure can increase the frequency at which petite cells arise, pick multiple colonies that have undergone 3 passages on GdnHCl and check for functional mitochondrial respiration by examining their ability to grow into visible colonies on YP-Glycerol plates (10 g of yeast extract, 20 g of peptone, 20 mL of glycerol, and 20 g of agar per 1 L) after 7 days.
- Pick multiple colonies from the YP-Glycerol plates and test for the maintenance of stable phenotypic states using growth in the presence of the stressor compared to non-"cured" and naïve (i.e., isogenic cells that do not harbor the phenotypic state, such as BY4741 MATa haploids) controls in a similar manner as described in step 1.13.
- Test Hsp70 chaperone dependence.
- "Cure" via a plasmid.
- Pick single transformants for each, streak to single colonies on an SD-URA plate (0.74 g of CSM-URA, 6.7 g of yeast nitrogen base without amino acids, 20 g of glucose, and 20 g of agar per 1 L), and grow for 3 days at 30 °C and atmospheric CO2.
- Repeat this passaging 2 more times on SD-URA plates.
- Pick multiple colonies and streak to single colonies on 5-FOA plates to eliminate the plasmid.
- Pick multiple single colonies for each isolate and test for retention (or loss) of the stable phenotypic states in a similar manner as described in step 1.13.
- "Cure" via genetic crossing.
- Cross BY4741 MATa haploid strains harboring stable phenotypic states and naïve strains of the opposite mating type (in this case, BY4742 MATα) harboring genetic deletions in two of the four yeast Hsp70 paralogs (ssa1Δ ssa2Δ). NOTE: This strain is deficient in Hsp70 chaperone function.
- Select for the growth of diploids by streaking to single colonies on SD-LYS-MET (0.74 g of CSM-LYS-MET, 6.7 g of yeast nitrogen base without amino acids, 20 g of glucose, and 20 g of agar per 1 L) double-dropout medium.
- Pick single diploid colonies and grow for 24 h at 30 °C in 8 mL of pre-sporulation medium (0.8% yeast extract, 0.3% peptone, 10% dextrose, and 100 mg/L adenine sulfate).
- Spin down the cultures for 3 min at 3,000 xg, aspirate the pre-sporulation (pre-SPO) medium, and wash the pellets once with sterile water.
- Re-suspend the pellets in 2 mL of sporulation medium (1% potassium acetate, 0.1% yeast extract, 0.05% glucose, and 0.01% amino acid add-back mix (2 g of histidine, 10 g of leucine, 2 g of lysine, and 2 g of uracil)). Grow at 25°C for 5 days.
- Move the cultures to 30 °C and incubate for an additional 48 h.
- Assess the efficiency of sporulation by looking for the presence of tetrads using a light microscope32.
- Take 40 µL of culture, spin down for 1 min at 3,000 xg in a 1.5-mL tube, and re-suspend in an equal volume of zymolyase enzyme mix (1 M sorbitol, 0.1 M EDTA, and 10 mg/mL zymolyase 100T)
- Incubate at 25 °C for 5 min.
- Apply 10 µL of digested culture to a very dry, very level YPD agar plate and allow the culture to slowly spread down the plate in a straight line.
- Allow the plates to dry for at least 30 min in a laminar flow hood.
- Using a dissection microscope, separate the spores into tetrads and array them on the plate.
- Allow the individual haploid spores to grow into colonies and test for their ability to grow on SD-HIS-LEU (0.67 g of CSM-HIS-LEU, 6.7 g of yeast nitrogen base without amino acids, 20 g of glucose, and 20 g of agar per 1L) plates, indicating that they harbor both genetic deletions (ssa1Δ ssa2Δ).
- Test each for the maintenance of the phenotypic state in the Hsp70-deficient background in a similar manner as described in step 1.13. NOTE: Crossing these spores back to a wildtype strain with restored Hsp70 function should not restore the prion phenotype.
- Testing for non-Mendelian inheritance.
- Cross BY4741 MATa haploid strains harboring stable phenotypic states and isogenic naïve controls to an isogenic naïve strain of the opposite mating type (in this case, BY4742 MATα).
- Select for the growth of diploids by streaking to single colonies on SD-LYS-MET double-dropout medium.
- Pick single diploid colonies and grow for 24 h at 30 °C in 8 mL of pre-sporulation medium.
- Repeat steps 2.2.2.3-2.2.2.12, as described above.
- Allow the individual haploid spores to grow into colonies and test each for maintenance of the phenotypic state through meiosis using the growth assay, as in step 1.13. Note: Another defining feature of prions is that they can be heritably eliminated through the removal of the original casual protein. Thus, analogously crossing a prion strain with a strain harboring a genetic deletion of the original induced protein will abrogate prion phenotypes in the spores that inherit the deletion. Also note that is possible for the prion to be maintained in a "cryptic" state in such a mutant if the deleted gene is required to manifest the prion phenotype but not to propagate the prion itself. To distinguish between these two possibilities, cross the spores back to a naïve wildtype strain and test whether the prion phenotype re-emerges in a diploid genetic background, where the function of the gene has been restored.
- Cytoduction.
- Generate the initial BY4742 recipient strain.
- Through transformation, introduce a defective KAR allele (kar1-15) that prevents nuclear fusion during mating23.
- Make this strain "petite" (i.e., incompetent for mitochondrial respiration) by inoculating a single colony in YPD broth with 0.25% ethidium bromide.
- Grow the culture at 30 °C until late exponential/stationary phase (OD600 ~1).
- Dilute 1:1,000 in fresh YPD with 0.25% ethidium bromide and repeat twice.
- Once the culture reaches late exponential/early stationary phase (OD600 0.8-1.2), plate to single colonies by diluting 1:10,000 in sterile water and plating 50 µL on a YPD plate. After growing for 3 days at 30 °C, pick multiple colonies and test each for respiration incompetence by examining their ability to grow into visible colonies on YP-Glycerol plates after 7 days.
- Perform initial cytoduction into BY4742.
- Mix cells of the donor BY4741 strain harboring a stable phenotypic state with cells of the naïve BY4742 kar1-15 recipient strain on the surface of a YPD agar plate.
- Grow for 24 h at 30 °C and atmospheric CO2 (407.05 ppm) and transfer to methionine dropout medium containing glycerol (SGly-MET: 0.74 g of CSM-MET, 6.7 g of yeast nitrogen base without amino acids, 20 mL of glycerol, and 20 g of agar per 1 L). NOTE: This selects for both BY4742 nuclear markers, along with allowing the restoration of functional mitochondria via cytoplasmic exchange.
- After 3-5 days, pick multiple single colonies and perform another round of selection on SD-MET (0.74 g of CSM-MET, 6.7 g of yeast nitrogen base without amino acids, 20 g of glucose, and 20 g of agar per 1 L).
- In parallel, confirm that the colonies are not diploids by passaging on SD-LYS-MET medium.
- Perform reverse cytoductions.
- Mix the new donor strains (this time, the successful BY4742 kar1-15 cytoductants) with petite naïve BY4741 cells (generated as above) on YPD agar.
- Repeat cytoductions, as described previously (steps 2.3.2.1-2.3.2.4), except selecting for BY4741 recipient nuclear markers on glycerol medium lacking lysine (SGly-LYS: 0.74 g of CSM-LYS, 6.7 g of yeast nitrogen base without amino acids, 20 mL of glycerol, and 20 g of agar per 1 L).
- Pick multiple cytoductants and test for the maintenance of stable phenotypic states using the growth assay, as before in step 1.13.
- Protein transformation.
- Prepare the lysate.
- Grow 50 mL of cells harboring stable phenotypic states in YPD for 18 h at 30 °C.
- Pellet the cultures at 3,000 x g for 4 min and then wash twice: once in autoclaved H2O and then in 1 M sorbitol.
- Re-suspend the cells in 200 mL of SCE buffer (1 M sorbitol, 10 mM EDTA, 10 mM DTT, 100 mM sodium citrate, and 1 mini-EDTA-free protease inhibitor tablet per 50 mL, pH 5.8) with 50 U/mL of yeast lytic enzyme 100T.
- Incubate the mix at 35 °C for 30 min.
- Sonicate the cells at 20 kHz and 20% intensity for 10 s on ice with a sonic dismembrator.
- Remove the cell debris by centrifugation at 10,000 x g for 15 min at 4 °C.
- Move the supernatants to a clean tube and add RNase I and biotinylated DNase at a 3-fold excess (as determined by units of activity) and incubate at 37 °C for 1 h.
- Remove the DNase I by adding excess streptavidin-agarose beads, incubating for 5 min, and pelleting by centrifugation at 10,000 x g for 1.5 min.
- Transfer the supernatant to a fresh tube.
- Prepare the recipient spheroplast.
- Grow 5 mL of naïve recipient cells in YPD to mid-exponential phase (OD600 ranging from 0.5-1).
- Harvest by centrifugation (3,000 x g for 4 min) and wash 4 times: twice in H2O and twice in 1 M sorbitol.
- Re-suspend the cells in 1 M sorbitol containing 200 U/mL yeast lytic enzyme and incubate for 15 min at 35 °C to digest the cell walls.
- Harvest the resulting spheroplasts with gentle centrifugation at 600 x g for 5 min and wash with 1 mL of 1 M sorbitol and then again with 1 mL of STC buffer (1 M sorbitol, 10 mM Tris pH 7.5, and 10 mM CaCl2).
- Resuspend the washed spheroplasts in 50 µL of STC buffer. NOTE: Use scissors to cut off the last ~0.7 cm of a pipette tip; this will create a wider mouth and prevent the lysis of the spheroplasts.
- Transform the naïve cells with protein.
- Incubate the mix at 25 °C for 30 min.
- Centrifuge for 5 min at 600 x g to collect the spheroplasts and re-suspend in 150 µL of SOS-buffer (1 M sorbitol, 7 mM CaCl2, 0.25% yeast extract, and 0.5% peptone).
- Recover for 30 min at 30 °C.
- Plate the entire culture on SD-URA plates.
- Quickly overlay the plated culture with warm (~45 °C) SD-URA containing 0.8% agar. Note: This will prevent the spheroplasts from bursting
- Incubate the transformation plates for 2-3 days at 30 °C.
- Using a toothpick, pick successfully growing cells from the overlaid plate and re-streak to single colonies on SD-URA plates.
- Pick dozens of colonies for each and streak on 5-FOA plates to eliminate URA3-marked carrier plasmid.
- Pick colonies that grow on the 5-FOA plates and test for the transfer of stable phenotypic states using the growth assay, as before (step 1.13).
Representative Results
Protein overexpression is known to dramatically alter cellular phenotypes27. Indeed, with an initial screening approach, hundreds of new phenotypes were reproducibly recovered from the overexpression of clones from the yeast ORFeome using just ten stressors. However, the assays described above allow for the assessment of whether cells retain any long-term stable phenotypes following this overexpression. One protein capable of encoding such a state is Psp1. Psp1 is a protein of unknown function that can suppress mutants in the replication machinery (e.g., in pol alpha34). Overexpression of the PSP1 ORF modulated cellular phenotypes in a variety of stressors. Strikingly, one phenotype (resistance to manganese chloride) was maintained for hundreds of generations in cells long after overexpression had ceased (i.e., progeny who had never experienced Psp1 induction directly, but whose ancestors had) (Figure 1A). The half-life of Psp1 is ~5 h35, making it extremely unlikely that the resistance phenotype is due to the propagation of long-lived, stable Psp1 protein from mothers to their daughters over this timescale (as the original protein would be degraded within a handful of generations).
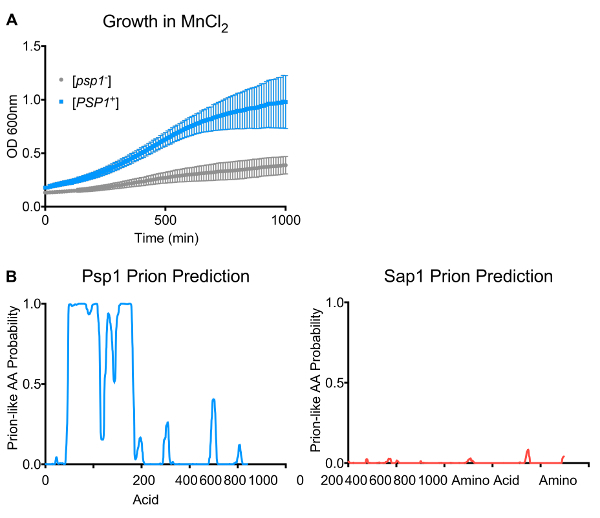
Because the MnCl2 resistance phenotype arose at a high frequency and in multiple independent transient overexpression experiments, it is extremely unlikely that this phenotype was due to de novo mutation. A possible explanation for the heritable nature of this phenotype is the induction of a self-templating element (i.e., a prion). Indeed, the Psp1 amino acid content harbors some small stretches of asparagine and glutamine, reminiscent of canonical prions such as [PSI+], which are driven by proteins that contain long stretches of these amino acids. Predictive algorithms36 score the N-terminus of Psp1 as moderately "prion-like" (Figure 1B, Rank 173 in the yeast proteome). However, unlike canonical prion proteins, Psp1 protein from cells harboring the stable phenotypic state did not form amyloid fibers, as judged by semi-denaturing agarose gel electrophoresis28,37. To determine whether Psp1 formed a bona fide prion, we tested whether the pattern of inheritance of the Psp1-induced phenotypic state was consistent with hallmarks of prion biology: reliance on protein homeostasis machinery for faithful propagation and non-Mendelian inheritance in meiosis. Finally, a "gold standard" test of transmission was performed using only assembled protein.
Unlike mutations or other forms of epigenetic inheritance, prions are uniquely reliant on the function of molecular chaperones and other arms of the protein homeostasis network that regulate protein folding in the cell. Most canonical prions require the Hsp104 disaggregase to act on the amyloid fibers that they adopt. This process generates heritable "seeds" that transmit the prion conformation from mothers to daughters13. One notable exception to this requirement is the [ISP+] prion, which is curable via Hsp104 inhibition but does not require its disaggregase activity for propagation38. Furthermore, we and others have recently reported many examples of prions that are Hsp104-independent and are instead regulated by other molecular chaperones (primarily Hsp70 and, in rare cases, Hsp90)25,28. We first tested whether the inheritance of the Psp1-induced MnCl2 resistance depended on Hsp104. Cells harboring this state were passaged 3 times on rich medium (YPD) supplemented with a low dose of guanidine hydrochloride (an Hsp104 inhibitor). We then returned the cells to standard rich medium to restore full Hsp104 function. This regimen did not affect the heritability of the Psp1-induced MnCl2 resistance, establishing that the cellular state was Hsp104-independent (Figure 2).
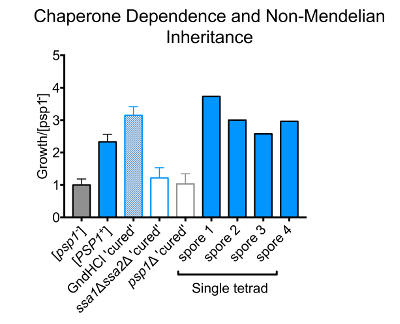
Next, a crossing approach was employed to test whether the Psp1-induced phenotypic state depended on Hsp70. The MnCl2 resistant cells were mated to an Hsp70-deficient strain, with genetic deletions in two of the four yeast Hsp70 paralogs (ssa1Δ ssa2Δ). Diploids were selected and then sporulated to generate haploid spores. We tested for the maintenance of the Psp1-dependent state and found that the Psp1-induced MnCl2 resistance phenotype had been completely eliminated in spores harboring the ssa1Δ ssa2Δ double deletion (Figure 2). Thus, the Psp1-induced phenotypic state requires the activity of Hsp70 to pass from one generation to the next.
Another defining characteristic of prions is a non-Mendelian pattern of inheritance. Traits encoded by mutations in DNA segregate 2:2 in meiotic crosses: half of the progeny inherit one parental allele and other half inherit the other parental allele. The inheritance of prions, in contrast, does not depend upon changes in DNA, but rather transmissible changes in protein conformation. Thus, they are inherited by most, if not all, meiotic progeny in genetic crosses (3:1 or 4:0 inheritance)39. We tested the inheritance pattern of the Psp1-induced MnCl2 resistance by crossing strains harboring the state to naïve controls of the opposite mating type. The resulting diploids were sporulated and tested for the presence of the Psp1 MnCl2 resistance phenotype within spores derived from the same tetrads. All spores from 2 separate tetrads in which one parent harbored the Psp1-dependent epigenetic state inherited the corresponding MnCl2 resistance phenotype (Figure 2). In contrast, no cells from naïve control crosses displayed the phenotype. These data establish that the Psp1-induced phenotypic state is not encoded by a DNA-based mutation. However, it is important to note that while the Psp1 state is not encoded in DNA, the state does require the continual presence of the PSP1 gene. Crossing the Psp1-dependent phenotypic state into a genetic background lacking the PSP1 gene (psp1Δ) heritably eliminated the MnCl2 resistance phenotype (Figure 2). Thus, overexpression of Psp1 does not simply orchestrate the formation of a highly stable feedback loop that functions autonomously of Psp1 once initiated.
The very definition of protein-based inheritance is the transmission of stable traits to progeny using only assembled protein as the heritable material. To carry out such "protein transformations," a method that is agnostic with respect to protein stoichiometry, modification, or interaction was used40,41. Separate cultures of cells harboring the Psp1-induced phenotypic state and naïve controls were grown to mid-exponential phase, and their cell walls were lysed to generate donor spheroplasts. These spheroplasts were then sonicated and spun down to pellet the resulting cell debris. Finally, the remaining lysates were subjected to over-digestion with both RNase and DNase to eradicate nucleic acid (to aid steps later in the protocol, the biotinylated DNase enzyme was subsequently removed via affinity purification).
These nucleic acid-depleted lysates were used to transform naïve cells. To aid in the uptake of heritable protein "seeds," the cell walls of the naïve recipients were enzymatically digested prior to transformation. A URA3-marked carrier plasmid with the protein donor lysates was also included to enable the selection of cells that were competent for the uptake of foreign material. The transformants were plated on medium lacking uracil, resistant colonies were selected, and the URA3-marked carrier plasmid was removed by growth on 5-FOA. The resistance of the resulting colonies to MnCl2 was then examined. Strikingly, over half of the transformants tested harbored heritable MnCl2 resistance (Figure 3). In contrast, no cells transformed with naïve lysates displayed any such phenotype. Importantly, these efficiencies were gleaned from cellular lysates expressing endogenous levels of Psp1 protein (~340 molecules per cell). This is remarkably more efficient than the transformation of other yeast prions, such as [PSI+]. These results establish that Psp1 is capable of forming a protein-based heritable element-a "prion"-henceforth known as [PSP1+]. Furthermore, the protein transformation technique described here is sufficient for screening for non-amyloid, Hsp104-independent, protein-based inheritance in a high-throughput manner.
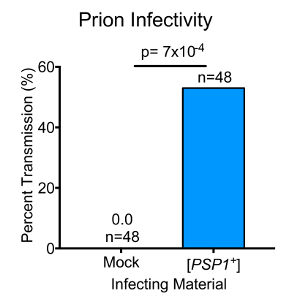
Figure 1. Transient overexpression of PSP1 drives heritable MnCl2 resistance. (A) Growth of uninduced ([psp1-]) and Psp1-induced ([PSP1+]) strains in SD-CSM with 10 mM MnCl2. The error bars represent the standard error of the mean from three biological replicates. (B) Left: The Psp1 primary amino acid sequence scores as highly prion-like in prion-predicting algorithms (PLAAC). Right: Representative PLAAC analysis for most inducing proteins recovered in our screen predicted virtually no significant prion-like sequence features. Please click here to view a larger version of this figure.
Figure 2. Psp1 chaperone dependence and pattern of inheritance. The growth of strains in SD-CSM with 10 mM MnCl2, normalized to a corresponding naïve [psp1-] control. The inhibition of Hsp104 (GdnHCl "cured") does not impair Psp1-dependent MnCl2 resistance, whereas the removal of the Hsp70 chaperone (ssa1Δ ssa2Δ) and the PSP1 gene (psp1Δ) heritably eliminates this phenotype. All spores from single tetrads of crosses between strains harboring the Psp1-dependent phenotypic state and naïve strains display MnCl2 resistance, indicating non-Mendelian (4:0) inheritance of the phenotype (n=2 tetrads). The error bars represent the standard error of the mean from three biological replicates. Please click here to view a larger version of this figure.
Figure 3. Transmission of Psp1-dependent phenotypic state via heritable protein "seeds." Rates of infectivity for protein transformation comparing mock ([psp1-] lysates) and [PSP1+]- harboring lysates as the transmissible material. Infectivity is calculated as the percent of transmission divided by the amount of seeded protein used. Over 53% of naïve cells transformed with seeded [PSP1+] received the corresponding MnCl2 resistance phenotype, whereas no cells transformed with [psp1-] displayed MnCl2 resistance (p=7.4 x 10-4 by Student's t-test). The infectivity rate of the previously characterized yeast prion [PSI+] is displayed via the dotted horizontal line40. Please click here to view a larger version of this figure.
Discussion
The first yeast prions were identified by their unusual phenotypes and perplexing patterns of inheritance. The characteristics of these prions were then used to build algorithms and computational tools to screen for additional prion proteins. The method described here, in contrast, is experimental and relies on transient overexpression to create a lasting change-a stable state-encoded in protein conformation. However, if the efficiency of "seeding" prion assembly by overexpression for any given protein is very low, that protein will continually emerge as a false negative in overexpression screens of this type. One such modification to correct this would be to use a 2-micron plasmid for protein overexpression in future experiments. Finally, each induced prion has its own unique set of growth phenotypes and will not be apparent in every condition assayed. Thus, the number of different conditions and doses tested limits the number of hits.
Importantly, not all types of protein-based inheritance will be equally recovered using this method. Proteins that cannot be overexpressed efficiently or without toxicity will obviously be continually missed. Mitotically unstable elements, such as "mnemons," would never propagate to daughters following the initial overexpression22. In contrast, other types of long-lived bistable switches could theoretically be induced via transient overexpression42,43. However, these states are typically not dependent on protein homeostatic machinery or transmissible via "seeded" proteins. In addition, prions that rely on other chaperones (outside of Hsp70 and Hsp104) or additional arms of the protein homeostasis network for propagation would fail the chaperone-dependency assays described here. Finally, a low abundance proteins that also form amyloid might have infectivity rates below the limit of detection in the protein transformation setup.
This protocol describes a technique for inducing stable protein-based epigenetic states via protein overexpression, as well as further downstream steps to validate whether each induced epigenetic state is a bona fide prion. The example presented in this paper, Psp1, is an example of a protein that displays a "prion-like" amino acid bias and could theoretically be recovered using previously developed bioinformatic algorithms. However, the inability of Psp1 to form amyloid and its unusual chaperone dependence (Hsp104) would have quickly disqualified it from further analyses and thus eliminated it from prion consideration. However, the screening techniques presented in this paper are agnostic to these assumptions and instead focus on the underlying patterns of inheritance and the sufficiency of protein alone to transmit the corresponding phenotypes. Indeed, the vast majority of protein-based inheritance recovered with this method was devoid of N/Q-rich sequence bias.
This method was used to probe the entire yeast ORFeome for its capacity to elicit protein-based inheritance in an unbiased fashion using only a small number of stressors (25 mM cadmium chloride, 1 mM cobalt chloride, 2 mM copper sulfate, 1 mM diamide, 0.2 mM fluconazole, 50 mM hydroxyurea, 20 mM manganese chloride, 0.75 mM paraquat, 50 mM radicicol, 80 J/m2 UV-irradiation, and 10 mM zinc sulfate). However, this approach could easily be modified to screen genetic networks or specific cellular responses in a more targeted manner. For example, functionally related proteins or all proteins regulated in a discrete signaling network could be induced via transient overexpression and screened with stressors related to their biological function. In contrast, a larger set of proteins could be screened with a more comprehensive set of stressors to investigate whether specific cellular responses have naturally evolved to harbor prion switches. Finally, although we have conducted these studies in yeast, many aspects of the experiments (e.g., transient protein expression, chaperone "curing," etc.) could be generalized to other model systems in the future. For example, mammalian tissue culture is amenable to overexpression via plasmid-based systems, and fluorescence foci could be also be used as a readout for heritable self-assembly, as described previously28. Furthermore, protein-coding sequences from other organisms could be expressed in yeast and tested for their ability to elicit prion-like inheritance using the methods described here.
Disclosures
The authors have nothing to disclose.
Acknowledgments
We thank Sohini Chakrabortee, Sandra Jones, David Garcia, Bhupinder Bhullar, Amelia Chang, Richard She, and Susan Lindquist for their assistance in developing the assays used in this paper, as well as the reviewers for their thoughtful comments.
References
- Halfmann R, Alberti S, Lindquist S. Prions, protein homeostasis, and phenotypic diversity. Trends Cell Biol. 2010;20(3):125–133. doi: 10.1016/j.tcb.2009.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byers JS, Jarosz DF. Pernicious pathogens or expedient elements of inheritance: the significance of yeast prions. PLoS Pathog. 2014;10(4):1003992. doi: 10.1371/journal.ppat.1003992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarosz DF, et al. Cross-kingdom chemical communication drives a heritable, mutually beneficial prion-based transformation of metabolism. Cell. 2014;158(5):1083–1093. doi: 10.1016/j.cell.2014.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- True HL, Lindquist SL. A yeast prion provides a mechanism for genetic variation and phenotypic diversity. Nature. 2000;407(6803):477–483. doi: 10.1038/35035005. [DOI] [PubMed] [Google Scholar]
- Suzuki G, Shimazu N, Tanaka M. A yeast prion, Mod5, promotes acquired drug resistance and cell survival under environmental stress. Science. 2012;336(6079):355–359. doi: 10.1126/science.1219491. [DOI] [PubMed] [Google Scholar]
- Hou F, et al. MAVS forms functional prion-like aggregates to activate and propagate antiviral innate immune response. Cell. 2011;146(3):448–461. doi: 10.1016/j.cell.2011.06.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai X, et al. Prion-like polymerization underlies signal transduction in antiviral immune defense and inflammasome activation. Cell. 2014;156(6):1207–1222. doi: 10.1016/j.cell.2014.01.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majumdar A, et al. Critical role of amyloid-like oligomers of Drosophila Orb2 in the persistence of memory. Cell. 2012;148(3):515–529. doi: 10.1016/j.cell.2012.01.004. [DOI] [PubMed] [Google Scholar]
- Khan MR, et al. Amyloidogenic Oligomerization Transforms Drosophila Orb2 from a Translation Repressor to an Activator. Cell. 2015;163(6):1468–1483. doi: 10.1016/j.cell.2015.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fioriti L, et al. The Persistence of Hippocampal-Based Memory Requires Protein Synthesis Mediated by the Prion-like Protein CPEB3. Neuron. 2015;86(6):1433–1448. doi: 10.1016/j.neuron.2015.05.021. [DOI] [PubMed] [Google Scholar]
- Derkatch IL, Bradley ME, Hong JY, Liebman SW. Prions affect the appearance of other prions: the story of [PIN(+)] Cell. 2001;106(2):171–182. doi: 10.1016/s0092-8674(01)00427-5. [DOI] [PubMed] [Google Scholar]
- Alberti S, Halfmann R, King O, Kapila A, Lindquist S. A systematic survey identifies prions and illuminates sequence features of prionogenic proteins. Cell. 2009;137(1):146–158. doi: 10.1016/j.cell.2009.02.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chernoff YO, Lindquist SL, Ono B, Inge-Vechtomov SG, Liebman SW. Role of the chaperone protein Hsp104 in propagation of the yeast prion-like factor [psi+] Science. 1995;268(5212):880–884. doi: 10.1126/science.7754373. [DOI] [PubMed] [Google Scholar]
- Patino MM, Liu JJ, Glover JR, Lindquist S. Support for the prion hypothesis for inheritance of a phenotypic trait in yeast. Science. 1996;273(5275):622–626. doi: 10.1126/science.273.5275.622. [DOI] [PubMed] [Google Scholar]
- Wickner RB. [URE3] as an altered URE2 protein: evidence for a prion analog in Saccharomyces cerevisiae. Science. 1994;264(5158):566–569. doi: 10.1126/science.7909170. [DOI] [PubMed] [Google Scholar]
- Sondheimer N, Lindquist S. Rnq1: an epigenetic modifier of protein function in yeast. Mol Cell. 2000;5(1):163–172. doi: 10.1016/s1097-2765(00)80412-8. [DOI] [PubMed] [Google Scholar]
- Balbirnie M, Grothe R, Eisenberg DS. An amyloid-forming peptide from the yeast prion Sup35 reveals a dehydrated beta-sheet structure for amyloid. Proc Natl Acad Sci U S A. 2001;98(5):2375–2380. doi: 10.1073/pnas.041617698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glover JR, et al. Self-seeded fibers formed by Sup35, the protein determinant of [PSI+], a heritable prion-like factor of S. cerevisiae. Cell. 1997;89(5):811–819. doi: 10.1016/s0092-8674(00)80264-0. [DOI] [PubMed] [Google Scholar]
- King CY, et al. Prion-inducing domain 2-114 of yeast Sup35 protein transforms in vitro into amyloid-like filaments. Proc Natl Acad Sci U S A. 1997;94(13):6618–6622. doi: 10.1073/pnas.94.13.6618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shorter J, Lindquist S. Hsp104 catalyzes formation and elimination of self-replicating Sup35 prion conformers. Science. 2004;304(5678):1793–1797. doi: 10.1126/science.1098007. [DOI] [PubMed] [Google Scholar]
- Ferreira PC, Ness F, Edwards SR, Cox BS, Tuite MF. The elimination of the yeast [PSI+] prion by guanidine hydrochloride is the result of Hsp104 inactivation. Mol Microbiol. 2001;40(6):1357–1369. doi: 10.1046/j.1365-2958.2001.02478.x. [DOI] [PubMed] [Google Scholar]
- Caudron F, Barral Y. A super-assembly of Whi3 encodes memory of deceptive encounters by single cells during yeast courtship. Cell. 2013;155(6):1244–1257. doi: 10.1016/j.cell.2013.10.046. [DOI] [PubMed] [Google Scholar]
- Wickner RB, Edskes HK, Shewmaker F. How to find a prion: [URE3], [PSI+] and [beta] Methods. 2006;39(1):3–8. doi: 10.1016/j.ymeth.2006.04.009. [DOI] [PubMed] [Google Scholar]
- Chernoff YO, Derkach IL, Inge-Vechtomov SG. Multicopy SUP35 gene induces de-novo appearance of psi-like factors in the yeast Saccharomyces cerevisiae. Curr Genet. 1993;24(3):268–270. doi: 10.1007/BF00351802. [DOI] [PubMed] [Google Scholar]
- Brown JC, Lindquist S. A heritable switch in carbon source utilization driven by an unusual yeast prion. Genes Dev. 2009;23(19):2320–2332. doi: 10.1101/gad.1839109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coustou V, Deleu C, Saupe S, Begueret J. The protein product of the het-s heterokaryon incompatibility gene of the fungus Podospora anserina behaves as a prion analog. Proc Natl Acad Sci U S A. 1997;94(18):9773–9778. doi: 10.1073/pnas.94.18.9773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sopko R, et al. Mapping pathways and phenotypes by systematic gene overexpression. Mol Cell. 2006;21(3):319–330. doi: 10.1016/j.molcel.2005.12.011. [DOI] [PubMed] [Google Scholar]
- Chakrabortee S, et al. Intrinsically Disordered Proteins Drive Emergence and Inheritance of Biological Traits. Cell. 2016;167(2):369–381. doi: 10.1016/j.cell.2016.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Y, et al. Approaching a complete repository of sequence-verified protein-encoding clones for Saccharomyces cerevisiae. Genome Res. 2007;17(4):536–543. doi: 10.1101/gr.6037607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swain PS, et al. Inferring time derivatives including cell growth rates using Gaussian processes. Nat Commun. 2016;7:13766. doi: 10.1038/ncomms13766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarosz DF, Lancaster AK, Brown JC, Lindquist S. An evolutionarily conserved prion-like element converts wild fungi from metabolic specialists to generalists. Cell. 2014;158(5):1072–1082. doi: 10.1016/j.cell.2014.07.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neiman AM. Sporulation in the budding yeast Saccharomyces cerevisiae. Genetics. 2011;189(3):737–765. doi: 10.1534/genetics.111.127126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gietz D, St Jean A, Woods RA, Schiestl RH. Improved method for high efficiency transformation of intact yeast cells. Nucleic Acids Res. 1992;20(6):1425. doi: 10.1093/nar/20.6.1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Formosa T, Nittis T. Suppressors of the temperature sensitivity of DNA polymerase alpha mutations in Saccharomyces cerevisiae. Mol Gen Genet. 1998;257(4):461–468. doi: 10.1007/s004380050670. [DOI] [PubMed] [Google Scholar]
- Christiano R, Nagaraj N, Frohlich F, Walther TC. Global proteome turnover analyses of the Yeasts S. cerevisiae and S. pombe. Cell Rep. 2014;9(5):1959–1965. doi: 10.1016/j.celrep.2014.10.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lancaster AK, Nutter-Upham A, Lindquist S, King OD. PLAAC: a web and command-line application to identify proteins with prion-like amino acid composition. Bioinformatics. 2014;30(17):2501–2502. doi: 10.1093/bioinformatics/btu310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halfmann R, Lindquist S. Screening for amyloid aggregation by Semi-Denaturing Detergent-Agarose Gel Electrophoresis. J Vis Exp. 2008. [DOI] [PMC free article] [PubMed]
- Rogoza T, et al. Non-Mendelian determinant [ISP+] in yeast is a nuclear-residing prion form of the global transcriptional regulator Sfp1. Proc Natl Acad Sci U S A. 2010;107(23):10573–10577. doi: 10.1073/pnas.1005949107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shorter J, Lindquist S. Prions as adaptive conduits of memory and inheritance. Nat Rev Genet. 2005;6(6):435–450. doi: 10.1038/nrg1616. [DOI] [PubMed] [Google Scholar]
- Tanaka M, Chien P, Naber N, Cooke R, Weissman JS. Conformational variations in an infectious protein determine prion strain differences. Nature. 2004;428(6980):323–328. doi: 10.1038/nature02392. [DOI] [PubMed] [Google Scholar]
- Tanaka M, Weissman JS. An efficient protein transformation protocol for introducing prions into yeast. Methods Enzymol. 2006;412:185–200. doi: 10.1016/S0076-6879(06)12012-1. [DOI] [PubMed] [Google Scholar]
- Roberts BT, Wickner RB. Heritable activity: a prion that propagates by covalent autoactivation. Genes Dev. 2003;17(17):2083–2087. doi: 10.1101/gad.1115803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozbudak EM, Thattai M, Lim HN, Shraiman BI, Van Oudenaarden A. Multistability in the lactose utilization network of Escherichia coli. Nature. 2004;427(6976):737–740. doi: 10.1038/nature02298. [DOI] [PubMed] [Google Scholar]


