ABSTRACT
The discovery of disease biomarkers, along with the use of “liquid biopsies” as a minimally invasive source of biomarkers, continues to be of great interest. In inflammatory diseases of the central nervous system (CNS), cerebrospinal fluid (CSF) is the most obvious biofluid source. Extracellular vesicles (EVs) are also present in CSF and are thought to be potential “biomarker treasure chests”. However, isolating these CSF-derived EVs remains challenging. This small-scale pilot study developed and tested a protocol to enrich for CSF-EVs, both in relapsing remitting multiple sclerosis (RRMS) CSF and controls. These were subsequently compared, using an aptamer based proteomics array, SOMAscan™. EVs were enriched from RRMS patient (n = 4) and non-demyelinating control (idiopathic intracranial hypertension (IIH) (n = 3)) CSF using precipitation and mini size-exclusion chromatography (SEC). EV-enriched fractions were selected using pre-defined EV characteristics, including increased levels of tetraspanins. EVs and paired CSF were analysed by SOMAscan™, providing relative abundance data for 1128 proteins. CSF-EVs were characterised, revealing exosome-like features: rich in tetraspanins CD9 and CD81, size ~100 nm, and exosome-like morphology by TEM. Sufficient quantities of, SOMAscan™ compatible, EV material was obtained from 5 ml CSF for proteomics analysis. Overall, 348 and 580 proteins were identified in CSF-EVs and CSF, respectively, of which 50 were found to be significantly (t-test) and exclusively enriched in RRMS CSF-EVs. Selected proteins, Plasma kallikrein and Apolipoprotein-E4, were further validated by western blot and appeared increased in CSF-EVs compared to CSF. Functional enrichment analysis of the 50 enriched proteins revealed strong associations with biological processes relating to MS pathology and also extracellular regions, consistent with EV enrichment. This pilot study demonstrates practicality for EV enrichment in CSF derived from patients with MS and controls, allowing detailed analysis of protein profiles that may offer opportunities to identify novel biomarkers and therapeutic approaches in CNS inflammatory diseases.
KEYWORDS: Extracellular vesicles, multiple sclerosis, cerebrospinal fluid, size exclusion chromatography, proteomics, biomarkers
Introduction
Selecting a relevant biological sample is a crucial step in protein biomarker candidate identification. The use of fluid biopsies, usually blood via venepuncture, is an attractive option, as they are minimally invasive. Blood has been used in numerous studies, predominantly in cancer, to detect and identify circulating biomarkers, utilising components such as circulating tumour cells and extracellular vesicles to help characterise tumours [1,2]. In diseases of the central nervous system (CNS), however, cerebrospinal fluid (CSF) may offer a more relevant biofluid. In this study, we were particularly interested in multiple sclerosis (MS); the most common cause of progressive neurological disability, in individuals between 20–40 years of age. To date, there has been limited success in identifying proteins in CSF with levels that appear to be altered in MS [3]. In order to optimise the utility of CSF as a biomarker, it may be most appropriate to remove or reduce the levels of the most abundant proteins to increase the probability of identifying less abundant proteins, which may be of most relevance in disease. An alternative approach to examining whole CSF is to study CSF derived extracellular vesicles (EVs), such as exosomes and microvesicles.
Due to their universal presence in biofluids, including CSF [4,5], EVs have become of considerable interest and have been referred to as possible biomarker “treasure chests” [6]. Enriching for EVs in CSF may help reduce the concentration of abundant proteins, but in turn also concentrate neurological disease biomarkers that associate with EVs. The enrichment of proteins and RNA linked to disease and cellular stress in EVs are well documented [7–9]. Furthermore, identification of proteins associated with processes such as cell signalling, inflammation, antigen presentation, complement modulation and neuronal viability [10], which may increase the sensitivity and specificity of clinical features in MS [11], have also been identified.
However, even with EV enrichment, significant challenges remain in investigating EVs as a potential source of biomarkers, including the removal of highly abundant non-EV associated proteins that are commonly present as principal components of biological fluids and can confound the identification of lower abundance proteins of interest in disease. However, new technologies are becoming available to overcome these hurdles and size exclusion chromatography (SEC) shows good utility in this regard [12–14], although it may not entirely eliminate non-vesicular material, which can still confound mass spectrometry-based analyses [13]. Recent use of protein-array methods offers an alternative approach for proteomics analysis, minimising the impact of high abundance contaminating proteins within the sample and enhancing the ability to identify relatively low abundance proteins of interest in disease. One such approach was used to examine human blood plasma and urine derived EVs isolated using SEC, is the SOMAscan® assay. This high throughput multiplex aptamer based array allowed simultaneous measurement and relative quantitative analysis of over 1000 proteins and has shown some promise in other diseases, including cancers [13].
In this proof of concept study we examined a novel method of CSF-EV enrichment using a precipitation and SEC methodology and subsequently examined and compared the proteome of CSF-EVs and CSF from relapsing remitting (RR) MS patients and non-demyelinating disease controls (Idiopathic intracranial hypertension; IIH). This was performed using the SOMAscan® biomarker discovery platform to overall assess this strategy for future use in the identification of novel protein biomarkers in CNS inflammatory diseases such as MS.
Experimental procedures
Cerebrospinal fluid samples were obtained from consented individuals (Wales REC 14/WA/0073) placed on ice and processed within 30 min. CSF was rendered acellular by centrifugation at 2000 × g, 10 min, 4°C and cell-free CSF supernatant stored at −80°C in 300 µl aliquots. Details of the patient-demographics are detailed in Supplementary Table 1.
CSF extracellular vesicle isolation
CSF (5 ml) was thawed at ambient temperature and vortexed for 20 s prior to the concentration of EVs using the Exo-Spin method (Cell GS, Cambridge, UK) following manufacturers protocol. Membrane precipitation buffer (Buffer A) was added and incubated at 4°C for 1 h, then centrifuged at 20,000 × g for 2 h at 4°C. The subsequent pellets were re-suspended in a total of 100 µl PBS per sample and further purified using mini pre-packed size exclusion columns (Exo-Spin™, Cell GS). All size-exclusion was performed under gravity. The sample was added to the column and eight fractions of 100 µl were collected and their protein and particle concentration determined, by NanoDrop™ and NanoSight™ analysis respectively. Fractions or pooled fractions were stored at −80°C prior to further analysis.
Throughout the rest of the report the different samples will be referred to as follows:
RRMS-EV: Relapsing remitting multiple sclerosis enriched cerebrospinal fluid extracellular vesicles.
RRMS-CSF: Relapsing remitting multiple sclerosis cell free cerebrospinal fluid.
IIH-EV: Idiopathic intracranial hypertension enriched cerebrospinal fluid extracellular vesicles.
IIH-CSF: Idiopathic intracranial hypertension cell free cerebrospinal fluid.
Nanoparticle tracking analysis (NanoSight™)
Nanoparticle tracking analysis (NanoSight™) was performed as previously described [13,15], with some modifications. Three videos of 30 s were taken under controlled fluid flow with a pump speed set to 80. Videos were analysed using the batch analysis tool of NTA 2.3 software (version 2.3 build 2.3.5.0033.7-Beta7), where minimum particle size, track length and blur were set to “automatic”. The area under the histogram for each triplicate measurement was averaged and used in further analysis.
Plate based immuno-assay for tetraspanin proteins
Column fractions were bound to protein-binding ELISA plates (at a dilution of 1:4). After overnight coupling and blocking (with 1% (w/v) BSA in PBS for 2 h at room temperature (RT)), the bound material were labelled with primary antibodies against proteins including CD9 (R&D systems) and CD81 (AbD serotec) or HSA (human serum albumin) (250 ng/ml) (R&D systems) was added for 2 h at RT on a plate shaker. After three washes, goat anti-mouse-biotinylated antibody (Perkin Elmer) diluted 1:2500 was added for 1.5 h. After three washes, Europium-conjugated streptavidin was added for 45 min. After a final six washes, a signal was obtained using time-resolved fluorometry, measured using a Wallac Victor-II multi-label plate reader (PerkinElmer) [13].
SDS-PAGE and immunoblotting
Cell-free CSF or EV enriched isolates were boiled in SDS sample buffer containing 20 mM DTT as previously described [16] briefly samples were separated using NuPAGE precast 4–20% gel (Invitrogen) and transferred and probed as described previously [15]. Membranes were probed with antibodies against KLKB1 (Plasma Kallikrein; Merck Millipore), ApoE4 and DKK3 (ThermoFisher Scientific), C6, TSG101 (SantaCruz Biotechnology), S100A9 (R&D Systems).
Transmission electron microscopy
CSF-EVs, from both RRMS patients and IIH controls, isolated by precipitation and mini-SEC, were stored at −80°C prior to transmission electron microscopy (TEM). The EVs were thawed on ice and negatively stained, as previously described by Connolly et al. [17].
Preparation of samples for the SOMAscan™ array
CSF and CSF-EVs were prepared for the SOMAscan® array as previously described [13]. Samples were diluted to 200 µg/ml in buffer (1x SomaLogic SB17, 1&NP40 and 0.5% (w/v) sodium deoxycholate). The subsequent sample supernatant was mixed with the SOMAmer® reagents for binding, at a sample concentration of 20 µg/ml), prior to a series of washing steps, followed by quantification on a custom Agilent hybridisation array. The relative fluorescence unit (RFU) for each SOMAmer® measured is proportional to the original protein concentration.
Data handling and presentation
The RFU output from the array was normalised using quantile normalisation and any significant differences between the cell-free CSF and RRMS CSF and their respective CSF derived EVs was assessed using row-by-row t-test, correcting for multiple testing using the Benjamini-Hochberg (BH) procedure. A conservative RFU cut-off value of 200 was subsequently chosen to distinguish between absent and present, based on previous studies using the platform [15,18], and was used in all subsequent analyses.
Graphs were generated using R in RStudio version 0.99.483 for Windows (RStudio, Inc., Boston, MA) or GraphPad Prism version 5.01 for Windows (GraphPad Software, San Diego, CA). For Gene Ontology analysis using Gprofiler, all reported genes for each SOMAmer®, because of protein complex recognition, were included as previously published [13]. For Gprofiler, the input and background gene list was converted into ENSEMBL codes with the following options: Gene Ontology, Reactome and KEGG selected, p-value adjustment = Benjamini-Hochberg. The resulting file was loaded into the enrichment map plugin on the Cytoscape (version 3.3.0) software, where the node and edge settings were adjusted to reflect q-value, gene number and significance [19].
Results
Isolation and characterisation of vesicles from CSF
Extracellular vesicles were enriched using a combination of precipitation with mini-SEC. Sample characterisation was undertaken on post-precipitation mini-SEC fractions for both RRMS and control (IIH) CSF vesicle preparations (RRMS-EV or IIH-EV, respectively) (Figure 1(a, b)). The results for the RRMS-EV enrichment (Figure 1(a)) suggested a higher concentration of particles in relation to protein in fractions 4–7, with fractions 5 and 6 coinciding the highest levels of both CD9 and CD81 (Figure 1(a), green and blue line). HSA was also present in these fractions, but it is not known whether this was a contaminant or HSA-associated with the vesicles. Due to limited sample availability, the presence of other non-EV associated proteins could not be investigated here. The results for the IIH-EV samples (Figure 1(b )) were different, demonstrating much lower levels of protein and EV associated protein markers, suggesting less EVs in the non-demyelinating disease controls (IIH). Overall protein concentrations for fraction 6 (Figure 1(c)) demonstrated a significant difference (p = 0.0015; Mann-Whitney U-test; n = 4) between RRMS-EV and control IIH-EVs used for the proteomics analysis.
Figure 1.
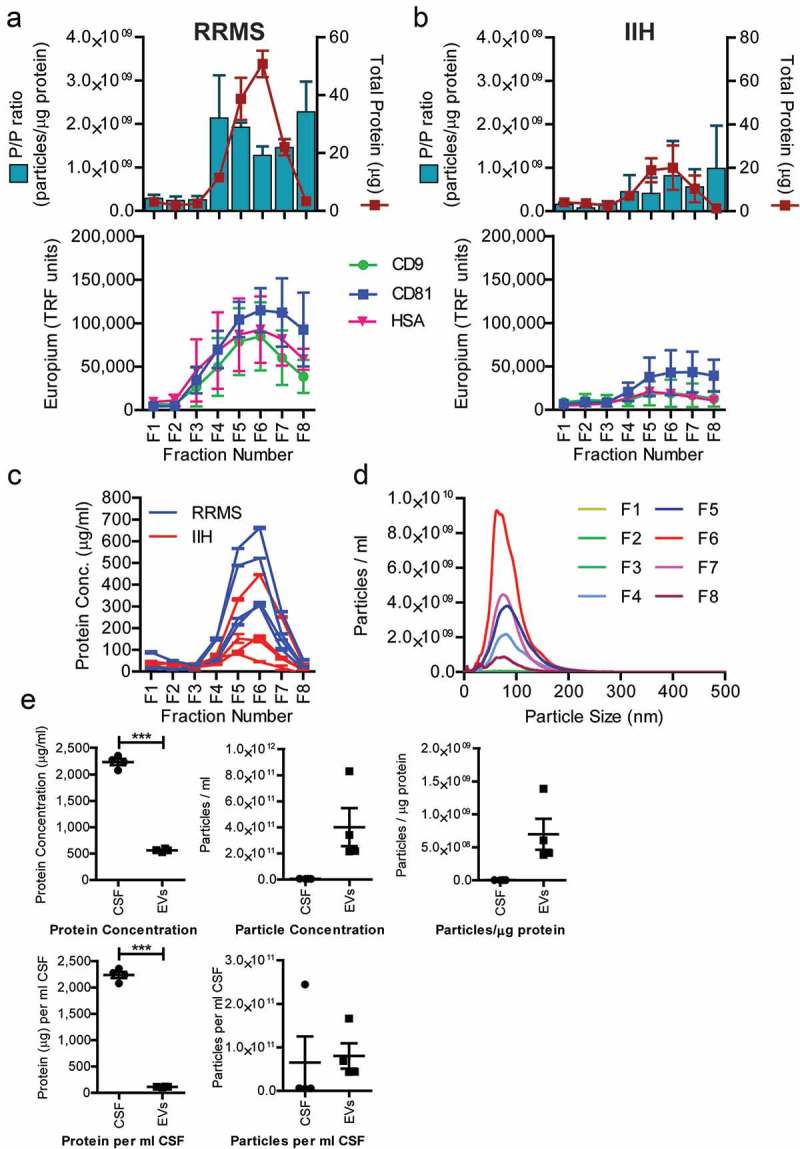
EV isolate sample characterisation. CSF size-exclusion fractions post-EV precipitation, for RRMS patients (a) and non-demyelinating disease controls (IIH) (b). Pelleted material from the precipitation procedure was subject to mini-SEC and eight serial fractions were collected and analysed. The protein concentration was estimated by NanoDrop™ (absorbance at 280 nm) and the particle concentration measured by nanoparticle tracking analysis (NanoSight™). The ratio of particles to protein (particles/µg) was calculated and plotted (left axis: blue bars), with total protein (µg/ml) on the right axis (red line) (±SEM). A proportion of each fractions was also immobilised onto high-protein-binding microplates. After blocking, the wells were stained with primary antibodies against CD9, CD81 or HSA and detected using TRF as a readout (arbitrary TRF units shown) (a, b). The protein concentration for all eight fractions for each of the samples (RRMS – blue lines; IIH – red lines, ±SEM) used for subsequent proteomics analysis was measured (c). A proportion of the eight fractions was used to examine the size distribution of particles during enrichment method development. Mean particle size and distribution from NanoSight™ analysis of RRMS-EV mini-SEC isolation is shown (d). Fractions 5 and 6 were identified as vesicle enriched and were pooled. Total protein and particle concentration were measured, in addition to protein and particles per millilitre of originating CSF. Particle-to-protein ratios were calculated (particle/ml) as a sample purity estimation. These RRMS-EV sample data (filled squares) are plotted as dot-plots and compared to RRMS-CSF (filled circles) values (e). (a), (b) and (d) were performed on method development patient CSF (n = 3) and (c) and (e) are from CSF used for the proteomics experiments (n = 4).
Particle size profiles for the mini-SEC fractions were also assessed by nanoparticle tracking analysis (NTA), demonstrating the presence of particles with a mean and mode particle size of around 100 nm, consistent with that of exosome-like sized EVs (Figure 1(d )). Furthermore, pooled fractions (5 and 6) were examined by TEM to assess their morphology, revealing the EV population to be heterogeneous and of exosome-like size (range = ~ 30–100 nm; Figure 2, main image) in both RRMS and IIH, consistent with the NanoSight™ analysis (Figure 2).
Figure 2.
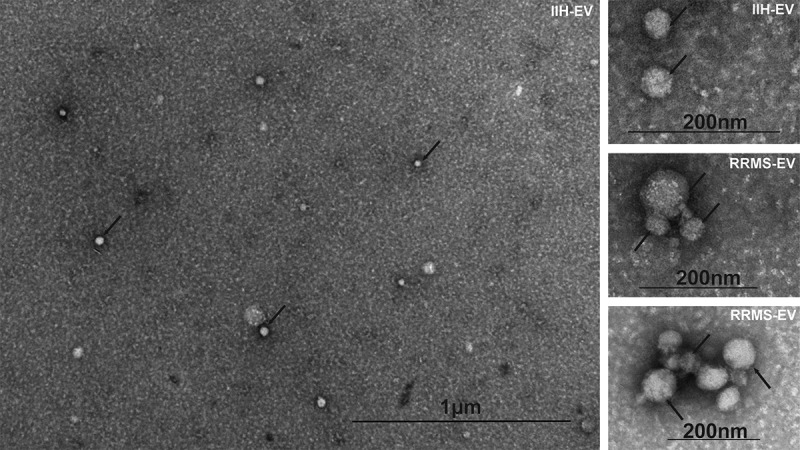
CSF-EV imaging by transmission electron microscopy. A proportion of both RRMS and IIH EV isolates were examined using transmission electron microscopy, as indicated (top right of each image), and representative fields are shown including a wide-field view (main image). Representatives of the heterogeneous EV populations are highlighted with black arrows and the scale indicated on each image.
Pooled fractions 5 and 6 underwent further analysis, including examining RRMS-EV yield and purity, by NanoDrop™ and NanoSight™, respectively (Figure 1(e )). A significant difference was observed between the total protein concentrations, and protein per millilitre of CSF, of the pooled EV fractions compared to the originating CSF (n = 4; p ≤ 0.0001; paired t-test), showing a major loss of total protein upon vesicle concentration. There was also a trend towards an increase in particle/protein ratio (p = 0.0725), indicating an increase in vesicle purity, as an intended consequence of the vesicle enrichment strategy.
Overall, the protocol implemented to isolate EVs from CSF has been partially successful, focusing in on fractions containing tetraspanins, nanoparticles and vesicles and yielding sufficient material for the RRMS specimens for protein profiling. Vesicle preparations from IIH; however, were significantly more difficult, due to the relative scarcity of vesicular material in this specimen type.
Protein profiling of CSF vesicles and matched cell-free CSF
The SOMAscan® platform was tested for compatibility with the EV-enriched sample prepared using the precipitation buffer, paired pooled cell-free CSF and CSF-EVs (n = 4 RRMS, 3 IIH) samples were examined. Pooled samples (four samples per pool) were used in this initial discovery phase in order to maximise the amount of information from limited CSF volumes and to also increase the chances of identifying proteins elevated across all samples. Overall, 346/1128 proteins were identified in CSF EVs (both RRMS and IIH controls) and 580/1128 were identified in cell-free CSF. These proteins all demonstrated mean relative fluorescence unit (RFU) values over 200, used as a cut-off value for presence vs absence of a protein. This data is summarised in Figure 3 and appended in full in Supplementary Table 2.
Figure 3.
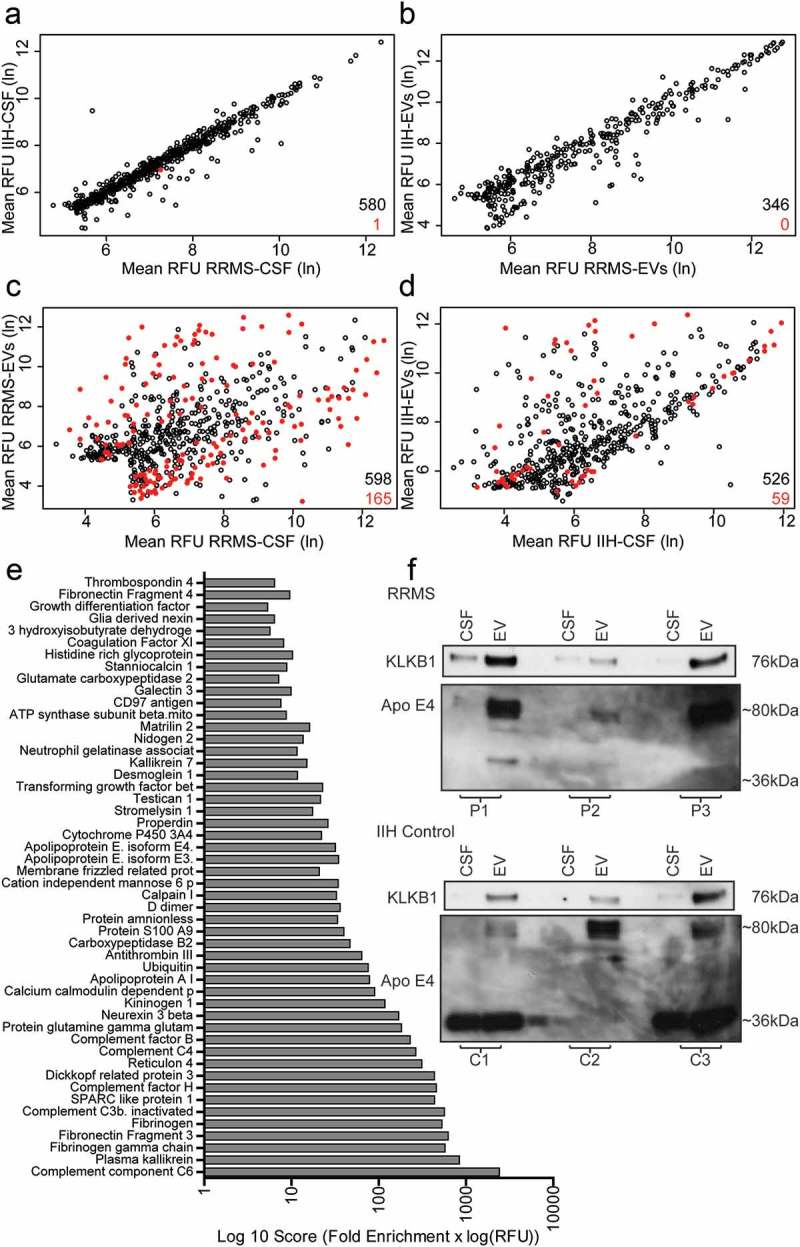
RRMS EV enriched protein identification and validation. The scatterplots shown in (a–d) give an overview of the results of the SOMAscan® array signifying all of the SOMAmer® bound proteins identified in the study. Comparisons were made between RRMS-CSF and IIH-CSF, a total of 580 proteins with RFUs > 200, (a) and corresponding CSF-EVs, 346 proteins, (b) only a single protein, vascular cell adhesion protein 1, is shown to be significantly different (p = 0.007) between RRMS-CSF and the IIH-CSF samples. In order to identify proteins that were uniquely enriched in RRMS-EVs, the CSF samples were compared with their corresponding EVs (c, d) Of these proteins, 71 were significantly higher in the CSF EVs. Fifty proteins were identified as uniquely enriched in the RRMS CSF EVs compared to whole CSF. These proteins are shown in the bar graph (e), ranked by an arbitrary scoring mechanism (fold enrichment × log(RFU)). From this list, several proteins were chosen for validation by western blot using completely new validation samples. The western blots show staining for KLKB1 (Plasma Kallikrein) and Apo E4 for matching CSF and CSF EVs for each of three RRMS patients (P1–3) and three control patients (C1–3). KLKB1 was observed at the expected molecular weight of ~ 76 kDa at higher levels in the EVs than the CSF in all instances. ApoE4 was primarily detected at ~ 80 kDa in the RRMS samples, with higher levels observed in the EV preparations. In the IIH controls, staining can be seen at the expected ~ 36 kDa for C1 and C3 and the 80 kDa form appears to be enriched in the EVs only. (a–d) Open black circles represent the mean RFU of SOMAmer® bound proteins with a p-value > 0.05. Red filled circles represent significant differences (p ≤ 0.05) in the levels of the SOMAmer® bound proteins. All statistical test were t-tests with Benjamini-Hochberg correction for multiple comparisons. The total number of proteins is indicated in black and the number of significantly different proteins in red.
We initially compared RRMS-CSF vs IIH-CSF and RRMS-EVs vs IIH-EVs. Only a single significantly different protein (Vascular cell adhesion protein 1; p <0.01; t-test with Benjamini–Hochberg procedure) was identified in the CSF and no significant differences were identified between EV sources (Figure 3(a,b)). This was not entirely unexpected, due to the limited number of samples that could be analysed in this study.
Subsequently, RRMS-CSF and RRMS-EVs were compared (Figure 3(c)), revealing 165 significantly different proteins, with 71 being increased in RRMS-EVs (t-test with Benjamini–Hochberg procedure). Comparing IIH-CSF and IIH-EVs flagged 59 significantly different proteins (t-test with Benjamini–Hochberg procedure), of which 38 were significantly increased proteins (Figure 3(d)). In these comparisons, all identifications that were not significantly different (i.e. p >0.05) were discarded. In addition, those identifications found in the IIH-EV specimens and also in the RRMS-EV specimens were removed. By this process of elimination, 50 significantly different proteins unique to the RRMS-EVs were revealed. These proteins were subsequently ranked using a scoring system previously used by Webber and Clayton are shown in Figure 3(e) [20].
Data analysis validation of proteins specifically enriched in RRMS EVs
In order to examine the 50 RRMS-EV enriched proteins, manual interrogation was used to discover any previous associations with vesicles or MS research publications in the PubMed database, as shown in full in Supplementary Table 3. Research publications (PubMed searches performed 29 September 2015) for 30 of these proteins were identified relating to MS, including numerous complement components, fibrinogen and apolipoproteins. Proteins not previously identified in association with MS included plasma kallikrein (KLKB1), Neurexin-3-beta, protein amnionless, testican-1, kallikrein-7 and thrombospondin-4.
From the information obtained from these searches, five proteins (KLKB1, ApoE4, DKK3, C6 and S100A9) were chosen for subsequent validation by western blot. For the validation, six samples from six different individuals (three RRMS, three IIH controls) were used and the cell-free CSF and CSF-EVs compared (Figure 3(f )).
KLKB1 was selected as it has been associated with EVs and other neurological conditions, but not MS. Interestingly, KLKB1 was found to be present not just in RRMS-EVs, but also the IIH-EVs. Overall, KLKB1 was enriched in the EV fraction compared to the cell-free CSF. This protein is, therefore, not able to distinguish MS patients from IIH controls.
Apo E4 was selected as it is present in EVs and neurological conditions including MS and Alzheimer’s disease. The APOE4 allele of this gene is known to increase the risk of Alzheimer’s disease [21]. It was also possible to see differences between the RRMS samples and the IIH control. For the IIH control bands were observed at the expected molecular weight of ~ 36 kDa in two out of three controls, but this 36 kDa band was completely absent from the RRMS patient samples. There was, however, another band present at ~ 80 kDa, which was uniquely present in the EV enriched fraction of all of the CSF samples. Unfortunately, we were unable to resolve DKK3, C6 or S100A9 by western blot. TSG101 was also not detectable with the amount of protein obtained.
Protein functional enrichment analysis
The associations of the 50 RRMS enriched proteins were also explored on a more global level, using functional enrichment analysis. This allowed the identification of statistically relevant associations of proteins that were over-represented in the dataset, and that may have associations with functional processes. The results of this are shown in Figure 4, where the larger the circle is the more gene terms are associated with that node and the deeper the colour the more significant the associations are. The network shows that the majority of these larger nodes are associated with vesicles, for example: extracellular vesicle, vesicle, membrane-bounded vesicles and extracellular membrane-bounded organelle. This, along with the other evidence presented, is again suggestive of the successful enrichment of vesicles by the isolation technique developed. There are also numerous smaller but significant nodes present in the network including nodes associated with wound healing, regulatory processes and clot formation.
Figure 4.
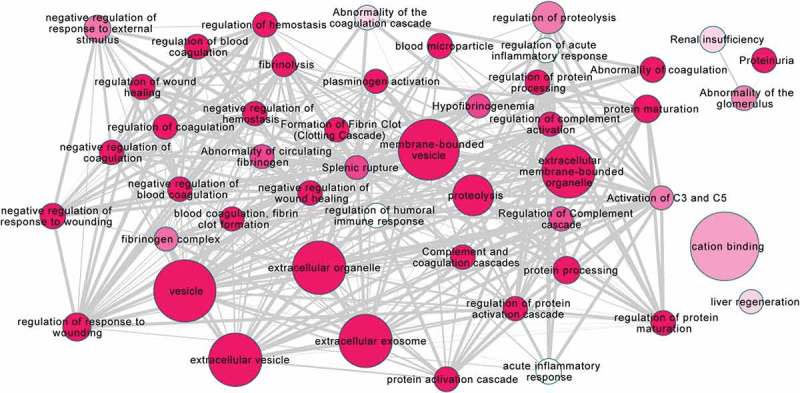
Functional enrichment analysis of RRMS EV unique proteins using Gprofiler.
Functional enrichment analysis was performed on the 50 RRMS EV unique proteins using Gprofiler. The size of each node demonstrates more gene terms associated with that node. The darker the colour of the node the more significant these associations are. The line thickness is the ratio of overlap from small to large terms. The majority of the larger nodes are associated with vesicles or extracellular organelles. There are numerous smaller nodes present with associations with processes such as wound healing, regulatory processes and clot formation.
Discussion
In this study, we present a novel practical approach for the enrichment of EVs from CSF for non-mass spectrometry based proteomics analysis. As well as demonstrating this simple quick (< 3 h) enrichment approach, we present a semi-quantitative approach for discovering novel proteins of potential interest as biomarkers and/or as mechanistic features of MS, using aptamer based protein array technology.
It remains challenging to isolate CSF-EVs due to their concentration and the volume of CSF available from individual donors. However, the enrichment method employed in this study was effective in concentrating and isolating EVs from volumes of 5 ml and differs from serial ultracentrifugation, which provides only a crude EV enriched pellet [4,5,22,23] and allows enrichment through the use of the membrane specific precipitant and subsequent size exclusion chromatography. The use of a mini-SEC column, post-precipitation, also means the method is amenable to handling lots of samples at once and quickly (< 10 min/sample), unlike ultracentrifugation and some of the larger SEC columns available. Saman et al. examined CSF-EVs in relation to Alzheimer’s disease using a more rigorous approach utilising a sucrose gradient approach [24]; however, this approach is lengthy (> 16 h) and is unsuitable for the analysis of multiple clinical samples. Using the precipitation with mini-SEC approach, we were able to isolate EV enriched fractions, as demonstrated by the presence of EV associated markers, tetraspanins CD9 and CD81. The size profile of the particles within these tetraspanin rich fractions was typical of EVs of exosome-size (~ 100 nm). This was supported by TEM images, showing a heterogeneous population of EVs consistent with exosomes. Due to the limitations of sample availability, we were unable to confirm the presence of endosome specific markers TSG101, Alix or LAMP-2 or the absence of contaminating proteins, such as Gp96. Our particles, based on the characterisation presented, are, thus, described as exosome-like EVs. Furthermore, the feasibility nature of the study did not allow us to investigate the effects of freeze thawing on the EVs contained within the CSF samples; however, we have demonstrated that it is possible to enrich EVs present in stored CSF. As CSF is not routinely taken, the ability to study EVs from archived patient samples is important.
The origin of the HSA present, in our samples, as mentioned in the results, is not known. This may be a soluble contaminant, among potentially many other non-vesicular proteins in these preparations, which we were unable to explore further due to the limited sample availability. Determining whether or not a discovered protein is genuinely vesicular would require more detailed follow-up investigations on a protein-by-protein basis.
In relation to the use of the aptamer based proteomics array, it does not appear to be impinged by the presence of PEG-like substances that would normally be a difficult problem to overcome with a traditional liquid chromatography/mass spectrometry based analysis. The SOMAscan® array is, nevertheless, a closed platform with a finite menu of detectable analytes, unlike mass spectrometry.
We were able to identify 348 CSF derived EV proteins and 580 cell free CSF proteins using this approach. Proteins identified in CSF-EVs include lactate dehydrogenase and GAPDH, reporting with high RFUs consistent with those reported previously by the group [15]. Out of those 348 proteins identified in CSF-EVs, 50 proteins demonstrated a significant enrichment (t-test with Benjamini–Hochberg procedure) in RRMS-EVs compared to RRMS-CSF, and these were not enriched in the control specimens. We, therefore, highlight both; the potential of our approach for CSF-EV enrichment and analysis, and the identification of several proteins hitherto not identified in the context of EVs or RRMS, which warranted further investigation.
In relation to these proteins, one of the most interesting observations was the presence of what appeared to be a dimer or ApoE4 containing complex found enriched in all of the EV samples, by Western blot at ~ 80 kDa. Furthermore, the natural monomer of 36 kDa was surprisingly not detected in the context of RRMS-EV. Unlike ApoE3 and ApoE2, ApoE4 is not thought to form disulphide-linked dimers in plasma or CSF, due to the lack of a cysteine residue. A study by Elliott et al. [25] examined apolipoprotein-E dimers in human frontal cortex and hippocampus, which also demonstrated a lack of ApoE4 dimers in human brain tissue. In fact, in the case of ApoE3/4 heterodimers these were also only present at very low levels. An interesting report by van Neil et al. [26] highlights ApoE interaction with exosomes in relation to amyloid-related diseases such as Alzheimer’s disease and their potential role in neutralising toxic PMEL-derived amyloid peptides. Van Neil et al. [26] demonstrated cryo-EM ApoE specific regions on exosomes that could cluster important cofactors of fibrillation. This association with exosomes and intraluminal vesicles demonstrates a new pathway for ApoE secretion and extends the properties of exosomes in pigment cell pathologies.
We also investigated the serine protease protein, KLKB1, which did not appear to be RRMS specific, but its presence was elevated in the EV enriched fractions compared to the originating CSF, consistent with the SOMAscan™ data. KLKB1 is thought to be a secreted protein, but has been previously identified in association with EVs in other biological fluids including blood, neutrophils and urine [27–29]. Based on these observations it is unlikely that this enrichment is CNS or disease specific.
We also wanted to investigate the data from our pilot study in more detail, exploring whether from just a few samples we would be able to identify biological associations of interest in MS. We, therefore, looked more broadly at the 50 enriched proteins using functional enrichment analysis. We revealed associations with vesicles, with six out of the seven largest nodes including vesicle relevant terms supporting the EV origin of the samples analysed, further supporting our CSF-EV enrichment strategy. Amongst the smaller nodes there were many significant links to wound healing and coagulation, processes which have previously been investigated in association with MS. For example, blood coagulation protein fibrinogen, deposited in the CNS after blood–brain barrier disruption, was identified by Ryu et al. [30] to promote autoimmunity and induce inflammatory demyelination. Regulation of complement activation also features in the analysis and complement proteins have been demonstrated to be differentially expressed in MS patient plasma compared to neuromyelitis optica spectrum disorder [31]. To date, it is not known what impact EV associated complement components have on MS pathology.
The overall picture from this analysis is that the 50 proteins identified as enriched specifically in RRMS CSF-EVs by using the SOMAscan® array may be indicative of involvement in processes such as the complement pathway, coagulation and wound healing, which have all been demonstrated to be altered in MS, and that our CSF-EV enrichment method does enrich for vesicle associated proteins. In subsequent higher powered studies using our approach it may be possible to tease out more MS specific proteins and perhaps EV proteins specific to the CNS. This would allow the specific pull out of CNS related EVs found in the blood circulation, opening yet more possibilities for studying EVs in CNS diseases. Currently research into EVs in relation to MS is limited, but various aspects of their relevance to the disease have been studied, including; their potential role in remyelination [32], as markers for therapy response/disease activity [33–35], therapeutic target [35], disease mechanism [36] and their role in pregnancy related reduced CNS pathology in EAE mouse models [37,38].
In summary, we highlight a simple method for the enrichment of smaller EVs (~ 100 nm) from human CSF using a precipitation step coupled with mini size exclusion chromatography. The subsequent selected fractions are high in EV associated markers and their size and morphology are exosome-like. The analysis of a very small number of samples of RRMS-EVs and controls, along with the originating CSF, has highlighted that there is the potential to identify EV associated proteins specifically enriched in RRMS. These EV-enriched proteins are otherwise missed when analysing the whole cell-free CSF and there is, therefore, value in concentrating this particular fraction of the CSF-proteome for discovery of disease relevant markers. The limited data from this proof of concept study demonstrates the potential for the application of these tools to identify proteins of interest as both biomarkers and potential therapeutic targets for neurological diseases such as multiple sclerosis.
Supplementary Material
Funding Statement
This work was supported by the Multiple Sclerosis Society [Grant reference 13].
Disclosure statement
No potential conflict of interest was reported by the authors.
Supplementary data
Supplemental data for this article can be accessed here.
References
- [1]. Jia S, Zhang R, Li Z, et al. Clinical and biological significance of circulating tumor cells, circulating tumor DNA, and exosomes as biomarkers in colorectal cancer. Oncotarget. 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2]. Perakis S, Speicher MR.. Emerging concepts in liquid biopsies. BMC Med. 2017;15(1):75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3]. Farias AS, Santos LM. How can proteomics elucidate the complexity of multiple sclerosis? Proteomics Clin Appl. 2015;9:844–847. [DOI] [PubMed] [Google Scholar]
- [4]. Akers JC, Ramakrishnan V, Kim R, et al. miRNA contents of cerebrospinal fluid extracellular vesicles in glioblastoma patients. J Neurooncol. 2015;123(2):205–216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5]. Lee J, McKinney KQ, Pavlopoulos AJ, et al. Exosomal proteome analysis of cerebrospinal fluid detects biosignatures of neuromyelitis optica and multiple sclerosis. Clin Chim Acta. 2016;462:118–126. [DOI] [PubMed] [Google Scholar]
- [6]. Duijvesz D, Burnum-Johnson KE, Gritsenko MA, et al. Proteomic profiling of exosomes leads to the identification of novel biomarkers for prostate cancer. PLoS One. 2013;8(12):e82589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7]. Riazifar M, Pone EJ, Lötvall J, et al. Stem cell extracellular vesicles: extended messages of regeneration. Annu Rev Pharmacol Toxicol. 2017;57:125–154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8]. Kadota T, Yoshioka Y, Fujita Y, et al. Extracellular vesicles in lung cancer-from bench to bedside. Semin Cell Dev Biol. 2017;67:39–47. [DOI] [PubMed] [Google Scholar]
- [9]. Quek C, Hill AF. The role of extracellular vesicles in neurodegenerative diseases. Biochem Biophys Res Commun. 2017;483(4):1178–1186. [DOI] [PubMed] [Google Scholar]
- [10]. van der Pol E, Böing AN, Harrison P, et al. Classification, functions, and clinical relevance of extracellular vesicles. Pharmacol Rev. 2012;64(3):676–705. [DOI] [PubMed] [Google Scholar]
- [11]. Raphael I, Webb J, Stuve O, et al. Body fluid biomarkers in multiple sclerosis: how far we have come and how they could affect the clinic now and in the future. Expert Rev Clin Immunol. 2015;11(1):69–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12]. Böing AN, van der Pol E, Grootemaat AE, et al. Single-step isolation of extracellular vesicles by size-exclusion chromatography. J Extracell Vesicles. 2014;3:10.3402/jev.v3.23430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13]. Welton JL, Brennan P, Gurney M, et al. Proteomics analysis of vesicles isolated from plasma and urine of prostate cancer patients using a multiplex, aptamer-based protein array. J Extracell Vesicles. 2016;5:31209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14]. Welton JL, Webber JP, Botos L-A, et al. Ready-made chromatography columns for extracellular vesicle isolation from plasma. J Extracell Vesicles. 2015;4:27269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15]. Webber J, Stone TC, Katilius E, et al. Proteomics analysis of cancer exosomes using a novel modified aptamer-based array (SOMAscan™) platform. Mol Cell Proteomics. 2014;13(4):1050–1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16]. Welton JL, Khanna S, Giles PJ, et al. Proteomics analysis of bladder cancer exosomes. Mol Cell Proteomics. 2010;9(6):1324–1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17]. Connolly KD, Guschina IA, Yeung V, et al. Characterisation of adipocyte-derived extracellular vesicles released pre- and post-adipogenesis. J Extracell Vesicles. 2015;4:29159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18]. Gold L, Ayers D, Bertino J, et al. Aptamer-based multiplexed proteomic technology for biomarker discovery. PLoS One. 2010;5(12):e15004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19]. Cline MS, Smoot M, Cerami E, et al. Integration of biological networks and gene expression data using cytoscape. Nat Protoc. 2007;2(10):2366–2382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20]. Webber J, Clayton A. How pure are your vesicles? J Extracell Vesicles. 2013;2:19861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21]. Farrer LA, Cupples LA, Haines JL, et al. Effects of age, sex, and ethnicity on the association between apolipoprotein E genotype and Alzheimer disease. A meta-analysis. APOE and Alzheimer Disease Meta Analysis Consortium. JAMA. 1997;278(16):1349–1356. [PubMed] [Google Scholar]
- [22]. Akers JC, Ramakrishnan V, Kim R, et al. MiR-21 in the extracellular vesicles (EVs) of cerebrospinal fluid (CSF): a platform for glioblastoma biomarker development. PLoS One. 2013;8(10):e78115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23]. Shi R, Wang P-Y, Li X-Y, et al. Exosomal levels of miRNA-21 from cerebrospinal fluids associated with poor prognosis and tumor recurrence of glioma patients. Oncotarget. 2015;6(29):26971–26981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24]. Saman S, Kim W, Raya M, et al. Exosome-associated tau is secreted in tauopathy models and is selectively phosphorylated in cerebrospinal fluid in early Alzheimer disease. J Biol Chem. 2012;287(6):3842–3849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25]. Elliott DA, Halliday GM, Garner B. Apolipoprotein-E forms dimers in human frontal cortex and hippocampus. BMC Neurosci. 2010;11:23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26]. van Niel G, Bergam P, Di Cicco A, et al. Apolipoprotein E regulates amyloid formation within endosomes of pigment cells. Cell Rep. 2015;13(1):43–51. [DOI] [PubMed] [Google Scholar]
- [27]. Principe S, Jones EE, Kim Y, et al. In-depth proteomic analyses of exosomes isolated from expressed prostatic secretions in urine. Proteomics. 2013;13(10–11):1667–1671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28]. Bosman GJ, Lasonder E, Luten M, et al. The proteome of red cell membranes and vesicles during storage in blood bank conditions. Transfusion. 2008;48(5):827–835. [DOI] [PubMed] [Google Scholar]
- [29]. Dalli J, Montero-Melendez T, Norling LV, et al. Heterogeneity in neutrophil microparticles reveals distinct proteome and functional properties. Mol Cell Proteomics. 2013;12(8):2205–2219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30]. Ryu JK, Petersen MA, Murray SG, et al. Blood coagulation protein fibrinogen promotes autoimmunity and demyelination via chemokine release and antigen presentation. Nat Commun. 2015;6:8164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31]. Hakobyan S, Luppe S, Evans DR, et al. Plasma complement biomarkers distinguish multiple sclerosis and neuromyelitis optica spectrum disorder. Mult Sclerosis J. 2016. ;23(7):946–955. [DOI] [PubMed] [Google Scholar]
- [32]. Pusic AD, Pusic KM, Clayton BL, et al. IFNγ-stimulated dendritic cell exosomes as a potential therapeutic for remyelination. J Neuroimmunol. 2014;266(1–2):12–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33]. Lowery-Nordberg M, Eaton E, Gonzalez-Toledo E, et al. The effects of high dose interferon-β1a on plasma microparticles: correlation with MRI parameters. J Neuroinflammation. 2011;8:43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34]. Sheremata WA, Jy W, Horstman LL, et al. Evidence of platelet activation in multiple sclerosis. J Neuroinflammation. 2008;5:27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35]. Verderio C, Muzio L, Turola E, et al. Myeloid microvesicles are a marker and therapeutic target for neuroinflammation. Ann Neurol. 2012;72(4):610–624. [DOI] [PubMed] [Google Scholar]
- [36]. Marcos-Ramiro B, Oliva Nacarino P, Serrano-Pertierra E, et al. Microparticles in multiple sclerosis and clinically isolated syndrome: effect on endothelial barrier function. BMC Neurosci. 2014;15:110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37]. Gatson NN, Williams JL, Powell ND, et al. Induction of pregnancy during established EAE halts progression of CNS autoimmune injury via pregnancy-specific serum factors. J Neuroimmunol. 2011;230(1–2):105–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38]. Williams JL, Gatson NN, Smith KM, et al. Serum exosomes in pregnancy-associated immune modulation and neuroprotection during CNS autoimmunity. Clin Immunol. 2013;149(2):236–243. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


