Abstract
Root hairs increase root surface area for better water uptake and nutrient absorption by the plant. Because they are small in size and often obscured by their natural environment, root hair morphology and function are difficult to study and often excluded from plant research. In recent years, microfluidic platforms have offered a way to visualize root systems at high resolution without disturbing the roots during transfer to an imaging system. The microfluidic platform presented here builds on previous plant-on-a-chip research by incorporating a two-layer device to confine the Arabidopsis thaliana main root to the same optical plane as the root hairs. This design enables the quantification of root hairs on a cellular and organelle level and also prevents z-axis drifting during the addition of experimental treatments. We describe how to store the devices in a contained and hydrated environment, without the need for fluidic pumps, while maintaining a gnotobiotic environment for the seedling. After the optical imaging experiment, the device may be disassembled and used as a substrate for atomic force or scanning electron microscopy while keeping fine root structures intact.
Keywords: Bioengineering, Issue 126, roots, microfluidics, plant-on-a-chip, Arabidopsis thaliana, root hair, SEM, AFM, organelle, high-resolution, imaging, treatment
Introduction
Fine root features increase water and nutrient acquisition for the plant, exploring new soil spaces and increasing the total root surface area. The turnover of these fine root features plays a major role in stimulating the underground food chain1 and the number of fine roots in certain plant species is expected to double under elevated atmospheric carbon dioxide2. Fine roots are generally defined as those smaller than 2 mm in diameter, although new definitions advocate for characterizing fine roots by their function3. Like many fine roots, root hairs provide the function of uptake and absorption but occupy a much smaller space with diameters on the order of microns. Because of their small size, root hairs are difficult to image in situ and are often overlooked as a part of the overall root architecture in field scale experiments and models.
Ex terra root hair studies, such as from seedlings grown on agar plates, have provided the scientific community with valuable information on cellular growth and transport4,5. While agar plates allow root systems to be imaged non-destructively and in real time, they do not offer high environmental control for the addition of experimental treatments such as nutrients, plant hormones, or bacteria. An emerging solution to facilitating high resolution imaging while also affording dynamic environmental control has been the advent of microfluidic platforms for plant studies. These platforms have enabled the non-destructive growth and visualization of several plant species for high throughput phenotyping6,7,8,9, isolated chemical treatments10, force measurements11,12, and the addition of microorganisms13. Microfluidic platform designs have focused on the use of single open space fluidic layers in which the roots may propagate, permitting the root hairs to drift in and out of optical focus during growth or treatment.
Here we present a procedure for developing a two-layer microfluidic platform using photo and soft-lithography methods that builds upon previous plant-on-a-chip designs by confining the seedling root hairs to the same imaging plane as the main root. This allows us to track root hair development in real time, at high resolution, and throughout the experimental treatment process. Our culturing methods allow Arabidopsis thaliana seedlings to be germinated from seed within the platform and cultured for up to a week in a hydrated and sterile environment that does not require the use of syringe pump equipment. Once the time-lapse imaging experiment has concluded, the platform presented here can be opened without disturbing the position of the finer root features. This allows the use of other high resolution imaging methods. Here we provide representative results for the quantification and visualization of root hair morphology in this platform by optical, scanning electron microscopy (SEM), and atomic force microscopy techniques (AFM).
Protocol
1. Two-layer Platform Fabrication
- Fabrication of multilayer masters
- Spin coat epoxy-based negative photoresist (~63.45% solids, 1,250 cSt) according to the manufacturer's specifications (2,000 rpm for 45 s) onto a 4 inch diameter silicon wafer to obtain the desired height of 20 µm for the first design layer.
- Soft-bake the resist coated wafers for 4 min at 95 °C. Allow wafer to cool for 5 min. Expose the wafer to UV light for 15 s (~150 mJ/cm2 at 365 nm) through a photomask in a UV contact aligner to define the geometry of the bottom layer.
- Post-exposure bake the wafer for 5 min at 95 °C. Without developing, return the wafer to the spin coater and spin on a second layer of epoxy-based negative photoresist (~76.75% solids, 80,000 cSt) at 3,000 rpm to achieve a second layer height of 150 - 200 µm.
- Allow the wafer to rest for 5 min before transferring to a 95 °C hotplate for 45 min. After baking on the hotplate, remove the wafer and allow the resist to cool and harden at room temperature for another 5 min on a flat level surface. Align and expose the wafer to the second layer photomask for 30 s in a UV contact aligner (dose of approximately 300 mJ/cm2).
- Perform a post-exposure bake by placing the wafer on a hotplate for 15 min at 95 °C, and then allow the wafer to cool on a flat level surface for 5 min before developing.
- Develop both resist layers simultaneously by placing the wafer into a plastic dish and submerging the wafer in the appropriate developer (see Materials Table). Gently rock the dish occasionally to wash fresh developer over the wafer.
- After 17 min, rinse the wafer with isopropanol (IPA). If a white film appears, continue to iterate between rinsing the wafer with developer and IPA until the film disappears. Dry the patterned silicon wafer with nitrogen.
- Polydimethylsiloxane Soft-lithography
- Expose the silicon wafer to an air plasma for 30 s in a plasma cleaner set on high (see Materials Table) to clean. Silanize the wafer with trichloro (1H,1H,2H,2H-perfluoro-n-octyl) silane in a chemical hood on a hotplate below the flashpoint of the silane (85 °C) for 2 h.
- Pour a 10:1 ratio of polydimethylsiloxane (PDMS) to curing agent onto the silicon master wafer. Degas the mixed PDMS in a vacuum chamber and then cure the polymer in a 70 °C oven for 1 h or until the PDMS is fully cured.
- Using a scalpel, cut the PDMS devices and peel them from the silicon master wafer. Use a 1.5 mm biopsy punch to create seed and treatment inlets, punching the seed inlet at a 45° angle to encourage root growth into the main channel.
- Use clear adhesive tape to remove debris from the PDMS device and place the device design side down on a glass coverslip. Autoclave the assembled devices.
2. Planting Devices
- Arabidopsis thaliana seed preparation
- Surface sterilize A. thaliana seeds in a microfuge tube with a solution of 30% commercially available bleach diluted in de-ionized water and 0.1% Triton X detergent for 7 min. Wash seeds 4 times with sterile water.
- Stratify seeds by refrigerating microfuge tube overnight or up to a week at 4 °C.
- Device Preparation
- Place sterilized devices in a vacuum degassing chamber to remove air from the gas permeable PDMS and fluidic channels to improve ease of filling.
- Remove the devices from the vacuum chamber and immediately submerge the devices in a Petri dish filled with liquid ¼ strength plant based media at pH 5.7.
- Using a pipette, pull liquid through the inlets to fill the device. Make sure no air bubbles remain within channels by visual inspection.
- Transfer individual devices to new dry Petri dishes. Pour or pipette hot agar around the device until the agar level is almost flush with the top of the PDMS device. Allow agar to solidify. NOTE: Devices are now ready for planting seeds or can be stored at 4 °C until needed.
- Planting seeds within devices
- In a sterile environment, transfer one sterilized seed to the inlet of each device using a small pipette.
- Cover the Petri dish with wax film (see Materials Table) and place in a light/dark cycling growth chamber or windowsill at room temperature. Orient the Petri dish so that the devices are vertical and gravity will encourage the roots to grow through the channel.
3. Treatment
- Experimental Treatments
- At the desired time in the seedling's development, add the experimental treatment to the seedling by adding a prescribed amount of the treatment to each of the 8 side ports via pipette.
- Reseal the Petri dish and return to growth chamber or windowsill.
- Proceed with imaging samples at the desired time to capture individual time points or begin time lapse imaging.
4. Optical Imaging
- Lower Resolution (4 - 20X) Imaging
- Place the entire Petri dish containing the device and seedling under an inverted bright field microscope for lower resolution (4 - 20X) bright field or differential interference contrast (DIC) imaging. Optimize lighting conditions to elucidate morphological features of interest by adjusting exposure times, lamp brightness, aperture, and the polarity of light. Drive the stage to the desired area of the root and focus on the root or root hair(s) of interest.
- Acquire a single time point or a time series of images. To visualize the growth of root hairs, image growing root hairs once per min. To visualize the growth of the main root, acquire one image every 30 min. Return the Petri dish and the seedlings to growth chamber or windowsill in a vertical position after imaging is complete.
- Higher Resolution (20 - 63X) Imaging
- Once the seedling has grown for a desired time, use forceps to remove coverslip and device from the agar. Clean off the bottom of the coverslip using an ethanol wetted laboratory tissue.
- Apply the recommended immersion media to the objective as suggested by the microscope lens manufacturer. Place the coverslip down on the microscope stage and raise the stage to contact the immersion media on the objective. Optimize the lighting conditions by adjusting exposure times, lamp brightness, aperture, and the polarity of light. Drive the stage to the desired area and focus on root hair(s) of interest. NOTE: DIC is needed to visualize cytoplasmic streaming, and fluorescence markers are needed to visualize organelle movement.
- To quantify root hair growth over time, acquire one image per min. To visualize organelle movement, minimize the fluorescence exposure time while still retaining an identifiable fluorescence signal from the organelles. Acquire images of the organelles as quickly as the exposure time allows. For longer time-lapse imaging, use a live-cell imaging stage incubator to maintain ambient temperature and moisture. NOTE: A 63X oil objective is recommended for imaging root hair organelles.
5. Non-Optical Imaging
- Device preparation
- Once seedling has grown for desired time, remove the device and coverslip from the Petri dish.
- Turn device upside down and gently peel away the coverslip so that the root stays within the PDMS channel.
- Proceed to use the PDMS device as a substrate for the root in higher resolution atomic force or scanning electron microscopy.
- Scanning Electron Microscopy
- Deposit a thin (~20 nm) layer of chromium onto the root and surrounding PDMS using a Dual Gun Electron Beam Evaporation Chamber.
- Transfer the root and PDMS device to a scanning electron microscope chamber. NOTE: Imaging conditions, i.e. voltage, current and working distance will need to be optimized to achieve the desired resolution for a given application.
- Atomic Force Microscopy (AFM)
- Mount the PDMS device root side up on an AFM specimen holder. To increase contrast in the camera and more easily distinguish the root from the PDMS, place the PDMS device on a flat reflective substrate (such as mica coated in gold) before mounting the device on the AFM specimen holder.
- Secure a liquid well attachment on top of the PDMS device and fill it with water to keep the roots hydrated during imaging.
- Load the specimen holder into the AFM. Adjust the z-control for the thickness of the PDMS device and drive the cantilever to the region of interest on the root using a camera for guidance.
- Align the laser with the tip of the cantilever. For best results with contact mode imaging, use cantilevers with spring constants of 0.01 or 0.03 N/m to exert minimal force on the root during scanning.
- Slowly lower the scanning mechanism until the cantilever just makes contact with the sample. Adjust the scan size for the desired region and choose a scan speed of 1 line/s with 256 voltage points per line. Acquire the scan.
Representative Results
The two-layer PDMS microfluidic devices described here have a 200 µm high channel for the main Arabidopsis root and a 20 µm high chamber to confine laterally growing root hairs (Figure 1A). This design may be used for plant species with similar root diameters as Arabidopsis thaliana and can be readily modified to accommodate species of different sizes. The design incorporates an inlet for the plant as well as 8 side inlets for any desired chemical or biological treatment. Prefilling the devices with media and pouring agar around the device keeps the root environment hydrated for the duration of the experiment without the need for external fluidic equipment. (Figure 1B) The agar also stabilizes the device within the Petri dish, allowing the seedlings to be grown vertically to encourage root growth down the main channel. The Petri dish keeps the seedlings contained in a gnotobiotic environment and allows the root system to be imaged through the Petri dish to magnifications up to 20X. Another option would be to directly seal the PDMS device to a glass-bottom Petri dish for even higher optical magnifications.
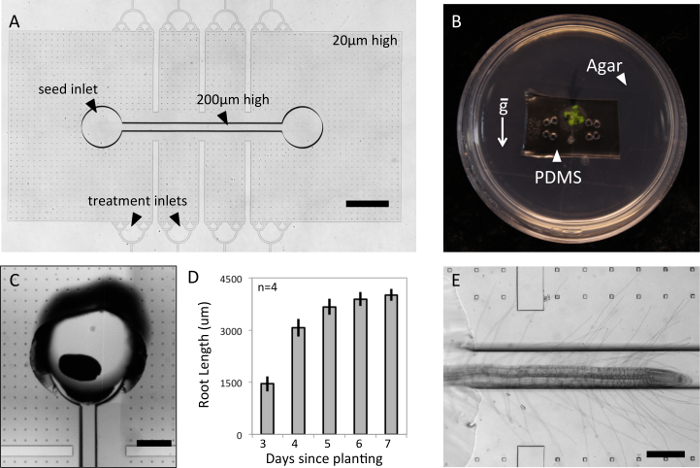
Arabidopsis thaliana seeds can be directly planted into the device inlet that is punched at a 45-degree angle to facilitate root growth into the main channel. (Figure 1C) The height of the PDMS device should not exceed a few millimeters, as the leaves must be able to grow out of the top of the platform. Because of their small size, Arabidopis seeds are very difficult to orient so that the emerging root radical faces the channel upon germination. Therefore, approximately half of the seeds planted will germinate with their leaves in the channels and cannot be used for root visualization experiments. Devices from these seedlings may be re-autoclaved and used again beginning from step 2.2. To encourage phototrophic growth of Arabidopsis thaliana shoots, the viewing chamber of the device may be covered in aluminum foil to block light and improve success rate. Arabidopsis thaliana roots have grown steadily in this platform for a week, at which point growth becomes directed toward lateral root formation. (Figure 1D) The growth rate of roots in this platform are comparable to growth rates of Arabidopsis thaliana roots in other microfluidic platforms.13 The two-layer design successfully confines the root hairs to the same imaging plane as the main root. (Figure 1E) However, some root hairs may continue growing in the main channel out of optical focus and future designs should aim to more gradually guide root hairs into the root hair chamber.
Figure 1: Growth of Arabidopsis thaliana in Two-layer Microfluidic Platform. (A) Device consists of two layers to confine root hairs to the same imaging plane as the main root (scale 1 mm). A. thaliana seedlings may be (B) contained in a gnotobiotic environment, (C) germinated from seed within the device, Scale bar = 400 µm, (D) and monitored for growth over the course of a week (error bars are standard deviation, SD). (E) A representative root is imaged in the platform, Scale bar = 200 µm. Please click here to view a larger version of this figure.
Within the microfluidic platform, root hairs can be quantified on a cellular level in terms of their length, density, and the angularity of their growth from the root. (Figure 2A) Root hair length and density of A. thaliana wild type seedlings grown in our platform are within range of seedlings grown vertically on agar plates.14 The angle of root hair growth is typically perpendicular to the surface of the root. In our platform, the angularity of root hair growth toward the tip may be an artifact of the confinement of the root hairs to a 2-dimensional plane. Mechanical stimulation, in this case by the walls of the microfluidic device, has been shown to induce changes in root growth and orientation which may explain the discrepancy between a confined microfluidic platform and an open agar plate.15 It is unlikely that this phenotype is the result of a nutrient stress in our platform as A. thaliana seedlings utilize stored food reserves from the seed in the first few days of growth. Although hypoxia may be a concern in fully saturated root systems, the oxygen permeability of PDMS has previously demonstrated the biocompatibility of the polymer with oxygen dependent tissues.16,17
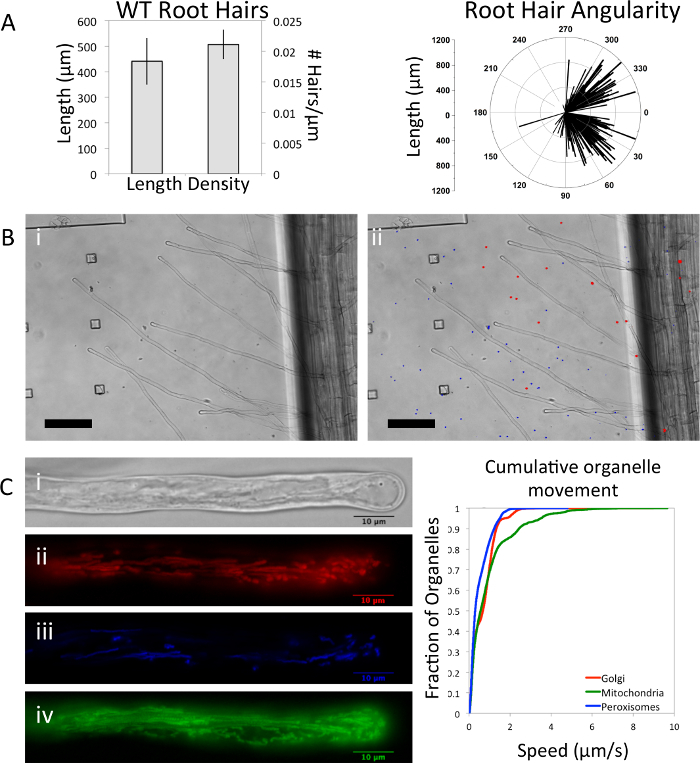
One of the strongest advantages of microfluidic platforms over agar plate methods is the ability to uniformly add precise concentrations of chemical treatments to the organisms. In single channel microfluidic platforms, the addition of treatments can potentially displace fine root hairs from optical focus. Here we demonstrate the maintenance of root hair optical focus in our two-layer microfluidic platform during the addition of fluorescent beads. In Figure 2B a seedling is imaged (i) before and (ii) after the addition of fluorescent carboxylated polystyrene beads (blue and red). The addition of this abiotic treatment did not disrupt the orientation or optical focus of the seedling's root hairs.
Higher magnification imaging and fluorescent markers may be used to image and quantify organelle level changes in root hair development (Figure 2C). Cytoplasmic streaming can be seen in a root hair from a (i) differential interference contrast image and, in another root hair from a seedling containing a triple organelle marker18, the trajectory and spatial distribution of three organelles ii) Golgi (mCherry), (iii) peroxisomes (CFP), and (iv) mitochondria (YFP) can be seen from the maximum intensity projection in each respective panel. The movement of these three organelles are captured in a cumulative distribution plot in Figure 2C.
Figure 2: Root Hair Characterization and Treatment. Root hairs were quantified on a cellular level by (A) length, density, and angularity for n= 4 wild type (WT) seedlings (error bars are SD). Angularity is determined as the angle between the main root tip and root hair tip with the main root tip defined as the 0° mark. (B) The optical focus of the root hairs remains unchanged (i) before and (ii) after treating them with polystyrene fluorescent beads (Scale bar = 100 µm). (C) Organelle level quantification is demonstrated by imaging the (i) cytoplasmic streaming from a differential interference contrast image and, in a separate root hair, the fluorescence of three organelles: (ii) Golgi (iii) peroxisomes, and (iv) mitochondria (Scale bar = 10 µm). Organelle trajectories were visualized by merging fluorescent images taken every 23 - 32 s for 20 s. The movements of the three organelles were tracked automatically and cumulative speed distributions plotted on the right. Please click here to view a larger version of this figure.
The PDMS device presented here has not been chemically bonded to a glass coverslip but rather forms a weaker physical bond that is established during the autoclaving step. This allows the device to be disassembled after the optical imaging experiment is complete. The PDMS device may be peeled from the glass, inverted, and used as a substrate for higher resolution non-optical imaging of the root morphology (Figure 3A). The platform conveniently holds the root in place during contact with a cantilever during atomic force microscopy (Figure 3B). Further, if care is taken during the disassembly process, root hairs will remain in position on the PDMS substrate. This is demonstrated from the optical image of the root before the disassembly (Figure 3C) and after disassembly of the platform and coating the seedling in a 20 nm conductive chromium layer for imaging with a scanning electron microscope (Figure 3D). To further preserve the seedling before deconstructing the platform, an aldehyde-based fixative may be injected into the platform to crosslink the plant tissue proteins in place.
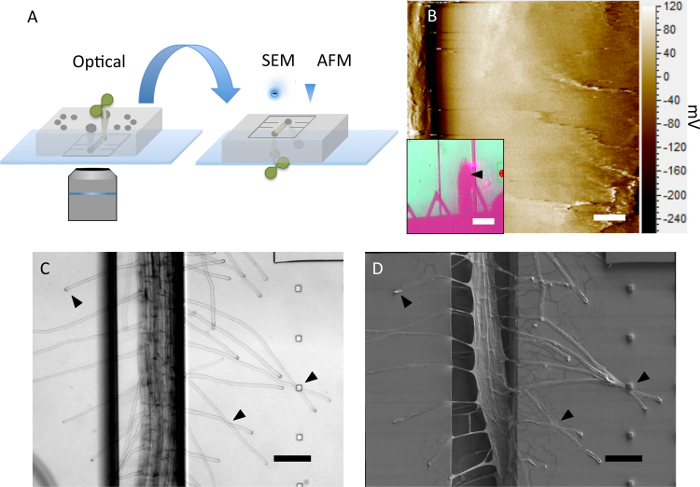
Figure 3: Device Deconstruction and Non-optical Imaging. (A) The microfluidic platform can be disassembled and used as a substrate for high-resolution non-optical imaging methods. (B) The surface topography (diffraction image) of an Arabidopsis thaliana root tip is imaged using contact mode atomic force microscopy, Scale bar = 2 µm. The location of the cantilever on the root is indicated by a black arrow in the inset, Scale bar = 100 µm. (C) Optical image and (D) corresponding scanning electron micrograph of the same seedling before and after device is disassembled. Arrows emphasize root hairs that remain undisturbed, Scale bar = 100 µm. Please click here to view a larger version of this figure.
Discussion
The method described in this article for creating a plant-on-a-chip platform is unique in that the two-layer design confines the root hairs to a single imaging plane and the platform may be deconstructed and used as a substrate for high resolution non-optical imaging. Using high-resolution non-optical imaging can provide valuable information about the plant tissue that could not be obtained from optical imaging alone. For example, AFM imaging can provide force measurements to calculate the elasticity of root tissues during development or after a specific chemical or biological treatment. Similarly, SEM imaging can provide high-resolution details of surface topography of the root tissue and, when coupled with chemical imaging, can provide information on elemental composition of the tissues.19 Future generations of this microfluidic platform will include optimization for compatibility with other chemical imaging systems such as matrix-assisted laser desorption/ionization- mass spectroscopy (MALDI-MS) and Coherent anti-Stokes Raman spectroscopy (CARS).
Critical steps in this method involve using agar to secure the device and provide hydration to the plant without the need for complicated fluid flow procedures. Care must also be taken when deconstructing the PDMS device from the glass substrate in order to keep the root morphology intact. If the experimental goal is solely for low to medium resolution optical imaging without the need for device deconstruction, the procedure may be modified to chemically bond the device to the underlying glass substrate using oxygen plasma. Higher resolution optical images can be obtained without removing the device from its gnotobiotic environment by chemically bonding the PDMS directly to a glass bottomed Petri dish and then pouring agar around the PDMS as previously described.
The design of 8 treatment side channels was an attempt to confine treatments to certain regions of the root. However, this design was unsuccessful in preventing the diffusion of the treatment to other areas of the root due to the conductance of the open area in the main root channel. In order to locally treat the root, the device architecture will either need to be redesigned to confine treatments or the treatment will need to be introduced via a viscous media to temporarily slow diffusion throughout the device.
If it is desirable for experimental treatments to be added via flow, modifications will also be required for this platform. Currently the addition of flow to any of the platform inlets results in the expulsion of the seed from its inlet and difficulty in controlling the location of the fluidic treatments. This method works well for seedlings up to one week of age. It is limited in how long seedlings may continue to grow in the platform, because of the length of the main channel. Future modifications will involve elongating the main channel and incorporating more 200 µm high channels for lateral roots while retaining the 20 µm tall chamber for root hair confinement. This modification will require knowledge on the anticipated location of lateral root emergence in order to design an appropriate device.
Disclosures
The authors have nothing to disclose.
Acknowledgments
This manuscript has been authored by UT-Battelle, LLC under Contract No. DE-AC05-00OR22725 with the U.S. Department of Energy. The United States Government retains and the publisher, by accepting the article for publication, acknowledges that the United States Government retains a non-exclusive, paid-up, irrevocable, world-wide license to publish or reproduce the published form of this manuscript, or allow others to do so, for United States Government purposes. The Department of Energy will provide public access to these results of federally sponsored research in accordance with the DOE Public Access Plan (http://energy.gov/downloads/doe-public-access-plan).
This work was supported in part by the Genomic Science Program, U.S. Department of Energy, Office of Science, Biological and Environmental Research, as part of the Plant Microbe Interfaces Scientific Focus Area (http://pmi.ornl.gov). The fabrication of the microfluidic platforms was carried out in the Nanofabrication Research Laboratory at the Center for Nanophase Materials Sciences, which is a DOE Office of Science User Facility. JAA is supported by an NSF graduate research fellowship DGE -1452154
References
- Pritchard SG. Soil organisms and global climate change. Plant Pathol. 2011;60(1):82–99. [Google Scholar]
- Norby RJ, Ledford J, Reilly CD, Miller NE, O'Neill EG. Fine-root production dominates response of a deciduous forest to atmospheric CO2 enrichment. Proc. Natll. Acad. Sci. USA. 2004;101(26):9689–9693. doi: 10.1073/pnas.0403491101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCormack ML, et al. Redefining fine roots improves understanding of below-ground contributions to terrestrial biosphere processes. New Phytol. 2015;207(3):505–518. doi: 10.1111/nph.13363. [DOI] [PubMed] [Google Scholar]
- Mangano S, Juarez SPD, Estevez JM. ROS regulation of polar-growth in plant cells. Plant Physiol. 2016;171(3):1593–1605. doi: 10.1104/pp.16.00191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ketelaar T, Emons AM. The Actin Cytoskeleton in Root Hairs: A Cell Elongation Device. Root Hairs. 2009. pp. 211–232.
- Grossmann G, et al. The RootChip: an integrated microfluidic chip for plant science. Plant Cell. 2011;23(12):4234–4240. doi: 10.1105/tpc.111.092577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grossmann G, et al. Time-lapse fluorescence imaging of Arabidopsis root growth with rapid manipulation of the root environment using the RootChip. J. Vis. Exp. 2012. [DOI] [PMC free article] [PubMed]
- Jiang H, Xu Z, Aluru MR, Dong L. Plant chip for high-throughput phenotyping of Arabidopsis. Lab Chip. 2014;14(7):1281. doi: 10.1039/c3lc51326b. [DOI] [PubMed] [Google Scholar]
- Busch W, et al. A microfluidic device and computational platform for high-throughput live imaging of gene expression. Nat. Methods. 2012;9(11) doi: 10.1038/nmeth.2185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meier M, Lucchettta E, Ismagilov R. Chemical Stimulation of the Arabidopsis thaliana Root using Multi-Laminar Flow on a Microfluidic Chip. Lab Chip. 2010;10(16):2147–2153. doi: 10.1039/c004629a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozoe K, Hida H, Kanno I, Higashiyama T, Notaguchi M. Early characterization method of plant root adaptability to soil environments. Proc. of 28th IEEE Interntl. Conf. Micro. Electro Mech. Syst. 2015.
- Sanati Nezhad A. Microfluidic platforms for plant cells studies. Lab on a chip. 2014. pp. 3262–3274. [DOI] [PubMed]
- Parashar A, Pandey S. Plant-in-chip: Microfluidic system for studying root growth and pathogenic interactions in Arabidopsis. App. Phys. Lett. 2011;98(26):2009–2012. [Google Scholar]
- Rigas S, et al. Root gravitropism and root hair development constitute coupled developmental responses regulated by auxin homeostasis in the Arabidopsis root apex. New Phytolol. 2013;197(4):1130–1141. doi: 10.1111/nph.12092. [DOI] [PubMed] [Google Scholar]
- Bengough AG, McKenzie BM, Hallett PD, Valentine TA. Root elongation, water stress, and mechanical impedance: A review of limiting stresses and beneficial root tip traits. J. Exp. Bot. 2011;62(1):59–68. doi: 10.1093/jxb/erq350. [DOI] [PubMed] [Google Scholar]
- Sia SK, Whitesides GM. Microfluidic devices fabricated in poly(dimethylsiloxane) for biological studies. Electrophor. 2003;24(21):3563–3576. doi: 10.1002/elps.200305584. [DOI] [PubMed] [Google Scholar]
- Millet LJ, Stewart ME, Sweedler JV, Nuzzo RG, Gillette MU. Microfluidic devices for culturing primary mammalian neurons at low densities. Lab chip. 2007;7(8):987–994. doi: 10.1039/b705266a. [DOI] [PubMed] [Google Scholar]
- Nelson BK, Cai X, Nebenführ A. A multicolored set of in vivo organelle markers for co-localization studies in Arabidopsis and other plants. Plant J. 2007;51(6):1126–1136. doi: 10.1111/j.1365-313X.2007.03212.x. [DOI] [PubMed] [Google Scholar]
- Talbot MJ, White RG. Cell surface and cell outline imaging in plant tissues using the backscattered electron detector in a variable pressure scanning electron microscope. Plant Methods. 2013;9(1):40. doi: 10.1186/1746-4811-9-40. [DOI] [PMC free article] [PubMed] [Google Scholar]


