Abstract
The purpose of this study was to investigate the distribution of Chlamydia trachomatis (CT) genotypes in infective diseases of the female lower genital tract, especially in cervical diseases.
This study included 128 CT-positive women. DNA was extracted from cervical swabs. Omp1 gene PCR-RFLP and sequencing were used to confirm the subtypes of CT. The association of subtypes with age, clinical symptoms, cervical cytology, and biopsy results was further analyzed.
Omp1 gene PCR-RFLP and sequencing showed that the order of prevalent CT genotypes in the female lower genital tract was D (n=38, 29.69%), followed by E (n=28, 21.88%), G (n=21, 16.41%), and F (n=16,12.50%). Genotypes J, H, and K were comparatively rare. Genotype I was not identified in our samples. Further analysis showed that patients with genotype G were more frequently co-infected with other bacteria. Genotype G was also associated with mucopurulent cervicitis (MPC) and cervical intraepithelial neoplasia (CIN). Patients with genotype E were commonly co-infected with HR-HPV. Although genotype D was the most prevalent, it was a relatively low-risk type.
These results provide information on distribution of CT genotypes in infective diseases of the female lower genital tract, which is instrumental to developing a vaccine for CT.
MeSH Keywords: Chlamydia trachomatis; Genotype; Sequence Analysis, Protein
Background
Chlamydia trachomatis (CT) is the most common sexually transmitted disease (STD), accounting for 40% of all STDs [1]. In 2011, an epidemiological survey in England showed that CT was the most prevalent STD, and exceeded gonorrhea and syphilis infection, especially in young people under the age of 25 [2]. In the United States, the infection rate of CT in 2006 was 362.3/100 000, and increased to 452.6/100 000 in 2010, which suggest that 3–4 million new cases were infected in 5 years [3]. After CT infection, males manifest nongonococcal urethritis, post-gonorrhea urethritis epididymitis, and prostatitis. For females, cervicitis, urethritis, and pelvic inflammation are commonly involved. In fact, for some patients there are no obvious symptoms or there are only mild symptoms after CT infection. If the patient does not get timely diagnosis and treatment, serious complication may follow, such as infertility, ectopic pregnancy, pneumonia, and neonatal conjunctivitism. It is also a risk factor for cervical cancer [4].
MOMP is the main antigen of CT. Studies showed that Ompl, the encoding gene of MOMP, has a different structure. According to the difference, CT was divided into 15 subtypes: A, B, Ba, C, D, E, F, G, H, I, J, K, L1, L2, and L3. Clinical studies showed that different subtypes have different histotropism and pathogenicity. Genotypes A–C are related to conjunctivitis, which is a cause of blindness. Genotypes D–K cause urogenital canal infection, such as urethritis, epididymitis in men, and cervicitis in women. Lymphogranuloma venereum is induced by genotypes L1–3 [5].
In the present study, we used PCR-RFLP and sequencing to examine the subtypes of CT in outpatients. The prevalence of different subtypes and the relationship with clinical characteristics were analyzed.
Material and Methods
Study population
From January 2010 to May 2015, 603 patients who received cervical cancer screening in Liuzhou People’s Hospital were examined for cytology, HPV, and CT. Ages ranged from 20 to 70 years old. Exclusion criteria were: 1) pregnant women, 2) taking sex hormone drugs during the most recent 6 months, 3) taking antibiotics in the most recent 1 month, and 4) pathological analysis showing cervical cancer. Finally, 128 CT-positive patients were recruited into this study and further analyzed. Informed consent was obtained from all participants and the study was approved by the Ethics Committee of Liuzhou People’s Hospital.
DNA extraction
The samples were centrifuged at 12 000 rpm for 30 min. The supernatant was removed and 20 μl protein K (TIANGEN, Beijing) was added, followed by incubation for 10 min at 37°C. TIAN amp Bacteria DNA Kit (TIANGEN, Beijing) was used to extract genomic DNA.
PCR
The following primers were used to amplify Ompl gene: Forward primer: 5′-GCCGCTTTGAGTTCTGCTTCCTC-3′, Reverse primer: 5′-ATTTACGTGAGCAGCTCTCTCAT-3′ (Invitrogen). PCR was performed in a 50 μl reaction volume containing 45 μL PCR Mix (Invitrogen), including 22 mM Tris-HCl (pH 8.4), 55 mM KCl, 1.65 mM MgCl2, 220 μM dGTP, 220 μM dATP, 220 μM dTTP, 220 μM dCTP, recombinant Taq DNA polymerase 22 U/ml, 2 μl primers, 1 μl dNTP, and 2 μl H2O. The thermal cycling conditions were 94°C for 2 min, followed by 35 cycles at 94°C for 30 s, 62°C for 30 s, 72°C for 1 min, and a final extension of 72°C for 5 min. We used 5 μl PCR product to run 1% agarose gel, and the size of Ompl gene was 1100 bp. The PCR product was used for sequencing.
Restricted fragment length polymorphisms (RFLP)
We digested 10 μl PCR product by AluI. A 20 μl reaction volume, containing 2 μl NEBuffer 4, 0.5 μl AluI, 10 μl PCR product, and 9.5 μl H2O, was incubated at 37°C for 3 h and 70°C for 10 min. HpaII+ EcoRI+ HinfI digestion system was also conducted, containing 2 μl NEBuffer 4, 0.5 μl HpaII/0.5 μl EcoRI/0.5 μl HinfI, 10 μl PCR product, and 7.5 μl H2O at 37°C for 3 h and 70°C for 10 min. We used 10 μl digestion product to run 2% agarose gel.
Statistical analysis
SPSS 17.0 statistical software was used to analyze the data. The chi-square test was used to analyze enumeration data. For other data, Fisher’s exact test was used. A significant difference was defined at p values less than 0.05.
Results
Age and clinical symptom distribution of patients
Analysis of the age distribution showed that CT-positivity was highest in patients 20–30 years old, followed by those 31–40 years old, and was low in patients who were under age 20 and those who were over age 40 (Table 1). Leucorrhea abnormality and contact bleeding were the main clinical symptoms (Table 2).
Table 1.
Age distribution of CT positive women.
| Age (yeas old) | Number | % |
|---|---|---|
| <21 | 7 | 5.47% |
| 20–30 | 57 | 44.53% |
| 31–40 | 43 | 33.59% |
| >40 | 21 | 16.41% |
Table 2.
The clinical symptom distribution in CT positive patients.
| Clinical symptom | Number | % |
|---|---|---|
| Leucorrhea abnormality | 42 | 32.81% |
| Contact bleeding | 16 | 12.50% |
| CT positive in sexual partner | 12 | 9.37% |
| Urinary tract infection | 7 | 5.47% |
| Abdominal pain | 9 | 7.03% |
| Physical examination | 35 | 27.34% |
| Others | 7 | 5.47% |
CT genotype distribution and association with clinical characteristics
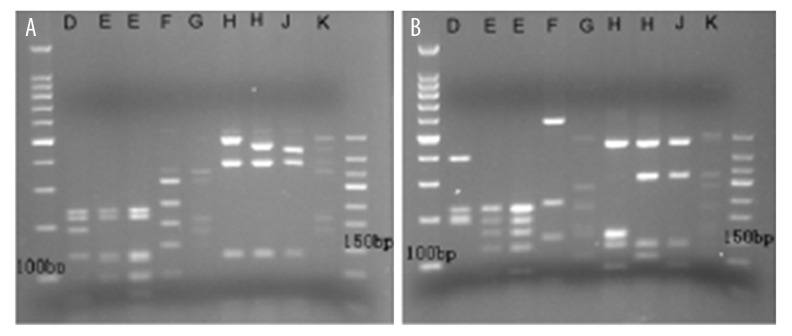
Through comparing the sequencing and RFLP, we determined the CT genotype for each patient. Figure 1 shows a typical image of RFLP. The analysis of Ompl genotype distribution showed that genotype D was the most common subtype in outpatients (n=38, 29.69%), followed by genotype E (n=28, 21.88%), G (n=21, 16.41%), and F (n=16, 12.50%). Genotypes J, H, and K were comparatively rare. Genotype I was not identified in our samples. There was no obvious difference among CT genotypes in clinical symptoms or age distribution. Further analysis showed that patients with genotype D were less commonly co-infected with other bacteria (11.76%/41.56%, P=0.000, OR 0.316, 95%CI 0.147–0.677) and that infection with genotype D was not related to cervical pathological changes (14.63%/36.78%, P=0.013, OR 0.406, 95%CI 0.186–0.884). However, patients with genotype G was more frequently co-infected with other bacteria (27.45%/9.09%, P=0.006, OR 1.928, 95%CI 1.293–2.874). Genotype G infection was also associated with MPC (27.45%/9.09%, P=0.045, OR 1.499, 95%CI 1.071–2.096). The cervical pathological changes induced by genotype G were also higher than with other genotypes (26.83%/11.49%, P=0.029, OR 1.868, 95%CI 1.124–3.106).
Figure 1.
RFLP restriction patterns of different CT genotypes. (A) RFLP restriction patterns of different CT genotypes after digestion of AluI. (B) RFLP restriction patterns of different CT genotypes after digestion with HpaII, EcoRI, and HinfI.
Patients with genotype F were also commonly co-infected with other bacteria (19.61%/7.79%, P=0.048, OR 1.707, 95%CI 1.087–2.680). Patients with genotype E were commonly co-infected with HR-HPV (35.19%/12.16%, P=0.002, OR 1.939, 95%CI 1.340–2.805) (Table 3).
Table 3.
CT genotype distribution and association with clinical characters.
| Number | CT genotype (%) | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| D | E | F | G | H | I | J | K | ||
| Age | |||||||||
| ≤30 years old | 64 | 31.58 | 22.81 | 10.53 | 15.79 | 3.51 | 0.00 | 10.53 | 5.26 |
| <30 years old | 64 | 33.33 | 19.61 | 13.73 | 11.76 | 9.8 | 0.00 | 5.88 | 7.84 |
| Clinical charactera | |||||||||
| No | 52 | 26.92 | 26.92 | 13.46 | 13.46 | 5.77 | 0.00 | 5.77 | 7.69 |
| Yes | 76 | 31.58 | 18.42 | 11.84 | 18.42 | 5.26 | 0.00 | 10.53 | 3.95 |
| Cervix | |||||||||
| Smooth | 62 | 33.87 | 24.19 | 11.29 | 9.68c | 8.06 | 0.00 | 8.06 | 4.84 |
| Mucopurulent | 66 | 25.76 | 19.7 | 13.64 | 22.73 | 3.03 | 0.00 | 9.09 | 6.06 |
| Infectionb | |||||||||
| No | 77 | 41.56d | 20.78 | 7.79e | 9.09e | 6.69 | 0.00 | 10.39 | 3.9 |
| Yes | 51 | 11.76 | 23.53 | 19.61 | 27.45 | 3.92 | 0.00 | 5.88 | 7.84 |
| Pathological change | |||||||||
| Nomal | 87 | 36.78f | 24.14 | 10.34 | 11.49f | 4.6 | 0.00 | 6.9 | 5.75 |
| CIN | 41 | 14.63 | 17.07 | 17.07 | 26.83 | 7.32 | 0.00 | 12.19 | 4.88 |
| HR-HPV | |||||||||
| Negative | 74 | 40.54g | 12.16g | 8.11 | 18.92 | 6.76 | 0.00 | 8.11 | 5.41 |
| Postive | 54 | 14.81 | 35.19 | 18.52 | 12.96 | 3.7 | 0.00 | 9.26 | 5.56 |
no clinical symptoms, no CT positive in sexual partner and no other lower genital tract infection;
co-infected with other bacteria and protozoa;
there was a distribution difference between genotype G and cervix change (27.45%/9.09%, P=0.045, OR 1.499, 95%CI 1.071–2.096);
genotype D was less co-infected with other bacteria (11.76%/41.56%, P=0.000, OR 0.316, 95%CI 0.147–0.677;
genotype G and F were frequently co-infected with bacteria (27.45%/9.09%, P=0.006,OR 1.928,95%CI 1.293–2.874) and (19.61%/7.79%, P=0.048, OR 1.707, 95%CI 1.087–2.680);
The cervical pathological changes induced by genotype G was also higher than other genotypes (26.83%/11.49%, P=0.029,OR 1.868, 95%CI 1.124–3.106); Genotype D was not related to the cervical pathological changes(14.63%/36.78%, P=0.013, OR 0.406, 95%CI 0.186–0.884);
Genotype E was frequently co-infected with HR-HPV.(35.19%/12.16%, P=0.002, OR 1.939, 95%CI 1.340–2.805).
Discussion
Chlamydia trachomatis infection in the genital tract is believed to be the most prevalent sexually transmitted disease. For females, occurrence of serious complications, such as infertility and ectopic pregnancy, is significantly associated with CT infection. Long-term CT infection is also a risk factor for genital tract tumors [6]. Comprehensive studies worldwide show there are different distributions of CT genotypes. There are differences in region, ethnicity, gender, and sexual orientation. However, genotype E is dominant, and the positive rates of D, F, and G are also high [7]. In the present study, we examined the genotype distribution of CT and found that D was the most prevalent CT genotype in the female lower genital tract, followed by E, G, and F, which is similar to results of a survey conducted in the United States [8].
In methodology, MOMP monoclonal antibody and polyclonal antibody and Ompl gene polymorphism are used to classify the subtype of CT [9]. The Ompl genotype examination is more sensitive and specific than serological typing. In fact, serological typing lags behind the reaction of the infection site [10]. There are also no commercial reagents available with which to perform CT typing. PCR-RFLP is widely used to analyze the gene sequence and gene polymorphism [8]. In the present study, we used PCR-RFLP and sequencing to confirm the genotype of CT, and found that it is a stable method for detecting the genotype of CT.
CT infection has different histotropism and clinical characteristics, depending on different genotypes. To analyze the association of subtypes with age, clinical symptoms, cervical cytology, and biopsy results, we observed the common clinical characteristics of CT-positive patients, fining that they included leucorrhea abnormality, contact bleeding, positive CT in a sexual partner, urinary tract infection, and abdominal pain. Physical examination showed purulent vaginal secretions, flush on cervical surface, and easy bleeding. Some patients did not show any signs and physical examination showed normal secretions and cervix. The different clinical characteristics may be related to the molecular structure of different genotypes [11].
We further analyzed the relationship between genotypes and clinical characteristics. We found that patients with genotype D were less likely to be co-infected with other bacteria, and having this genotype was not related to cervical pathological changes. However, patients with genotype G were more frequently co-infected with other bacteria. Genotype G infection was also associated with mucopurulent cervicitis. The cervical pathological changes induced by genotype G were also greater than in patients with other genotypes. Patients with genotype E were more likely to be co-infected with HR-HPV [12]. As HR-HPV is the most common cause of CIN and is major etiologic agent of cervical cancer, it seems that genotype E should be associated more frequently with CIN. In fact, the different subtypes of HPV had different relationships with cervical cancer. In the present study, we did not examine the subtype of HPV and did not analyze the influence of different subtypes. We found that the patients with genotype G infection commonly had cervical cytology abnormity (CIN/non-CIN=26.83%/11.49%, P=0.029, OR 1.868, 95%CI 1.124–3.106), suggesting that genotype G may be the pathogenic factor in HR-HPV-negative patients. Patients with genotype E were commonly co-infected with HR-HPV, but genotype E infection was not significantly related to the CIN, suggesting that genotype E may be not the pathogenic factor. We also found that patients with genotype F were also commonly co-infected with other bacteria. Some studies using serum antibodies found a high rate of infection with other bacteria and protozoa in CT-F-positive vaginal secretions. Thus, these results are also similar to those of our study.
In addition, we analyzed the age distribution of individuals with CT and found that that rates of infection were highest in those 20–30 years old, followed by those 31–40 years old. Rates of infection were low in those under age 20 and in those over age 40. Generally, people 20–40 years old are sexually active, so the prevalence of CT in this age group is in line with other sexually transmitted diseases. It is also consistent with the report by Detels [13]. There was no obvious genotype difference among different age groups.
Conclusions
Although antibiotics play an important role in treatment of CT infection, regional breakout frequently occurs, such as CT-L2 [14]. The development of a vaccine is a promising way to prevent CT infection [15]. This epidemiological investigation shows the distribution of different genotypes of CT and will be instrumental in screening and prevention.
Footnotes
Conflict of interest
The authors declare no conflict of interest.
Source of support: This work was supported by the Natural Science Foundation of Guangxi (Z20090289)
References
- 1.Stephens AJ, Aubuchon M, Schust DJ. Antichlamydial antibodies, human fertility, and pregnancy wastage. Infect Dis Obstet Gynecol. 2011;2011:525182. doi: 10.1155/2011/525182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Health Protection Agency. Health Protection Report. 2011. Sexually transmitted infections in England. [Google Scholar]
- 3.Centers for Disease Control and Prevention. Sexually Transmitted DiseaseSurveillance. Atlanta, GA: US Department of Health and Human Services; 2010. [Google Scholar]
- 4.Silva J, Cerqueira F, Medeiros R. Chlamydia trachomatis infection: implications for HPV status and cervical cancer. Arch Gynecol Obstet. 2014;289:715–23. doi: 10.1007/s00404-013-3122-3. [DOI] [PubMed] [Google Scholar]
- 5.Quint KD, Bom RJ, Bruisten SM, et al. Comparison of three genotyping methods to identify Chlamydia trachomatis genotypes in positive men and women. Mol Cell Probes. 2010;24:266–70. doi: 10.1016/j.mcp.2010.04.007. [DOI] [PubMed] [Google Scholar]
- 6.Alibek K, Karatayeva N, Bekniyazov I. The role of infectious agents in urogenital cancers. Infect Agent Cancer. 2012;7:35. doi: 10.1186/1750-9378-7-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gharsallah H, Frikha-Gargouri O, Sellami H, et al. Chlamydia trachomatis genovar distribution in clinical urogenital specimens from Tunisian patients: high prevalence of C. trachomatis genovar E and mixed infections. BMC Infect Dis. 2012;12:333. doi: 10.1186/1471-2334-12-333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gao X, Chen XS, Yin YP, et al. Distribution study of Chlamydia trachomatis serovars among high-risk women in China performed using PCR-restriction fragment length polymorphism genotyping. J Clin Microbiol. 2007;45:1185–89. doi: 10.1128/JCM.02076-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nunes A, Gomes JP. Evolution, phylogeny, and molecular epidemiology of Chlamydia. Infect Genet Evol. 2014;23:49–64. doi: 10.1016/j.meegid.2014.01.029. [DOI] [PubMed] [Google Scholar]
- 10.Silva J, Cerqueira F, Ribeiro J, et al. Is Chlamydia trachomatis related to human papillomavirus infection in young women of southern European population? A self-sampling study. Arch Gynecol Obstet. 2013;288:627–33. doi: 10.1007/s00404-013-2771-6. [DOI] [PubMed] [Google Scholar]
- 11.Nunes A, Borrego MJ, Gomes JP. Genomic features beyond Chlamydia trachomatis phenotypes: what do we think we know? Infect Genet Evol. 2013;16:392–400. doi: 10.1016/j.meegid.2013.03.018. [DOI] [PubMed] [Google Scholar]
- 12.Madeleine MM, Anttila T, Schwartz SM, et al. Risk of cervical cancer associated with Chlamydia trachomatis antibodies by histology, HPV type and HPV cofactors. Int J Cancer. 2007;120:650–55. doi: 10.1002/ijc.22325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Detels R, Green AM, Klausner JD, et al. The incidence and correlates of symptomatic and asymptomatic Chlamydia trachomatis and Neisseria gonorrhoeae infections in selected populations in five countries. Sex Transm Dis. 2011;38:503–9. [PMC free article] [PubMed] [Google Scholar]
- 14.Hafner LM, Wilson DP, Timms P. Development status and future prospects for a vaccine against Chlamydia trachomatis infection. Vaccine. 2014;32:1563–71. doi: 10.1016/j.vaccine.2013.08.020. [DOI] [PubMed] [Google Scholar]
- 15.Brunham RC, Rappuoli R. Chlamydia trachomatis control requires a vaccine. Vaccine. 2013;31:1892–97. doi: 10.1016/j.vaccine.2013.01.024. [DOI] [PMC free article] [PubMed] [Google Scholar]