Abstract
Bacterial biofilms are surface-associated communities that are vastly studied to understand their self-produced extracellular polymeric substances (EPS) and their roles in environmental microbiology. This study outlines a method to cultivate biofilm attachment to the System for Analysis at the Liquid Vacuum Interface (SALVI) and achieve in situ chemical mapping of a living biofilm by time-of-flight secondary ion mass spectrometry (ToF-SIMS). This is done through the culturing of bacteria both outside and within the SALVI channel with our specialized setup, as well as through optical imaging techniques to detect the biofilm presence and thickness before ToF-SIMS analysis. Our results show the characteristic peaks of the Shewanella biofilm in its natural hydrated state, highlighting upon its localized water cluster environment, as well as EPS fragments, which are drastically different from the same biofilm's dehydrated state. These results demonstrate the breakthrough capability of SALVI that allows for in situ biofilm imaging with a vacuum-based chemical imaging instrument.
Keywords: Chemistry, Issue 126, SALVI, ToF-SIMS, biofilm, in situ, molecular imaging, microfluidics
Introduction
Bacterial biofilms are surface-associated communities which have evolved over time as a defense for bacteria to survive varying adverse physical and mechanical stimuli, wherein cells are able to attach and survive in many possible environments.1,2 Biofilms are vastly investigated and have applications in many fields such as biomedicine, biomedical engineering, agriculture, and industrial research and development.1,2 Understanding the chemical mapping of these complex microbial communities, including their self-produced extracellular polymeric substances (EPS) and their local water-cluster environment, is essential to gaining an accurate and detailed depiction of their biological activities.2
Biofilms exist and grow within a highly hydrated state. This presents a great challenge in using vacuum-based surface analysis techniques such as time-of-flight secondary ion mass spectrometry (ToF-SIMS) due to the difficulty in studying volatile liquids in vacuum. As a result, vacuum-based surface analysis techniques have been limited almost exclusively to studying biofilm samples at only their dried state. However, studying a biofilm in its dried state inhibits the accurate investigation of its true biological microenvironment. It often causes drastic changes to the EPS integrity and biofilm morphology, which has been demonstrated after comparing dry biofilm mass spectral results to in situ liquid studies.3,4 This article presents a solution for studying biofilms within their natural hydrated state by employing the use of our System for Analysis at the Liquid Vacuum Interface (SALVI),5,6 a microfluidic reactor that contains liquid under its thin silicon nitride (SiN) membrane in a microchannel made of polydimethylsiloxane (PMDS), thus providing direct access to the secondary ion probe beam while still maintaining the structural integrity of the liquid matrix within a vacuum chamber.7,8
S. oneidensis MR-1 mutated to express green fluorescence protein (GFP) was chosen as a model organism for this biofilm procedure illustration due to its metabolic versatility and common use in environmental and applied microbiology, which was based heavily on its unique capability for metal reduction and extracellular electron transfer.9,10,11 Additionally, the presence of GFP allowed for easy continuous biofilm-thickness monitoring through fluorescence microscopy, using a fluorescein isothiocyanate (FITC) filter. Our previous studies have shown evidence of this bacteria favoring attachment to the SiN window using in operando fluorescence imaging for biofilm growth to a thickness of up to one hundred micrometers.4,12 While this paper will only discuss the confirmation of biofilm's presence through fluorescence microscopy, the SALVI is compatible with other optical imaging methods such as super-resolution fluorescence imaging (i.e., structured illumination microscopy (SIM)9) and confocal laser scanning microscopy (CLSM) imaging4). Optical imaging can serve to measure the biofilm thickness, and obtain a 3D image of, the shape of the biofilm as it appears, confirming its thickness and its attachment to the SiN window.9 While GFP was used in the SIMS analysis, S. oneidensis without GFP was used for the growth curve, as this only required measurement of optical density and did not require any fluorescent imaging. Generally, the difference between the GFP tagged and untagged species in the growth curve is insignificant. Additionally, while this protocol uses S. oneidensis MR-1 GFP as a model organism to describe the procedure, this procedure is designed for any bacterial strain that may be needed for cultivation within SALVI. Although, given knowledge of the bacterial strain needed, some growth conditions such as time, temperature, and oxygen environment may need to be modified to accommodate the strain of bacteria to be used. For growth medium, this procedure uses "nanowires" medium, tryptic soy broth (TSB) without dextrose, and tryptic soy agar (TSA) without dextrose for culturing. The composition of "nanowires" medium has been specially formulated for the growth and for monitoring of extensions of the membrane and periplasm of S. oneidensis that appear to take the shape of small wires, and the medium composition has been established within previous research.13,14
Our previous protocol on in situ liquid ToF-SIMS has illustrated the benefit that SALVI has to offer for protein immobilization and attachment to the SiN, as well as a detailed protocol on ToF-SIMS analysis and data reduction.12 Rather than reiterate data reduction steps, this paper will serve to instead focus on the unique approach of setting up and cultivating biofilms within our SALVI microchannel, as well as the imaging steps to detect biofilm presence and thickness prior to ToF-SIMS analysis. While biofilms have been previously limited to only dried samples within the chamber of vacuum-based surface analytical techniques, detailed EPS and biofilm chemical mapping of live biofilms can now be obtained in situ because of this new capability.
Protocol
1. Preparation of Materials
- Preparation of Medium Tubing
- Serum Bottles (one needed per biofilm culture and three needed per growth curve) Note: As mentioned in the introduction, any growth medium suitable to provide the nutrients needed for the strain of bacteria of interest can be utilized for this procedure; in this case, "nanowires" media and TSB without dextrose medium was used for the growth of S. oneidensis MR-1 GFP.13
- Deposit 20 mL of growth medium into one 70 mL serum bottle, cap the stopper and crimp the bottle. Cover the top with a piece of clean sterile aluminum foil.
- Autoclave the bottle with a liquid cycle for 30 min at 121 °C. Afterward, store the sterilized serum bottle at room temperature.
- Anaerobic Culture Tubes
- Deposit 5 mL of growth medium into an anaerobic culture tube with air headspace, put on the stopper and crimp the bottle. Cover the top with sterile foil.
- Preparing Bubble Trap Tubing
- Using a 22 gauge needle, carefully punch a hole through a serum bottle rubber stopper.
- Cut 20 in of 1/32" polytetrafluoroethylene (PTFE) tubing with a razor such that one end of the segment of tubing is pointed.
- Using the pointed end, thread the PTFE tubing through the hole in the top portion of the rubber stopper. Push the tube through until roughly 2 cm is through the stopper and cut the pointed end of the PTFE tube with a razor to have a flat end.
- Remove the plunger of a 5 mL syringe and fit the rubber stopper from step 1.2.3 into the open end of the syringe. The 2 cm end of PTFE tubing is within the barrel of the syringe. This is the bubble trap.
- Wrap the bubble trap (the PTFE tubing, rubber stopper, and syringe) made in step 1.2.4 with aluminum foil and secure with autoclave tape. Autoclave the foil package to sterilize the tubing with a gravity cycle at 121 °C for 30 min. Store the sealed foil package at room temperature until ready for use in step 2.4.
- Bacterial Growth Curve NOTE: This protocol uses S. oneidensis MR-1 GFP as a model organism for growing the biofilm in SALVI. However, this procedure can be adapted to accommodate other bacteria growing aerobically or anaerobically. Depending on which strain is used, growth time and environment may need to be adjusted accordingly. NOTE: -80 °C bacterial freezer stocks consisted of a 1:1 mixture of TSB without dextrose and glycerol. The growth curve example shown in Figure 1B was prepared using a starter culture of TSB with no dextrose, transferred in respective 0.2 mL quantities three times to 70 mL serum bottles with 20 mL "nanowires" medium to remove traces of TSB. However, this protocol assumes that only one growth medium will be used, and that subsequent transfers will not be conducted, as this was done with these particular medium solutions for S. oneidensis. Although not outlined in this protocol, extra transfers like this are recommended as initially depositing the bacteria to the rich TSB helps the frozen cells recover; and three subsequent transfers to "nanowires" ensures that all TSB is gone and that typical growth dynamics are occurring.
- Inoculating the Starter Culture
- Remove the bacterial glycerol stock from the -80 °C freezer and warm the stock with a gloved hand to thaw as quickly as possible. Move this freezer stock to the biological safety cabinet (BSC).
- Within the BSC, use a 1 mL syringe with a 22G needle attached to transfer 0.1 mL of freezer stock to a capped and crimped anaerobic culture tube containing 5 mL of growth medium with no dextrose, prepared in step 1.1.2. Dispose of the remaining freezer stock in the appropriate biological waste container, as freezing again could shock the cells.
- Incubate the starter culture in a shaker/incubator at 30 °C at 150 rotations per minute (rpm) and be sure to take an optical density at 600 nm (OD600) reading when ready for use. Record the time of growth and OD600 such that it can be done at that same time for every subsequent use, such that results are reproducible. As an example, this starter culture was allowed to grow for 15 h and used at an OD600 of approximately 1.0.
- Inoculating Growth Medium
- Take the three serum bottles prepared in step 1.1.1.2, and the starter culture prepared in 1.3.1.3, and bring both to the BSC.
- Within the BSC, using a sterile 1 mL syringe with a 22G needle attached, transfer 0.1 mL of solution from the starter culture to each of the serum bottles, respectively.
- Sterilize the top of the serum bottles with 70% ethanol and cover with sterilized aluminum foil, label appropriately, and transfer to an orbital shaker within an incubator. Set the orbital shaker to 150 rpm and set the incubator to 30 °C. Incubate until the first OD600 data point is taken, within step 1.3.3.
- Obtaining OD 600 Data Points
- Prepare a blank by depositing 100 µL of filter-sterilized distilled deionized (DI) water into a sterile cuvette. Wrap plastic paraffin film around the top of the cuvette so that the water will not be contaminated. Store at room temperature. Use this blank with the UV/Vis Spectrophotometer before taking any data points for the growth curve by inserting the cuvette and pressing "blank". NOTE: Every 48 h a new blank should be prepared to avoid incorrect reading, as regularly replacing a blank is good practice.
- For each OD600 data point, remove the serum bottles prepared in step 1.3.2.3 from the incubator and transfer to a sterilized BSC. Take 0.1 mL of the inoculated medium from each serum bottle and deposit into three separately labeled sterile cuvettes.
- After blanking, insert cuvette containing the culture into the ultraviolet visible (UV/Vis) Spectrophotometer and read the OD600 one at a time, and record all three readings for that time point. NOTE: For S. oneidensis MR-1, data points were taken at 1, 14, 28, 32, 41, 57, 81, and 105 h after inoculation. The growth is complete when the bacteria has completed the death phase. These points should be adjusted accordingly if there is lack of knowledge on the particular growth time and tendency of bacteria used in this protocol. If there is uncertainty about the growth tendency, data points should be collected more frequently, for example, every three h for the first 12 h, every six h for subsequent 36 h, at 12 h intervals for the following 100 h, and at 24 h intervals for the final 124 h.
- Using the OD600 data points as the y-axis and the time as the x-axis, graph a curve of the average of the three points taken with standard deviation error bars for each time point using a graphing software. The S. oneidensis MR-1 growth curve is displayed in Figure 1B in the representative results section.
- Using the graph prepared in step 1.3.3.4, identify the timeframe of the log-phase section of the growth curve. For Shewanella, this is between 12 and 33 h of growth; therefore it can be inferred that at 24 h of growth, Shewanella will be within its log-phase of growth, assuming that the bacteria will be cultured using the same media and treatment before use that was used for the growth curve.
2. Culturing the Bacteria
- Day One: Inoculating the Agar Plate NOTE: This section of the procedure was used with an agarose plate to take one colony-forming unit (CFU) at log-phase, instead of the liquid starter culture used for the growth curve. This could be assumed to reproduce the same growth conditions, due to the fact that TSA without dextrose was used and subsequently transferred to "nanowires" medium in the growth curve procedure, and TSA has the same ingredients that comprise TSB.
- Remove S. oneidensis MR-1 GFPbacteria stock from -80 °C freezer and place into an ice bucket, place this bucket inside of sterilized BSC.
- Within the BSC, use a sterile 1 µL inoculation loop to scrape the surface of the frozen bacteria stock and use the loop to T-streak the surface of an agarose plate.
- After sealing the sides of the plate with plastic paraffin film, invert the plate and store in a 30 °C incubator for 24 h, until individual colonies appear.
- Day Two: Inoculating the Serum Bottle
- Remove the plate from the incubator and open within the BSC.
- Within the BSC, clean the surface of the rubber stopper on a prepared serum bottle from step 1.1.1 with 70% ethanol in DI water.
- Select an individual CFU from the agar plate and, using a sterile syringe with an attached 22 gauge needle, deposit enough growth media to dislodge the colony from the plate without touching any neighboring colonies and mix the colony with the medium with the tip of the needle. NOTE: A singly colony should be selected that is far enough away from other bacteria on the plate, such that medium can be deposited onto it without touching any other colonies.
- Using the same syringe, extract the liquid from the surface of the plate.
- After tapping out bubbles within the syringe, inject the liquid into the serum bottle, and place onto an orbital shaker within 30 °C set at 150 rpm for 24 h. NOTE: Tapping the bubbles out of the syringe is important in order to avoid introducing more oxygen to the serum bottle, as introducing more oxygen to the bottle can change experimental conditions and reduce consistency between growth times for bacteria.
- Day Two: Sterilization of the SALVI Microchannel NOTE: To promote sterility of the tubing system, steps that require detaching the syringe from the tubing and replacement with a new syringe should be done within the BSC. To do this, simply detach the syringe from the syringe pump by unscrewing the metal syringe holder, and bring the syringe with tubing attached to a sterile BSC. When tubing system is mentioned in the procedures, this refers to the syringe containing the liquid reservoir, attached with a polyetheretherketon (PEEK) injector to the PTFE tubing, attached to the drip chamber, which has a PEEK injector fitting attached to the SALVI inlet tubing, as well as the outlet container that the SALVI outlet tubing is sealed to.
- Take 2 mL of 70% ethanol into a syringe and connect to the PEEK fitting on the SALVI. After connecting the syringe to a syringe pump and attaching the outlet of the SALVI to a waste bottle, allow the ethanol DI water solution to run through the SALVI at 20 µL/min for 1.5 h.
- Take 4 mL of sterilized DI water into a syringe and connect to the inlet of the same SALVI. After connecting the syringe to a syringe pump, allow the water to run through the SALVI at 20 μL/min for at least 3 h.
- Within a BSC, open the foil packet prepared in step 1.2.5 and connect a sterile PEEK injection fitting to the end of the 5 mL syringe and sterile PEEK fittings to the end of the TFE tubing. Take ~ 3 mL of sterile medium into a syringe and attach to the TFE tube. Invert the 5 mL drip chamber and, using a sterile syringe, inject the growth medium into the drip chamber until it reaches a total volume of 1 mL.
- Connect the end of the 5 mL syringe drip chamber to the inlet of the SALVI.
- Take 10 mL of growth medium into a sterile syringe and connect to the inlet of the drip tubing. After connecting the syringe to a syringe pump, allow the medium to run through the SALVI at 20 µL/min for 12 h (or overnight). Use adhesive tape to secure these parts. Cover the outlet bottle with foil or plastic paraffin film to minimize the probability of dust particles and organisms contained therein to contaminate the medium. A detailed depiction of how this setup should look can be found in Figure 1A.
- Allow the drip tubing to be vertical, meaning that the syringe pump should be placed on an elevated surface with drip tubing secured with tape, and with the SALVI secured on a flat surface below.
- Run medium through the SALVI for 12 h to ensure that all traces of ethanol have been removed from the microchannel and tubing of the SALVI before inoculating with bacteria.
- For added protection, keep the SALVI microchannel chamber within a sterilized Petri dish, with the sides cut to fit the inlet and outlet tubing. Additionally, keep tape always on the window to protect the SiN membrane and channel prior to imaging analysis.
- Day Three: Inoculation of the SALVI Microchannel
- Remove the whole tubing system (syringe connected to drip chamber connected to SALVI connected to the waste bottle) from the syringe pump and bring it to a sterilized BSC. Additionally, remove the serum vial from step 2.2.5 from the incubator and bring it to the BSC.
- Clean the surface of the serum bottle stopper with 70% ethanol to prevent any contamination, and then use a sterile syringe with an attached sterile 22G needle to extract 4 mL of bacteria from the bottle.
- Detach the SALVI from the 5 mL chamber of the drip tubing, and connect the syringe with inoculated medium directly to the inlet of the SALVI. Leave the drip tubing within a sterile aluminum foil packet or within the BSC.
- Attach the syringe with SALVI and outlet bottle to the syringe pump to run at 20 µL/min for 3 h to inoculate the channel of the SALVI, in order to allow for multiple volume changes of the liquid contained within SALVI.
- After inoculation, disconnect the syringe with SALVI from the syringe pump and bring to the BSC. After attaching the inlet of the SALVI back to the 5 mL drip chamber from step 2.4.3, take 20 mL of growth medium into a sterile syringe and attach to the inlet of the drip tubing.
- Bring the tubing attached to the SALVI from the BSC to the syringe pump and allow the medium to pass through the tubing at a rate of 2 µL/min for six to ten days, or until fluorescence imaging (discussed in step 3) shows favorable visible biofilm growth for ToF-SIMS analysis. Refill fresh medium as it runs out within the BSC by filling a new sterile syringe with 10mL of growth medium and attaching to the SALVI after the previous growth medium has run out. NOTE: After beginning inoculation at 2 µL/min, the flow rate should never be increased or decreased, as changing the flow rate would create shear-stress within the channel and detach the biofilm. It is critical that this flow rate should never be changed; to prepare for this, ~24 h before SIMS, observe the tubing closely to make sure there are no bubbles so that there is no need to push more medium at a faster rate throughout the tubing.
- Avoid bubbles in the microchannel, as they can push the biofilm out of the channel. It is important to be cautious of bubbles within the bottom of the 5 mL drip tubing where it connects to the SALVI tubing. Fresh medium can be injected into the end of the PEEK injector to ensure that no air will be forced into the SALVI.
3. Optical Imaging of the Biofilm within the SALVI Microchannel
- Fluorescence Microscopy Imaging NOTE: The Shewanella used as a model organism within this protocol is mutated to express GFP; as such, it does not require staining. If the bacteria require staining, this should always be done at the same flowrate (e.g., 2 µL/min) to avoid biofilm detachment. Additionally, when cells go without nutrient availability, they will detach. Therefore, if any additional staining is required, the stain should be injected to the growth medium rather than water before supplying to stain the cells.
- Detach the SALVI from the drip tubing inside the BSC and close the SALVI by screwing the PEEK fittings to finger-tighten the PEEK union.
- Tape the tubing of the SALVI microchannel chamber securely onto a glass slide, such that the window is completely flat and facing upward. Remove the protective tape from the window. Clamp the slide to the stage of the microscope by placing the glass slide between the stage clamps on the platform.
- Lower the stage of the microscope such that the top of the SALVI is positioned close enough to the end of the 10X objective, where adjustment of the focus will not cause the lens to touch the window.
- Switch on the backlight and find the channel using the 10x microscope objective by adjusting the focus. At this time, make sure the presence of bacterial attachment to the SiN window of the SALVI is apparent in comparison to the window of a control channel with no inoculated bacteria. For closer imaging of cells, switch to the 20x objective. When compared to an empty SALVI channel, the presence of bacteria attached to the window should have a strong green fluorescence.
- Turn off the backlight and turn on the microscope mercury source. Wait 2-3 min and switch to the FITC filter set on the microscope to view and capture images of the biofilm attached to the window. As an example of what the matured biofilm will look like when ready, refer to Figure 1C.
- Turn off the mercury source and detach the SALVI from the slide. If needed, attach to 2 µL/min medium flow once again to allow for more growth before imaging again, or transfer the SALVI to secondary containment use for ToF-SIMS analysis. This is needed if biofilm thickness is concluded to not be thick enough, and more time for growth is required before ToF-SIMS analysis. To attach to medium flow once again, refer to step 2.3.6. NOTE: If put back under medium flow, fluorescence imaging should be done again before ToF-SIMS analysis.
- Give the biofilm growth medium to feed it until ToF-SIMS analysis can be conducted. While not recommended, if needed, the closed SALVI can be stored within 4 °C overnight, but should be allowed to warm to room temperature before inserting to the vacuum chamber of the ToF-SIMS. However, analyze the biofilm immediately after detached from medium flow to reduce biofilm detachment.
4. ToF-SIMS Data Acquisition
NOTE: This section serves as an overview, and is discussed in greater detail within our earlier protocol describing adsorbed protein molecule liquid SIMS analysis.12
- Install SALVI into the ToF-SIMS Loadlock Chamber NOTE: Gloves should be worn at all times when handling the SALVI device and installing it onto the ToF-SIMS stage to avoid potential contaminations during surface analysis.
- Mount SALVI onto the stage and fix it with several screws. Make sure that the SiN window is flat by adjusting screw tightness and inserting silicon wafer pieces at the bottom of the PDMS microchannel chamber. Remove the protective tape from the window, then load the stage into the ToF-SIMS loadlock chamber.
- Open the ToF-SIMS loadlock, hold and set the stage horizontally onto the loading platform, and close the loadlock door. Initiate the vacuum pump and wait to ensure that suitable vacuum is reached.
- Move the SALVI into the main chamber once the vacuum is stabilized to 1x10-7 mbar.
- Depth Profiling and Collecting Data Points
- Find the microfluidic channel using the optical microscope equipped in ToF-SIMS.
- Select positive or negative mode before data acquisition. Use the 25 keV Bi3+ beam as the primary ion beam in all measurements, and use the electron flood gun to neutralize the surface charging during all measurements.
- Scan the Bi3+ beam with 150 ns pulse width on a round area with a diameter of ~2 µm with 64 pixels by 64 pixels resolution. After punching through the 100 nm SiN window, continue to scan for another 150 s to collect high intensity data for imaging. After punch-through, the counts will increase significantly within the depth profiling region. After stabilizing, this can be referred to as the high-intensity region.
- Reduce the pulse width to 50 ns for data collection to acquire spectra with better mass resolution. Continue this acquisition for about another 200 s.
- Repeat these steps for collecting at least three positive and three negative data points. NOTE: Be sure to space the punch-through areas such that the SiN window will not break and leak into the chamber of the vacuum from compromised window integrity.
5. ToF-SIMS Data Analysis
- Mass Calibration using IonToF Software
- Open the analysis software of ToF-SIMS (Measurement Explorer) and then click the "profiles", "spectra" and "image" buttons, respectively, to process depth profile, m/z spectrum, and image data.
- Open the data files obtained throughout ToF-SIMS data acquisition.
- Select the depth profiling data of interest and reconstruct the spectra according to the depth profile temporal series.
- Press "F3" to open the "mass calibration" window. Choose peaks for calibration based on chemical compounds which are expected to exist in the specific sample.
- Peaks of Interest Selection
- As peak selection is necessary for sample analysis, determine characteristic peaks of the sample according to literature or other previous findings, if any. Compare the intensity of peaks of interest in the m/z spectra. If necessary, add new peaks or delete interference peaks in the peak list.
- Click on a peak, find the red lines at the left bottom window. Move the red lines to enclose the whole peak.
- Select a peak matching the chemical formula in the main spectrum window, and then to click the "add peak" button above the window.
- Export Mass Calibrated Data
- To export a peak list, in the "Peak List" menu, select "Save…" the peak list in the "itmil" format.
- To export a depth profile, in the "Profile" window, click on the "File" menu, then select "Export", and then select "Save as" a ".txt" file.
- To export an m/z spectrum file, in the "Spectra" window, click on the "File" menu, then select "Export", and then select "Save as" a ".txt" file.
- To export an image file, in the "Image" window, print screen and save as the image file.
6. ToF-SIMS Data Plotting and Presentation
Use a graphic tool to import the data.
Make a plot using the m/z as the x-axis and peak intensity as the y-axis to show the reconstructed spectrum. An example is given in Figure 2A.
Combine reconstructed two-dimensional (2D) images of different m/z and form a matrix to show either positive or negative ion mapping. An example is given in Figure 2B.
Representative Results
These representative results serve to show how the chemical profile of the attached biofilm can be identified and interpreted, as obtained through ToF-SIMS. After plotting mass spectra from ToF-SIMS data acquisition, highlighted briefly in the procedures section, peak identification should be conducted in order to assign identities to each respective m/z value. This can be done through extensive literature review on mass spectrometry on bacteria and specific chemical fragments that are expected to be present within the bacteria studied, such as various water clusters, fatty acids, and protein fragments.16,17,18,19,20 These representative results only show the negative mass spectra as obtained by the S. oneidensis MR-1 biofilm and a DI water sample. Interpretation of positive spectra follow a similar procedure.
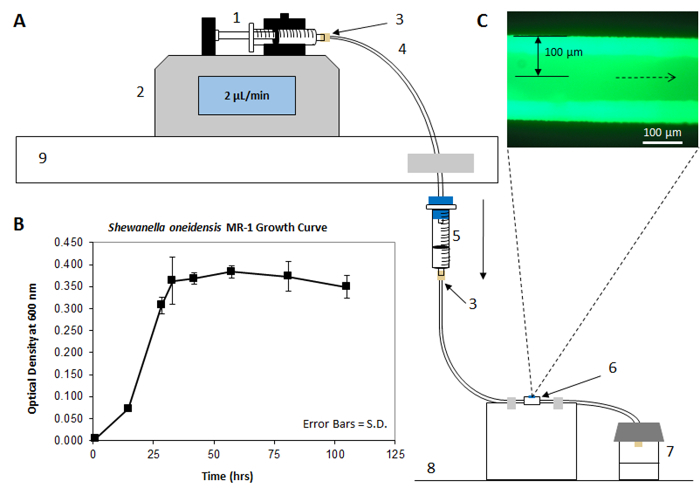
Figure 1A shows the schematic setup according to this protocol when cultivating a biofilm in the SALVI microchannel. On the top left, the syringe is attached to a syringe pump, which keeps medium flowing at a constant rate throughout the PFTE tubing system and inside the microchannel. The syringe pump is placed above the drip chamber, which prevents backwards contamination to the medium reservoir to prevent motile organisms from swimming up-stream. The medium flows from the drip chamber reservoir directly into the PFTE tubing of the SALVI, which passes through the microchannel and through the outlet tubing to a sealed outlet bottle. The dotted lines point from the SiN window to an image of a matured S. oneidensis MR-1 GFP cells captured by fluorescence microscopy, detailed in step 3.1. Figure 1B shows an example of a growth curve established using the model organism for this protocol, S. oneidensis MR-1. From the growth phases in this graph, it can be concluded that the log-phase of growth occurs between 15-32 h in these specific cultivation conditions. Additional information from this graph shows that 0-15 h is the lag phase of growth, and 33-105 h represents the stationary phase of growth. Figure 1C is an image acquired with fluorescence microscopy showing an example of a matured biofilm.
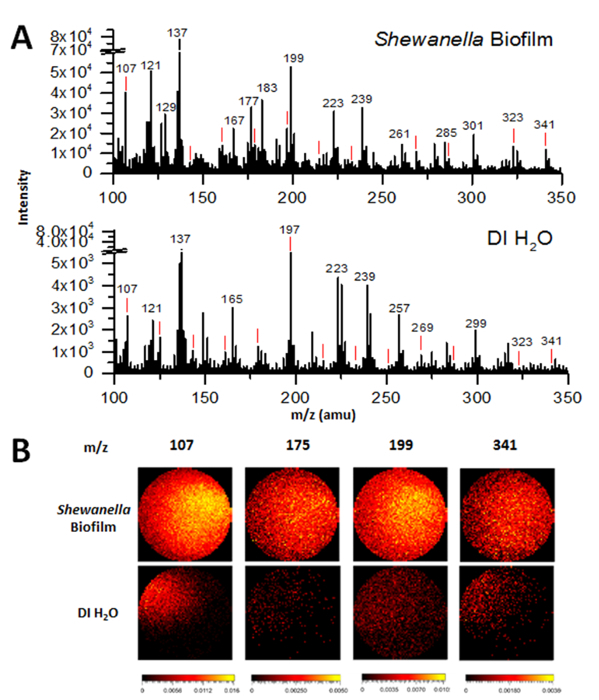
Figure 2A shows the negative mass spectra of a hydrated S. oneidensis MR-1 biofilm within the microfluidic channel, as obtained by in situ liquid ToF-SIMS. This graph shows an example of the selected mass range (m/z 100-350) comparison between the biofilm of S.oneidensis MR-1 and DI water, the latter was obtained as a system control. Figure 2B displays a comparison of 2D images of peaks of interest in the MR-1 biofilm and DI water obtained by ToF-SIMS. Once interesting m/z values can be identified from studying the mass spectra, potential peak identification of m/z values are properly attributed to chemical fragments, as supported by the IonToF SIMS software library and literature survey. Peaks of interest in Figure 2A include water clusters (i.e., m/z 107 (H2O)5OH-, 125 (H2O)6OH-, 143 (H2O)7OH-, 161 (H2O)8OH-, 179 (H2O)9OH-, 197 (H2O)10OH-, 215 (H2O)11OH-, 233 (H2O)12OH-, 251 (H2O)13OH-, 269 (H2O)14OH-, 287 (H2O)15OH-, 323 (H2O)17OH-, 341 (H2O)19OH-))9, as denoted by red lines above respective peaks, quorum sensing related compounds biomarkers (i.e., m/z 175 C10H9NO2-), EPS byproducts (m/z 123 C2H4PO4-, 159 P2O8H-, 216 Cr2O7-, 285 octadecanoethiols, 325 polar compounds, C12 Polar compounds)15,16,18, and fatty acid chain fragments (i.e. m/z 127 [C2H3(CH2)3COO]-, 255 C16:0 fatty acid, 279 linoleic acid, C18H31O2-, 325 C21H40O2-, 341 surface lipids, 18-MEA bound through thioester linkage, stearic acid C18H35O2-, ions of monoacylglyceryls of palmitic acid C19H17O6-).16,19
Since the biofilm is hydrated, it is not surprising that some peaks seen in the biofilm sample are similar to those found in DI water. These water cluster peaks are marked with a red line above each peak in Figure 2A, including m/z 107, 125, 143, 161, 179, 197, 215, 233, 251, 269, 287, 323, and 341 and corresponding to (H2O)5OH- (H2O)6OH- (H2O)7OH- (H2O)8OH- (H2O)9OH- (H2O)10OH- (H2O)11OH- (H2O)12OH- (H2O)13OH- (H2O)14OH- (H2O)15OH- (H2O)17OH- (H2O)19OH-, respectively.9 As there is prevalence of many characteristic water clusters in this mass spectrum, this results provides a strong evidence of the chemical water cluster environment present in a hydrated biofilm. Additionally, non-water cluster related peaks observed in both spectra can commonly be attributed as interference peaks, typically from the PDMS, a component of the microfluidic channel. For example, one common known interference peak from PDMS is the m/z 137 in the negative ion mode.
When comparing the biofilm SIMS mass spectra to that of DI water, as shown in Figure 2A, some peaks are not present in the DI water, yet they appear in the biofilm. For example, the peak at m/z 255 ([CH3(CH2)14COO]-, palmitic acid) is present at high intensity in the biofilm sample, but not at all in the DI water sample. This would suggest that the signals come from in the biofilm, most likely the biofilm and/or its self-generated EPS. Many interesting peaks were identified in Figure 2A. For example, EPS related peaks include m/z 123-, found to be C2H4PO4-, 159 (P2O8H-), 216 (Cr2O7-), 285 (octadecanoethiols), and 325 (polar compounds, C12 Polar compounds).17,18,20 Peaks associated with fatty acids were found to be 127 ([C2H3(CH2)3COO]-), 255 ([CH3(CH2)14COO]-, palmitic acid), 279 ([CH3(CH2)15COO]-, linoleic acid), 317 (C21H33O2-), 325 (C21H40O2-), and 341 (Surface lipids, 18-MEA bound through thioester linkage, stearic acid C18H35O2-, ions of monoacylglyceryls of palmitic acid C19H17O6-).16,19 Lastly, a quinolone signal quorum sensing signal related peak of C10H9NO2- was observed at m/z 175.
Figure 2B displays 2D images of the distribution of four different interesting ions between the S. oneidensis MR-1 biofilm (top row) and DI water (bottom row). The m/z value 107 ((H2O)5OH-) shows a water cluster of significant intensity within the samples, the m/z value 175 (C10H9NO2-) depicts a quorum sensing related signal, m/z 255 is a fatty acid ([CH3(CH2)14COO]-, palmitic acid), and m/z 341 (C21H41O2-) may represent both lipid fragments and water clusters due to the 1 amu mass accuracy.9 Brighter regions of the 2D pictures indicate a higher count of the molecular ion present in the image. As expected, signals for the QS compound, and lipids were far greater in quantity within the biofilm sample. While the signal for the water cluster (m/z 107) was stronger throughout the biofilm sample, it was also present in the DI water sample, as expected. Lastly, the signal at m/z 341 was found to be homogeneous within the biofilm sample, but very weakly within the DI water sample. While m/z 341 was a water cluster peak, it was also representative of several different fatty acid and surface lipids.16 Its stronger presence within the biofilm sample after normalization of data suggests that the presence of lipid fragments far outweighs the presence of water clusters.
Figure 1: Experimental Schematic. (A) Displays the experimental schematic of the biofilm setup as it appears within the laboratory. Starting at the top left, medium flows from the syringe (1), which is attached to the syringe pump (2), controlling the rate at which liquid moves through. Arrows indicate the flow direction throughout the system. The syringe (1) is connected via a PEEK injector fitting (3) to the TFE tubing (4). Medium flows through a drip chamber (5) to avoid contamination, and eventually moves through the SALVI (6) to the sealed outlet container (7). The syringe pump is positioned on an elevated surface (9) such that the drip chamber (5) can be perpendicular to the flat surface (8) that the SALVI (6) is placed upon. (B) Growth curve for Shewanella oneidensis MR-1 in "nanowires" medium. Time 0-15 h represents lag phase, time 15-33 h represents log phase, time 33-105 h represents stationary phase, and time 105 h or longer represents death phase. Error bars represent standard deviation. (C) A picture of a matured biofilm acquired with fluorescence microscopy. Please click here to view a larger version of this figure.
Figure 2: Comparisons of ToF-SIMS negative spectra of the hydrated Shewanella oneidensis biofilm and MR-1 medium in the SALVI microchannel. (A) The m/z SIMS mass spectra of the Shewanella oneidensis biofilm in MR-1 medium and DI water. The latter was used as a control. Red lines indicate locations of characteristic water cluster peaks. (B) 2D image comparison of peaks of interest between the biofilm and DI water. From left to right, images display a water cluster (m/z 107 (H2O)5OH-), a quorum sensing/hormone signal (m/z 175, C10H9NO2-), a fatty acid (m/z 255, [CH3(CH2)14COO]-, palmitic acid), and surface lipids/EPS fragments (m/z 341, 18-MEA, C21H41O2-). Please click here to view a larger version of this figure.
Discussion
After inoculating at log-phase, it is important to test the number of days and temperature at which the biofilm should grow before it is healthy and thick enough for imaging, as described in step 3.1. This procedure specifically covers culturing a S. oneidensis MR1 biofilm at room temperature; however different room temperatures can influence the rate of growth. Therefore, it is critical to use optical imaging to understand whether the biofilm is ready before proceeding to ToF-SIMS analysis. Similarly, different strains of bacteria require different growth conditions and length to achieve a desirable thickness for analysis. While the growth curve in Figure 1b depicts log phase 200 of the bacteria to be 12-32 h, and this was used at 24 h throughout the procedure, it is important to note that this time cannot be used as log-phase for other strains of bacteria without establishing the growth curve independently. While the log phase was between 12 and 33 h of growth for S. oneidensis, 24 h was chosen due to the fact that it was in the portion of growth where oxygen was starting to limit the growth-rate of the organism, towards the end of log-phase. Experiments showed that this was the time cells began to produce nanowires, thus primed to form successful biofilms that could survive in low oxygen environments.3,13 Consequently, the time chosen to use the bacteria during log phase should be influenced by research experience and knowledge gained from literature on experimental study of the bacteria being studied. Additionally, a separate growth curve must be established to understand the log-phase for all different strains of bacteria that will be used for biofilm growth using this approach. The growth rate of 2 µL/min was calculated so as not to exceed the maximum growth-rate of S. oneidensis MR-1, and the total time was chosen to get a mature biofilm that was not overgrown and prone to detaching. These rates can be adjusted accordingly to knowledge of different bacteria to be used with this setup.
Some limitations to using SALVI for biofilm growth include biofouling within the channel as well as the probability of bacteria to attach to the flat walls of the channel. Despite growth at a constant rate, recommended to be 2 µL/min within this protocol, biofouling can still occur if the biofilm is allowed to grow for too long. In such a case and without warning, the biofilm can detach from the window and exit the SALVI to the outlet containment. Biofouling can also occur simply by adding mechanical force (i.e., moving) the SALVI, which cannot be avoided. However, this can be kept in check by viewing the attachment of the biofilm to the SiN window before ToF-SIMS analysis under a light microscope. One other limitation includes the smoothness of the PDMS microchannel which can make it difficult for some bacteria to attach. However, this can be changed in the design of the SALVI to accommodate by creating ribbed edges to the channel, if attachment of the bacteria is an issue. Lastly, cell counts can be done in lieu of OD600 readings for growth curve determination. For example, studies have shown direct and consistent correlation of cell counts to OD600 readings.21 Therefore, OD600 is deemed sufficient in evaluating biofilm growth.
Limitations to this technique are few, as the SALVI essentially acts as a culture dish which has much versatility for use in many applications, such as anaerobically within a glove box, or within any temperature-controlled environmental chamber. Some minor limitations include uncontrollable room conditions such as relative humidity, temperature, placement near sunlight, which are still possible to be accommodated for, for each specific bacteria's optimal growth conditions. In addition, some of these technical challenges can be overcome with an incubator with temperature or RH mediation features in the biofilm setup.
SALVI is a vacuum-compatible microfluidic interface. In this work, a 200 x 300 µL microfluidic channel was used to culture biofilms followed with optical and liquid SIMS imaging. Liquids present a technical challenge to study using vacuum based techniques, because they are volatile and difficult to retain in the liquid phase in vacuum. Thus vacuum techniques such as ToF-SIMS have been traditionally restricted to only dry and cryogenic samples.22 Additionally, SALVI can be used for diverse imaging and spectroscopy using a variety of microscopy and spectroscopy techniques.4,9 Our group has worked to continually expand various SALVI applications in in situ analysis of liquids and solid-liquid interfaces.15,23,24 Our most recent effort has effectively shown bacteria attachment to the SiN membrane.9 After initial attachment, continual flow of medium over time has been shown to successfully cultivate a biofilm directly on the SIN window of the SALVI. During ToF-SIMS, the primary ion beam is used to bombard holes of 2 µm in diameter. This dimension is similar to the cell length of the biofilm attached to the SiN window, thus providing a chemical profile of the biofilm in its naturally hydrated microenvironment. This is highly important due to the difference of a biofilm's chemical identity, presence of EPS, and water cluster environment when comparing the biofilms in its hydrated state with dried samples.9 Without the use of SALVI, ToF-SIMS could only be conducted on dry and cryogenic samples, providing chemical mapping which fails to capture the chemical profile of a sample in its natural hydrated state.
As shown in Figure 1A, it is important to keep the syringe pump above the drip tubing in order to allow for the drip tubing to be perpendicular to the SALVI microchannel. Additionally, bubbles will be less likely to become trapped within the tubing of the device, if the outlet PFTE tubing (within the outlet bottle) is lower than the inlet tubing (attached to the drip chamber). Another critical step of cultivating bacteria for SALVI growth is to first obtain a growth curve of the particular bacteria strain that will be used, as described within step 1.3. This can be done by obtaining growth curves with optical density measurements. A growth curve, as shown in Figure 1B, is important for understanding the timeframe at which bacteria will be within the log-phase of growth, when it can be assumed that the bacteria are healthiest. Utilizing bacteria within this phase of growth facilitates the creation of a healthy biofilm microenvironment and ensures that the biofilm will grow at its expected rate. The SALVI microchannel is sterilized in room temperature with 70% ethanol and DI water, as it cannot be autoclaved for sterilization prior to use with ToF-SIMS. This is due to the fact that autoclaving can both damage the window of the SALVI and introduce water vapor to the channel. After several days of growth, the microchannel should be checked with fluorescence microscopy to validate bacterial attachment. An example can be found in Figure 1C, this image was taken to show a matured biofilm. Due to the flow path of the medium, the bacteria more favorably attaches and grows along the edges of the microchannel, as this location is where flow and shear-forces are lowest.
In summary, SALVI is an excellent method for which to culture and study biofilms using in situ chemical mapping, as it allows for providing the living biofilm in its natural hydrated state to the vacuum-based imaging mass spectrometer. This unique approach can provide more information on a biofilm's water cluster microenvironment, as well as to gain a deeper understanding of its EPS byproducts. This information could be used to understand a biofilm's biological activities, and can be utilized in various applications such as in biomedicine, biomedical engineering, agriculture, and industrial research and development.
Disclosures
The authors have nothing to disclose.
Acknowledgments
We are grateful to the Pacific Northwest National Laboratory (PNNL) Earth and Biological Sciences (EBD) mission seed Laboratory Directed Research and Development (LDRD) fund for support. Instrumental access was provided through a W. R. Wiley Environmental Molecular Sciences Laboratory (EMSL) General User Proposal. EMSL is a national scientific user facility sponsored by the Office of Biological and Environmental Research (BER) at PNNL. The authors thank Dr. Yuanzhao Ding for proof reading the manuscript and providing useful feedback. PNNL is operated by Battelle for the DOE under Contract DE-AC05-76RL01830.
References
- Renner LD, Weibel DB. Physicochemical regulation of biofilm formation. MRS Bull. 2011;36(5):347–355. doi: 10.1557/mrs.2011.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flemming HC, Wingender J. The biofilm matrix. Nat Rev Microbiol. 2010;8(9):623–633. doi: 10.1038/nrmicro2415. [DOI] [PubMed] [Google Scholar]
- Aldeek F, et al. Patterned hydrophobic domains in the exopolymer matrix of Shewanella oneidensis MR-1 biofilms. Appl Environ Microbiol. 2013;79(4):1400–1402. doi: 10.1128/AEM.03054-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hua X, et al. In situ molecular imaging of a hydrated biofilm in a microfluidic reactor by ToF-SIMS. Analyst. 2014;139(7):1609–1613. doi: 10.1039/c3an02262e. [DOI] [PubMed] [Google Scholar]
- Yu XY, Yang L, Cowin JP, Iedema M, Zhu Z. Systems and methods for analyzing liquids under vacuum. US Patent. 2013.
- Yu XY, Liu B, Yang L, Zhu Z, Marshall MJ. Microfluidic electrochemical device and process for chemical imaging and electrochemical analysis at the electrode-liquid interface in situ. US Patent. 2014.
- Yang L, Yu XY, Zhu ZH, Thevuthasan T, Cowin JP. Making a hybrid microfluidic platform compatible for in situ imaging by vacuum-based techniques. J Vac Sci Technol A. 2011;29(6) [Google Scholar]
- Yang L, Yu XY, Zhu ZH, Iedema MJ, Cowin JP. Probing liquid surfaces under vacuum using SEM and ToF-SIMS. Lab Chip. 2011;11(15):2481–2484. doi: 10.1039/c0lc00676a. [DOI] [PubMed] [Google Scholar]
- Ding Y, et al. In Situ Molecular Imaging of the Biofilm and Its Matrix. Anal Chem. 2016. [DOI] [PubMed]
- Yang J, Ghobadian S, Montazami R, Hashemi N. Proceedings of the Asme 11th Fuel Cell Science, Engineering, and Technology Conference, 2013. 2013.
- Yu F, Wang CX, Ma J. Applications of Graphene-Modified Electrodes in Microbial Fuel Cells. Materials. 2016;9(10) doi: 10.3390/ma9100807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J, Zhou Y, Hua X, Zhu Z, Yu XY. In Situ Characterization of Hydrated Proteins in Water by SALVI and ToF-SIMS. J Vis Exp. 2016. p. e53708. [DOI] [PMC free article] [PubMed]
- Hill EA. Effects of Electron-Transport-System Impairment on Hydrogen Gas Production by the Bacterium Shewanella oneidensis MR-1. Washington State University; 2007. Master's thesis. [Google Scholar]
- McCormick AJ, et al. Biophotovoltaics: oxygenic photosynthetic organisms in the world of bioelectrochemical systems. Energy Environ Sci. 2015;8(4):1092–1109. [Google Scholar]
- Liu B, et al. In situ chemical probing of the electrode-electrolyte interface by ToF-SIMS. Lab Chip. 2014;14:855–859. doi: 10.1039/c3lc50971k. [DOI] [PubMed] [Google Scholar]
- Keune K, Hoogland F, Boon JJ, Peggie D, Higgitt C. Evaluation of the "added value" of SIMS: A mass spectrometric and spectroscopic study of an unusual Naples yellow oil paint reconstruction. Int J mass Spectrom. 2009;284(1-3):22–34. [Google Scholar]
- Lee M. Mass Spectrometry Handbook. John Wiley & Sons Inc; 2012. p. 988. [Google Scholar]
- Petrovic M, Barcelo D. Determination of anionic and nonionic surfactants, their degradation products, and endocrine-disrupting compounds in sewage sludge by liquid chromatography/mass spectrometry. Anal Chem. 2000;72(19):4560–4567. doi: 10.1021/ac000306o. [DOI] [PubMed] [Google Scholar]
- Vickerman JC. Molecular Imaging and Depth Profiling by Mass Spectrometry--Sims, MALDI or DESI. Analyst. 2011;136(11) doi: 10.1039/c1an00008j. [DOI] [PubMed] [Google Scholar]
- Weng LT, Bertrand P, Stonemasui JH, Stone WEE. Tof Sims Study of the Desorption of Emulsifiers from Polystyrene Latexes. Surf Interface Anal. 1994;21(6-7):387–394. [Google Scholar]
- Peñuelas-Urquides K, et al. Measuring of Mycobacterium tuberculosis crowth. A correlation of the optical measurements with colony forming units. Braz J Microbiol. 2013;44(1):287–289. doi: 10.1590/S1517-83822013000100042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dohnalkova AC, et al. Imaging hydrated microbial extracellular polymers: comparative analysis by electron microscopy. Appl Environ Microbiol. 2011;77(4):1254–1262. doi: 10.1128/AEM.02001-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L, et al. In situ SEM and ToF-SIMS analysis of IgG conjugated gold nanoparticles at aqueous surfaces. Surf Interface Anal. 2013;46:224–228. [Google Scholar]
- Yu JC, et al. Capturing the transient species at the electrode-electrolyte interface by in situ dynamic molecular imaging. Chem Commun. 2016;52(73):10952–10955. doi: 10.1039/c6cc02893d. [DOI] [PubMed] [Google Scholar]


