Abstract
Many cancer types, including pancreatic cancer, have a dense fibrotic stroma that plays an important role in tumor progression and invasion. Activated cancer associated fibroblasts are a key component of the tumor stroma that interact with cancer cells and support their growth and survival. Models that recapitulate the interaction of cancer cells and activated fibroblasts are important tools for studying the stromal biology and for development of antitumor agents. Here, a method is described for the rapid generation of robust 3-dimensional (3D) spheroid co-culture of pancreatic cancer cells and activated pancreatic fibroblasts that can be used for subsequent biological studies. Additionally, described is the use of 3D spheroids in carrying out functional metabolic assays to probe cellular bioenergetics pathways using an extracellular flux analyzer paired with a spheroid microplate. Pancreatic cancer cells (Patu8902) and activated pancreatic fibroblast cells (PS1) were co-cultured and magnetized using a biocompatible nanoparticle assembly. Magnetized cells were rapidly bioprinted using magnetic drives in a 96 well format, in growth media to generate spheroids with a diameter ranging between 400-600 µm within 5-7 days of culture. Functional metabolic assays using Patu8902-PS1 spheroids were then carried out using the extracellular flux technology to probe cellular energetic pathways. The method herein is simple, allows consistent generation of cancer cell-fibroblast spheroid co-cultures and can be potentially adapted to other cancer cell types upon optimization of the current described methodology.
Keywords: Cancer Research, Issue 126, Pancreatic cancer, 3-dimensional spheroids, co-culture, extracellular flux, bioenergetics, mitochondrial stress
Introduction
In the last decade, numerous in vitro 3D models have been developed to recapitulate and investigate the in vivo tumor biology, microenvironment and growth conditions of cancer cells1,2. Two-dimensional (2D) monolayer cell culture systems with uniform exposure to biochemical factors and investigational compounds fail to replicate the native 3D tumor-stromal interactions exposed to a gradient of compounds diffusing through the extracellular matrix proteins (ECM)3,4. Thus, in comparison to 2D tissue culture models, 3D cancer models have emerged to show better potential at simulating the tumor microenvironment and served as important tools to better understand in vivo tumor characteristics, such as hypoxia, desmoplasia, dormancy, drug penetrance, toxicity and therapeutic resistance5,6. To this end, 3D models have potential to bridge the gap between 2D cell culture and whole animal models by mimicking in vivo tumor features, while being relatively inexpensive and optimized for rapid generation and consistency. These advantages are being exploited to accelerate translational research in many areas including cancer biology, morphogenesis and tissue engineering7,8.
In a surge of evolving 3D tissue culture methods, magnetic levitation techniques have recently been developed and described for growth, assaying and imaging of spheroids derived from various cell types9,10,11,12. Magnetic 3D bioprinting exploits the use of magnetic forces to engineer tissues by magnetizing cells with biocompatible nanoparticles and printing them in multi-well formats. This allows rapid production of consistent, near identical 3D spheroids, which can be harnessed and employed for a plethora of downstream applications for biochemical and biophysical investigation10. Here we have adapted the magnetic bioprinting technique using a biocompatible material called Nanoshuttle (NS) composed of iron oxide, poly L-lysine and gold nano particles to label pancreatic cancer cells and fibroblasts. NS attaches electrostatically to the plasma membrane, is not known to bind to any specific receptors, and releases off the cell surface within a week. It requires very low magnetic forces (30pN), enough to aggregate but not harm cells and does not affect cell viability, metabolism or proliferation to make it extremely biocompatible for 3D cultures10,13,14,15.
In this study, using pancreatic cancer as an example and model, we describe the generation and metabolic assay of 3D cancer cell-fibroblast spheroids. Starting from cells cultured in 2D vessels, we illustrate the culture and growth conditions of pancreatic tumor-fibroblast co-culture spheroids using magnetic bioprinting. Cultured spheroids were then used in functional metabolic assays using an extracellular flux analyzer, a technology demonstrated to simultaneously measure the two major energy producing pathways, glycolysis and mitochondrial respiration, in a variety of live cells and tissues16,17,18,19,20. Glycolysis was measured as a change in the extracellular acidification rate (ECAR), while mitochondrial respiration or oxidative phosphorylation was measured as oxygen consumption rate (OCR). We propose that this method developed for pancreatic tumor spheroids can serve as a backbone for optimizing and translating 3D tumor spheroid generation and assay to other cell/tissue types.
Protocol
1. Culture of Pancreatic Cancer 3D Spheroids using Magnetic Bioprinting
Using standard aseptic tissue culture technique, culture cells of interest in a T75 flask to a confluency of 70-80% in appropriate growth media. NOTE: Typically, 5-7 × 106 cells from a 70-80% confluent T75 flask were harvested. Two different cell types were used in this study - Patu8902 (pancreatic tumor cells) and PS1 cells (activated pancreatic stellate cells21). Both cell lines were cultured in Roswell Park Memorial Institute media 1640 (RPMI-1640) supplemented with 10% (v/v) fetal bovine serum(FBS), 1× penicillin-streptomycin (p/s).
Wash cells once with 5 mL of Dulbecco's phosphate buffered saline(DPBS).
Detach cells from plastic surface by trypsinizing with 2 mL of 0.05% Trypsin-EDTA for 5-10 min at 37 °C. Check for cell detachment under a microscope.
Deactivate trypsin by addition of 8 mL growth media to detached cells. Avoid over trypsinization, as this can adversely affect the health/viability of cells.
Pipet cells up and down to generate a single cell suspension and count cell density using an automated cell counter or a hemocytometer.
In the meantime, equilibrate a vial of NS to ambient temperature.
Calculate the amount of cells required for seeding the desired number of spheroids (wells) and aliquot to a new 15 mL conical tube. For example, to seed an entire 96 well plate with 5,000 cells/well, aliquot at least 5.5 × 105 cells.
Collect cells by centrifuging at 500 x g for 3 min. Carefully aspirate or decant the supernatant and resuspend the cells at a concentration of 1.0 x 106 cells/mL in growth media. Pipette gently using a wide bored pipet tip to avoid shearing. For example, resuspend 5.5 x 105 cells in 550 µl of growth media.
- Magnetize the desired amount of cells using the equilibrated NS (step 1.6).
- Directly add NS to the cell suspension in growth media and agitate gently. For efficient magnetic labeling of cells, use 10 µL NS per 100 µL cells (resuspended at 1 x 106 cells/mL). NOTE: For a typical experiment, label 5.5×105 Patu8902 cells in 550 µL with 55 µL NS and 1.1×106 PS1 cells in 1100 µL, (separately) with 110 µL NS.
- Gently invert tube a few times to ensure cell suspension. Temporarily, transfer the cell-NS mix into a single well of a 24-well plate. Do this separately for each cell type to be magnetized. Cover plate and incubate cells at room temperature for 2 h with gentle shaking to allow NS binding to cell surface. NOTE: Culture and assay of spheroids starting either from Patu8902 or PS1 alone cells, in addition to co-culture spheroids starting with both cell types premixed before printing on magnetic drives was successfully achieved using this method in independent experiments. A method for generating co-culture spheroids from Patu8902 and PS2 cells is described below in the subsequent steps.
Pipet up and down the magnetized cells gently to mix evenly and prepare a master mix containing the desired cell number. For seeding spheroids, use a total of 15,000 cells (5,000 Patu8902 + 10,000 PS1) and adjust to a total volume of 150 µL per well of a 96-well plate. NOTE: The final volume dispensed into each well must be the same regardless of the number of cells used.
Place a cell repellent 96-well plate atop the 96-well magnetic spheroid drive and dispense 150 µL of cell mix into each well. Observe the cells slowly collect to the center of the well above the magnet. Cover and incubate the plate overnight in a 5% CO2 incubator set at 37 °C with the magnetic spheroid drive still attached. Remove the magnetic drive and incubate for further growth for up to 7 days. NOTE: It is critical to use cell repellent growth plates for spheroid printing using magnetic spheroid drives. Ensure that the spheroid drive with thinner smaller magnets is used, not the holding drive.
Monitor the growth of spheroids every day. Replenish with fresh growth media every three days by placing the growth plate mounted on the magnetic holding drive at an angle and using a multichannel pipet to draw out spent media. NOTE: Patu8902 and PS1 cells co-seeded at 15,000 cells/well will grow into 3D spheroids with a diameter of 400-600 µm within 5-7 days. At this point spheroids are ready for metabolic characterization, drug evaluation, and other downstream applications.
2. Metabolic Assay of 3D Pancreatic Tumor Spheroids for Bioenergetics Pathways using the Extracellular Flux Analyzer
Note: In this section, the analysis of the metabolic functions of the spheroids using an extracellular flux analyzer with assay microplates specifically designed for 3D spheroids is described.
- Washing and Transfer of spheroids from growth plates to spheroid assay microplates. NOTE: Spheroids may be used for metabolic assays when they reach a diameter of ~500 µm.
- The day prior to performing the assay, hydrate the probe or sensor cartridge with assay calibrant (200 µL/well) in the provided utility plate and incubate overnight (4-18 h) in a humidified incubator (non-CO2) set at 37 °C.
- Prepare the appropriate assay medium for the desired metabolic assay by supplementing the base media (see the Table of Materials) with either 2 mM L-glutamine for glycolysis stress test (GST) assay or with 1 mM pyruvate, 2 mM L-glutamine and 10 mM glucose for a mitochondrial stress test (MST) assay. Note: The extracellular flux analyzer measures both mitochondrial respiration (OCR) and glycolysis (ECAR) of live cells in a 96-well plate format. These rates provide an overview of cellular metabolic function in samples under investigation. Please refer to the manual in the assay kits for detailed instructions.
- After adding assay specific supplements (refer Step 2.1.1), warm the supplemented assay medium in a 37 °C water bath and adjust the pH to 7.4 ± 0.1 with 0.1N NaOH. Keep the medium warm until use.
- Reconstitute assay kit reagents in the calibrated assay medium to recommended stock concentrations. NOTE: These are made at 10× the final concentration desired in the wells. Stock concentrations for glucose, oligomycin and 2-deoxy-glucose (2-DG) are 100 mM, 100 µM and 500 mM, respectively.
- Observe spheroids in the growth plate under a light microscope to check for spheroid morphology and overall uniformity; in terms of the shape and structure of the spheroids.
- Transfer the growth plate onto a magnetic holding drive and carefully aspirate ~120 µL growth media. Gently wash spheroids with ~120 µL of warmed and pre-calibrated assay media, aspirate and repeat the wash twice. Inspect the spheroids under the microscope again to ensure that spheroids are not washed away.
- Prepare the spheroid assay microplate by adding 180 µL warm assay media to each well.
- Set a P20 micropipette to 10 µL and fit a wide bored tip to it. If wide bored tips are not available, cut off the tips of regular pipette tips with a clean scissors or scalpel to widen the bore. Note: This should reduce shearing and preserve the overall morphology of the spheroids while being transferred to assay plates.
- Use a warm (37 °C) surface to carry out transfer of spheroids. Use an X-ray film viewer surface warmed with a white-light lamp to enhance contrast and aid visualization of spheroids during the transfer process.
- Carefully aspirate a single pre-washed spheroid from the cell repellent growth plate using a P20 micropipette fitted with a wide bore tip and gently transfer spheroid directly into the center of each well of a spheroid assay plate for carrying out the metabolic assay. Allow each spheroid to gently fall into the central micro-chamber of each well by pure gravity to populate the assay plate. NOTE: It usually takes 5-8 s for each spheroid to fall into the center of the well by gravity. Do not pull out the pipette until the spheroids have settled down in the micro-chamber. Do not deposit spheroids in the 4 corner wells of the assay plate (A1, A12, H1, H12) since these will be used as background wells.
- Monitor the morphology and position of each spheroid to ensure that each spheroid is in the center of each well. If necessary, reposition to the center of well by aspirating out spheroid using a wide bore pipet tip, and subsequent deposition by gravity.
- Once all spheroids have been transferred, place the assay plate in a 37 °C humidified air incubator (non-CO2) for one hour prior to assay.
- Loading sensor cartridge with compounds and performing the assay
- Prepare 10× concentrated assay specific reagents in the assay specific media described in step 2.1.2. For example, to carry out GST, reconstitute glucose, oligomycin, and 2-DG in recommended volumes referred to in the assay manual. Both GST and MST assays were carried out using Patu8902-PS1 spheroids.
- Properly orient the sensor cartridge with rows labeled A-H on the left hand side and place a loading guide on top of the sensor cartridge. Ensure correct loading of desired port by orienting the loading guide so that the letter corresponding to the port to be loaded is at the upper left hand corner.
- Using a multichannel pipette, dispense reagents directly into injection ports. Hold loading guides in position using fingertips throughout the procedure. Note: Each reagent will be injected in a recommended port referred in the assay manual. Avoid creating air bubbles but do not tap any part of the cartridge to remove air bubbles.
- Remove loading guide and position at eye level with cartridge to visually inspect injection ports for even loading.
- Create the experimental assay design/instrument protocol using the instrument controller Wave software (henceforth, analysis software) as described in section 2.3.
- Assay design and execution using the analysis software
- Create an assay template for the experiment
- Open the analysis software and click "Templates" or choose from an existing assay template. Double click on "Blank Template".
- Define experimental groups and conditions.
- Define Injection strategies for ports A, B, and C. Leave port D empty. NOTE: For GST assay, only these ports will be loaded with Glucose, Oligomycin and 2-DG. In case of MST assay, oligomycin, FCCP and rotenone will be loaded.
- Define pre-treatments if spheroids were treated with one or more test compounds and the control group. Define the assay media to include source, supplements, and other information.
- Define one or more cell type (e.g. Patu8902, PS1) seeing the density and passage number information.
- Click "Generate Groups".
- Click "Plate Map" tab and assign groups to the assay plate corresponding to the experimental design.
- Select the four corner wells as background wells. Ensure that there are no spheroids in these wells.
- Run assay on the analyzer
- Click on the Instrument Protocol tab. Keep the default protocol commands by checking "Calibrate", "Equilibrate" and "Baseline Measurement Cycle".
- Click "Injections" and define compound for each port. Edit measurement cycles to 6 cycles from the default 3 cycles for spheroids. Keep the default 3 min mix, 0 min wait, and 3 min measure times. Note: Default mix-wait-measure times are 3 min, 0 min, 3 min, respectively. Three basal rate measurements are typically taken prior to injection of first assay compound. These measurements can be calibrated and readjusted as per the experimental design.
- Review Protocol and Group Summary.
- Save assay design template.
- Proceed to pre-assay calibration once injection ports have been loaded with assay substrates. Transfer the cartridge with the utility plate to the extracellular flux analyzer. NOTE: This usually completes within 15-20 min and calibrates the sensors to conduct OCR and ECAR measurements.
- After calibration of the cartridge, replace the utility plate with the pre-warmed assay plate containing 3D spheroids and initiate the assay. After the measurements are completed, export the data to spreadsheet or graphing and statistical software to analyze the data.
Representative Results
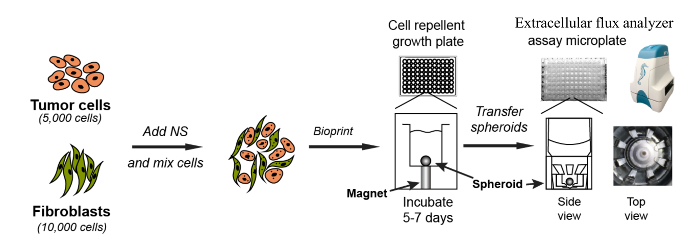
The general workflow for magnetic bioprinting and extracellular flux analysis of 3D spheroidsFigure 1 depicts an overview of the entire process described in this protocol. Pancreatic cancer cells (Patu8902) and activated stellate cells (PS1) were cultured in tissue culture flasks containing appropriate growth media to a confluency of 70-80%. Cells were detached from the 2D culture vessel and washed, cell density was determined and cells finally resuspended in growth media at 1×106 cells/mL. NS, a nanoparticle assembly consisting of gold, iron oxide, and poly-L-lysine was used to magnetize cells. Individually magnetized Patu8902 and PS1 cells were then mixed and printed onto 96-well cell repellent growth plates mounted on a magnetic spheroid drive. We have optimized the 15,000 to be the total number of cells for seeding a single well (5,000 Patu8902 cells mixed with 10,000 PS1 cells to generate a single spheroid). After incubation under appropriate tissue culture conditions for 5-7 days, 3-dimensional spheroids were obtained, during which time the growth of these spheroids was monitored and recorded. After washing with assay media, spheroids were physically transferred onto a spheroid microplate to carry out metabolic assays using an extracellular flux analyzer for simultaneous measurement of glycolysis and mitochondrial respiration.
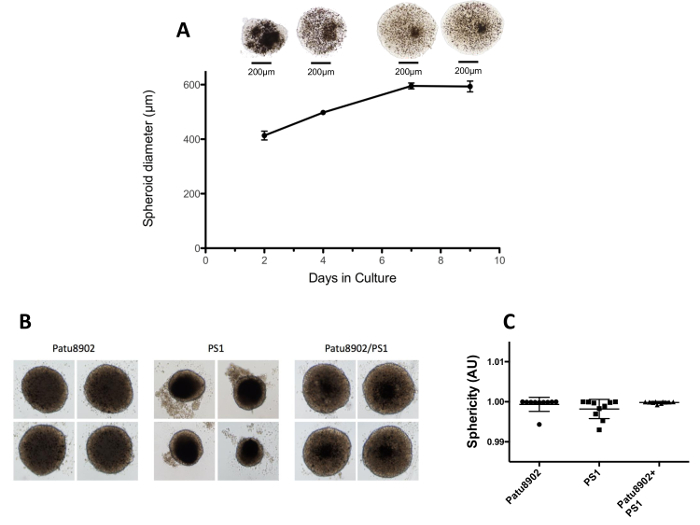
Culture and growth of pancreatic tumor-fibroblast spheroids To obtain functional and robust spheroids, the epithelial cancer cell line Patu8902 and activated stellate cells PS1 were cultured in RPMI-1640 supplemented with fetal bovine serum. Pat8902 and PS1 cells magnetized with NS were then mixed together in a ratio of 1:2, so as to seed a total of 15,000 cells per well in a growth plate. Within 24 h of printing the cells onto the magnetic drives, cells focalized to the center of the well exactly on the top of each magnetic pin under each well. Growth of Patu8902-PS1 spheroids was monitored by imaging on days 2, 4, 7 and 9 after seeding cells. Figure 2 shows quantification of the average diameter of representative spheroids at various time points above. The diameter of spheroids increases from 414 µm on day 2 to 596 µm on day 7 post printing. There was no significant difference between growth on days 7 and 9, and thus all further experimentation was limited to 7 days of spheroid growth. We noticed that the spheroids acquire a sharper morphology especially around the outer edges and the NS is progressively more evenly diffused throughout the volume of the spheroid with more days in culture.We also estimated the sphericity of 10 spheroids based on the length of their major and minor axes. The average sphericity is 0.92 ± 0.04 for Patu8902 alone (tumor), 0.95 ± 0.04 for PS1 (stromal) alone and 0.94 ± 0.02 for co-culture spheroids.
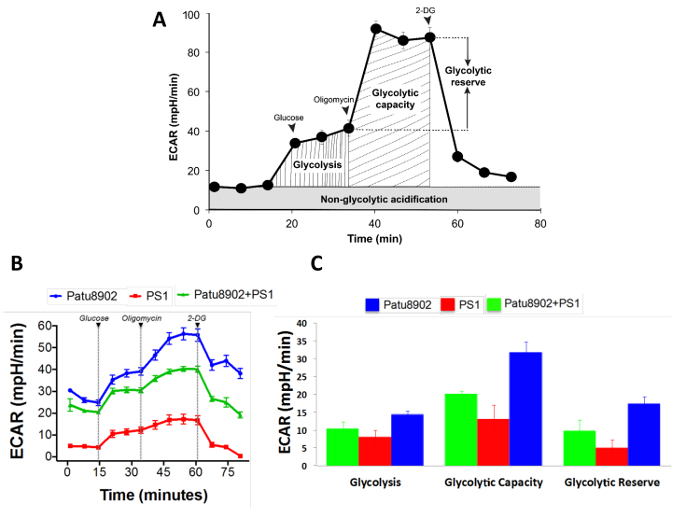
Functional metabolic assay of pancreatic 3D spheroids using the extracellular flux analyzer To test the functionality of the spheroids, we employed metabolic and bioenergetics analysis to probe the energy pathways in spheroids. We carried out a glycolysis stress test on spheroids cultured as described above. To this end, spheroids generated from either Patu8902 or PS1 cells in addition to those resulting from a co-culture of the two cell types were subjected to the assay. Interestingly, all three types of spheroids, whether originating from distinct or mixed cells, exhibited a biologically functional response to the GST assay. Upon injecting a saturating concentration of glucose, the spheroids types tested show increases in the glycolytic pathway as evidenced by an increase in ECAR (Figure 3). When, mitochondrial ATP production was inhibited using oligomycin, energy production shifts to glycolysis and pushed cells to maximal glycolytic capacity, seen as further increase in ECAR. Inhibition of glycolysis using 2-DG, a glucose analog that competitively binds glucose hexokinase, resulted in a decrease in ECAR, confirming that the prior increase in ECAR was due to glycolytic activity. We noticed that the spheroids tested in this study responded in this fashion, although the PS1 alone spheroids (average diameter 250 µm) had a relatively lower signal, possibly due to their smaller size and lower glycolytic capacity compared to the Patu8902 cancer cells (average diameter 600 µm). In general, a higher ECAR signal from tumor cell derived spheroids compared to those from relatively smaller PS1 fibroblast derived spheroids, could possibly be attributed to the cancer cells' inherent metabolic inclination towards glycolysis22,23. All three types of spheroids were derived from seeding the same number of cells, although we noticed spheroid size variation among each type, with PS1 alone spheroids being the smallest, followed by co-culture spheroids, and Patu8902 alone spheroids being the largest.
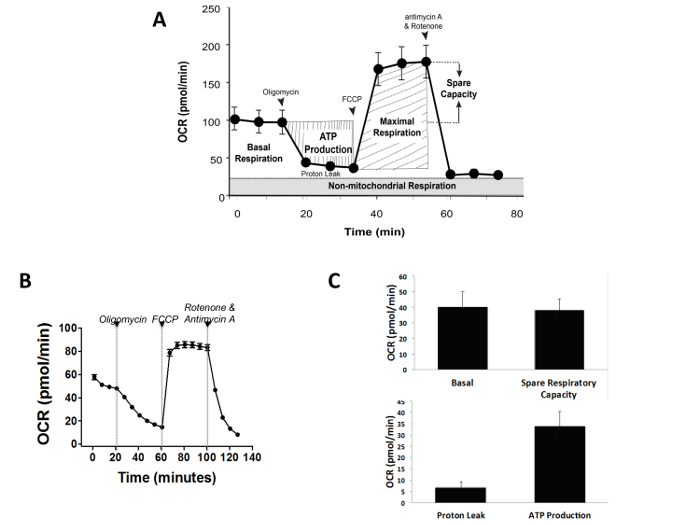
To test mitochondrial function in 3D spheroids, we subjected co-culture spheroids to a MST assay. The MST measures key parameters of mitochondrial function by directly measuring OCR of cells (Figure 4A and 4B). A serial injection with compounds (oligomycin, FCCP, and a mix of rotenone and antimycin A) was used to measure key mitochondrial parameters such as, ATP production, maximal respiration, and non-mitochondrial respiration. These parameters and basal respiration rates were further used to calculate proton leak and spare respiratory capacity (Figure 4C). A decrease in OCR in response to oligomycin, an inhibitor of ATP synthase (complex V) correlates to the mitochondrial respiration associated with cellular ATP production. Spheroids were incubated with oligomycin for a longer duration to allow maximal penetration and efficient response time. A second injection with the uncoupler FCCP stimulates maximal oxygen consumption and a corresponding increase in OCR. The FCCP-stimulated OCR was used to calculate spare respiratory capacity, a measure of the ability of the cell to respond to increased energy demand. The third injection; a mix of rotenone, (a Complex I inhibitor), and antimycin A (a Complex III inhibitor) blocks mitochondrial respiration, seen as a sharp decrease in OCR (Figure 4B).
Overall, our observations confirm the functionality of cultured spheroids using an extracellular flux analyzer as a measure of their metabolic footprint/activity both in terms of the glycolytic and mitochondrial potential as described above.
Figure 1. Schematic representation of the methodology for the culture and assay of pancreatic cancer cells/fibroblast spheroids. Patu8902 and PS1cells were cultured, trypsinized, washed and counted. For seeding each spheroid, Patu8902 (5,000 cells/well) and PS1 fibroblasts (10,000 cells/well) were magnetized using NS and printed on a cell repellent growth plate on day 1. After seeding, the growth plate was mounted on a magnetic spheroid drive (with one magnet under each well of the 96 well format growth plate) and allowed to culture for 2-7 days to obtain 3D spheroids. The resulting spheroids were washed and transferred to the spheroid assay microplates to carry out the metabolic assays on the extracellular flux instrument. Please click here to view a larger version of this figure.
Figure 2. Growth of pancreatic tumor-fibroblast spheroids. (A) A representative image of Patu8902-PS1 co-culture spheroid corresponding to the day in culture is shown on the top and the size in diameter of the spheroid (µm) is quantified below. Progressive increase in spheroid size and diffusion of NS particles (dark dots) throughout its volume is evident between days 2 and 7. Spheroids were kept in culture for an additional 2 days after this, which did not result in a significant increase in spheroid diameter. (B) Representative pictures of spheroids derived from tumor cells (Patu8902), stroma cells (PS1) and co-cultured cells (Patu8902 + PS1) is shown. (C) Sphericity data is depicted from 10 individual spheroids from each group. Error bars represent standard deviation (SD). Please click here to view a larger version of this figure.
Figure 3. Glycolysis Stress Test (GST) with pancreatic spheroids. (A) Schematic representation of a typical glycolytic function assay with various assay parameters depicted. (B) An example of a GST test performed on the pancreatic spheroids cultured using the methodology described. Glucose injection ramps up glycolysis seen as an increase in ECAR, with further increase on addition of oligomycin to shut off mitochondrial respiration. ECAR falls in response to 2-DG, a competitive analog of glucose, by blocking glycolysis. All spheroids are noted to exhibit a functional response to the GST assay. (C) An output of the GST assay using the manufacturer's report generator depicting the three major parameters of the GST assay. Error bars represent standard error of mean (SEM). Please click here to view a larger version of this figure.
Figure 4. Mitochondrial Stress Test (MST) with Patu8902-PS1 spheroids. (A) Schematic representation of a typical mitochondrial function assay with various assay parameters is depicted. (B) An example of a MST on the pancreatic tumor-fibroblast spheroids is shown. (C) Co-culture 3D spheroids exhibit a functional response to the MST assay. As expected, mitochondrial function measured as a decline in OCR was noticed when spheroids were challenged with oligomycin, an ATP synthase inhibitor. Using FCCP to ramp up electron flow resulted in maximal OCR, which immediately collapsed upon inhibition of mitochondrial respiration using a -cocktail of mitochondrial complex inhibitors. Error bars represent standard error of mean (SEM). Please click here to view a larger version of this figure.
Discussion
Using pancreatic cancer, the 3rd leading cause of cancer related deaths in the United States24, as an example and model, a rapid and consistent method was developed to grow and assay 3D spheroids using the co-culture of pancreatic cancer cells and activated pancreatic fibroblasts. Using magnetic bioprinting, spheroids ranging in size from 400-600 µm in diameter were obtained. These were subjected to metabolic assays to successfully test the functionality of the cultured 3D spheroids. This method is simple, consistent and can be easily adapted to other cell types.
While this methodology will accelerate and add to the translational value of the assays performed with 3D spheroids, it can be improved by developing techniques that allow multiplexing of spheroid manipulation and handling. This should further reduce the overall time, cost and labor required to carry out the assay. Additionally, spheroid volumes may be used to normalize OCR signals as reported by others6. Spheroid volume normalized by average volume of a cell could be used to determine OCR per cell, but since growth of spheroids will not be identical, calculating an absolute OCR per cell might be challenging. Currently, this methodology is limited by the lack of an efficient tool to multiplex spheroid handling and transfer from growth plates to assay environment.
The method described is modified and adapted from the magnetic bioprinting technology, and further validated to show the functional relevance by applying the cultured spheroids to a downstream metabolic assay. The method provides an important tool to generate robust, viable and functional spheroids that contain the two major cell types found in most tumor tissues, cancer cells and cancer associated fibroblasts, which can be employed for other investigational assays, such as drug screens. The relative ease and efficiency of the NS methodology is a major improvement over other methods25, such as the hanging drop method2,26 of spheroid generation.
Robust spheroids generated as such provide an in vitro tumor model that includes stromal cells, which are often missing from many 3D culture studies. The possibilities for application of 3D spheroid models generated as such are vast. For example, such models can be employed to investigate cell-cell interactions, bioenergetics shifts and to test genetic susceptibility and dependence of various therapeutic regimens. 3D spheroids that recapitulate the in vivo tumor microenvironment serve as highly relevant and useful tool for drug screening studies and those investigating the mechanisms of action of current and novel therapeutics. Metabolic assays probing energy pathways using spheroids cultures with mutant cells may help shed new light into the role of those genes and related pathways in the pathobiology in a relevant 3D model of cancer, like the spheroids described in this study.
The successful application of this method relies foremost on the health of cells required for magnetic printing, which is why cell growing in logarithmic phase should be harvested and magnetized. It is critical to use the magnetic spheroid drive to print magnetized cells and not the holding drive, which should be utilized only during the spheroid washing and media replenishing steps of the described method. The other most critical aspect for successful readout of metabolic assays is to maintain the morphology of the spheroids during the transfer process from the growth plate to the assay plate.
Overall, this protocol has been established for generation of 3D spheroids that contain both cancer and stromal cells, following the subsequent metabolic assay of the spheroids using the extracellular flux analyzer. The key features of this method are that it is quick, easily adaptable and consistent, relevant to and mimics the in vivo tumor microenvironment, is comparatively inexpensive and may serve as a new tool for studying tumor biology and developing anticancer agents.
Disclosures
The authors George W. Rogers and Andrew Neilson are employees/shareholders of Agilent Technologies that produces reagents and instruments used in this Article. All other authors declare that they have no competing financial interests.
Acknowledgments
We would like to thank Samuel Zirbel for assistance in optimizing culture conditions and monitoring spheroid growth. We are grateful to Agilent technologies for providing the SeaHorse XFe96 analyzer, assay reagents and technical insights. We also thank Dr. Hemant Kocher at Barts Cancer Institute in United Kingdom for providing the PS1 cells. This work was supported in part by the Seena Magowitz Foundation for Pancreatic Cancer Research and a Stand Up To Cancer-Cancer Research UK-Lustgarten Foundation Pancreatic Cancer Dream Team Research Grant (Grant Number: SU2C-AACR-DT-20-16). Stand Up To Cancer is a program of the Entertainment Industry Foundation. Research grants are administered by the American Association for Cancer Research, the scientific partner of SU2C.
References
- Nyga A, Cheema U, Loizidou M. 3D tumour models: Novel in vitro approaches to cancer studies. J Cell Commun Signal. 2011;5(3):239–248. doi: 10.1007/s12079-011-0132-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ware MJ, et al. Generation of an in vitro 3D PDAC stroma rich spheroid model. Biomaterials. 2016;108:129–142. doi: 10.1016/j.biomaterials.2016.08.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S. Beyond the Petri dish. Nat Biotechnol. 2004;22(2):151–152. doi: 10.1038/nbt0204-151. [DOI] [PubMed] [Google Scholar]
- Cukierman E, Pankov R, Stevens DR, Yamada KM. Science. Vol. 294. New York, N.Y: 2001. Taking cell-matrix adhesions to the third dimension; pp. 1708–1712. November. [DOI] [PubMed] [Google Scholar]
- Imamura Y, et al. Comparison of 2D- and 3D-culture models as drug-testing platforms in breast cancer. Oncol Rep. 2015;33(4):1837–1843. doi: 10.3892/or.2015.3767. [DOI] [PubMed] [Google Scholar]
- Jiang L, et al. Reductive carboxylation supports redox homeostasis during anchorage-independent growth. Nature. 2016;532(7598):255–258. doi: 10.1038/nature17393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada KM, Cukierman E. Modeling Tissue Morphogenesis and Cancer in 3D. Cell. 2007;130(4):601–610. doi: 10.1016/j.cell.2007.08.006. [DOI] [PubMed] [Google Scholar]
- Pampaloni F, Reynaud EG, Stelzer EHK. The third dimension bridges the gap between cell culture and live tissue. Nat Rev Mol Cell Biol. 2007;8(10):839–845. doi: 10.1038/nrm2236. [DOI] [PubMed] [Google Scholar]
- Souza GR, et al. Three-dimensional tissue culture based on magnetic cell levitation. Nat Nanotech. 2010;5(4):291–296. doi: 10.1038/nnano.2010.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haisler WL, Timm DM, Gage JA, Tseng H, Killian TC, Souza GR. Three-dimensional cell culturing by magnetic levitation. Nat Protoc. 2013;8(10):1940–1949. doi: 10.1038/nprot.2013.125. [DOI] [PubMed] [Google Scholar]
- Tseng H, et al. A spheroid toxicity assay using magnetic 3D bioprinting and real-time mobile device-based imaging. Sci Rep. 2015;5:13987. doi: 10.1038/srep13987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tseng H, et al. Luminescent Viability Assays in Magnetically Bioprinted 3D Cultures. Promega Corporation; 2015. pp. 1–9. Updated (Figure 1) [Google Scholar]
- Tseng H, et al. Assembly of a three-dimensional multitype bronchiole coculture model using magnetic levitation. Tissue Eng Part C Methods. 2013;19(9):665–675. doi: 10.1089/ten.TEC.2012.0157. [DOI] [PubMed] [Google Scholar]
- Daquinag AC, Souza GR, Kolonin MG. Adipose tissue engineering in three-dimensional levitation tissue culture system based on magnetic nanoparticles. Tissue Eng Part C, Methods. 2013;19(5):336–344. doi: 10.1089/ten.tec.2012.0198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tseng H, et al. A three-dimensional co-culture model of the aortic valve using magnetic levitation. Acta Biomat. 2013;10:173–182. doi: 10.1016/j.actbio.2013.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholls DG, Darley-Usmar VM, Wu M, Jensen PB, Rogers GW, Ferrick D. a Bioenergetic profile experiment using C2C12 myoblast cells. J Vis Exp. 2010. pp. e3–e7. [DOI] [PMC free article] [PubMed]
- Wang R, et al. The Acute Extracellular Flux (XF) Assay to Assess Compound Effects on Mitochondrial Function. J Biomol Screening. 2015;20(3):422–429. doi: 10.1177/1087057114557621. [DOI] [PubMed] [Google Scholar]
- Gibert Y, McGee SL, Ward AC. Metabolic profile analysis of zebrafish embryos. J Vis Exp. 2013. p. e4300. [DOI] [PMC free article] [PubMed]
- Mookerjee SA, Brand MD. Measurement and Analysis of Extracellular Acid Production to Determine Glycolytic Rate. J Vis Exp. 2015. p. e53464. [DOI] [PMC free article] [PubMed]
- Traba J, Miozzo P, Akkaya B, Pierce SK, Akkaya M. An Optimized Protocol to Analyze Glycolysis and Mitochondrial Respiration in Lymphocytes. J Vis Exp. 2016. p. e54918. [DOI] [PMC free article] [PubMed]
- Froeling FEM, et al. Organotypic culture model of pancreatic cancer demonstrates that stromal cells modulate E-cadherin, beta-catenin, and Ezrin expression in tumor cells. Am J Pathol. 2009;175(2):636–648. doi: 10.2353/ajpath.2009.090131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JW, Dang CV. Cancer's molecular sweet tooth and the warburg effect. Cancer Res. 2006;66(18):8927–8930. doi: 10.1158/0008-5472.CAN-06-1501. [DOI] [PubMed] [Google Scholar]
- Holley AK, Dhar SK, St. Clair DK. Curbing cancer's sweet tooth: Is there a role for MnSOD in regulation of the Warburg effect. Mitochondrion. 2013;13(3):170–188. doi: 10.1016/j.mito.2012.07.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegel RL, Miller KD, Jemal A. Cancer statistics, 2017. CA: Cancer J Clinicians. 2017;67(1):7–30. doi: 10.3322/caac.21387. [DOI] [PubMed] [Google Scholar]
- Lin RZ, Chang HY. Recent advances in three-dimensional multicellular spheroid culture for biomedical research. Biotechnol J. 2008;3(9-10):1172–1184. doi: 10.1002/biot.200700228. [DOI] [PubMed] [Google Scholar]
- Kelm JM, Timmins NE, Brown CJ, Fussenegger M, Nielsen LK. Method for generation of homogeneous multicellular tumor spheroids applicable to a wide variety of cell types. Biotechnol Bioeng. 2003;83(2):173–180. doi: 10.1002/bit.10655. [DOI] [PubMed] [Google Scholar]


