ABSTRACT
Background: Biogenic amines (BAs) are metabolites produced by the decarboxylation of amino acids with significant physiological functions in eukaryotic and prokaryotic cells. BAs can be produced by bacteria in fermented foods, but little is known concerning the potential for microbes within the human gut microbiota to produce or degrade BAs.
Objective: To isolate and identify BA-producing and BA-degrading microbes from the human gastrointestinal tract.
Design: Fecal samples from human volunteers were screened on multiple growth media, under multiple growth conditions. Bacterial species were identified using 16S rRNA sequencing and BA production or degradation was assessed using ultra-performance liquid chromatography.
Results: In total, 74 BA-producing or BA-degrading strains were isolated from the human gut. These isolates belong to the genera Bifidobacterium, Clostridium, Enterococcus, Lactobacillus, Pediococcus, Streptococcus, Enterobacter, Escherichia, Klebsiella, Morganella and Proteus. While differences in production or degradation of specific BAs were observed at the strain level, our results suggest that these metabolic activities are widely spread across different taxa present within the human gut microbiota.
Conclusions: The isolation and identification of microbes from the human gut with BA-producing and BA-degrading metabolic activity is an important first step in developing a better understanding of how these metabolites influence health and disease.
KEYWORDS: Biogenic amines, UPLC, gut ecology, bacterial metabolites
Introduction
The gut microbiome is composed of up to 1000 different bacterial species that encode for 100 times more genes than those of the human genome [1]. This community of microorganisms is seen as a virtual organ, responsible for important functions that contribute to the overall health of the host [2]. An imbalance between protective and pathogenic microbes in the gut microbial ecosystem, termed dysbiosis, has often been linked with various disorders, such as inflammatory bowel disease, asthma, obesity and diabetes [3,4].
The importance of the microbiota is related not only to its taxonomic diversity, but also to the metabolites that the bacteria produce and degrade. So far, only a few microbial metabolites have been assessed for their contribution to human health, with the exception being short-chain fatty acids [5]. However, the human gut microbiota produces many other metabolites that may influence the host. In this study, we focused on identifying the human gut-derived bacteria that produce biogenic amines (BAs), including histamine, tyramine, cadaverine, spermine, spermidine and putrescine. BAs are metabolites produced by the decarboxylation of amino acids (AAs), with significant physiological functions in eukaryotic cells as they are precursors for the synthesis of hormones, alkaloids, nucleic acids and proteins [6]. BAs such as histamine are immune mediators and neurotransmitters, whereas others, such as putrescine, spermidine and spermine, are needed for optimal cell growth and differentiation, stabilization of the DNA negative charge, RNA transcription, protein synthesis, apoptosis and regulation of the immune response [7–12]. In bacteria, BAs are also essential for growth and proliferation [13], but the concentrations produced and secreted are far higher than those of eukaryotic cells. This difference could represent, at least in part, a defense mechanism used by bacteria to withstand acidic stress conditions, in addition to many other, so far unknown, cellular and immune regulatory properties [14,15].
The direct influence of this microbial-associated metabolic activity on the human host, i.e. production or degradation of BAs, has been poorly evaluated. Spermine has been previously demonstrated to inhibit pro-inflammatory cytokine secretion by lipopolysaccharide-stimulated peripheral blood mononuclear cells [16] and its mucosal concentration appeared to be reduced in patients with ulcerative colitis compared to healthy controls [17]. Histamine can have both pro-inflammatory and anti-inflammatory effects on immunoregulatory processes, depending on which histamine receptor is activated [18,19]. We previously demonstrated that Lactobacillus saerimneri 30a (which secretes histamine and cadaverine) induces rapid weight loss and immunological effects, dependent on activation of histamine receptor-2 within the mucosa [20]. In addition, the levels of histamine-secreting bacteria were shown to be increased in the gut of asthma patients compared to healthy volunteers, and the level of one histamine-secreting microbe positively correlated with the severity of disease [21]. In murine models, microbiota-derived taurine, histamine and spermine were shown to influence host–microbiome interactions by co-modulating NLRP6 inflammasome signaling, epithelial interleukin-18 secretion and downstream anti-microbial peptide secretion [22].
However, other than histamine, it is unknown which BAs can be produced or degraded by bacteria derived from the human gut. This study aims to isolate and identify some of these microorganisms, so that a better insight can be developed into the biological importance of BA-producing and BA-degrading bacteria from the human gut in the host, as well as of the microbiota itself. Using a rapid and simple screening approach, numerous taxonomically different strains were isolated and identified, and their capacities to produce or degrade BAs were assessed.
Materials and methods
Isolation of bacterial strains
Human adult volunteers (n = 14) collected a single fecal sample at their workplace or home residence using a fecal collection kit (containing plastic gloves, a cardboard seat cover, a plastic container and a disposal bag) and the fecal sample was immediately sealed in the airtight plastic container. The sealed plastic container was placed on ice and transported to the study laboratory within 4 h. Samples were then immediately divided into 1 g aliquots and stored frozen until analysis at −80°C, under ambient air. Bacteria were isolated from frozen fecal samples by adding 2 ml of phosphate-buffered saline (Life Technologies, Grand Island, NY, USA) to 1 g of feces and the obtained suspension was vortexed until homogeneity. The solution was centrifuged at 4°C at 1000 rpm for 3 min to pelletize the fecal material. Then, 100 µl of supernatant was plated on brain–heart infusion agar (BHI; Oxoid, Basingstoke, UK), tryptic soy agar (TSA; Oxoid), de Man–Rogosa–Sharpe agar (MRS; Becton Dickinson, Sparks, MD, USA) and reinforced clostridial medium agar (RCM; Oxoid). Plates were incubated at 37°C under anaerobic conditions using an anaerobic pouch (Oxoid). Random colonies were selected and after three successive transfers on to the corresponding solid medium, pure cultures were obtained. Isolates were stocked in 40% glycerol (Sigma-Aldrich, St. Louis, MO, USA) at −80°C. For clarity, Gram-positive and Gram-negative bacteria are separated in all the tables.
16S rRNA identification of the isolates
Molecular identification of the isolates was performed by sequencing the 16S ribosomal RNA (rRNA) regions without a prior DNA extraction step, by colony polymerase chain reaction (PCR). A single colony was isolated and DNA amplified via PCR using primers 27F and 1492R and GoTaq Green Master Mix polymerase (Promega Corporation, Madison, WI, USA) under the following conditions: 95°C for 4 min; 95°C for 40 s, 54°C for 45 s and 72°C for 1 min (32 cycles); and 72°C for 5 min. The 16S rRNA genes were sequenced by GATC Biotech (Constance, Germany) and subsequently compared with all GenBank entries by a BLASTN search.
Quantification of BAs
To assess BA production by the isolated bacteria, all strains were grown in liquid medium (BHI, TSB, MRS and RCM) containing pyridoxal-5-phosphate at 37°C and under anaerobic conditions in 14 ml round-bottomed polystyrene tubes. In addition, medium was supplemented with an AA cocktail (0.1% final concentration each of histidine, lysine, arginine, ornithine and tyrosine) or a BA cocktail (0.03% final concentration each of histamine, cadaverine, spermine, spermidine, putrescine and tyramine). Bacterial supernatants were collected after 48 h culture by centrifugation at 14,000 rpm for 10 min. Samples were stored at −20°C until derivatization.
Standard chemicals for spermidine trihydrochloride, histamine dihydrochloride, putrescine dihydrochloride (Acros Organics, Morris, NJ, USA), cadaverine dihydrochloride, spermine tetrahydrochloride and tyramine hydrochloride (Sigma-Aldrich, St. Louis, MO, USA) were of analytical grade. Standard stock solutions were prepared at 1000 mg/l in distilled water, and were further diluted to generate standard curves.
In the next step, 475 μl of bacterial supernatant or standard solution was mixed with 25 μl of 5 g/l 1,7-diaminoheptane (Internal standard; Sigma-Aldrich, St. Louis, MO, USA), 100 µl of 2 M NaOH, 150 µl of saturated sodium bicarbonate solution and 1 ml of dansyl chloride (Sigma-Aldrich) solution (10 mg/ml in acetone), and then incubated at 40°C, 200 rpm, for 45 min. Residual dansyl chloride was removed by adding 50 µl of 25% ammonium hydroxide (Merck, Darmstadt, Germany). After 30 min at 25°C, the volume was adjusted to 2.5 ml with acetonitrile (Biosolve Chimie, Dieuze, France) and centrifuged at 3500 rpm for 5 min, and supernatants were filtered (0.22 µm) before ultra-performance liquid chromatography (UPLC) analysis. Duplicate samples were analyzed in parallel.
Separation was carried out by UPLC on an ACQUITY UPLC H-Class Bio System (Waters Corp., Milford, MA, USA) equipped with a quaternary solvent manager, sample manager with flow-through needle, column manager and diode array detector. Data processing was performed using MassLynx v. 4.1 (Waters Corp.). Based on their different hydrophobicities, the dansylated BAs were separated on an ACQUITY UPLC BEHC18 column (1.7 µm particle size, 2.1 mm x 50 mm; Waters Corp.) and the samples were eluted with a gradient elution of (A) acetonitrile (100%), (B) acetonitrile (50%) as follows: 0–0.72 min, A 40%, B 60%; 0.72–1.07 min, A 40–80%, B 60–20%; 1.07–1.42 min, A 80–90%, B 20–10%; 1.42–2.11 min, A 90–95%, B 10–5%; 2.11–2.46 min, A 95–40%, B 5–60%, 2.46–4.20 min, A 40%, B 60%. The flow rate was kept at 0.6 ml/min, column temperature at 25°C and injection 1 µl and the detection wavelength was 217 nm.
Results
Isolation of gut bacterial strains
Extensive literature reviews suggested that the BA-producing bacteria isolated from food were mostly facultative anaerobes and generally belong to the phyla Firmicutes and Proteobacteria. Therefore, we decided to search for new isolates using the media MRS, RCM, TSB and BHI, under anaerobic conditions generated by anaerobic pouches.
From the 14 volunteers [seven male and seven female, all Caucasian, with a median age of 39 years (range 22–60 years) and median body mass index of 23.2 kg/m2 (range 20.2–24.902 kg/m2)], a total of 74 presumptive BA-producing strains was isolated. The 16S rRNA gene sequences of the isolates were determined and compared with those deposited in Genbank. The results showed that the strains were closely related (similarity of >99% for all strains) to the genera Bifidobacterium, Clostridium, Enterococcus, Lactobacillus, Pediococcus, Streptococcus, Enterobacter, Escherichia, Klebsiella, Morganella and Proteus. In total, 54 isolates were Gram positive (Table 1) and 20 isolates were Gram negative (Table 2).
Table 1.
Gram-positive isolates, medium used for their isolation and 16S rRNA Genbank accession number.
| Strain | Medium | Genbank accession no. | |
|---|---|---|---|
| Bifidobacterium adolescentis | A.1 | BHI | KX674032 |
| Bifidobacterium longum | A.2 | RCM | KX674040 |
| A.3 | RCM | KX673988 | |
| Bifidobacterium pseudocatenulatum | A.4 | RCM | KX674010 |
| A.5 | RCM | KX673984 | |
| A.6 | BHI | KX674008 | |
| A.7 | RCM | KX673991 | |
| A.8 | RCM | KX673986 | |
| Clostridium perfringens | B.1 | BHI | KX674025 |
| B.2 | BHI | KX674031 | |
| B.3 | BHI | KX674026 | |
| Enterococcus avium | C.1 | TSB | KX673997 |
| C.2 | BHI | KX674028 | |
| Enterococcus faecalis | C.3 | TSB | KX674015 |
| C.4 | TSB | KX674016 | |
| C.5 | TSB | KX674019 | |
| C.6 | TSB | KX674022 | |
| C.7 | TSB | KX674023 | |
| C.8 | TSB | KX674046 | |
| C.9 | TSB | KX674051 | |
| C.10 | TSB | KX674048 | |
| C.11 | TSB | KX674045 | |
| C.12 | TSB | KX674012 | |
| C.13 | TSB | KX674013 | |
| C.14 | TSB | KX674014 | |
| Enterococcus faecium | C.15 | MRS | KX674033 |
| Enterococcus gallinarum | C.16 | BHI | KX674030 |
| Enterococcus sp. | C.17 | RCM | KX673985 |
| Lactobacillus crispatus | D.1 | MRS | KX674034 |
| D.2 | MRS | KX674035 | |
| Lactobacillus fermentum | D.3 | MRS | KX674011 |
| D.4 | MRS | KX674003 | |
| D.5 | MRS | KX674004 | |
| D.6 | MRS | KX674002 | |
| D.7 | RCM | KX674007 | |
| Lactobacillus gasseri | D.8 | MRS | KX673989 |
| D.9 | MRS | KX673998 | |
| D.10 | MRS | KX673995 | |
| Lactobacillus salivarius | D.11 | MRS | KX674005 |
| D.12 | MRS | KX673996 | |
| D.13 | MRS | KX674006 | |
| D.14 | MRS | KX673990 | |
| D.15 | MRS | KX673994 | |
| D.16 | RCM | KX674009 | |
| D.17 | RCM | KX673980 | |
| D.18 | RCM | KX673983 | |
| D.19 | RCM | KX673981 | |
| D.20 | MRS | KX673993 | |
| D.21 | MRS | KX674037 | |
| Lactobacillus vaginalis | D.22 | MRS | KU612267 |
| Pediococcus pentosaceus | E.1 | MRS | KX674036 |
| E.2 | MRS | KX674038 | |
| Streptococcus salivarius | F.1 | MRS | KX674039 |
| Streptococcus vestibularis | F.2 | MRS | KX673987 |
BHI, brain–heart infusion; RCM, reinforced clostridial medium; TSB, tryptic soy broth; MRS, de Man–Rogosa–Sharpe.
Table 2.
Gram-negative isolates, medium used for their isolation and 16S rRNA Genbank accession number.
| Strain | Medium | Genbank accession no. | |
|---|---|---|---|
| Enterobacter cloacae | G.1 | TSB | KX674047 |
| Escherichia coli | H.1 | TSB | KX673982 |
| H.2 | TSB | KX674041 | |
| H.3 | TSB | KX674042 | |
| H.4 | TSB | KX673999 | |
| H.5 | TSB | KX674000 | |
| H.6 | TSB | KX674001 | |
| H.7 | TSB | KX674043 | |
| H.8 | TSB | KX674044 | |
| H.9 | TSB | KX674017 | |
| H.10 | TSB | KX674018 | |
| H.11 | TSB | KX674020 | |
| H.12 | TSB | KX674021 | |
| H.13 | BHI | KX674027 | |
| H.14 | BHI | KX674029 | |
| Escherichia fergusonii | H.15 | TSB | KX673992 |
| Klebsiella pneumoniae | I.1 | TSB | KX674052 |
| I.2 | TSB | KX674024 | |
| Morganella morganii | J.1 | TSB | KU6122676 |
| Proteus mirabilis | K.1 | TSB | KX674049 |
TSB, tryptic soy broth; BHI, brain–heart infusion.
Bacterial production of BAs in the presence of AAs
To assess the production of BAs, strains were grown for 48 h in the presence of an AA cocktail (containing histidine, lysine, arginine, ornithine and tyrosine) and supernatants were subsequently assessed by UPLC. All isolates produced at least one BA; however, significant differences between species were observed. A representative UPLC plot is shown in supplementary Figure S1.
In general, Gram-negative bacteria seemed to generate more BAs than Gram-positive bacteria (Figure 1). Putrescine and cadaverine were the main BAs produced by the majority of Gram-negative bacteria, but were not usually secreted by Gram-positive bacteria (Figure 1). The only exceptions were the two strains of Enterococcus faecalis (C.10 and C.11) that were capable of producing cadaverine. Histamine secretion was rarely observed, with only one Gram-positive strain (Lactobacillus vaginalis, D.22) and one Gram-negative strain (Morganella morganii, J.1) showing high levels of histamine secretion (Figure 1).
Figure 1.
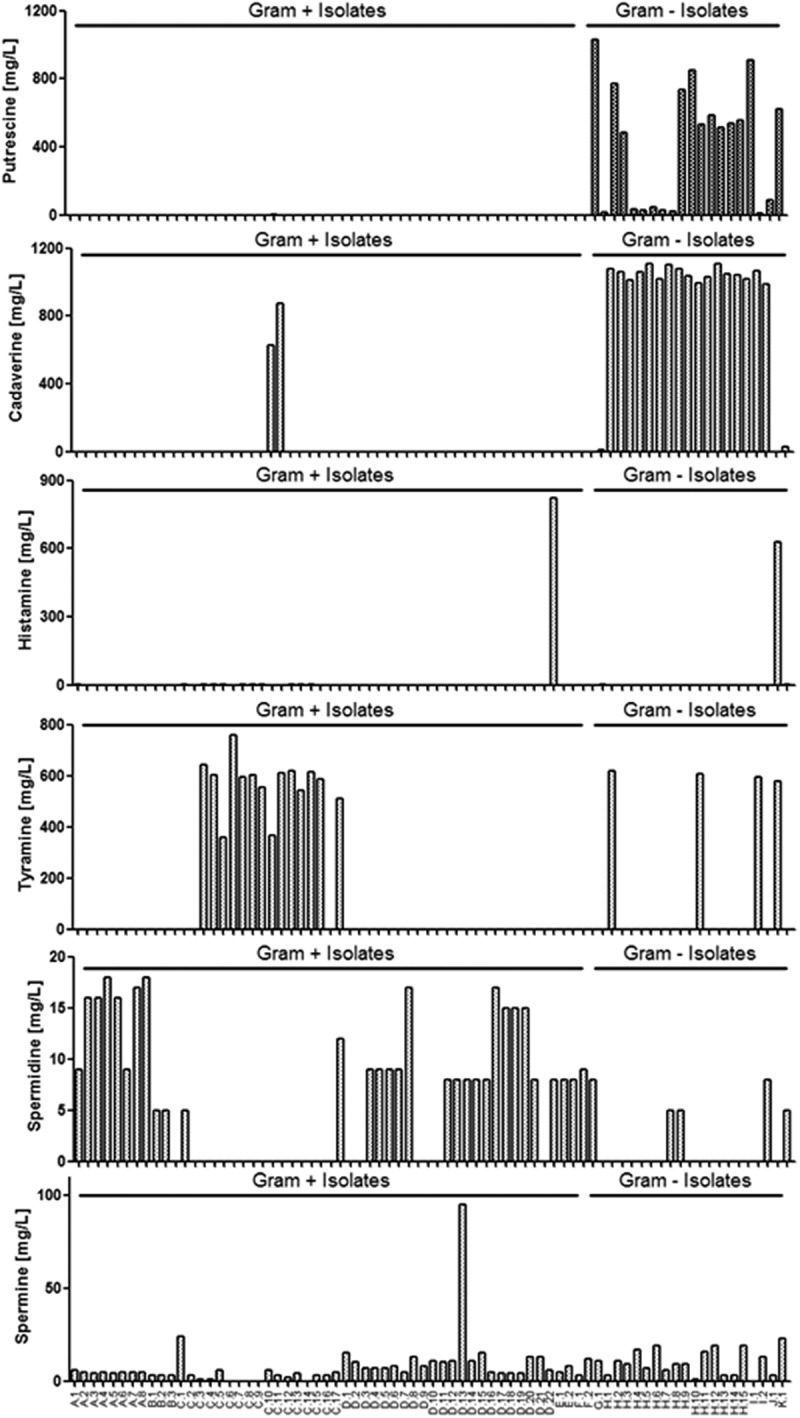
Gut-derived bacteria produce biogenic amines from amino acids (AAs). Production of putrescine, cadaverine, histamine, tyramine, spermidine and spermine (mg/l) in the presence of AAs by isolated strains is illustrated. Results are expressed as the mean of two independent experiments. In total, 54 isolates are Gram positive (Table 1) and 20 isolates are Gram negative (Table 2).
Tyramine was produced by the majority of Enterococcus strains, but in some species specificity was observed since tyramine was not produced by the E. avium or E. gallinarum isolates (Figure 1). Tyramine was produced by only four Gram-negative isolates: two Escherichia coli strains (H.1 and H.10), one Klebsiella pneumoniae strain (I.1) and Morganella morganii (J.1).
Spermidine was produced by all strains from the genera Bifidobacterium, by two of the three Clostridium isolates (B.1 and B.2), but by only two of the 17 Enterococcus strains (C.1 and C.17). Lactobacillus crispatus and Lactobacillus gasseri strains were not able to produce spermidine, but Lactobacillus fermentum, Lactobacillus salivarius (except D.21) and Lactobacillus vaginalis were capable of producing spermidine (Figure 1). The Pediococcus and Streptococcus isolates also secreted spermidine (Figure 1). For Gram-negative bacteria, only two E. coli strains (H.7 and H.8), one K. pneumoniae strain (I.2) and Proteus mirabilis (K.1) were capable of secreting spermidine under these culture conditions (Figure 1).
Spermine was secreted by all Gram-positive strains, except for five Enterococcus isolates (C.6, C.7, C.8, C.9 and C.14). Similarly, spermine was secreted by all Gram-negative isolates, except for K. pneumoniae I.1 (Figure 1).
Tabulated data for Gram-positive and Gram-negative bacteria can be observed in supplementary Tables S1 and S2, respectively.
Bacterial production and degradation of BAs in the presence of BAs
In vivo, bacteria are exposed to multiple substrates and may not produce BAs in the sole presence of AAs. Thus, we determined whether these isolated strains could produce or degrade BAs in the presence of other BAs (putrescine, cadaverine, histamine, tyramine, spermidine and spermine). Strains were grown for 48 h in the presence of the BA cocktail and supernatants were quantified by UPLC.
Putrescine levels were decreased following incubation with the majority of Gram-positive strains (Figure 2), other than the Enterococcus strains C.1, C.10 and C.11. Gram-negative strains increased or decreased putrescine levels independently of species, with some strains increasing putrescine levels and other strains decreasing levels.
Figure 2.
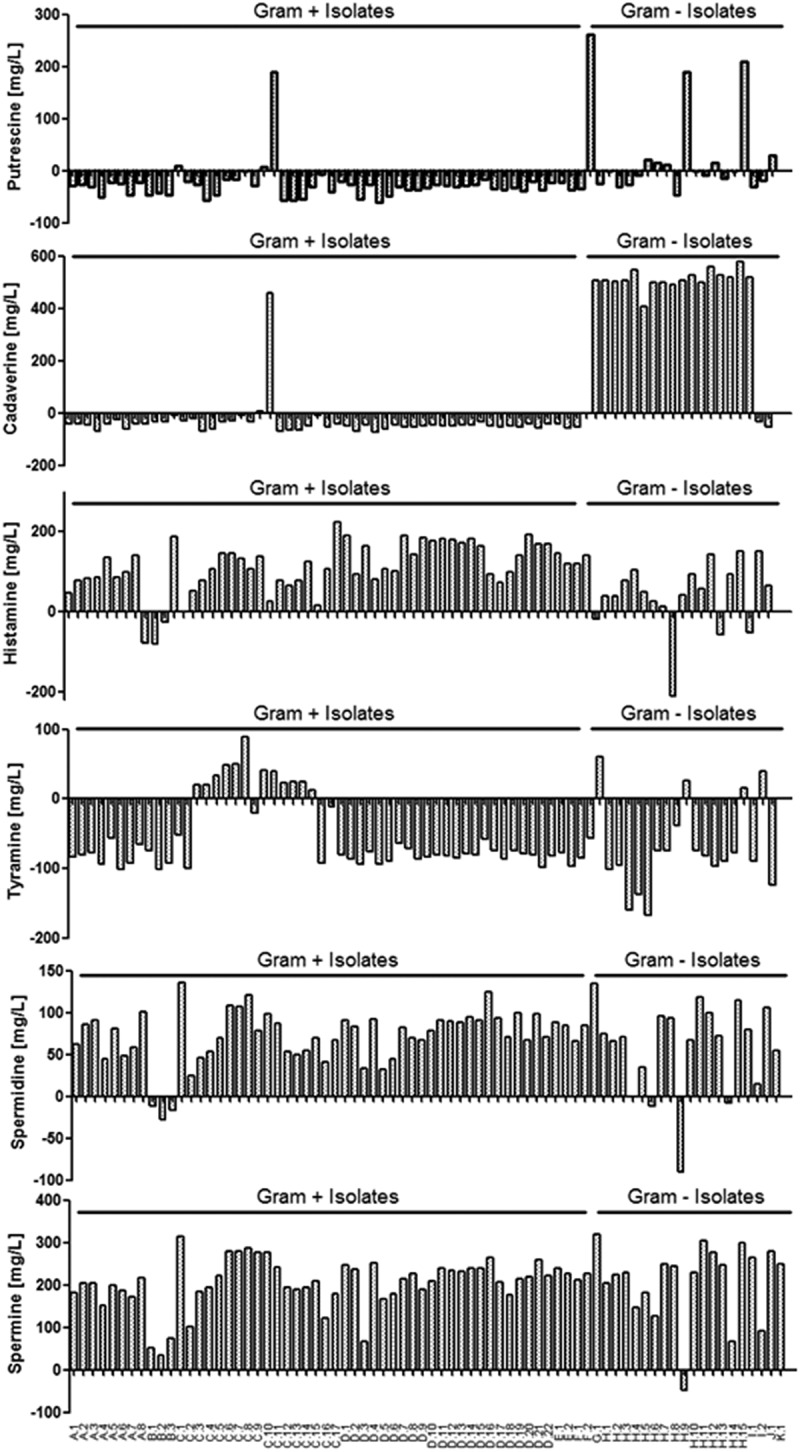
Gut-derived bacterial biogenic amine (BA) metabolism is altered in the presence of BAs. Production and degradation of putrescine, cadaverine, histamine, tyramine, spermidine and spermine (mg/l) in the presence of BAs by isolated strains is illustrated. A positive number represents an increase, while a negative number represents a decrease in BA concentration. Results are expressed as the mean of two independent experiments. In total, 54 isolates are Gram positive (Table 1) and 20 isolates are Gram negative (Table 2).
Similarly to changes in putrescine levels, the majority of Gram-positive strains utilized or degraded cadaverine that led to reduced cadaverine levels (Figure 2). Two of the three Enterococcus isolates that had increased putrescine levels also increased cadaverine levels (C.10 and C.11). Co-incubation of BAs with Gram-negative microbes resulted in increased levels of cadaverine for the majority of isolates tested (Figure 2). The only exceptions were Enterobacter cloacae (G.1), M. morganii (J.1) and P. mirabilis (K.1).
In contrast, the vast majority of both Gram-positive and Gram-negative isolates increased the level of histamine when co-cultured in the presence of the BA cocktail (Figure 2). However, a small number of strains was able to degrade and/or utilize histamine. All of the Gram-positive Clostridium perfringens isolates (B.1, B.2 and B.3) reduced histamine levels, while the Gram-negative strains E. coli (H.1, H.9 and H.14) and K. pneumoniae (I.2) also reduced histamine levels. Of note, E. coli H.9 completely removed all histamine that was added to the culture. The UPLC plot for E. coli H.9 illustrating the removal of histamine can be observed in supplementary Figure S2.
Tyramine levels were reduced by all Gram-positive isolates, except for Enterococcus species (Figure 2). As described above, tyramine was not produced by the E. avium or E. gallinarum isolates in the presence of AAs and also was not produced in the presence of BAs (Figure 2). Similarly to the Gram-positive isolates, tyramine levels were reduced by the majority of Gram-negative bacteria, except for E. coli (H.1 and H.10), M. morganii (J.1) and K. pneumoniae (I.2).
Spermidine levels were increased following co-incubation with BAs for the majority of both Gram-positive and Gram-negative bacteria (Figure 2). However, all the Gram-positive C. perfringens isolates (B.1, B.2 and B.3) reduced spermidine levels, while the Gram-negative strains E. coli (H.6, H.9 and H.14) also reduced spermidine levels. This metabolic activity is similar to that described above for the reduction of histamine levels.
Finally, spermine levels were increased following co-incubation of BAs with all microbes tested, with the exception of the Gram-negative strain E. coli (H.9), which seems to have the unusual ability to degrade and/or utilize the majority of BAs tested.
Tabulated data for differences in BA concentrations compared to the uninoculated control media (supplemented only with the BA cocktail) are shown in supplementary Tables S3 and S4 for Gram-positive and Gram-negative strains, respectively.
Discussion
The in vivo influence of microbial-derived BAs on the host is still poorly understood. Nonetheless, with the high level of BA monitoring in food products [23], it is equally important to determine the identity of bacterial strains and influence of the production of BAs by the resident microbiota in the human gut, independently from consumption of these BAs. Our data suggest that many different microbial species can produce, utilize or degrade BAs.
Of the 74 gut isolates we tested, the production or degradation of BAs appeared to be a common, rather than a rare, microbial metabolic activity, with some important differences between species and strains. For example, some strains could produce histamine and/or tyramine at levels above the maximum limit tolerated in fish products (histamine 200 mg/kg; tyramine 100 mg/kg), as required by the US Food and Drug Administration (FDA) and the World Health Organization (WHO) [24,25]. Tyramine secretion was observed for most of the Enterococcus species, as reported before for Enterococcus strains isolated from meat, cheese, fish and wine [26]. The secretion of tyramine from Gram-negative bacteria could not be associated with a specific species; rather, it was a strain-specific trait (observed for two E. coli strains, one K. pneumoniae strain and M. morganii). Potentially, horizontal gene transfer could represent a putative mechanism for dissemination of tyramine-related metabolic genes across different taxa [27,28].
Putrescine and cadaverine were secreted primarily by Gram-negative bacteria, as previously reported by other research groups [29,30]. The production of cadaverine by two strains of E. faecalis is, however, more surprising, since previous reports did not detect cadaverine production by this species [31]. Although cadaverine is only toxic at high doses, it was previously shown to potentiate histamine toxicity in rats and guinea pigs [32]. More studies are needed to determine the synergic effects between BAs, but an in vivo imbalance of BAs could result in damage to the host.
An increased level of histamine in the gastrointestinal tract is associated with a range of mucosal inflammatory disorders [20,22]. In this study, elevated histamine production was observed in the presence of AAs only with strains M. morganii and L. vaginalis, as previously reported by others [33,34]. More surprisingly, an increase in histamine concentration was observed for almost all strains when incubated with BAs.
Spermidine and spermine are usually formed from putrescine, which probably explains the significant increase in the secretion of these two BAs when the bacterial isolates were incubated with the BA cocktail, compared to the AAs cocktail. However, many of the Gram-positive strains were able to secrete both spermidine and spermine in the presence of AA alone, even though no putrescine was secreted by these microbes. This suggests that all the putrescine formed by these strains was immediately converted, or that spermidine and spermine were formed from an alternative substrate. Spermidine and spermine are molecules with known regulatory functions in prokaryotic and eukaryotic cells [13], and their concentration appeared to be normally tightly controlled in bacteria [35]. That drastic difference in histamine, spermidine and spermine concentrations may represent a system of regulation in the presence of one or several BAs. Very few data to date have suggested that BAs possess self-regulatory activities; however, it was demonstrated that the protein AtoC/Az (an ornithine decarboxylase inhibitor) from E. coli was regulated at a transcriptional level in the presence of BAs [36].
The in vivo impact of this drastic change in BAs levels is difficult to interpret at this stage, but taken together, these data emphasize the importance of further exploring the regulatory mechanisms behind the production of BAs by bacteria, as well as the synergistic effects of BAs from a toxicological point of view. One obvious limitation of our current study is that we did not examine these isolates for their ability to secrete catecholamines and serotonin, which are important neurotransmitters. However, recent studies by others have clearly shown the presence and importance of these metabolites from gut-associated microbes, including the bidirectional character of microbiota–host interactivity via neuromediators including BAs [37,38].
A small number of bacteria isolated from food has been described as BA degraders, including Micrococcus varians [39], Natrinema gari [40], Brevibacterium linen [41], Vergibacillus sp. SK33 [42], Lactobacillus sakei, Lactobacillus curvatus [43] and Staphylococcus xylosus [44]. To our knowledge, there is no information available about the in vivo presence of BA-degrading bacteria within the gut of humans. In this study, we observed that all isolated strains of C. perfringens had the highest overall BA-degrading capacities, with decreased concentrations of putrescine, cadaverine, histamine, tryramine and spermidine, and increased concentration of spermine. Previous reports have shown that C. perfringens was a BA producer rather than a BA degrader, with secretion of histamine [45], cadaverine and putrescine being described [46]. The strains isolated in this study did not display this trend even when grown in the presence of AAs. One could speculate that the BA degraders might be used as potential beneficial strains to counteract an unbalanced gut microbiota consisting of harmful BA producers. Of note, the L. fermentum and B. pseudocatenulatum strains displayed good overall degradative capabilities and are known as potential probiotic species. On the other hand, increased exposure to BA degradation products may be a contributory factor to the development of various serious chronic and acute diseases [47]. More studies are therefore required to determine the impact and role of BA degraders in host–microbiota interactions.
Finally, one BA-degrading bacterium that caught our attention was E. coli H.9. Besides being able to utilize putrescine, tyramine and spermidine, E. coli H.9 is the only isolate that was able to remove 100% of the histamine present in the medium. Further physiological and genomic analyses are currently underway with this strain to determine the molecular mechanisms underpinning this activity, and further studies will examine the functional ability of this microbe to reduce the severity of allergic responses mediated by histamine.
In this study, we report for the first time the diversity of human gut bacteria producing and/or degrading BAs. These isolates were taxonomically heterogeneous and produced dissimilar levels of BAs depending on the strains and the culture conditions used. Additional studies are needed to determine how these BAs interact and are regulated in vivo, which, in turn, would help us to understand the complex mechanisms of BA toxicity occurring between host and microbiome. Our data also emphasize that studies on host–microbiota interaction relying solely on 16S rRNA sequencing may lack essential metabolomic information needed to fully understand the metabolic and immunological impact of an altered gut microbiota composition.
Supplementary Material
Funding Statement
This work was supported by the Swiss National Science Foundation (SNSF) [CRSII3_154488].
Disclosure statement
Liam O’Mahony has received research support from GSK and consulted for Alimentary Health Ltd. Cezmi Akdis has received research support from Novartis and Stallergenes and consulted for Actellion, Aventis and Allergopharma. The remaining authors have no potential conflicts of interest.
Supplemental data
Supplemental data for this article can be accessed here.
References
- [1].Ley RE, Turnbaugh PJ, Klein S, et al. Microbial ecology: human gut microbes associated with obesity. Nature. 2006;444:1022–9. [DOI] [PubMed] [Google Scholar]
- [2].Frei R, Lauener RP, Crameri R, et al. Microbiota and dietary interactions – an update to the hygiene hypothesis? Allergy. 2012;67:451–461. [DOI] [PubMed] [Google Scholar]
- [3].Carding S, Verbeke K, Vipond D, et al. Dysbiosis of the gut microbiota in disease. Microb Ecol Health D. 2015;26:26191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Huang YJ, Marsland BJ, Bunyavanich S, et al. The microbiome in allergic disease: current understanding and future opportunities-2017 PRACTALL document of the American academy of allergy, asthma & immunology and the european academy of allergy and clinical immunology. J Allergy Clin Immunol. 2017;139:1099–1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Barcik W, Untersmayr E, Pali-Schöll I, et al. Influence of microbiome and diet on immune responses in food allergy models. Drug Discov Today Dis Models. 2016;17-18:71–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Spano G, Russo P, Lonvaud-Funel A, et al. Biogenic amines in fermented foods. Eur J Clin Nutr. 2010;64:S95–S100. [DOI] [PubMed] [Google Scholar]
- [7].Igarashi K, Kashiwagi K.. Modulation of cellular function by polyamines. Int J Biochem Cell Biol. 2010;42:39–51. [DOI] [PubMed] [Google Scholar]
- [8].Tsavkelova EA, Botvinko IV, Kudrin VS, et al. Detection of neuromediator amines in microorganisms by high-performance liquid chromatography. Dokl Akad Nauk. 2000;372:840–842. [PubMed] [Google Scholar]
- [9].Shishov VA, Kirovskaya TA, Kudrin VS, et al. Amine neuromediators, their precursors, and oxidation products in the culture of Escherichia coli K-12. Appl Biochem Micro. 2009;45:494–497. [PubMed] [Google Scholar]
- [10].Yunes RA, Poluektova RU, Dyachkova MS, et al. GABA production and structure of gadB/gadC genes in Lactobacillus and Bifidobacterium strains from human microbiota. Anaerobe. 2016;42:1–8. [DOI] [PubMed] [Google Scholar]
- [11].Özogul F. Production of biogenic amines by Morganella morganii, Klebsiella pneumonia and Hafnia alvii using a rapid HPLC method. Eur Food Res Technol. 2004;219:465–469. [Google Scholar]
- [12].Oleskin AV, Zhilenkova OG, Shenderov BA, et al. Lactic-acid bacteria supplement fermented dairy products with human behavior-modifying neuroactive compounds. J Pharm Nutrit Sci. 2014;4P:199–206. [Google Scholar]
- [13].Shah P, Swiatlo E. A multifaceted role for polyamines in bacterial pathogens. Mol Microbiol. 2008;68:4–16. [DOI] [PubMed] [Google Scholar]
- [14].Rhee JE, Rhee JH, Ryu PY, et al. Identification of the cadBA operon from Vibrio vulnificus and its influence on survival to acid stress. FEMS Microbiol Lett. 2002;208:245–251. [DOI] [PubMed] [Google Scholar]
- [15].Lee YH, Kim BH, Kim JH, et al. CadC has a global translational effect during acid adaptation in Salmonella enterica serovar typhimurium. J Bacteriol. 2007;189:2417–2425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Zhang M, Caragine T, Wang H, et al. Spermine inhibits proinflammatory cytokine synthesis in human mononuclear cells: a counterregulatory mechanism that restrains the immune response. J Exp Med. 1997;185:1759–1768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Weiss TS, Herfarth H, Obermeier F, et al. Intracellular polyamine levels of intestinal epithelial cells in inflammatory bowel disease. Inflamm Bowel Dis. 2004;10:529–535. [DOI] [PubMed] [Google Scholar]
- [18].Frei R, Ferstl R, Konieczna P, et al. Histamine receptor 2 modifies dendritic cell responses to microbial ligands. J Allergy Clin Immunol. 2013;132:194–204. [DOI] [PubMed] [Google Scholar]
- [19].Smolinska S, Jutel M, Crameri R, et al. Histamine and gut mucosal immune regulation. Allergy. 2014;69:273–281. [DOI] [PubMed] [Google Scholar]
- [20].Ferstl R, Frei R, Schiavi E, et al. Histamine receptor 2 is a key influence in immune responses to intestinal histamine-secreting microbes. J Allergy Clin Immunol. 2014;134:744–746. [DOI] [PubMed] [Google Scholar]
- [21].Barcik W, Pugin B, Westermann P, et al. Histamine-secreting microbes are increased in the gut of adult asthma patients. J Allergy Clin Immunol. 2016;138:1491–1494. [DOI] [PubMed] [Google Scholar]
- [22].Levy M, Thaiss CA, Zeevi D, et al. Microbiota-modulated metabolites shape the intestinal microenvironment by regulating NLRP6 inflammasome signaling. Cell. 2015;163:1428–1443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Ladero V, Calles-Enriquez M, Fernandez M, et al. Toxicological effects of dietary biogenic amines. Curr Nutr Food Sci. 2010;6:145–156. [Google Scholar]
- [24].US Food & Drug Administration report Processing parameters needed to control pathogens in cold smoked fish; Chapter IV. Potential hazards in cold-smoked fish: biogenic amines. Available from: http://www.fda.gov/Food/FoodScienceResearch/SafePracticesforFoodProcesses/ucm094576.htm.
- [25].Joint FAO/WHO food standards programme codex committee on fish and fishery products. Available from: ftp://ftp.fao.org/codex/meetings/ccffp/ccffp33/fp33_12e.pdf.
- [26].Ladero V, Fernández M, Calles-Enríquez M, et al. Is the production of the biogenic amines tyramine and putrescine a species-level trait in enterococci? Food Microbiol. 2012;30:132–138. [DOI] [PubMed] [Google Scholar]
- [27].Coton E, Coton M. Evidence of horizontal transfer as origin of strain to strain variation of the tyramine production trait in Lactobacillus brevis. Food Microbiol. 2009;26:52–57. [DOI] [PubMed] [Google Scholar]
- [28].Lucas PM, Wolken WAM, Claisse O, et al. Histamine-producing pathway encoded on an unstable plasmid in Lactobacillus hilgardii 0006. Appl Environ Microbiol. 2005;71:1417–1424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Buňková L, Buňka F, Klčovská P, et al. Formation of biogenic amines by Gram-negative bacteria isolated from poultry skin. Food Chem. 2010;121:203–206. [Google Scholar]
- [30].Pircher A, Bauer F, Paulsen P. Formation of cadaverine, histamine, putrescine and tyramine by bacteria isolated from meat, fermented sausages and cheeses. Eur Food Res Technol. 2007;226:225–231. [Google Scholar]
- [31].Sarantinopoulos P, Andrighetto C, Georgalaki MD, et al. Biochemical properties of enterococci relevant to their technological performance. Int Dairy J. 2001;11:621–647. [Google Scholar]
- [32].Lyons DE, Beery JT, Lyons SA, et al. Cadaverine and aminoguanidine potentiate the uptake of histamine in vitro in perfused intestinal segments of rats. Toxicol Appl Pharmacol. 1983;70:445–458. [DOI] [PubMed] [Google Scholar]
- [33].Klausen NK, Huss HH. Growth and histamine production by Morganella morganii under various temperature conditions. Int J Food Microbiol. 1987;5:147–156. [Google Scholar]
- [34].Diaz M, del Rio B, Ladero V, et al. Isolation and typification of histamine-producing Lactobacillus vaginalis strains from cheese. Int J Food Microbiol. 2015;215:117–123. [DOI] [PubMed] [Google Scholar]
- [35].Tabor H, Tabor CW. Biosynthesis and metabolism of 1,4-diaminobutane, spermidine, spermine, and related amines. Adv Enzymol Relat Areas Mol Biol. 1972;36:203–268. [DOI] [PubMed] [Google Scholar]
- [36].Filippou PS, Lioliou EE, Panagiotidis CA, et al. Effect of polyamines and synthetic polyamine-analogues on the expression of antizyme (AtoC) and its regulatory genes. BMC Biochem. 2007;8:1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Patterson E, Cryan JF, Fitzgerald GF, et al. Gut microbiota, the pharmabiotics they produce and host health. Proc Nutr Soc. 2014;73:477–789. [DOI] [PubMed] [Google Scholar]
- [38].Wall R, Cryan JF, Ross RP, et al. Bacterial neuroactive compounds produced by psychobiotics. Adv Exp Med Biol. 2014;817:221–239. [DOI] [PubMed] [Google Scholar]
- [39].Leuschner RGK, Hammes WP. Tyramine degradation by micrococci during ripening of fermented sausage. Meat Sci. 1998;49:289–296. [DOI] [PubMed] [Google Scholar]
- [40].Tapingkae W, Tanasupawat S, Parkin KL, et al. Degradation of histamine by extremely halophilic archaea isolated from high salt-fermented fishery products. Enzyme Microbial Technol. 2010;46:92–99. [Google Scholar]
- [41].Leuschner RGK, Hammes WP. Degradation of histamine and tyramine by Brevibacterium linens during surface ripening of Munster cheese. J Food Prot. 1998;61:874–878. [DOI] [PubMed] [Google Scholar]
- [42].Yongsawatdigul J, Rodtong S, Raksakulthai N. Acceleration of Thai fish sauce fermentation using proteinases and bacterial starter cultures. J Food Sci. 2007;72:M382–M390. [DOI] [PubMed] [Google Scholar]
- [43].Dapkevicius MLNE, Nout MJR, Rombouts FM, et al. Biogenic amine formation and degradation by potential fish silage starter microorganisms. Int J Food Microbiol. 2000;57:107–114. [Google Scholar]
- [44].Mah JH, Hwang HJ. Inhibition of biogenic amine formation in a salted and fermented anchovy by Staphylococcus xylosus as a protective culture. Food Control. 2009;20:796–801. [Google Scholar]
- [45].Yoshinaga DH, Frank HA. Histamine-producing bacteria in decomposing skipjack tuna (Katsuwonus pelamis). Appl Environ Microbiol. 1982;44:447–452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Allison C, Macfarlane GT. Influence of pH, nutrient availability, and growth rate on amine production by bacteroides fragilis and clostridium perfringens. Appl Environ Microbiol. 1989;55:2894–2898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Pegg EA. Toxicity of polyamines and their metabolic products. Chem Res Toxicol. 2013;26:1782–1800. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


