Abstract
For years, distinctions between skeletal muscle fiber types were best visualized by myosin-ATPase staining. More recently, immunohistochemical staining of myosin heavy chain (MyHC) isoforms has emerged as a finer discriminator of fiber-type. Type I, type IIA, type IIX and type IIB fibers can now be identified with precision based on their MyHC profile; however, manual analysis of these data can be slow and down-right tedious. In this regard, rapid, accurate assessment of fiber-type composition and morphology is a very desirable tool. Here, we present a protocol for state-of-the-art immunohistochemical staining of MyHCs in frozen sections obtained from mouse hindlimb muscle in concert with a novel semi-automated algorithm that accelerates analysis of fiber-type and fiber morphology. As expected, the soleus muscle displayed staining for type I and type IIA fibers, but not for type IIX or type IIB fibers. On the other hand, the tibialis anterior muscle was composed predominantly of type IIX and type IIB fibers, a small fraction of type IIA fibers and little or no type I fibers. Several image transformations were used to generate probability maps for the purpose of measuring different aspects of fiber morphology (i.e., cross-sectional area (CSA), maximal and minimal Feret diameter). The values obtained for these parameters were then compared with manually-obtained values. No significant differences were observed between either mode of analysis with regards to CSA, maximal or minimal Feret diameter (all p > 0.05), indicating the accuracy of our method. Thus, our immunostaining analysis protocol may be applied to the investigation of effects on muscle composition in many models of aging and myopathy.
Keywords: Medicine, Issue 126, Skeletal muscle, fiber-type, histology, immunohistochemistry, myosin heavy chain, automated analysis
Introduction
It has been known for some time that skeletal muscle is composed of single fibers of many types1. Initially, two groups of fibers were characterized based on their contractile properties and named, appropriately, slow-twitch (type I) and fast-twitch (type II). These categories were further distinguished on the basis of fiber metabolism. Since type I fibers are rich in mitochondria and reliant on oxidative metabolism, they were elucidated by robustly positive nicotinamide adenine dinucleotide-tetrazolium reductase (NADH-TR) diaphorase2 or succinate dehydrogenase (SDH)3 staining. By contrast, type II fibers exhibited lesser and variable degrees of NADH-TR diaphorase or SDH staining and were divided into two fast-twitch subgroups (type IIA and type IIB) somewhat crudely based on their relative oxidative capacities. These distinctions between fibers have been visualized more effectively by myosin-ATPase staining where type I fibers stain dark after a pre-incubation at pH 4.0 and type IIB fibers absorb precipitate following pre-incubation at pH 10.0 with type IIA fibers staining intermediately4.
More recently, immunohistochemical staining of myosin heavy chain (MyHC) isoforms has emerged as a finer discriminator of fiber-type5. Type I, type IIA and type IIB fibers can be all identified with precision based on their MyHC profile. In addition, another fast-twitch metabolically-intermediate fiber type, type IIX, has been identified6. Hybrid fibers expressing more than one MyHC have also been confirmed5,7,8. Some species such as the cat and the baboon are known not to express Type IIB MyHCs6. Though MyHC immunostaining is currently the state-of-the-art assessment of muscle composition, the analysis of the data obtained via this technique is cumbersome and time-consuming without automated assistance. To this end, a handful of semi-automated methods to analyze these data have been developed5,9,10. Here, we present a relatively standard protocol for immunohistochemical identification of muscle fiber-type5,7,8,10 along with a novel semi-automated algorithm that accelerates analysis of fiber-type and fiber morphology with accuracy.
Protocol
All procedures involving mice were approved by the University of Colorado-Anschutz Medical Campus Institutional Animal Care and Use Committee (91813(05)1D).
1. Day 1: Primary (1°) Immunostaining with Bovine Serum Albumin (BSA) Blocking
Air-dry frozen sections of mouse hindlimb muscle (e.g., tibialis anterior, soleus) mounted on charged slides for ~ 30 min11. Draw a border around the sections using a hydrophobic barrier PAP pen.
Place ~ 250 µL of 5% BSA/phosphate buffered saline (BSA/PBS) on each slide to block non-specific antibody binding. Incubate slides at room temperature for 1 h.
While blocking, prepare 1:50 dilutions of the 1° antibody supernatant in 5% BSA/PBS (all are mouse monoclonal antibodies; see Table 16,12); prepare 250 µL of solution per slide. Keep 1° antibody stocks and dilutions on ice.
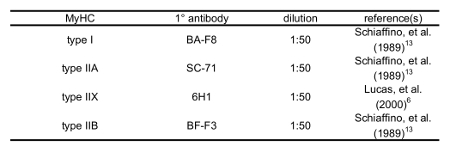 Table 1: Primary Antibodies used to Distinguish MyHCs.
Table 1: Primary Antibodies used to Distinguish MyHCs.
Following aspiration of the 5% BSA/PBS blocking solution, add 250 µL of 1° antibody dilution to the appropriate slides. Add 250 µL of 5% BSA/PBS to secondary (2°) antibody-only negative control slides.
Incubate at 4 °C for 24 - 48 h in a humid environment. NOTE: A Petri dish covered in aluminum foil with dampened filter paper may serve as a suitable incubation chamber.
2. Day 2: 2° Immunostaining with Fluorescent Antibodies
Aspirate 1° antibody solution or control 5% BSA/PBS solution. Wash all slides three times, 10 min with 5% BSA.
While washing, prepare 1:200 dilutions of the purified, fluorophore-conjugated 2° antibodies in 5% BSA/PBS (see Table 2, all are goat anti-mouse). Prepare 250 µL of solution per slide. Keep 2° fluorophore-conjugated antibodies in the dark on ice.
 Table 2: Fluorophore-conjugated Secondary Antibodies used to Visualize Primary Antibody Recognition of MyHCs.
Table 2: Fluorophore-conjugated Secondary Antibodies used to Visualize Primary Antibody Recognition of MyHCs.
Following aspiration of the third application of 5% BSA/PBS blocking solution, add 250 µL of 2° antibody dilution to all slides. Incubate for 90 min at room temperature in a dark, humid environment.
Aspirate 2° antibody solution. Wash all slides three times, 10 min with 5% BSA/PBS.
Rinse with PBS. Dry for 10 min.
Mount the coverglass with a non-permanent, low-viscosity aqueous mounting medium.
When dry, seal the slide edges with nail polish. Dry and store in a dark slidebox.
3. Imaging Slides with Epifluorescence Microscopy
Clean the slides with a small, 70% ethanol-soaked lab-wipe.
- Obtain digital images of immunostained muscle sections using an epifluorescence microscope equipped with a photographic apparatus (see the Table of Materials).
- Select an area of approximately 1 mm2 (10X objective, 1.4 NA).
- View Alexa 488-conjugated 2° antibody fluorescence via a 505 nm long-pass filter. View the fluorescence generated by excitation of Alexa 594-conjugated 2° antibodies via a 595 nm long-pass filter. Digitize images with a PC computer equipped with a compatible imaging software. Include the scale bar from imaging software for later analysis.
4. Analysis of Images with Fiji
Note: Before proceeding with analysis, Fiji (available freely from the National Institutes of Health, Bethesda, MD) must be installed via https://imagej.net/Fiji/Downloads. Also, the macro used in this process is provided in the Supplemental File. Place the macro file in an easily accessible directory.
Load a brightfield image of a selected section in Fiji. Navigate to Plugins → Segmentation → Trainable Weka Segmentation.
- In order to stipulate the cellular areas of the section, draw a line on a fiber, and click "Add to class 1". Then, draw a line on the space between fibers, and click "Add to class 2" to mark the extracellular domains. Repeat until there are 5-10 labels in each class.
- Click Train Classifier. On the resulting red and green overlay, add more labels manually (see step 4.2) to incorrectly segmented pieces of the image, if necessary. Repeat until fibers (red) are appropriately separated from the space between fibers (green).
Click "Get Probability" and save the image in a newly labeled folder.
Repeat the above procedure (steps 4.2 - 4.3) with the fluorescent image taken from the same field. Label immunostained fibers as "class 1" and all non-fluorescent regions as "class 2". Save the resulting probability map to the same folder where the processed brightfield image is stored.
Open one of the previously saved probability maps in Fiji. Draw a straight line on the scale bar provided by the imaging software. Go to Analyze → Set Scale. Change the parameters (i.e., known distance, unit of length) to their appropriate values as given by the scale bar, then check the "Global" box to standardize the scale for each image.
Navigate to Plugins → Macros → Run → Tyagi et al. fiber quantification macro.ijm (see the Supplemental File). Immediately, a navigation pane will appear. Open the image's host folder; a dialog box will appear. For each dialog box, change the "Background" dropdown value to "Light." NOTE: The folder will now be populated with images of the fiber outlines and spreadsheet software files of the results (CSA and Feret diameter are most pertinent to quantifying fiber morphology; measured parameters can be adjusted in Analyze Set Measurements). Fiber morphology data generated by the pore-analysis function of Fiji will be automatically transferred to spreadsheet software files for both fluorescent and bright field images.
Calculate the fraction of fibers expressing a given MyHC in a field by dividing the number of fluorescent fibers in the field by the number of total fibers in the corresponding bright field image.
Representative Results
Hindlimb muscles (i.e., tibialis anterior, soleus) dissected from a male C57BL/6 mouse of unknown age were flash frozen by sinking a plastic mold containing the muscle in OCT compound in liquid nitrogen-cooled isopentane. Then, using a cryotome, 8-10 µm serial sections were cut at -20 °C and transferred to different positively-charged glass slides12.
We chose tibialis anterior and soleus because these muscles are predominantly composed of fast- and slow-twitch fibers, respectively13,14. Following immunohistochemical staining for individual type I, IIA, IIX and IIB MyHCs, brightfield and fluorescent images were captured. The left panels of Figure 1 show sections of a soleus muscle immunostained with the type I, IIA, IIX and IIB MyHC-specific antibodies listed in Table 1.
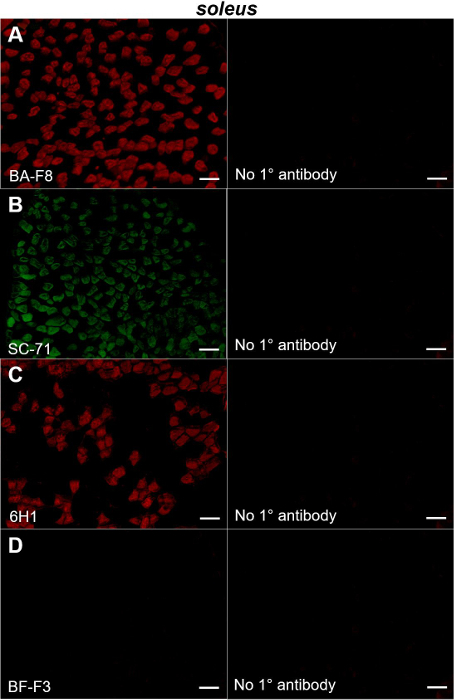
Figure 1: Immunohistochemical staining of MyHC type I, IIA, IIX and IIB in mouse soleus muscle. Left panels show fluorescent images obtained from sections of a mouse soleus muscle probed with primary antibodies directed to type I (BA-F8; A), type IIA (SC-71; B), type IIX (6H1; C) and type IIB (BF-F3; D) MyHCs. Right panels show the sections in which the primary antibody used in the experiment depicted in the corresponding left panel was omitted. Bars= 100 µm. Please click here to view a larger version of this figure.
Specifically, we observed substantial fractions of fibers expressing type I (Figure 1A, left) and type IIA MyHCs (Figure 1B, left) with a fair amount of the remaining fibers displaying positive staining for type IIX MyHCs (Figure 1C, left). By contrast, virtually no type IIB fibers were evident (Figure 1D, left). These observations are consistent with earlier reports that soleus muscles are composed mostly of type I, type IIA and type IIX fibers (less so) with almost no type IIB fibers5,10,12. Importantly, no staining was observed in sections in which the primary antibody had been omitted from the protocol, demonstrating the specificity of the antibodies (Figure 1A-D, right panels).
Figure 2: Immunohistochemical staining of MyHC type IIB, IIX, IIA and I in mouse tibialis anterior muscle. Left panels show fluorescent images obtained from sections of a mouse tibialis anterior muscle probed with primary antibodies directed to type IIB (A), type IIX (B), type IIA (C) and type I (D) MyHCs. Right panels show the sections in which the primary antibody used in the experiment depicted in the adjacent left panel was omitted. Bars = 100 µm. Please click here to view a larger version of this figure.
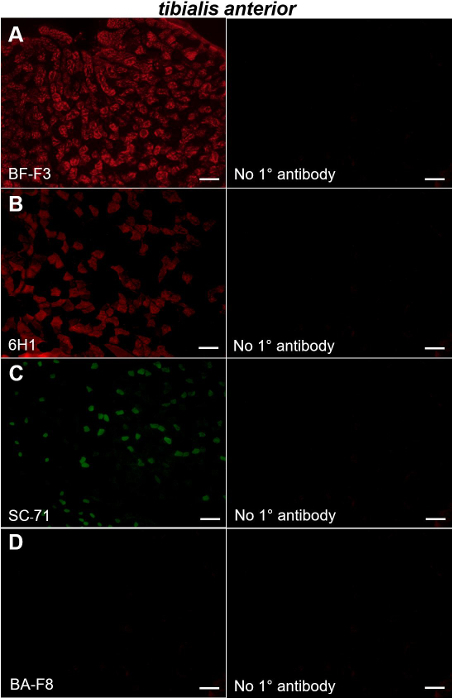
We also stained tibialis anterior muscles with the aforementioned antibodies. Tibialis anterior muscles were composed mostly of type IIB and type IIX MyHC-positive fibers (Figure 2A-B, left panels) with smaller contributions from fibers expressing type IIA MyHCs (Figure 2C, left); virtually no fibers expressing type I MyHC were observed (Figure 2D, left). Our data indicating that the mouse tibialis anterior is composed predominantly of fast twitch type IIA, type IIB and type IIX fibers are consistent with earlier studies5,10,13. Again, no staining was observed in the sections in which the primary antibody had been omitted from the protocol (Figure 2A-D, right panels).
Figure 3: Iterated computerized fiber segmentation. Brightfield image of a mouse tibialis anterior section (A). Transformation of the same image using our semi-automated algorithm (B). Fluorescent image of the same section showing immunostaining of type IIA MyHC (C). Transformation image showing only type IIA MyHC-expressing fibers (D). Bars = 100 µm. Please click here to view a larger version of this figure.
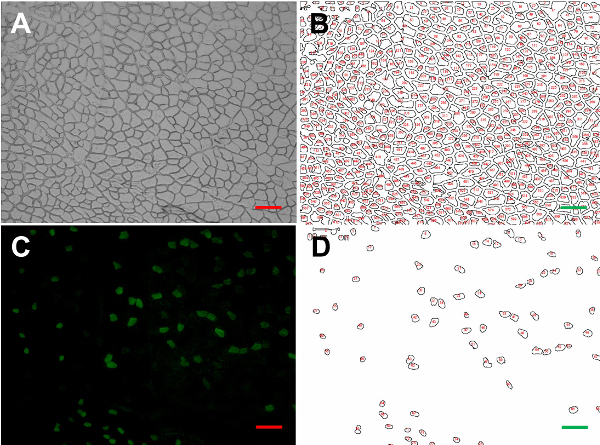
For the purpose of morphometric analysis, brightfield images of the immunostained fields were also obtained. The example shown in Figure 3A is from a tibialis anterior muscle section probed with SC-71 antibodies directed to type IIA MyHCs13. Using our original semi-automated algorithm, several image transformations were used to generate probability maps from the brightfield images. A typical image transformation derived from the brightfield image shown in Figure 3A is presented in Figure 3B. Using the transformation and the pore analysis function of Fiji, we measured the mean CSA (891.4 ± 45.0 µm2), maximum (46.9 ± 1.3 µm) and minimum (26.0 ± 0.8 µm) mean Feret diameters of each fiber in the field. To test the fidelity of our measurements, we also assessed these parameters in the brightfield image manually. The mean CSA (867.0 ± 52.0 µm2), maximum (45.7 ± 3.5 µm) and minimum (25.6 ± 1.9 µm) mean Feret diameters obtained using the more tedious manual acquisition method were not significantly different from the semi-automated method (all p > 0.05, unpaired t-tests; Table 3).
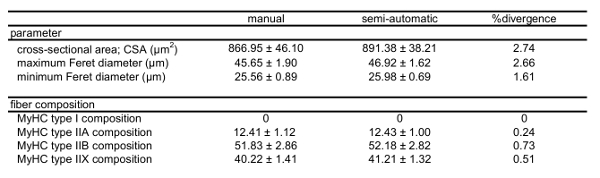 Table 3: Fiber morphology and composition of tibialis anterior muscle immunostained for type I, IIA, IIB and IIX MyHCs as determined by manual and semi-automatic analytic methods. Mean fiber CSA, mean Feret diameters and MyHC type in a mouse tibialis anterior quantified via traditional manual evaluation versus the presented algorithm. Please note that combined fractions exceed a value of 1 due to the presence of fibers expressing more than one MyHC type.
Table 3: Fiber morphology and composition of tibialis anterior muscle immunostained for type I, IIA, IIB and IIX MyHCs as determined by manual and semi-automatic analytic methods. Mean fiber CSA, mean Feret diameters and MyHC type in a mouse tibialis anterior quantified via traditional manual evaluation versus the presented algorithm. Please note that combined fractions exceed a value of 1 due to the presence of fibers expressing more than one MyHC type.
Figure 3C shows a fluorescent image of the same field immunostained with antibodies directed to type IIA MyHCs. Positively-stained fibers in the field were selected by number (Figure 3D) and used to determine the fraction of type IIA MyHC-expressing fibers by dividing by the total number of fibers in the brightfield transformation field (Table 3). The average CSA, maximum and minimum Feret diameters of type IIA fibers were then assessed (Table 3). The proportion of type I, type IIX and type IIB fibers, as well as their dimensions, were determined by repeating this protocol in serial sections immunostained with BA-F8, 6H1 and BF-F3 antibodies, respectively6,12(Table 4; Figure 4A-D).
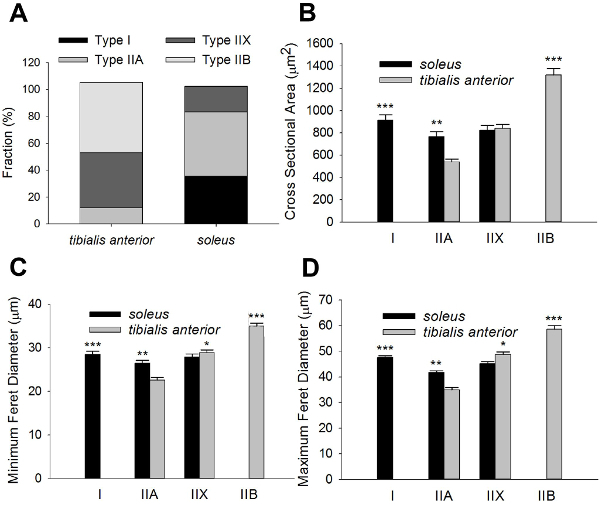
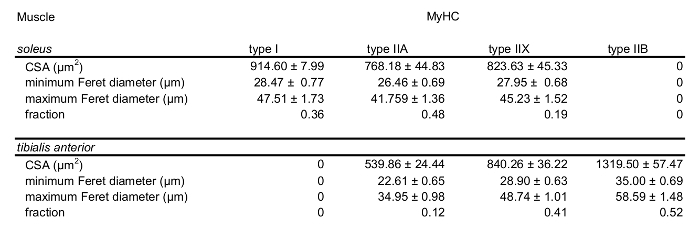 Table 4: MyHC content and morphology in mouse soleus and tibialis anterior. Fiber type compositions of representative sections of the soleus and tibialis anterior. Quantification of average CSA, minimal and maximal Feret diameters of type I, type IIA, type IIX and type IIB in both the soleus and tibialis anterior are shown. Data are given as mean ±SEM.
Table 4: MyHC content and morphology in mouse soleus and tibialis anterior. Fiber type compositions of representative sections of the soleus and tibialis anterior. Quantification of average CSA, minimal and maximal Feret diameters of type I, type IIA, type IIX and type IIB in both the soleus and tibialis anterior are shown. Data are given as mean ±SEM.
We also assessed these parameters in a field obtained from the soleus muscle (Table 4; Figure 4A-D). 3,052 and 2,876 individual fibers were counted for the tibialis anterior and soleus muscles, respectively. Taken together, these data demonstrate the feasibility of our method in fiber-type and morphometric analysis.
Figure 4: Morphometric analysis data summary. Fiber-type compositions of representative sections of the tibialis anterior and soleus muscles (A). Quantification of average CSA, minimimal and maximal Feret diameters of type I, type IIA, type IIX and type IIB fivers in both the tibialis anteriort and soleus are shown in (B-D). Data are given as mean ±SEM. Significant differences between the tibialis anterior and soleus are indicated (*denotes p < 0.05: ** denotes p < 0.01; *** denotes p < 0.005; t-test). 3,052 and 2,876 individual fibers were counted for the tibialis anterior and soleus muscles, respectively. Please click here to view a larger version of this figure.
 Supplemental File: Fiber quantification macro.
Please click here to download this file.
Supplemental File: Fiber quantification macro.
Please click here to download this file.
Discussion
Here, we have provided useful direction for the identification of skeletal muscle fiber types. In doing so, we describe a novel algorithm for analysis of the data.
Since our results largely confirm those of previous reports5,8,10 and reflect our own manual measurements, the algorithm appears to be accurate. Still, we encountered some infrequent experimental pitfalls including unclear fiber borders resulting from sectioning artifact. The impact of these artifacts can be minimized through careful sectioning and vigilant examination of the processed section prior to analysis. In particular, less-than-optimal image segmentation may be circumvented through multiple rounds of reclassification on the same image to achieve sharper contrast.
The most crucial step in our protocol is step 4.2 in which we stipulate the cellular and extracellular areas of the section by drawing a line on a given fiber, thereby distinguishing a "class 1" or "class 2" area. Obviously, this manual distinction is critical because it determines the boundaries of the muscle fibers; an error in this step will adversely affect the assessment of the fiber dimensions.
In this study, we probed sections for only one MyHC in a given serial section. However, some fibers, such as hybrid fibers, express two types of MyHC, indicative of overlapping metabolic/functional profiles (e.g., type IIX and type IIB). Thus, the fractions of fiber-type summed to a value > 1 for both the soleus and tibialis anterior (Figure 4A). Hybrid fibers can be identified with double-, or even triple-, labelling protocols such as those described by Lee et al.7and Bloemberg and Quadrilatero5, respectively. Importantly, our method of analysis can be applied to these more complex staining protocols. Another limitation of our protocol is that it is not fully automatic; however, we believe that this minor concession ensures that our method does not compromise rigor for the sake of speed.
While the MyHC compositions of both soleus and tibialis anterior muscles observed in this study reflect the results of previous studies5,6,7,10,14, there are some contrasts with earlier studies regarding the assessment of CSA. For example, our measurements of average CSA are somewhat smaller than those reported by either Bloemberg and Quadrilatero5 and Allen et al.15 In particular, our CSA measurements ranged from 539.86 ± 24.44 µm2 to 1319.50 ± 57.47 µm2 across fiber types whilst theirs spanned roughly 735.7 ± 31.2 µm2 to 3073.8 ± 51.3 µm2. However, our values more closely resemble those observed by Augusto et al.13 and Lucas et al.6 who reported average CSAs ranging from 436 ± 605 µm2 to 2404 ± 412.5 µm2 and 815 ± 221 µm2 to 1904 ± 570 µm2, respectively. Thus, some variability in morphometric analysis of mouse hindlimb fibers exists across the literature.
Immunohistochemistry presents an efficient way to label and classify muscle fibers on the basis of their MyHC content. Accurate quantification of immunohistochemical experiments is essential to making meaningful conclusions about the data; to this end, a method which can automate the tedious parts of analysis is a useful one, especially when hundreds, or even thousands, of fibers need to be measured and classified. In addition to providing measurements similar to manual observations, our algorithm provides an accurate assessment of these data without bias; this advantage over manual measurements ensures scientific rigor. In this regard, our methodology may be useful in the characterization of the effects of genetic and experimental manipulations that impact muscle integrity. Specifically, MyHC immunohistochemistry coupled with our method of rapid analysis may be employed to assess morphological and metabolic changes in muscle of disease models such as muscular dystrophies, ALS and cancer cachexia. Moreover, the efficacy of interventions which prevent atrophy or cause hypertrophy may also be assessed.
Disclosures
The authors have no conflicting interests to disclose.
Acknowledgments
We are grateful to the Boettcher Foundation and the Amyotrophic Lateral Sclerosis Association (#17-II-344) for their support of this research.
References
- Engel WK. The essentiality of histo- and cytochemical studies of skeletal muscle in the investigation of neuromuscular disease. Neurol. 1962;12:778–784. [PubMed] [Google Scholar]
- Dobrowolny G, et al. Skeletal muscle is a primary target of SOD1G93A-mediated toxicity. Cell Metab. 2008;8:425–436. doi: 10.1016/j.cmet.2008.09.002. [DOI] [PubMed] [Google Scholar]
- Sultana N, et al. Restricting calcium currents is required for correct fiber type specification in skeletal muscle. Development. 2016;143(9):1547–1559. doi: 10.1242/dev.129676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guth L, Samaha FJ. Procedure for the histochemical demonstration of actomyosin ATPase. Exp Neurol. 1970;28(2):365–367. [PubMed] [Google Scholar]
- Bloemberg D, Quadrilatero J. Rapid determination of myosin heavy chain expression in rat, mouse, and human skeletal muscle using multicolor immunofluorescence analysis. PLoS One. 2012;7(4) doi: 10.1371/journal.pone.0035273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucas CA, Kang LH, Hoh JF. Monospecific antibodies against the three mammalian fast limb myosin heavy chains. Biochem Biophys Res Commun. 2000;272(1):303–308. doi: 10.1006/bbrc.2000.2768. [DOI] [PubMed] [Google Scholar]
- Lee CS, et al. Ca2+ permeation and/or binding to Cav1.1 fine-tunes skeletal muscle Ca2+ signaling to sustain muscle function. Skelet Muscle. 2015;5(4) doi: 10.1186/s13395-014-0027-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawano S, et al. A one-step immunostaining method to visualize rodent muscle fiber type within a single specimen. PLoS One. 2016;11(11) doi: 10.1371/journal.pone.0166080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meunier B, Picard B, Astruc T, Labas R. Development of image analysis tool for the classification of muscle fibre type using immunohistochemical staining. Histochem Cell Biol. 2010;134(3):307–317. doi: 10.1007/s00418-010-0733-7. [DOI] [PubMed] [Google Scholar]
- Kammoun M, Casser-Malek I, Meunier B, Picard B. A simplified immunohistochemical classification of skeletal muscle fibres in mouse. Eur J Histochem. 2014;58(2):2254. doi: 10.4081/ejh.2014.2254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar A, Accorsi A, Rhee Y, Girgenrath M. Do's and don'ts in the preparation of muscle cryosections for histological analysis. J Vis Exp. 2015. [DOI] [PMC free article] [PubMed]
- Schiaffino S, et al. Three myosin heavy chain isoforms in type 2 skeletal muscle fibres. J Muscle Res Cell Motil. 1989;10(197) doi: 10.1007/BF01739810. [DOI] [PubMed] [Google Scholar]
- Augusto V, Padovani CR, Campos GER. Skeletal muscle fiber types in C57Bl6j mice. Braz J Morphol Sci. 2004;21(2):89–94. [Google Scholar]
- Schiaffino S, Reggiani C. Fiber types in mammalian skeletal muscles. Physiol Rev. 2011;91(4):1447–1531. doi: 10.1152/physrev.00031.2010. [DOI] [PubMed] [Google Scholar]
- Allen DL, Harrison BC, Maass A, Bell ML, Byrnes WC, Leinwand LA. Cardiac and skeletal muscle adaptations to voluntary wheel running in the mouse. J Appl Physiol. 2001;90(5):1900–1908. doi: 10.1152/jappl.2001.90.5.1900. [DOI] [PubMed] [Google Scholar]


