Summary
The relapsing fever agent Borrelia hermsii undergoes multiphasic antigenic variation through gene conversion of a unique expression site on a linear plasmid by an archived variable antigen gene. To further characterize this mechanism we assessed the repertoire and organization of archived variable antigen genes by sequencing ~85% of plasmids bearing these genes. Most archived genes shared with the expressed gene a ≤ 62 nucleotide (nt) region, the upstream homology sequence (UHS), that surrounded the start codon. The 59 archived variable antigen genes were arrayed in clusters with 13 repetitive, 214 nt long downstream homology sequence (DHS) elements distributed among them. A fourteenth DHS element was downstream of the expression locus. Informative nucleotide polymorphisms in UHS regions and DHS elements were applied to the analysis of the expression site of relapse serotypes from 60 infected mice in a prospective study. For most recombinations, the upstream cross-over occurred in the UHS’s second half, and the downstream cross-over was in the DHS’s second half. Usually the closest archival DHS element was used, but occasionally a more distant DHS was employed. The upstream extragenic crossover site in B. hermsii contrasts with the downstream extragenic crossover site for antigenic variation in African trypanosomes.
Keywords: antigenic variation, gene conversion, relapsing fever, spirochete, lipoprotein
Introduction
The spirochetal infection relapsing fever has a distinctive clinical course in untreated patients and in experimental animals: up to 13 febrile episodes are separated by periods of well-being. This arthropod-borne disease is caused by several Borrelia species, which are grouped according to their geographic ranges: Old World (Palearctic or Afro-tropical) species, such as B. duttonii, B. hispanica, and B. crocidurae, and New World (Nearctic or Neotropic) species, such as B. hermsii, B. turicatae, and B. venezuelensis (Barbour, 2005). With the exception of the louse-borne B. recurrentis, relapsing fever spirochetes are transmitted by soft ticks, such as Ornithodoros hermsi in the case of B. hermsii. Natural reservoirs for relapsing fever species include a variety of mammals and birds, but most commonly are rodents (Dworkin et al., 2002; Schwan and Piesman, 2002).
Being dependent on hematophagous arthropods for transmission between vertebrates, relapsing fever agents prolong their residence in the blood by sequential evasion of the host’s adaptive immune responses (Barbour et al., 2000). This evasion is accomplished by true antigenic variation, that is, within a clonal population. One surface-exposed lipoprotein is replaced by another of sufficient antigenic distance that current antibodies are ineffective against cells expressing the second antigen (Barbour, 2002). During this relapse of disease, the newly-emerged variants, each designated as a different “serotype”, proliferate in a vertebrate host until the next wave of antibodies clears it in turn. Meanwhile, other serotypes have spontaneously appeared in the population at an estimated frequency of 10−4 to 10−3, and these constitute the predominant antigenic types for subsequent relapses. For the HS1 strain of B. hermsii 25 unique serotypes had been identified prior to the present study (Barbour, 1985; Restrepo et al., 1992; Stoenner et al., 1982).
The serotype-specific lipoproteins of relapsing fever Borrelia species are of two types that are not discernibly homologous: Variable Large Proteins (Vlp) of about 36 kDa and Variable Small Proteins (Vsp) of about 20 kDa (Hinnebusch et al., 1998; Restrepo et al., 1992). Vsp and Vlp proteins are encoded by vsp or vlp genes, which are numbered according to serotype; for example, vlp7 encodes Vlp7, which confers serotype 7 identity (Burman et al., 1990; Carter et al., 1994; Restrepo et al., 1992). The vlp genes are further categorized into four sub-families, α-vlp, β-vlp, γ-vlp, and δ-vlp, with less than 60% sequence identity between them (Restrepo et al., 1992; Hinnebusch et al., 1998). In species that have been examined to date, the vsp and vlp genes are located on linear plasmids of 28–32 kb (Kitten and Barbour, 1990; Plasterk et al., 1985). The vsp and vlp genes in the genome of a Borrelia spirochete are transcriptionally silent with the exception of one locus: a duplicated vsp or vlp gene positioned immediately downstream of a σ70-type prokaryotic promoter and near the telomere of a 28 kb linear plasmid (Barbour et al., 1991; Kitten and Barbour, 1990). When a particular vsp or vlp gene at the expression site is replaced by another gene in what appears to be a non-reciprocal recombination, it is lost but the original archived copy is retained intact (Plasterk et al., 1985; Restrepo et al., 1992).
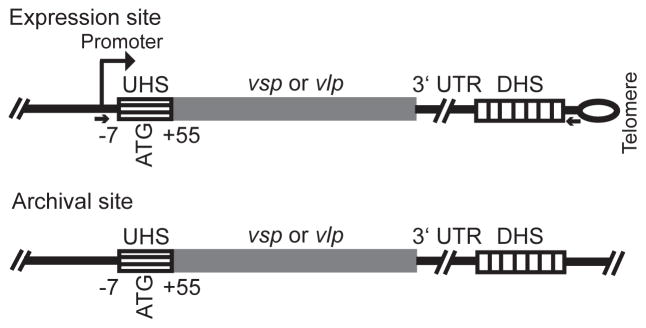
These features of antigenic variation by B. hermsii are consistent with gene conversion. Figure 1 schematically represents the substrates for this recombination. A previous study of switches between serotypes 7 and 21, both of the α-vlp sub-family, indicated possible boundaries for these recombinations (Barbour et al., 1991; Kitten and Barbour, 1990). The upstream crossover point appeared to be within a ~60 nucleotide (nt) region comprising the interval between the start codon and the transcriptional start site and the coding sequence for part of the lipoprotein’s signal peptide (Barbour et al., 1991). This region was called the “UHS” for upstream homology sequence. The downstream boundary for the recombination in these switches appeared to be a 214 nt non-coding sequence downstream of the expressed allele on one plasmid and the silent vlp7 and vlp21 genes on other plasmids and was called the “DHS” for downstream homology sequence (Burman et al., 1990; Kitten and Barbour, 1990). Sequences of the expression locus for 25 different vsp or vlp genes provided further circumstantial evidence on the DHS sequence’s role in the recombination (Restrepo et al., 1992). The 3′ untranslated region (3′ UTR) between the end of the vsp or vlp gene and the DHS at the expression site was associated with the unique vsp or vlp rather than expression locus itself, thus pointing to the more distal DHS as the site of the downstream cross-over.
Figure 1.
Schematic representation of expression and archival sites for vsp and vlp genes of Borrelia hermsii. The drawing is not to scale. The locations of the UHS regions that surround the start codon (ATG), the 3′ untranslated region (UTR), and DHS elements are given. By the numbering system, +1 is the transcriptional start position at the expression site. The UHS regions at the archival sites varied in length to the extent that they were >90% identical to the UHS at the expression site. The expression site is adjacent to a hair-pin telomere, indicated by the loop. The small arrows give the location of PCR primers (see text) for amplification of the expression site but not the silent site vsp or vlp gene.
To examine the hypothesis that the UHS and DHS are the cross-over points for most gene conversions at the expression site, we more fully characterized the repertoire of vsp and vlp alleles, the organization of archived vsp and vlp genes, and locations of additional DHS elements in B. hermsii by large-scale sequencing of B. hermsii’s plasmids in the genome. We also carried out prospective studies of the first relapses of infections of mice infected with one of two different serotypes representing different vlp sub-families. Our aim was to examine relapse populations with lineages that were better defined than what was previously available. In the course of the study, we discovered additional polymorphisms in the UHS and DHS sequences that allowed fine mapping of the recombination points and established the role of the extragenic DHS elements in antigenic variation during relapsing fever.
Results
Organization of vsp and vlp genes and DHS elements on B. hermsii plasmids
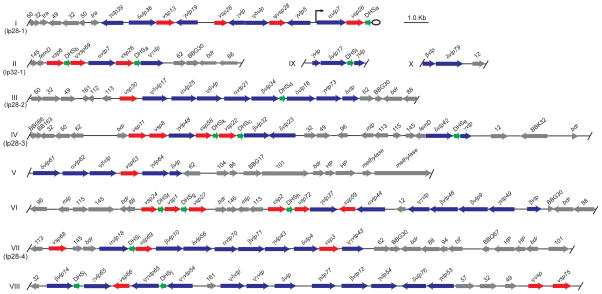
Ten large sequence fragments of plasmids, numbered I through X, each had at least one vsp, vlp, or DHS sequence. They ranged in length from 2,583 bp to 23,848 bp and in total comprised 152,569 bp of nonredundant sequence, which was 34% of all the plasmid sequences in fragments of at least 2.0 kb; 36 other sequence fragments, ranging in size from 2,054 to 47,233 bp, contained sequences that were homologous to B. burgdorferi or B. hermsii genes known to be on circular plasmids (Casjens et al., 2000; Simpson et al., 1990b; Stevenson et al., 2000) or were in the 53 kb linear plasmid (Carter et al., 1994; Porcella et al., 2005) or 180 kb linear plasmid (Zhong et al., 2006; Schwan et al., manuscript in preparation). Physical maps of fragments I – X are shown in the Figure 2, and details on the ORFs and other features of these sequence contigs are given in the Table S1 of supplementary materials. Fragments I, II, and III were previously identified as partial sequences of three linear plasmids, now named lp28-1, lp32-1, and lp28-2, respectively (Kitten and Barbour, 1990; Plasterk et al., 1985). Pulsed field gel electrophoresis and Southern blot analysis mapped fragments IV and VII to linear plasmids of about 28 kb (data not shown). Addition of fragments IV or VII to each other or to fragments I, II, or III would have produced a combined fragment of ≥31 kb in length, and, accordingly, fragments IV and VII were provisionally designated as parts of plasmids lp28-3 and lp28-4, respectively (Figure 2).
Figure 2.
Physical maps of 10 (I – X) fragments of B. hermsii linear plasmids that contain vsp genes (red arrows), vlp genes (blue arrows), and/or DHS elements (green arrows). Also shown are the locations of other open reading frames (ORF), which are indicated by gene names (e.g. femD and bdr) or by Borrelia burgdorferi gene names (e.g. BBG30) or paralogous family numbers (e.g. 50) (Casjens et al., 2000; Fraser et al., 1997). When an ORF had no discernible homology with a protein in the GenBank database, it was designated a hypothetical protein (HP). The arrowheads indicate either the direction of transcription in the case of vsp and vlp genes and other ORFs or the orientation with respect to the expression site for the DHS elements. The start and stop positions for each open reading frame or sequence element are given in Table S1 of supplementary materials. The vsp and vlp genes are further distinguished by the number of the serotype they specify (e.g. vsp6) and, in the case of vlp genes, by appending their membership in vlp sub-families α, β, γ, and δ. Pseudo genes are indicated by Ψ, and truncated or otherwise incomplete vsp or vlp sequences were not assigned a serotype number. The 12 different genotypes of the near-identical DHS elements are indicated by a letter (a – l) subscript (see Figure 5). Some fragments were identified with linear plasmids of known sizes: lp28-1, lp28-2, lp28-3, lp28-4, and lp32-1. The expression site in this example of serotype 7 is adjacent the right telomere of plasmid lp28-1; the expression site promoter is indicated by the raised arrow. Serotypes 7 and 21 are exceptional in having a silent vsp or vlp downstream of the active vlp at the expression site (Kitten and Barbour, 1990); in other serotypes only a single vsp or vlp gene is between the promoter and the sub-telomeric DHS (Restrepo et al., 1992).
In the physical maps of fragments I – X the expressed vsp or vlp gene on plasmid lp28-1 by convention is on the plus strand, and transcription is directed toward the right telomere (Kitten and Barbour, 1990; Meier et al., 1985; Plasterk et al., 1985). This was also the orientation for the silent vlp7 and vlp21 on plasmids lp32-1 and lp28-2, respectively (Barbour et al., 1991; Kitten and Barbour, 1990), and, accordingly, the 7 fragments were oriented so that the majority of archived vsp and vlp genes were on the plus strand. The 10 fragments contained 21 vsp alleles and 38 vlp alleles, including 7 α-vlp genes, 8 β-vlp genes, 13 γ-vlp genes, and 10 δ-vlp genes. The archived versions of the 25 previously identified vsp and vlp genes were located in these sequence fragments, and the current study identified 34 more.
To estimate the extent to which the 10 sequence fragments included the entire vsp and vlp repertoire in the genome, we used another approach to identify vsp and vlp genes, namely PCR amplification with vsp family- and vlp subfamily-specific primers of B. hermsii HS1 genomic DNA (Hinnebusch et al., 1998). The PCR products were ligated into plasmid vectors, and the inserts of individual transformant colonies were sequenced. There were 37 unique vsp and vlp genes among 296 clones whose inserts were sequenced. Of these, 34 (92%) were present in the 10 fragments. (The 3 additional genes were designated vlp47, vlp51, and vlp55.) From this sampling, we concluded that shotgun sequencing and sequence assembly into fragments captured about 90% of the vsp and vlp repertoires in B. hermsii. For another measure of the completeness of the sequencing, the overall total of 444,430 bp of non-redundant plasmid sequence available for this study was compared to the estimated 550 kb of plasmid DNA in a genome of B. hermsii: one 180 kb, one 53 kb, approximately five 28–32 kb, and one 18 kb linear plasmids, as well as five 32 kb circular plasmids (Barbour et al., 2000; Ferdows et al., 1996; Kitten and Barbour, 1992; Stevenson et al., 2000). By this criterion, we cumulatively determined 81% of the plasmid sequences in the B. hermsii genome.
Besides the 59 ORFs identified as vsp and vlp genes, we found 14 pseudogenes and 9 partial fragments of vsp or vlp genes among the large sequence fragments (Figure 2). The vsp and vlp genes, pseudogenes, and fragments were grouped in arrays, usually but not always in the same orientation. There were a total of 13 DHS elements, each 214 nt, within or adjacent to a cluster of vsp and/or vlp genes. The DHS elements were ≤ 117 bp downstream from the stop codon for a vsp or vlp gene. The fourteenth DHS element was near the telomere of the expression plasmid lp28-1 (Barbour et al., 1991; Kitten and Barbour, 1990). Between the clusters of vsp and vlp genes and their DHS elements were several ORFs that were orthologous to plasmid genes of the Lyme disease agents B. burgdorferi and B. garinii (Casjens et al., 2000; Glockner et al., 2004). These included ORFs similar to bdr genes (Zückert et al., 1999), mlp genes, and members of B. burgdorferi paralogous families 32, 62, 88, 50, 49, 145, 113, 13, 96, 116, 161, and 101, in decreasing order of frequency. A few ORFs were unique to B. hermsii among proteins in the GenBank database and labeled as hypothetical proteins. There were degraded fragments of a transposase (tra) sequence, similar to U85588 of B. burgdorferi, but not a full-length ORF with detectable homology to a known transposase or recombinase in any of the plasmid sequences.
Mouse infections and relapses
For the prospective study we chose serotypes 7 and 17, because they represented two different vlp sub-families, α and δ. In the first set of experiments 19 mice were infected with serotype 7, and 18 were infected with serotype 17. In the second set of experiments 11 mice were infected with serotype 7, and 12 were infected with serotype 17. In both sets of experiments relapses of spirochetes in the blood occurred between days 7–9 after initial inoculation. DNA was extracted from the plasma and then subjected to PCR with a forward primer for the promoter at the expression site and a reverse primer from the sequence between the DHS element and plasmid’s right telomere (Figure 1). The resultant PCR products ranged in size from 1.0 to 2.3 kb and were cloned and sequenced over their lengths.
Among the 60 infected mice, 83 relapse serotypes were identified. These comprised 16 different serotypes: 1–2, 4, 6–7, 13–14, 16–18, 24, 26–27, 42, 46, and 58. The last three had not previously been observed. All the relapse serotypes were accounted for by the vsp and vlp genes in fragments I through X. The distribution of different serotypes by gene family among the relapse populations did not differ significantly (p > 0.05) by goodness-of-fit between sets of experiments, and, accordingly, for subsequent analyses the two sets’ results were combined. Analysis of the frequencies of different serotypes during relapses will be reported elsewhere (Barbour et al., submitted for publication). For 70 of the 83 relapses, there were available sequences for the UHS regions at both the expression site and the archival site for the incoming vsp or vlp gene (Table S2). For 68 relapses there were available sequences for the DHS elements at both the expression site and archival locus (Table S3).
Recombination within the UHS sequence
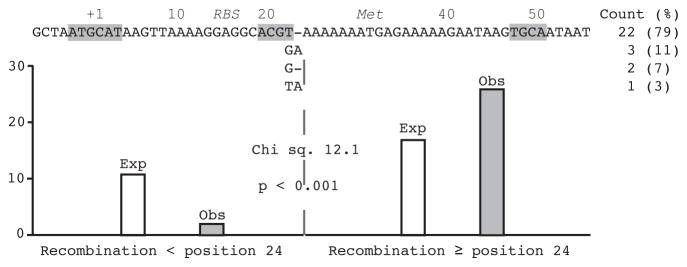
A UHS at its greatest length of 62 bp stretches from 7 nucleotides upstream of the transcriptional start nucleotide C at position +1, through the ribosomal binding sequence (positions +13–17) and start codon (positions +29–30), and then to the T at position +55, 26 nucleotides into the signal peptide coding region for the Vsp or Vlp lipoprotein (Figures 1 and 3). We aligned the UHS sequences at expression sites for serotypes 1–14, 16–19, and 21–27 with 28 UHS sequences of ≥ 33 nt at the archival sites (Table S2). Four different UHS sequences were found; the polymorphisms were at positions 22 and 23 (Figure 3). Two of the polymorphisms, a T- or GA, were found at expression sites as well as archived site, while the other two, a G- or TA, were noted only in archival sites. Most UHS regions had T- at positions 22–23. All 28 UHS sequences contain the palindrome TGCA, and 23 of 28 have the palindrome ACGT.
Figure 3.
UHS regions for 28 archival vsp and vlp genes of B. hermsii. Numbering of the positions is according to the transcriptional start position (+1) at the expression site; also shown are the start codon (Met) and presumed ribosomal binding sequence (RBS). Differences between the 4 sequence variants at positions +22 and 23 are shown; the counts of each variant are indicated on the right. The identification of a given archival vsp or vlp with a UHS sequence variant is given in Table S1 of supplementary materials. The 6-mer and 4-mer palindromes in the UHS are highlighted by gray. Shown below the sequence are expected (Exp) and observed (Obs) cross-over points with respect to position +24 for 28 relapses involving an infection with serotype 17, which has a GA for the expression site UHS, and relapse to a serotype whose archival vsp or vlp has another UHS sequence variant (see text). The goodness-of-fit analysis of observed and expected results with Chi square value and 2-tailed p value for 1 degree of freedom is shown. The relapse isolates for this analysis are indicated by italicized names in Table S2.
Another rationale for using serotype 7 and serotype 17 in the prospective study was the difference between the two serotypes in their UHS sequences at the expression site: serotype 7 had a T- at positions +22–23, whereas serotype 17 had a GA. Of the 40 relapse populations following serotype 7 infections, 38 (95%) had the same expression site UHS genotype as that of serotype 7, i.e. T-. Similarly, of the 30 relapse serotypes after serotype 17 infections, 27 (90%) had the same expression site UHS type as that of serotype 17, i.e. GA. The expression site UHS of the relapse serotypes was most often that of the infecting strain, which suggested that the upsteam boundary for the recombination was usually 3′ to the UHS polymorphism site.
To confirm this, we examined 28 relapses from serotype 17 in which the presumptive donor site for the recombination had an informative T- at the UHS’s polymorphic site instead of the GA found at the expression site for serotype 17 (Tables S1 and S2). The findings for these relapses are shown in Figure 3. In 26 out of these 28 cases the expression site UHS had type GA instead of T-. If we limit the length of DNA available for recombination to 52 nt, the average length for archival UHS regions (Table S2), then the proportion of the UHS sequence distal to polymorphism site is 0.62. If the crossover point occurred randomly over the length of the UHS, then one would expect that only 17, instead of the observed 26 relapses, would have a GA out of the 28 relapses examined (Figure 3). When we examined each of the 28 relapses and used the length of the UHS at the archival site to specifically set the expectation of a cross-over distal to the polymorphic site, the 1-tailed probability of the observed or a more extreme outcome was <0.001. The results of this further analysis confirmed that the crossover predominantly occurred to the 3′ side of the polymorphism site. The prior existence of the GA in the original serotype 17’s expression site, as well as the 2 exceptions among the 28 relapses in this study, indicated that the recombination was not invariably site-specific. The 62 and 61 nt long UHS sequences may differ in phenotype if the increase from 11 to 12 nt in the spacing of the presumed ribosomal binding sequence and start codon affects expression levels.
We examined 70 relapses for which we had sufficient sequence to fully examine their UHS regions (Table S2). In only one (1.4%) of these relapses was the upstream cross-over point for the recombination beyond the initial codons for the signal peptide and within the coding sequence for a processed Vsp or Vlp lipoprotein. In this case the relapse isolate from a mouse originally infected with serotype 7 had an α-vlp that was a chimeric gene of vlp7 for 252 bp at the 5′ end and vlp18 for 825 bp at the 3′ end (accession number DQ355027). In the only other known example of intragenic recombination at the expression site, a chimeric α-vlp gene was also found (Kitten et al., 1993). We have not yet observed at the expression site a chimeric vsp or vlp that derived its 5′ end from the archival site and retained the original vsp/vlp at its 3′ end.
Recombination within the 3′ UTR
Another possible region for recombination was the 3′ UTR, located between most vsp/vlp genes and the DHS at the expression site and ranging in length from 41 to 117 nt (Figure 1) (Kitten et al., 1990; Restrepo et al., 1992). If the 3′ UTR at the expression site was largely the same as that flanking the vsp or vlp donor at the archival site, this would indicate that the gene conversion extended beyond the end of the vsp or vlp gene itself. To determine this, we examined the 3′ UTR sequences of the expression sites of the previously identified serotypes and relapse isolates, as well as the 3′ UTR’s of the archival sites (Table S4).
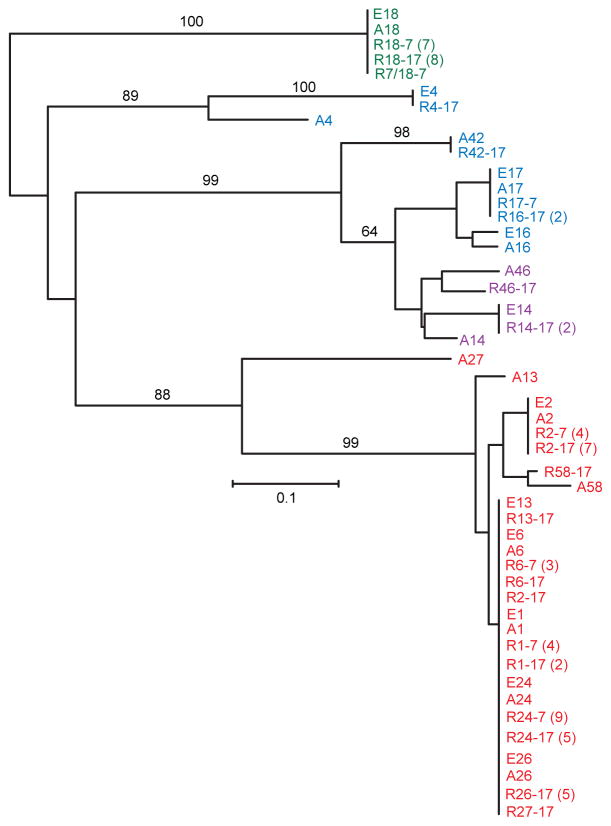
Figure 4 shows a phylogram of the 3′ UTR sequences of the relapses (labeled “R”) of the prospective study, the expressed genes (labeled “E”) from the original collection of 25 serotypes (Barbour et al., 2000; Restrepo et al., 1992), and those archival genes (labeled “A”) represented among the relapse populations. The different text colors represent the vsp, α-vlp, β-vlp, γ-vlp, and δ-vlp families. Because of the differing lengths of the 3′ UTR sequences, the alignments were made from the 5′ end and were limited to the first 43 nucleotides. The branch lengths correspond to nucleotide distance. In 59 (87%) of 68 cases, the 3′ UTRs of the expressed genes were identical or highly similar to those of the corresponding vsp or vlp gene at the archival site, as would be predicted for recombinations extending downstream of the stop codon of the vsp or vlp gene.
Figure 4.
Neighbor-joining distance phylogram of aligned 3′ UTR sequences of expressed and archived vsp and vlp genes of B. hermsii. The sequences are given in Table S4. The sequences from expressed genes are distinguished between those serotypes (E) previously characterized (Burman et al., 1990; Restrepo et al., 1992) and those of relapses (R) from the current study. 3′ UTR sequences for archival genes are designated by “A”; in cases without an adjacent DHS element a maximum of 117 nucleotides was included in the alignment. The first number in the sequence name after E, R, or A indicates the serotype and, for relapses, the second number indicates the infecting serotype. Sequences with ≤ 2 nucleotide differences were grouped together; the numbers in each group of two or more are given in parentheses. The text color indicates the vsp or vlp family: red, vsp; green, α-vlp; purple, β-vlp; and blue, δ-vlp. There were no instances of a γ-vlp among the relapses. The black numbers along the branches indicate the % support from 500 bootstraps, if greater than 60%. The size marker (0.1) for the branch lengths represents nucleotide distance.
In 8 out of 68 cases the 3′ UTR of the expressed gene in the relapse isolate was different from the 3′ UTR for the archived gene (Figure 4 and Table S4): 2 cases of serotype 24 (R24-7-17 and R24-17-12), 1 case of serotype 2 (R2-17-93), 1 case of serotype 13 (R13-17-89), 1 case of serotype 46 (R46-17-94), 2 cases of serotype 16 (R16-17-96 and R16-17-107), and 1 case of serotype 27 (R27-17-109). In addition, there was a ninth relapse (R4-17-115), in which the difference of the expression site 3′ UTR from the archival site was only apparent beyond the first 34 nt and, thus, not shown in Figure 4. In only 3 of these 9 cases was the 3′ UTR of the newly expressed vsp or vlp wholly or partially like that of the original expression site: two serotype 16 relapses from serotype 17 (R16-17-96 and R16-17-107) and one serotype 4 from serotype 17 (R4-17-115). The observed 65 relapses with 3′ UTR replacement at the expression was twice the expected 34 cases of replacement, if a priori there was an equal likelihood of retention or replacement (Chi square 37.7 1 d.f., p < 0.00001).
Recombination within the DHS elements
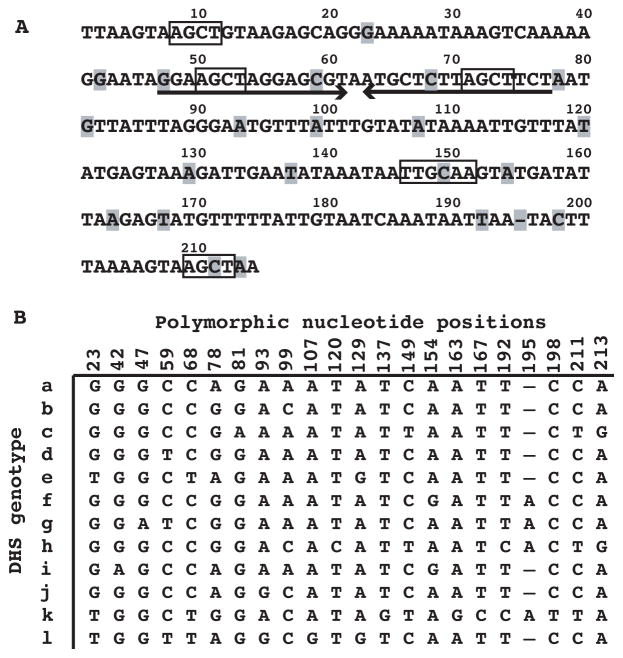
We previously noted differences at a few positions between expression site DHS sequences (Kitten and Barbour, 1990). In the present study we identified several additional polymorphic positions, for a total 22, in the DHS at the expression site for 16 serotypes and among the 13 DHS elements at the archival loci (Figure 5A). The 22 polymorphic positions occurred over the length of the element, beginning at position 23, and were a mean of 8 nucleotides (range of 1–24) apart. Figure 5B is an alignment of a total 12 unique sequences found among the 13 DHS elements at the archival sites; these were designated DHSa through DHSl. The DHS also contains an inverted repeat from positions 47 to 77 with a predicted 15 bp stem loop (ΔG = −18.0 kcal/mol). As in the UHS, there were both 6-base (TTGCAA) and 4-base (AGCT) palindromes in the DHS sequences. The two AGCT palindromes located in the putative stem loop structure were present in all DHS elements.
Figure 5. DHS elements of B. hermsii.
A. The nucleotide sequence of DHSa with positions marked on the top and polymorphic positions indicated by highlighting. A large inverted repeat element is indicated by arrows, and 4-mer and 6-mer palindromes are boxed.
B. Alignment of nucleotides at 22 polymorphic positions for genotypes a through l.
Serotypes 7 and 17 had DHSa and DHSl genotypes, respectively, at the expression site; these sequences differed at 7 (32%) of the 22 polymorphic positions: 23, 59, 68, 93, 99, 107, and 129. We compared these sequences of the infecting serotype with those subsequently observed in the relapse serotypes (Table S3). If the DHS sequences of the relapse serotypes were the same as that of the infecting serotypes, then this would be evidence that the downstream recombination boundary was either 5′ to DHS or no more than 23 nucleotides into the DHS. In fact, we found that among 29 relapse isolates after serotype 7 infections (Table S3), none had the same DHS type as that of the infecting serotype (Chi square 29.0, 1 d.f., p < 0.0001), and among 39 relapse isolates after serotype 17 infections (Table S3), 36 (92%) had a different DHS genotype from that of the infecting serotype (Chi square 27.9, 1 d.f., p < 0.0001).
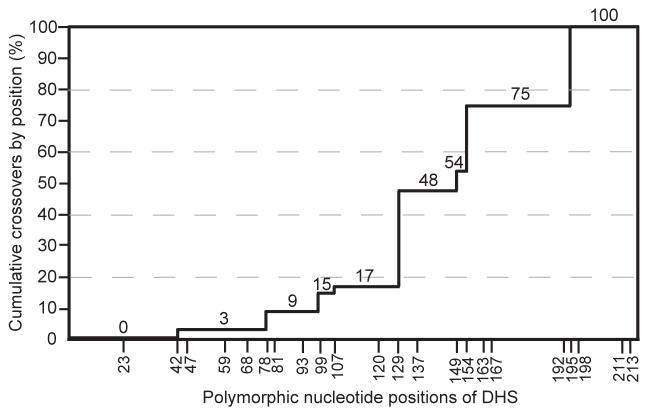
We next studied the sources of genetic novelty of the expression site DHS elements among the relapses in comparison to the DHS elements at the corresponding archival site. There were 68 relapse isolates for this analysis; 29 relapses were from serotype 7 infection and 39 were from serotype 17 infection (Table S3). The expression site’s DHS of infecting serotypes and relapse isolates, along with the donor DHS elements on the archival plasmids, were aligned. In 3 of the 68 relapses, the presumptive donor DHS had the same sequence as the DHS at the original expression site, and, thus, these were not informative for analysis. Among the remaining 65 relapses, 43 had a DHS type that was identical in sequence to the archival site donor, and 22 relapse expression sites had a DHS that was a chimera of the archival site DHS donor at the 5′ end and the infecting serotype’s expression site DHS at the 3′ end (Table S3). Since both the archival and expression site DHS sequences were identical in some parts of the element, we could not map the crossover point to a single or few nucleotides in most cases. We instead determined the inclusive ranges in which cross-overs possibly occurred. Figure 6 summarizes this data as of the cumulative crossovers by the position of the most proximal possible cross-over point. In 54 (83%) out of 65 relapses the crossover was after position 129. If the crossover point occurred randomly over the length of a DHS bounded by informative polymorphic sites, then one would expect that 33 relapses, instead of the observed 54 relapses, would have a crossover after position 129 for the 65 relapses examined (Chi square 27.1, 1 d.f., 2-tailed p < 0.001). In no cases was there evidence of a cross-over after position 211. All the relapse expression site DHS sequences had a C at position 211 and A at position 213, like the infecting serotype. These included 11 cases in which the presumed donor had different nucleotides at positions 211 and 213. In 3 cases (R18-7-20, R18-7-23, and R1-17-3 of Table S3) the relapse isolate had an adenine at position 195 that was not present in either the infecting serotype’s expression site or the presumed DHS donor. Whether this was templated from a DHS element with the A instead of a gap at position 195 or a base duplication during recombination or replication is not known.
Figure 6.
Percent cumulative cross-overs by polymorphic position (x-axis) of the DHS elements for 65 relapse isolates. The percent values are indicated at each level as well as on the y-axis. The sequences are given in Table S3.
In 55 (85%) of the 65 informative relapses, the 5′ end of the new expression site DHS was most similar in sequence to the DHS that was closest downstream of the donor vsp or vlp at the archival locus. However, in 10 (15%) cases the new expression site DHS was instead more similar to that of a more distantly placed DHS element at the archival site.
Origin of DHS elements
To investigate the origin of the DHS elements, we determined whether a similar sequence was downstream of vsp genes in another relapsing fever agent, B. turicatae (AF129737, AF129434, and AF130429), and examined the downstream flanking sequence for the vtp gene at its expression site on a 53 kb linear plasmid within B. hermsii (accession number L24911). An alignment of these sequences with the DHSa element is shown in Figure 7 and suggests that these sequences have a common lineage. The 3 sequences were identical at 43 (61%) out of 70 aligned positions. Moreover, each of the 3 sequences has a potential stem loop structure of ≥ 27 nucleotides in length and with direct repeats of AGCT in each of the arms.
Figure 7.
Alignment of partial nucleotide sequences of DHSa of B. hermsii, DHS-like sequence in B. turicatae, and 3′ flanking region of vtp (formerly vsp33) gene of B. hermsii. Accession numbers are provided in the text. The consensus sequence is shown at the bottom. The positions are numbered according to the B. hermsii DHSa element. Long inverted repeats are indicated by arrows, and 4-mer palindromes are highlighted with gray.
Discussion
In this prospective study of experimental relapsing fever we examined 83 relapse serotypes from infected mice. We determined the sequences of each of the serotypes’ expression sites from the UHS regions that surround the start of the vsp or vlp gene and then through the DHS elements in the subtelomeric region of a linear plasmid (Figure 1). These sequences were compared to previously identified expression sites and to homologous sequences at the archival sites for vsp and vlp genes. To achieve a more comprehensive comparison, we determined the DNA sequence of 152,559 bp, an estimated ~85% of the linear plasmids that bear archival vsp and vlp genes. For all the relapses in this study we located and characterized the archival site for the vsp or vlp gene that was duplicated for the expression site. Rather than depending on a single serotype to initiate infection, we used one of two serotypes, 7 or 17, which represented different vlp families. Except for the serotype-specifying genes at the expression site, the populations were isogenic (Barbour et al., 1991; Kitten and Barbour, 1990; Plasterk et al., 1985).
In previous studies of infections in mice the vsp or vlp gene at the expression site was almost always replaced completely by a different vsp or vlp gene in the relapse serotype (Kitten and Barbour, 1990; Plasterk et al., 1985; Restrepo et al., 1992). In the present study there was only one example of recombination within a vlp or vsp itself: a chimera of vlp7 and vlp18. Consequently, for further defining the boundaries of recombination our attention was on the UHS region, the 3′ UTR downstream of vsp or vlp gene, and the DHS element. Archival vsp and vlp genes generally were located in arrays of at least two genes, and differed in the lengths of their associated UHS sequences and in the distances to the nearest DHS element downstream (Figure 2). We used sequence polymorphisms in the UHS regions and in the DHS elements, as well as the linkages of 3′ UTR sequences to their corresponding vsp and vlp genes, to map the cross-over points for recombinations resulting in the relapse serotypes. Following the principle of parsimony, we assumed that a single recombination event with two cross-over points accounted for the placement of a new vsp or vlp next to the promoter at the expression site, but, as discussed below, this assumption does not rule out subsequent rearrangements at the expression site.
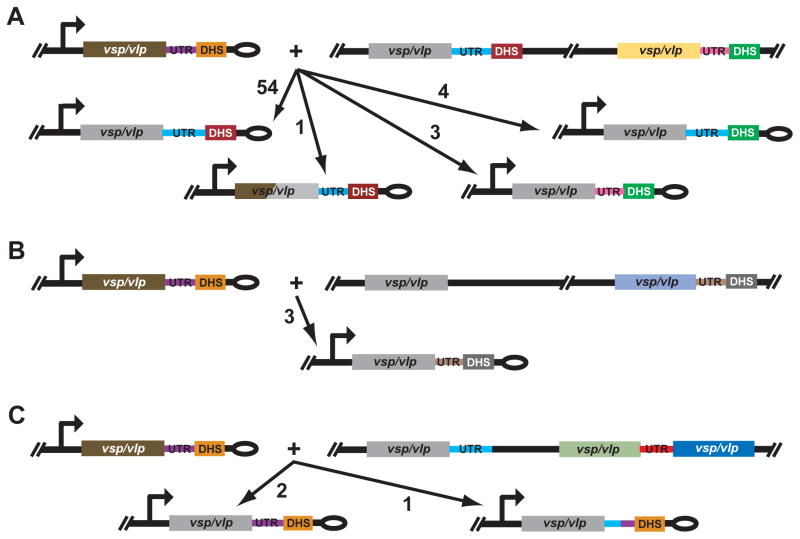
A schematic summary of the recombinations that occurred in 68 independent and informative cases of relapse is shown in Figure 8. In 62 (91%) of the 68 cases the donor vsp or vlp was adjacent to a DHS element, separated only by the 3′ UTR, at its archival site. In 55 (81%) of these 62 cases, the adjacent 3′ UTR and DHS at the expression site were those linked to the vsp or vlp at its archival site (panel A). With one exception, a relapse with a chimera of vlp7 and vlp18 at the expression site, the replacement extended to within the UHS. In 4 of the 62 cases, the 3′ UTR sequence was the one linked to the vsp or vlp at the archival site but a more distantly located DHS element was utilized for the recombination, and in 3 of the 62 cases both the 3′ UTR and the DHS were those located at a distance from the donor vsp or vlp at its archival site (panel A). Panels B and C of Figure 8 show the reactants for 6 recombinations in which the donor vsp or vlp either was not close to a DHS at its archival site (panel B) or did not have a detectable DHS element downstream of it in the sequenced fragment (panel C). When a DHS distant from the donor vsp or vlp was involved in the switch, that DHS and the 3′ UTR in front of it, even if linked to another vsp and vlp, could juxtapose at the expression site with the newly activated vsp or vlp (panel B). In some other cases, the expression site DHS was fully retained, and the expression site 3′ UTR was either fully retained or was a chimera with 3′ UTR linked to the donor vsp or vlp at the archival site (panel C).
Figure 8.
Schematic representations of recombination outcomes for 68 relapse isolates of B. hermsii. The drawing is not to scale. The infecting serotype’s expressed vsp or vlp is shown as brown in panel A, B, and C; the direction of transcription is indicated by the arrow. The hair-pin telomere of the expression plasmid is denoted by an ellipse. The archival vsp or vlp gene that is the donor for each recombination is shown as gray for each recombination. Other vsp/vlp genes, their accompanying 3′ UTR sequences, and different DHS elements in the figure as denoted by other colors. The 3 panels differ in the characteristics of the archival site for the recombination. In panel A there is a DHS element adjacent to the 3′ untranslated region (UTR) for the donor archival gene and a more distant DHS downstream of another vsp or vlp and its 3′ UTR. In panel B the donor archival gene is at a distance from nearest DHS. In panel C there was not a downstream DHS element on the sequence fragment. In each panel the numbers next to the arrows with long arrowheads indicate the frequencies of each type of recombination event.
These findings established the importance of the UHS region of the vsp or vlp gene and the non-coding extragenic DHS element for the recombination. In only 4% of the cases did the antigen switch occur without the apparent involvement of a DHS element at the archival site. In the minority of cases when the nearest DHS element was not employed, a more distant one at the archival site was used. The polymorphisms in the UHS and DHS sequences allowed for identification of the sources for recombinations at the expression site and for mapping of the cross-over points for the majority of recombinations. For the UHS these were most commonly within its second or 3′ half, which encodes the start of the signal peptide, and for the DHS this was usually between positions 129 and 211.
B. burgdorferi also manifests antigenic variation during infections, but the conversion of its expression site by a silent locus is partial and the boundaries for the recombination are entirely intragenic (Zhang and Norris, 1998). The antigenic variation of relapsing fever Borrelia spp. instead most closely resembles that of African trypanosomes, such as Trypanosoma brucei, in its biological and genetic features (Donelson, 1995; Barbour and Restrepo, 2000; Borst, 2002). Both the prokaryotic B. hermsii and the eukaryotic T. brucei employ gene conversion for replacement of a full-length or near full-length variant gene at a telomeric expression site, on a linear plasmids in B. hersmii and a chromosome in T. brucei. The variable antigen repertoires of both pathogens are extensive: 60–70 for B. hermsii, as the present study establishes, and upwards of a thousand for T. brucei (Vanhamme et al., 2001). In both organisms there are repetitive sequences flanking or surrounding the 5′ and 3′ ends of the expression site gene and some of the variant genes at archival sites, and these are the boundaries for the recombination. In the African trypanosomes the upstream recombination locus is a set of imperfect 70-bp repeats that are not transcribed and are 5′ to the variable antigen gene. The downstream recombination locus for trypanosomes is a conserved sequence that comprises the end of coding region and the 3′ UTR (Aline and Stuart, 1989; Liu et al., 1983). Thus, both B. hermsii and T. brucei have an intragenic recombination region on one flank of the variant gene and an extragenic one on the other flank. The extragenic element is upstream of the variant gene in the trypanosome and downstream in the bacterium.
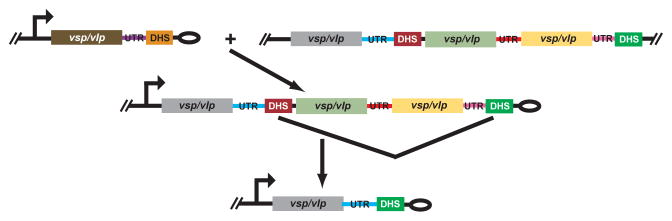
Our current model for the mechanism of antigenic variation in B. hermsii is as follows: Recombination is initiated by a break in the DHS element at the telomeric expression site. The telomeres of Borrelia linear plasmids are inherently recombinogenic (Chaconas, 2005), and this tendency may be exacerbated by the inverted repeat within the DHS. There may also be the action of an undefined recombinase or endonuclease at the inverted repeat or one or other of the shorter 4-bp and 6-bp palindromes within the DHS. In any case, another DHS element in the genome provides a template for the repair of the break, and the resultant heteroduplex extends over the length of archival donor, including the 3′ UTR and vsp or vlp itself, and branch migration terminates within the UHS region. This appears to be the most common type of event. Occasionally, the recombination ends within the vsp or vlp itself when they are the same family, as was the case for the vlp7/18 chimera observed in the present study. At other times, a more distal DHS element is used for the strand invasion and repair, either because there is no DHS adjacent to the archival vsp or vlp or because the most proximal one has inexplicably been skipped. For the latter case, we predict that there is at least one intermediate form of the expression plasmid, such as the one shown in Figure 9. This would be a longer plasmid than what one usually encounters, and the expression site promoter would no longer be telomeric. The greater separation of the expression site promoter and vsp or vlp from the telomere may also be deleterious, as indicated by a mutant in which the expression site was silent when it was no longer near the telomere (Barbour et al., 2000). This circumstance could provide a selection for cells in which deletions occurred between the DHS elements, thus shortening the plasmid and bringing the promoter and vsp/vlp close to a sub-telomeric location again. A precedence in B. hermsii for this proposed mechanism is the documented deletion between short repeats in bringing about the activation of the silent vsp26 gene in a population of serotype 7 cells (Restrepo et al., 1994). Finally, as the examples in panel C of Figure 8 indicate, there are some infrequent cases in which a DHS element at an archival site may not be involved. In such cases the DHS element at the expression site is unchanged.
Figure 9.
Proposed model of recombination events when a distal instead of the most proximal DHS is used. See legend for Figure 8 for description of schematic features. The model shows a two-step process in which the intervening sequence between two DHS elements in the intermediate form is deleted, thus yielding a new expressed gene near the telomere.
The DHS sequence is repeated several times in the genome, albeit with small differences in sequence between the repeats, but the DHS does not have the features of a transposable element. There is an inverted repeat, but it is in the middle of sequence and does not constitute the ends. Highly similar sequences to B. hermsii’s DHS elements were found downstream of expressed and silent genes in another relapsing fever agent, B. turicatae. A possible origin for the element is suggested by the similarity of both the B. hermsii and B. turicatae DHS sequences to the putative rho-independent terminator following the vtp (formerly vsp33) gene on another plasmid in B. hermsii (Carter et al., 1994) (Figure 7). The vtp gene and its promoter are homologous to the ospC genes of B. burgdorferi (Carter et al., 1994). There is only one vtp or ospC gene in the genome of B. hermsii or B. burgdorferi, unlike the multiplicity of alleles of the vsp and vlp genes. We postulate that in the relapsing fever Borrelia spp. lineage there were duplications of the vtp gene over time, resulting in the large repertoire of variable antigen genes we now observe in B. hermsii and other relapsing fever species (Rich et al., 2001). We further propose that one of these duplications established an alternative expression site for these genes. While the vsp (as well as vlp) genes diversified, presumably under both immune and niche selections in their vertebrate hosts (Barbour, 2002), DHS elements minimally differ from each other, thereby providing for a set of substrates for future recombinations and rearrangements. The DHS elements may serve a transcription terminator function in some serotypes but probably not all. Indeed, the DHS element at the expression site in serotypes 7 and 21 seems not to function as the primary terminator. There are sequences with the features of a terminator in the 3′ UTR for vlp7 and vlp21 (Burman et al., 1990; Kitten and Barbour, 1990), and transcription appears to stop before the DHS element is reached in these serotypes (Meier et al., 1985). Thus, while the DHS elements trace a lineage back to a transcriptional control sequence in Borrelia, they now may primarily be “hot-spots” for recombinations, upon which these pathogens depend for persistence in the host.
Experimental Procedures
Strains and culture conditions
The origins of B. hermsii isolates HS1 and DAH were described by Stoenner et al. (Stoenner et al., 1982) and Schwan et al. (Schwan et al., 1996), respectively. By the criterion of the sequences of their vtp, 16S rRNA, flaB, gyrB, and glpQ genes (Porcella et al., 2005), as well as other sequences (this study), HS1 and DAH are different isolates of the same strain enzootic in the northwestern United States (Schwan et al., 1996; Thompson et al., 1969). Frozen stocks of serotypes 7 and 17 of isolate HS1 in mouse plasma were at least 98% pure in serotype by immunoflourescence assay with serotype-specific antisera (Stoenner et al., 1982). For the present study serotypes 7 and 17 were cloned again by limiting dilution in adult female CB17 scid mice (Charles River Laboratories), and the identities of the serotypes was confirmed by sequencing of the expression site (Restrepo et al., 1992). Cells were counted in a Petroff-Hausser counting chamber by phase contrast microscopy. B. hermsii cells were cultivated in BSK II medium at 34°C (Barbour, 1984). Escherichia coli strains INVF′ and Top10F′ (Invitrogen) were grown in Mueller-Hinton or Luria-Bertani medium (Difco).
Mouse infections
In two sets of experiments, “1” and “2”, we identified and isolated the serotypes of the first relapses in mice infected initially with either serotype 7 or 17. Groups of 4–6 wk old female BALB/c mice (Charles River Laboratories) were inoculated intraperitoneally on day 0 with 0.1 ml phosphate-buffered saline, pH 7.4 (PBS) with 5 mM MgCl2 and 1–3 viable spirochetes in set 1 and 0.3–0.6 spirochetes in set 2. The mice were monitored daily for the presence and density of spirochetes by phase-contrast microscopy of a wet mount of tail vein blood. Between 90–100% of mice in set 1 and 30–70% in set 2 experiments were infected. A relapse was the reappearance of spirochetes in blood under microscopy (400X) after absence ≥ 1 day; relapses generally were first detected on days 7, 8, or 9. At that time mice were terminally exsanguinated under anesthesia. Infected plasma was frozen at −76°C in plasma with 10% DMSO (v/v).
DNA methods
Genomic DNA from B. hermsii isolate HS1 was extracted using the DNeasy Tissue Kit (Qiagen), and genomic DNA of B. hermsii isolate DAH was extracted as previously described (Simpson et al., 1990a). For plasmid-enriched DNA, spirochetes were harvested by centrifugation from plasma and resuspended in 50 mM Tris, pH 8.0–50 mM EDTA-15% [w/v] sucrose (TES) with 4 mg/ml of proteinase K. After incubation for 15 min at 37°C, cells were lysed by adding a 1.25X volume of 1% sodium deoxycholate in TES and then incubating at 65°C for 15 min. Thereafter, treatment with diethyl pyrocarbonate, precipitation of the proteins with ammonium acetate, and then precipitation of nucleic acids with isopropanol were carried out as described (Barbour, 1988). Plasmid DNA from E. coli was extracted by the alkaline lysis method or with the High Pure Plasmid Isolation Kit (Roche). DNA was subjected to electrophoresis in a 1.0% agarose gel with a buffer of 90 mM Tris, pH8.3–90 mM boric acid-2mM EDTA (TBE); fragments were cut from the gel and purified by Perfectprep Gel Cleanup Kit (Eppendorf). Isolated products were cloned into the plasmid pCRII in E. coli INVαF′ or the plasmid pCR2.1 in E. coli Top10F′ (Invitrogen). Custom oligonucleotides for primers were synthesized on an Applied Biosystems DNA synthesizer or obtained commercially.
Polymerase chain reactions
For all reactions the sample was subjected to an initial denaturation at 94°C for 5 min and a final extension at 72°C for 7 min. For each of the intervening 30 to 40 cycles, the denaturation step was 94°C for 1 min and extension was 72°C for 1 min, except as noted. The preparation of samples and amplification were performed in separate rooms, and a negative control was included with each set of reactions. The vsp or vlp gene at the expression site on the linear plasmid lp28-1 was amplified as described (Restrepo and Barbour, 1994). The forward and reverse primers (annealing conditions) were, respectively 5′-TAAACTTTGAAAGTTGAGGTATAATGC-3′ and 5′-TAGTACAAATCCCCTTGCCGCTTC-3′ (60° C for 1 min), and for each cycle the extension was 2 min. DNA was first treated with mung bean nuclease to facilitate denaturation of the plasmid telomeres (Kitten and Barbour, 1990). For amplification of vsp and the four vlp sub-families (Hinnebusch et al., 1998), the following sets of forward and reverse primers (and annealing temperatures) were used: vsp, 5-′AAGTCTGAYGGAACAGTRC-3; and 5′-TTATTKTGAGAAGGTTTYTC-3′ (43°C); α-vlp, 5′-AGTGCKGAGAATGCYTTT-3′ and 5′-AWCATTCTTTACTGTCTTYT-3′ (39°C); β-vlp, 5′-CAAGGATTYCAAGATATWT-3′ and 5′-ATAYCTTATTTACWGCACTT-3′ (37°C); γ-vlp, 5-′AATAGACTTAGGTAATGATT-3′ and 5′-GCAATAGTTARTGTATCTARTG-3′ (37°C); and δ-vlp, 5′-ATACTAAGAAAAGTGATATAGG-3′ and 5′-CTTGTTTAACTKTAGCWAG-3′ (37°C). For amplification of probes for size standards, the pairs of forward and reverse primers (and annealing conditions) were the following: 5′-AGCTAAGAGTAATGATGGCAAT-3′ and 5′-ATTTATCACCTTTAGCCATTCT-3′ (55°C for 1 min) for the expression site on the lp28-1 plasmid (accession number DQ218042); 5′-CAGATGGTCTTACTGCTGAAGC-3′ and 5′-CAGCAACAACCTTTTCCTTTAG-3′ (55°C for 1 min) for vtp (formerly vsp33; L24911) on a 53 kb linear plasmid (Carter et al., 1994); and 5′-ACTTGCTGTTCAATCTGGTAATGG-3′ and 5′-GTTGATTTCATCTGTAAGTTGCTCAATTT-3′ (60°C for 1 min) for flaB (X53940) on the 950 kb linear chromosome (Ferdows et al., 1996).
Pulsed-field gel electrophoresis and Southern blot analysis
109 B. hermsii cells were embedded in 80 μl 0.5% (w/v) low melting temperature agarose (BioWhittaker Molecular Applications) and treated with 1 mg/ml proteinase K in 50 mM Tris (pH8.0)-50mM EDTA-1% sodium dodecyl sulfate (SDS) at 50°C for 24 h (Ferdows et al., 1996). Washed agarose blocks were loaded into wells of a 1% agarose gel, and pulsed-field gel electrophoresis was performed with a CHEF Mapper apparatus (Bio-Rad) at 14°C for 21 h in 0.5X TBE with the following settings: 6 V/cm with a pulsed-field angle of 120°, initial switch of 1 second, and final switch of 6 second. The gels were stained with ethidium bromide, and Southern blot analysis was carried out as described (Zhong and Barbour, 2004). The probes were 100 nucleotides or longer, and hybridization was carried out at 60°C in 0.12 M Na2HPO4, pH 7.2–0.25 M NaCl-7% [w/v)] SDS; the final wash was with 0.15 M NaCl-0.015 M sodium citrate-0.1% SDS at 60°C.
DNA sequencing and curation
For isolate HS1, primer-directed, dye-termination sequencing of PCR products or recombinant plasmids was done manually as described (Barbour et al., 1996) or by capillary electrophoresis on Applied Biosystems 373A or Beckman Coulter CEQ 3000 automated DNA sequencers. Partial sequences of linear plasmids lp28-1 (formerly called lp7E), lp32-1 (lp7S), and lp28-2 (lp21S) were previously determined and had the following accession numbers (with fragment number and range of positions corresponding to Figure 2 and Table S1): DQ218042 (entire fragment I), DQ166207 (positions 1–5485 of fragment II), and DQ172919 (1-10900 of fragment III). Additional sequences were from a B. hermsii genomic library in pUC18 (Putteet-Driver et al., 2004). Clones of interest were identified by colony hybridization with radiolabeled PCR-amplified fragments for selected vsp or vlp genes, the plasmid inserts were sequenced, and the sequence assemblies were confirmed by PCR across the inserts of different clones (Barbour et al., 1996). The additional HS1 sequences were assigned accession numbers AY840995 (13086-22593 of fragment IV), AY838879 (11124-22769 of fragment VII) and DQ173930 (entire fragment X). Newly-identified vsp or vlp sequences of HS1 from PCR-based studies (see above) in this study were assigned the following accession numbers: DQ423795 (vlp47), DQ423796 (vlp51), and DQ423797 (vlp55), DQ423793 (vlp42), DQ423792 (vlp46), and DQ423794 (vsp58).
A genomic DNA library of B. hermsii isolate DAH was constructed with DNA that had been sheared by nebulization (http://www.genome.ou.edu/protocol_book/protocol_partII.html). In brief, 25 μg of DNA were suspended in 500 ul 10 mM Tris, pH 8.0–1 mM EDTA and 25% glycerol. The mixture was forced by nitrogen gas through a plastic nebulizer (No. 4101, IPI Medical Products) to create DNA fragments between 2 and 3 kb. The sheared DNA was gel-purified, concentrated, and treated with Klenow and T4 DNA polymerase, and ligated into the vector pCR4Blunt-TOPO (Invitrogen). Nucleotide sequence data from colony-PCR-amplified clones were obtained with Big-Dye terminator chemistry and an ABI3700 automated capillary electrophoresis sequencer (Applied Biosystems) as described (Smoot et al., 2002). Sequence data were stored with the FINCH data management system (Geospiza) and assembled with SPS-PHRAP (Southwest Parallel Software). Sequence and physical gaps were analyzed with CROSS_MATCH (http://www.phrap.org) and SEQUENCHER4.1 (Gene Codes). Primers were designed with CONSED (http://www.phrap.com/consed/) and obtained from Sigma-Genosys (Sigma-Genosys) to initiate gap closures and re-sequence regions of low quality. The nine fragments of DAH sequence used for the present study were assigned the following GenBank accession numbers (with fragment number and range of positions corresponding to Figure 2 and Table S1): CP000273 (683-12119 of fragment I), CP000274 (1-8913 of fragment II), CP000275 (1563-16331 of fragment III), CP000276 (1-21928 of fragment IV), CP000277 (entire fragment V), CP000278 (entire fragment VI), CP000279 (1-15901 of fragment VII), CP000280 (entire fragment VIII), and CP000281 (entire fragment IX).
A total of 54,474 bp of nonredundant DNA sequence of plasmids were determined for isolate HS1, and 429,868 bp were determined for isolate DAH. A sequence fragment was identified as plasmid in origin if it had previously been shown to be a plasmid (Carter et al., 1994; Kitten and Barbour, 1992), contained a vsp or vlp gene (Carter et al., 1994; Ferdows et al., 1996; Plasterk et al., 1985), or if it contained a sequence homologous to B. burgdoreri genes known to be restricted to plasmids (Casjens et al., 2000; Fraser et al., 1997). Nine of the fragments (139,478 bp) of DAH sequences and 6 fragments (54,474 bp) of HS1 sequence contained at least one vsp or vlp sequence (see below). The sequences of isolates HS1 and DAH coincided over 5 fragments of 4,778 to 11,433 nucleotides for a total 39,912 nucleotides, of which only 22 (0.06%) differed between the sequencing projects and isolates.
The sequences were the queries for BLASTN, BLASTX, and TBLASTX searches of a local database of known vsp and vlp genes and DHS elements (http://spiro.mmg.uci.edu/blast) and the GenBank database. An open reading frame (ORF) with homology to vsp or vlp genes was assigned a new allele designation, e.g. vsp28 or vlp42, if the nucleotide sequence was <90% identical to a previously numbered vsp or vlp gene over its ORF length. A vsp- or vlp-like sequence missing more than 100 bp at its 5′ end and/or with at least one nonsense mutation or confirmed frame shift was called a pseudogene. If the pseudogene was >90% identical in nucleotide sequence to a previously identified vsp or a vlp gene, it was also assigned a number. A vsp- or vlp-like sequence less than 70% of its usual length was designated a vsp or vlp sub-family fragment but not assigned a unique allele name. Other ORFs with E-values of <10−4 by BLAST searches against bacterial sequences were named according to presumptive function or according to B. burgdorferi genome nomenclature for chromosomal genes or for plasmid-borne paralogous gene families (Casjens et al., 2000; Fraser et al., 1997). Nucleotide sequences were aligned with Clustalx version 1.83 (http://www.embl.de/~chenna/clustal/darwin). Phylograms by the neighbor-joining distance criterion were produced with Phylo_Win software (http://pbil.univ-lyon1.fr/software/phylowin.html) (Galtier et al., 1996).
Supplementary Material
Table S1. Open reading frames (ORF) and DHS elements on 10 fragments of linear plasmids of Borrelia hermsii with vsp and vlp genes shown in physical maps of Figure 2.
Table S2. Polymorphic sites (positions 22–23) of UHS regions of relapse expression sites and archival sites.
Table S3. DHS elements at expression sites for infecting and relapse isolates and the presumptive donor at archival site.
Table S4. Alignment of nucleotide sequences of the 3′ UTR for vsp and vlp genes at expression sites for previously-identified serotypes (E) and relapse isolates (R) and at archival sites (A)
Acknowledgments
We thank Carol Carter, Hany Mattaous, and Merry Schrumpf and for technical assistance. This work was supported by NIH grant AI24424 and by the Intramural Research Program of NIAID, NIH.
References
- Aline RF, Jr, Stuart K. Trypanosoma brucei: conserved sequence organization 3′ to telomeric variant surface glycoprotein genes. Exp Parasitol. 1989;68:57–66. doi: 10.1016/0014-4894(89)90008-8. [DOI] [PubMed] [Google Scholar]
- Barbour AG. Isolation and cultivation of Lyme disease spirochetes. Yale J Biol Med. 1984;57:521–525. [PMC free article] [PubMed] [Google Scholar]
- Barbour AG. Plasmid analysis of Borrelia burgdorferi, the Lyme disease agent. J Clin Microbiol. 1988;26:475–478. doi: 10.1128/jcm.26.3.475-478.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbour AG. Antigenic variation by relapsing fever Borrelia species and other bacterial pathogens. In: Craig NL, Craigie R, Gellert M, Lambowitz A, editors. Mobile DNA II. Washington, D.C: ASM Press; 2002. pp. 972–994. [Google Scholar]
- Barbour AG, Burman N, Carter CJ, Kitten T, Bergström S. Variable antigen genes of the relapsing fever agent Borrelia hermsii are activated by promoter addition. Mol Microbiol. 1991;5:489–493. doi: 10.1111/j.1365-2958.1991.tb02132.x. [DOI] [PubMed] [Google Scholar]
- Barbour AG, Carter CJ, Bundoc V, Hinnebusch J. The nucleotide sequence of a linear plasmid of Borrelia burgdorferi reveals similarities to those of circular plasmids of other prokaryotes. J Bacteriol. 1996;178:6635–6639. doi: 10.1128/jb.178.22.6635-6639.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbour AG, Carter CJ, Sohaskey CD. Surface protein variation by expression site switching in the relapsing fever agent Borrelia hermsii. Infect Immun. 2000;68:7114–7121. doi: 10.1128/iai.68.12.7114-7121.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbour AG, Restrepo BI. Antigenic variation in vector-borne pathogens. Emerg Infect Dis. 2000;6:449–457. doi: 10.3201/eid0605.000502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbour AG. Relapsing fever. In: Goodman Jesse L, DTD, Sonenshine Daniel E., editors. Tick-Borne Diseases of Humans. Washington, D.C: American Society for Microbiology Press; 2005. pp. 268–291. [Google Scholar]
- Barbour AG, Stoenner HG. Antigenic variation of Borrelia hermsii. In: Herskowitz I, Simon M, editors. Genome Rearrangement. Vol. 20. New York: UCLA Symposia on Molecular and Cellular Biology, Alan R. Liss, Inc; 1985. pp. 123–135. Vol. New Series. [Google Scholar]
- Borst P. Antigenic variation in eukaryotic parasites. In: Craig Nancy L, RC, Gellert Martin, Lambowitz Alan M., editors. Mobile DNA II. Washington, D.C: ASM press; 2002. pp. 953–971. [Google Scholar]
- Burman N, Bergström S, Restrepo BI, Barbour AG. The variable antigens Vmp7 and Vmp21 of the relapsing fever bacterium Borrelia hermsii are structurally analogous to the VSG proteins of the African trypanosome. Mol Microbiol. 1990;4:1715–1726. doi: 10.1111/j.1365-2958.1990.tb00549.x. [DOI] [PubMed] [Google Scholar]
- Carter CJ, Bergstrom S, Norris SJ, Barbour AG. A family of surface-exposed proteins of 20 kilodaltons in the genus Borrelia. Infect Immun. 1994;62:2792–2799. doi: 10.1128/iai.62.7.2792-2799.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casjens S, Palmer N, van Vugt R, Huang WM, Stevenson B, Rosa P, Lathigra R, Sutton G, Peterson J, Dodson RJ, Haft D, Hickey E, Gwinn M, White O, Fraser CM. A bacterial genome in flux: the twelve linear and nine circular extrachromosomal DNAs in an infectious isolate of the Lyme disease spirochete Borrelia burgdorferi. Mol Microbiol. 2000;35:490–516. doi: 10.1046/j.1365-2958.2000.01698.x. [DOI] [PubMed] [Google Scholar]
- Chaconas G, Stewart PE, Tilly K, Bono JL, Rosa P. Telomere resolution in the Lyme disease spirochete. EMBO J. 2001;20:3229–3237. doi: 10.1093/emboj/20.12.3229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donelson JE. Mechanisms of antigenic variation in Borrelia hermsii and African trypanosomes. J Biol Chem. 1995;270:7783–7786. doi: 10.1074/jbc.270.14.7783. [DOI] [PubMed] [Google Scholar]
- Dworkin MS, Schwan TG, Anderson DE., Jr Tick-borne relapsing fever in North America. Med Clin North Am. 2002;86:417–433. viii–ix. doi: 10.1016/s0025-7125(03)00095-6. [DOI] [PubMed] [Google Scholar]
- Ferdows MS, Serwer P, Griess GA, Norris SJ, Barbour AG. Conversion of a linear to a circular plasmid in the relapsing fever agent Borrelia hermsii. J Bacteriol. 1996;178:793–800. doi: 10.1128/jb.178.3.793-800.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraser CM, Casjens S, Huang WM, Sutton GG, Clayton R, Lathigra R, White O, Ketchum KA, Dodson R, Hickey EK, Gwinn M, Dougherty B, Tomb JF, Fleischmann RD, Richardson D, Peterson J, Kerlavage AR, Quackenbush J, Salzberg S, Hanson M, van Vugt R, Palmer N, Adams MD, Gocayne J, Weidman J, Utterback T, Watthey L, McDonald L, Artiach P, Bowman C, Garland S, Fuji C, Cotton MD, Horst K, Roberts K, Hatch B, Smith HO, Venter JC. Genomic sequence of a Lyme disease spirochaete, Borrelia burgdorferi. Nature. 1997;390:580–586. doi: 10.1038/37551. [DOI] [PubMed] [Google Scholar]
- Galtier N, Gouy M, Gautier C. SEAVIEW and PHYLO_WIN: two graphic tools for sequence alignment and molecular phylogeny. Comput Appl Biosci. 1996;12:543–548. doi: 10.1093/bioinformatics/12.6.543. [DOI] [PubMed] [Google Scholar]
- Glockner G, Lehmann R, Romualdi A, Pradella S, Schulte-Spechtel U, Schilhabel M, Wilske B, Suhnel J, Platzer M. Comparative analysis of the Borrelia garinii genome. Nucleic Acids Res. 2004;32:6038–6046. doi: 10.1093/nar/gkh953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinnebusch BJ, Barbour AG, Restrepo BI, Schwan TG. Population structure of the relapsing fever spirochete Borrelia hermsii as indicated by polymorphism of two multigene families that encode immunogenic outer surface lipoproteins. Infect Immun. 1998;66:432–440. doi: 10.1128/iai.66.2.432-440.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitten T, Barbour AG. Juxtaposition of expressed variable antigen genes with a conserved telomere in the bacterium Borrelia hermsii. Proc Natl Acad Sci USA. 1990;87:6077–6081. doi: 10.1073/pnas.87.16.6077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitten T, Barbour AG. The relapsing fever agent Borrelia hermsii has multiple copies of its chromosome and linear plasmids. Genetics. 1992;132:311–324. doi: 10.1093/genetics/132.2.311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitten T, Barrera AV, Barbour AG. Intragenic recombination and a chimeric outer membrane protein in the relapsing fever agent Borrelia hermsii. J Bacteriol. 1993;175:2516–2522. doi: 10.1128/jb.175.9.2516-2522.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu AY, Van der Ploeg LH, Rijsewijk FA, Borst P. The transposition unit of variant surface glycoprotein gene 118 of Trypanosoma brucei. Presence of repeated elements at its border and absence of promoter-associated sequences. J Mol Biol. 1983;167:57–75. doi: 10.1016/s0022-2836(83)80034-5. [DOI] [PubMed] [Google Scholar]
- Meier JT, Simon MI, Barbour AG. Antigenic variation is associated with DNA rearrangements in a relapsing fever Borrelia. Cell. 1985;41:403–409. doi: 10.1016/s0092-8674(85)80013-1. [DOI] [PubMed] [Google Scholar]
- Plasterk RH, Simon MI, Barbour AG. Transposition of structural genes to an expression sequence on a linear plasmid causes antigenic variation in the bacterium Borrelia hermsii. Nature. 1985;318:257–263. doi: 10.1038/318257a0. [DOI] [PubMed] [Google Scholar]
- Porcella SF, Raffel SJ, Anderson DE, Jr, Gilk SD, Bono JL, Schrumpf ME, Schwan TG. Variable tick protein in two genomic groups of the relapsing fever spirochete Borrelia hermsii in western North America. Infect Immun. 2005;73:6647–6658. doi: 10.1128/IAI.73.10.6647-6658.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Putteet-Driver AD, Zhong J, Barbour AG. Transgenic expression of RecA of the spirochetes Borrelia burgdorferi and Borrelia hermsii in Escherichia coli revealed differences in DNA repair and recombination phenotypes. J Bacteriol. 2004;186:2266–2274. doi: 10.1128/JB.186.8.2266-2274.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Restrepo BI, Kitten T, Carter CJ, Infante D, Barbour AG. Subtelomeric expression regions of Borrelia hermsii linear plasmids are highly polymorphic. Mol Microbiol. 1992;6:3299–3311. doi: 10.1111/j.1365-2958.1992.tb02198.x. [DOI] [PubMed] [Google Scholar]
- Restrepo BI, Barbour AG. Antigen diversity in the bacterium B. hermsii through “somatic” mutations in rearranged vmp genes. Cell. 1994;78:867–876. doi: 10.1016/s0092-8674(94)90642-4. [DOI] [PubMed] [Google Scholar]
- Restrepo BI, Carter CJ, Barbour AG. Activation of a vmp pseudogene in Borrelia hermsii: an alternate mechanism of antigenic variation during relapsing fever. Mol Microbiol. 1994;13:287–299. doi: 10.1111/j.1365-2958.1994.tb00423.x. [DOI] [PubMed] [Google Scholar]
- Rich SM, Sawyer SA, Barbour AG. Antigen polymorphism in Borrelia hermsii, a clonal pathogenic bacterium. Proc Natl Acad Sci USA. 2001;98:15038–15043. doi: 10.1073/pnas.071042098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwan TG, Schrumpf ME, Hinnebusch BJ, Anderson DE, Jr, Konkel ME. GlpQ: an antigen for serological discrimination between relapsing fever and Lyme borreliosis. J Clin Microbiol. 1996;34:2483–2492. doi: 10.1128/jcm.34.10.2483-2492.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwan TG, Piesman J. Vector interactions and molecular adaptations of Lyme disease and relapsing fever spirochetes associated with transmission by ticks. Emerg Infect Dis. 2002;8:115–121. doi: 10.3201/eid0802.010198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson WJ, Garon CF, Schwan TG. Analysis of supercoiled circular plasmids in infectious and non-infectious Borrelia burgdorferi. Microb Pathog. 1990a;8:109–118. doi: 10.1016/0882-4010(90)90075-2. [DOI] [PubMed] [Google Scholar]
- Simpson WJ, Garon CF, Schwan TG. Borrelia burgdorferi contains repeated DNA sequences that are species specific and plasmid associated. Infect Immun. 1990b;58:847–853. doi: 10.1128/iai.58.4.847-853.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smoot JC, Barbian KD, Van Gompel JJ, Smoot LM, Chaussee MS, Sylva GL, Sturdevant DE, Ricklefs SM, Porcella SF, Parkins LD, Beres SB, Campbell DS, Smith TM, Zhang Q, Kapur V, Daly JA, Veasy LG, Musser JM. Genome sequence and comparative microarray analysis of serotype M18 group A Streptococcus strains associated with acute rheumatic fever outbreaks. Proc Natl Acad Sci USA. 2002;99:4668–4673. doi: 10.1073/pnas.062526099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevenson B, Porcella SF, Oie KL, Fitzpatrick CA, Raffel SJ, Lubke L, Schrumpf ME, Schwan TG. The relapsing fever spirochete Borrelia hermsii contains multiple, antigen-encoding circular plasmids that are homologous to the cp32 plasmids of Lyme disease spirochetes. Infect Immun. 2000;68:3900–3908. doi: 10.1128/iai.68.7.3900-3908.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoenner HG, Dodd T, Larsen C. Antigenic variation of Borrelia hermsii. J Exp Med. 1982;156:1297–1311. doi: 10.1084/jem.156.5.1297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson RS, Burgdorfer W, Russell R, Francis BJ. Outbreak of tick-borne relapsing fever in Spokane County, Washington. JAMA. 1969;210:1045–1050. [PubMed] [Google Scholar]
- Vanhamme L, Pays E, McCulloch R, Barry JD. An update on antigenic variation in African trypanosomes. Trends Parasitol. 2001;17:338–343. doi: 10.1016/s1471-4922(01)01922-5. [DOI] [PubMed] [Google Scholar]
- Zhang JR, Norris SJ. Genetic variation of the Borrelia burgdorferi gene vlsE involves cassette-specific, segmental gene conversion. Infect Immun. 1998;66:3698–704. doi: 10.1128/iai.66.8.3698-3704.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong J, Barbour AG. Cross-species hybridization of a Borrelia burgdorferi DNA array reveals infection- and culture-associated genes of the unsequenced genome of the relapsing fever agent Borrelia hermsii. Mol Microbiol. 2004;51:729–748. doi: 10.1046/j.1365-2958.2003.03849.x. [DOI] [PubMed] [Google Scholar]
- Zhong J, Skouloubris S, Dai Q, Myllykallio H, Barbour AG. Function and evolution of plasmid-borne genes for pyrimidine biosynthesis in Borrelia spp. J Bacteriol. 2006;188:909–918. doi: 10.1128/JB.188.3.909-918.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zückert WR, Meyer J, Barbour AG. Comparative analysis and immunological characterization of the Borrelia Bdr protein family. Infect Immun. 1999;67:3257–3266. doi: 10.1128/iai.67.7.3257-3266.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Open reading frames (ORF) and DHS elements on 10 fragments of linear plasmids of Borrelia hermsii with vsp and vlp genes shown in physical maps of Figure 2.
Table S2. Polymorphic sites (positions 22–23) of UHS regions of relapse expression sites and archival sites.
Table S3. DHS elements at expression sites for infecting and relapse isolates and the presumptive donor at archival site.
Table S4. Alignment of nucleotide sequences of the 3′ UTR for vsp and vlp genes at expression sites for previously-identified serotypes (E) and relapse isolates (R) and at archival sites (A)