Summary
Cyclic GMP-AMP synthase (cGAS) is a cytosolic DNA sensor that induces the IFN antiviral response. However, the regulatory mechanisms that mediate cGAS-triggered signaling have not been fully explored. Here, we show the involvement of a small GTPase RAB2B and its effector protein Golgi-associated RAB2B interactor-like 5 (GARIL5) in the cGAS-mediated IFN response. RAB2B-deficiency affects the IFN response induced by cytosolic DNA. Consistent with this, RAB2B-deficiency enhances replication of vaccinia virus, a DNA virus. After DNA stimulation, RAB2B colocalizes with stimulator of interferon genes (STING), the downstream signal mediator of cGAS, on the Golgi apparatus. GTP-binding activity of RAB2B is required for its localization on the Golgi apparatus and for recruitment of GARIL5. GARIL5 deficiency also affects the IFN response induced by cytosolic DNA and enhances replication of vaccinia virus. These findings indicate that the RAB2B-GARIL5 complex promotes IFN responses against DNA viruses by regulating the cGAS-STING signaling axis.
Keywords: Host defense, Innate Immunity, Antiviral response, Interferon, Rab GTPase, Organelle
eTOC Blurb
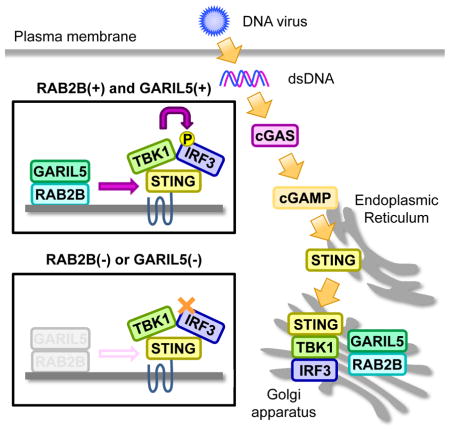
Takahama et al. show that RAB2B GTPase recruits its effector protein GARIL5 into the Golgi apparatus to positively regulate cytosolic DNA-triggered activation of the cGAS-STING signaling axis and promotes type I IFN-mediated host defense response to DNA virus.
Introduction
Innate immunity functions as the first line of defense against a variety of invading pathogens. The recognition of pathogen-associated molecular patterns by germline-encoded pattern-recognition receptors (PRRs), such as Toll-like receptors (TLRs), retinoic acid-inducible gene- I (RIG-I)-like receptors, nucleotide binding oligomerization domain (NOD)-like receptors, and cyclic GMP-AMP (cGAMP) synthase (cGAS), initiates innate immune responses (Palm and Medzhitov, 2009; Beutler, 2009; Kawai and Akira, 2009; Schroder and Tschopp, 2010; Cai et al., 2014). Upon recognition, these receptors trigger signal transduction pathways that induce type I interferon (IFN) and proinflammatory cytokines, which are essential to generate an innate immune response, as well as in subsequent adaptive immune responses.
DNA derived from bacteria, DNA viruses, and dead host cells trigger an innate immune response (Ishii et al., 2006; Stetson and Medzhitov, 2006; Ishii et al., 2008; Charrel-Dennis et al., 2008; Schroder et al., 2009; Hornung et al., 2009). Recent studies have identified cGAS as a cytosolic DNA sensor (Sun et al., 2013; Wu et al., 2013). Upon DNA binding, cGAS synthesizes cGAMP from ATP and guanosine triphosphate (GTP) (Ablasser et al., 2013; Gao et al., 2013; Diner et al., 2013). In turn, cGAMP functions as a second messenger and is recognized by stimulator of interferon genes (STING), an endoplasmic reticulum (ER)-resident protein. After cGAMP recognition, STING moves from the ER to the Golgi apparatus and finally reaches cytoplasmic punctate structures to assemble with TANK-binding kinase 1 (TBK1) (Ishikawa and Barber, 2008; Ishikawa et al., 2009; Saitoh et al., 2009). Subsequently, TBK1 phosphorylates the transcription factor interferon regulatory factor 3 (IRF3) to activate the transcription of type I IFN and IFN-inducible genes. Consistent with this, the cGAS-STING-TBK1 signaling axis plays an essential role in antiviral IFN responses against DNA viruses (Li et al., 2013; Schoggins et al., 2013). Therefore, clarification of the molecular mechanisms underlying cGAS-triggered signaling is needed to better understand the process of innate immunity.
The Rab family belongs to the Ras superfamily of small GTPases. Approximately 60 Rab genes are present in the human genome, and a number of these are conserved from yeast to mammals. The Rab family regulates various cellular events such as vesicle formation, vesicle movement along the cytoskeleton, and membrane fusion that occurs on organelles (Stenmark, 2009). Comprehensive screening experiments have revealed that Rab GTPases and their regulatory molecules, GDP-GTP exchange factors and GTPase-activating proteins, control various biological processes such as autophagy, exocytosis, cell migration, and primary cilium formation (Matsui and Fukuda, 2013; Zografou et al., 2012; Linford et al., 2012; Yoshimura et al., 2007). Importantly, Rab GTPases are also involved in TLR-mediated immune responses. RAB11A regulates the recruitment of TLR4 and TRIF-related adaptor molecule (TRAM) to the phagosome, a process requiring TLR4 signaling (Husebye et al., 2010). Proper intracellular trafficking of TLR4 and TRAM by RAB11A is required for maximal activation of IRF3, and RAB7B limits activation of the TLR4 signaling pathway by promoting lysosomal degradation of TLR4 (Wang et al., 2007). Therefore, Rab family GTPases might play a critical role in regulating innate immune responses mediated by other PRRs. Although previous studies have shown that the ER and Golgi apparatus are involved in STING-mediated signaling, little is known about a regulator of STING that acts on these organelles. Hence, we focused on the Rab family and identified a positive regulator of the cGAS-STING signaling axis. Here, we show that RAB2B is required for antiviral responses against DNA viruses.
Results
RAB2B regulates DNA-induced innate immune responses
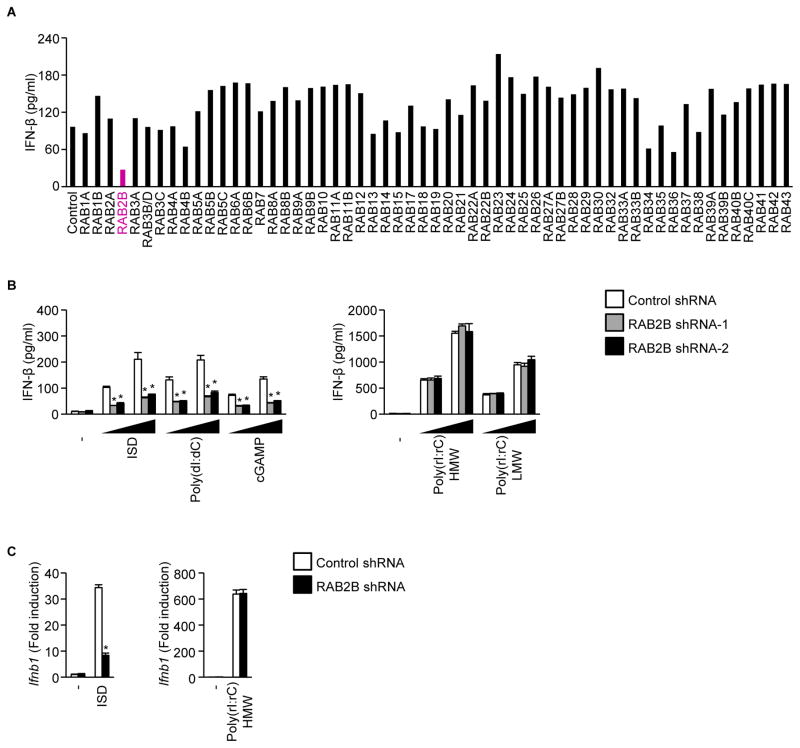
We first performed small interfering RNA (siRNA)-based screening to search for Rab GTPases whose knockdown affected cytosolic DNA-induced production of IFN-β in immortalized mouse embryonic fibroblasts (MEFs). Knockdown of Rab2b, Rab34, and Rab36 mRNA inhibited IFN-β production induced by DNA (Figure 1A). In the present study, we focused on RAB2B, which was the enzyme that showed the strongest effect on IFN-β production. We next examined the involvement of RAB2B in a DNA-induced innate immune response in primary MEFs stably expressing short hairpin RNA (shRNA) against RAB2B or control shRNA. Knockdown of Rab2b mRNA inhibited production of IFN-β and CXCL10 induced by interferon stimulatory DNA (ISD), poly (dI:dC), and cGAMP (Figures 1B, S1, and S2A). Knockdown of Rab2b mRNA inhibited upregulation of Ifnb1 and Cxcl10 mRNA by ISD (Figures 1C and S2B). However, knockdown of Rab2b mRNA did not affect the innate immune response induced by RNA such as high- and low-molecular weight poly (rI:rC) (Figures 1B, 1C, S2A, and S2B). These findings indicated that RAB2B promotes DNA-induced immune responses.
Figure 1. RAB2B regulates DNA-induced innate immune responses.
(A) Immortalized wild-type MEFs treated with siRNA against the indicated Rab GTPases were stimulated with poly (dI:dC) for 24 h. The levels of IFN-β in the culture supernatants were measured by ELISA.
(B) Primary MEFs stably expressing the indicated shRNAs were stimulated with the indicated ligands (500 ng/ml or 1000 ng/ml) for 24 h. The levels of IFN-β in the supernatants were measured by ELISA. The results shown are means ± SD (n = 3); *P < 0.01.
(C) Primary MEFs stably expressing the indicated shRNAs were stimulated with the indicated ligands for 6 h. The levels of Ifnb1 mRNA were measured by quantitative RT-PCR. The results shown are means ± SD (n = 3); *P < 0.01.
GTP-binding activity of RAB2B is involved in DNA-induced immune responses
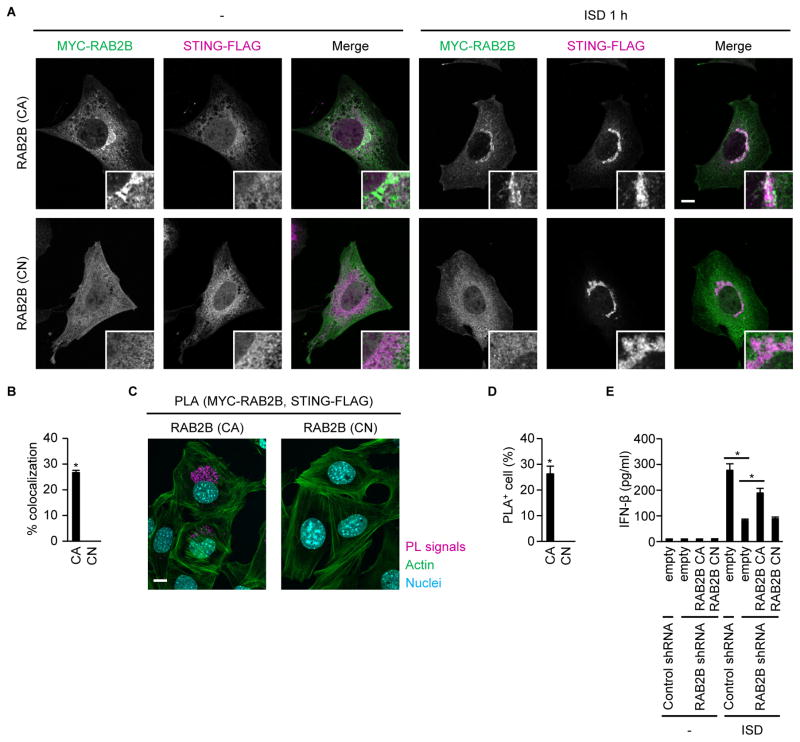
Rab GTPases cycle between a GTP-bound active form and a guanosine diphosphate (GDP)-bound negative form, and function as molecular switches (Stenmark, 2009). Therefore, we examined the importance of the GTP-binding activity of RAB2B in regulating DNA-induced immune responses. Constitutively active RAB2B (Q65L) colocalized with STING (Figures 2A and 2B). However, constitutively negative RAB2B (S20N) did not localize on membrane-bound compartments or colocalize with STING (Figures 2A and 2B). Consistent with this, the spatial approximation of STING to constitutively active RAB2B, but not constitutively negative RAB2B, occurred after DNA stimulation (Figures 2C and 2D). Furthermore, complementation of constitutively active RAB2B, but not constitutively negative RAB2B, promoted IFN-β production induced by ISD in MEFs expressing shRNA that targeted the 3′-untranslated region of Rab2b mRNA (Figure 2E). These findings indicated that RAB2B regulation of STING-mediated innate immune responses was dependent on its GTP-binding activity.
Figure 2. GTP-binding activity of RAB2B is required for a DNA-induced innate immune response.
(A and B) Primary MEFs stably expressing shRNA against RAB2B, together with STING-FLAG and constitutively active MYC-RAB2B or constitutively negative MYC-RAB2B, were established by retroviral transduction and transfected with ISD. Colocalization of MYC-RAB2B and STING-FLAG was observed under a confocal laser scanning microscope and fluorescence microscope (A) (scale bars, 10 μm). The frequency of colocalization of MYC-RAB2B with STING-FLAG in ISD-stimulated MEFs was determined (B). The graph shows means ± SD (n = 3); *P < 0.01.
(C and D) Proximity-ligation assay (PLA) of MYC-RAB2B and STING-FLAG in primary MEFs stably expressing shRNA against RAB2B, together with STING-FLAG and constitutively active MYC-RAB2B or constitutively negative MYC-RAB2B, after stimulation with ISD (C) (scale bars, 10 μm). The frequency of cells with proximity ligation-positive signals was determined (D). The graph shows means ± SD (n = 3); *P < 0.01.
(E) Primary MEFs stably expressing the indicated shRNAs, together with constitutively active RAB2B or constitutively negative RAB2B, were transfected with ISD. The levels of IFN-β in the supernatants were measured by ELISA. The results shown are means ± SD (n = 3); *P < 0.01.
RAB2B-GARIL5 complex regulates DNA-induced innate immune responses
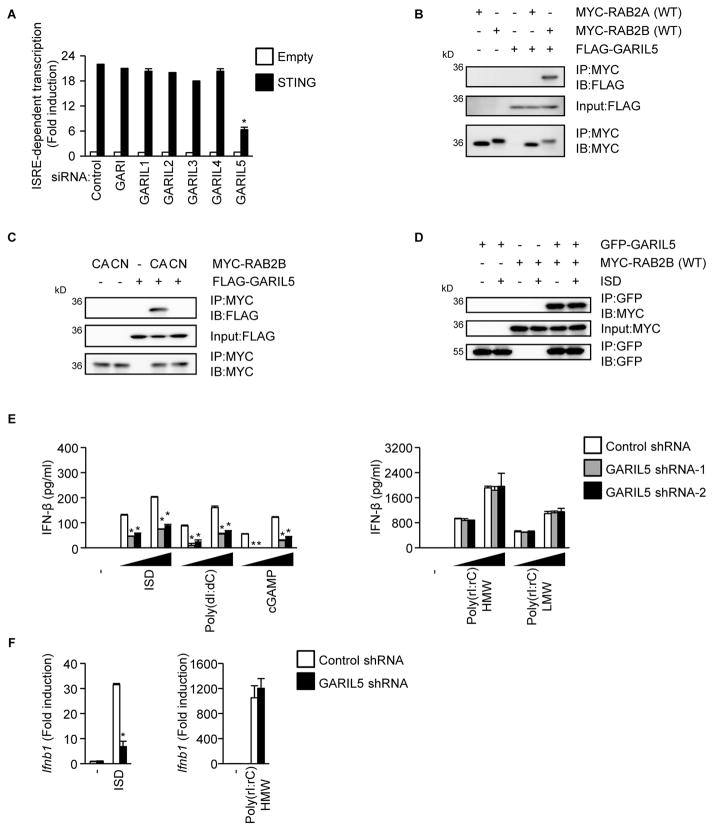
Rab GTPases perform their regulatory function by recruiting specific effector molecules. Golgi-associated RAB2B interactor (GARI) was identified as a candidate effector molecule of RAB2B (Fukuda et al., 2008). In addition to GARI, GARI-like 1 (GARIL1), GARIL2, GARIL3, GARIL4, and GARIL5 each harbor a putative RAB2B-binding domain (Fukuda et al., 2008). Thus, we examined the role of GARI family members in DNA-induced RAB2B-mediated responses. Knockdown of GARIL5 mRNA, but not other GARI family members, inhibited interferon-stimulated response element (ISRE)-dependent transcriptional activation induced by ectopic expression of STING (Figure 3A). Immunoprecipitation followed by immunoblot analysis showed that GARIL5 interacted with RAB2B, but not RAB2A (Figure 3B). GARIL5 interacted with constitutively active RAB2B, but not constitutively negative RAB2B (Figure 3C). Furthermore, GARIL5 interacted with wild-type RAB2B, regardless of ISD stimulation (Figure 3D). Knockdown of Garil5 mRNA inhibited production of IFN-β and CXCL10 induced by ISD, poly (dI:dC), and cGAMP, but not poly (rI:rC) (Figures 3E, S1, and S3A). Knockdown of Garil5 mRNA also inhibited upregulation of Ifnb1 and Cxcl10 mRNA by ISD, but not poly (rI:rC) (Figures 3F and S3B). These findings indicated that GARIL5 binds to the GTP-bound active form of RAB2B and regulates DNA-induced IFN responses.
Figure 3. GARIL5 regulates DNA-induced innate immune responses.
(A) HEK293 cells treated with the indicated siRNAs were transfected with the STING expression vector, along with pISRE-Luc and pRL-TK. The cell lysates were examined for ISRE-dependent transcriptional activity using a luciferase assay. The results shown are means ± SD (n = 3); *P < 0.01.
(B) HEK293 cells were transfected with FLAG-GARIL5 expression vector, together with the wild-type MYC-RAB2A or wild-type MYC-RAB2B expression vector. Whole cell lysates were immunoprecipitated and immunoblotted with the indicated antibodies.
(C) HEK293 cells were transfected with the FLAG-GARIL5 expression vector, together with the constitutively active MYC-RAB2B or constitutively negative MYC-RAB2B expression vector. Whole cell lysates were immunoprecipitated and immunoblotted with the indicated antibodies.
(D) Primary MEFs stably expressing the indicated proteins were established by retroviral transduction and transfected with ISD. Whole cell lysates were immunoprecipitated and immunoblotted with the indicated antibodies.
(E) Primary MEFs stably expressing the indicated shRNAs were stimulated with the indicated ligands (500 ng/ml or 1000 ng/ml) for 24 h. The levels of IFN-β in the supernatants were measured by ELISA. The results shown are means ± SD (n = 3); *P < 0.01.
(F) Primary MEFs stably expressing the indicated shRNAs were stimulated with the indicated ligands for 6 h. The levels of Ifnb1 mRNA were measured by quantitative RT-PCR. The results shown are means ± SD (n = 3); *P < 0.01.
RAB2B-GARIL5 complex regulates the phosphorylation of IRF3
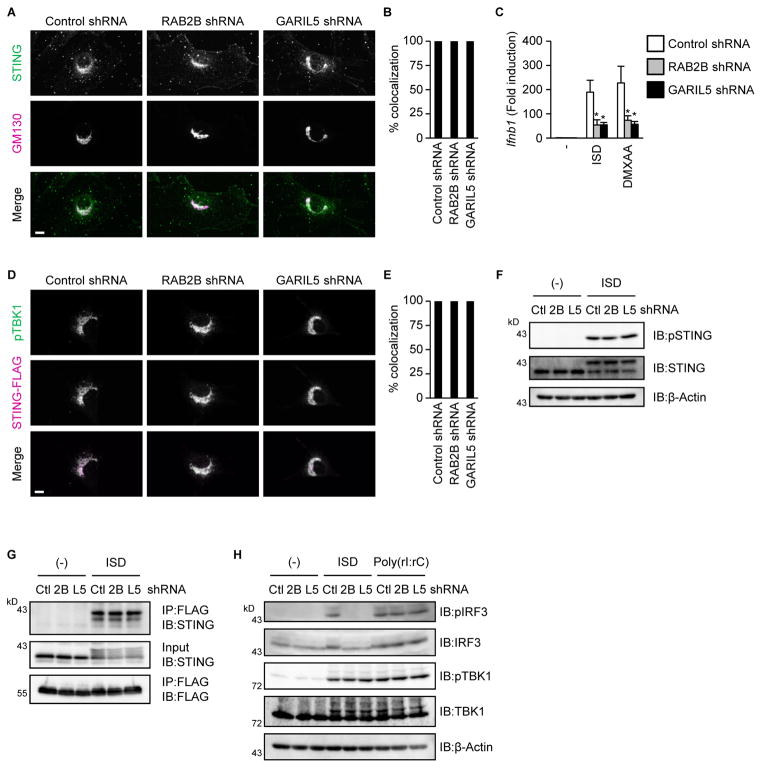
The translocation of STING from the ER to the Golgi apparatus facilitates cGAS-STING signaling (Ishikawa et al., 2009; Saitoh et al., 2009). Therefore, we examined whether RAB2B and GARIL5 regulates STING trafficking. STING moved from the ER to the Golgi apparatus 1 h after stimulation with 5,6-dimethylxanthenone-4-acetic acid (DMXAA), a direct activator of mouse STING. Knockdown of Rab2b or Garil5 mRNA did not inhibit DMXAA-induced STING trafficking (Figures 4A and 4B). However, knockdown of Rab2b or Garil5 mRNA inhibited upregulation of Ifnb1 mRNA by DMXAA (Figure 4C). These findings suggested that RAB2B and GARIL5 do not regulate STING trafficking; rather, they function downstream of STING on the Golgi apparatus.
Figure 4. RAB2B-GARIL5 complex regulates the phosphorylation of IRF3.
(A and B) Primary MEFs stably expressing the indicated shRNAs were established by retroviral transduction and stimulated with DMXAA (100 μg/ml) for 1 h. Colocalization of endogenous STING and GM130 was observed under a fluorescence microscope (A) (scale bars, 10 μm). The frequency of colocalization of endogenous STING with GM130 in DMXAA-stimulated MEFs was determined (B). The graph shows means ± SD (n = 3); *P < 0.01.
(C) Primary MEFs stably expressing the indicated shRNAs were stimulated with ISD (1000 ng/ml) or DMXAA (100 μg/ml) for 1 h. The levels of Ifnb1 mRNA were measured by quantitative RT-PCR. The results shown are means ± SD (n = 3); *P < 0.01.
(D and E) Primary MEFs stably expressing the indicated shRNAs, together with STING-FLAG, were stimulated with DMXAA (100 μg/ml) for 1 h. The localization of phosphorylated TBK1 and STING-FLAG was observed by a fluorescence microscope (D) (scale bars, 10 μm). The frequency of colocalization of phosphorylated TBK1 with STING-FLAG in DMXAA-stimulated MEFs was determined (E). The graph represents means ± SD (n = 3); *P < 0.01.
(F) Primary MEFs stably expressing the indicated shRNAs were stimulated with ISD (1000 ng/ml) for 1 h. Whole cell lysates were subjected to immunoblot analysis using anti-phospho STING, anti-STING, and anti-β-actin.
(G) Primary MEFs stably expressing the indicated shRNAs, together with STING-MYC and FLAG-IRF3-S385A/S386A, were stimulated with ISD (1000 ng/ml) for 1 h. Whole cell lysates were immunoprecipitated and immunoblotted with the indicated antibodies.
(H) Primary MEFs stably expressing the indicated shRNAs were stimulated with ISD (1000 ng/ml) or poly (rI:rC) (1000 ng/ml) for 1 h. Whole cell lysates were subjected to immunoblot analysis using anti-phospho IRF3, anti-IRF3, anti-phospho TBK1, anti-TBK1, and anti-β-actin.
After recruitment to ligand-binding STING, TBK1 phosphorylates itself and STING to facilitate the activation of IRF3 (Liu et al., 2015). Thus, we assessed the involvement of RAB2B and GARIL5 in spatial regulation of phospho-TBK1 and phosphorylation of STING. Knockdown of Rab2b or Garil5 mRNA did not inhibit colocalization of STING with phosphorylated TBK1 (Figures 4D and 4E). Furthermore, knockdown of Rab2b or Garil5 mRNA did not inhibit phosphorylation of STING (Figure 4F). We next examined whether RAB2B and GARIL5 regulate the association of IRF3 with STING and phosphorylation of IRF3 by TBK1. We utilized a well-established experimental system that enabled detection of the complex containing STING and IRF3 (Liu et al., 2015). Consistent with previous data, retrovirally transduced IRF3 containing the substitutions S385A and S386A associated with STING after ISD stimulation (Figure 4G). Knockdown of Rab2b or Garil5 did not inhibit interactions between STING and IRF3-S385A/S386A (Figure 4G). On the other hand, knockdown of Rab2b or Garil5 mRNA inhibited phosphorylation of IRF3 by ISD, but not by double-stranded RNA (dsRNA) (Figure 4H). These findings indicated that RAB2B and GARIL5 regulate phosphorylation of IRF3 by TBK1 through an unknown mechanism.
RAB2B promotes colocalization of GARIL5 with STING
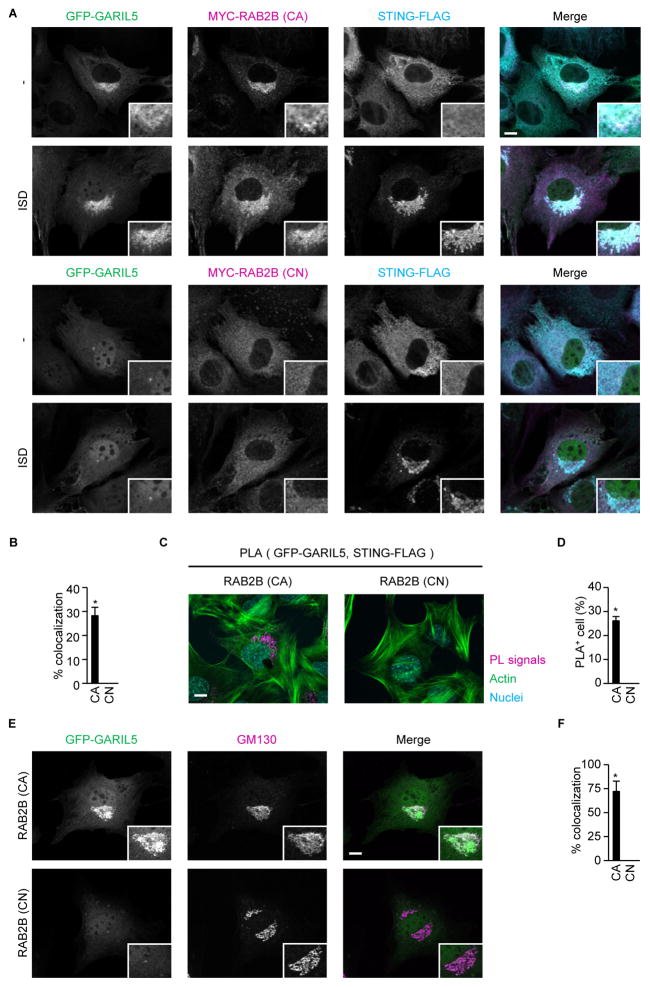
The involvement of GARIL5 in DNA-induced IFN responses prompted us to examine the subcellular localization of GARIL5. RAB2B recruited GARIL5 to STING-positive compartments in a manner that was dependent on RAB2B GTP-binding activity (Figures 5A and 5B). Consistent with this, GTP-binding activity of RAB2B promoted spatial approximation of GFP-GARIL5 and STING-FLAG (Figures 5C and 5D). Furthermore, GARIL5 colocalized with GM130-positive Golgi apparatus, also in a RAB2B GTPase-dependent manner (Figures 5E and 5F). These findings indicated that RAB2B recruits GARIL5 to modulate STING-triggered signaling.
Figure 5. GTP-binding activity of RAB2B promotes spatial approximation of GARIL5 and STING<.
(A and B) Primary MEFs stably expressing shRNA against RAB2B, together with GFP-GARIL5, STING-FLAG, and constitutively active MYC-RAB2B or constitutively negative MYC-RAB2B, were established by retroviral transduction and transfected with ISD. The localization of GFP-GARIL5, STING-FLAG, and MYC-RAB2B was observed under a confocal laser scanning microscope and fluorescence microscope (A) (scale bars, 10 μm). The frequency of colocalization of GFP-GARIL5 with STING-FLAG in ISD-stimulated MEFs was determined (B). The graph represents means ± SD (n = 3); *P < 0.01.
(C and D) Proximity-ligation assay (PLA) of GFP-GARIL5 and STING-FLAG in primary MEFs stably expressing shRNA against RAB2B, together with GFP-GARIL5, STING-FLAG, and constitutively active MYC-RAB2B or constitutively negative MYC-RAB2B, after stimulation with ISD (C) (scale bars, 10 μm). The frequency of cells with proximity ligation-positive signals was determined (D). The graph shows means ± SD (n = 3); *P < 0.01.
(E and F) Primary MEFs stably expressing shRNA against RAB2B, together with GFP-GARIL5 and constitutively active MYC-RAB2B or constitutively negative MYC-RAB2B, were transfected with ISD. The localization of GFP-GARIL5 and GM130 was observed under a confocal laser scanning microscope and fluorescence microscope (E) (scale bars, 10 μm). The frequency of colocalization of GFP-GARIL5 with GM130 in ISD-stimulated MEFs was determined (F). The graph shows means ± SD (n = 3); *P < 0.01.
RAB2B and GARIL5 are involved in antiviral responses to DNA viruses
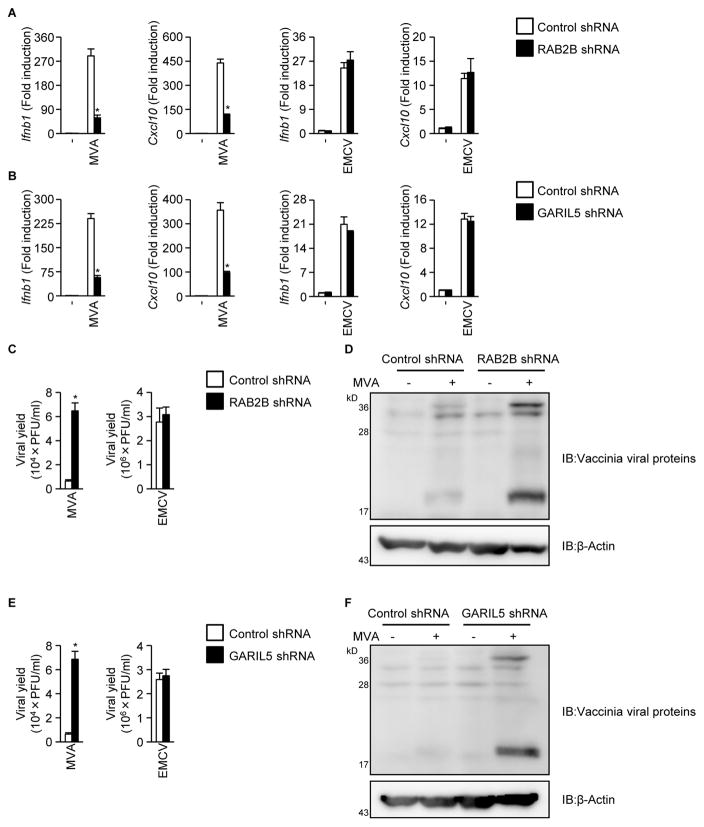
Because the cGAS-STING signaling axis contributes to the establishment of an antiviral state against DNA viruses (Li et al., 2013; Schoggins et al., 2013), we examined the possible involvement of RAB2B and GARIL5 in antiviral response to DNA viruses. Knockdown of Rab2b or Garil5 mRNA inhibited transcription of Ifnb1 and Cxcl10 induced by modified vaccinia virus Ankara strain (MVA) (Figures 6A and 6B). Knockdown of Rab2b or Garil5 mRNA also inhibited production of IFN-β induced by baculovirus, which triggers the cGAS-STING signaling axis (Figure S4; Ono et al., 2014). However, knockdown of Rab2b or Garil5 mRNA did not affect transcription of Ifnb1 and Cxcl10 induced by encephalomyocarditis virus (EMCV), an RNA virus that is recognized by MDA5 (Figures 6A and 6B; Kato et al., 2006). Consistent with this, knockdown of Rab2b or Garil5 mRNA greatly enhanced the replication efficiency of MVA (Figures 6C–6F). However, knockdown of Rab2b or Garil5 mRNA did not affect the replication efficiency of EMCV (Figures 6C and 6E). These findings indicated that the RAB2B-GARIL5 complex promotes IFN-dependent antiviral responses to DNA viruses that are recognized by cGAS.
Figure 6. RAB2B and GARIL5 limit replication of vaccinia virus by promoting the cGAS-STING signaling axis.
(A) Primary MEFs stably expressing the indicated shRNAs were established by retroviral transduction and infected with vaccinia virus (MOI = 1) or EMCV (MOI = 0.02). The levels of Ifnb1 and Cxcl10 mRNA were determined by quantitative RT-PCR. The results shown are means ± SD (n = 3); *P < 0.01.
(B) Primary MEFs stably expressing the indicated shRNAs were infected with vaccinia virus (MOI = 1) or EMCV (MOI = 0.02). The levels of Ifnb1 and Cxcl10 mRNA were determined by quantitative RT-PCR. The results shown are means ± SD (n = 3); *P < 0.01.
(C) Primary MEFs stably expressing the indicated shRNAs were infected with vaccinia virus (MOI = 1) or EMCV (MOI = 0.02). The viral titers in the culture supernatants were determined by TCID50 assay. The results shown are means ± SD (n = 3); *P < 0.01.
(D) Primary MEFs stably expressing the indicated shRNAs were infected with vaccinia virus (MOI = 1). Whole cell lysates were subjected to immunoblot analysis using anti-MVA antibody.
(E) Primary MEFs stably expressing the indicated shRNAs were infected with vaccinia virus (MOI = 1) or EMCV (MOI = 0.02). The viral titers in the culture supernatants were determined by TCID50 assay. The results shown are means ± SD (n = 3); *P < 0.01.
(F) Primary MEFs stably expressing the indicated shRNAs were infected with vaccinia virus (MOI = 1). Whole cell lysates were subjected to immunoblot analysis using anti-MVA antibody.
Discussion
In the present study, we showed that the RAB2B-GARIL5 complex promotes double-stranded DNA (dsDNA)-induced antiviral innate immune responses by regulating phosphorylation of IRF3 by TBK1. However, how the RAB2B-GARIL5 complex facilitates the phosphorylation of IRF3 by TBK1 has not been completely elucidated. The RAB2B-GARIL5 complex colocalizes with STING on Golgi apparatus, and neither RAB2B nor GARIL5 interacts with STING, TBK1, or IRF3 (data not shown). Hence, the RAB2B-GARIL5 complex indirectly promotes dsDNA-induced phosphorylation of IRF3 by TBK1 by recruiting an additional positive regulator of STING on the Golgi apparatus. The molecular functions of RAB2B-binding GARI family members remain unknown at present. Because GARI family members are involved in antiviral innate immune responses and maintenance of Golgi morphology (Aizawa and Fukuda, 2015), their targets and mode of action should be clarified. In future studies, we will address these points to increase our understanding of RAB2B-dependent biological processes that occur on the Golgi apparatus.
The mechanism contributing to STING trafficking is still unclear. Although the RAB2B-GARIL5 complex promotes STING-dependent IFN response, it is not involved in STING trafficking. RAB1A, RAB1B and RAB2A are known to be involved in ER-to-Golgi trafficking. However, knockdown of RAB1A, RAB1B, or RAB2Adoes not affect the production of IFN-β induced by dsDNA (Figure 1A). Hence, these RabGTPases might compensate for each other in driving ER-to-Golgi trafficking of STING. Alternatively, an unconventional trafficking system that does not depend on these Rab GTPases might drive STING trafficking. It would be important to clarify the precise mechanism of STING trafficking for a better understanding of cytosolic DNA-induced innate immune response.
It has become clear that organelles play pivotal roles in signal transduction from nucleic acid-sensing PRRs. As shown in the present study, cGAS and its downstream regulator STING utilize the Golgi apparatus for signal transduction. RIG-I-like receptors detect cytosolic RNA and mediate this signal with an adaptor protein, IPS-1 (also known as MAVS/VISA/CARDIF), which is expressed on mitochondria (Kawai et al., 2005; Seth et al., 2005; Xu et al, 2005). Importantly, Rab family members regulate multiple events that occur on organelles. Although we showed that RAB2B regulates cytosolic DNA-induced innate immune responses, Rab family members capable of regulating cytosolic RNA-induced innate immune responses have not been identified. Therefore, the role of Rab GTPases in signal transduction from these nucleic acid-sensing PRRs must be clarified in future studies.
Pathogenic microorganisms have evolved to counteract host defenses. In particular, many viruses can suppress expression of type I IFN and IFN-stimulated genes. Influenza A virus NS1 interacts with RIG-I and disrupts RIG-I-triggered IFN responses (Pichlmair et al., 2006; Mibayashi et al., 2007). Hepatitis C virus NS3-4A induces degradation of IPS-1 to shut down the RIG-I-IPS-1 signaling axis (Li et al., 2005; Meylan et al., 2005). Human herpesvirus-8 interferon regulatory factor (IRF) competes with IRF3 to bind to the promoters of type I IFN and IFN-stimulated genes (Burýsek et al., 1999). Hence, disruption of the cGAS-STING signaling axis by DNA viruses has been speculated. Because vaccinia virus encodes proteins that suppress the expression of type I IFN and IFN-stimulated genes (Smith et al., 2001), it would be interesting to assess whether vaccinia virus proteins target RAB2B and GARIL5 to disrupt the cGAS-STING signaling axis.
Experimental Procedures
Reagents
ISD, cGAMP, high molecular weight poly (rI:rC), and low molecular weight poly (rI:rC) were purchased from InvivoGen. Poly (dI:dC) was purchased from Sigma. The enzyme-linked immunosorbent assay (ELISA) kit used to detect mouse IFN-β was purchased from PBL Biomedical Laboratories. The ELISA kit to identify mouse CXCL10 was purchased from R&D Systems. The following commercial antibodies were used: anti-IRF3 (#4302, Cell Signaling), anti-phospho-IRF3 (Ser396) (#4947, Cell Signaling), anti-STING (#3337, Cell Signaling; 19851-1-AP, Proteintech), anti-phospho-STING (Ser366) (#85735, Cell Signaling), anti-TBK1 (ab40676, Abcam), anti-phospho-TBK1 (#5483, Cell Signaling), anti-GM130 (610822, BD Biosciences), anti-FLAG (F1804, Sigma), horse radish peroxidase (HRP)-labeled anti-FLAG (A8592, Sigma), anti-MYC (A190-103A, Bethyl Laboratories), HRP-labeled anti-MYC (#2040, Cell Signaling), HRP-labeled β-actin (sc-1615, Santa Cruz), HRP-labeled anti-rabbit IgG (GE Healthcare), Alexa Fluor 488-conjugated anti-mouse IgG (A11029, Life Technologies), Alexa Fluor 488-conjugated anti-chicken IgG (A11039, Life Technologies), Alexa Fluor 568-conjugated anti-mouse IgG (A11031, Life Technologies), Alexa Fluor 568-conjugated anti-chicken IgG (A11041, Life Technologies), and Alexa Fluor 647-conjugated anti-mouse IgG (A21236, Life Technologies). Can Get Signal immunostain Solution A was purchased from TOYOBO. An siRNA library targeting mouse Rab GTPase family members was synthesized at Nippon EGT (Matsui and Fukuda, 2013). Unless otherwise noted, reagents were purchased from Nacalai Tesque.
Cells and viruses
Primary MEFs were prepared from pregnant female mice on embryonic day 13.5 as described previously (Kato et al., 2006). Immortalized wild-type MEFs and HEK293 cells were previously characterized (Saitoh et al., 2009). Plat-E cells (Morita et al., 2000) were kindly donated by Dr. T. Kitamura (The University of Tokyo). Cells were cultured in Dulbecco’s modified Eagle’s medium supplemented with 10% fetal calf serum (Life Technologies) in a 5% CO2 incubator. Modified vaccinia virus Ankara strain was purchased from the American Type Culture Collection. EMCV and baculovirus has been described previously (Kato et al., 2006; Ono et al., 2014). The viral titers were determined with 50% tissue culture infectious dose (TCID50) assays as described previously (Saitoh et al., 2009).
Plasmids
The reporter plasmids pISRE-Luc and pRL-TK were purchased from Stratagene and Promega, respectively. The retroviral cDNA expression plasmids pMRX-ires-puro and pMRX-ires-bsr were kindly donated by Dr. S. Yamaoka (Tokyo Medical and Dental University), and the previously characterized plasmid pcDNA3 STING-MYC was used (Saitoh et al., 2009). Complementary DNA encoding RAB2A was inserted into pcDNA3-MYC-MCS, generating pcDNA-MYC-RAB2A. Complementary DNA encoding RAB2B was inserted into pcDNA3-MYC-MCS and pMRX-MYC-MCS-ires-puro, generating pcDNA-MYC-RAB2B and pMRX-MYC-RAB2B-ires-puro, respectively. Plasmids derived from pEGFP-C1 encoding constitutively active and constitutively negative mutants of RAB2B were described previously (Fukuda et al., 2008). Complementary DNA encoding GARIL5 was inserted into pcDNA3-FLAG-MCS and pMRX-GFP-MCS-ires-puro, generating pcDNA3-FLAG-GARIL5 and pMRX-GFP-GARIL5-ires-puro, respectively. FLAG-tagged STING was inserted into pMRX-ires-bsr, generating pMRX-STING-FLAG-ires-bsr. The retroviral shRNA expression plasmid pSuper-retro-puro was characterized in an earlier report (Saitoh et al., 2006). Complementary DNA sequences inserted immediately downstream of the H1 promoter of pSuper-retro-puro were as follows (only the sense strand sequence is shown): specific to RAB2B, 5′-GTCATGTCTCCTCCTTCAG-3′ and 5′-GTGATTTCATTGCGTGTAT-3′; specific to GARIL5, 5′-GACTCAGACAAGATCCTTC-3′ and 5′-GTAAAGTCACAAGCTCTAG-3′; an unrelated control was used (Misawa et al., 2013).
Quantitative real-time (RT)-PCR
Total RNA was isolated using an RNA Microprep kit according to the manufacturer’s instructions (Zymo Research). Reverse transcription was performed using ReverTra Ace in accordance with the manufacturer’s instructions (Toyobo). For quantitative PCR, cDNA fragments were amplified with Real-Time PCR Master Mix in accordance with the manufacturer’s instructions (Toyobo). Fluorescence from the TaqMan probe was detected using a 7500 Real-Time PCR System (Applied Biosystems), and the expression levels of Ifnb, Cxcl10, Rab2b, and Garil5 mRNA were normalized to that of Actb mRNA.
ELISA
The levels of IFN-β and CXCL10 in culture supernatants were measured using an ELISA in accordance with the manufacturer’s instructions.
Immunocytochemistry
Cells cultured on coverslips were fixed with 3% paraformaldehyde and then processed for immunocytochemistry as previously described (Saitoh et al., 2012). Samples were examined with an LSM 780 confocal laser-scanning microscope (Carl Zeiss) and DMI6000B fluorescence microscope (Leica Microsystems).
Immunoblotting
Cells were washed with ice-cold phosphate-buffered saline and then lysed in lysis buffer (1% Nonidet P-40, 50 mM Tris-HCl (pH 7.4), 150 mM NaCl) supplemented with a complete protease inhibitor cocktail tablet (Roche) and a phosphatase inhibitor cocktail tablet (Roche). Cell lysates were incubated for 15 min at 4°C, then centrifuged at 14,000 × g for 15 min at 4°C. The supernatants were boiled in 2-mercaptoethanol-containing sample buffer, subjected to sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) and transferred to polyvinylidene difluoride membrane (Millipore). The membranes were then blocked with Tris-buffered saline containing 20 mM Tris-HCl (pH 7.4), 135 mM NaCl, 0.05% Tween 20, and 5% skim milk and incubated with primary antibody at room temperature for 1 h or overnight at 4°C and then with HRP-conjugated secondary antibody at room temperature for 1 h. The immune complexes and cell lysates were visualized using a Luminata Forte Western HRP Substrate (Millipore), ImageQuant LAS-4000 (GE Healthcare), and FUSION-Solo 7S (Vilber-Lourmat).
Immunoprecipitation
HEK293 cells seeded on 100-mm dishes were transiently transfected with a total of 10 μg of various plasmids. At 24 h after transfection, the cells were lysed in lysis buffer and centrifuged at 14,000 × g for 15 min at 4°C. The supernatants were incubated with antibody for 1 h at 4°C, and Protein G-Sepharose 4B Fast Flow beads (GE Healthcare) were added. After 1 h of incubation at 4°C, the beads were washed four times with lysis buffer. The immunoprecipitates were boiled in sample buffer and subjected to SDS-PAGE. The immunoprecipitation assay for STING-IRF3 interactions was performed as described previously (Liu et al., 2015).
Reporter assay
HEK293 cells plated on 24-well plates were transfected with the indicated siRNAs, and then transiently transfected with 90 ng of pISRE-Luc, 10 ng of pRL-TK, and 400 ng of expression plasmid by Lipofectamine 2000 transfection reagent (Life Technologies). At 24 h after transfection, luciferase activities in total cell lysates were measured using a Dual-Luciferase Reporter Assay System (Promega).
In situ proximity ligation assay
In situ proximity ligation assays were performed using a Duolink In Situ PLA Kit (Olink Bioscience) according to the manufacturer’s instructions. In brief, MEFs expressing the indicated genes were transfected with ISD, fixed, and then permeabilized with digitonin. The cells were incubated with primary antibodies in antibody buffer for 1 h at 37°C and then washed in wash buffer A. Next, the cells were incubated with PLA Probes from Duolink anti-mouse PLA MINUS and anti-rabbit PLA PLUS in antibody buffer for 1 h at 37°C and then washed in wash buffer A. Thereafter, the ligation reagent was added to the cells and incubated for 30 min at 37°C, followed by washing in wash buffer A. For amplification, the cells were incubated with amplification-polymerase solution for 100 min at 37°C and washed in wash buffer B. After the final wash in 0.01× wash buffer B for 1 min, the cells were mounted with phalloidin and Hoechst 33342 to stain actin filaments and nuclei. Samples were examined with an LSM 780 confocal laser-scanning microscope and a DMI6000B fluorescence microscope.
Statistical analysis
Student’s t test or ANOVA plus post hoc Tukey testing was used to determine statistical significance. A P-value of < 0.05 was considered significant.
Supplementary Material
Highlights.
RAB2B recruits its effector protein GARIL5 into the Golgi apparatus.
RAB2B and GARIL5 regulate cytosolic DNA-induced IFN response.
RAB2B and GARIL5 do not regulate cytosolic RNA-induced IFN response.
RAB2B and GARIL5 limit replication of DNA virus.
Acknowledgments
We thank T. Kitamura (The University of Tokyo) for Plat-E packaging cells and S. Yamaoka (Tokyo Medical and Dental University) for the plasmids pMRX-ires-puro and pMRX-ires-bsr. We also thank the members of the Division of Inflammation Biology for their assistance. This work was partly supported by the Japan Society for the Promotion of Science KAKENHI (grant numbers 26713005 and 23659231 to T.S.; 26111501 and 16H01189 to M.F.); the Ministry of Education, Culture, Sports, Science, and Technology KAKENHI (grant number 17H06009 and 17H06415 to T.S.); a research grant from the Takeda Science Foundation (to T.S.); a research grant from the Japan Foundation for Pediatric Research (to T.S.); and a National Institutes of Health (grant PO1-AI070167 to S.A.).
Footnotes
Author Contributions
M.T. performed the experiments and analyzed the data. T.K. and T.M. provided assistance in carrying out experiments. M.F, N.O, T.O., and Y.M contributed material support. S.A. supervised the project. M.T. and T.S. designed the experiments and wrote the manuscript.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Ablasser A, Goldeck M, Cavlar T, Deimling T, Witte G, Röhl I, Hopfner KP, Ludwig J, Hornung V. cGAS produces a 2′-5′-linked cyclic dinucleotide second messenger that activates STING. Nature. 2013;498:380–384. doi: 10.1038/nature12306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aizawa M, Fukuda M. Small GTPase Rab2B and Its Specific Binding Protein Golgi-associated Rab2B Interactor-like 4 (GARI-L4) Regulate Golgi Morphology. Journal of Biological Chemistry. 2015;290:22250–22261. doi: 10.1074/jbc.M115.669242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beutler B. Microbe sensing, positive feedback loops, and the pathogenesis of inflammatory diseases. Immunological Reviews. 2009;227:248–263. doi: 10.1111/j.1600-065X.2008.00733.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burýsek L, Yeow WS, Lubyová B, Kellum M, Schafer SL, Huang YQ, Pitha PM. Functional analysis of human herpesvirus 8-encoded viral interferon regulatory factor 1 and its association with cellular interferon regulatory factors and p300. Journal of Virology. 1999;73:7334–7342. doi: 10.1128/jvi.73.9.7334-7342.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai X, Chiu YH, Chen ZJ. The cGAS-cGAMP-STING Pathway of Cytosolic DNA Sensing and Signaling. Molecular Cell. 2014;54:289–296. doi: 10.1016/j.molcel.2014.03.040. [DOI] [PubMed] [Google Scholar]
- Charrel-Dennis M, Latz E, Halmen KA, Trieu-Cuot P, Fitzgerald KA, Kasper DL, Golenbock DT. TLR-Independent Type I Interferon Induction in Response to an Extracellular Bacterial Pathogen via Intracellular Recognition of Its DNA. Cell Host & Microbe. 2008;4:543–554. doi: 10.1016/j.chom.2008.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diner ECAJ, Burdette DL, Wilson SC, Monroe KM, Kellenberger CA, Hyodo M, Hayakawa Y, Hammond MC, Vance RE. The Innate Immune DNA Sensor cGAS Produces a Noncanonical Cyclic Dinucleotide that Activates Human STING. Cell Reports. 2013;3:1355–1361. doi: 10.1016/j.celrep.2013.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukuda M, Kanno E, Ishibashi K, Itoh T. Large Scale Screening for Novel Rab Effectors Reveals Unexpected Broad Rab Binding Specificity. Molecular & Cellular Proteomics. 2008;7:1031–1042. doi: 10.1074/mcp.M700569-MCP200. [DOI] [PubMed] [Google Scholar]
- Gao P, Ascano M, Wu Y, Barchet W, Gaffney BL, Zillinger T, Serganov AA, Liu Y, Jones RA, Hartmann G, et al. Cyclic [G(2′,5′)pA(3′,5′)p] Is the Metazoan Second Messenger Produced by DNA-Activated Cyclic GMP-AMP Synthase. Cell. 2013;153:1094–1107. doi: 10.1016/j.cell.2013.04.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hornung V, Ablasser A, Charrel-Dennis M, Bauernfeind F, Horvath G, Caffrey DR, Latz E, Fitzgerald KA. AIM2 recognizes cytosolic dsDNA and forms a caspase-1-activating inflammasome with ASC. Nature. 2009;458:514–518. doi: 10.1038/nature07725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Husebye H, Aune MH, Stenvik J, Samstad E, Skjeldal F, Halaas Ø, Nilsen NJ, Stenmark H, Latz E, Lien E, et al. The Rab11a GTPase Controls Toll-like Receptor 4-Induced Activation of Interferon Regulatory Factor-3 on Phagosomes. Immunity. 2010;33:583–596. doi: 10.1016/j.immuni.2010.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishii KJ, Coban C, Kato H, Takahashi K, Torii Y, Takeshita F, Ludwig H, Sutter G, Suzuki K, Hemmi H, et al. A Toll-like receptor–independent antiviral response induced by double-stranded B-form DNA. Nature Immunology. 2006;7:40–48. doi: 10.1038/ni1282. [DOI] [PubMed] [Google Scholar]
- Ishii KJ, Kawagoe T, Koyama S, Matsui K, Kumar H, Kawai T, Uematsu S, Takeuchi O, Takeshita F, Coban C, et al. TANK-binding kinase-1 delineates innate and adaptive immune responses to DNA vaccines. Nature. 2008;451:725–729. doi: 10.1038/nature06537. [DOI] [PubMed] [Google Scholar]
- Ishikawa H, Barber GN. STING is an endoplasmic reticulum adaptor that facilitates innate immune signalling. Nature. 2008;455:674–678. doi: 10.1038/nature07317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishikawa H, Ma Z, Barber GN. STING regulates intracellular DNA-mediated, type I interferon-dependent innate immunity. Nature. 2009;461:788–792. doi: 10.1038/nature08476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato H, Takeuchi O, Sato S, Yoneyama M, Yamamoto M, Matsui K, Uematsu S, Jung A, Kawai T, Ishii KJ, et al. Differential roles of MDA5 and RIG-I helicases in the recognition of RNA viruses. Nature. 2006;441:101–105. doi: 10.1038/nature04734. [DOI] [PubMed] [Google Scholar]
- Kawai T, Akira S. The roles of TLRs, RLRs and NLRs in pathogen recognition. International Immunology. 2009;21:317–337. doi: 10.1093/intimm/dxp017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawai T, Takahashi K, Sato S, Coban C, Kumar H, Kato H, Ishii KJ, Takeuchi O, Akira S. IPS-1, an adaptor triggering RIG-I- and Mda5-mediated type I interferon induction. Nature Immunology. 2005;6:981–988. doi: 10.1038/ni1243. [DOI] [PubMed] [Google Scholar]
- Li K, Foy E, Ferreon JC, Nakamura M, Ferreon ACM, Ikeda M, Ray SC, Gale M, Lemon SM. Immune evasion by hepatitis C virus NS3/4A protease-mediated cleavage of the Toll-like receptor 3 adaptor protein TRIF. Proceedings of the National Academy of Sciences. 2005;102:2992–2997. doi: 10.1073/pnas.0408824102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li XD, Wu J, Gao D, Wang H, Sun L, Chen ZJ. Pivotal Roles of cGAS-cGAMP Signaling in Antiviral Defense and Immune Adjuvant Effects. Science. 2013;341:1390–1394. doi: 10.1126/science.1244040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linford A, Yoshimura SI, Bastos RN, Langemeyer L, Gerondopoulos A, Rigden DJ, Barr FA. Rab14 and Its Exchange Factor FAM116 Link Endocytic Recycling and Adherens Junction Stability in Migrating Cells. Developmental Cell. 2012;22:952–966. doi: 10.1016/j.devcel.2012.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S, Cai X, Wu J, Cong Q, Chen X, Li T, Du F, Ren J, Wu Y-T, Grishin NV, Chen ZJ. Phosphorylation of innate immune adaptor proteins MAVS, STING, and TRIF induces IRF3 activation. Science. 2015;347:aaa2630. doi: 10.1126/science.aaa2630. [DOI] [PubMed] [Google Scholar]
- Matsui T, Fukuda M. Rab12 regulates mTORC1 activity and autophagy through controlling the degradation of amino-acid transporter PAT4. EMBO reports. 2013;14:450–457. doi: 10.1038/embor.2013.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meylan E, Curran J, Hofmann K, Moradpour D, Binder M, Bartenschlager R, Tschopp J. Cardif is an adaptor protein in the RIG-I antiviral pathway and is targeted by hepatitis C virus. Nature. 2005;437:1167–1172. doi: 10.1038/nature04193. [DOI] [PubMed] [Google Scholar]
- Mibayashi M, Martinez-Sobrido L, Loo YM, Cardenas WB, Gale M, Garcia-Sastre A. Inhibition of Retinoic Acid-Inducible Gene I-Mediated Induction of Beta Interferon by the NS1 Protein of Influenza A Virus. Journal of Virology. 2007;81:514–524. doi: 10.1128/JVI.01265-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misawa T, Takahama M, Kozaki T, Lee H, Zou J, Saitoh T, Akira S. Microtubule-driven spatial arrangement of mitochondria promotes activation of the NLRP3 inflammasome. Nature Immunology. 2013;14:454–460. doi: 10.1038/ni.2550. [DOI] [PubMed] [Google Scholar]
- Morita S, Kojima T, Kitamura T. Plat-E: an efficient and stable system for transient packaging of retroviruses. Gene Therapy. 2000;7:1063–1066. doi: 10.1038/sj.gt.3301206. [DOI] [PubMed] [Google Scholar]
- Ono C, Ninomiya A, Yamamoto S, Abe T, Wen X, Fukuhara T, Sasai M, Yamamoto M, Saitoh T, Satoh T, Kawai T, Ishii KJ, Akira S, Okamoto T, Matsuura Y. Innate immune response induced by baculovirus attenuates transgene expression in mammalian cells. J Virol. 2014;88:2157–2167. doi: 10.1128/JVI.03055-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palm NW, Medzhitov R. Pattern recognition receptors and control of adaptive immunity. Immunological Reviews. 2009;227:221–233. doi: 10.1111/j.1600-065X.2008.00731.x. [DOI] [PubMed] [Google Scholar]
- Pichlmair A, Schulz O, Tan CP, Naslund TI, Liljestrom P, Weber F, Sousa CRE. RIG-I-Mediated Antiviral Responses to Single-Stranded RNA Bearing 5′-Phosphates. Science. 2006;314:997–1001. doi: 10.1126/science.1132998. [DOI] [PubMed] [Google Scholar]
- Saitoh T, Fujita N, Hayashi T, Takahara K, Satoh T, Lee H, Matsunaga K, Kageyama S, Omori H, Noda T, et al. Atg9a controls dsDNA-driven dynamic translocation of STING and the innate immune response. Proceedings of the National Academy of Sciences. 2009;106:20842–20846. doi: 10.1073/pnas.0911267106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saitoh T, Komano J, Saitoh Y, Misawa T, Takahama M, Kozaki T, Uehata T, Iwasaki H, Omori H, Yamaoka S, et al. Neutrophil Extracellular Traps Mediate a Host Defense Response to Human Immunodeficiency Virus-1. Cell Host & Microbe. 2012;12:109–116. doi: 10.1016/j.chom.2012.05.015. [DOI] [PubMed] [Google Scholar]
- Saitoh T, Tun-Kyi A, Ryo A, Yamamoto M, Finn G, Fujita T, Akira S, Yamamoto N, Lu KP, Yamaoka S. Negative regulation of interferon-regulatory factor 3–dependent innate antiviral response by the prolyl isomerase Pin1. Nature Immunology. 2006;7:598–605. doi: 10.1038/ni1347. [DOI] [PubMed] [Google Scholar]
- Schoggins JW, Macduff DA, Imanaka N, Gainey MD, Shrestha B, Eitson JL, Mar KB, Richardson RB, Ratushny AV, Litvak V, et al. Pan-viral specificity of IFN-induced genes reveals new roles for cGAS in innate immunity. Nature. 2013;505:691–695. doi: 10.1038/nature12862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroder K, Muruve DA, Tschopp J. Innate Immunity: Cytoplasmic DNA Sensing by the AIM2 Inflammasome. Current Biology. 2009:19. doi: 10.1016/j.cub.2009.02.011. [DOI] [PubMed] [Google Scholar]
- Schroder K, Tschopp J. The Inflammasomes. Cell. 2010;140:821–832. doi: 10.1016/j.cell.2010.01.040. [DOI] [PubMed] [Google Scholar]
- Seth RB, Sun L, Ea CK, Chen ZJ. Identification and Characterization of MAVS, a Mitochondrial Antiviral Signaling Protein that Activates NF-κB and IRF3. Cell. 2005;122:669–682. doi: 10.1016/j.cell.2005.08.012. [DOI] [PubMed] [Google Scholar]
- Smith EJ, Marie I, Prakash A, Garcia-Sastre A, Levy DE. IRF3 and IRF7 Phosphorylation in Virus-infected Cells Does Not Require Double-stranded RNA-dependent Protein Kinase R or I B Kinase but Is Blocked by Vaccinia Virus E3L Protein. Journal of Biological Chemistry. 2001;276:8951–8957. doi: 10.1074/jbc.M008717200. [DOI] [PubMed] [Google Scholar]
- Stenmark H. Rab GTPases as coordinators of vesicle traffic. Nature Reviews Molecular Cell Biology. 2009;10:513–525. doi: 10.1038/nrm2728. [DOI] [PubMed] [Google Scholar]
- Stetson DB, Medzhitov R. Recognition of Cytosolic DNA Activates an IRF3-Dependent Innate Immune Response. Immunity. 2006;24:93–103. doi: 10.1016/j.immuni.2005.12.003. [DOI] [PubMed] [Google Scholar]
- Sun L, Wu J, Du F, Chen X, Chen ZJ. Cyclic GMP-AMP Synthase Is a Cytosolic DNA Sensor That Activates the Type I Interferon Pathway. Science. 2013;339:786–791. doi: 10.1126/science.1232458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Chen T, Han C, He D, Liu H, An H, Cai Z, Cao X. Lysosome-associated small Rab GTPase Rab7b negatively regulates TLR4 signaling in macrophages by promoting lysosomal degradation of TLR4. Blood. 2007;110:962–971. doi: 10.1182/blood-2007-01-066027. [DOI] [PubMed] [Google Scholar]
- Wu J, Sun L, Chen X, Du F, Shi H, Chen C, Chen ZJ. Cyclic GMP-AMP Is an Endogenous Second Messenger in Innate Immune Signaling by Cytosolic DNA. Science. 2013;339:826–830. doi: 10.1126/science.1229963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu LG, Wang YY, Han KJ, Li LY, Zhai Z, Shu HB. VISA Is an Adapter Protein Required for Virus-Triggered IFN-β Signaling. Molecular Cell. 2005;19:727–740. doi: 10.1016/j.molcel.2005.08.014. [DOI] [PubMed] [Google Scholar]
- Yoshimura SI, Egerer J, Fuchs E, Haas AK, Barr FA. Functional dissection of Rab GTPases involved in primary cilium formation. The Journal of Cell Biology. 2007;178:363–369. doi: 10.1083/jcb.200703047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zografou S, Basagiannis D, Papafotika A, Shirakawa R, Horiuchi H, Auerbach D, Fukuda M, Christoforidis S. A complete Rab screening reveals novel insights in Weibel-Palade body exocytosis. Journal of Cell Science. 2012;125:4780–4790. doi: 10.1242/jcs.104174. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.