Abstract
The heat shock protein, Hsp60, is one of the most abundant proteins in Helicobacter pylori. Given its sequence homology to the Escherichia coli Hsp60 or GroEL, Hsp60 from H. pylori would be expected to function as a molecular chaperone in this organism. H. pylori is an organism that grows on the gastric epithelium, where the pH can fluctuate between neutral and 4.5 and the intracellular pH can be as low as 5.0. This study was performed to test the ability of Hsp60 from H. pylori to function as a molecular chaperone under mildly acidic conditions. We report here that Hsp60 could suppress the acid-induced aggregation of alcohol dehydrogenase (ADH) in the 7.0–5.0 pH range. Hsp60 was found to undergo a conformational change within this pH range. It was also found that exposure of hydrophobic surfaces of Hsp60 is significant and that their exposure is increased under acidic conditions. Although, alcohol dehydrogenase does not contain exposed hydrophobic surfaces, we found that their exposure is triggered at low pH. Our results demonstrate that Hsp60 from H. pylori can function as a molecular chaperone under acidic conditions and that the interaction between Hsp60 and other proteins may be mediated by hydrophobic interactions.
Keywords: Hsp60, Molecular chaperone, Protein aggregation, Acid stress, Conformational changes
Highlights
-
•
Hsp60 of H. pylori could suppress the acid-induced aggregation of alcohol dehydrogenase (ADH) at low pH.
-
•
Exposure of hydrophobic surfaces of Hsp60 and ADH is triggered under acidic conditions.
-
•
The interaction between Hsp60 of H. pylori and other proteins may be mediated by hydrophobic interactions.
1. Introduction
Helicobacter pylori, is a Gram-negative, microaerophilic bacterium present in the stomach of approximately half of the human population [1]. Chronic infection by this microorganism can, in certain individuals, give rise to gastric and duodenal ulcers, gastric adenocarcinoma, and mucosa-associated lymphoid tissue lymphoma [2], [3]. H. pylori, survives transient exposure to extreme acid prior to adherence and growth on the gastric epithelium where the pH can fluctuate between neutral and 4.5 and the intracellular pH can be as low as 5.0 [4], [5]. Urease is the key enzyme of H. pylori responsible for its survival in the stomach [4]. This enzyme hydrolyzes urea into ammonia, neutralizing the gastric acid [6]. Under neutral and moderately acidic conditions, urease and the heat shock protein, Hsp60, are the most abundant proteins in H. pylori.[4], [7]. Given its sequence homology to the E. coli Hsp60 or GroEL [8], Hsp60 from H. pylori would be expected to function as a molecular chaperone in this organism. Their amino acid sequence identity and similarity are 61.5% and 78.6%, respectively (see Appendix A). The molecular chaperones are a class of proteins that have been shown to facilitate the folding of nascent polypeptides, activation of other proteins, and protection of native proteins against the effects of heat, and oxidative and acid stress [9], [10], [11], [12], [13], [14]. The co-expression of H. pylori urease, Hsp60, and Hsp10 in E. coli was shown to substantially increase the activity of urease [15], which suggested that urease activity was protected by the heat shock proteins. The role of Hsp60 as an extracellular antigen of H. pylori has been extensively reported [16], [17] and the H. pylori Hsp10 protein has been shown to contain a unique His- and Cys-rich domain at the C terminus and to function as a specialized nickel chaperone through its unique C-terminal extension [18], [19]. Interestingly, Hsp60 from H. pylori has been reported to exist as a mixture of dimers and tetramers [20] which is in contrast to that of GroEL, a tetradecamer of 14 identical subunits of 58 kDa arranged in a double-stacked ring structure [9], [21]. The x-ray structure of Hsp60 of H. pylori has not been determined. Thus, a model of the monomeric structure of this protein was generated with the RaptorX software (see Appendix B). The modeled structure of Hsp60 of H. pylori displayed a striking similarity with the structure of GroEL of E. coli. Moreover, analysis of the hydrophobicity of the amino acid sequences of Hsp60 of H. pylori and GroEL of E. coli gave similar hydrophobic profiles (see Appendix C). No reports, however, have been made on the potential chaperone activity of Hsp60 from H. pylori. It is well known that protein denaturation can be induced under acidic conditions [22]. During moderately acidic conditions, such as those experienced by H. pylori, partially unfolded proteins may exist [23] and molecular chaperones; such as Hsp60 and Hsp10 may stabilize them against acid-induced aggregation.
This study was performed to determine if Hsp60 could act as a molecular chaperone under moderately acidic conditions. The model protein alcohol dehydrogenase (ADH) was used as a substrate protein for Hsp60 given its lower stability in the absence of its cofactor, Zn2+[24], [25]. Here, we report that Hsp60 prevented the acid-induced aggregation of ADH in the 7.0–5.0 pH range where the enzyme alone was observed to aggregate. The aggregation of Hsp60 was not detected under these conditions. It is also reported here that Hsp60 undergoes a significant conformational change within this pH range. It was also found that exposure of hydrophobic surfaces of Hsp60 is significant at neutral pH and that their exposure is further increased by moderately acidic conditions. Although, ADH does not contain exposed hydrophobic surfaces at neutral pH, it was found that their exposure is triggered by moderately acidic conditions. Thus, it is demonstrated here, that Hsp60 from H. pylori can function as a molecular chaperone under moderately acidic conditions. Our results also show that the interaction between Hsp60 from H. pylori and other proteins, under moderately acidic conditions, may be mediated by hydrophobic interactions.
2. Materials and methods
2.1. Reagents and proteins
All the reagents used here were of analytical grade. Isopropyl-β-D-thiogalactopyranoside (IPTG) was purchased from Gold Biotechnology and benzonase nuclease from Novagen. The protease inhibitor cocktail and horse liver ADH were from Sigma. The H. pylori Hsp60 gene was synthesized by GenScript (New Jersey) and inserted into the expression vector pET-22b(+). The resulting plasmid pET-Hsp60 was transformed into competent E. coli BL21 (DE3) cells using ampicillin resistance for selection. The Hsp60 site-directed mutant Y359W, in which Tyr was replaced with Trp at the 359 amino acid residue position was also made by GenScript. The expressed proteins were purified using His GraviTrap chromatography (GE Healthcare) followed by dialysis. Purification of Hsp60 was confirmed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) [26] and its concentration determined by using the Bradford assay (BioRad).
2.2. Light scattering of acid-induced aggregation of ADH
The aggregation of ADH was monitored by using light scattering. Measurement of the light scattering of ADH alone or in the presence of Hsp60 was made with a Fluoromax-3 spectrophotometer using an excitation wavelength of 350 nm. The fluorescence emission was recorded at the same wavelength. The appropriate blanks were subtracted. Samples were incubated in either 50 mM Tris-HCl (pH 7.5-6.5) or 50 mM sodium citrate (pH 6.0-5.0).
2.3. Measurement of tryptophan fluorescence
Hsp60 Y359W was incubated in 50 mM Tris-HCl (pH 7.5) or 50 mM sodium citrate (pH 5.5). Trp fluorescence emission was recorded at 25 °C between 330 and 400 nm with a Fluoromax-3 spectrofluorometer using an excitation wavelength of 295 nm.
2.4. Measurement of protein surface hydrophobicity
Changes on surface hydrophobicity of Hsp60 or ADH after incubation in 50 mM Tris-HCl (pH 7.5) or 50 mM sodium citrate (pH 5.5) were determined by measuring the fluorescence of bisANS at 25 °C between 410 and 600 nm with a Fluoromax-3 spectrofluorometer using an excitation wavelength of 394 nm.
3. Results and discussion
3.1. Aggregation of ADH at low pH
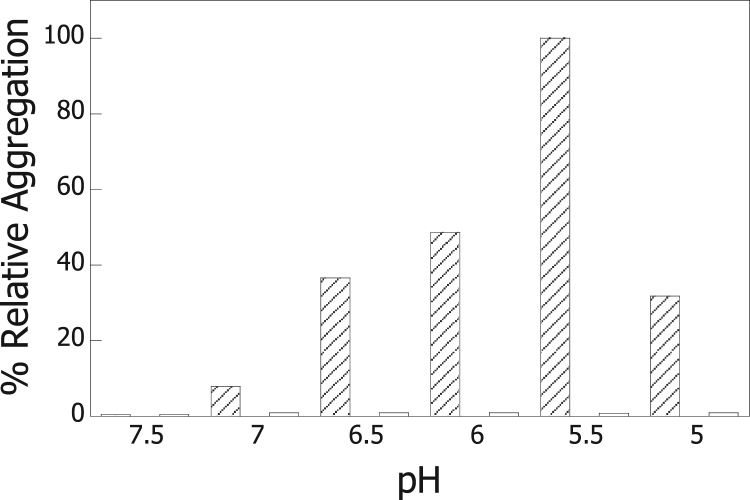
Given the sequence homology of H. pylori Hsp60 to the E.coli Hsp60 protein or GroEL [8], [27], Hsp60 from H. pylori would be expected to function as a molecular chaperone. Here, we have tested the ability of Hsp60 from H. pylori to prevent the acid-induced aggregation of a native protein. The model protein alcohol dehydrogenase (ADH) from horse liver is a dimer composed of two equal subunits of 40 kDa each that has been reported to be stabilized by its cofactor, Zn2+[24], [25]. Thus, the potential aggregation of ADH at moderately acidic conditions was monitored by incubating the enzyme without Zn2+ in a series of buffers of decreasing pH (pH 7.5-5.0) and measuring the light scattering of the samples with time. Light scattering measurements showed that ADH aggregation occurred in the 7.0–5.0 pH range. Fig. 1 shows the aggregation of ADH relative to that observed for the sample at pH 5.5 (100%) for which the highest light scattering signal was detected. The results in the figure show that an increased aggregation of ADH was observed when the enzyme was incubated in buffers of lower pH values (diagonal bars). Light scattering for all samples was detected to maximally increase 20–30 min after the addition of ADH to the buffers. The potential aggregation of Hsp60 alone was monitored by incubating the protein in the same pH range and measuring the light scattering of the samples with time. No significant changes in light scattering were detected (open bars) that would indicate either Hsp60 aggregation or disassembly by an increase or decrease in the light scattering, respectively.
Fig. 1.
Aggregation of ADH and Hsp60 monitored by using light scattering. ADH (1 μM) or Hsp60 (1 μM) was incubated in 50 mM Tris-HCl (pH 7.5-6.5) or 50 mM sodium citrate (pH 6.0-5.0) at 25 °C. The figure shows the % aggregation of ADH (diagonal bars) and Hsp60 (open bars) relative to the maximal aggregation (100%) corresponding to the highest scattering signal detected for ADH (pH 5.5).
3.2. Hsp60 suppresses the aggregation of ADH at low pH
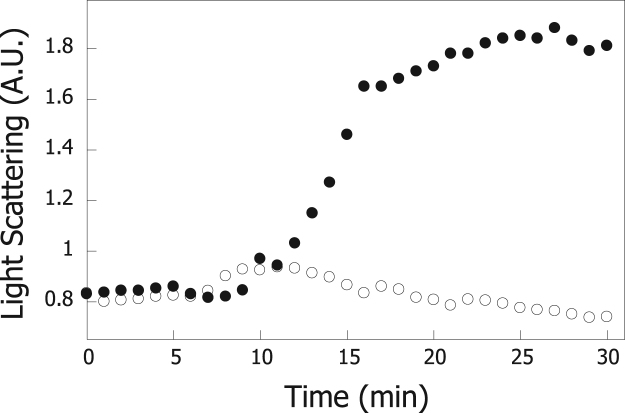
Fig. 2 shows the time course of aggregation of ADH in the absence (closed circles) or presence (open circles) of Hsp60 at pH 5.5 for which maximum aggregation was observed for ADH alone. The results in the figure show that ADH did not aggregate in the presence of Hsp60 at pH 5.5 (open circles).
Fig. 2.
Time course of the light scattering of ADH. ADH (1 μM) was added after 5 min to 50 mM sodium citrate, pH 5.5 at 25 °C without (closed circles) or with 1 μM Hsp60 (open circles).
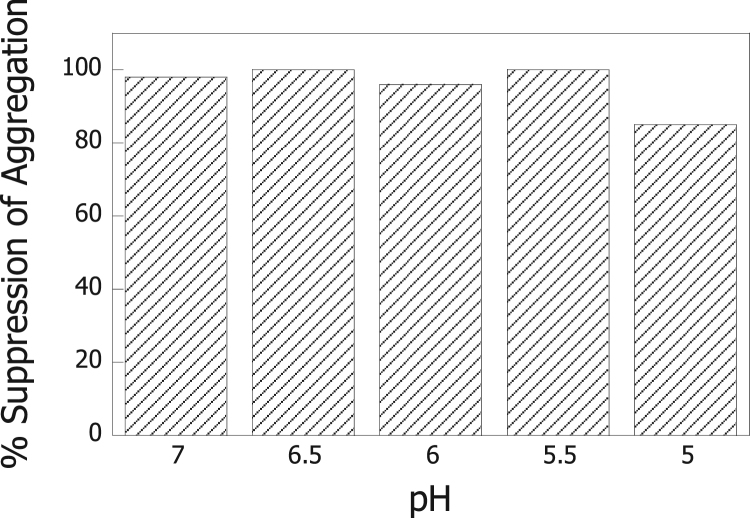
The effect of Hsp60 in suppressing the aggregation of ADH was then tested at the other pH values (7.0-5.0) at which ADH was also observed to aggregate. As shown in Fig. 3, Hsp60 was also able to suppress aggregation of ADH in these samples. Therefore, the aggregation of ADH was prevented by Hsp60 over the entire 7.0–5.0 pH range.
Fig. 3.
Suppression of aggregation of ADH by Hsp60 monitored by using light scattering. Hsp60 (1 μM) was incubated in 50 mM Tris-HCl (pH 7.5-6.5) or 50 mM sodium citrate (pH 6.0-5.0) at 25 °C. ADH (1 μM) was added after 5 min and light scattering was recorded for 30 min after the addition of ADH to the buffers.
Although, ADH exists as a dimer [25], and Hsp60 from H. pylori has been reported to exist as a mixture of dimers and tetramers [20], in these aggregation assays, ADH and Hsp60 were mixed in a 1:1 monomeric molar ratio. Interestingly, the suppression of aggregation of ADH by Hsp60 over the entire 7.0–5.0 pH range reported here was observed using that molar ratio. Sub-stoichiometric amounts of Hsp60, however, did not suppress completely the observed aggregation of ADH (not shown). Thus, given that Hsp60 from H. pylori apparently does not form a double ring; such as that of GroEL, where the binding protein is encapsulated inside the double donut cavity, the interaction of Hsp60 from H. pylori with other proteins may be different under moderately acidic conditions.
3.3. Changes in tryptophan environment of Hsp60 at low pH
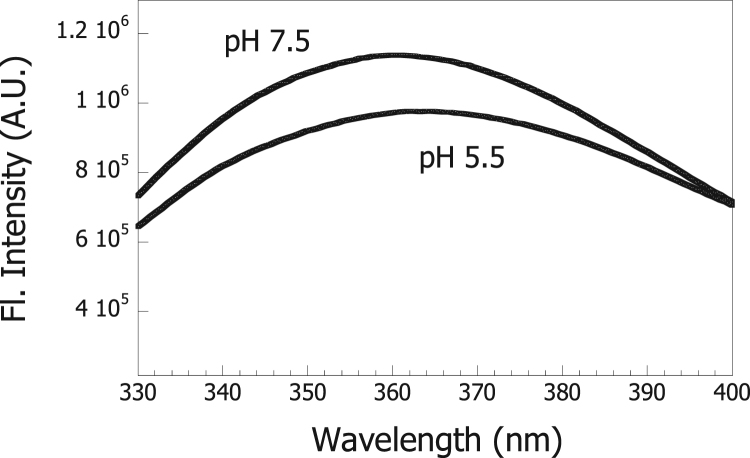
Hsp60 from H. pylori does not contain any Trp residues. Therefore, for these measurements, we made use of a site-directed mutant of Hsp60, Y359W, in which a Tyr residue was replaced with a Trp residue at the 359 amino acid residue position. This position is highly conserved in the sequences of Hsp60 proteins [27]. Fig. 4 shows the Trp fluorescence spectrum of the Y359W mutant which displayed a maximal emission centered at ~360 nm at pH 7.5, characteristic of a Trp residue that is significantly exposed to the polar solvent.
Fig. 4.
Tryptophan fluorescence of the Y359W mutant of Hsp60. The Y359W mutant was incubated with 50 mM Tris-HCl, pH 7.5 or 50 mM sodium citrate pH 5.5 and fluorescence recorded as described in “Materials and methods”. The final concentration of the Hsp60 mutant was 2.0 μM (monomers).
As shown in Fig. 4, the fluorescence emission of this mutant decreased slightly at pH 5.5, indicating that changes in the environment of Trp-359 occurred under acidification. The fluorescence spectrum of the Y359W mutant showed a maximal emission at 362 nm at pH 5.5. The observed red shift in the fluorescence spectrum for the mutant at pH 5.5 indicates that Trp-359 was even more exposed to the polar solvent than at pH 7.5.
Interestingly, Hsp60 from H. pylori has nine His residues in contrast to GroEL that has only one [27]. Thus, given that His is the only amino acid residue with a pKa close to 6.5, it is possible that protonation of His residues is involved in structural changes of Hsp60. The observed red shift in the fluorescence spectra of the Hsp60 mutant at pH 5.5 suggests that quenching of the fluorescence intensity is not likely to be due to a pH-induced conformational change involving approximation between the Trp residue and a His residue, but rather to an increased exposure of the Trp residue to the aqueous medium.
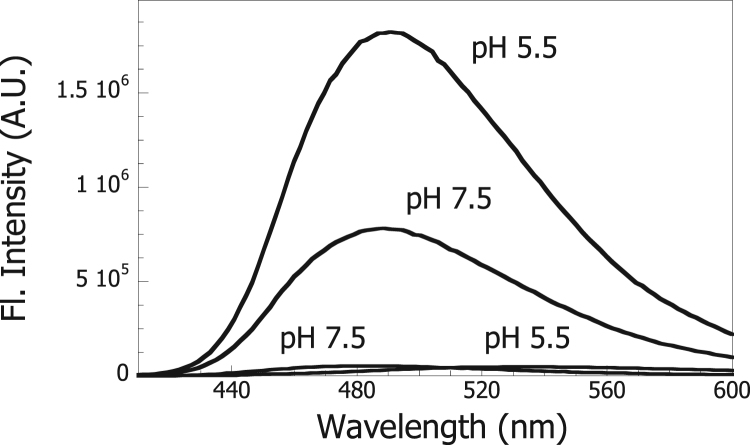
3.4. Changes in exposure of hydrophobic surfaces of Hsp60 at low pH
Changes in protein conformation induced by acidic conditions are often associated with the change in surface hydrophobicity [28], [29], [30]. Also, it is generally believed that molecular chaperones interact with their client substrate proteins via hydrophobic interactions. Here, we used the hydrophobic reporter probe, 1,1’-bis(4-anilino) naphthalene-5,5’-disulfonic acid (bisANS), to detect the exposure of hydrophobic surfaces of Hsp60. BisANS is virtually non-fluorescent in an aqueous buffer, but becomes highly fluorescent when it is bound to hydrophobic surfaces on proteins that are exposed to the solvent [31], [32]. As shown in Fig. 5, bisANS alone displayed a very weak fluorescence (lowest spectra). Fluorescence of bisANS, however, increased when this was incubated in the presence of Hsp60 (middle spectrum) at pH 7.5, showing that Hsp60 contains a significant exposure of hydrophobic surfaces. As shown in Fig. 5, the bisANS fluorescence increased even more when this was incubated in the presence of Hsp60 (highest spectrum) at pH 5.5. The hydrophobicity of the Y359W mutant, that was previously used to detect pH-induced conformational changes of Hsp60, was found to be virtually the same than that of wild type Hsp60 in the 7.5–5.5 pH range.
Fig. 5.
Measurement of exposure of hydrophobic surfaces of Hsp60. Fluorescence emission spectra were recorded for bisANS alone in 50 mM Tris-HCl, pH 7.5 or 50 mM sodium citrate, pH 5.5 (lowest spectra). Hsp60 was added to bisANS in 50 mM Tris-HCl, pH 7.5 (middle spectrum) or to bisANS in 50 mM sodium citrate, pH 5.5 (highest spectrum). The final concentrations of bisANS and Hsp60 were 10 μM and 1 μM (monomers), respectively.
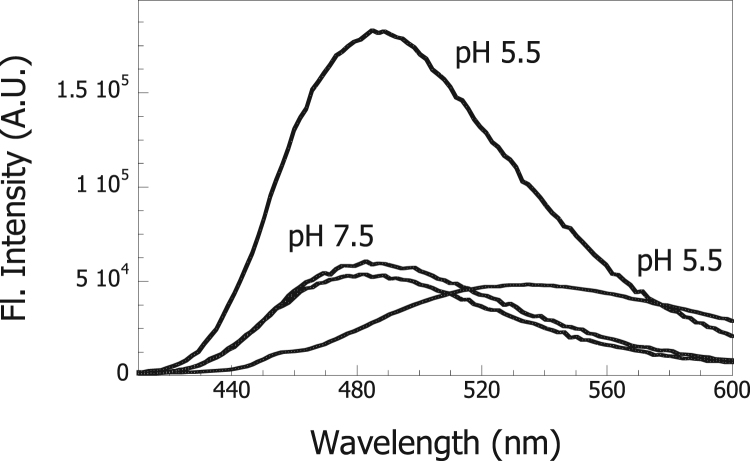
3.5. Changes in exposure of hydrophobic surfaces of ADH at low pH
BisANS fluorescence was also measured in the presence of ADH. As shown in Fig. 6, the bisANS fluorescence (lowest spectra) displayed a very small increase in the presence of ADH (middle spectrum) at pH 7.5, indicating the absence of exposed hydrophobic surfaces in the protein. The fluorescence of bisANS, however, increased when this was incubated with ADH (highest spectrum) at pH 5.5. Altogether, these results show that moderately acidic conditions trigger conformational changes of Hsp60 leading to an increase in the exposure of hydrophobic surfaces allowing it to bind other proteins also exposing hydrophobic surfaces under these conditions.
Fig. 6.
Measurement of exposure of hydrophobic surfaces of ADH. Fluorescence emission spectra were recorded for bisANS alone in 50 mM Tris-HCl, pH 7.5 or 50 mM sodium citrate, pH 5.5 (lowest spectra). ADH was added to bisANS in 50 mM Tris-HCl, pH 7.5 (middle spectrum) or to bisANS in 50 mM sodium citrate, pH 5.5 (highest spectrum). The final concentrations of bisANS and Hsp60 were 10 μM and 1 μM (monomers), respectively.
The experiments reported here have established that Hsp60 from H. pylori can function as a molecular chaperone, under moderately acidic conditions, by preventing the acid-induced aggregation of a protein. Our findings are consistent with an expected chaperone role for Hsp60 given its high degree of homology to GroEL [8], [27]. Interestingly, while GroEL consists of a double ring in which each ring has seven equal subunits [9], [21], Hsp60 from H. pylori exists as a mixture of dimers and tetramers [20]. Our light scattering measurements of Hsp60 could not detect any structural changes in the tested pH range suggesting that Hsp60 retained its quaternary structure during these experiments. Functional lower oligomers have been reported for the Hsp60 protein from M. tuberculosis[33]. The unusual quaternary structure of H. pylori Hsp60 raises the question if, under in vivo or certain in vitro conditions, Hsp60 could undergo oligomerization like the Hsp60 from M. tuberculosis into a double ring structure like that of GroEL [34] or if Hsp60 could undergo disassembly into monomers under more acidic conditions like the E. coli chaperones HdeA and HdeB [13]. Whereas, in the experiments reported here, Hsp60 by itself was an effective chaperone, it has been shown that the folding process of newly synthesized proteins in E. coli involves several molecular chaperones [9]. Thus, it remains to be seen whether Hsp60 from H. pylori can support the folding of other proteins and interact with other chaperones. Also, the nature of the complexes detected here and the mechanism of release of the bound proteins remains to be elucidated. Although, GroEL has the ability to hydrolyze ATP, other molecular chaperones cannot. We have found that ATP binds Hsp60 from H. pylori causing a significant conformational change (unpublished results). Thus, it is possible that ATP may be involved in the release of bound proteins from Hsp60 like in the GroEL-mediated protein folding [9]. The neutralization of the acidic environment may also be required to facilitate protein folding like in the process mediated by the molecular chaperones HdeA and HdeB which bind denatured proteins in an extremely acidic environment [13]. All these questions await further experimentation.
4. Conclusions
In summary, our results demonstrate that Hsp60 from H. pylori can function as a molecular chaperone under moderately acidic conditions. Our results strongly suggest that the interaction between Hsp60 and other proteins undergoing acid-induced denaturation is mediated by hydrophobic interactions. The ability of Hsp60 to protect a protein exposed to moderately acidic conditions is significant in light of the fact that under in vivo conditions of moderate acid stress, like those in which H. pylori grows, partially denatured proteins may actually exist in the conformations we have duplicated here for ADH.
Acknowledgments
This research was supported by a development grant to J.A.M. from the California State University Program for Education and Research in Biotechnology (CSUPERB).
Footnotes
Supplementary data associated with this article can be found in the online version at http://dx.doi.org/10.1016/j.bbrep.2016.11.011.
Appendix A. Supplementary material
Supplementary material
Supplementary material
Supplementary material
References
- 1.Covacci A., Telford G., Del Giudice J., Parsonnet J., Rappuoli R. Helicobacter pylori virulence and genetic geography. Science. 1999;284:1328–1333. doi: 10.1126/science.284.5418.1328. [DOI] [PubMed] [Google Scholar]
- 2.Kusters J.G., van Vliet A.H., Kuipers E.J. Pathogenesis of Helicobacter pylori infection. Clin. Microbiol. Rev. 2006;19:449–490. doi: 10.1128/CMR.00054-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ferreira A.C., Isomoto H., Moriyama M., Fujioka T., Machado J.C., Yamaoka Y. Helicobacter and gastric malignancies. Helicobacter. 2008;13:28–34. doi: 10.1111/j.1523-5378.2008.00633.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Slonczewski J.L., McGee D.J., Phillips J., Kirkpatrick C., Mobley H.L.T. pH-Dependent protein profiles of helicobacter pylori analyzed by two-dimensional gels. Helicobacter. 2000;5:240–247. doi: 10.1046/j.1523-5378.2000.00037.x. [DOI] [PubMed] [Google Scholar]
- 5.Sachs G., Weeks D.L., Wen Y., Marcus E.A., Scott D.R. Acid acclimation by helicobacter pylori. Physiology. 2005;20:429–438. doi: 10.1152/physiol.00032.2005. [DOI] [PubMed] [Google Scholar]
- 6.Khan J., Karim A., Iqbal S. Helicobacter urease: niche construction at the single molecule level. J. Biosci. 2009;34:503–511. doi: 10.1007/s12038-009-0069-4. [DOI] [PubMed] [Google Scholar]
- 7.Jungblut P.R., Bumann D., Haas G., Zimny-Arndt U., Holland P., Lamer S., Siejak F., Aebischer A., Meyer T.F. Comparative proteome analysis of helicobacter pylori. Mol. Microbiol. 2000;36:710–725. doi: 10.1046/j.1365-2958.2000.01896.x. [DOI] [PubMed] [Google Scholar]
- 8.Macchia G., Massone A., Burroni D., Covacci A., Censini S., Rappuoli R. The Hsp60 protein of Helicobacter pylori: structure and immune response in patients with gastroduodenal diseases. Mol. Microbiol. 1993;9:645–652. doi: 10.1111/j.1365-2958.1993.tb01724.x. [DOI] [PubMed] [Google Scholar]
- 9.Fink A.L. Chaperone-mediated protein folding. Physiol. Rev. 1999;79:425–449. doi: 10.1152/physrev.1999.79.2.425. [DOI] [PubMed] [Google Scholar]
- 10.Sun Y., MacRae T.H. Small heat shock proteins: molecular structure and chaperone function. Cell. Mol. Life Sci. 2005;62:2460–2476. doi: 10.1007/s00018-005-5190-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Muga A., Moro F. Thermal adaptation of heat shock proteins. Curr. Protein Pept. Sci. 2008;9:552–566. doi: 10.2174/138920308786733903. [DOI] [PubMed] [Google Scholar]
- 12.Kumsta C., Jakob U. Redox-regulated chaperones. Biochemistry. 2009;48:4666–4676. doi: 10.1021/bi9003556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hong W., Wu Y.E., Fu X., Chang Z. Chaperone-dependent mechanisms for acid resistance in enteric bacteria. Trends Microbiol. 2012;20:328–335. doi: 10.1016/j.tim.2012.03.001. [DOI] [PubMed] [Google Scholar]
- 14.Li J., Soroka J., Buchner J. The Hsp90 chaperone machinery: conformational dynamics and regulation by co-chaperones. Biochim. Biophys. Acta. 2012;1823:624–635. doi: 10.1016/j.bbamcr.2011.09.003. [DOI] [PubMed] [Google Scholar]
- 15.Suerbaum S., Thiberge J.M., Kansau I., Ferrero R.L., Labigne A. Helicobacter pylori hspA-hspB heat-shock gene cluster: nucleotide sequence, expression, putative function and immunogenicity. Mol. Microbiol. 1994;14:959–974. doi: 10.1111/j.1365-2958.1994.tb01331.x. [DOI] [PubMed] [Google Scholar]
- 16.Kansau I., Labigne A. Heat shock proteins of Helicobacter pylori. Aliment. Pharmacol. Ther. 1996;10:51–56. doi: 10.1046/j.1365-2036.1996.22164005.x. [DOI] [PubMed] [Google Scholar]
- 17.Bai Y., Li L.R., Wang J.D., Chen Y., Jin J.F., Zhang Z.S., Zhou D.Y., Zhang Y.L. Expression of helicobacter pylori Hsp60 protein and its immunogenicity. World J. Gastroenterol. 2003;9:2711–2714. doi: 10.3748/wjg.v9.i12.2711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cun S., Li H., Ge R., Lin M.C.M., Sun H. A histidine-rich and cysteine-rich metal-binding domain at the C terminus of heat shock protein A from Helicobacter pylori. J. Biol. Chem. 2008;283:15142–15151. doi: 10.1074/jbc.M800591200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schauer K., Muller C., Carrière M., Labigne A., Cavazza C., De Reuse H. The Helicobacter pylori GroES cochaperonin HspA functions as a specialized nickel chaperone and sequestration protein through its unique C-terminal extension. J. Bacteriol. 2010;192:1231–1237. doi: 10.1128/JB.01216-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lin C.Y., Huang Y.S., Li C.H., Hsieh Y.T., Tsai N.M., He P.J., Hsu W.T., Yeh Y.C., Chiang F.H., Wu M.S., Chang C.C., Liao K.W. Characterizing the polymeric status of Helicobacter pylori heat shock protein 60. Biochem. Biophys. Res. Commun. 2009;388:283–289. doi: 10.1016/j.bbrc.2009.07.159. [DOI] [PubMed] [Google Scholar]
- 21.Braig K., Otwinowski Z., Hegde R., Boisvert D.C., Joachimiak A., Horwich A.L., Sigler P.B. The crystal structure of the bacterial chaperonin GroEL at 2.8 A. Nature. 1994;371:578–586. doi: 10.1038/371578a0. [DOI] [PubMed] [Google Scholar]
- 22.Goto Y., Takahashi N., Fink A.L. Mechanism of acid-induced folding of proteins. Biochemistry. 1990;29:3480–3488. doi: 10.1021/bi00466a009. [DOI] [PubMed] [Google Scholar]
- 23.Uversky V.N., Goto Y. Acid denaturation and anion-induced folding of globular proteins: multitude of equilibrium partially folded intermediates. Curr. Protein Pept. Sci. 2009;10:447–455. doi: 10.2174/138920309789352029. [DOI] [PubMed] [Google Scholar]
- 24.Brändén C.-I., Eklund H., Nordström B., Boiwe T., Söderlund G., Zeppezauer E., Ohlsson I., Åkeson Å. Vol. 70. 1973. Structure of liver alcohol dehydrogenase at 2.9-angstrom resolution; pp. 2439–2442. (Proc. Natl. Acad. Sci. USA). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schneider G., Eklund H., Cedergren-Zeppezauer E., Zeppezauer M. Structure of the complex of active site metal-depleted horse liver alcohol dehydrogenase and NADH. EMBO J. 1983;2:685–689. doi: 10.1002/j.1460-2075.1983.tb01485.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Laemmli U.K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 27.Brocchieri L., Karlin S. Conservation among HSP60 sequences in relation to structure, function, and evolution. Protein Sci. 2000;9:476–486. doi: 10.1110/ps.9.3.476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Qa'Dan M., Spyres L.M., Ballard J.D. pH-Induced conformational changes in clostridium difficile. Toxin B Infect. Immun. 2000;68:2470–2474. doi: 10.1128/iai.68.5.2470-2474.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dollery S.J., Delboy M.G., Nicola A.V. Low pH-Induced conformational change in herpes simplex virus glycoprotein B. J. Virol. 2010;84:3759–3766. doi: 10.1128/JVI.02573-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tutar Y., Arslan D., Tutar L. Heat, pH induced aggregation and surface hydrophobicity of S. cerevesiae Ssa1 Protein. Protein J. 2010;29:501–508. doi: 10.1007/s10930-010-9280-2. [DOI] [PubMed] [Google Scholar]
- 31.Rosen C.G., Weber G. BisANS dimer formation from 1-anilino-8-naphthalene sulfonate catalyzed by bovine serum albumin. fluorescent molecule with exceptional binding properties. Biochemistry. 1969;8:3915–3920. doi: 10.1021/bi00838a006. [DOI] [PubMed] [Google Scholar]
- 32.Hawe A., Sutter M., Jiskoot W. Extrinsic fluorescent dyes as tools for protein characterization. Pharm. Res. 2008;25:1487–1499. doi: 10.1007/s11095-007-9516-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Qamra R., Srinivas V., Mande S.C. Mycobacterium tuberculosis GroEL homologues unusually exist as lower oligomers and retain the ability to suppress aggregation of substrate proteins. Mol. Biol. 2004;342:605–617. doi: 10.1016/j.jmb.2004.07.066. [DOI] [PubMed] [Google Scholar]
- 34.Fan M., Rao T., Zacco E., Ahmed M.T., Shukla A., Ojha A., Freeke J., Robinson C.V., Benesch J.L., Lund P.A. The unusual mycobacterial chaperonins: evidence for in vivo oligomerization and specialization of function. Mol. Microbiol. 2012;85:934–944. doi: 10.1111/j.1365-2958.2012.08150.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material
Supplementary material
Supplementary material