Abstract
Legumain (EC 3.4.22.34) is an asparaginyl endopeptidase. Legumain activity has been detected in various mouse tissues including the kidney, spleen and epididymis. Legumain is overexpressed in the majority of human solid tumors and transcription of the legumain gene is regulated by the p53 tumor suppressor in HCT116 cells. The legumain activity is also increased under acid conditions in Alzheimer's disease brains. DJ-1/PARK7, a cancer- and Parkinson's disease-associated protein, works as a coactivator to various transcription factors, including the androgen receptor, p53, PSF, Nrf2, SREBP and RREB1. Recently, we found that legumain expression, activation and cleavage of annexin A2 are regulated by DJ-1 through p53. In this study, we found that the expression levels of legumain mRNA were increased in the cerebrum, kidney, spleen, heart, lung, epididymis, stomach, small intestine and pancreas from DJ-1-knockout mice, although legumain activity levels were decreased in the cerebrum, spleen and heart from DJ-1-knockout mice. Furthermore, we found that cystatin E/M expression was increased in the spleen, cerebrum and heart from DJ-1-knockout mice. These results suggest that reduction of legumain activity is caused by an increase of cystatin E/M expression in the spleen, cerebrum and heart from DJ-1-knockout mice.
Abbreviations: MHC, major histocompatibility complex; AMC, 7-amino-4-methylcoumarin; Z, benzyloxycarbonyl; MCA, methylcumarinamide
Keywords: Legumain, DJ-1, Cystatin E/M
Highlights
-
•
Legumain is strongly activated in the epididymis from DJ-1-knockout mice.
-
•
Expression level of legumain mRNA is increased but activity is decreased in the spleen, cerebrum and heart from DJ-1-knockout mice.
-
•
Expression level of cystatin E/M is increased in the spleen, cerebrum and heart from DJ-1-knockout mice.
1. Introduction
Legumain (EC 3.4.22.34) is an asparaginyl endopeptidase belonging to the cysteine peptidase C13 family [1]. Legumain activity has been detected in a number of mouse tissues, including the kidney, placenta, spleen, liver, testis and thymus [2]. Legumain degrades annexin A2 [3], tetanus toxin C-terminal fragment [4], cathepsins B, H and L [4], (pro)legumain [5], [6], [7], [8], [9] and cystatins C and E/M [10]. Cystatins are reversible competitive inhibitors of C1 cysteine proteases [11]. Plant-derived papain and mammalian cathepsins B, H and L interact with cystatins [12]. Legumain cleaves cystatin C in specific positions, and legumain activity is competitively inhibited by other legumain substrates [13]. Legumain is also overexpressed in the majority of human solid tumors, including breast cancer [14], prostate cancer [15], gastric cancer [16], colorectal cancer [17] and ovarian cancer [18]. Legumain activity is increased under acid conditions in Alzheimer's disease brains [19], [20], [21]. Legumain is also a key enzyme in the processing of foreign- and self-antigens for MHC class II antigen presentation [22], [23]. However, Rene Maehr et al. reported that legumain is not essential for MHC class II antigen presentation [5]. Legumain activity is inhibited by type 2 cystatins, and cystatin E/M is the most potent legumain inhibitor [24], [25]. Legumain is mainly expressed in proximal tubules of the rat kidney [26], and legumain might have an important role in remodeling of the extracellular matrix through degradation of fibronectin in renal proximal tubular cells [27]. Legumain protease activity is regulated by pH [28]. Legumain activity is rapidly destroyed at neutral pH [6], [7], and neutral pH destabilizes legumain or change it from a protease to a ligase [10]. Legumain is also responsible for catalyzing both cleavage of peptide bond and ligation of cyclotides in a single processing event [29].
DJ-1 has been identified by us as a novel oncogene [30] and was also later identified as a causative gene for a familial form of Parkinson's disease, park7 [31]. DJ-1 has a function of transcriptional regulation [32], [33], [34], [35], [36], [37], [38], [39], [40], [41], [42], [43], and it acts as a coactivator that binds to p53 [34], [39].
Recently, we reported that the expression and protease activity of legumain are regulated by p53 through its binding to intron 1 of the legumain gene [44] and that legumain expression and protease activity are regulated by p53 through DJ-1 on intron 1 of the mouse legumain gene after analyses using DJ-1-knockout cells [45].
In this study, to confirm that legumain expression and protease activity are also regulated by DJ-1 in vivo, we examined the expression level and activity of legumain in various tissues of DJ-1-knockout mice. We found that the expression level of legumain mRNA was increased in a number of tissues from DJ-1-knockout mice, including the cerebrum, spleen and heart. However, legumain activity was decreased in these tissues. We also found that cystatin E/M expression was increased in these tissues from DJ-1-knockout mice. The results suggest that reduction of legumain activity is caused by an increase of cystatin E/M expression in the cerebrum, spleen and heart from DJ-1-knockout mice.
2. Materials and methods
2.1. Materials
PrimeScript RT Master Mix and Z-Ala-Ala-Asn-MCA were obtained from Takara (Shiga, Japan) and Peptide Institute (Osaka, Japan), respectively. RNAlater and RNeasy Mini were purchased from Qiagen (Venlo, Netherlands). All other chemicals were of analytical grade and were purchased from Wako Pure Chemicals (Osaka, Japan).
2.2. Animals
Wild-type and DJ-1-knockout mice were described previously [28]. DJ-1-knockout mice with a C57BL/6 background and C57BL/6 mice that were used as control mice with DJ-1 (+/+) were housed under SPF conditions. Tissues were isolated from wild-type and DJ-1-knockout mice at 23 weeks of age, and their total RNAs and protein were extracted for further analyses. The four mice of each genotype were used in a pool of samples for each experiment, and experiments were performed in duplicates.
2.3. Reverse transcription and quantitative PCR
Total RNAs were prepared from wild-type and DJ-1-knockout mice tissues using an RNeasy mini kit. Reverse transcription was carried out in a mixture containing 500 ng of total RNAs using PrimeScript RT Master Mix (Takara, Shiga, Japan) with manufacture's protocol. The specific primers under the conditions of 95 °C for 30 s, 39 cycles of 95 °C for 10 s, and 60 °C for 30 s by using an SYBR Premix Ex Taq II (Takara) and a quantitative PCR system (MiniOpticon, Bio-Rad, Hercules, CA, USA). β-actin (ACTB) mRNA was also amplified as a reference gene. Nucleotide sequences of oligonucleotides used for quantitative PCR primers were as follows: mLgmn-F: 5′-CTTCCGCACACACTGCTTTA-3′, mLgmn-R: 5′-CTTTGTCCATGGCCATCTCT −3′, mCystatin E/M-F: 5′-CACCAGTCTCCAACCTCCAC-3′ and mCystatin E/M-R: 5′-CACAGTGGGACACAATGGGA-3′. Each experiment was performed in duplicates. To demonstrate that the differential expression is not result of primer bias, the efficiency was evaluated for each primer using a Bio-Rad CFX Manager 3.1 software, and stability of the valuable reference gene β-actin was also demonstrated.
2.4. Western blotting and antibodies
To examine the expression levels of cystatin E/M in tissues from wild-type and DJ-1-knockout mice, proteins were extracted from the cerebrum, spleen, heart and epididymis in a solution containing 50 mM sodium citrate (pH 5.0). The proteins were then separated on a 12.5% polyacrylamide gel and subjected to Western blotting with respective antibodies. The antibodies used were anti-Actin (1:1000, Santa Cruz Biotechnology, Santa Cruz, CA, USA), anti-legumain (1:1000, R&D Systems, Minneapolis, MN, USA), anti-cystatin E/M (1:1000, Aviva Systems Biology, San Diego, CA, USA) and rabbit anti-DJ-1 (1:1000) antibodies. Proteins on the membrane were reacted with an Alexa Fluor 680-conjugated secondary antibody (Molecular Probes, Eugene, OR, USA) or IRDye 800- (Rockland, Philadelphia, PA, USA) and visualized by using an infrared imaging system (Odyssey, LI-COR, Lincoln, NE, USA). The relative expression level of Cystatin E/M was detected by measuring the band intensity using a densitometry.
2.5. Proteolytic activity of legumain
Enzyme activity of legumain was examined by measuring the fluorometrical number (excitation, 380 nm; emission, 440 nm) of liberation of AMC in a mixture containing 10 μl of 10 mM Z-Ala-Ala-Asn-MCA, 100 μl of 0.5 M sodium citrate buffer (pH 5.0), 5 μl of 1 M 2-mercaptoethanol, 20 μl of enzyme solution and water (18 mΩ) in a total volume of 1 ml. After incubation of the mixture at 37 °C for 30 min, 2 ml of 0.2 M acetic acid was added to the mixture to stop the reaction. Enzyme assays were carried out using 100 µg of total protein from each tissue. The enzyme concentration was quantified using fluorescence intensity of AMC. One unit (U) of activity was defined as the amount of enzyme that hydrolyzed 1 μmol of the substrate per min.
2.6. Statistical analyses
Data are expressed as means±S.E. Statistical analyses were performed using analysis of variance (one-way ANOVA) followed by unpaired Student's t-test.
2.7. Ethics statement
All animal experiments were carried out in accordance with the National Institutes of Health Guide for the Care and Use of Laboratory Animals, and the protocols were approved by the Committee for Animal Research at Hokkaido University (permit number 08–0467).
3. Results
3.1. Legumain expression in wild-type and DJ-1-knockout mice
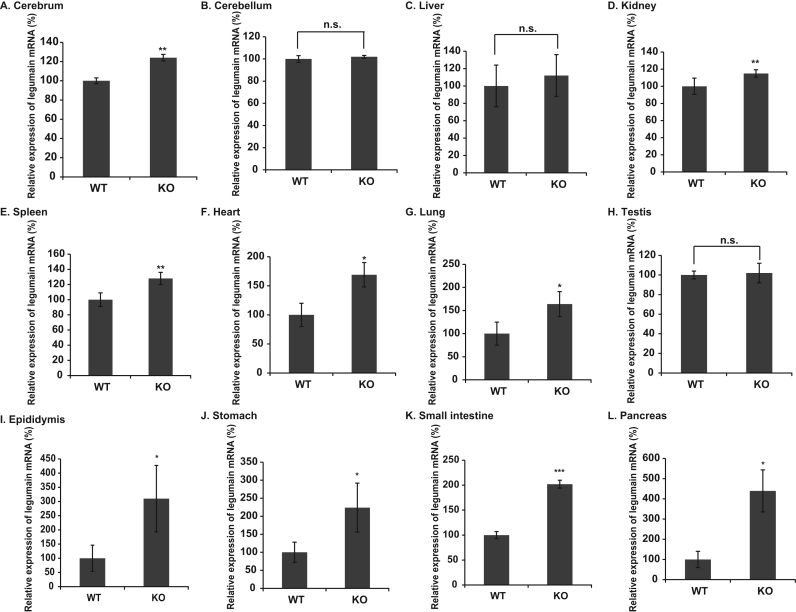
To explore the regulation of legumain gene expression in wild-type and DJ-1-knockout mice, the expression levels of legumain and actin mRNA were examined by RT-qPCR. Actin mRNA was used as a reference gene. As shown in Fig. 1A, D-G and I-L, the expression levels of legumain mRNA in the cerebrum, kidney, spleen, heart, lung, epididymis, stomach, small intestine and pancreas from DJ-1-knockout mice were increased by about 20%, 25%, 28%, 70%, 65%, 210%, 120%, 100% and 340% of those in wild-type tissues, respectively. The expression levels of legumain mRNA were not increased in the cerebellum, liver and testis from DJ-1-knockout mice (Fig. 1B, C and H). The expression of the β-actin gene was stable between tissues, and the reaction efficiency was between 1.8 and 2.0.
Fig. 1.
Expression levels of legumain mRNA in wild-type and DJ-1-knockout tissues. Expression levels of legumain mRNA in wild-type and DJ-1-knockout tissues were examined by RT-qPCR (real-time PCR). Actin mRNA was used as a reference gene. The relative expression level of each mRNA toward actin mRNA is shown. A. cerebrum, B. cerebellum, C. liver, D. kidney, E. spleen, F. heart, G. lung, H. testis, I. epididymis, J. stomach, K. small intestine, L. pancreas. Statistical analyses were carried out using Student's t-test. The number of experiments (n) was 4. Statistically significant: *p<0.05, **p<0.01, **p<0.001. Not significant: n.s.
3.2. Legumain activity in wild-type and DJ-1-knockout mice
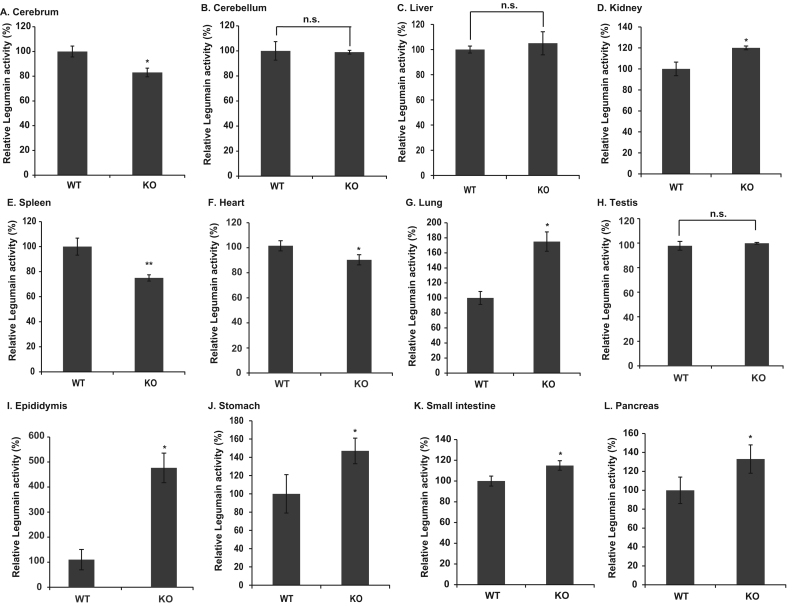
To examine the regulation of legumain activity in wild-type and DJ-1-knockout mice, the levels of legumain activity were measured using Z-Ala-Ala-Asn-MCA as a substrate. As shown in Fig. 2D, G and I-L, the levels of legumain activity in the kidney, lung, epididymis, stomach, small intestine and pancreas from DJ-1-knockout mice were increased by about 20%, 75%, 330%, 47%, 15% and 33% of those in wild-type tissues, respectively. The levels of legumain activity were not increased in the cerebellum, liver and testis from DJ-1-knockout mice (Fig. 2B, C and H). Interestingly, the levels of legumain activity in the cerebrum, spleen and heart from DJ-1-knockout mice were decreased by about 17%, 25% and 11% of those in wild-type tissues, respectively (Fig. 2A, E and F). For example, the enzyme unit of legumain was 0.68 and 0.50 mU in the spleen, and was 0.25 and 1.2 mU in the epididymis from wild-type and DJ-1-knockout mice, respectively.
Fig. 2.
Protease activities of legumain in wild-type and DJ-1-knockout tissues. The levels of legumain activity in wild-type and DJ-1-knockout tissues were examined by a proteolytic assay as described in Materials and methods. The relative legumain activity is shown. A. cerebrum, B. cerebellum, C. liver, D. kidney, E. spleen, F. heart, G. lung, H. testis, I. epididymis, J. stomach, K. small intestine, L. pancreas. Statistical analyses were carried out using Student's t-test. The number of experiments (n) was 4. Statistically significant: *p<0.05, **p<0.01. Not significant: n.s.
3.3. Cystatin E/M expression in the cerebrum, spleen, heart and epididymis from wild-type and DJ-1-knockout mice
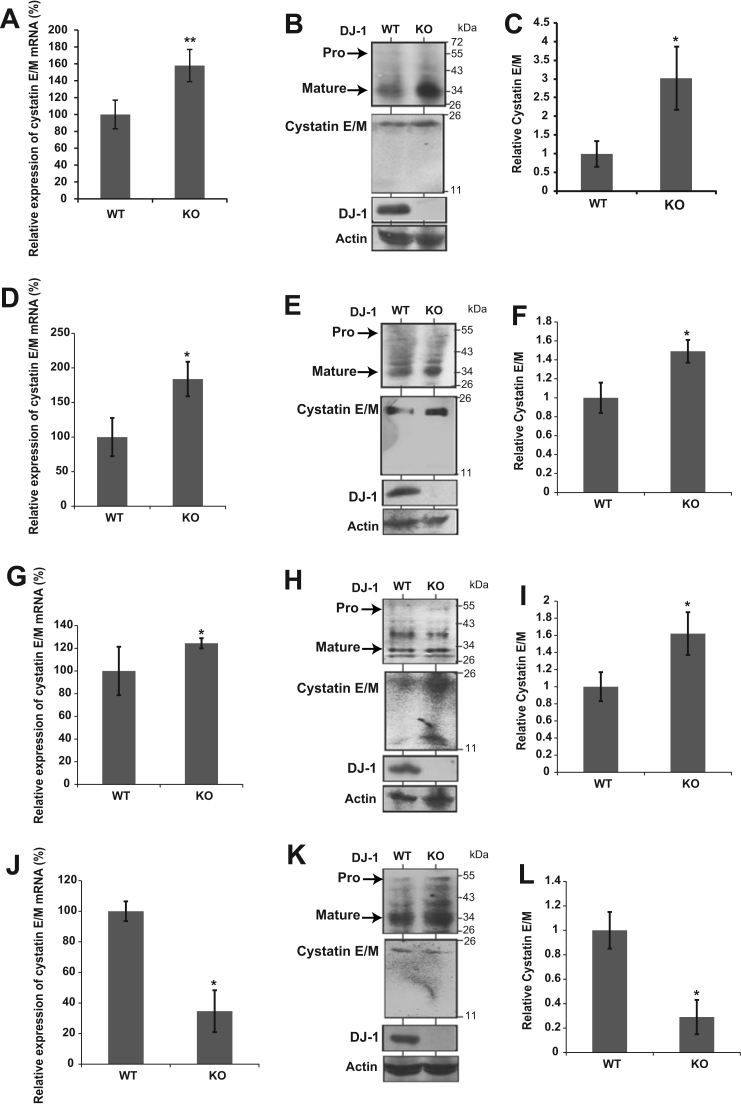
Since reduction of legumain activity was found in the cerebrum, spleen and heart from DJ-1-knockout mice, the expression level of cystatin E/M was measured. Since the greatest increase of legumain activity was found in the epididymis from DJ-1-knockout mice, the expression level of cystatin E/M was also measured. The expression levels of cystatin E/M and actin mRNA were examined by RT-qPCR. Actin mRNA was used as a reference gene. As shown in Fig. 3A, D and G, the expression levels of cystatin E/M mRNA were increased in the spleen, cerebrum and heart from DJ-1-knockout mice. On the other hand, the expression level of cystatin E/M mRNA was decreased in the epididymis from DJ-1-knockout mice (Fig. 3J). Western blotting was carried out to examine the expression levels of cystatin E/M in the spleen, cerebrum, heart and epididymis from DJ-1-knockout mice. As shown in Fig. 3B, E and H, the expression levels of cystatin E/M were increased in the spleen, cerebrum and heart from DJ-1-knockout mice, and quantification of cystatin E/M is shown in Fig. 3C, F and I. On the other hand, the expression level of cystatin E/M was decreased in the epididymis from DJ-1-knockout mice (Fig. 3K), and quantification of cystatin E/M is shown in Fig. 3L.
Fig. 3.
Effect of cystatin E/M expression on inhibition of legumain activity. Relative mRNA level of cystatin E/M was examined by RT-qPCR (real-time PCR) in the spleens (A), cerebrums (D), hearts (G) and epididymis (J) from wild-type and DJ-1-knockout mice. Actin mRNA was also amplified by real-time PCR as a reference gene. Statistical analyses were carried out using Student's t-test. The number of experiments (n) was 4. Statistically significant: *p<0.05, **p<0.01. Proteins extracted from wild-type and DJ-1-knockout mice spleens (B), cerebrums (E), hearts (H) and epididymis (K) were analyzed by Western blotting with anti-cystatin E/M, anti-legumain, anti-DJ-1 and anti-β-actin antibodies. β-actin was used as a reference protein. The expression level of cystatin E/M was examined by Western blotting. “Pro” and “Mature” indicate active and inactive forms of legumain, respectively. The relative expression level of cystatin E/M toward actin is shown in (C) spleen, (F) cerebrum, (I) heart and (L) epididymis. Statistical analyses were carried out using Student's t-test. The number of experiments (n) was 4. Statistically significant: *p<0.05.
4. Discussion
In this study, we first found that expression of the legumain gene was increased in the cerebrum, kidney, spleen, heart, lung, epididymis, stomach, small intestine and pancreas from DJ-1-knockout mice. The greatest increase in the activity of legumian was detected in the epididymis from DJ-1-knockout mice. Prolegumain is activated by the auto-catalysis under acidic pH [7], [9]. Prolegumain and DJ-1 are secreted to seminal fluid [46]. DJ-1 is related to infertility of rats and mice and it takes part in fertilization for sperm to penetrate into the zonae pellucida of eggs [47], [48], [49], [50], [51], [52]. The level of DJ-1 was shown to be significantly lower in seminal plasma from asthenozoospermia patients than in seminal plasma from healthy donors [46]. Prolegumain secretion into epididymal fluid may be increased and prevent maturation of sperm. Interestingly, we also found that legumain activity was decreased in the cerebrum, spleen and heart from DJ-1-knockout mice. Cystatins are reversible competitive inhibitors of C1 cysteine proteases [11]. Plant derived papain and mammalian cathepsins B, H and L interact with cystatins [12]. Legumain activity is inhibited by cystatin superfamily type 2 members, including cystatins C, F and E/M, and cystatin E/M has strong affinity to legumain [24]. Secreted prolegumain and cystatin E/M are able to be internalized, and cystatin E/M inhibits legumain auto-activation intra- and extracellularly [53]. Legumain activity is also regulated by pH, and neutral pH destabilizes legumain or changes it from a protease to a ligase [10], [28]. For example, the expression level of legumain mRNA was increased about 4-fold in the pancreas from DJ-1-knockout mice. However, legumain activity was increased only 1.35-fold in the pancreas from DJ-1-knockout mice. These results indicate that legumain may be destabilized and that its activity is inhibited under a neutral pH condition in the pancreas of DJ-1-knockout mice. Since legumain activity was strongly inhibited in the spleen in comparison with the activity in the cerebrum and heart from DJ-1-knockout mice, expression levels of cystatin E/M were measured. Cystatin E/M mRNA and protein levels were increased in the spleen, cerebrum and heart from DJ-1-knockout mice. On the other hand, cystatin E/M mRNA and protein levels were decreased in the epididymis. These results suggest that increased cystatin E/M inhibited legumain activity in the spleen, cerebrum and heart but not in the epididymis of DJ-1-knockout mice. However, since we did not measure the functional activity in vivo of legumain and since relationship between legumain activity and difference in its sub-cellular localization and subtle pH-change in vivo was not investigated, the current study has limitations and further study is needed.
In conclusion, legumain is strongly activated in the epididymis of DJ-1-knockout mice, and legumain activities are decreased in the cerebrum, spleen and heart of DJ-1-knockout mice, whereas expression levels of mRNA are increased in those tissues. Cystatin E/M mRNA and protein levels are increased in the spleen of DJ-1-knockout mice. It would therefore be interesting to further analyze the association between legumain and DJ-1 in transcriptional regulation of cystatin E/M in the spleen, cerebrum and heart.
Competing interests
The authors have declared that no competing interests exist.
Footnotes
Supplementary data associated with this article can be found in the online version at doi:10.1016/j.bbrep.2016.12.010.
Appendix A. Transparency document
Supplementary material
.
References
- 1.Ishii S. Legumain: asparaginyl endopeptidase. Methods Enzymol. 1994;244:604–615. doi: 10.1016/0076-6879(94)44044-1. [DOI] [PubMed] [Google Scholar]
- 2.Chen J.M., Dando P.M., Stevens R.A.E. Cloning and expression of mouse legumain, a lysosomal endopeptidase. Biochem. J. 1998;335:111–117. doi: 10.1042/bj3350111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yamane T., Hachisu R., Yuguchi M. Knockdown of legumain inhibits cleavage of annexinA2 in the mouse kidney. Biochem. Biophys. Res. Commun. 2013;430:482–487. doi: 10.1016/j.bbrc.2012.12.010. [DOI] [PubMed] [Google Scholar]
- 4.Manoury B., Hewitt E.W., Morrice N., Dando P.M., Barrett A.J., Watts C. An asparaginyl endopeptidase processes a microbial antigen for class II MHC presentation. Nature. 1998;396:695–699. doi: 10.1038/25379. [DOI] [PubMed] [Google Scholar]
- 5.Maehr R., Hang H.C., Mintern J.D., Kim Y.M., Cuvillier A., Nishimura M., Yamada K., Shirahama-Noda K., Hara-Nishimura I., Ploegh H.L. Asparagine endopeptidase is not essential for class II MHC antigen presentation but is required for processing of cathepsin L in mice. J. Immunol. 2005;174:7066–7074. doi: 10.4049/jimmunol.174.11.7066. [DOI] [PubMed] [Google Scholar]
- 6.Dall E., Brandstetter H. Activation of legumain involves proteolytic and conformational events, resulting in a context- and substrate-dependent activity profile. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 2012;68:24–31. doi: 10.1107/S1744309111048020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen J.M., Fortunato M., Barrett A.J. Activation of human prolegumain by cleavage at a C-terminal asparagine residue. Biochem. J. 2000;352(Pt 2):327–334. [PMC free article] [PubMed] [Google Scholar]
- 8.Li D.N., Matthews S.P., Antoniou A.N., Mazzeo D., Watts C. Multistep autoactivation of asparaginyl endopeptidase in vitro and in vivo. J. Biol. Chem. 2003;278:38980–38990. doi: 10.1074/jbc.M305930200. [DOI] [PubMed] [Google Scholar]
- 9.Halfon S., Patel S., Vega F., Zurawski S., Zurawski G. Autocatalytic activation of human legumain at aspartic acid residues. FEBS Lett. 1998;438:114–118. doi: 10.1016/s0014-5793(98)01281-2. [DOI] [PubMed] [Google Scholar]
- 10.Dall E., Fegg J.C., Briza P., Brandstetter H. Structure and mechanism of an aspartimide-dependent peptide ligase in human legumain. Angew. Chem. Int. Ed. Engl. 2015;54:2917–2921. doi: 10.1002/anie.201409135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ochieng J., Chaudhuri G. Cystatin superfamily. J. Health Care Poor Underserved. 2010;21:51–70. doi: 10.1353/hpu.0.0257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Turk V., Bode W. The cystatins: protein inhibitors of cysteine proteinases. FEBS Lett. 1991;285:213–219. doi: 10.1016/0014-5793(91)80804-c. [DOI] [PubMed] [Google Scholar]
- 13.Zhao L., Hua T., Crowley C., Ru H., Ni X., Shaw N., Jiao L., Ding W., Qu L., Hung L.W., Huang W., Liu L., Ye K., Ouyang S., Cheng G., Liu Z.J. Structural analysis of asparaginyl endopeptidase reveals the activation mechanism and a reversible intermediate maturation stage. Cell Res. 2014;24:344–358. doi: 10.1038/cr.2014.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lewen S., Zhou H., Hu H.D. A Legumain-based minigene vaccine targets the tumor stroma and suppresses breast cancer growth and angiogenesis. Cancer Immunol. Immunother. 2008;57:507–515. doi: 10.1007/s00262-007-0389-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ohno Y., Nakashima J., Izumi M. Association of legumain expression pattern with prostate cancer invasiveness and aggressiveness. World J. Urol. 2013;31:359–364. doi: 10.1007/s00345-012-0977-z. [DOI] [PubMed] [Google Scholar]
- 16.Li N., Liu Q., Su Q. Effects of legumain as a potential prognostic factor on gastric cancers. Med. Oncol. 2013;30:621. doi: 10.1007/s12032-013-0621-9. [DOI] [PubMed] [Google Scholar]
- 17.Haugen M.H., Johansen H.T., Pettersen S.J. Nuclear legumain activity in colorectal cancer. PLoS One. 2013;8:e52980. doi: 10.1371/journal.pone.0052980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang L., Chen S., Zhang M. Legumain: a biomarker for diagnosis and prognosis of human ovarian cancer. J. Cell Biochem. 2012;113:2679–2686. doi: 10.1002/jcb.24143. [DOI] [PubMed] [Google Scholar]
- 19.Lillico S.G., Mottram J.C., Murphy N.B. Characterisation of the QM gene of Trypanosoma brucei. FEMS Microbiol. Lett. 2002;211:123–128. doi: 10.1111/j.1574-6968.2002.tb11213.x. [DOI] [PubMed] [Google Scholar]
- 20.Pivtoraiko V.N., Stone S.L., Roth K.A. Oxidative stress and autophagy in the regulation of lysosome-dependent neuron death. Antioxid. Redox Signal. 2009;11:481e496. doi: 10.1089/ars.2008.2263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Castellani R.J., Honda K., Zhu X. Contribution of redox-active iron and copper to oxidative damage in Alzheimer disease. Ageing Res. Rev. 2004;3:319e326. doi: 10.1016/j.arr.2004.01.002. [DOI] [PubMed] [Google Scholar]
- 22.Manoury B., Hewitt E.W., Morrice N. An asparaginyl endopeptidase processes a microbial antigen for class II MHC presentation. Nature. 1998;396:695–699. doi: 10.1038/25379. [DOI] [PubMed] [Google Scholar]
- 23.Burster T., Beck A., Tolosa E. Cathepsin G, and not the asparagine-specific endoprotease, controls the processing of myelin basic protein in lysosomes from human B lymphocytes. J. Immunol. 2004;172:5495–5503. doi: 10.4049/jimmunol.172.9.5495. [DOI] [PubMed] [Google Scholar]
- 24.Rawlings N.D., Barrett A.J., Bateman A. MEROPS: the database of proteolytic enzymes, their substrates and inhibitors. Nucleic Acids Res. 2012;40:D343–D350. doi: 10.1093/nar/gkr987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Alvarez-Fernandez M., Barrett A.J., Gerhartz B. Inhibition of mammalian legumain by some cystatins is due to a novel second reactive site. J. Biol. Chem. 1999;274:19195–19203. doi: 10.1074/jbc.274.27.19195. [DOI] [PubMed] [Google Scholar]
- 26.Yamane T., Takeuchi K., Yamamoto Y. Legumainfrom bovine kidney: its purification, molecular cloning, immunohistochemical localization and degradation of annexin II andvitamin D-binding protein. Biochim. Biophys. Acta. 2002;1596:108–120. doi: 10.1016/s0167-4838(02)00209-1. [DOI] [PubMed] [Google Scholar]
- 27.Morita Y., Araki H., Sugimoto T. Legumain/asparaginyl endopeptidase controls extracellular matrix remodeling through the degradation of fibronectin in mouse renal proximal tubular cells. FEBS Lett. 2007;581:1417–1424. doi: 10.1016/j.febslet.2007.02.064. [DOI] [PubMed] [Google Scholar]
- 28.Dall E., Brandstetter H. Structure and function of legumain in health and disease. Biochimie. 2016;122:126–150. doi: 10.1016/j.biochi.2015.09.022. [DOI] [PubMed] [Google Scholar]
- 29.Saska I., Gillon A.D., Hatsugai N., Dietzgen R.G., Hara-Nishimura I., Anderson M.A., Craik D.J. An asparaginyl endopeptidase mediates in vivo protein backbone cyclization. J. Biol. Chem. 2007;282:29721–29728. doi: 10.1074/jbc.M705185200. [DOI] [PubMed] [Google Scholar]
- 30.Nagakubo D., Taira T., Kitaura H. DJ-1, a novel oncogene which transforms mouse NIH3T3 cells in cooperation with ras. Biochem. Biophys. Res. Commun. 1997;231:509–513. doi: 10.1006/bbrc.1997.6132. [DOI] [PubMed] [Google Scholar]
- 31.Bonifati V., Rizzu P., van Baren M.J. Mutations in the DJ-1 gene associated with autosomal recessive early-onset parkinsonism. Science. 2003;299:256–259. doi: 10.1126/science.1077209. [DOI] [PubMed] [Google Scholar]
- 32.Takahashi K., Taira T., Niki T. DJ-1 positively regulates the androgen receptor by impairing the binding of PIASx alpha to the receptor. J. Biol. Chem. 2001;276:37556–37563. doi: 10.1074/jbc.M101730200. [DOI] [PubMed] [Google Scholar]
- 33.Niki T., Takahashi-Niki K., Taira T. DJBP: a novel DJ-1-binding protein, negatively regulates the androgen receptor by recruiting histone deacetylase complex, and DJ-1 antagonizes this inhibition by abrogation of this complex. Mol. Cancer Res. 2003;1:247–261. [PubMed] [Google Scholar]
- 34.Shinbo Y., Taira T., Niki T. DJ-1 restoresp53 transcription activity inhibited by Topors/p53BP3. Int. J. Oncol. 2005;26:641–648. [PubMed] [Google Scholar]
- 35.Xu J., Zhong N., Wang H. The Parkinson's disease-associated DJ-1 protein is a transcriptional co-activator that protects against neuronal apoptosis. Hum. Mol. Genet. 2005;14:1231–1241. doi: 10.1093/hmg/ddi134. [DOI] [PubMed] [Google Scholar]
- 36.Zhong N., Kim C.Y., Rizzu P. DJ-1 transcriptionally up-regulates the human tyrosine hydroxylase by 11 inhibiting the sumoylation of pyrimidine tract-binding protein associated splicing factor. J. Biol. Chem. 2006;281:20940–20948. doi: 10.1074/jbc.M601935200. [DOI] [PubMed] [Google Scholar]
- 37.Clements C.M., McNally R.S., Conti B.J. DJ-1, acancer- and Parkinson's disease-associated protein, stabilizes the antioxidant transcriptional master regulator Nrf2. Proc. Natl. Acad. Sci. USA. 2006;103:15091–15096. doi: 10.1073/pnas.0607260103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tillman J.E., Yuan J., Gu G. DJ-1 binds androgen receptor directly and mediates its activity inhormonally treated prostate cancer cells. Cancer Res. 2007;67:4630–4637. doi: 10.1158/0008-5472.CAN-06-4556. [DOI] [PubMed] [Google Scholar]
- 39.Fan J., Ren H., Jia N. DJ-1decreases Bax expression through repressing p53 transcriptional activity. J. Biol. Chem. 2008;283:4022–4030. doi: 10.1074/jbc.M707176200. [DOI] [PubMed] [Google Scholar]
- 40.Ishikawa S., Taira T., Takahashi-Niki K. Human DJ-1-specific transcriptional activation of tyrosine hydroxylase gene. J. Biol. Chem. 2010;285:39718–39731. doi: 10.1074/jbc.M110.137034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yamaguchi S., Yamane T., Takahashi-Niki K. Transcriptional activation of low-density lipoprotein receptor gene by DJ-1 and effect of DJ-1 on cholesterol homeostasis. PLoS One. 2012;7:e38144. doi: 10.1371/journal.pone.0038144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yamane T., Suzui S., Kitaura H. Transcriptional activation of the cholecystokinin gene by DJ-1 through interactionof DJ-1 with RREB1 and the effect of DJ-1 on the cholecystokinin level in mice. PLoS One. 2013;8:e78374. doi: 10.1371/journal.pone.0078374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yamane T., Sugimoto N., Maita H. Identification of DJ-1-associated regions on human genes from SH-SY5Y cells using chromatin immunoprecipitation sequence technique. Mol. Biol. 2013;3:15. [Google Scholar]
- 44.Yamane T., Murao S., Kato-Ose I. Transcriptional regulation of the legumain gene by p53 in HCT116 cells. Biochem. Biophys. Res. Commun. 2013;438:613–618. doi: 10.1016/j.bbrc.2013.08.007. [DOI] [PubMed] [Google Scholar]
- 45.Yamane T., Yamamoto Y., Nakano Y. Expression and protease activity of mouse legumain are regulated by the oncogene/transcription co-activator, DJ-1, through p53 and cleavage of annexin A2 is increased in DJ-1-knockout cells. Biochem. Biophys. Res. Commun. 2015;467:472–477. doi: 10.1016/j.bbrc.2015.10.032. [DOI] [PubMed] [Google Scholar]
- 46.Wang J., Wang J., Zhang H.R. Proteomic analysis of seminal plasma from asthenozoospermia patients reveals proteins that affect oxidative stress responses and semen quality. Asian J. Androl. 2009;11:484–491. doi: 10.1038/aja.2009.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wagenfeld A., Yeung C.H., Strupat K. Shedding of a rat epididymal sperm protein associated with infertility induced by ornidazole and alpha-chlorohydrin. Biol. Reprod. 1998;8:1257–1265. doi: 10.1095/biolreprod58.5.1257. [DOI] [PubMed] [Google Scholar]
- 48.Welch J.E., Barbee R.R., Roberts N.L. SP22: a novel fertility protein from a highly conserved gene family. J. Androl. 1998;19:385–393. [PubMed] [Google Scholar]
- 49.Wagenfeld A., Yeung C.H., Shivaji S. Expression and cellular localization of contraception associated protein. J. Androl. 2000;21:954–963. [PubMed] [Google Scholar]
- 50.Okada M., Matsumoto K., Niki T. A target protein for an endocrine disrupter, participates in the fertilization in mice. Biol. Pharm. Bull. 2002;25:853–856. doi: 10.1248/bpb.25.853. [DOI] [PubMed] [Google Scholar]
- 51.Klinefelter G.R., Welch J.E., Perreault S.D. Localization of the sperm protein SP22 and inhibition of fertility in vivo and in vitro. Andrology. 2002;23:48–63. doi: 10.1002/jand.2002.23.1.48. [DOI] [PubMed] [Google Scholar]
- 52.Yoshida K., Sato Y., Yoshiike M. Immunocytochemical localization of DJ-1 in human male reproductive tissue. Mol. Reprod. Dev. 2003;66:391–397. doi: 10.1002/mrd.10360. [DOI] [PubMed] [Google Scholar]
- 53.Smith R., Johansen H.T., Nilsen H. Intra- and extracellular regulation of activity and processing of legumain by cystatin E/M. Biochimie. 2012;94:2590–2599. doi: 10.1016/j.biochi.2012.07.026. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material