Abstract
Small cell lung cancer (SCLC) is an aggressive neuroendocrine tumor characterized by rapid progression. The mechanisms that lead to a shift from initial therapeutic sensitivity to ultimate therapeutic resistance are poorly understood. Although the SCLC genomic landscape led to the discovery of promising agents targeting genetic alterations that were already under investigation, results have been disappointing. Achievements in targeted therapeutics have not been observed for over 30 years. Therefore, the underlying disease biology and novel targets urgently require a better understanding. Epigenetic regulation is deeply involved in the cellular plasticity that could shift tumor cells to the malignant phenotype. We have focused on a histone modifier, LSD1, that is overexpressed in SCLC and is a potent therapeutic target. Interestingly, the LSD1 splice variant LSD1+8a, the expression of which has been reported to be restricted to neural tissue, was detected and was involved in the expression of neuroendocrine marker genes in SCLC cell lines. Cells with high expression of LSD1+8a were resistant to CDDP and LSD1 inhibitor. Moreover, suppression of LSD1+8a inhibited cell proliferation, indicating that LSD1+8a could play a critical role in SCLC. These findings suggest that LSD1+8a should be considered a novel therapeutic target in SCLC.
Abbreviations: LSD1, lysine specific demethylase 1; KDM, lysine demethylase; SCLC, small cell lung cancer; NSCLC, non-small cell lung cancer
Keywords: LSD1, LSD1+8a, KDM, SCLC, Neuroendocrine marker, Resistance to chemotherapy
Highlights
-
•
LSD1+8a, which was reported to be restricted to neural tissue, is expressed in SCLC.
-
•
LSD1+8a is involved in the expression of neuroendocrine marker genes.
-
•
SCLC cell lines with high expression of LSD1+8a were resistant to chemotherapy.
1. Introduction
Small cell lung cancer (SCLC), accounting for approximately 15% of all lung cancers, is a neuroendocrine carcinoma that is clinically characterized by aggressive behavior, rapid growth, spread to distant organs, and poor prognosis [1]. Although the gold standard treatment of SCLC is platinum-based chemotherapy that achieves robust responses in initial treatment, the development of resistance and eventual relapse arises rapidly in the majority of patients [2]. Moreover, there have been no significant advances in the treatment of this disease during the past 30 years. Consequently, the prognosis of SCLC remains poor, with an overall 5-year survival rate of less than 10% [3].
Research over the last several decades has been focused on genetic regulation, as DNA sequencing based strategies have led to the discovery of genetic alterations in lung cancer. In fact, treatments against genetic alterations such as EGFR, EML-ALK, RET, and ROS all associated with non-small cell lung cancer (NSCLC), have been developed and have demonstrated dramatic clinical response [4], [5], [6], [7], [8]. Despite the fact that large scale genetic analyses have been performed in SCLC, numerous molecular approaches targeting genetic alterations in SCLC have failed to demonstrate clinical impact [2], [9], [10], [11], suggesting that a different point of focus should be considered to obtain a better understanding of SCLC. The focus on biological pathways that drive proliferation, survival, and resistance to chemotherapy has recently shifted from genetic to epigenetic regulation. Epigenetic regulation is responsible for the cellular plasticity that could be associated with metastatic ability and acquired resistance to chemotherapy [12]. Therefore, epigenetic dysregulation, resulting in dramatic changes in gene expression, could push tumor cells towards a malignant phenotype.
Histone methylation, which is tightly regulated by lysine methyltransferase (KMT) and lysine demethylase (KDM), is part of a major class of epigenetic mechanism and is critical for gene expression, cell cycle, and differentiation [13]. Lysine-specific demethylase 1 (LSD1)/KDM1 is a flavin adenine dinucleotide-dependent KDM that can remove mono- and dimethylation from histone H3 at lysine 4 (H3K4me1/2), leading to gene suppression [14]. LSD1 has also been recognized as a co-activator mediated by demethylation of histone H3 at lysine 9 (H3K9), leading to gene activation [15], [16]. LSD1 is ubiquitously expressed and recognized as an essential epigenetic regulator of pluripotency in ESCs [17]. However, the molecular mechanism of dual substrate specificity (to H3K4 and H3K9) has remained unknown because structural studies do not support the possibility that LSD1 can mediate H3K9 demethylation [18]. It has recently been discovered that LSD1+8a, which is an alternative LSD1 splice variant, containing a 12-bp exon E8a, is responsible for demethylation of H3K9 [19]. Interestingly, LSD1+8a was reported to be restricted to neural tissues where it plays an important role in neural differentiation [20], [21].
LSD1 is overexpressed in several human cancers including SCLC [22]. However, the expression of LSD1+8a in human cancer has not been reported. Although mechanisms of neural differentiation mediated by LSD1+8a have been investigated, the epigenetic role of LSD1+8a in SCLC has not been elucidated. Proof of neuroendocrine differentiation by immunohistochemistry is not required for the diagnosis of SCLC. However, the majority of these cases display a neural differentiation phenotype, suggesting that genes involved in neural differentiation must have a critical role in SCLC; this could be mediated by LSD1+8a. Herein, we report that LSD1+8a plays an important role in neural differentiation in SCLC.
2. Materials and methods
2.1. Cell culture
Small cell lung cancer (SCLC) cell lines, SBC3 and SBC5, were obtained from the Japan Health Sciences Foundation, Health Science Resources Bank (HSRB; JCRB0819 and JCRB0818) and PC9 was kindly provided by Dr. Kazuto Nishio (Department of Genome Biology, School of Medicine, Kinki University, Osaka). SCLC (H69, H446, H187, H82), NSCLC (A549, HCC827, Calu-3, H2170), and cervical cancer cell lines (Hela) were purchased from the American Type Culture Collection. All cells were cultured in RPMI 1640 supplemented with 10% heat-inactivated fetal bovine serum (FBS) and 1% (v/w) penicillin/streptomycin and incubated at 37 °C in a 5% CO2 atmosphere. The cells were routinely tested for Mycoplasma contamination using the MycoAlert Mycoplasma Detection Kit (Cambrex, Rockland, ME, USA).
2.2. Quantitative real-time PCR
Total RNA was extracted from cell lines using miRvana miRNA Isolation kit (Ambion, Austin, TX, USA) according to the manufacturer's instructions. Total RNA (500 ng) was reverse-transcribed to cDNA using the Revertra cDNA synthesis kit (Toyobo, Osaka, Japan). Quantitative real-time PCR (qPCR) was performed using SYBR Green Master Mix (Applied Biosystems, Foster City, CA, USA). Cycling conditions were as follows: denature hold at 95 °C for 20 s, 40 cycles of amplification (denature at 95 °C for 30 s, annealing and extension at 60 °C for 30 s), and melting-curve analysis. qPCR was performed in triplicate and the expression level of β-actin was used as an internal control. The primer sequences used to analyze the gene expression using qPCR are provided below.
LSD1
Forward, 5′ - TCGGGGCTCTTATTCCTATG - 3′
Reverse, 5′ - ATCGTATGTTCTCCCGCAAA - 3′
LSD1+8a (Ref. [19])
Forward, 5′ - GCTGTGGTCAGCAAACAAG - 3′
Reverse, 5′ - CTCTTTAGGAACCTTGACAGTGTC - 3′
CHGA
Forward, 5′ - TAAAGGGGATACCGAGGTGATG - 3′
Reverse, 5′ - TCGGAGTGTCTCAAAACATTCC - 3′
NCAM
Forward, 5′ - GGCATTTACAAGTGTGTGGTTAC - 3′
Reverse, 5′ - TTGGCGCATTCTTGAACATGA - 3′
SYP
Forward, 5′ - CTCGGCTTTGTGAAGGTGCT - 3′
Reverse, 5′ - CTGAGGTCACTCTCGGTCTTG - 3′
ENO2
Forward, 5′ - AGGTGCAGAGGTCTACCATAC - 3′
Reverse, 5′ - AGCTCCAAGGCTTCACTGTTC - 3′
GRP
Forward, 5′ - ACCGTGCTGACCAAGATGTA - 3′
Reverse, 5′ - TCAGGCTCCCTCTCTCAGAA - 3′
B3CAT1
Forward, 5′ - CTCCTTCGAGAACTTGTCACC - 3′
Reverse, 5′ - GGGTCAGTGAAGCCCTTCTT - 3′
SOX2
Forward, 5′ - TACAGCATGTCCTACTCGCAG - 3′
Reverse, 5′ - GAGGAAGAGGTAACCACAGGG - 3′
POU5F1B
Forward, 5′ - GAGTGAGAGGCAACCTGGAG - 3′
Reverse, 5′ - GCCGGTTACAGAACCACACT - 3′
KLF4
Forward, 5′ - CCCAATTACCCATCCTTCCT - 3′
Reverse, 5′ - ACGATCGTCTTCCCCTCTTT - 3′
MYC
Forward, 5′ - TTCGGGTAGTGGAAAACCAG - 3′
Reverse, 5′ - CACCGAGTCGTAGTCGAGGT - 3′
CD44
Forward, 5′ - TCCCAGACGAAGACAGTCCCTGGAT - 3′
Reverse, 5′ - CACTGGGGTGGAATGTGTCTTGGTC - 3′
PROM1
Forward, 5′ - GGCCCAGTACAACACTACCAA - 3′
Reverse, 5′ - CGCCTCCTAGCACTGAATTGATA - 3′
Actin
Forward, 5′ - CTCTTCCAGCCTTCCTTCCT - 3′
Reverse, 5′ - AGCACTGTGTTGGCGTACAG - 3′
2.3. RNA interference assay
siLSD1+8a #1 sequence (5ʹ - AACCUUGACAGUGUCAGCUUGUCCG - 3ʹ), siLSD1+8a #2 sequence (5ʹ - CAAGCUGACACUGUCAAGGUUCCUA - 3ʹ), and control siRNA (50 nM) were transfected into cells using Lipofectamine RNAiMAX (Life Technologies, Inc. Grand Island, NY, USA) according to the manufacturer's instructions.
2.4. Western blot analysis
The antibodies used for western blot analysis were anti-LSD1 antibody (1:1000, Cell Signaling Technology, Beverly, MA, USA) and mouse anti-GAPDH monoclonal antibody (1:3000, Wako, Osaka, Japan). Cells were lysed in M-PER (Thermo Scientific, Rockford, IL, USA) supplemented with protease inhibitors (Roche) and phosphatase inhibitors (Sigma-Aldrich, St. Louis, MO). Proteins were separated on a 4–20% polyacrylamide gradient-SDS gel, transferred on polyvinylidene difluoride membranes, and blocked in TBST (Tris-buffered saline and Tween20; 25 mM Tris, pH 7.4, 136 mM NaCl, 5 mM KCl, and 0.1% Tween) containing 5% milk. The antibodies were used at each dilution, as described above, in 5% milk/TBST. Blots were incubated with primary antibodies for 12 h at room temperature. Blots were washed (three times) with 5% milk/TBST and were incubated with the appropriate horseradish peroxidase-conjugated antibodies. Bound antibodies were detected with the ECL Prime Western Blotting System (RPN2232; GE Healthcare, Little Chalfont, Buckinghamshire, UK), and luminescent images were analyzed using a lumino imager (LAS-4000 mini; Fuji Film Inc. Tokyo, Japan).
2.5. Cell proliferation assay
Cells (1×103) were seeded onto 96-well microtiter plates followed by transfection with 50 nM siLSD1+8a or control siRNA. Cell proliferation assays were performed at 24, 48, and 72 h using WST-8 (cell counting kit-8, Dojindo Laboratories, Kumamoto, Japan). After 4 h, the optical density was measured at 450 nm with a microplate reader (Bio-Rad Laboratories, Hercules, CA, USA).
2.6. Chemosensitivity assay
Cisplatin (CDDP) was purchased from Sigma-Aldrich (St. Louis, MO). The LSD1 inhibitor, S2101, was purchased from Calbiochem (EMD-Millipore, Billerica, MA, USA). Cells (1×103) were seeded in 96-well microtiter plates in the absence or the presence of various concentrations of CDDP and S2101. After 72 h or 7 days of incubation, 10 μl of WST-8 (cell counting kit-8, Dojindo Laboratories, Kumamoto, Japan) was added to each well. After 4 h, the optical density was measured at 450 nm using a microplate reader (Bio-Rad Laboratories, Hercules, CA, USA). The results are expressed as the percentage of cell viability.
2.7. Patient sample collection
Tumor samples were obtained from SCLC patients who visited the Juntendo University Hospital. Patients gave consent before enrolment in the study under the Juntendo University Institutional Review Board (IRB)-approved protocols. All experiments conformed to the principles set out in the WMA Declaration of Helsinki.
2.8. Expression analysis of LSD1 splicing isoforms using public RNA-sequencing datasets
To examine RNA expression of LSD1 and its splicing isoforms, we downloaded RNA-sequencing (RNA-seq) data from the Cancer Cell Line Encyclopedia (CCLE) using GeneTorrent software (https://cghub.ucsc.edu/datasets/ccle.html). We obtained binary alignment/map (BAM) format files of the following 14 NSCLC cell lines: A549, DMS53, DMS79, DMS114, DMS153, H69, H82, H209, H446, H510, H596, H1155, SBC5, and SHP-77. The BAM files were visualized and analyzed by the Integrative Genomic Viewer (IGV: https://www.broadinstitute.org/igv/). Given that LSD1+8a is located from base pair 23,392,553 to 23,392,564 (12 base pairs) on chromosome 1, we counted the number of reads containing this region with exact splicing junctions at both ends. Cell lines that expressed more than 10 reads of LSD1+8a were allocated to the "LSD1+8a expression +" group (DMS79, H446, H1155, and SHP-77), and those without any reads of LSD1+8a were labeled "LSD1+8a expression -" group (A549, DMS114, H69, H209, and SBC5). We then compared these two groups using the Cufflinks (http://cole-trapnell-lab.github.io/cufflinks/) analysis software suite. Transcripts were assembled by the Cufflinks program for each cell line, and the obtained transcript information was merged into one file using Cuffmerge software. The Cuffdiff program was used to examine differentially expressed genes and their isoforms, comparing LSD1+8a expression + and - groups.
2.9. Statistical analysis
Before analyzing correlations between neuroendocrine markers, the Smirnov-Grubbs test was used to eliminate outliers. Statistical analysis was performed using a two-tailed Student's t-test. To compare multiple groups, a one-way analysis of variance (ANOVA) test was applied. Differences between means were considered statistically significant at P<0.05.
3. Results
3.1. Expression of LSD1 and variant isoform LSD1+8a
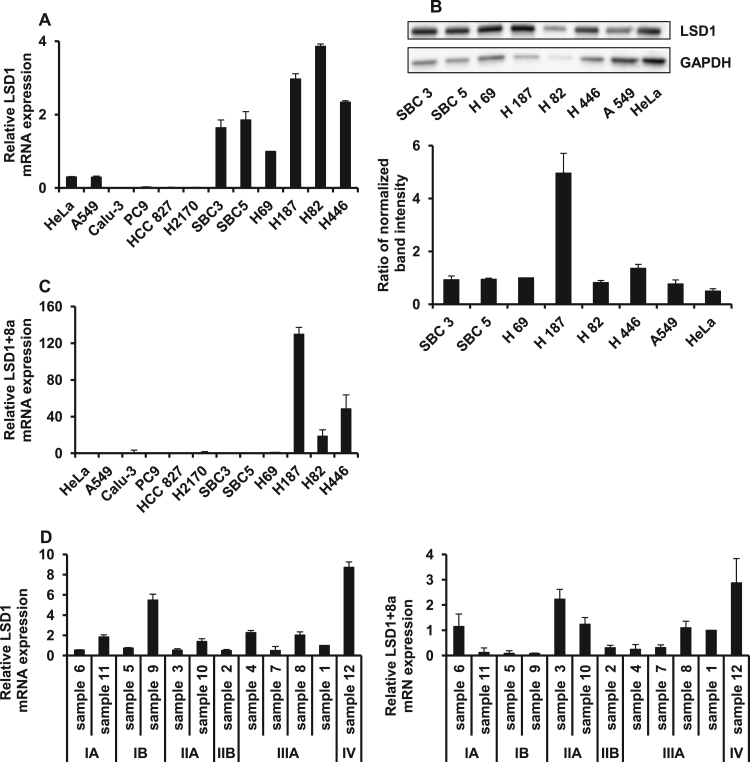
LSD1+8a has been reported to be restricted to neural tissue; to investigate if it is expressed in cancer cells, a database search was performed using the Cancer Cell Line Encyclopedia (CCLE). RNA sequencing data was analyzed in several human lung cancer cell lines, revealing that LSD1+8a was also expressed in a small number of lung cancer cell lines. Although the differences did not reach statistical significance, the expression of neuroendocrine marker genes tended to be higher in cell lines expressing LSD1+8a (Table 1). In contrast, the expression of stem cell marker genes tended to be lower in these cells (Table 2), suggesting that LSD1+8a expressing cells could exhibit neural differentiation. To investigate the expression of LSD1 and LSD1+8a in cancer cell lines, qPCR was performed for 12 human cancer cell lines (Fig. 1A and C). LSD1 expression was also validated using western blotting (Fig. 1B). The expression of LSD1 was higher in SCLC cell lines compared to that in NSCLC cell lines and a cervical cancer cell line. Interestingly, LSD1+8a was expressed in only a small subset of SCLC cell lines characterized by a neural differentiation phenotype (Fig. 1C). LSD1 and LSD1+8 expression was also investigated in 12 human surgical samples from patients with SCLC (Fig. 1D). Although there was no correlation between LSD1+8a expression and clinical stage in SCLC, LSD1+8a was detected in a small portion of clinical samples.
Table 1.
Neuroendocrine marker gene expression from RNA sequencing data in the Cancer Cell Line Encyclopedia (CCLE).
| Gene | LSD1+8a expression – | LSD1+8a expression+ | Log2 | P-value |
|---|---|---|---|---|
| (H69, SBC5, H209, DMS114, A549) | (H446, SHP-77, H1155, DMS79) | (fold change) | ||
| CHGA | 17.40 | 70.22 | 2.01 | 0.020 |
| NCAM1 | 19.44 | 52.94 | 1.45 | 0.189 |
| ENO2 | 0.55 | 0.90 | 0.71 | 0.529 |
| SYP | 17.78 | 55.16 | 1.63 | 0.077 |
| GRP | 80.94 | 107.83 | 0.41 | 0.802 |
| B3GAT1 | 1.79 | 16.42 | 3.20 | 0.033 |
RNA sequencing data in CCLE was analyzed for neuroendocrine marker genes in several human lung cancer cell lines including A549, DMS79, DMS114, H1155, H209, H446, H69, SBC5, and SHP-77.
Table 2.
Stem cell marker gene expression from RNA sequencing data in CCLE.
| Gene | LSD1+8a expression – | LSD1+8a expression+ | Log2 | P-value |
|---|---|---|---|---|
| (H69, SBC5, H209, DMS114, A549) | (H446, SHP-77, H1155, DMS79) | (fold change) | ||
| SOX2 | 65.15 | 10.47 | −2.64 | 0.134 |
| POU5F1B | 0.88 | 0.63 | −0.49 | 0.636 |
| KLF4 | 5.14 | 0.80 | −2.68 | 0.088 |
| MYC | 29.44 | 35.58 | 0.27 | 0.727 |
| CD44 | 25.66 | 7.94 | −1.69 | 0.096 |
| PROM1 | 17.11 | 1.55 | −3.46 | 0.003 |
RNA sequencing data in CCLE was analyzed for stem cell marker genes in several human lung cancer cell lines including A549, DMS79, DMS114, H1155, H209, H446, H69, SBC5, and SHP-77.
Fig. 1.
Expression of LSD1 and LSD1+8a in cancer cell lines and human samples. A) LSD1 mRNA expression in SCLC, NSCLC, and cervical cancer cell lines was analyzed by qPCR. B) LSD1 protein expression in SCLC, NSCLC, and cervical cancer cell lines was analyzed by western blotting. GAPDH was used as loading control. The bar graph on the bottom shows relative abundance of LSD1, which was determined by calculating the ratio of LSD1 intensity to that of GAPDH. C) LSD1+8a mRNA expression in SCLC cell lines was analyzed by qPCR. D) LSD1 and LSD1+8a mRNA expression in surgical specimens in patients with SCLC was analyzed by qPCR. Data are presented as the mean±S.D. in triplicate experiments.
3.2. Correlation of LSD1+8a and neuroendocrine markers
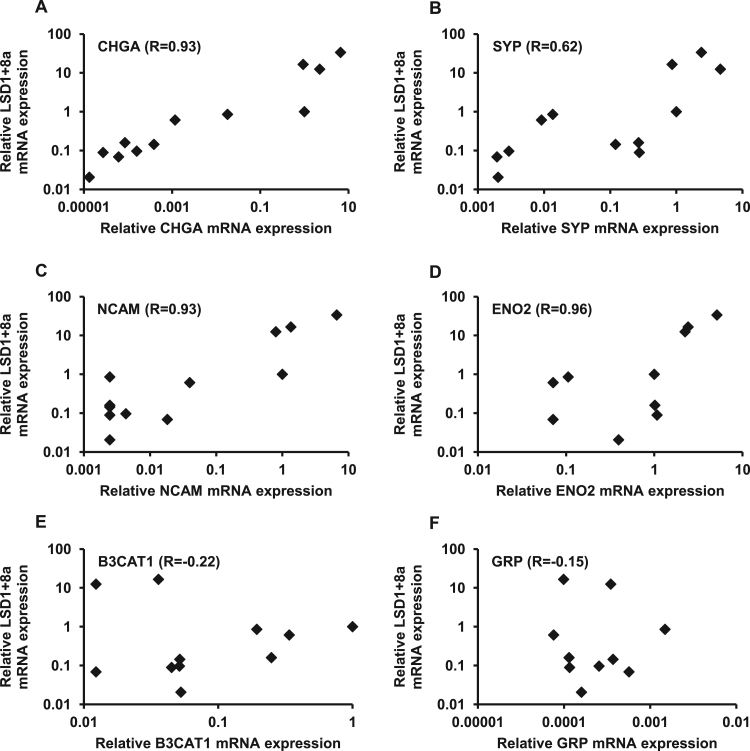
To validate the database search results, the expression of genes involved in neural differentiation, including those for chromogranin A (CHGA), synaptophysin (SYP), neural cell adhesion molecule (NCAM), enolase 2 (ENO2), beta-1,3-glucuronyltransferase 1 (B3CAT1), and gastrin-releasing peptide (GRP), was analyzed by qPCR. The correlation between the expression of these neural markers and LSD1+8a was then investigated in human cancer cell lines (Fig. 2). The Pearson correlation coefficients were significant between LSD1+8a and the expression of CHGA (Fig. 2A, R=0.93, p<0.05), SYP (Fig. 2B, R=0.62, p<0.05), NCAM (Fig. 2C, R =0.93, p<0.05), and ENO2 (Fig. 2D, R=0.93, p<0.05) but not B3CAT1 (Fig. 2E, R=−0.22) and GRP (Fig. 2F, R=−0.15). Thus, a significant positive correlation was observed between LSD1+8a and neuroendocrine marker genes in human cancer cell lines, suggesting that LSD1+8a could mediate the expression of neuroendocrine marker genes in cancer cell lines.
Fig. 2.
Pearson correlation of LSD1+8a and neuroendocrine marker gene expression in human cancer cell lines. A) Chromogranin A (CHGA), B) synaptophysin (SYP), C) neural cell adhesion molecule (NCAM), D) enolase 2 (ENO2), E) beta-1,3-glucuronyltransferase 1 (B3CAT1), and F) gastrin-releasing peptide (GRP) mRNA expression in 12 human cancer cell lines was analyzed by qPCR. Pearson's correlation coefficient is displayed in the upper left corner.
3.3. Suppression of LSD1+8a repressed the expression of neuroendocrine markers
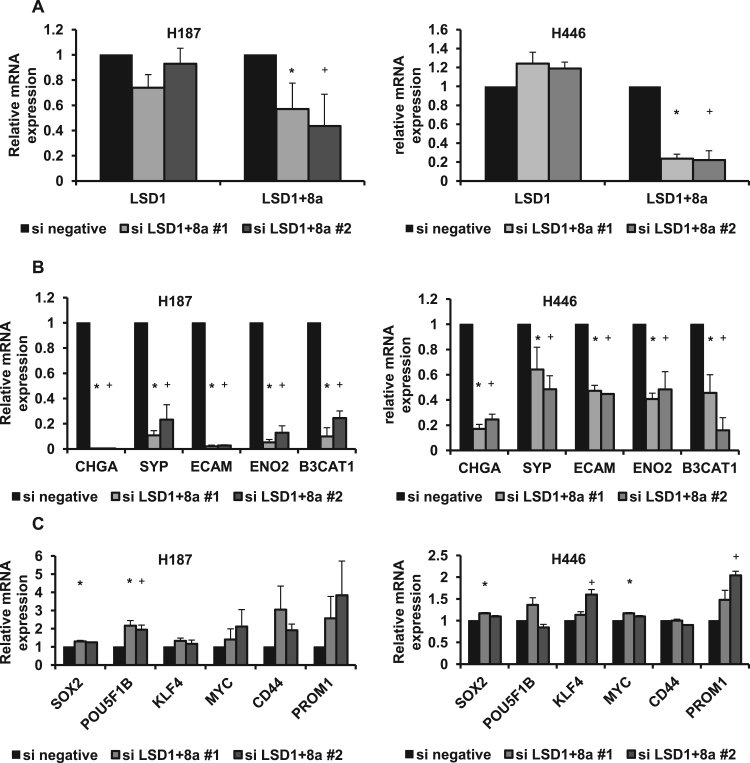
To investigate whether LSD1+8a is involved in the expression of neuroendocrine marker genes in SCLC, an siRNA targeting LSD1 exon 8a was designed to specifically knockdown the LSD1+8a isoform. H187 and H446 cells were transfected with specific siRNA targeting LSD1 exon 8a or control siRNA, and qPCR was performed (Fig. 3A and B). As expected, suppression of LSD1+8a led to a decrease in LSD1+8a mRNA expression but not LSD1 mRNA expression. This knockdown also led to a significant decrease in the expression of CHGA, ENO2, NCAM, SYP, and B3CAT1, suggesting that LSD1+8a could regulate the expression of neuroendocrine marker genes in SCLC. On the other hand, the expression of stem cell marker genes tended to be higher in these cells transfected with siLSD1+8a (Fig. 3C). Although the differences in some of stem cell marker genes did not reach statistical significance, the trend is just the opposite of the neuroendocrine marker genes, suggesting that LSD1+8a could play an important role in neural differentiation in SCLC.
Fig. 3.
Effect of LSD1+8a suppression by siRNA on expression of neuroendocrine marker genes. A) LSD1 and LSD1+8a mRNA expression was analyzed by qPCR in H187 and H446 cells transfected with specific siRNA against LSD1 exon 8a or control siRNA. The expression of each control mRNA was set to one. B) Chromogranin A (CHGA), synaptophysin (SYP), neural cell adhesion molecule (NCAM), enolase 2 (ENO2), and beta-1,3-glucuronyltransferase 1 (B3CAT1) mRNA expression was analyzed by qPCR in H187 and H446 transfected with specific siRNA against LSD1 exon 8a. Expression of each control mRNA was set to one. C) SOX2, POU5F1B, KLF4, MYC, CD44, and PROM1 mRNA expression was analyzed by qPCR in H187 and H446 transfected with specific siRNA against LSD1 exon 8a. Expression of each control mRNA was set to one. Data are presented as the mean±S.D. in triplicate experiments. *P<0.05 vs siLSD1+8a # 1. +P<0.05 vs siLSD1+8a # 2.
3.4. Suppression of LSD1+8a repressed the cell viability
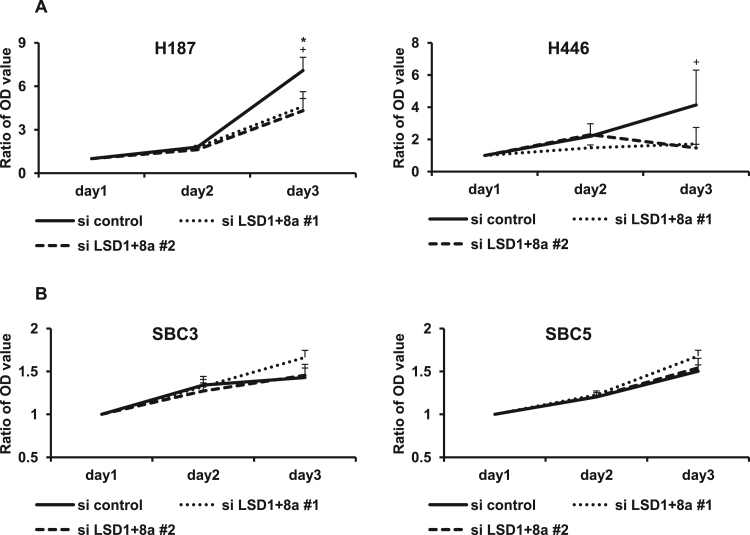
To investigate how suppression of LSD1+8a expression affects SCLC cell viability, a proliferation assay was performed following transfection with siLSD1+8a or control siRNA (Fig. 4A and B). Interestingly, compared to in control conditions, siLSD1+8a significantly repressed cell viability in H187 and H446 cells with LSD1+8a expression. However, there were no effects on SBC3 and SBC5 cells without LSD1+8a expression, suggesting that suppression of LSD1+8a inhibits cell viability in SCLC with LSD1+8a expression.
Fig. 4.
Effect of LSD1+8a suppression by siRNA on the cell viability. A) A cell proliferation assay was performed using H187 and H446 cells transfected with siLSD1+8a or control siRNA at 24, 48, and 72 h. The optical density at 24 h was set to one. B) A cell proliferation assay was performed using SBC3 and SBC5 cells transfected with siLSD1+8a or control siRNA at 24, 48, and 72 h. The optical density at 24 h was set to one. Data are presented as the mean±S.D. in triplicate experiments. *P<0.05 vs siLSD1+8a # 1. +P<0.05 vs siLSD1+8a # 2.
3.5. LSD1+8a induced resistance to chemotherapeutic agents
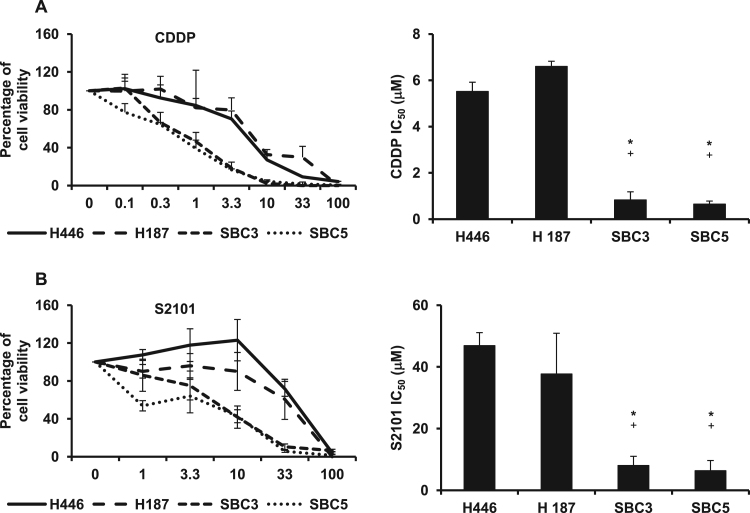
To evaluate differences in sensitivity to chemotherapeutic agents between SCLC cell lines with high and those with low LSD1+8a expression, a chemosensitivity assay was performed using CDDP and LSD1 inhibitor, S2101, which was developed as a TCP-analogue with LSD1 inhibitory activity (Fig. 5A and B). The IC50 values of CDDP against H446 and H187 were 5.51±0.40 μM and 6.59±0.22 μM, respectively, whereas those against SBC3 and SBC5 were 0.82±0.35 μM and 0.64±0.13 μM, respectively. The IC50 of S2101 against H446 and H187 were 46.87±4.23 μM and 37.69±13.21 μM, respectively, whereas those against SBC3 and SBC5 were 8.01±3.00 μM and 6.34±3.31 μM, respectively. These results indicated that SCLC cells with high LSD1+8a isoform expression were more resistant to CDDP and S2101. Taken together, neural differentiation induced by LSD1+8a in SCLC could lead to CDDP and LSD1 inhibitor resistance.
Fig. 5.
In vitro chemosensitivity assay in SCLC cell lines. SCLC cell lines including H446, H187, SBC3, and SBC5 were seeded in 96-well flat bottom plates and cultured for 72 h or 7 days in the absence or presence of various concentrations of cisplatin (CDDP; A) and LSD1 inhibitor (S2101; B). IC50 values are presented as the mean±S.D. in triplicate experiments. *P<0.05 vs H446. +P<0.05 vs H187.
4. Discussion
In this study, we detected LSD1+8a in SCLC cell lines and in clinical specimens; LSD1+8a mRNA has been reported to be restricted to neural tissues. There were significant positive correlations between LSD1+8a and the expression of neuroendocrine differentiation genes in human cancer cell lines. The suppression of LSD1+8a by siRNA led to a decrease in the expression of neuroendocrine marker genes and inhibited cell proliferation in SCLC cell lines. Moreover, cells with high expression of LSD1+8a were resistance to CDDP and LSD1 inhibitor, suggesting that LSD1+8a could play a critical role in SCLC.
LSD1+8a has been reported to have a key role in differentiation in neural tissues [20], [21]. In our study, neuroendocrine marker genes were downregulated by suppression of LSD1+8a in SCLC cell lines. This finding indicates that LSD1+8a is involved in neural differentiation in SCLC. However, it is unclear how LSD1+8a can regulate neuroendocrine marker expression. Benoit et al. revealed that LSD1+8a is essential for neural maturation by activating target gene expression though H3K9me2 demethylation [19]. The simplest hypothesis is that LSD1+8a regulates specific subsets of target genes such as CHGA, SYP, and NCAM by demethylation of the H3K9, which is associated with gene activation. However, Toffolo et al. demonstrated that phosphorylation of the second residue encoded by exon 8a converts LSD1+8a into a dominant negative enzyme isoform that is unable to repress gene transcription though dissociation of CoREST and the HDAC1/2 corepressor complex [21]. Future studies are required to elucidate the mechanisms of neural differentiation induced by LSD1+8a in SCLC.
In general, SCLC is considered to be the most aggressive form of lung cancer and is associated with a less favorable outcome [1]. However, there are certain cases that appear sensitive to chemotherapy and irradiation, which is generally associated with a favorable outcome. It is difficult to distinguish SCLC subtypes according to prognosis by histopathological findings. Expression profiling has been reported to have the ability to predict prognosis more accurately. Large-scale gene expression analyses revealed that neuroendocrine gene expression is associated with a less favorable outcome in lung cancer [23], [24]. It has also been recently reported that patients with SCLC with low neuroendocrine marker expression, including for CHGA, SYP, and NCAM, comprise an identified subset with good prognosis [25]. We demonstrated significant positive correlations between LSD1+8a expression and neuroendocrine genes, including those of CHGA, SYP, and NCAM. Moreover, SCLC cell lines with high LSD1+8a isoform expression were resistant to LSD1 inhibitor, which is a novel target for cancer therapy [26], [27], [28]. These findings suggested that high expression of LSD1+8a could be considered not only one of the candidates of surrogate marker for the prediction of prognosis but also a novel target that should be focused to improve outcome in patients with SCLC.
The most fascinating aspect of this study is that it is the first report showing that LSD1+8a is expressed in SCLC, wherein it plays a critical role. These results contribute to the understanding of the epigenetic modifier, LSD1+8a, which should be considered a novel surrogate marker and novel therapeutic target for SCLC.
Acknowledgements
This study was supported by Grants-in-Aid for Scientific Research No. 24591176 (Kazuhisa Takahashi) from the Ministry of Education, Culture, Sports, Science, and Technology of Japan.
Footnotes
Transparency document associated with this article can be found in the online version at doi:10.1016/j.bbrep.2016.11.015.
Appendix A. Transparency document
Supplementary material
.
Supplementary material
.
Supplementary material
.
Supplementary material
.
Supplementary material
.
Supplementary material
.
Supplementary material
.
Supplementary material
.
Supplementary material
.
Supplementary material
.
Supplementary material
.
Supplementary material
.
Supplementary material
.
Supplementary material
.
Supplementary material
.
Supplementary material
.
Supplementary material
.
Supplementary material
.
Supplementary material
.
References
- 1.Semenova E.A., Nagel R., Berns A. Origins, genetic landscape, and emerging therapies of small cell lung cancer. Genes Dev. 2015;29:1447–1462. doi: 10.1101/gad.263145.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sgambato A., Casaluce F., Maione P., Rossi A., Sacco P.C., Panzone F., Ciardiello F., Gridelli C. Medical treatment of small cell lung cancer: state of the art and new development. Expert Opin. Pharm. 2013;14:2019–2031. doi: 10.1517/14656566.2013.823401. [DOI] [PubMed] [Google Scholar]
- 3.Shepherd F.A., Crowley J., Van Houtte P., Postmus P.E., Carney D., Chansky K., Shaikh Z., Goldstraw P. The International Association for the Study of Lung Cancer lung cancer staging project: proposals regarding the clinical staging of small cell lung cancer in the forthcoming (seventh) edition of the tumor, node, metastasis classification for lung cancer. J. Thorac. Oncol. 2007;2:1067–1077. doi: 10.1097/JTO.0b013e31815bdc0d. [DOI] [PubMed] [Google Scholar]
- 4.Mok T.S., Wu Y.L., Thongprasert S., Yang C.H., Chu D.T., Saijo N., Sunpaweravong P., Han B., Margono B., Ichinose Y., Nishiwaki Y., Ohe Y., Yang J.J., Chewaskulyong B., Jiang H., Duffield E.L., Watkins C.L., Armour A.A., Fukuoka M. Gefitinib or carboplatin-paclitaxel in pulmonary adenocarcinoma. N. Engl. J. Med. 2009;361:947–957. doi: 10.1056/NEJMoa0810699. [DOI] [PubMed] [Google Scholar]
- 5.Maemondo M., Inoue A., Kobayashi K., Sugawara S., Oizumi S., Isobe H., Gemma A., Harada M., Yoshizawa H., Kinoshita I., Fujita Y., Okinaga S., Hirano H., Yoshimori K., Harada T., Ogura T., Ando M., Miyazawa H., Tanaka T., Saijo Y., Hagiwara K., Morita S., Nukiwa T. Gefitinib or chemotherapy for non-small-cell lung cancer with mutated EGFR. N. Engl. J. Med. 2010;362:2380–2388. doi: 10.1056/NEJMoa0909530. [DOI] [PubMed] [Google Scholar]
- 6.Kwak E.L., Bang Y.J., Camidge D.R., Shaw A.T., Solomon B., Maki R.G., Ou S.H., Dezube B.J., Janne P.A., Costa D.B., Varella-Garcia M., Kim W.H., Lynch T.J., Fidias P., Stubbs H., Engelman J.A., Sequist L.V., Tan W., Gandhi L., Mino-Kenudson M., Wei G.C., Shreeve S.M., Ratain M.J., Settleman J., Christensen J.G., Haber D.A., Wilner K., Salgia R., Shapiro G.I., Clark J.W., Iafrate A.J. Anaplastic lymphoma kinase inhibition in non-small-cell lung cancer. N. Engl. J. Med. 2010;363:1693–1703. doi: 10.1056/NEJMoa1006448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Drilon A., Wang L., Hasanovic A., Suehara Y., Lipson D., Stephens P., Ross J., Miller V., Ginsberg M., Zakowski M.F., Kris M.G., Ladanyi M., Rizvi N. Response to Cabozantinib in patients with RET fusion-positive lung adenocarcinomas. Cancer Discov. 2013;3:630–635. doi: 10.1158/2159-8290.CD-13-0035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.A.M.Kamat, Commentary on "Phase II trial of cetuximab with or without paclitaxel in patients with advanced urothelial tract carcinoma." Y.N. Wong, S. Litwin, D. Vaughn, S. Cohen, E.R. Plimack, J. Lee, W. Song, M. Dabrow, M. Brody, H. Tuttle, G. Hudes, University of Pennsylvania, Philadelphia, PA: J Clin Oncol 2012;30(28):3545-51 [Epub 2012 Aug 27], Urol Oncol, 31, 2013, 719. [DOI] [PMC free article] [PubMed]
- 9.Peifer M., Fernandez-Cuesta L., Sos M.L., George J., Seidel D., Kasper L.H., Plenker D., Leenders F., Sun R., Zander T., Menon R., Koker M., Dahmen I., Muller C., Di Cerbo V., Schildhaus H.U., Altmuller J., Baessmann I., Becker C., de Wilde B., Vandesompele J., Bohm D., Ansen S., Gabler F., Wilkening I., Heynck S., Heuckmann J.M., Lu X., Carter S.L., Cibulskis K., Banerji S., Getz G., Park K.S., Rauh D., Grutter C., Fischer M., Pasqualucci L., Wright G., Wainer Z., Russell P., Petersen I., Chen Y., Stoelben E., Ludwig C., Schnabel P., Hoffmann H., Muley T., Brockmann M., Engel-Riedel W., Muscarella L.A., Fazio V.M., Groen H., Timens W., Sietsma H., Thunnissen E., Smit E., Heideman D.A., Snijders P.J., Cappuzzo F., Ligorio C., Damiani S., Field J., Solberg S., Brustugun O.T., Lund-Iversen M., Sanger J., Clement J.H., Soltermann A., Moch H., Weder W., Solomon B., Soria J.C., Validire P., Besse B., Brambilla E., Brambilla C., Lantuejoul S., Lorimier P., Schneider P.M., Hallek M., Pao W., Meyerson M., Sage J., Shendure J., Schneider R., Buttner R., Wolf J., Nurnberg P., Perner S., Heukamp L.C., Brindle P.K., Haas S., Thomas R.K. Integrative genome analyses identify key somatic driver mutations of small-cell lung cancer. Nat. Genet. 2012;44:1104–1110. doi: 10.1038/ng.2396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.George J., Lim J.S., Jang S.J., Cun Y., Ozretic L., Kong G., Leenders F., Lu X., Fernandez-Cuesta L., Bosco G., Muller C., Dahmen I., Jahchan N.S., Park K.S., Yang D., Karnezis A.N., Vaka D., Torres A., Wang M.S., Korbel J.O., Menon R., Chun S.M., Kim D., Wilkerson M., Hayes N., Engelmann D., Putzer B., Bos M., Michels S., Vlasic I., Seidel D., Pinther B., Schaub P., Becker C., Altmuller J., Yokota J., Kohno T., Iwakawa R., Tsuta K., Noguchi M., Muley T., Hoffmann H., Schnabel P.A., Petersen I., Chen Y., Soltermann A., Tischler V., Choi C.M., Kim Y.H., Massion P.P., Zou Y., Jovanovic D., Kontic M., Wright G.M., Russell P.A., Solomon B., Koch I., Lindner M., Muscarella L.A., la Torre A., Field J.K., Jakopovic M., Knezevic J., Castanos-Velez E., Roz L., Pastorino U., Brustugun O.T., Lund-Iversen M., Thunnissen E., Kohler J., Schuler M., Botling J., Sandelin M., Sanchez-Cespedes M., Salvesen H.B., Achter V., Lang U., Bogus M., Schneider P.M., Zander T., Ansen S., Hallek M., Wolf J., Vingron M., Yatabe Y., Travis W.D., Nurnberg P., Reinhardt C., Perner S., Heukamp L., Buttner R., Haas S.A., Brambilla E., Peifer M., Sage J., Thomas R.K. Comprehensive genomic profiles of small cell lung cancer. Nature. 2015;524:47–53. doi: 10.1038/nature14664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Metro G., Duranti S., Fischer M.J., Cappuzzo F., Crino L. Emerging drugs for small cell lung cancer – an update. Expert Opin. Emerg. Drugs. 2012;17:31–36. doi: 10.1517/14728214.2012.656588. [DOI] [PubMed] [Google Scholar]
- 12.Petronis A. Epigenetics as a unifying principle in the aetiology of complex traits and diseases. Nature. 2010;465:721–727. doi: 10.1038/nature09230. [DOI] [PubMed] [Google Scholar]
- 13.Black J.C., Van Rechem C., Whetstine J.R. Histone lysine methylation dynamics: establishment, regulation, and biological impact. Mol. Cell. 2012;48:491–507. doi: 10.1016/j.molcel.2012.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shi Y., Lan F., Matson C., Mulligan P., Whetstine J.R., Cole P.A., Casero R.A. Histone demethylation mediated by the nuclear amine oxidase homolog LSD1. Cell. 2004;119:941–953. doi: 10.1016/j.cell.2004.12.012. [DOI] [PubMed] [Google Scholar]
- 15.Metzger E., Wissmann M., Yin N., Muller J.M., Schneider R., Peters A.H., Gunther T., Buettner R., Schule R. LSD1 demethylates repressive histone marks to promote androgen-receptor-dependent transcription. Nature. 2005;437:436–439. doi: 10.1038/nature04020. [DOI] [PubMed] [Google Scholar]
- 16.Perillo B., Ombra M.N., Bertoni A., Cuozzo C., Sacchetti S., Sasso A., Chiariotti L., Malorni A., Abbondanza C., Avvedimento E.V. DNA oxidation as triggered by H3K9me2 demethylation drives estrogen-induced gene expression. Science. 2008;319:202–206. doi: 10.1126/science.1147674. [DOI] [PubMed] [Google Scholar]
- 17.Whyte W.A., Bilodeau S., Orlando D.A., Hoke H.A., Frampton G.M., Foster C.T., Cowley S.M., Young R.A. Enhancer decommissioning by LSD1 during embryonic stem cell differentiation. Nature. 2012;482:221–225. doi: 10.1038/nature10805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Forneris F., Binda C., Adamo A., Battaglioli E., Mattevi A. Structural basis of LSD1-CoREST selectivity in histone H3 recognition. J. Biol. Chem. 2007;282:20070–20074. doi: 10.1074/jbc.C700100200. [DOI] [PubMed] [Google Scholar]
- 19.Laurent B., Ruitu L., Murn J., Hempel K., Ferrao R., Xiang Y., Liu S., Garcia B.A., Wu H., Wu F., Steen H., Shi Y. A specific LSD1/KDM1A isoform regulates neuronal differentiation through H3K9 demethylation. Mol. Cell. 2015;57:957–970. doi: 10.1016/j.molcel.2015.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zibetti C., Adamo A., Binda C., Forneris F., Toffolo E., Verpelli C., Ginelli E., Mattevi A., Sala C., Battaglioli E. Alternative splicing of the histone demethylase LSD1/KDM1 contributes to the modulation of neurite morphogenesis in the mammalian nervous system. J. Neurosci. 2010;30:2521–2532. doi: 10.1523/JNEUROSCI.5500-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Toffolo E., Rusconi F., Paganini L., Tortorici M., Pilotto S., Heise C., Verpelli C., Tedeschi G., Maffioli E., Sala C., Mattevi A., Battaglioli E. Phosphorylation of neuronal lysine-specific demethylase 1LSD1/KDM1A impairs transcriptional repression by regulating interaction with CoREST and histone deacetylases HDAC1/2. J. Neurochem. 2014;128:603–616. doi: 10.1111/jnc.12457. [DOI] [PubMed] [Google Scholar]
- 22.Hayami S., Kelly J.D., Cho H.S., Yoshimatsu M., Unoki M., Tsunoda T., Field H.I., Neal D.E., Yamaue H., Ponder B.A., Nakamura Y., Hamamoto R. Overexpression of LSD1 contributes to human carcinogenesis through chromatin regulation in various cancers. Int. J. Cancer. 2011;128:574–586. doi: 10.1002/ijc.25349. [DOI] [PubMed] [Google Scholar]
- 23.Jones M.H., Virtanen C., Honjoh D., Miyoshi T., Satoh Y., Okumura S., Nakagawa K., Nomura H., Ishikawa Y. Two prognostically significant subtypes of high-grade lung neuroendocrine tumours independent of small-cell and large-cell neuroendocrine carcinomas identified by gene expression profiles. Lancet. 2004;363:775–781. doi: 10.1016/S0140-6736(04)15693-6. [DOI] [PubMed] [Google Scholar]
- 24.Bhattacharjee A., Richards W.G., Staunton J., Li C., Monti S., Vasa P., Ladd C., Beheshti J., Bueno R., Gillette M., Loda M., Weber G., Mark E.J., Lander E.S., Wong W., Johnson B.E., Golub T.R., Sugarbaker D.J., Meyerson M. Classification of human lung carcinomas by mRNA expression profiling reveals distinct adenocarcinoma subclasses. Proc. Natl. Acad. Sci. USA. 2001;98:13790–13795. doi: 10.1073/pnas.191502998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hamanaka W., Motoi N., Ishikawa S., Ushijima M., Inamura K., Hatano S., Uehara H., Okumura S., Nakagawa K., Nishio M., Horai T., Aburatani H., Matsuura M., Iwasaki A., Ishikawa Y. A subset of small cell lung cancer with low neuroendocrine expression and good prognosis: a comparison study of surgical and inoperable cases with biopsy. Hum. Pathol. 2014;45:1045–1056. doi: 10.1016/j.humpath.2014.01.001. [DOI] [PubMed] [Google Scholar]
- 26.Arrowsmith C.H., Bountra C., Fish P.V., Lee K., Schapira M. Epigenetic protein families: a new frontier for drug discovery. Nat. Rev. Drug Discov. 2012;11:384–400. doi: 10.1038/nrd3674. [DOI] [PubMed] [Google Scholar]
- 27.Mohammad H.P., Smitheman K.N., Kamat C.D., Soong D., Federowicz K.E., Van Aller G.S., Schneck J.L., Carson J.D., Liu Y., Butticello M., Bonnette W.G., Gorman S.A., Degenhardt Y., Bai Y., McCabe M.T., Pappalardi M.B., Kasparec J., Tian X., McNulty K.C., Rouse M., McDevitt P., Ho T., Crouthamel M., Hart T.K., Concha N.O., McHugh C.F., Miller W.H., Dhanak D., Tummino P.J., Carpenter C.L., Johnson N.W., Hann C.L., Kruger R.G. A DNA hypomethylation signature predicts antitumor activity of LSD1 inhibitors in SCLC. Cancer Cell. 2015;28:57–69. doi: 10.1016/j.ccell.2015.06.002. [DOI] [PubMed] [Google Scholar]
- 28.Zhang X., Lu F., Wang J., Yin F., Xu Z., Qi D., Wu X., Cao Y., Liang W., Liu Y., Sun H., Ye T., Zhang H. Pluripotent stem cell protein Sox2 confers sensitivity to LSD1 inhibition in cancer cells. Cell Rep. 2013;5:445–457. doi: 10.1016/j.celrep.2013.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material
Supplementary material
Supplementary material
Supplementary material
Supplementary material
Supplementary material
Supplementary material
Supplementary material
Supplementary material
Supplementary material
Supplementary material
Supplementary material
Supplementary material
Supplementary material
Supplementary material
Supplementary material
Supplementary material
Supplementary material
Supplementary material