Abstract
Pepsin is generally used in the preparation of F(ab)2 fragments from antibodies. The antibodies that are one of the largest and fastest growing categories of bio- pharmaceutical candidates. Differential scanning calorimetric is principally suitable method to follow the energetics of a multi-domain, fragment to perform a more exhaustive description of the thermodynamics in an associating system. The thermodynamical models of analysis include the construction of a simultaneous fitting of a theoretical expression. The expression depending on the equilibrium unfolding data from multimeric proteins that have a two-state monomer. The aim of the present study is considering the DSC data in connection with pepsin going through reversible thermal denaturation. Afterwards, we calculate the homology modeling identification of pepsin in complex multi-domain families with varied domain architectures. In order to analyze the DSC data, the thermal denaturation of multimer proteins were considered, the “two independent two-state sequential transitions with domains dissociation model” was introduced by using of the effective ΔG concept. The reversible unfolding of the protein description was followed by the two-state transition quantities which is a slower irreversible process of aggregation. The protein unfolding is best described by two non-ideal transitions, suggesting the presence of unfolding intermediates. These evaluations are also applicable for high throughput investigation of protein stability.
Abbreviations: DSC, Differential Scanning Calorimetry; Tm, Thermal midpoint; ΔHcal, Calorimetric enthalpies; ΔHVH, Van’t Hoff enthalpy
Keywords: Multimeric proteins, Thermal denaturation, Protein unfolding, Differential Scanning Calorimetric, Sequential transition
Highlights
-
•
The “two independent two-state transitions model” was used for analyzing of thermal denaturation of multi-domain proteins.
-
•
ΔGDeff is a useful parameter that gives an intuitive appreciation of the stability of multimeric proteins.
-
•
All experimental data suggests that at pH 4 the stability of pepsin is enhanced, the positive big value at pH 4 in Ts=298.15 K.
-
•
The corresponding that is a well-defined function for thermodynamic stability of multi-domain proteins reversible thermal denaturation.
1. Introduction
Pepsin is commonly used in the preparation of F(ab)2 fragments from antibodies. Antibodies and antibody-derived molecules are one of the largest and fastest growing classes of bio- pharmaceutical candidates and products. Differential scanning calorimetry (DSC) is the most direct experimental technique to resolve the energies out of conformational transitions of biological macromolecules. The stability of multi-domain proteins is commonly investigated by DSC. One of the great advantages of DSC is that it can detect fine-tuned of interactions between the individual domains of a protein. Followed by measuring the temperature dependence of the partial heat capacity, a basic thermodynamic property, DSC gives immediate access to the thermodynamic mechanism that governs a conformational equilibrium. The way proteins work imposes constraints on their function. Knowing the sources of the protein stability is essential to recognize their structure and function. One of the method for quantifying the stability of a protein is to populate the native and unfolded states by physical and chemical means. Then, the transitions measured by DSC or fluorescence, and absorption spectroscopy were evaluated [1], [2]. During the last two decades DSC has significantly contributed to the development of our current understanding of the energetics and thermodynamic properties of protein folding-unfolding transitions [3]. By scanning microcalorimetry, it was shown that thermal transition is connected purely to the denaturation of protein molecules in the crystal and it is not accompanied by the crystal disintegration into separate molecules [4].
At critical temperatures and higher, along with typical loosening of the protein globuleis is observed along with it oligomeric molecules undergo dissociation process into subunits. Subsequently these smaller components may associate with each other and oligomeric structures. This association may result in irreversible denaturation and deviant physicochemical behavior [5]. The denaturation unfolding process is strongly dependent on the heating rate. As it is expected, the unfolding process is kinetically controlled by the presence of an irreversible reaction. CD signal on heating of proteins, constructing ellipticity quantities at wavelength distinct is very applicable for following the denaturation process [6]. A large body of research on the thermodynamics of small monomeric and single domain proteins has indicated that the hydrophobic effect and loss of conformational entropy are the major determinants of stability in the native state [7]. The unfolding transitions of several small globular proteins are usually highly cooperative. They closely follow a two-state mechanism under equilibrium conditions [8], [9]. However, in some cases, stable intermediates have been detected and partially characterized [10], [11]. Much of the disagreement between theoretical and experimental results of thermal denaturation of proteins derives from the necessity of using some models to interpret the thermodynamic data for proteins [1]. In these cases that have no easy experimental methods, applying an empirical formula can be a rational. For overcoming to the above difficulties, the present study uses of the effective ΔG concept, the thermodynamics of a multi-domain protein which undergo thermal denaturation is studied as a function of protein concentration. ΔGeff is a valuable factor that provides an natural increase of the stability of multimeric proteins; one can determine the section of unfolded protein from ΔGeff of a multimeric protein as easily as doing so from the ΔG° of a monomeric protein [8]. In this regards, we try to discuss and introduce “the two independent two-state transitions with a domain dissociation model”.
2. Methods
DSC measurements were carried out on a MicroCall “MC-2” Differential Scanning Calorimeter (Micro All Inc., Northampton, MA) with cell volumes 1.14 mL, at heating rates 1.5 °C min−1. DSC scans were obtained in a temperature range from 283.15 to 373.15 K. During the measurements, the protein concentrations was 30 µM, and pH ranged from 1.0 to 4.0 [12]. The buffers used for acid denaturation of pepsin are at different pH values KCl/HCl (pH 0.8–1.4), Gly/HCl (pH 1.6–3.0), sodium acetate (pH 3.5–5.0) at a concentration of 20 mM [13]. Pepsin, lyophilized powder (≥2500 units mg−1 protein) as well as Aldrich. Degassing during the calorimetric measurements was prevented by additional constant pressure of 1 atm over the liquids in the cells. At first, the solvent was placed in both the sample and reference compartments. A DSC curve corresponding to solvent vs. solvent run was used as an instrumental baseline. The calorimetric data was corrected for calorimetric baseline (by subtracting solvent-solvent scan).
In this study we used the Swiss-Model template library (SMTL) [14], [15] database to analyze the domain organization of proteins. The SCOP [16] and CATH [17] families corresponding to multi-domain proteins have been used to identify single domain homologues. A control dataset of non-redundant single domain proteins obtained from completely different families has also been formed. All the structures have been optimized at 298 K by using the optimizing tool available in FoldX (version 3.0) [18]. Then free energy computations were done.
3. Results and discussion
3.1. The thermodynamic models
We clearly pointed out that this manuscript is the continuation of previously published work, as described in the framework of this model. The previously published paper were also clearly identified (reference [12]) at the manuscript.
The framework used for fitting DSC data offers four models. They all of them use the Levenberg-Marquardt non-linear least-square method [19], there was a conflict with the number of parameters involved:
1) Two-state with zero ΔCp ( parameters: Thermal midpoint (Tm), Calorimetric enthalpies (ΔHcal)); 2) Non-two state with zero ΔCp (parameters: Tm, ΔHcal, Van’t Hoff enthalpy (ΔHVH)); 3) Two-state with non-zero ΔCp (parameters: Tm, ΔHcal, ΔCp, BL0, BL1) and 4) Dissociation with non-zero ΔCp (parameters: Tm, ΔHcal, ΔCp, BL0, BL1, n- number of multimers).
With exception of parameter number (4), one or more transitions can be used to fit the models, each transition has a relevant parameter set, for example in the case of two overlying transitions can be used to fit to one or more transitions. In the case of multiple transitions, each transition has its own complete parameter set, e.g., if pattern 1 is used to fit two overlapping transitions there will be two independent parameters sets (Tm1, ΔHcal1) and (Tm2, ΔHcal2). These parameters specify the thermal midpoint (Tm) and heat change (ΔHcal) for each transition. BL0 and BL1 parameters define the slope and intercept of the low-temperature baseline segment, they are not repeated and appears only once in each model. While all four models use calorimetric heat change, only non-two state with zero ΔCp model has a van’t Hoff heat change (ΔHVH). The ΔHcal is determined only by the area under transition peak, while the van’t Hoff heat is determined only by the shape (ΔCpmax at transition midpoint). The transition sharpness is associated with ΔHVH largernes. The relationship between ΔHcal and ΔHVH can sometimes provide insights not accessible from ΔHcal model alone. If a protein is composed of two identical domains, which unfold independently with the same Tm and ΔHcal, then the ratio of Hcal/HVH, will be 2, while it would be 1 if the protein had a single domain. If, on the other hand, the protein dimerized and dimer underwent only a single coupled transition then, the ratio will be 0.5, etc.
In the model 1, it is possible to define that overlapping transitions are either independent or sequential in nature, for example, if two architectural domains are interacting strongly, it is possible to assume that their transitions will be coupled in a sequential manner. However, the independent model might better describe two transitions that are thoroughly uncoupled from one another. In practice, this option is often not critical because the sequential and independent models lead essentially identical effects whenever the Tm's of two transitions are separated by a couple of degrees or more. The mathematical derivations for each pattern has been introduced in the previous article [19]. Generally is the aim, the objective is using the simplest model which produces a good framework for the data. Therefore, if data is described by two-state model using two transitions, it would be preferred over a two-state model using three transitions or a non-two state model having two transitions. In this study, a noble two independent two-state transitions with domains dissociation model is introduced, as the fifth fitting model.
3.2. The two independent two-state transitions with domains dissociation model
First, for investigating of thermal multi-domains protein stability, the modified Gibss-Helmholts equation is determined. The fundamental assumption in this model is the equilibrium reversibility of thermal denaturation process in multimeric protein between the folded and unfolded states. As the inherent difficulty in the treatment and analysis of their equilibrium behavior in experimental scope, the use of the Gibss-Helmholts empirical modified function in terms of the number of domains can be a cross cut to thermodynamic purposes.
Eq. (1) shows the equilibrium between n-identical domain protein and its unfolded monomer without any intermediate state is shown in:
| (1) |
The reaction equilibrium constant is as follows:
| (2) |
The definition of unfolded protein fraction, fD is expressed as follow:
| (3) |
Where, Pt is the total protein concentration in domain units. As fallow and ΔG° can be expressed as functions of fD:
| (4) |
| (5) |
| (6) |
In order to determine the modified ΔHD and ΔSD functions in terms of the temperature, applying corresponding prameters (,) will be a useful approach. In the following way:
| (7) |
and
| (8) |
| (9) |
The values of and at TG (Temperature where ) ( and , respectively), can be obtained as follow:
| (10) |
| (11) |
The following equation can be easily extracted by comparing Eqs. (10), (11).
| (12) |
Substitution of the above mentioned equation into Eq. (7), the modified function for in terms of the number of domains can be obtained by:
| (13) |
By following the similar trend for , the corresponding statement for can be obtained as follow:
| (14) |
On the other hand, it can be rewritten as:
| (15) |
Subsequently, it can be shown that:
| (16) |
and
| (17) |
so
| (18) |
and
| (19) |
Therefore:
| (20) |
The modified function of in terms of the number of domains (Eq. (21)) derived by substitution of Eq. (20) into Eq. (15).
can be defined as follow:
| (21) |
The modified function of in terms of the number of domains (Eq. (23)) can be obtained by substitution of Eq. (22) into Eq. (6).
| (22) |
The modified Gibss-Helmholtz equation (Eq. (23)) for multi-domain proteins undergo thermal denaturation can be obtained by substitution of corresponding equation for (Eq. (13)), (Eq. (20)) and (Eq. (21)), into famous Gibbs Eq. ().
| (23) |
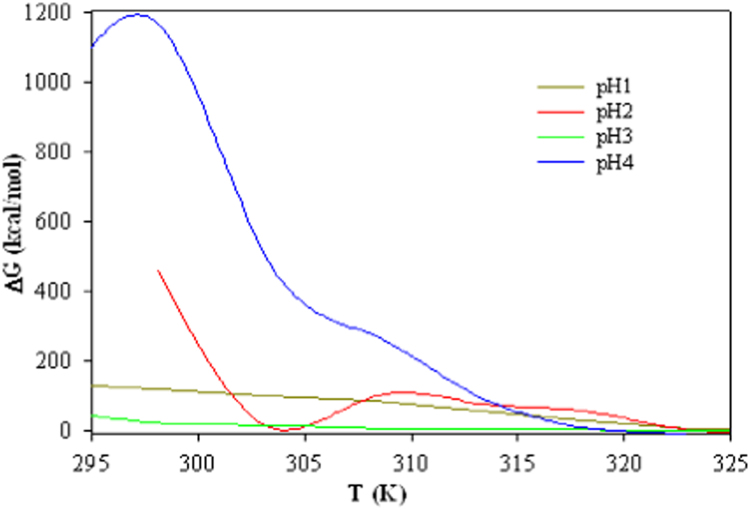
This is a well-defined function for thermodynamic stability of multi-domain proteins that undergo reversible thermal denaturation. The theoretical results of and from in this study (Eqs. (8), (9), (10), (11), (12), (13), (14), (15), (16), (17), (18), (19), (20), (21), (22), (23)) provided in Table 1 to be compared with experimental data [12]. As mentioned earlier, the values of arising from present model are in good agreement with the calorimetric values. They can fit a two-state transition process. According to the DSC thermogram, it was also determined that the low pH-induced denaturation of pepsin affects the domains of the protein differently in comparison with heat induced denaturation. This difference indicates the presence of thermal fluctuations in the native conformation of pepsin [12]. It is also consistent with the non-ideal unfolding observed in DSC experiments. The thermal denaturation determinations of would confirm the assumption that DSC transition peaks can indeed be evaluated by thermodynamic transition patterns. Moreover, it allows the analysis of protein unfolding transitions by the use of thermodynamic variation in state models. The analysis of the transitions by a two-state transition model, requires that the DSC results, in terms of Tm and the unfolding enthalpy, as is indeed observed for the multimeric proteins. Extensive calorimetric studies on small globular domains have demonstrated that these proteins typically show an ideal two-state behavior. The exothermic enthalpy variation observed in a DSC experiment is accredit to unfolding of a part of the protein molecule. In base of the theoretical values of in this study, the stability of arrangement in pepsin can be predicted at four mentioned pH (1−4) can be predicted. All experimental data suggests that at pH 4 the stability of pepsin is enhanced, as illustrated in Fig. 1, the positive big value at pH 4 in T=298.15 K is evident on this suggestion. At the temperature of the first peak observed (and for low heating rates), it was observed that the concentration of unfolded pepsin is high; accordingly, so is the rate of aggregation was also high. At those situations, most pepsin molecules will be thoroughly unfolded before being incorporated in the aggregations. At higher temperatures (low heating rates) the unfolding takes place at a low speed, which leads to a low concentration of (partially) unfolded pepsin molecules. The rate of aggregation is respectively slower and it could well be that at such low aggregation rates pepsin molecules are incorporated in the aggregation before they obtain efficient time for complete unfolding.
Table 1.
The theoretical thermodynamic parameters at TG for the thermal unfolding of pepsin at different pH values within the framework of this study.
| pH | TG (K) | ΔHcal (kcal mol−1) | ΔHDeffmodel (kcal mol−1) | ΔGDeffmodel (kcal mol−1) |
|---|---|---|---|---|
| The first transition | ||||
| 1.0 | — | — | — | — |
| 2.0 | 305.55±0.2 | 128±5 | 105.7 | 117.96 |
| 3.0 | 322.15±0.2 | 76±4 | 70.12 | –0.9 |
| 4.0 | 320.05±0.2 | 44±3 | 24.43 | 45.8 |
| The second transition | ||||
| 1.0 | 332.65±0.2 | 210±5 | 149.48 | 0.006 |
| 2.0 | 339.85±0.2 | 133±3 | 128.52 | –2.2 |
| 3.0 | 348.55±0.2 | 117±4 | 43.45 | –49.43 |
| 4.0 | 349.85±0.2 | 108±2 | 80.28 | –56.8 |
Fig. 1.
Temperature dependence of the Gibbs energy difference in the native and denatured states of Pepsin at various pH values as a function of temperature.
3.3. Multi-domain homology
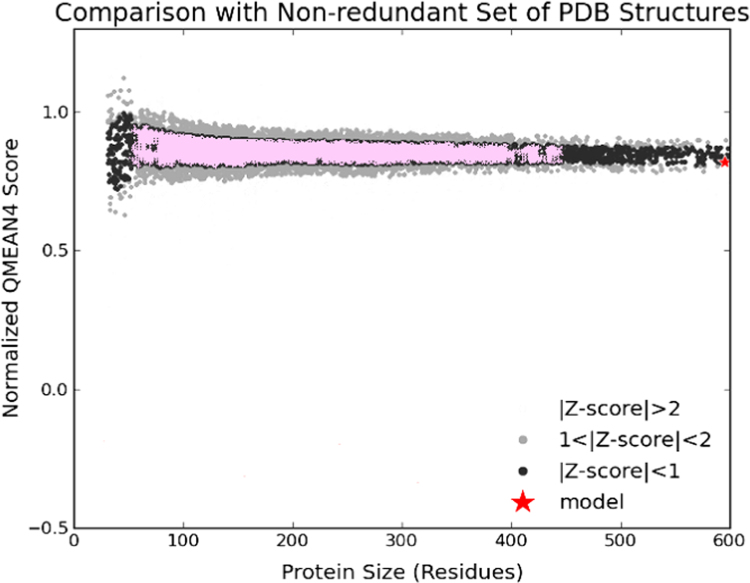
The route and method of denaturation affect the structure of the formed aggregates. We comput the problem of homology modeling identification is computed in complex multi-domain families that have varied domain architectures. The Swiss-Model template library (SMTL) was performed along with Blast [14] and HHBlits [15] for evolutionary related structures matching of target sequence Sus scrofa (Pig) Pepsin A. Overall 549 templates were found, some of them are mentioned in Table 2. The templates with the highest quality have then been selected for building a model. Models are built based on the target-template alignment using Promod-II (Fig. 2). Coordinates are first coneserved between the target and the template, are copied from the template to the model. Insertions and deletions are remodeled using a fragment library (Fig. 3). Side chains are then reconstructed. Eventually, the geometry of the resulted model is equalized by using a force field. In case loop modeling with ProMod-II [20] does not give gratifying results, an alternative model is built with Modeller [21]. The global and per-residue model quality is appraised using the QMEAN scoring function [22]. For enhancing the performance, weights of the individual QMEAN terms have been trained specifically for Swiss Model. Ligands present in the template structure (N – Ethoxycarbonyl - L- leucyl-N-[(1R,2S,3S)-1-(cyclohexylmethyl) - 2,3- dihydroxy-5-methylhexyl] -L- leucinamide) are transferred by homology to the model. When the following criteria are met (Gallo -Casserino, to be published): (a) The ligands are annotated as biologically relevant in the template library, (b) the ligand is in contact with the model, (c) the ligand is not clashing with the protein, (d) the residues in contact with the ligand are conserved between the target and the template. If any of these four principal is not satisfied, a certain ligand will not be included in the model. The model summary contains some information on why and which ligand has not been covered (Fig. 4). Homo oligomeric structure of the target protein is predicted based on the analysis of pairwise interfaces of the identified template structures. For each relevant interface between polypetide chains (interfaces with more than 10 residue-residue interactions), the Qscore Oligomer [23] is predicted by considering features such as similarity to target and frequency of observing this interface in the identified templates (Kiefer, Bertoni, Biasini, to be published). The prediction is performed with a random forest regressor using these features as input parameters to predict the probability of conservation for each interface (Fig. 5). The Qscore Oligomer of the whole complex is then calculated as the weight averaged Qscore Oligomer of the interfaces. The oligomeric state of the target is predicted to be the same as in the template. The similarity is when Qscore Oligomer is predicted to be higher or equal to 0.5.
Table 2.
The templates for the homology modeling of Pepsin A.
| Template | Seq identity | Oligo-state | Found | Method | Resolution | Seq similarity | Coverage |
|---|---|---|---|---|---|---|---|
| 2PSG.1.A | 99.46 | homo-dimer | HHblits | X-ray | 1.80 Å | 0.61 | 0.96 |
| 1PSA.1.A | 100.00 | homo-dimer | BLAST | X-ray | 2.90 Å | 0.61 | 0.85 |
| 1YX9.1.A | 100.00 | monomer | BLAST | X-ray | 3.00 Å | 0.61 | 0.85 |
| 1F34.1.A | 100.00 | hetro-oligomer | BLAST | X-ray | 2.45 Å | 0.61 | 0.85 |
| 3PEP.1.A | 99.69 | monomer | BLAST | X-ray | 2.30 Å | 0.61 | 0.85 |
| 4PEP.1.A | 99.69 | monomer | BLAST | X-ray | 1.80 Å | 0.61 | 0.85 |
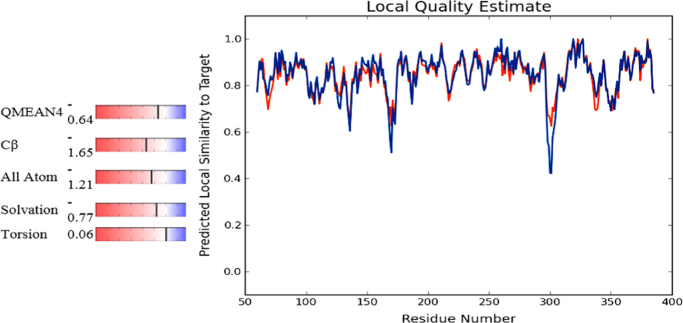
Fig. 2.
Local similarity quality estimate in target vs, residue number.
Fig. 3.
Fragment library of coordinates which are conserved between the target and the template.
Fig. 4.
The geometry of the model with ligand by using a force field.
Fig. 5.
Normalize scores for a number of residues non-redundant set of PDB structure.
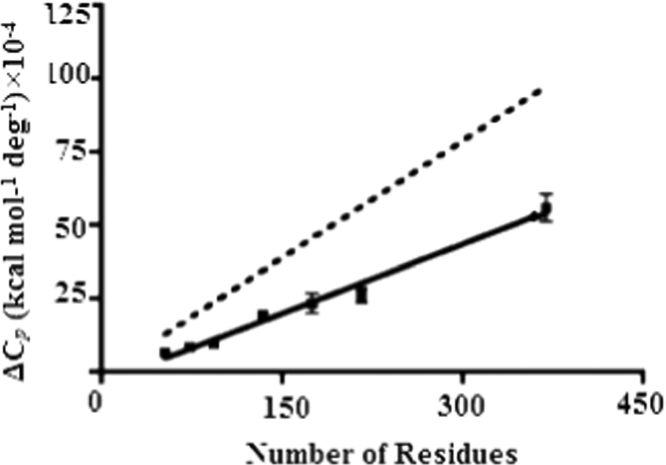
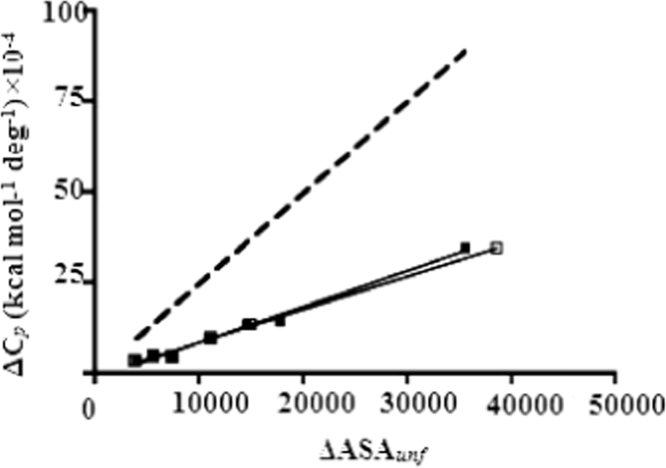
Furthermore, it can be estimate the correlation between the change in heat capacity (ΔCp) and the surface area, depends on unfolding ΔASAunf for a set of domain proteins. It is widely accepted that the value of is dependent on the change in solvent accessible to non polar surface area upon unfolding (ΔASAunf), which is directly proportional to the number of residues in a protein. Such data has been assembled for a variety of template domain proteins, and the slope of the plot of ∆Cp vs. number of residues is 0.020 kcal mol−1 deg−1 residues−1. Interestingly, the slope of a plot of ∆Cp vs. number of residues for the set of globular proteins is 0.015 kcal mol−1 deg−1 residues−1, which is significantly smaller than for template domain proteins (Fig. 6). This observation implies that the surface area exposed to upon unfolding of a globular protein is smaller than expected from the number of residues in the protein. It can be rationalized by two unusual features of domain proteins: 1) their unfolded state is more compact than predicted by the typical self avoiding random walk treatment of the denatured state, and 2) their elongated native state results in a greater surface area to volume ratio than for a globular domain protein. These effects both will contribute to the observed deviation from the behavior of domain proteins. The change in accessible surface area/energy while going from folded to compact unfolded is less than that in going from folded to extended unfolded. We observe a small deviation for domain proteins with 20 or fewer repetitions (Fig. 7). Therefore, it can be concluded that the predominant cause of the deviation in the ∆Cp values is the nature of the unfolded state.
Fig. 6.
Linear relationship between ∆Cp and the number of residues in the protein. ∆Cp is plotted versus number of residues for the proteins acid protease (solid symbols). The solid black line is linear fit of the data, giving a slope of 0.020 kcal mol−1 deg−1 residues−1. The dotted black shows the collected data for ∆Cp versus number of residues for many globular domain proteins. The slope of this line is 0.015 kcal mol−1 deg−1 residues−1.
Fig. 7.
Correlation between the change in heat capacity (ΔCp) and the surface area exposed to unfolding ΔASAunf for a set of globular domain proteins (dashed line) and for acid protease proteins (symbols). The filled squares represent the ΔASAunf calculated using ASAN from the acid protease crystal structures in the unfolded state. The empty squares represent the ΔASAunf calculated using ASAN from the acid protease number of residues and the surface area exposed expected for globular domain protein of that size in the unfolded state. The solid black lines show the linear regression fit to the data.
3.4. Thermodynamic domain interfaces
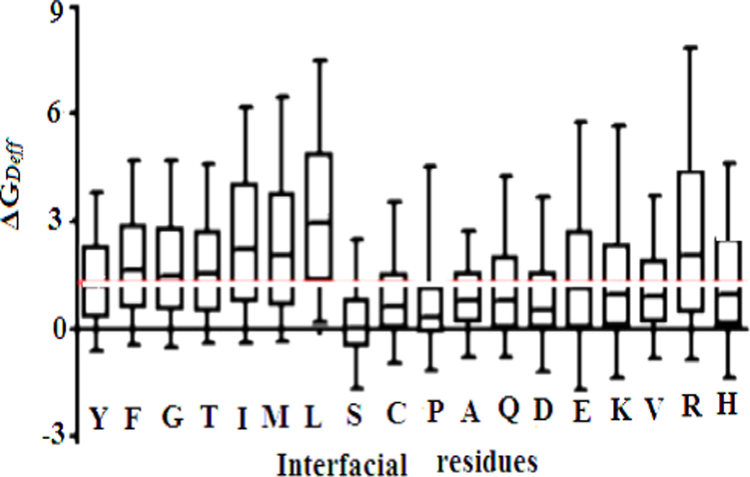
Domain interfaces in several multi-domain proteins are involved in some important function, i.e., they take part in binding or catalysis or act as hinges to facilitate conformational transitions and it may not be possible to tune the functional interfaces to promote folding. We computationally investigated the role of interface domain interaction in full length protein that contains two domain of protein Sus scrofa (Pig) Pepsin A and we found that an altogether domain insertion, promotes unfolding. Strong interface domain interaction is involved in unfolding of multi-domain protein. The simulations were performed close to the unfolding temperature, TG, where multiple transitions occur between the equally populated folded and unfolded ensembles. As a result the best sampling of the transition region is achieved. The presence of a single free energy barrier separating the native and unfolded ensembles at TG implies that the protein unfolds. If the different domains of a multi-domain protein fold at different TGs, partially unfolded states get populated at temperatures between the lowest and the highest domain specific TGs. Upon mutation, a domain specific decrease of TG can result in the incomplete folding of that domain at the TG of the whole protein and the population of partially unfolded states in the folded ensemble. This process in reduced folding cooperativity. Unfolding is usually deduced from the heat capacity curve using the ratio of the van’t Hoff enthalpy (ΔHVH) to the calorimetric enthalpy (ΔHcal). We computed the stability (ΔGDeff) of each of the proteins/domains in these templates using the energy function (Fig. 8). It was found that the domains of multi-domain proteins considered in isolation (7.65±0.86 Kcal.mol-1). Examination of unfolding with DSC indicated that at least two unfolding transitions exist. At and the higher temperature (and lower enthalpy) transition correlated with low aggregation. This result revealed that the least conformationally stable regions of a multi-domain protein are not necessarily the most aggregation.
Fig. 8.
Effect of interfacial residues on the energy of association between the interacting domains. Each of the interfacial residues of the 450 interfaces was mutated to Alanine and the change in the interaction energies between the two domains was computed. All residues except Glycines and Alanines were mutated in this analysis.. It is observed that the medians values of ΔG for these mutations are above the 1.5 kcal mol−1 threshold.
We make changes to Pepsin A which alters the free energetic balance between domains and in turn the unfolding. However, since the domains of Pepsin A are of unequal in size. The largest contribution to Cv(TG) comes from the unfolding of core which is not perturbed much in simulations. Thus, the ΔHVH/ΔHcal is not a sensitive measure of the unfolding of Pepsin A and its mutants. Therefore, we use the height of the free energy barrier was used at TG and the ‘‘unfoldedness’’ of the protein in this fold ensemble to infer the degree of unfolding. We define unfoldedness is defined as the ratio of the population of a mutant at the value of the reaction coordination where Pepsin A is unfolded to the population of Pepsin A at the value of the reaction coordination where Pepsin A is unfolded. This definition intrinsically assumes that the value of the reaction coordination where Pepsin A is unfolded is greater than or equal to the amount of the reaction coordinate where the mutants are unfolded.
The presence of multiple domains in proteins can cause interactions between partially unfolded domains and in turn to increment unfolding. However, several multi-domain proteins unfold unstablility in vitro. Unfolding, the all or no folding of a protein with the population of few intermediates, diminishes partly folded states.
4. Conclusions
It was shown that the use of effective ΔG provides some valuable information about the stability of multimeric proteins. ΔGDeff is a useful parameter that gives an intuitive appreciation of the stability of multimeric proteins, one can calculate the fraction of unfolded protein from ΔGDeff of a multimeric protein can be calculated by the ΔG° of a monomeric protein. The DSC thermogram can be fit to the for a two-state process, and the unfolding process. The reversible unfolding of the protein described by the two-state transition quantities is followed by a slower irreversible process of aggregation.
This model can be used to analyze the changes in the structural and functional properties of a number of large multimeric proteins subjected to broad range temperature variations. The description of the thermal denaturation of multimeric proteins may demand more complex models.
Acknowledgments
The authors are greatfully acknowledge from financial support by Isfahan University and Falavarjan Branch, Islamic Azad University (Grant no. 51723890822022). Authors also appreciate the Swiss- Model for the homology modeling project "PEPA_PIG P00791 Pepsin A(3.4.23.1)", and for help in evolutionary related structures matching the target sequence.
Footnotes
Transparency document associated with this article can be found in the online version at http://dx.doi.org/10.1016/j.bbrep.2017.01.005.
Contributor Information
Ali Asghar Rastegari, Email: aarastegari@gmail.com, rastegari@iaufala.ac.ir.
Abdol-Khalegh Bordbar, Email: bordbar@chem.ui.ac.ir.
Appendix A. Transparency document
Transparency document
References
- 1.Panse V.G., Swaminathan C.P., Aloor J.J., Surolia A., Varadarajan R. Unfolding thermodynamics of the tetrameric chaperone. SecB, Biochem. 2000;39:2362–2369. doi: 10.1021/bi992484l. [DOI] [PubMed] [Google Scholar]
- 2.Privalov P.L., Putekhin S.A. Scanning microcalorimetry in studying temperature- induced changes in proteins. Methods Enzymol. 1986;131:4–51. doi: 10.1016/0076-6879(86)31033-4. [DOI] [PubMed] [Google Scholar]
- 3.Manetto G.D., La Rosa C., Grasso D.M., Milardi D. Evaluation of thermodynamic properties of irreversible protein thermal unfolding measured by DSC. J. Therm. Anal. Cal. 2005;80:263–270. [Google Scholar]
- 4.Makarov A.A., Esipova N.G., Monaselidze D.R., Mgeladze G.N., Madzhagaladze G.V., Kuranova L.P., Grebenko A.L. Microcalorimetry of thermal denaturation of pepsin and trypsin in the crystal state. Biochim. Biophys. Acta. 1976;434:286–289. doi: 10.1016/0005-2795(76)90060-x. [DOI] [PubMed] [Google Scholar]
- 5.Artiukhov V.G., Basharina O.V., Vashanov G.A., Nakvasina M.A., Putintseva O.V. Thermal denaturation of oligomeric proteins: structural and functional modifications, the sequence of steps. Biofizika. 2004;49(4):617–630. [PubMed] [Google Scholar]
- 6.Tello-Solis S.R., Hernández-Arana A. Effect of irreversibility on the thermodynamic characterization of the thermal denaturation of Aspergillus saitoi acid proteinase. Biochem. J. 1995;311:969–974. doi: 10.1042/bj3110969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ramprakash J., Doseeva V., Galkin A., Krajewski W., Muthukumar L., Pullalarevu S., Demirkan E., Herzberg O., Moult J., Schwarz F.P. Comparison of the chemical and thermal denaturation of proteins by a two-state transition model. Anal. Biochem. 2008;374:221–230. doi: 10.1016/j.ab.2007.10.005. [DOI] [PubMed] [Google Scholar]
- 8.Park C., Marqusee S. Analysis of the stability of multimeric proteins by effective ΔG and effective m-values. Protein Sci. 2004;13:2553–2558. doi: 10.1110/ps.04811004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kim P.S., Baldwin R.L. Specific intermediates in the folding reactions of small proteins and the mechanism of protein folding. Annu. Rev. Biochem. 1982;51:459–489. doi: 10.1146/annurev.bi.51.070182.002331. [DOI] [PubMed] [Google Scholar]
- 10.Privalov P.L. Stability of proteins: small globular proteins. Adv. Protein Chem. 1979;33:167–241. doi: 10.1016/s0065-3233(08)60460-x. [DOI] [PubMed] [Google Scholar]
- 11.Hernández-Arana A., Soriano-Garcia M. Detection and characterization by circular dichroism of a stable intermediate state formed in the thermal unfolding of papain. Biochim. Biophys. Acta. 1988;954:170–175. doi: 10.1016/0167-4838(88)90068-4. [DOI] [PubMed] [Google Scholar]
- 12.Rastegari A.A., Behnaz Buzari B., Pavelkic V., Gopcevic K., Petkovic M., Bordbar A.K. Thermal denaturation of pepsin at acidic media: using DSC, MALDI TOF MS and PAGE techniques. Thermochim. Acta. 2013;568:165–170. [Google Scholar]
- 13.Fiorentino G., Giudice I.D., Petraccone L., Bartolucci S., Vecchio P.D. Conformational stability and ligand binding properties of BldR, a member of the MarR family, from Sulfolobus solfataricus. Bioch. Bioph. Acta. 2014;1844:1167–1172. doi: 10.1016/j.bbapap.2014.03.011. [DOI] [PubMed] [Google Scholar]
- 14.Altschul S.F., Madden T.L., Schaffer A.A., Zhang J., Zhang Z., Miller W., Lipman D.J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25(17):3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Remmert M., Biegert A., Hauser A., Soding J. HHblits: lightning-fast iterative protein sequence searching by HMM-HMM alignment. Nat. Methods. 2012;9:173–175. doi: 10.1038/nmeth.1818. [DOI] [PubMed] [Google Scholar]
- 16.Andreeva A., Howorth D., Chandonia J.M., Brenner S.E., Hubbard T.J., Chothia C., Murzin A.G. Data growth and its impact on the SCOP database: new developments. Nucleic Acids Res. 2008;36:D419–D425. doi: 10.1093/nar/gkm993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Greene L.H., Lewis T.E., Addou S., Cuff A., Dallman T., Dibley M., Redfern O., Pearl F., Nambudiry R., Reid A., Sillitoe I., Yeats C., Thornton J.M., Orengo C.A. The CATH domain structure database: new protocols and classification levels give a more comprehensive resource for exploring evolution. Nucleic Acids Res. 2007;35:D291–D297. doi: 10.1093/nar/gkl959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schymkowitz J., Borg J., Stricher F., Nys R., Rousseau F., Serrano L. The FoldX web server: an online force field. Nucleic Acids Res. 2005;33:W382–W388. doi: 10.1093/nar/gki387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.DSC Data Analysis in Origin, Tutorial Guide. The Calorimetry Experts, Version 5.0, 1998, p. 66.
- 20.Guex N., Peitsch M.C. SWISS-model and the Swiss-PdbViewer: an environment for comparative protein modeling. Electrophoresis. 1997;18:2714–2723. doi: 10.1002/elps.1150181505. [DOI] [PubMed] [Google Scholar]
- 21.Sali A., Blundell T.L. Comparative protein modelling by satisfaction of spatial restraints. J. Mol. Biol. 1993;234:779–815. doi: 10.1006/jmbi.1993.1626. [DOI] [PubMed] [Google Scholar]
- 22.Benkert P., Biasini M., Schwede T. Toward the estimation of the absolute quality of individual protein structure models. Bioinformatics. 2011;27:343–350. doi: 10.1093/bioinformatics/btq662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mariani V., Kiefer F., Schmidt T., Haas J., Schwede T. Assessment of template based protein structure predictions in CASP9. Proteins. 2011;79(Suppl 10):S37–S58. doi: 10.1002/prot.23177. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Transparency document