Abstract
Basophils have been erroneously considered as minor relatives of mast cells, due to some phenotypic similarity between them. While recent studies have revealed non-redundant roles for basophils in various immune responses, basophil-derived effector molecules, including lipid mediators, remain poorly characterized, compared to mast cell-derived ones. Here we analyzed and compared eicosanoids produced by mouse basophils and mast cells when stimulated with IgE plus allergens. The production of 5-LOX metabolites such as LTB4 and 5-HETE was detected as early as 0.5 h post-stimulation in both cell types, even though their amounts were much smaller in basophils than in mast cells. In contrast, basophils and mast cells showed distinct time course in the production of COX metabolites, including PGD2, PGE2 and 11-HETE. Their production by mast cells was detected at both 0.5 and 6 h post-stimulation while that by basophils was detectable only at 6 h. Of note, mast cells showed 8–9 times higher levels of COX-1 than did basophils at the resting status. In contrast to unaltered COX-1 expression with or without stimulation, COX-2 expression was up-regulated in both cell types upon activation. Importantly, when activated, basophils expressed 4–5 times higher levels of COX-2 than did mast cells. In accordance with these findings, the late-phase production of the COX metabolites by basophils was completely ablated by COX-2 inhibitor whereas the early-phase production by mast cells was blocked by COX-1 but not COX-2 inhibitor. Thus, the production of COX metabolites is differentially regulated by COX-1 and COX-2 in basophils and mast cells.
Abbreviations: BMBAs, bone marrow derived basophils; BMMCs, bone marrow derived mast cells; COX, cyclooxygenase; LOX, lipoxygenase; TNP, 2,4,6-trinitrophenyl; OVA, Ovalbumin; HETE, hydroxyeicosatetraenoic acid; PGD2, prostaglandin D2; PGE2, prostaglandin E2; LTA4, leukotriene A4; LTB4, leukotriene B4; LTC4, leukotriene C4; LTD4, leukotriene D4
Chemical compounds: BW-A4C (PubChem CID: 6438354), SC-560 (PubChem CID: 4306515), Celecoxib (PubChem CID: 2662)
Keywords: Basophils, Mast cells, Eicosanoids, Prostaglandins, COX-2, LC-MS/MS
Highlights
-
•
Basophils and mast cells show distinct time course of COX metabolite production.
-
•
Basophils and mast cells show differential expression and induction of COX isoforms.
-
•
COX metabolite production by basophils but not mast cells is mediated by COX-2.
1. Introduction
Basophils have been sometimes mixed up with mast cells in spite of distinct cell lineages. Basophils are the rarest granulocytes, representing less than 1% of blood-circulating leukocytes, and share some phenotypic features with tissue-resident mast cells. Both types of cells possess basophilic granules in the cytoplasm, express high-affinity IgE receptor FcεRI on the cell surface, and release proinflammatory mediators such as histamine and proteases upon activation [1]. Therefore, basophils have often considered erroneously as minor and non-redundant relatives or even blood-circulating precursors of tissue-resident mast cells. Although the research on basophils had long been hampered by their paucity and the absence of physiologically-relevant cell lines, recent studies utilizing novel analytical tools, including basophil-deficient engineered mice, revealed crucial and non-redundant roles for basophils in various immune responses such as allergy and protective immunity against parasitic infections [2], [3], [4]. Nevertheless, basophil-derived effector molecules involved in immune responses, including lipid mediators, remains ill-defined, compared to those derived from mast cells.
Eicosanoids, lipid mediators generated by the metabolism of arachidonic acid, play important roles in a series of biological responses including allergic inflammation and immunomodulation [5], [6], [7], [8]. The first step of eicosanoid biosynthesis is the release of arachidonic acid from membrane phospholipids through the action of phospholipases. The next step bifurcates into two major pathways among others, mediated by either lipoxygenase (LOX) or cyclooxygenase (COX). 5-LOX converts membrane-derived arachidonic acid to leukotriene A4 (LTA4) and 5-HETE. LTA4 is then catalyzed by LTA4 hydrolase to form LTB4 while it is conjugated with glutathione by LTC4 synthase to form LTC4 that is further converted into LTD4 and LTE4 by extracellular metabolism [8], [9]. In the other pathway mediated by COX, two isoforms of the COX enzyme, COX-1 and COX-2, have been identified [10], [11], [12]. While COX-1 is constitutively expressed in most cells, COX-2 expression can be up-regulated in response to a variety of stimuli. Both isoforms display the identical biochemical function in spite of relatively low homology (60–65%) at the amino acid level, and convert arachidonic acid to prostaglandin H2 (PGH2) [13]. PGH2 is subsequently catalyzed to PGD2, PGE2, PGI2, PGF2α and thromboxanes by prostaglandin synthase specific to each of them [8], [10].
Eicosanoids released from mast cells have been extensively studied by using bone marrow-derived mast cells (BMMCs). When stimulated with IgE and corresponding antigens, BMMCs quickly produce and release LTB4, LTC4, 5-HETE and PGD2 [14], [15]. In contrast, basophils were thought for many years to release a much narrower range of eicosanoids, such as LTC4 alone [16], [17], [18]. Although recent study expanded the range of eicosanoids released by basophils [19], systematic and comparable studies on the spectrum and quantity of eicosanoids released by activated basophils and mast cells remain to be done. In the present study, we analyzed and compared the time course, repertoire and quantity of eicosanoids produced by mouse basophils and mast cells when stimulated with IgE plus allergens, by using liquid chromatography-tandem mass spectrometry (LC-MS/MS). We also determined and compared the catalytic enzymes involved in eicosanoid production by basophils and mast cells, and demonstrated for the first time that COX-1 and COX-2 were differentially used in the production of prostaglandins and 11-HETE by basophils and mast cells.
2. Material and methods
2.1. Mice
C57BL/6 J mice were purchased from CLEA Japan. All animal studies were approved by Institutional Animal Care and Use Committee of Tokyo Medical and Dental University and the Ethics Committee for Animal Experiments of Ono Pharmaceutical Co., Ltd.
2.2. Generation and activation of BMBAs and BMMCs
Bone marrow-derived basophils (BMBAs) and bone marrow-derived mast cells (BMMCs) were generated as described previously [20], [21] with minor modification. In brief, mouse bone marrow cells were cultured with 10 ng/mL murine IL-3 and 2 ng/mL murine SCF for 40 days to obtain BMMCs while they were cultured with 0.1 ng/mL IL-3 for 7 days to obtain BMBAs. In case of BMBAs, the basophil fraction in cultured bone marrow cells was purified with positive sorting of CD49b+ cells by using MACS pro system (Miltenyi Biotec). The purity of basophils (CD49b+CD200R3+cKit-) in the BMBA preparation and mast cells (CD200R3+cKit+) in the BMMC preparation was >90%. Identity of basophils and mast cells was verified by their selective expression of Mcpt8 and Mcpt6 mRNAs encoding serine proteases mMCP-8 and mMCP6, respectively [20], [22], [23]. BMBAs and BMMCs were sensitized overnight with 1 μg/mL of hapten 2,4,6-trinitrophenyl (TNP)-specific IgE (IGELb4, ATCC-TIB141), and then incubated with 10 ng/mL TNP-conjugated ovalbumin (TNP12-OVA, Biosearch Tech.) or control ovalbumin (OVA, Invivogen) for indicated time periods. In some experiments, before stimulation, cells were pretreated for 10 min with of 3 μmol/L of inhibitors, including BW-A4C (5-LOX inhibitor, Sigma), SC-560 (COX-1 inhibitor, Cayman Chemicals), and celecoxib (COX-2 inhibitor, Sigma).
2.3. Preparation and activation of primary basophils
CD49b+ cells were enriched from mouse bone marrow cells by using MACS pro system, sensitized for 3 h with 1 μg/mL of anti-TNP-IgE, and then incubated with 10 ng/mL of TNP-OVA or control OVA for 30 min. After treatment with 0.1%NaN3, CD49b+CD200R3+c-Kit- basophils were sorted by FACS AriaIII (BD biosciences), and subjected to quantitative RT-PCR analysis.
2.4. Quantitative RT-PCR
Total RNA was prepared using RNeasy™ Mini Kit (Qiagen), followed by cDNAs synthesis with Superscript VILO™ Master Mix (Thermofisher). Quantitative PCR of the cDNA was performed on Agilent Mx3005P system using a Taqman Fast Universal PCR Master Mix (Applied Biosystems) and Taqman gene expression assay primer and probe mix as follows: Gapdh (Mm_99999915_g1), Mcpt6 (Mm 01301240_g1), Mcpt8 (Mm_00484935_g1), Alox5 (Mm_01182747_m1), Ptgs1 (Mm00477214_m1), Ptgs2 (Mm_00478374_m1). Gene expression levels were normalized by Gapdh expression levels.
2.5. Quantitative analysis of eicosanoids
Eicosanoids were extracted in the presence of deuterated internal standard (LTB4-6,7,14,15-d4 and 15(S)-HETE-5,6,8,9,11,12,14,15-d8, Cayman chemicals) by using CHCl3/methanol/acetic acid (49.5:49.5:1). For LC-MS/MS analysis, a 5500 QTRAP triple quadrupole mass spectrometer (Sciex Co., Ltd.) equipped with an UFLC Poroshell 120 SB-C18 column (2.7 µm particle size, 2.1×150 mm; Agilent) was used. Samples were eluted with mobile phase A consists of ultra-pure water with 0.1% acetic acid, and B of acetonitrile/methanol (4:6 v/v) with 0.1% acetic acid. Gradient separations were performed from 27% of B for 5 min, ramped to 70% after 15 min, ramped to 85% after 25 min, and ramped to 100% after 30 min and held for 10 min, with flow rates of 0.2 mL/min. MS/MS analyses were conducted in negative ion mode. Eicosanoids were identified by multiple reaction monitoring (MRM) using transitions of a set of parent ion mass (m/z) > daughter ion mass (m/z) (Table 1). Concentration of each eicosanoid was carried out in a five-point calibration from the peak area. Eicosanoids for LC-MS/MS standards (LTB4, 5-HETE, PGD2, PGE2, 11-HETE, and 12-HETE) were purchased from Cayman Chemicals.
Table 1.
Parameters for identification of eicosanoids.
| Targets | IS | RT (min) | Transition (m/z) Parent > Daughter | LLOQ (ng/mL) |
|---|---|---|---|---|
| LTB4 | LTB4-d4 | 19.72 | 335.0>195.0 | 0.1 |
| 5-HETE | 15-HETE-d8 | 24.88 | 319.0>115.0 | 0.05 |
| PGD2 | LTB4-d4 | 16.70 | 351.0>189.0 | 0.5 |
| PGE2 | LTB4-d4 | 16.50 | 351.0>271.0 | 0.05 |
| 11-HETE | 15-HETE-d8 | 23.94 | 319.0>167.0 | 0.05 |
IS; Internal standard, RT; Column retention time, LLOQ; Lower limit of quantification.
3. Results and discussion
3.1. Basophils release 5-LOX metabolites, LTB4 and 5-HETE at the early phase after stimulation
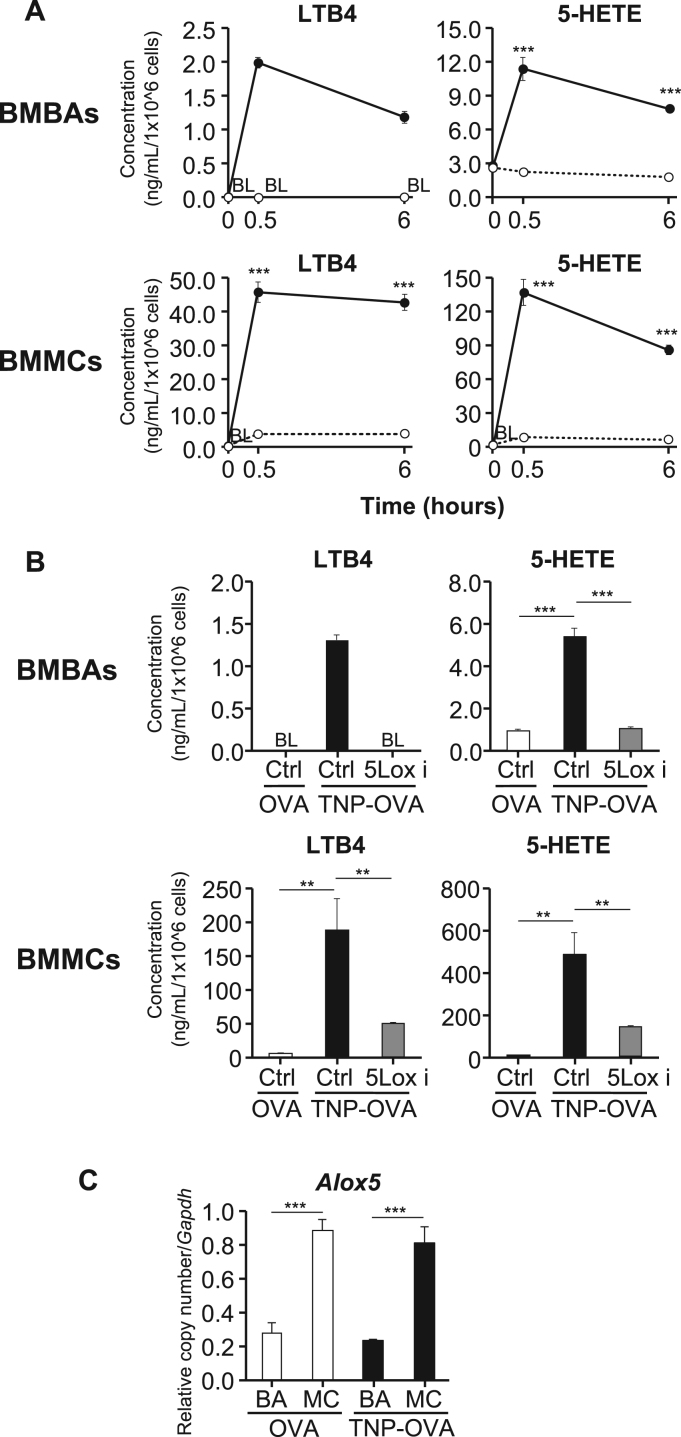
We first analyzed and compared the time course of production and repertoire of 5-LOX metabolites released from activated basophils and mast cells. To this end, bone marrow-derived basophils (BMBAs) and bone marrow-derived mast cells (BMMCs) were generated from mouse bone marrow cells, and stimulated with TNP-specific IgE plus the corresponding antigen TNP-OVA or control OVA. Culture supernatants were collected at 0, 0.5 and 6 h post-stimulation, and subjected to LC-MS/MS analysis. The production of LTB4 and 5-HETE was detected in culture supernatants of activated BMBAs as early as 0.5 h post-stimulation as in the case of BMMCs, even though their amounts were 10–20 times less in BMBAs than in BMMCs (Fig. 1A).
Fig. 1.
Comparative analysis of time-course and quantity of LOX-metabolite production by basophils and mast cells. (A) BMBAs and BMMCs were sensitized with TNP-specific IgE and then incubated with TNP-OVA (closed circles) or control OVA (open circles) for the indicated time periods. LOX-metabolites in culture supernatants were quantified by LC-MS/MS (mean±SEM, n=3). Statistical analysis was performed by using student's t-test. (B) IgE-sensitized BMBAs and BMMCs were pretreated with BW-A4C (5-LOX inhibitor) or control vehicle (DMSO) and subsequently incubated with TNP-OVA or OVA for 0.5 h. LTB4 and 5-HETE in culture supernatants were quantified as in A (mean±SEM, n=3). BL indicates below lower limit of quantification. Statistical analysis was performed by using student's t-test. (C) IgE-sensitized BMBAs (BA) and BMMCs (MC) were treated as in A for 0.5 h and subjected to quantitative RT-PCR analysis for the Alox5 expression. The relative amount of Alox5 mRNAs encoding 5-LOX was calculated in comparison to that of Gapdh mRNAs in each cell type (mean±SEM, n=3). Statistical analysis was performed by using 2-way ANOVA and student's t-test. All data shown are representative of at least three independent experiments. **;p<0.01; ***;p<0.001.
LTB4 and 5-HETE are known to be generated from arachidonic acid through either non-enzymatic oxidation or 5-LOX-mediated enzymatic process [24], [25]. As shown in Fig. 1B, the pretreatment of cells with BW-A4C (5-LOX inhibitor [26]) impaired the production of both LTB4 and 5-HETE by BMBAs and BMMCs, indicating that the 5-LOX-mediated enzymatic process is predominantly involved in their production by both cell types. Of note, quantitative RT-PCR analysis revealed that the expression of Alox5 mRNAs encoding 5-LOX remained unaltered even after stimulation in both cell types while the level of expression was 3 times lower in BMBAs than in BMMCs (Fig. 1C). This may account for the observations that the production of LTB4 and 5-HETE was detected as early as 0.5 h post-stimulation (without up-regulating the 5-LOX expression) in both cell types while BMBAs released less amounts of them compared to BMMCs. Taken together, both basophils and mast cells appeared to utilize 5-LOX as a key enzyme to produce LTB4 and 5-HETE at the early phase after stimulation, although their amounts released from basophils and mast cells were not comparable.
3.2. Basophils and mast cells show distinct time course of COX metabolite production upon stimulation
We next analyzed and compared the time course of production and repertoire of COX metabolites among eicosanoids released by activated BMBAs and BMMCs. In accordance with previous studies [15], the production of PGD2, PGE2 and 11-HETE by BMMCs was detected as early as 0.5 h post-stimulation (Fig. 2). In sharp contrast, those products in culture supernatants of BMBAs was detected only at 6 h but not 0.5 h post-stimulation (Fig. 2), suggesting that the production of COX metabolites by BMBAs started at the later phase after stimulation, unlike in BMMCs. Despite of the difference in the time course of production, the amounts of PGE2 and 11-HETE released by BMBAs and BMMCs appeared to be comparable (Fig. 2) although BMBAs released 10 times less amounts of PGD2 than did BMMCs as observed in the production of LTB4 and 5-HETE. This is the first report, to our knowledge, that basophils release 11-HETE upon stimulation. Previous study reported the detection of PGD2 and PGE2 in culture supernatants of IgE-antigen-stimulated BMBAs at 0.5 h post-stimulation even though the analysis at the later phase was not performed [19]. It remains to be clarified what makes this difference in the detection at 0.5 h post-stimulation, but it should be noted that BMBAs were stimulated with IgE plus antigens in the presence of small amount (0.1 ng/mL) of IL-3 to keep them alive in our experiments whereas 100 times more (10 ng/mL) of IL-3 was added to the culture in the previous study [19].
Fig. 2.
Distinct time course of COX-metabolite production by basophils and mast cells. IgE-sensitized BMBAs and BMMCs were stimulated with TNP-OVA (closed circles) or control OVA (open circles) as in Fig. 1A. COX-metabolites in culture supernatants were quantified by LC-MS/MS (mean±SEM, n=3). Statistical analysis was performed by using student's t-test. All data shown are representative of at least three independent experiments. ***;p<0.001.
3.3. Differential expression and induction of COX-1 and COX-2 in basophils and mast cells
Two distinct isoforms, COX-1 and COX-2, are involved in the COX pathway [10]. The distinct time course of COX metabolite production by BMBAs and BMMCs prompted us to examine the expression profile of the Ptgs1 and Ptgs2 genes encoding COX-1 and COX-2, respectively [11], [12]. Quantitative RT-PCR analysis revealed that BMMCs had 8–9 times higher levels of Ptgs1 mRNA expression than did BMBAs at the resting status (Fig. 3A, left). The stimulation with IgE plus antigens did not significantly alter the level of the Ptgs1 mRNA expression in both cell types (Fig. 3A, left). In contrast to this homeostatic expression of the Ptgs1 gene, the Ptgs2 expression was up-regulated in both cell types upon stimulation (Fig. 3A, right). Importantly, after stimulation, BMBAs expressed 4–5 times higher levels of Ptgs2 mRNAs than did BMMCs (Fig. 3A, right). Thus, BMBAs and BMMCs displayed the difference in not only the level of the homeostatic COX-1 expression but also that of the induced COX-2 expression, suggesting that this difference may account for the distinct time course of COX metabolite production. Of note, primary basophils displayed the property similar to that of BMBAs, namely the low level of Ptgs1 mRNA expression regardless of stimulation while highly up-regulated Ptgs2 expression upon stimulation (Fig. 3B).
Fig. 3.
Differential expression and induction of COX-1 and COX-2 in basophils and mast cells. (A) BMBAs (BA) and BMMCs (MC) were stimulated with TNP-OVA or control OVA as in Fig. 1C, and subjected to quantitative RT-PCR analysis for the Ptgs1 and Ptgs2 expression. The relative amount of Ptgs1 and Ptgs2 mRNAs encoding COX-1 and COX-2, respectively, was calculated in comparison to that of Gapdh mRNAs in each cell type (mean±SEM, n=3). (B) Primary basophils isolated from the bone marrow were stimulated and analyzed for the Ptgs1 and Ptgs2 expression as in A (mean±SEM, n=3). All data shown are representative of at least three independent experiments. Statistical analysis was performed by using 2-way ANOVA and student's t-test.***;p<0.001; n.s., no significant difference.
3.4. Differential usage of COX-1 and COX-2 by basophils and mast cells for the COX metabolite production at the different timing after stimulation
To compare the relative contribution of COX-1 and COX-2 to the production of PGD2, PGE2, and 11-HETE in BMBAs and BMMCs, cells were pretreated with a selective inhibitor of each COX isoform prior to the stimulation. Notably, celecoxib, a COX-2 inhibitor [27], [28], completely abolished the late-phase production of all three COX metabolites by BMBAs (Fig. 4A), in accordance with the highly up-regulated COX-2 in activated BMBAs (Fig. 3A, right). By contrast, celecoxib showed no significant inhibitory effect on the early-phase production of any of those COX metabolites by BMMCs (Fig. 4B). Instead, SC-560, a COX-1 inhibitor [29], completely abrogated this production (Fig. 4B), in accordance with previous reports [30]. These results clearly demonstrated that the late-phase production of PGD2, PGE2 and 11-HETE by basophils is mediated by COX-2, in contrast to their early-phase production by mast cells via COX-1. This differential usage of COX-1 and COX-2 by basophils and mast cells at different time points after the stimulation appeared to be correlated well with the differential expression and induction of COX-1 and COX-2, namely the much stronger up-regulation of COX-2 in basophils upon stimulation while the higher COX-1 expression at the basal level in mast cells.
Fig. 4.
Differential usage of COX-1 and COX-2 in the production of PGD2, PGE2 and 11-HETE by mast cells and basophils. (A) IgE-sensitized BMBAs were pretreated with celecoxib (COX-2 inhibitor; Cox-2i) or control vehicle (DMSO, Ctrl), and then incubated with TNP-OVA or control OVA for 6 h. PGD2, PGE2 and 11-HETE in culture supernatants were quantified by LC-MS/MS (mean±SEM, n=3). BL indicates below lower limit of quantification. Statistical analysis was performed by using student's t-test. (B) IgE-sensitized BMMCs pretreated with SC-560 (COX-1 inhibitor; Cox-1i), celecoxib (COX-2 inhibitor; Cox-2i) or control vehicle (DMSO, Ctrl), and then incubated with TNP-OVA or control OVA for 0.5 h. PGD2, PGE2 and 11-HETE in culture supernatants were quantified by LC-MS/MS (mean±SEM, n=3). Statistical analysis was performed by using student's t-test for comparison of Ctrl/OVA group and Ctrl/TNP-OVA group and Dunnett's test for comparison of Ctrl/TNP-OVA group and COX2i/TNP-OVA or COX1i/TNP-OVA groups. All data shown are representative of at least three independent experiments. **p<0.01, ***p<0.001; n.s., no significant difference.
Basophils had often been considered erroneously as minor and non-redundant relatives of mast cells because of some phenotypic similarity between them [2]. In the present study, we demonstrated that basophils behave differently from mast cells in terms of the production of eicosanoids, particularly COX metabolites. Basophils released 5-LOX metabolites at the early phase post-stimulation as observed in mast cells whereas basophils released COX metabolites only at the late phase in contrast to mast cells (Supplementary Fig. 1). It is well documented that macrophages produce COX metabolites in a COX-2-dependent manner at the late phase post-stimulation [10], [31], [32]. Therefore, basophils appear to behave like macrophages rather than mast cells in the production of COX metabolites. This may be associated with the anatomical and functional differences between basophils and mast cells. Mast cells reside within peripheral tissues such as the skin and mucosa, and quickly respond to pathogens and allergens that penetrate the skin and mucosal barrier [33]. In contrast, basophils circulate in the peripheral blood under homeostatic conditions, and migrate into peripheral tissues only when inflammation occurs there. Thus, mast cells appear to contribute to the early phase response to pathogens or allergens while basophils contribute to the later phase [34]. Differential timing of COX metabolite production by tissue-resident mast cells and tissue-infiltrating basophils may also contribute to the maintenance of inflammatory milieu for a longer period. This would be beneficial for elimination of pathogens while deleterious in allergic responses to allergens.
In conclusion, we demonstrated in the present study that the production of COX metabolites by mast cells and basophils was differentially regulated by COX-1 and COX-2, in association with differential expression and induction of these two isoforms in mast cells and basophils. This results in the different timing of the COX metabolite production, namely at the early phase post-stimulation in mast cells while at the late phase in basophils.
Author contributions
T.B., M.M. and H.K. conceived the work; T.B. designed and did most experiments; S.F. and N.N. did LC/MS/MS analysis of eicosanoids. M.M., S.Y. and Y.Y. provided helpful discussions; T.B. and H.K. wrote the manuscript.
Competing financial interests
T.B., S.F., N.N. and M.M. are employees of Ono pharmaceutical co., ltd.
Acknowledgements
We thank for R. Matsunaga for technical assistance, A. Hayashi and K. Kishikawa for helpful advice, the members of the Karasuyama laboratory for helpful discussions and M. Miki for secretary assistance. This work was supported by the research grant from the Japanese Ministry of Education, Culture, Sports, Science and Technology (to H.K).
Footnotes
Supplementary data associated with this article can be found in the online version at doi:10.1016/j.bbrep.2017.03.004.
Contributor Information
Masashi Minami, Email: minami@ono.co.jp.
Hajime Karasuyama, Email: karasuyama.mbch@tmd.ac.jp.
Appendix A. Supplementary material
Supplementary material Supprimentaly Fig. 1. Summary of eicosanoid production and enzyme expressin by basophils and mast cells.
.
References
- 1.Galli S.J. Mast cells and basophils. Curr. Opin. Hematol. 2000;7:32–39. doi: 10.1097/00062752-200001000-00007. [DOI] [PubMed] [Google Scholar]
- 2.Karasuyama H., Mukai K., Obata K., Tsujimura, Wada T. Nonredundant roles of basophils in immunity. Annu Rev. Immunol. 2011;29:45–69. doi: 10.1146/annurev-immunol-031210-101257. [DOI] [PubMed] [Google Scholar]
- 3.Siracusa M.C., Kim B.S., Spergel J.M., Artis D. Basophils and allergic inflammation. J. Allergy Clin. Immunol. 2013;789–801:788. doi: 10.1016/j.jaci.2013.07.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Eberle J.U., Voehringer D. Role of basophils in protective immunity to parasitic infections. Semin Immunopathol. 2016 doi: 10.1007/s00281-016-0563-3. [DOI] [PubMed] [Google Scholar]
- 5.Boyce J.A. Eicosanoids in asthma, allergic inflammation, and host defense. Curr. Mol. Med. 2008;8:335–349. doi: 10.2174/156652408785160989. [DOI] [PubMed] [Google Scholar]
- 6.Nicolaou A., Mauro C., Urquhart P., Marelli-Berg F. Polyunsaturated Fatty Acid-derived lipid mediators and T cell function. Front Immunol. 2014;5:75. doi: 10.3389/fimmu.2014.00075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hohjoh H., Inazumi T., Tsuchiya S., Sugimoto Y. Prostanoid receptors and acute inflammation in skin. Biochim. 2014;107:78–81. doi: 10.1016/j.biochi.2014.08.010. [DOI] [PubMed] [Google Scholar]
- 8.Liu M., Yokomizo T. The role of leukotrienes in allergic diseases. Allergol. Int. 2015;64:17–26. doi: 10.1016/j.alit.2014.09.001. [DOI] [PubMed] [Google Scholar]
- 9.Murphy R.C., Gijón M.A. Biosynthesis and metabolism of leukotrienes. Biochem. J. 2007;405:379–395. doi: 10.1042/BJ20070289. [DOI] [PubMed] [Google Scholar]
- 10.Smith W.L., DeWitt D.L., Garavito R.M. Cyclooxygenases: structural, cellular, and molecular biology. Annu. Rev. Biochem. 2000;69:145–182. doi: 10.1146/annurev.biochem.69.1.145. [DOI] [PubMed] [Google Scholar]
- 11.O'Banion M.K., Winn V.D., Young D.A. cDNA cloning and functional activity of a glucocorticoid-regulated inflammatory cyclooxygenase. Proc. Natl. Acad. Sci. USA. 1992;89:4888–4892. doi: 10.1073/pnas.89.11.4888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hla T., Neilson K. Human cyclooxygenase-2 cDNA. Proc. Natl. Acad. Sci. USA. 1992;89:7384–7388. doi: 10.1073/pnas.89.16.7384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tanabe T., Tohnai N. Cyclooxygenase isozymes and their gene structures and expression. Prostaglandins Other Lipid Mediat. 2002;68–69:95–114. doi: 10.1016/s0090-6980(02)00024-2. [DOI] [PubMed] [Google Scholar]
- 14.Mencia-Huerta J.M., Razin E., Ringel E.W., Corey E.J., Hoover D., Austen K.F., Lewis R.A. Immunologic and ionophore-induced generation of leukotriene B4 from mouse bone marrow-derived mast cells. J. Immunol. 1983;130:1885–1890. [PubMed] [Google Scholar]
- 15.Murakami M., Matsumoto R., Urade Y., Austen K.F., Arm J.P. c-kit ligand mediates increased expression of cytosolic phospholipase A2, prostaglandin endoperoxide synthase-1, and hematopoietic prostaglandin D2 synthase and increased IgE-dependent prostaglandin D2 generation in immature mouse mast cells. J. Biol. Chem. 1995;270:3239–3246. doi: 10.1074/jbc.270.7.3239. [DOI] [PubMed] [Google Scholar]
- 16.Warner J.A., Peters S.P., Lichtenstein L.M., Hubbard W., Yancey K.B., Stevenson H.C., Miller P.J., MacGlashan D.W. Differential release of mediators from human basophils: differences in arachidonic acid metabolism following activation by unrelated stimuli. J. Leukoc. Biol. 1989;45:558–571. doi: 10.1002/jlb.45.6.558. [DOI] [PubMed] [Google Scholar]
- 17.Moqbel R., MacDonald A.J., Kay A.B. Enhanced granulocyte cytotoxicity by mediators derived from anti-IgE-stimulated human leucocytes. Immunology. 1986;59:87–93. [PMC free article] [PubMed] [Google Scholar]
- 18.Marone G., Kagey-Sobotka A., Lichtenstein L.M. Effects of arachidonic acid and its metabolites on antigen-induced histamine release from human basophils in vitro. J. Immunol. 1979;123:1669–1677. [PubMed] [Google Scholar]
- 19.Ugajin T., Satoh T., Kanamori T., Aritake K., Urade Y., Yokozeki H. FcεRI, but not FcγR, signals induce prostaglandin D2 and E2 production from basophils. Am. J. Pathol. 2011;179:775–782. doi: 10.1016/j.ajpath.2011.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ugajin T., Kojima T., Mukai K., Obata K., Kawano Y., Minegishi Y., Eishi Y., Yokozeki H., Karasuyama H. Basophils preferentially express mouse Mast Cell Protease 11 among the mast cell tryptase family in contrast to mast cells. J. Leukoc. Biol. 2009;86:1417–1425. doi: 10.1189/jlb.0609400. [DOI] [PubMed] [Google Scholar]
- 21.Dvorak A.M., Seder R.A., Paul W.E., Morgan E.S., Galli S.J. Effects of Interleukin 3 with or without the c-kit ligand. Stem Cell Factor, Surviv. cytoplasmic granule Form. mouse basophils Mast. Cells Vitr., Am. J. Pathol. 1994;144:160–170. [PMC free article] [PubMed] [Google Scholar]
- 22.Poorafshar M., Helmby H., Troye-Blomberg M., Hellman L. MMCP-8, the first lineage-specific differentiation marker for mouse basophils. Elev. Numbers potent IL-4-Prod. MMCP-8-Posit. Cells spleens Malar.-infected mice, Eur. J. Immunol. 2000;30:2660–2668. doi: 10.1002/1521-4141(200009)30:9<2660::AID-IMMU2660>3.0.CO;2-I. [DOI] [PubMed] [Google Scholar]
- 23.Reynolds D.S., Gurley D.S., Austen K.F., Serafin W.E. Cloning of the cDNA and gene of mouse mast cell protease-6. Transcr. Progenit. Mast. Cells Mast. Cells Connect. Tissue subclass, J. Biol. Chem. 1991;266:3847–3853. [PubMed] [Google Scholar]
- 24.Waddington E.I., Croft K.D., Sienuarine K., Latham B., Puddey I.B. Fatty acid oxidation products in human atherosclerotic plaque: an analysis of clinical and histopathological correlates. Atherosclerosis. 2003;167:111–120. doi: 10.1016/s0021-9150(02)00391-x. [DOI] [PubMed] [Google Scholar]
- 25.Razin E., Romeo L.C., Krilis S., Liu F.T., Lewis R.A., Corey E.J., Austen K.F. An analysis of the relationship between 5-lipoxygenase product generation and the secretion of preformed mediators from mouse bone marrow-derived mast cells. J. Immunol. 1984;133:938–945. [PubMed] [Google Scholar]
- 26.Payne A.N., Garland L.G., Lees I.W., Salmon J.A. Selective inhibition of arachidonate 5-lipoxygenase by novel acetohydroxamic acids: effects on bronchial anaphylaxis in anaesthetized guinea-pigs. Br. J. Pharm. 1988;94:540–546. doi: 10.1111/j.1476-5381.1988.tb11558.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.FitzGerald G.A., Patrono C. The coxibs, selective inhibitors of cyclooxygenase-2. N. Engl. J. Med. 2001;345:433–442. doi: 10.1056/NEJM200108093450607. [DOI] [PubMed] [Google Scholar]
- 28.Mardini I.A., FitzGerald G.A. Selective inhibitors of cyclooxygenase-2: a growing class of anti-inflammatory drugs. Mol. Inter. 2001;1:30–38. [PubMed] [Google Scholar]
- 29.Smith C.J., Zhang Y., Koboldt C.M., Muhammad J., Zweifel B.S., Shaffer A., Talley J.J., Masferrer J.L., Seibert K., Isakson P.C. Pharmacological analysis of cyclooxygenase-1 in inflammation. Proc. Natl. Acad. Sci. USA. 1998;95:13313–13318. doi: 10.1073/pnas.95.22.13313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Reddy S.T., Tiano H.F., Langenbach R., Morham S.G., Herschman H.R. Genetic evidence for distinct roles of COX-1 and COX-2 in the immediate and delayed phases of prostaglandin synthesis in mast cells. Biochem Biophys. Res Commun. 1999;265:205–210. doi: 10.1006/bbrc.1999.1658. [DOI] [PubMed] [Google Scholar]
- 31.Thivierge M., Rola-Pleszczynski M. Up-regulation of inducible cyclooxygenase gene expression by platelet-activating factor in activated rat alveolar macrophages. J. Immunol. 1995;154:6593–6599. [PubMed] [Google Scholar]
- 32.Lee S.H., Soyoola E., Chanmugam P., Hart S., Sun W., Zhong H., Liou S., Simmons D., Hwang D. Selective expression of mitogen-inducible cyclooxygenase in macrophages stimulated with lipopolysaccharide. J. Biol. Chem. 1992;267:25934–25938. [PubMed] [Google Scholar]
- 33.Galli S.J., Tsai M. Mast cells in allergy and infection: versatile effector and regulatory cells in innate and adaptive immunity. Eur. J. Immunol. 2010;40:1843–1851. doi: 10.1002/eji.201040559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mukai K., Matsuoka K., Taya C., Suzuki H., Yokozeki H., Nishioka K., Hirokawa K., Etori M., Yamashita M., Kubota T., Minegishi Y., Yonekawa H., Karasuyama H. Basophils play a critical role in the development of IgE-mediated chronic allergic inflammation independently of T cells and mast cells. Immunity. 2005;23:191–202. doi: 10.1016/j.immuni.2005.06.011. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material Supprimentaly Fig. 1. Summary of eicosanoid production and enzyme expressin by basophils and mast cells.