Abstract
Humans and mice differ substantially in their bile acid profiles as mice in addition to cholic acid (CA) predominantly synthesize 6β-hydroxylated muricholic acids (MCAs) whereas humans produces chenodeoxycholic acid (CDCA) and CA as primary bile acids. Identifying the gene performing 6β-hydroxylation would be useful for ‘humanizing’ the bile acid profile in mice for studies of the interaction between bile acids, gut microbiota, and host metabolism. We investigated the formation of MCAs in primary murine hepatocytes and found that αMCA is synthesized from CDCA and βMCA from UDCA. It is commonly assumed that the P450-enzyme CYP3A11 catalyzes 6β-hydroxylation of bile acids, thus we hypothesized that mice without the Cyp3a11 gene would lack MCAs. To test this hypothesis, we analyzed bile acid profiles in Cyp3a deficient mice, which lack 7 genes in the Cyp3a gene cluster including Cyp3a11, and compared them with wild-type littermate controls. Bile acid composition in liver, gallbladder, caecum and serum from Cyp3a knock out mice and wild-type littermate controls was analyzed with UPLC-MS/MS and revealed no major differences in bile acid composition. We conclude that Cyp3a11 is not necessary for 6β-hydroxylation and the formation of MCAs.
Keywords: P450 enzymes, Mouse models, FXR, 6-Hydroxylation
1. Introduction
Bile acids are important signalling molecules that bind to and activate nuclear and membrane receptors involved in a large number of metabolic processes [1], [2], [3], [4]. Farnesoid X receptor (FXR) is the most studied bile acid nuclear receptor and is efficiently activated by chenodeoxycholic acid (CDCA) and cholic acid (CA). It regulates metabolism of bile acids, lipids, and glucose and is therefore an attractive target for the treatment of metabolic diseases [5].
Bile acid-dependent regulation of metabolism via FXR has been widely studied in various mouse models but substantial differences between murine and human bile acid profiles limit the translation of findings into human pathophysiology. Specifically, besides different preferences in the formation of glycine or taurine conjugates, mice synthesize alpha- and betamuricholic acids (α/βMCA) in addition to chenodeoxycholic acid and cholic acid (CDCA and CA), which are the primary bile acids found in humans. The taurine conjugates of the MCAs (Tα/βMCA) have been identified by us and others as potent FXR antagonists [6], [7]. Germ-free mice, which can be used to study the interaction between human bacteria and bile acids when colonized with human microbiota, have higher levels of MCAs (specifically TβMCA) than conventionally raised mice. Ideal mouse models for the study of human gut microbiota and bile acid-regulated metabolism and diseases would be mice with a human bile acid profile, i.e. with TCDCA instead of Tα/βMCA.
It has been suggested that the mouse liver generates αMCA by 6β-hydroxylation of CDCA and that βMCA is generated by further 7β-epimerization of αMCA [8], [9], [10]. Another possible pathway for βMCA formation could be 6β-hydroxylation of ursodeoxycholic acid (UDCA), which is the 7β-isomer of CDCA and a primary bile acid in mice [6]. The P450-enzyme CYP3A11 is commonly presented as bile acid 6β-hydroxylase [11], [12], [13]; however, its involvement in the synthesis of murine bile has never been proven. In this study, we hypothesized that mice without Cyp3a11 would lack murine bile acids and thus could potentially be an improved model for studies of bile acid-dependent metabolism. To investigate if Cyp3a11 is required for the synthesis of murine bile acids we used a commercially available mouse strain with deletion of 7 genes in the Cyp3a gene cluster including Cyp3a11.
2. Materials and methods
2.1. Isolation of primary hepatocytes
Primary murine hepatocytes were isolated using a modified protocol from Zhang at al. [14]. Twelve week-old female C57BL/6 mice were anesthetized with isoflurane and the liver was perfused through the portal vein first with wash buffer (HBSS, with 0.5 mM EGTA and 25 mM HEPES) and then with digestion buffer (DMEM supplemented with 100 U/mL Penicillin, 0.1 mg/mL Streptomycin, 15 mM HEPES and 100 U/mL of collagenase Type IV, Sigma Aldrich) for 15 min.
After sufficient digestion, the gallbladder was removed and the liver cells were liberated in a petri dish containing digestion medium. The cell suspension was then filtered through a 70 µm cell strainer, washed 3 times by centrifugation at 50g for 3 min, and re-suspended in culture medium (DMEM/F12 with 5% Penicillin and Streptomycin and 10% FBS). Viability was assessed by trypan blue; viability was >90% for all cell preparations. Hepatocytes were plated on collagen-coated (Type I collagen; BD) plates and allowed to attach for 4 h at 37 °C in a humidified 5% CO2 incubator. Cells were washed once with culture medium and then switched to an FBS-free culture medium.
2.2. Bile acid experiments in vitro
Isolated primary murine hepatocytes were plated in 12-well plates (400,000 cells/mL). After 2 h, when the cells were attached, the medium was removed and 2 mL fresh medium together with 20 µl deuterium (d4)-labeled CDCA or UDCA (d4-CDCA or d4-UDCA, solved in DMSO) or DMSO alone were added to the cells. The final concentration of d4-bile acids in the wells were 25 µM for CDCA and 100 µM for UDCA. The cells were incubated for 24 h and then 100 µl of the medium was taken for bile acid analysis to determine the profile of d4-labeled bile acids.
2.3. Design of mouse experiments
Mice with a deletion of 7 genes (Cyp3a11, Cyp3a16, Cyp3a25, Cyp3a41, Cyp3a44, Cyp3a57, and Cyp3a59) in the Cyp3a gene cluster (Cyp3a KO) were purchased from Taconic, Model 8841. The mice were bred with C57/Bl6 wild-type mice for at least two generations to obtain homozygous Cyp3a KO mice and wild-type littermate controls. All mice were 10–14 weeks at the time of the analysis. First, blood was taken from portal and caval veins under deep isoflurane anesthesia. Next, the mice were killed and tissues (liver, gallbladder and caecum) were harvested. Whole body, liver, gallbladder, and caecum were weighed and all tissues were frozen in liquid nitrogen and stored at −80 °C until further analyzed. All experiments were performed using protocols approved by the University of Gothenburg Animal Studies Committee.
2.4. Bile acid analysis
Bile acids in liver, gallbladder, caecum and serum from portal and caval veins were analyzed using ultra-performance liquid chromatography-tandem mass spectrometry (UPLC-MS/MS) and quantified using a combination of unlabeled standards and d4-labeled internal standards as previously described [15]. This methodology was also used for measurement of hepatic 7alpha-hydroxy-4-cholesten-3-one (C4) as marker of bile acid synthesis. The whole gallbladder, approximately 50 mg of liver and caecum, and 25 µl serum was used for the analyses.
2.5. Quantitative real-time PCR
Approximately 30 mg of liver was homogenized using TissueLyzer (Qiagen) and total RNA was isolated using RNeasy kit (Qiagen). Conditions for qPCR and primer sequences were previously described [6]. Assays were performed in a 7900HT Fast Real-Time PCR System (Applied Biosystems) or CFX96 Real-Time System (Bio-Rad Laboratories). The reactions were analyzed using the ΔΔCT analysis method.
2.6. Preparation of liver microsomes
Liver biopsies were homogenized in 800 µl lysis buffer (100 mM Triethylammoinium bicarbonate (TEAB), 255 mM Sucrose, 10 mM EDTA) with a Tissuelyser for 3 min (25 Hz). The homogenate were first centrifuged in +4 °C for 5 min (500g) and the supernatants were transferred to new tubes and centrifuged for 20 min (20 000g) in +4 °C. 500 µl of the supernatants were then transferred to a Thick walled Beckman Coulter centrifuge tube (No 343778) prewashed with TEAB and ultra-centrifuged for 60 min (160,000g). The supernatants were discarded and the pellets were washed 3 times with TEAB (100 mM) and then resuspended in 200 µl lysis buffer (200 mM TEAB, 8 M Urea, 4% CHAPS, 0.1%SDS).
2.7. Proteomics analysis of liver microsomal samples
2.7.1. Sample preparation and digestion
The samples were homogenized by sonication in 200 µl lysis buffer (200 mM Triethylammoinium bicarbonate (TEAB), 8 M Urea, 4% CHAPS, 0.1%SDS). Total protein concentration was determined with Pierce™ Pierce 660 nm Protein Assay Reagent (Thermo Scientific). Samples from the different groups were pooled three and three resulting in two samples for each group. Aliquots containing 100 μg of each sample and 100 µg of a reference pool containing equal amounts of all samples were in solution digested with trypsin. Briefly, TEAB (100 mM) was added to a final volume of 90 µl, samples were reduced with 1 mM TCEP at 37 °C for 60 min and alkylated with 2 mM methyl methane thiosulfonate in RT for 20 min. Trypsin (Promega, Sequencing grade) was added in a ratio of 1:20 relative to protein amount and incubated overnight at 37 °C. Digested peptides were labeled using TMT 6-plex isobaric mass tagging reagents (Thermo Scientific) according to the manufacturer instructions. In a set, samples from all groups and the reference, were labeled with a unique tag from a TMT 6plex isobaric mass tag labeling kit. After TMT labeling, the samples in a set were pooled resulting in two independent sets in total to cover all samples.
The peptides were further purified and fractionated into seventeen fractions by Strong Cation Exchange Chromatography (ÄKTA-system, Amersham-Pharmacia) on a PolySULFOETHYL A™ column (100×2.1 mm, 5 µm 300 Å, PolyLC inc.) from 20% to 40% B over 40 min. Solvent A was 25 mM ammonium formate, pH 2.8 and solvent B was 500 mM ammonium formate, pH 2.8.
2.7.2. LC-MS/MS analysis and database search
Each fraction was desalted using PepClean C18 spin columns (Thermo Fisher Scientific) according to the manufacturer's guidelines. Samples were analyzed on an LTQ-Orbitrap Velos mass spectrometer interfaced to an Easy-nLC (Thermo Fisher Scientific). Peptides were separated on a C18 analytical column (200×0.075 mm I.D, 3 µm Dr. Maisch, Germany) over a 80 min gradient from 5% to 40% acetonitrile in 0.1% formic acid. The MS scans was performed at the resolution 30,000 with a mass range of m/z 400–1800. MS/MS analysis was performed in a data-dependent mode at 7500 in resolution and m/z 100–2000, with the top ten most abundant doubly or multiply charged precursor ions in each MS scan selected for MS/MS fragmentation.
For relative quantification, identification and annotation the MS raw data files for each TMT set were merged in the search using Proteome Discoverer version 1.4 (Thermo Fisher Scientific). The database search was performed with the Mascot search engine (Matrix Science) against Mus musculus Swissprot Database version November 2012 (Swiss Institute of Bioinformatics, Switzerland). The data was searched with MS peptide tolerance of 10 ppm and MS/MS tolerance for identification of 0.5 Da. Tryptic peptides were accepted with 1 missed cleavage and variable modifications of methionine oxidation, cysteine methylthiolation and fixed modifications of N-terminal TMT6plex and lysine TMT6plex were selected. The detected peptide threshold in the software was set to 1% False Discovery Rate by searching against a reversed database and identified proteins were grouped by sharing the same sequences to minimize redundancy. Only peptides unique for a given protein were considered for relative quantitation, excluding those common to other isoforms or proteins of the same family.
3. Results and discussion
3.1. Formation of αMCA from CDCA and βMCA from UDCA in primary murine hepatocytes
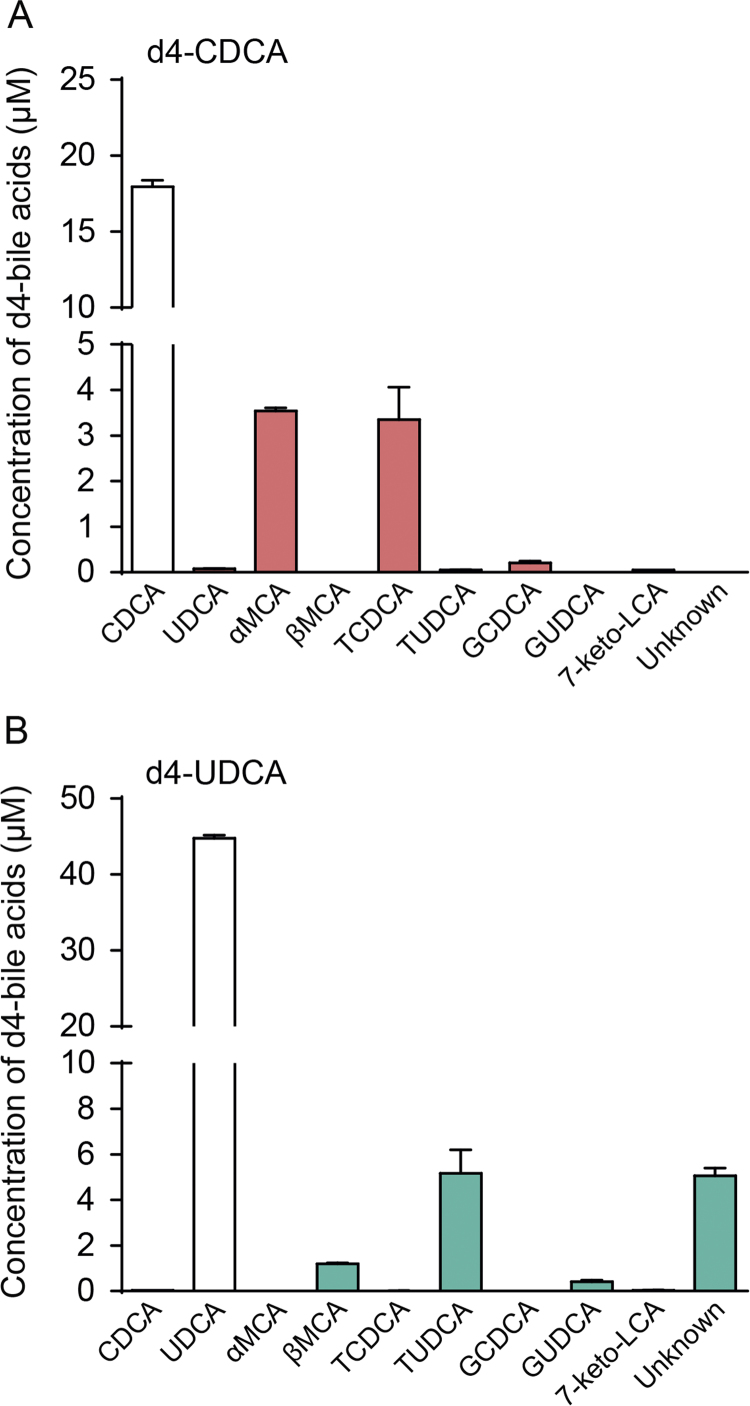
To investigate if MCAs are synthesized from CDCA or UDCA we incubated primary murine hepatocytes isolated from wild-type C57Bl6/J mice with d4-CDCA or d4-UDCA, and analyzed d4-labeled bile acids in the culture media after 24 h. We found that the media from hepatocytes incubated with d4-CDCA contained significant amounts of d4-αMCA and d4-TCDCA but no βMCA (Fig. 1A). In contrast, incubation with d4-UDCA generated d4-βMCA and d4-TUDCA but no d4-αMCA. Thus, βMCA is generated, at least in part, by 6β-hydroxylation of UDCA and not from CDCA, which commonly has been suggested [11], [12], [13].
Fig. 1.
Formation of murine bile acids in primary murine hepatocytes. (A-B) Concentrations of d4-labeled bile acids (µM) in medium collected from primary hepatocytes incubated with d4-CDCA (25 µM) (A) and d4-UDCA (100 µM) (B) for 24 h. Primary hepatocytes were isolated from two individual mice and plated in triplicates; CDCA, chenodeoxycholic acid; MCA, muricholic acid; UDCA, ursodeoxycholic acid; G, glycine-conjugated species; T, taurine-conjugated species.
3.2. Cyp3aKO mice have a largely normal phenotype and produce murine bile acids
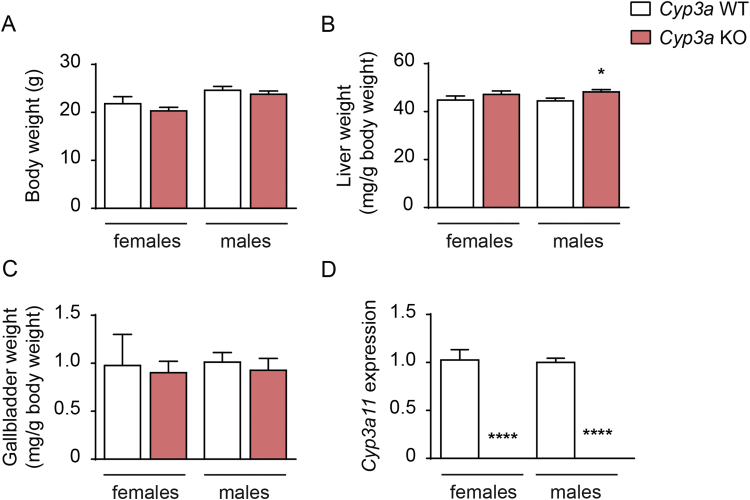
There were no apparent differences in the phenotype between the Cyp3a KO mice and their wild-type littermates. Body weight and gallbladder weights did not differ but relative liver weights were slightly increased in the male Cyp3a KO mice compared with their wild-type controls (48.2±1.0 versus 44.5±1.2 mg/g body weight, p=0.04; Fig. 2A-C). Analysis of Cyp3a11 gene expression in liver showed that Cyp3a11 was absent, verifying that Cyp3a11 was successfully deleted in the Cyp3a KO mice (Fig. 2D).
Fig. 2.
Characterization ofCyp3aKO mice. (A-C) Body weight (A) and relative liver (B) and gallbladder (C) weight in female and male Cyp3a KO and wild-type mice. (D) Gene expression of Cyp3a11 in livers from female and male Cyp3a KO and wild-type mice. Mean values±SEM are plotted; n=5–11 mice or samples/group; * P<0.05, ** P<0.01, *** P<0.001**** P<0.0001 indicate differences between female Cyp3a KO and wild-type or male Cyp3a KO and wild-type mice analyzed with unpaired t-test.
To determine if deletion of the Cyp3a gene cluster would affect bile acid composition we analyzed bile acid levels in liver, gallbladder, caecum and serum from portal and caval veins. Total bile acid levels were unchanged in all the analyzed compartments and there were only minor changes in the amounts of specific bile acids (Table 1, Supplemental Tables 1–3). In male Cyp3a KO mice, we found a slight decrease of TαMCA in the gallbladder whereas the levels of TβMCA were unchanged (Table 1). In liver and caecum there were no differences in the amount of TαMCA or TβMCA (Table 1, Supplemental Table 1). In serum from portal and caval veins, there were no significant changes in any bile acid, and there was rather a tendency towards higher levels of both TαMCA and TβMCA in Cyp3a KO mice (Supplemental Tables 2–3). These findings show that Cyp3a-deficient mice still synthesize normal amounts of murine bile acids.
Table 1.
Bile acid composition in liver and gallbladder. Liver (pmol/mg tissue) and gallbladder (nmol/mg tissue) bile acids in female and male Cyp3a KO and wild type mice.
| Liver bile acids (pmol/mg tissue) | Cyp3a KO females | Cyp3a WT females | Cyp3a KO males | Cyp3a WT males |
|---|---|---|---|---|
| TαMCA | 13.6 ± 3.9 | 8.4 ± 1.2 | 7.2 ± 1.1 | 8.0 ± 1.3 |
| TβMCA | 38.2 ± 9.4 | 26.8 ± 6.1 | 13.4 ± 2.6 | 18.6 ± 3.2 |
| TωMCA | 19.8 ± 3.9 | 11.4 ± 1.9 | 11.7 ± 1.4 | 11.4 ± 1.6 |
| TCA | 73.2 ± 16.1 | 69.1 ± 12.7 | 34.5 ± 2.5 | 54.8 ± 9.4 |
| TCDCA | 2.7 ± 0.5 | 2.0 ± 0.1 | 1.7 ± 0.2 | 1.5 ± 0.2 |
| TDCA | 5.3 ± 0.7 | 7.5 ± 1.3 | 6.6 ± 1.3 | 7.3 ± 1.3 |
| TUDCA | 2.1 ± 0.5 | 1.6 ± 0.16 | 1.6 ± 0.6 | 1.2 ± 0.30 |
| TLCA | traces | traces | 0.2 ± 0.0 | traces |
| αMCA | traces | traces | traces | |
| βMCA | 0.4 ± 0.1 | 0.3 ± 0.1 | 0.2 ± 0.1 | 0.2 ± 0.0 |
| ωMCA | 0.4 ± 0.1 | traces | 0.3 ± 0.0 | 0.2 ± 0.0 |
| CA | 1.2 ± 0.4 | 0.7 ± 0.2 | 0.6 ± 0.1 | 0.6 ± 0.1 |
| Total bile acids | 156.8 ± 35.7 | 127.9 ± 23.8 | 77.9 ± 10.1 | 103.8 ± 17.6 |
| C4 | 0.5 ± 0.1 | 0.4 ± 0.1 | 0.2 ± 0.0 | 0.2 ± 0.0 |
| total DCA/total CA ratio | 0.1 ± 0.0 | 0.1 ± 0.0 | 0.2 ± 0.0 | 0.2 ± 0.0 |
| Gallbladder bile acids (nmol/mg tissue) | Cyp3a KO females | Cyp3a WT females | Cyp3a KO males | Cyp3a WT males |
|---|---|---|---|---|
| TαMCA | 9.7 ± 1.6 | 7.5 ± 1.5 | 12.0 ± 1.3 | 7.4 ± 0.5 ** |
| TβMCA | 18.8 ± 2.2 | 17.7 ± 2.7 | 15.3 ± 2.4 | 12.5 ± 1.1 |
| TωMCA | 9.8 ± 1.0 | 6.3 ± 0.7 | 10.4 ± 1.4 | 7.5 ± 0.6 |
| TCA | 67.1 ± 4.3 | 55.2 ± 7.3 | 58.6 ± 6.7 | 56.7 ± 3.6 |
| TCDCA | 1.3 ± 0.2 | 0.8 ± 0.2 | 1.3 ± 0.2 | 0.7 ± 0.1 |
| TDCA | 1.5 ± 0.2 | 1.7 ± 0.2 | 2.2 ± 0.1 | 2.3 ± 0.2 |
| TUDCA | 1.0 ± 0.2 | 0.9 ± 0.2 | 1.1 ± 0.1 | 0.6 ± 0.1 ** |
| CA | 0.9 ± 0.3 | 0.6 ± 0.2 | 1.1 ± 0.1 | 1.1 ± 0.3 |
| Total bile acids | 110.0 ± 10.0 | 90.7 ± 12.9 | 102.0 ± 12.4 | 88.9 ± 6.5 |
Data are presented as mean values±SEM; n=5–11 mice/group; * P<0.05, ** P<0.01, *** P<0.001**** P<0.0001 indicate differences between female Cyp3a KO and wild type or male Cyp3a KO and wild type mice SEM analyzed with multiple t-test with correction using the Holm-Sidak method.; CA, cholic acid; CDCA, chenodeoxycholic acid; DCA, deoxycholic acid; LCA, lithocholic acid; MCA, muricholic acid; UDCA, ursodeoxycholic acid; T, taurine-conjugated species; C4, 7α-hydroxy-cholestene-4-one.
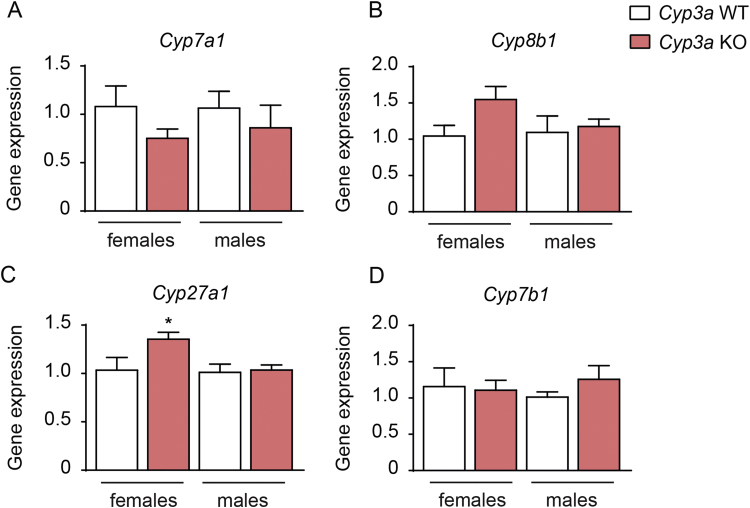
It has been described that mouse liver in contrast to human liver can re-hydroxylate DCA into CA [16]. To investigate if the livers of Cyp3a-deficient mice have reduced capacity to re-hydroxylate DCA we calculated the hepatic ratio between total DCA (TDCA+DCA) and total CA (TCA+CA). The Cyp3a KO mice had similar hepatic total DCA/total CA ratios as the wild-type control mice, which indicates that deletion of the genes in the Cyp3a gene cluster does not affect 7α-rehydroxylation of DCA (Table 1). In line with the minor changes in bile acid levels we found similar expression of the bile acid synthetic genes involved in the classical (neutral) and alternative (acidic) pathways of bile acid synthesis (Cyp7a1, Cyp8b1, Cyp27a1 and Cyp7b1) in Cyp3a KO and wild-type control mice (Fig. 3A-D), which is also reflected by similar hepatic C4 levels (Table 1). The only significant difference was a small increase in Cyp27a1 expression in the female Cyp3a KO mice (Fig. 3C).
Fig. 3.
Expression of genes involved in bile acid synthesis. (A-D) Expression of cytochrome P450 genes Cyp7a1 (A), Cyp8b1 (B), Cyp27a1 (C) and Cyp7b1 (D) in livers from female and male Cyp3a KO and wild-type mice. Mean values±SEM are plotted; n=5–11 mice or samples/group; * P<0.05, ** P<0.01, *** P<0.001**** P<0.0001 indicate differences between female Cyp3a KO and wild-type or male Cyp3a KO and wild-type mice analyzed with unpaired t-test.
Our study show that Cyp3a11 is not essential for the formation of murine bile acids. This is in agreement with a very recent study by Takahashi et al., which showed that mice without the Cyp3a gene cluster produce normal amounts of MCAs and suggested that Cyp2c70 is responsible for the species differences in bile acid metabolism between mice and humans [17]. To investigate if the expression of Cyp2c70 is elevated in germ-free mice, since they have increased levels of MCAs, we evaluated expression data from a previous microarray study performed in our group. Indeed, germ-free mice show higher expression of the Cyp2c70 gene than conventionally raised mice [18]. In contrast, the expression of Cyp3a11 was significantly lower in the germ-free mice supporting the dispensability of CYP3A11 in MCA formation. We also found a similar trend towards lower CYP3A11 and higher CYP2C70 levels in germ-free mice using proteomics (Fig. S1A-B).
Furthermore, we show that βMCA is generated from 6β-hydroxylation of UDCA in vitro and that synthesis of αMCA does not need to precede βMCA formation (Fig. 1A-B). This is an important finding and provides a new insight about the formation of bile acids. Takahashi et al. indicated that epimerization of αMCA to βMCA might occur in the mouse intestine by gut microbes [17] but our previous data suggest that at least some βMCA is produced in the liver since germ-free mice have higher levels of TβMCA compared with conventionally raised mice [6].
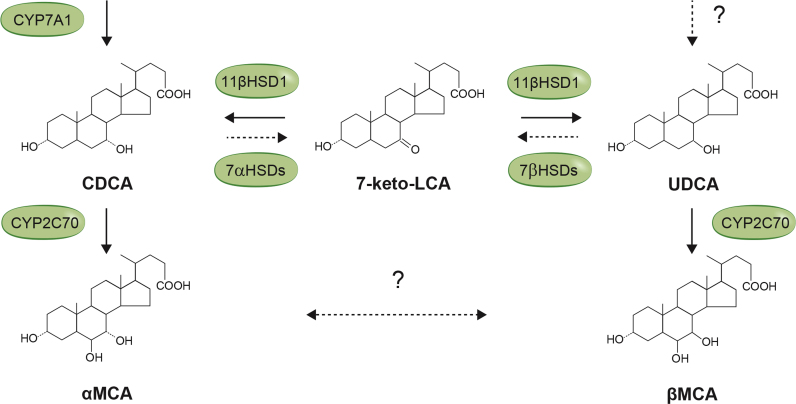
UDCA can be synthesized in the liver with 7-keto-LCA as a potential intermediate (Fig. 4). Several studies have shown that 7-keto-LCA is formed by gut bacteria with 7-hydroxysteroid dehydrogenase activity [19], [20], [21]. Other studies have shown that 7-keto-LCA is converted to CDCA and UDCA [22], [23] and studies with human and murine liver microsomes characterized 11β-hydroxysteroid dehydrogenase 1 (11β-HSD1) as the responsible enzyme [24], [25]. The conversion of CDCA to UDCA might occur in the mouse liver and this reaction is then likely to precede the formation of βMCA (Fig. 4). However, our in vitro experiments with primary murine hepatocytes did not show conversion from CDCA to UDCA and only traces of 7-keto-LCA (Fig. 1A). This, together with our previous findings that UDCA is a primary bile acid in mice and can be produced in the absence of bacteria, suggests that UDCA might be formed by an alternative pathway in the mouse liver (Fig. 4).
Fig. 4.
Synthesis of murine bile acids in mice. 7-Keto-LCA is an intermediate between CDCA and UDCA produced by bacteria with 7α/β-hydroxysteroid dehydrogenases (7α/βHSDs). 7-Keto-LCA can be converted to CDCA or UDCA by 11β-hydroxysteroid dehydrogenase1 (11βHSD1) in the liver. Our in vitro experiments showed that αMCA and βMCA can be synthesized from CDCA and UDCA, respectively. Further experiments are required to investigate if UDCA can be produced by an alternative pathway and if epimerization between αMCA and βMCA can occur in the liver and/or in the intestine by the gut microbiota.
In the future, Cyp2c70 KO mice may become an attractive “humanized” animal model for studies of bile acid-regulated metabolism and it would be important to investigate if germ-free Cyp2c70 KO mice also lack the murine bile acids and have a human bile acid profile.
Conflict of interest
The authors declare no competing financial interests of relevance for this manuscript.
Acknowledgements
We are grateful to Antonio Molinaro for isolation of primary hepatocytes, Anna Hallén for excellent technical assistance and Zakarias Gulic for superb mouse husbandry. The Proteomics Core Facility at Sahlgrenska Academy, Gothenburg University, performed the analysis for protein quantification. This study was supported by the Swedish Research Council and the regional agreement on medical training and clinical research (ALF) between Region Västra Götaland and Sahlgrenska University Hospital.
Footnotes
Supplementary data associated with this article can be found in the online version at doi:10.1016/j.bbrep.2017.02.011.
Appendix A. Supplementary material
Supplementary material Relative protein levels in liver microsomes. Relative protein level of CYP3A11 (A) and CYP2C70 (B) in liver microsomal preparations from germ-free (GF) and conventionally raised (CONV-R) mice. Each bar respresents n=2 samples/group and each sample contains proteins pooled from three different mice.
.
Supplementary material
.
Supplementary material
.
Supplementary material
.
References
- 1.Lefebvre P., Cariou B., Lien F., Kuipers F., Staels B. Role of bile acids and bile acid receptors in metabolic regulation. Physiol. Rev. 2009;89:147–191. doi: 10.1152/physrev.00010.2008. [DOI] [PubMed] [Google Scholar]
- 2.Ma H., Patti M.E. Bile acids, obesity, and the metabolic syndrome. Best. Pract. Res. Clin. Gastroenterol. 2014;28:573–583. doi: 10.1016/j.bpg.2014.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sato H., Macchiarulo A., Thomas C., Gioiello A., Une M., Hofmann A.F., Saladin R., Schoonjans K., Pellicciari R., Auwerx J. Novel potent and selective bile acid derivatives as TGR5 agonists: biological screening, structure-activity relationships, and molecular modeling studies. J. Med. Chem. 2008;51:1831–1841. doi: 10.1021/jm7015864. [DOI] [PubMed] [Google Scholar]
- 4.Wahlstrom A., Sayin S.I., Marschall H.U., Backhed F. Intestinal crosstalk between bile acids and microbiota and its impact on host metabolism. Cell Metab. 2016;24:41–50. doi: 10.1016/j.cmet.2016.05.005. [DOI] [PubMed] [Google Scholar]
- 5.Kuipers F., Bloks V.W., Groen A.K. Beyond intestinal soap--bile acids in metabolic control. Nat. Rev. Endocrinol. 2014;10:488–498. doi: 10.1038/nrendo.2014.60. [DOI] [PubMed] [Google Scholar]
- 6.Sayin S.I., Wahlstrom A., Felin J., Jantti S., Marschall H.U., Bamberg K., Angelin B., Hyotylainen T., Oresic M., Backhed F. Gut microbiota regulates bile acid metabolism by reducing the levels of tauro-beta-muricholic acid, a naturally occurring FXR antagonist. Cell Metab. 2013;17:225–235. doi: 10.1016/j.cmet.2013.01.003. [DOI] [PubMed] [Google Scholar]
- 7.Li F., Jiang C., Krausz K.W., Li Y., Albert I., Hao H., Fabre K.M., Mitchell J.B., Patterson A.D., Gonzalez F.J. Microbiome remodelling leads to inhibition of intestinal farnesoid X receptor signalling and decreased obesity. Nat. Commun. 2013;4:2384. doi: 10.1038/ncomms3384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hsla S.L., Matschiner J.T., Mahowald T.A., Elliott W.H., Doisy E.A., Jr., Thayer S.A., Doisy E.A. Bile acids. VI. The structure and synthesis of acid II. J. Biol. Chem. 1957;226:667–671. [PubMed] [Google Scholar]
- 9.Mahowald T.A., Matschiner J.T., Hsia S.L., Richter R., Doisy E.A., Jr., Elliott W.H., Doisy E.A. Bile acids. II. metabolism of deoxycholic acid-24-C14 and chenodeoxycholic acid-24-C14 in the rat. J. Biol. Chem. 1957;225:781–793. [PubMed] [Google Scholar]
- 10.Botham K.M., Boyd G.S. The metabolism of chenodeoxycholic acid to beta-muricholic acid in rat liver. Eur. J. Biochem. 1983;134:191–196. doi: 10.1111/j.1432-1033.1983.tb07550.x. [DOI] [PubMed] [Google Scholar]
- 11.Marschall H.U., Wagner M., Bodin K., Zollner G., Fickert P., Gumhold J., Silbert D., Fuchsbichler A., Sjovall J., Trauner M. Fxr(-/-) mice adapt to biliary obstruction by enhanced phase I detoxification and renal elimination of bile acids. J. Lipid Res. 2006;47:582–592. doi: 10.1194/jlr.M500427-JLR200. [DOI] [PubMed] [Google Scholar]
- 12.Cuesta S., de Juan, Monte M.J., Macias R.I., Wauthier V., Calderon P.B., Marin J.J. Ontogenic development-associated changes in the expression of genes involved in rat bile acid homeostasis. J. Lipid Res. 2007;48:1362–1370. doi: 10.1194/jlr.M700034-JLR200. [DOI] [PubMed] [Google Scholar]
- 13.Li T., Chiang J.Y. Nuclear receptors in bile acid metabolism. Drug Metab. Rev. 2013;45:145–155. doi: 10.3109/03602532.2012.740048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang W., Sargis R.M., Volden P.A., Carmean C.M., Sun X.J., Brady M.J. PCB 126 and other dioxin-like PCBs specifically suppress hepatic PEPCK expression via the aryl hydrocarbon receptor. PLoS One. 2012;7:e37103. doi: 10.1371/journal.pone.0037103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tremaroli V., Karlsson F., Werling M., Stahlman M., Kovatcheva-Datchary P., Olbers T., Fandriks L., le Roux C.W., Nielsen J., Backhed F. Roux-en-Y gastric bypass and vertical banded gastroplasty induce long-term changes on the human gut microbiome contributing to fat mass regulation. Cell Metab. 2015;22:228–238. doi: 10.1016/j.cmet.2015.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hofmann A.F., Hagey L.R. Bile acids: chemistry, pathochemistry, biology, pathobiology, and therapeutics. Cell. Mol. Life Sci. 2008;65:2461–2483. doi: 10.1007/s00018-008-7568-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Takahashi S., Fukami T., Masuo Y., Brocker C.N., Xie C., Krausz K.W., Wolf C.R., Henderson C.J., Gonzalez F.J. Cyp2c70 is responsible for the species difference in bile acid metabolism between mice and humans. J. Lipid Res. 2016 doi: 10.1194/jlr.M071183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Larsson E., Tremaroli V., Lee Y.S., Koren O., Nookaew I., Fricker A., Nielsen J., Ley R.E., Backhed F. Analysis of gut microbial regulation of host gene expression along the length of the gut and regulation of gut microbial ecology through MyD88. Gut. 2012;61:1124–1131. doi: 10.1136/gutjnl-2011-301104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hirano S., Masuda N. Epimerization of the 7-hydroxy group of bile acids by the combination of two kinds of microorganisms with 7 alpha- and 7 beta-hydroxysteroid dehydrogenase activity, respectively. J. Lipid Res. 1981;22:1060–1068. [PubMed] [Google Scholar]
- 20.MacDonald I.A., Rochon Y.P., Hutchison D.M., Holdeman L.V. Formation of ursodeoxycholic acid from chenodeoxycholic acid by a 7 beta-hydroxysteroid dehydrogenase-elaborating Eubacterium aerofaciens strain cocultured with 7 alpha-hydroxysteroid dehydrogenase-elaborating organisms. Appl. Environ. Microbiol. 1982;44:1187–1195. doi: 10.1128/aem.44.5.1187-1195.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fromm H., Sarva R.P., Bazzoli F. Formation of ursodeoxycholic acid from chenodeoxycholic acid in the human colon: studies of the role of 7-ketolithocholic acid as an intermediate. J. Lipid Res. 1983;24:841–853. [PubMed] [Google Scholar]
- 22.Fromm H., Carlson G.L., Hofmann A.F., Farivar S., Amin P. Metabolism in man of 7-ketolithocholic acid: precursor of cheno- and ursodeoxycholic acids. Am. J. Physiol. 1980;239:G161–G166. doi: 10.1152/ajpgi.1980.239.3.G161. [DOI] [PubMed] [Google Scholar]
- 23.Amuro Y., Yamade W., Kudo K., Yamamoto T., Hada T., Higashino K. Reduction of 7-ketolithocholic acid by human liver enzyme preparations in vitro. Am. J. Physiol. 1989;256:G67–G71. doi: 10.1152/ajpgi.1989.256.1.G67. [DOI] [PubMed] [Google Scholar]
- 24.Odermatt A., Da Cunha T., Penno C.A., Chandsawangbhuwana C., Reichert C., Wolf A., Dong M., Baker M.E. Hepatic reduction of the secondary bile acid 7-oxolithocholic acid is mediated by 11beta-hydroxysteroid dehydrogenase 1. Biochem. J. 2011;436:621–629. doi: 10.1042/BJ20110022. [DOI] [PubMed] [Google Scholar]
- 25.Penno C.A., Morgan S.A., Vuorinen A., Schuster D., Lavery G.G., Odermatt A. Impaired oxidoreduction by 11beta-hydroxysteroid dehydrogenase 1 results in the accumulation of 7-oxolithocholic acid. J. Lipid Res. 2013;54:2874–2883. doi: 10.1194/jlr.M042499. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material Relative protein levels in liver microsomes. Relative protein level of CYP3A11 (A) and CYP2C70 (B) in liver microsomal preparations from germ-free (GF) and conventionally raised (CONV-R) mice. Each bar respresents n=2 samples/group and each sample contains proteins pooled from three different mice.
Supplementary material
Supplementary material
Supplementary material