Abstract
The liver plays a central role in metabolism. Although many studies have described in vitro liver models for drug discovery, to date, no model has been described that can stably maintain liver function. Here, we used a unique, scaffold-free 3D bio-printing technology to construct a small portion of liver tissue that could stably maintain drug, glucose, and lipid metabolism, in addition to bile acid secretion. This bio-printed normal human liver tissue maintained expression of several kinds of hepatic drug transporters and metabolic enzymes that functioned for several weeks. The bio-printed liver tissue displayed glucose production via cAMP/protein kinase A signaling, which could be suppressed with insulin. Bile acid secretion was also observed from the printed liver tissue, and it accumulated in the culture medium over time. We observed both bile duct and sinusoid-like structures in the bio-printed liver tissue, which suggested that bile acid secretion occurred via a sinusoid-hepatocyte-bile duct route. These results demonstrated that our bio-printed liver tissue was unique, because it exerted diverse liver metabolic functions for several weeks. In future, we expect our bio-printed liver tissue to be applied to developing new models that can be used to improve preclinical predictions of long-term toxicity in humans, generate novel targets for metabolic liver disease, and evaluate biliary excretion in drug development.
Abbreviations: NASH, Non-alcoholic steatohepatitis; Dex, Dexamethasone; 8CPT-cAMP, 8-(4-Chlorophenylthio)adenosine 3′,5′-cyclic monophosphate; HE, hematoxylin and eosin; MT, Masson's trichrome; TUNEL, TdT-mediated dUTP nick end labeling; MRP2, multidrug resistance-associated protein 2; OAT, organic anion-transporting; NAFLD, Non-alcoholic fatty liver disease; ECM, Extracellular matrix
Keywords: Scaffold-free, 3D, Bio-printing, Liver, Drug discovery, Metabolism
Highlights
-
•
We constructed a unique human mini-liver model with 3D bio-printing technology.
-
•
Liver model maintained the drug-metabolizing functions for at least 7 weeks.
-
•
Liver model cultured for 11 weeks could regulate glucose production by insulin.
-
•
Liver model maintained NAFLD disease pathology for at least 3 weeks.
-
•
Liver model cultured for at least 3 weeks could secrete bile acid in the medium.
1. Introduction
The liver plays a central role in metabolism. Most metabolic functions are performed by hepatic parenchymal cells, which comprise about 78% of the liver [1]. Many previous studies have described in vitro human liver models [2] that comprised two-dimensional (2D) or suspension cultures of human primary hepatocytes. However, those models typically lost function rapidly [3]; therefore, drug discovery research is lacking a model capable of sustaining liver function over a long period. In particular, there is an extremely urgent need for models that can stably sustain drug metabolism function to enable in vitro predictions of drug hepatotoxicity and metabolite production in humans, which occur with long-term compound exposures [4], [5].
Several models capable of maintaining drug metabolism function for long times are known, including a micropatterned model [6], a 3D model based on transwell cultures [7], a spheroid 3D model [8], a model constructed with scaffold-free 3D bio-printing [9]. Among these models, the micropatterned model could recapitulate, to some extent, the appearance of liver toxicity [10] and metabolite [11] predicted in clinical trials conducted in collaboration with pharmaceutical companies. Thus, compared to other existing technologies, the micropatterned model is considered to represent a relatively high level of technology.
Glucose and lipid metabolism are two important metabolic functions in the liver. Diabetes, obesity, and non-alcoholic steatohepatitis (NASH) are known metabolic diseases that involve the liver. A human liver model that maintains the functions related to glucose and lipid metabolism for long times could be used to construct an in vivo-like human disease model. That model would be expected to contribute to the generation of novel drug discovery targets. Among existing technologies, the micropatterned model can maintain regulated glucose production for a long period [12]. In addition, it was used to construct an insulin resistance model with long-term glucose stimulation [13]. As described above, the ability of the micropatterned model to sustain glucose and lipid metabolism functions for a long time have made it a useful technique for constructing a human disease model.
Another role of the liver is bile secretion. Although a sandwich 2D model is often used to predict biliary excretion, its function is only stable for short durations [6], [14]. Moreover, its predictions of human hepatotoxicity [4] and metabolite [5] production are inferior to those of the micropatterned model. Yet another model has combined the micropattern and sandwich model approaches, it is not quantitatively accurate [15]. Therefore, a novel human 3D biliary excretion model capable of quantifying metabolites in secreted bile is highly desirable.
In the present study, we aimed to establish a method for stably producing small portions of human liver tissue with 3D bio-printing technology. In future, this unique human liver model could be widely useful in drug discovery research applications, such as safety, pharmacokinetics, identification of new drug discovery targets.
2. Materials and methods
2.1. Spheroid formation
Cryopreserved hepatocytes (HMCS1SA, Life Technologies or HUCSD, Triangle Research Laboratories) were used for this study. Spheroid formation was performed with commercially-available, U-bottom-type, low-binding 96-well plates. Primary hepatocytes (104 cells) and mouse fibroblasts (104 cells) were seeded in 96-well plates and cultured in the medium including Williams’ Medium E (Thermo Fisher Scientific), Primary Hepatocyte Maintenance Supplements (CM4000, Thermo Fisher Scientific), DMEM (Thermo Fisher Scientific), and FBS for 3 days.
2.2. 3D bio-printing and culture
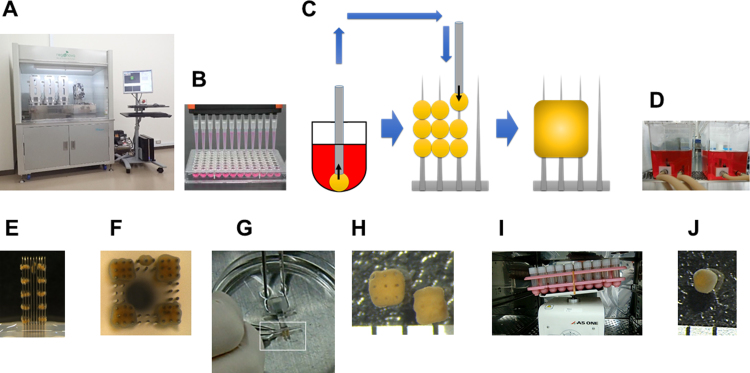
To produce functional 3D bio-printed liver tissue, we used a scaffold-free 3D bio-printer, Regenova® (Fig. 1) [16]. We constructed liver tissue with 9 spheroids, which were automatically selected and skewered onto a 9×9 needle array (NA1002, Cyfuse Biomedical). Spheroids skewered on the needle array were cultured in a perfusion chamber (VS0101, Cyfuse Biomedical) for 4 days. After the spheroids had fused to form liver tissue, the tissues were removed. Each of the tissues produced was placed in a screw-cap vial with 1 ml of culture medium and cultured with moderate rocking or shaking for more than 2 weeks.
Fig. 1.
Materials and process for manufacturing 3D bio-printed human liver tissue. (A) Photograph of the Regenova® apparatus. (B) Photograph of spheroids cultured in a U-bottom-type low-binding 96-well plate. (C) Schematic shows (left) collecting a spheroid, (middle) placing spheroids on skewers, and (right) tissue formation due to spheroid fusion on the 9×9 needle array. (D) Photograph of the perfusion chambers used as circulation cultures for spheroids that are skewered onto the needle array. (E and F) Photographs show the (E) side view and (F) top view of bio-printed liver tissues after 4 days of circulation culture, which promoted spheroid fusion on the needle array. (G) Photograph shows the removal of a printed liver tissue from the needle array. (H) Photograph shows bio-printed liver tissues after removal. Tissues are cubical in shape, with nine holes created by the needles. Scale=1 mm. (I) Photograph of rocking cultures for maintaining printed liver tissues. (J) After 60 days of culture, the shape of the printed liver tissue is roughly spherical, with a diameter of ~1 mm. Scale=1 mm.
2.3. Microarray analyses
Microarray (Agilent array) analyses were performed at Takara Bio Inc.
2.4. Gene expression analyses
Total RNA was extracted from tissue with TRIzol reagent (Thermo Fisher Scientific) and purified with the RNeasy 96 Kit (Qiagen). cDNA was synthesized with the PrimeScript RT reagent kit (Perfect Real Time, Takara Bio). Real-time PCR was carried out with the Power SYBR Green PCR Master Mix (Thermo Fisher Scientific), according to the manufacturer's instructions. PCR amplification was conducted on a StepOnePlus Real Time PCR System (Applied Biosystems). The primer sequences for human CYP3A4 were: 5′-TTCAGCAAGAAGAACAAGGACAA-3′ (forward) and 5′-GGTTGAAGAAGTCCTCCTAAGC-3′ (reverse). The primer sequences for human GAPDH were: 5′-AGAAGGCTGGGGCTCATTT-3′ (forward) and 5′-TGGACTGTGGTCATGAGTCCTT-3′ (reverse).
2.5. CYP3A4 activity determination
CYP3A4 activity in tissue was determined with a P450-Glo CYP3A4 Assay (Promega). A standard curve was generated with D-luciferin (Promega). The luminescence was measured with a Fluoroskan Ascent FL (ThermoScientific).
2.6. Glucose production assay
Bio-printed liver tissue (day 77) was washed three times with serum-free medium and incubated with the same medium at 37 °C in a CO2 incubator. After the tissue was washed three times with glucose-free medium, insulin (10 μM) was added to the tissue medium and incubated for 1 h at 37 °C in a CO2 incubator. In addition, tissue glucose production was measured by adding dexamethasone (Dex; 500 nM) and 8-(4-Chlorophenylthio)adenosine 3′,5′-cyclic monophosphate (8CPT-cAMP; 100 μM) to the medium and incubating for 24 h at 37 °C in a CO2 incubator. The glucose concentration in the medium was quantified with the Amplex Red Glucose/Glucose Oxidase Assay Kit (Molecular Probes). The absorbance at 492 nm was measured with a MULTISKAN FC (ThermoScientific).
2.7. Construction of bio-printed rat pathologic liver tissue
A Zucker fatty rat (male, 11 weeks) was purchased from Charles river. Construction of bio-printed rat liver tissue was basically carried out according to the methods described in Sections 2.1 and 2.2, Sections 2.1 and 2.2 above, except that the hepatocytes were derived from the Zucker fatty rat. On day 23, the structure was stained with hematoxylin and eosin (HE) and Oil-red O. Tissue staining was performed at Genostaff.
2.8. Bile acid secretion assay
The bile acids secretion in the medium were measured using the bio-printed liver tissues on day 24 for 3 days with the Total Bile Acid Assay Kit (Cell Biolabs, Inc.). The absorbance was measured at 405–620 nm with a MULTISKAN FC (ThermoScientific).
2.9. Tissue and immunochemical staining
Tissues were stained with HE, Masson's trichrome (MT), and oil-red O; labeled with TdT-mediated dUTP nick end labeling (TUNEL); and immunostained with antibodies against multidrug resistance-associated protein 2 (MRP2; Abcam), the endothelial marker, CD31 (Abcam), and organic anion-transporting polypeptides (OAT2/8; Novus Biologicals). All these assays were performed at Genostaff.
3. Results
3.1. Fabrication of 3D bio-printed human liver tissue
Fig. 1 is a schematic of the fabrication of 3D bio-printed human liver tissue. On day 0, primary hepatocytes were plated in low-binding, 96-well plates and cultured for 3 days. On day 3, spheroids that had formed were collected, and skewered onto a 9×9 needle array housed in the 3D bio-printer (Regenova). Skewered spheroids were cultured on the needle array in a perfusion chamber for 4 days. On day 7, spheroids had fused to form liver tissues; these tissues were removed from the needle array and cultured for about 2 weeks. Thus, at about 20 days after cell plating, 3D bio-printed liver tissues were ready for use in experiments.
3.2. 3D bio-printed liver tissue displayed a wide range of long-lasting liver functions
To test bio-printed liver tissue gene expression levels comprehensively, we performed microarray analyses. We compared gene expression in bio-printed liver tissues on day 22 with that of hepatocytes on day 0. These comparisons showed similar levels of expression for most genes related to key liver functions, including genes related to the metabolism of drugs, glucose, lipid, and urea, and genes that encoded transporter and serum proteins (Table 1). These results suggested that bio-printed liver tissue could maintain extensive liver functions for a long time (2 weeks).
Table 1.
Microarray analyses data about key liver function-related genes.
| Drug metabolizing enzyme | Drug transporter | ||||||||
| Reaction | Gene | Gene/GAPDH | Fold change | Distribution | Gene | Gene/GAPDH | Fold change | ||
| Day 0 | Day 29 | Day 0 | Day 29 | ||||||
| Phase I | CYP1A2 | 2.203 | 3.457 | 1.569 | Blood | OAT2 | 0.056 | 0.119 | 2.115 |
| CYP2B6 | 1.181 | 4.002 | 3.390 | OCT1 | 0.353 | 0.631 | 1.789 | ||
| CYP2C9 | 0.848 | 1.233 | 1.454 | OCTN2 | 0.014 | 0.017 | 1.193 | ||
| CYP2C19 | 0.246 | 0.256 | 1.037 | OATP1B1 | 0.122 | 0.157 | 1.283 | ||
| CYP2D6 | 0.082 | 0.145 | 1.771 | OATP1B3 | 0.014 | 0.001 | 0.032 | ||
| CYP3A4 | 0.168 | 2.712 | 16.10 | OATP2B1 | 0.047 | 0.196 | 4.222 | ||
| MAOA | 0.105 | 0.158 | 1.503 | NTCP | 0.170 | 0.210 | 1.238 | ||
| Phase II | UGT2B4 | 0.328 | 1.125 | 3.429 | MRP3 | 0.051 | 0.098 | 1.931 | |
| UGT1A6 | 0.833 | 1.204 | 1.444 | MRP6 | 0.047 | 0.087 | 1.838 | ||
| GSTT2 | 0.003 | 0.003 | 1.129 | Bile | MDR1 | 0.022 | 0.021 | 0.951 | |
| GSTA4 | 0.003 | 0.019 | 5.824 | MRP1 | 0.001 | 0.001 | 1.362 | ||
| SULT1A2 | 0.432 | 0.519 | 1.199 | MRP2 | 0.109 | 0.277 | 2.536 | ||
| COMT | 0.081 | 0.334 | 4.141 | BCRP | 0.001 | 0.002 | 1.844 | ||
| HNMT | 0.006 | 0.014 | 2.271 | MATE1 | 0.016 | 0.011 | 0.716 | ||
| NAT1 | 0.001 | 0.012 | 9.245 | BSEP | 0.013 | 0.013 | 1.075 | ||
| Metabolism related protein | |||||||||
| Function | Gene | Gene/GAPDH | Fold change | ||||||
| Day 0 | Day 29 | ||||||||
| Gluconeogenesis | GCGR | 0.222 | 0.143 | 0.647 | |||||
| PRKACA | 0.014 | 0.019 | 1.339 | ||||||
| G6PC | 0.204 | 0.202 | 0.991 | ||||||
| Glycolysis | INSR | 0.103 | 0.127 | 1.241 | |||||
| AKT1 | 0.137 | 0.181 | 1.320 | ||||||
| PDHA1 | 0.160 | 0.179 | 1.115 | ||||||
| TCA cycle | IDH1 | 0.074 | 0.199 | 2.692 | |||||
| Glycogen | GYS2 | 0.014 | 0.024 | 1.777 | |||||
| PYGL | 0.106 | 0.083 | 0.778 | ||||||
| Fatty acid synthesis | ACSL1 | 0.083 | 0.266 | 3.221 | |||||
| ACACA | 0.034 | 0.094 | 2.790 | ||||||
| SREBF1 | 0.542 | 0.560 | 1.033 | ||||||
| Cholesterol synthesis | SREBF2 | 0.017 | 0.028 | 1.632 | |||||
| HMGCR | 0.019 | 0.073 | 3.810 | ||||||
| Urea cycle | OTC | 0.065 | 0.152 | 2.330 | |||||
| ARG1 | 0.644 | 1.215 | 1.887 | ||||||
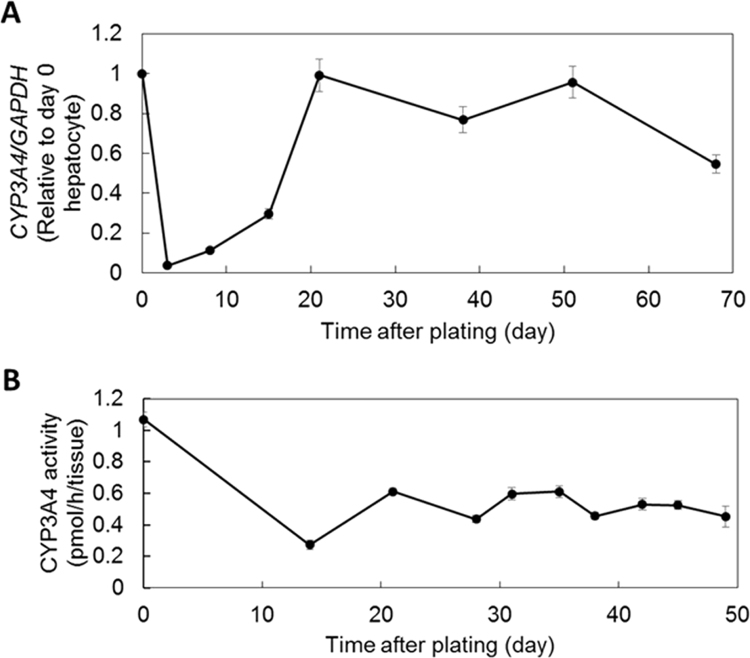
3.3. 3D bio-printed liver tissue showed long-lasting CYP3A4 expression and activity
To evaluate whether the detoxification function of bio-printed liver tissue was sustainable, we investigated the time course of CYP3A4 mRNA expression and CYP3A4 enzyme activity. Generally, it is well-known that, in 2D hepatocyte cultures, hepatic gene expression is lost rapidly, within the first few days. However, in bio-printed liver tissue, CYP3A4 expression had gradually increased from day 3 to day 20 (Fig. 2A). The maximum expression on day 20 was at the same level as the expression observed in hepatocytes on day 0. Furthermore, in bio-printed liver tissue, CYP3A4 activity was stably maintained after day 21 (Fig. 2B). However, the maximum activity was reduced to 57% of the activity observed in hepatocytes on day 0. These results suggested that bio-printed liver tissue could maintain drug metabolic functions for at least about 50 days.
Fig. 2.
3D bio-printed liver tissues showed sustained CYP3A4 expression and activity. Time courses of (A) CYP3A4 expression, relative to the reference gene (GAPDH) expression and (B) CYP3A4 activity in bio-printed liver tissues. All error bars represent SEM (n=3).
3.4. 3D bio-printed liver tissue maintained various long-lasting metabolic functions
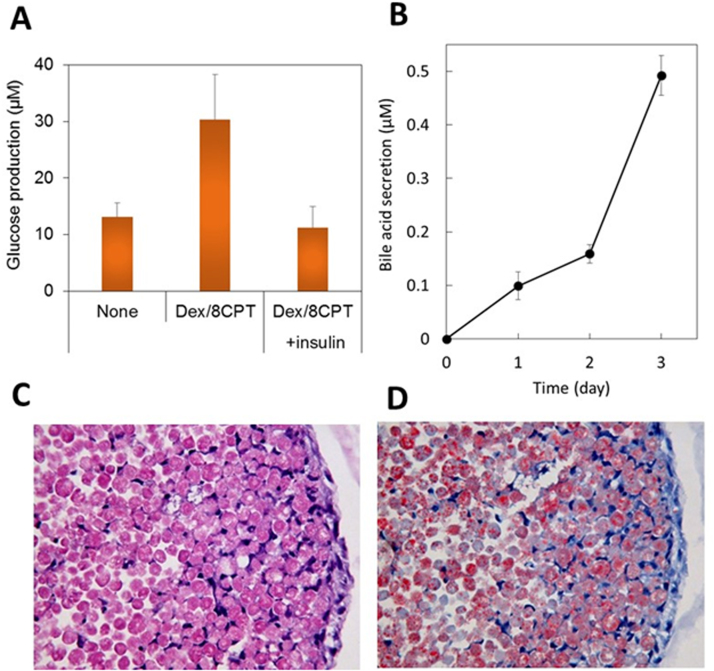
Bio-printed liver tissue showed relatively high expression levels of genes related to gluconeogenesis and insulin pathways (Table 1). Therefore, we investigated whether cAMP-induced gluconeogenesis was inhibited by insulin in bio-printed liver tissue. We found that insulin clearly inhibited cAMP-induced glucose production in the liver tissue (Fig. 3A).
Fig. 3.
3D bio-printed liver tissue maintained sugar and lipid metabolic functions for long periods. (A) Bio-printed human liver tissue glucose production was stimulated with 500 nM dexamethasone and 100 μM 8-(4-Chlorophenylthio)adenosine 3′,5′-cyclic monophosphate (Dex/8CPT); the increase was reduced with the addition of 10 μM insulin (day 77). Error bars represent SEM (n=3). (B) Time-dependent bile acid accumulation, measured in the medium of bio-printed human liver tissue (day 24–27). Error bars represent SEM (n=4). (C) Hematoxylin and eosin stained bio-printed liver tissue derived from a Zucker fatty rat (male, age 11 weeks) after 23 days of culture. (D) Oil-red O stained bio-printed liver tissue derived from a Zucker fatty rat shows lipid deposits after 23 days of culture.
Almost all bile acid transporters that we investigated showed high expression in bio-printed liver tissue (Table 1). Therefore, we investigated bile acid secretion from bio-printed liver tissue. We found that bio-printed liver tissue secreted bile acid into the medium, and the secreted bile acids accumulated over time (Fig. 3B).
We then investigated genes related to fatty acid and lipid synthesis. We found relatively high expression in bio-printed liver tissue (Table 1). We also investigated hepatocytes from Zucker fatty rats with non-alcoholic fatty liver disease (NAFLD) to determine whether bio-printed liver tissue could maintain a pathological condition for long periods. On day 23, we observed abundant oil-red O staining (Fig. 3D) among cells confirmed to be viable (Fig. 3C) in bio-printed liver tissue derived from Zucker rats with NAFLD. These results suggested that 3D bio-printed liver tissue could maintain for long periods the same glucose metabolism, bile acid secretion, and disease conditions that were present in the donor animal models or humans.
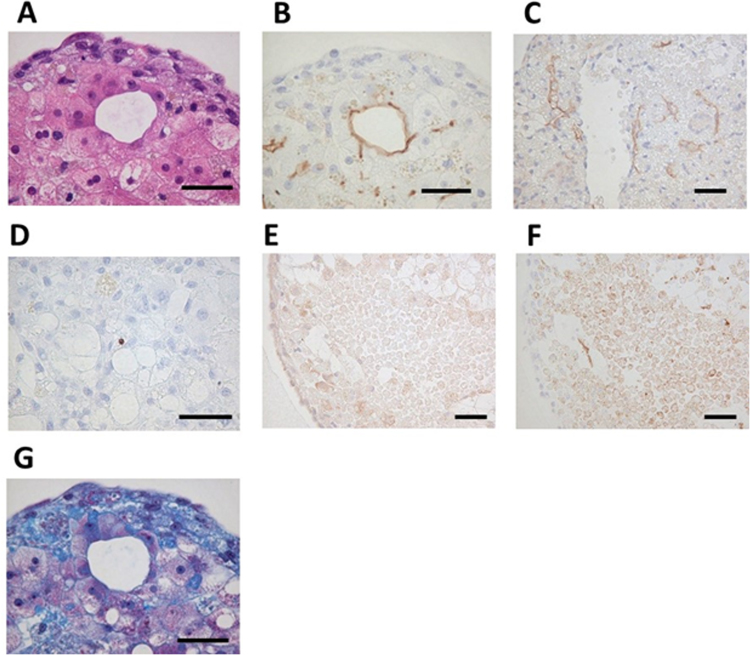
3.5. Scaffold-free 3D bio-printing induced self-organization in human hepatocytes
We performed histochemical staining in bio-printed human liver tissues. We identified liver-specific cord-like structures and two nuclei-containing hepatocytes on day 50 (Fig. 4A). Immunohistochemical studies revealed that these structures expressed a hepatic bile acid transporter known as MRP2, which is typically expressed on bile duct cell membranes (Fig. 4B). In addition, we observed blood capillary-like and sinusoidal-like structures that expressed CD31 (Fig. 4C). Little apoptosis was observed in bio-printed liver tissues, even on day 60 (Fig. 4D). The surface of the tissue exhibited strong positive signals for OATP1B1 and OATP1B3. These transporters are typically expressed on the sinusoidal membranes of hepatocytes (Fig. 4E). The MRP2-positive signals were found mostly inside the tissue spheres, but few signals were observed on the outer layer of the tissue (Fig. 4F). Also, abundant collagen deposits had accumulated in the tissue (Fig. 4G). These results suggested that this scaffold-free bio-printing technique performed with the Regenova and hepatocyte spheroids could produce liver structures with durable functions. Furthermore, these tissues exhibited both self organization and extracellular matrix (ECM) production (e.g., collagen), which are thought to contribute, at least in part, to effective liver function.
Fig. 4.
Self-organization in bio-printed human liver tissues. (A) HE staining shows structure of bio-printed liver tissue on day 50. (B) Immunostaining with MRP2 antibody detected bile acid transporters (day 50). (C) Immunostaining with CD31 antibody detected blood vessel-like and sinusoid-like structures (day 14). (D) TUNEL staining detected little apoptosis (day 60). (E) Immunostaining with OAT2/8 antibody detected drug uptake transporters (day 44). (F) Immunostaining with MRP2 antibody shows tissue distribution (day 44). (G) Masson's trichrome staining shows collagen accumulation (day 50). Black bars represent 50 µm.
4. Discussion
In this study, we used the scaffold-free 3D bio-printer, Regenova, to construct a novel liver tissue-like model with drug, glucose, lipid, and bile acid metabolism-related functions useful for drug discovery. Although many liver models have been described to date [2], our liver model is unique in its wide range of metabolic functions.
As shown in Table 1, the bio-printed liver tissue expressed transporters and enzymes related to drug metabolism at levels equivalent or higher than those observed in day-0 hepatocytes. Moreover, we estimated that these gene expression levels were similar to levels reported for existing technologies, such as the micropatterned model [6]. Currently, we are conducting drug toxicity tests and metabolite evaluations with commercially-available compounds. Moreover, we are also evaluating the ability of bio-printed liver tissue to reproduce clinically-confirmed hepatotoxicity and metabolite production, compared to existing technologies, such as the micropatterned model.
Among the genes listed in Table 1, we observed relatively high expression of glucose and lipid metabolism-related genes in bio-printed liver tissue. We then demonstrated that glucose production from bio-printed liver tissue was increased by enhancing the cAMP / PKA signaling and inhibited by enhancing insulin signaling. These results indicated that glucose metabolism was correctly regulated in fasting and satiety conditions. Generally, when fasting, glucagon stimulates the cAMP/PKA pathway in the liver to promote glucose production. On the other hand, after eating, insulin inhibits this pathway in the liver to reduce glucose production. When we evaluated liver tissue on day 77, we measured glucose released into the medium. The amount of glucose released increased with a cAMP-stimulating reagent and decreased to basal levels, when insulin was added with the cAMP-stimulating reagent. Thus, bio-printed liver tissue retained regulation of glucose production, even after culturing for 75 days or more.
When we tested pathologic NAFLD hepatocytes from a Zucker fatty rat, we found that bio-printed pathologic liver tissue retained the appropriate pathology. High lipid content was demonstrated in bio-printed pathologic liver tissue with oil-red O staining, and strong staining persisted at least until day 23. Therefore, the bio-printed liver tissues could maintain both normal functions and a pathological condition for long periods.
Currently, drug discovery studies in diabetes widely use primary cultured hepatocytes derived from normal or pathological animal models, However, a functional human liver tissue model that is sustainable long-term is likely to exhibit responses closer to those observed in the in vivo human environment. Therefore, the effective use of bio-printed liver tissues for drug discovery studies could contribute to the generation of new drug discovery targets and drugs.
As shown in Fig. 4B and C, bile duct-like structures and sinusoid vessel-like structures were found in the bio-printed liver tissue. We did not add sinusoidal endothelial cells or bile duct epithelial cells at the time of constructing the bio-printed structure; therefore, we assumed that these structures were derived from heterogeneities in the commercially-available primary cultured hepatocytes. Furthermore, our immunohistochemistry results showed that OATP1B1/1B3, which are known to localize on the vascular side of hepatocytes, were mainly localized to the surfaces of bio-printed structures (Fig. 4E). On the other hand, MRP2, which is known to localize on the bile duct side of hepatocytes, were mainly found in the internal layers of bio-printed structures (Fig. 4F). Interestingly, the bio-printed liver tissue could secrete bile acid into the medium, and it accumulated over time (Fig. 3B). These results suggested that bio-printed liver tissue might exert functions in the following order: (1) drug uptake, via OATP1B1/1B3, (2) drug metabolism with phase I and/or II enzymes, and (3) metabolite excretion, along with bile acid, via MRP2. In the latter stage, it remains unclear whether bile acid was secreted into the medium exactly through the bile duct. We plan to examine this point further in the near future.
In conclusion, we showed that bio-printed liver tissue could maintain functional metabolism of drugs, glucose, lipids, and bile acid for long periods. Little is known about the long-term maintenance of functions related to glucose production, lipid metabolism, and bile acid secretion. Bio-printed liver tissue is unique in its ability to maintain these features for several weeks. Therefore, in future, bio-printed liver tissue might be put into practical use as a preclinical model for hepatotoxicity predictions, in vitro NASH and/or diabetes studies, and biliary excretion studies. Further we also plan to develop two novel drug discovery models. One model will comprise bio-printed liver tissue composed of co-cultured human Kupffer cells and hepatic stellate cells. We expect that model to exhibit drug responsiveness close to that of human liver [17]. A second model, constructed by combining bio-printed liver tissue with a microfluidic system, will reflect the interactions between multiple organs [18], [19], [20], [21], [22].
Acknowledgments
We thank Mrs. Natsumi Mizoguchi for excellent technical support in the maintenance of bio-printed liver tissues and the biochemical assays including qPCR and enzyme activity. We thank Mr. Atushi Aburadani for excellent technical support in the maintenance of 3D bioprinter “Regenova”.
Footnotes
Transparency document associated with this article can be found in the online version at doi:10.1016/j.bbrep.2017.04.004.
Appendix A. Transparency document
Supplementary material
References
- 1.Si-Tayeb K., Lemaigre F.P., Duncan S.A. Organogenesis and development of the liver. Dev. Cell. 2010;18:175–189. doi: 10.1016/j.devcel.2010.01.011. [DOI] [PubMed] [Google Scholar]
- 2.Lin C., Khetani S.R. Advances in engineered liver models for investigating drug-induced liver injury. Biomed. Res. Int. 2016:1829148. doi: 10.1155/2016/1829148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Xu J.J., Henstock P.V., Dunn M.C., Smith A.R., Chabot J.R., de Graaf D. Cellular imaging predictions of clinical drug-induced liver injury. Toxicol. Sci. 2008;105:97–105. doi: 10.1093/toxsci/kfn109. [DOI] [PubMed] [Google Scholar]
- 4.Kaplowitz N. Idiosyncratic drug hepatotoxicity. Nat. Rev. Drug Discov. 2005;4:489–499. doi: 10.1038/nrd1750. [DOI] [PubMed] [Google Scholar]
- 5.Rawlins M.D. Cutting the cost of drug development? Nat. Rev. Drug Discov. 2004;3:360–364. doi: 10.1038/nrd1347. [DOI] [PubMed] [Google Scholar]
- 6.Khetani S.R., Bhatia S.N. Microscale culture of human liver cells for drug development. Nat. Biotechnol. 2008;26:120–126. doi: 10.1038/nbt1361. [DOI] [PubMed] [Google Scholar]
- 7.Kostadinova R., Boess F., Applegate D., Suter L., Weiser T., Singer T., Naughton B., Roth A. A long-term three dimensional liver co-culture system for improved prediction of clinically relevant drug-induced hepatotoxicity. Toxicol. Appl. Pharmacol. 2013;268:1–16. doi: 10.1016/j.taap.2013.01.012. [DOI] [PubMed] [Google Scholar]
- 8.Bell C.C., Hendriks D.F., Moro S.M., Ellis E., Walsh J., Renblom A., Fredriksson Puigvert L., Dankers A.C., Jacobs F., Snoeys J., Sison-Young R.L., Jenkins R.E., Nordling Å., Mkrtchian S., Park B.K., Kitteringham N.R., Goldring C.E., Lauschke V.M., Ingelman-Sundberg M. Characterization of primary human hepatocyte spheroids as a model system for drug-induced liver injury, liver function and disease. Sci. Rep. 2016;6:25187. doi: 10.1038/srep25187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nguyen D.G., Funk J., Robbins J.B., Crogan-Grundy C., Presnell S.C., Singer T., Roth A.B. Bioprinted 3D primary liver tissues allow assessment of organ-level response to clinical drug induced toxicity in vitro. PLoS One. 2016;11:e0158674. doi: 10.1371/journal.pone.0158674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Khetani S.R., Kanchagar C., Ukairo O., Krzyzewski S., Moore A., Shi J., Aoyama S., Aleo M., Will Y. Use of micropatterned cocultures to detect compounds that cause drug-induced liver injury in humans. Toxicol. Sci. 2013;132:107–117. doi: 10.1093/toxsci/kfs326. [DOI] [PubMed] [Google Scholar]
- 11.Wang W.W., Khetani S.R., Krzyzewski S., Duignan D.B., Obach R.S. Assessment of a micropatterned hepatocyte coculture system to generate major human excretory and circulating drug metabolites. Drug Metab. Dispos. 2010;38:1900–1905. doi: 10.1124/dmd.110.034876. [DOI] [PubMed] [Google Scholar]
- 12.Davidson M.D., Lehrer M., Khetani S.R. Hormone and drug-mediated modulation of glucose metabolism in a microscale model of the human liver. Tissue Eng. Part C. 2015;21:716–725. doi: 10.1089/ten.tec.2014.0512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Davidson M.D., Ballinger K.R., Khetani S.R. Long-term exposure to abnormal glucose levels alters drug metabolism pathways and insulin sensitivity in primary human hepatocytes. Sci. Rep. 2016;6:28178. doi: 10.1038/srep28178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bi Y.A., Kazolias D., Duignan D.B. Use of cryopreserved human hepatocytes in sandwich culture to measure hepatobiliary transport. Drug Metab. Dispos. 2006;34:1658–1665. doi: 10.1124/dmd.105.009118. [DOI] [PubMed] [Google Scholar]
- 15.S.C. Katherine, Hepatocytes in HepatoPac® function without the need for Matrigel®, Architecture is Important! The Hepregen Blog, 2014. 〈http://www.hepregen.com/hepregen-blog/hepatocytes-without-the-need-for-matrigel〉.
- 16.T. Koga, S. Nagasato, Y. Iwamoto, K. Nakayama, Method for production of three-dimensional structure of cells, Application No. 2009-509335, International application No.PCT/JP2008/056826, International Pub. No. WO2008/123614, 2008.
- 17.Rose K.A., Holman N.S., Green A.M., Andersen M.E., LeCluyse E.L. Co-culture of hepatocytes and Kupffer cells as an in vitro model of inflammation and drug-Induced hepatotoxicity. J. Pharm. Sci. 2016;105:950–964. doi: 10.1016/S0022-3549(15)00192-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ma C., Zhao L., Zhou E.M., Xu J., Shen S., Wang J. On-chip construction of liver lobule-like microtissue and Its application for adverse drug reaction assay. Anal. Chem. 2016;88:1719–1727. doi: 10.1021/acs.analchem.5b03869. [DOI] [PubMed] [Google Scholar]
- 19.Bhise N.S., Manoharan V., Massa S., Tamayol A., Ghaderi M., Miscuglio M., Lang Q., Shrike Zhang Y., Shin S.R., Calzone G., Annabi N., Shupe T.D., Bishop C.E., Atala A., Dokmeci M.R., Khademhosseini A. A liver-on-a-chip platform with bioprinted hepatic spheroids. Biofabrication. 2016;8:014101. doi: 10.1088/1758-5090/8/1/014101. [DOI] [PubMed] [Google Scholar]
- 20.Bavli D., Prill S., Ezra E., Levy G., Cohen M., Vinken M., Vanfleteren J., Jaeger M., Nahmias Y. Real-time monitoring of metabolic function in liver-on-chip microdevices tracks the dynamics of mitochondrial dysfunction. Proc. Natl. Acad. Sci. USA. 2016;113:E2231–E2240. doi: 10.1073/pnas.1522556113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rennert K., Steinborn S., Gröger M., Ungerböck B., Jank A.M., Ehgartner J., Nietzsche S., Dinger J., Kiehntopf M., Funke H., Peters F.T., Lupp A., Gärtner C., Mayr T., Bauer M., Huber O., Mosig A.S. A microfluidically perfused three dimensional human liver model. Biomaterials. 2015;71:119–131. doi: 10.1016/j.biomaterials.2015.08.043. [DOI] [PubMed] [Google Scholar]
- 22.Vernetti L.A., Senutovitch N., Boltz R., DeBiasio R., Shun T.Y., Gough A., Taylor D.L. A human liver microphysiology platform for investigating physiology, drug safety, and disease models. Exp. Biol. Med. 2016;241:101–114. doi: 10.1177/1535370215592121. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material