Abstract
Introduction:
Uganda has reported eight outbreaks caused by filoviruses between 2000 to 2016, more than any other country in the world. We used species distribution modeling to predict where filovirus outbreaks are likely to occur in Uganda to help in epidemic preparedness and surveillance.
Methods:
The MaxEnt software, a machine learning modeling approach that uses presence-only data was used to establish filovirus – environmental relationships. Presence-only data for filovirus outbreaks were collected from the field and online sources. Environmental covariates from Africlim that have been downscaled to a nominal resolution of 1km x 1km were used. The final model gave the relative probability of the presence of filoviruses in the study area obtained from an average of 100 bootstrap runs. Model evaluation was carried out using Receiver Operating Characteristic (ROC) plots. Maps were created using ArcGIS 10.3 mapping software.
Results:
We showed that bats as potential reservoirs of filoviruses are distributed all over Uganda. Potential outbreak areas for Ebola and Marburg virus disease were predicted in West, Southwest and Central parts of Uganda, which corresponds to bat distribution and previous filovirus outbreaks areas. Additionally, the models predicted the Eastern Uganda region and other areas that have not reported outbreaks before to be potential outbreak hotspots. Rainfall variables were the most important in influencing model prediction compared to temperature variables.
Conclusions:
Despite the limitations in the prediction model due to lack of adequate sample records for outbreaks, especially for the Marburg cases, the models provided risk maps to the Uganda surveillance system on filovirus outbreaks. The risk maps will aid in identifying areas to focus the filovirus surveillance for early detection and responses hence curtailing a pandemic. The results from this study also confirm previous findings that suggest that filoviruses are mainly limited by the amount of rainfall received in an area.
Keywords: disease model
Introduction
Uganda has experienced eight filovirus outbreaks; five Ebola Virus Disease (EVD) and three Marburg virus disease (MVD), between 2000 and 2016, more than any other country in the world.
The first outbreak in Uganda was caused by Ebolavirus of the species Sudan ebolavirus in 2000 in the Northern district of Gulu, where 425 cases were registered with a case fatality rate (CFR) of 53%1. The second outbreak was caused by Bundibugyo Ebolavirus in the western part of Uganda bordering with Democratic Republic of Congo (DRC), with 192 cases and a CFR of 34%2,3 . In 2011, another EVD outbreak occurred where only one case was involved in Luweero district Zirobwe village, 45 km North of Uganda’s Capital City Kampala4. Two more EVD outbreaks were observed in 2012, one in June in the Western District of Kibale and another in November, Luweero district in Central Uganda5.
Likewise, three outbreaks of MVD have occurred in Uganda; the first one was in Kamwenge district in 2007 associated with mining activity in the Kitaka gold mine that is occupied by bats6. This outbreak was later linked to cave-dwelling Egyptian fruit bats (Rousettus aegyptiacus) that occupy these mines, as they tested positive for Marburg virus by polymerase chain reaction (PCR)7,8. Another outbreak of MVD was in 2012 where several districts were involved with a CFR of 58% (15/26)9. This outbreak was also traced back to the same gold mines in Western Uganda, and subsequent testing of the bats in the mines revealed a spill over to human populations10. The latest MVD outbreak was in Kampala where the only fatal case was a health worker, and no other cases were identified11.
It is hypothesized that distribution of filoviruses is limited by the distribution of the bats, which are known probable reservoirs. All the filovirus outbreaks in humans have been reported to originate from Sub-Sahara Africa and only one species, Reston virus that is not known to infect humans was detected outside Sub-Sahara Africa in The Philippines12. It has been suggested that transmission from the natural reservoir occurs when humans get into contact with the reservoir or its body fluids such as feces, urine, and blood via activities such as hunting and consumption of bush meat13. Because previous outbreaks in Central Africa have been linked to reports of bush meat consumptions and deaths of wildlife14, many hypotheses have been put forward to suggest wildlife such as bats, primates, and antelopes as possible sources of infection. The debate on bats as potential reservoirs of Ebolaviruses is still not concluded, as no Ebolavirus has been isolated from bats despite finding some bats seropositive for Ebolavirus and others with viral RNA15. The role of non-human primates as reservoirs has been unconvincing since they do die from infection with filoviruses16,17,18,19. Other wildlife that has been reported to be infected by Ebolavirus was one duiker, whose bone tested positive by PCR in Republic of Congo bordering Gabon19. Dogs and pigs are the only domestic animals associated with ebolaviruses. Dogs were found to be IgG seropositive in Gabon20 whereas Reston virus has been reported in pigs and have shown potential for infection with Ebola virus21,22,23. Unlike EVD, there is progress in research in trying to describe the reservoirs of Marburg virus. Bats of species Rousettus aegyptiacus, found in Kitaka gold mine and Python cave from the Albertine region in Western Uganda have been described as potential reservoirs of Marburg virus in Uganda8,10,24. The bats in these caves have been linked to three MVD outbreaks, where artisanal gold miners got infected with Marburg virus6,9. Transmission of Marburg virus in human populations just like Ebolaviruses happens after a spillover event from the natural reservoir in wildlife. Lack of a clear reservoir and true source of infection or spill-overs into human populations has been a call for alternative methods of heightening surveillance and developing risk maps is one of them.
Situated in the rich and complex ecological systems with high biodiversity in East Africa, Uganda is not only affected directly by filovirus outbreaks but also vulnerable to outbreaks from neighboring countries such as DRC. For epidemic preparedness and response, Uganda’s health surveillance system needs to know where and when these epidemics are likely to occur. This will allow them to conduct active surveillance focusing in those areas for early detection to avoid pandemics and also focus research on reservoirs. This can be achieved by applying spatial epidemiology modeling techniques. One such technique is Ecological Niche Modeling (ENM) also known as Species Distribution Modeling (SDM), that has been used to establish the relationship between species and their environment25,26,27,28. ENM has also been used to predict the ecology and distribution of filoviruses before. Peterson et al (2014) used a Genetic Algorithm for Rule-Set Production (GARP) model to predict suitable environments for filoviruses as being in afro-tropics where EVD was being predicted more in the humid rain forest of Central and West Africa while MVD was more predicted to occur in the drier and more open areas of Central and East Africa29. More efforts were made to improve the spatial prediction model for MVD for Africa using a Bioclimatic variable (Bioclim)30, which predicted filoviruses mainly in Zimbabwe and abroad potential distribution across the arid woodland regions of Africa31. Furthermore, Pigott et al (2014) developed zoonotic niche maps for Marburg and Ebola viruses in Africa using species distribution models32,33. In these maps, they have predicted EVD at risk areas occupied by 22 million people while MVD is predicted to occur in 27 countries across Sub-Sahara Africa. Enhanced vegetation index which corresponds to high levels of rainfall was identified as the most important variable limiting the distribution of the Ebola virus in Africa32,33.
These predictions are not country specific, and they lack details of individual countries regarding vector and raster data. For example, they used online databases that are not accurate especially in estimating environmental covariates and getting coordinates of index cases, hence, affected countries find these maps limited for focused and targeted surveillance
A Maximum Entropy species distribution modeling environment (MaxEnt) has been used to predict the ecological niche for various species. The MaxEnt algorithm uses presence-only occurrence records to estimate the actual or potential geographic distribution of a species34 and has been known to outperform other species’ distribution modeling approaches such as Domain, Generalized Additive Models (GAM), Generalized Linear Models, Genetic Algorithm (GARP) and Bioclim35.
MaxEnt models have been used widely to predict ecological niches of different vectors and disease-causing organisms36,37,38,39,40,41,42,43, but it has not been used for prediction of filovirus outbreaks in Uganda. Briefly, MaxEnt is a multipurpose machine-learning technique and aims at estimating the probability of distribution of a species occurrence using the environmental features. Our major aim was to develop a country-specific risk map for Uganda using updated data on EVD/ MVD outbreaks and bat occurrence and environmental variables specific for Uganda using the MaxEnt modeling approach. The model outputs will improve filovirus epidemic preparedness, surveillance and response, and in the search for a reservoir especially in a disease prone country like Uganda
Materials and methods
EVD, MVD and Bat occurrence data
A total of 16 locations of the Ebolavirus outbreaks in Uganda since 2000 was obtained from published databases44. An additional 27 occurrence points for Ebola and Marburg virus diseases outbreaks were collected from the field where these outbreaks occurred especially for new outbreaks whose locations were not collected before. All locations where confirmed cases of Marburg or Ebola viruses were reported were collected with Global Positioning System (GPS) receiver and points were entered into an Excel spreadsheet. A total of 43 filovirus outbreak occurrence points (30 for EVD outbreak and 13 for MVD outbreak) were used for this prediction model (Supporting Information S1 File; see Appendix). These filovirus occurrence points represent households in villages where confirmed cases were residing. Due to the contagious nature of filoviruses, one household had more than one cases hence the reason for not using all the 562 EVD cases and 20 MVD cases. A fruit bat location survey was also done to determine the location of fruit bats in a cross-section of Uganda. We purposively selected districts to scout for bats based on previous filovirus outbreaks and anecdotal reports of bats in trees. Using a snowballing approach, we collected 84 fruit bat locations using a GPS receiver from different districts of the country. Here community members acted as informers of the roosting locations of fruit bats and caves that contain bats.
An additional, 517 bat locations from all over Uganda were generously provided by Kityo Robert (Department of Zoology, Makerere University Kampala Uganda) also published in Uganda Bat Atlas45, resulting in a total of 601 bat coordinates (Supporting Information S1 File; see Appendix).
Environmental covariates
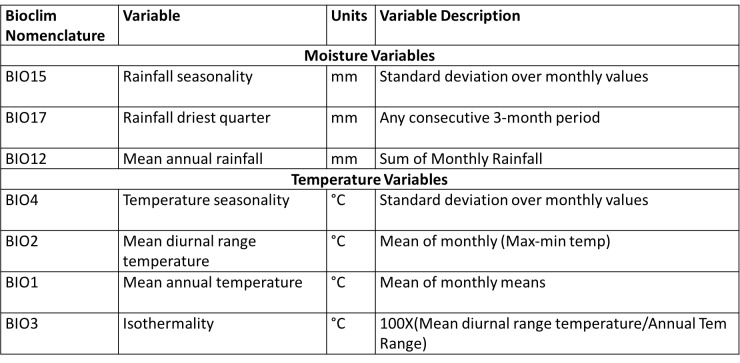
Ecologically suitable environmental covariates for filovirus outbreaks for Uganda were compiled from Africlim46, with a spatial resolution of 1 km. The environmental covariates considered were moisture (mean annual rainfall, rainfall wettest month, rainfall driest month, rainfall seasonality, rainfall wettest quarter, rainfall driest quarter, annual moisture index, moisture index arid quarter, number of dry months, length of longest dry season) and temperature variables (mean annual temperature, mean diurnal range in temperature, isothermality, temperature seasonality, maximum temperature warmest month, minimum temperature coolest month, annual temperature range, mean temperature warmest quarter, mean temperature coolest quarter, potential evapotranspiration). We used ENMTOOLs; a toolbox that facilitates quantitative comparisons of environmental niche models47 to test for multicollinearity between the predictor variables and we ran a pairwise Pearson correlation, and only variables with less than (+/-0.75) correlation were retained in the final prediction model (Supporting Information S2 File; see Appendix). After this test, only seven environmental variables were retained (Table 1); three moisture variables (Rainfall seasonality, Rainfall driest quarter, and mean annual rainfall) and four temperature variables (Temperature seasonality, Mean diurnal range in temperature, mean annual temperature and Isothermality).
Table 1: Environmental variables used in the models
Ecological Niche Model
We used MaxEnt Version 3.3k for modeling distribution of filovirus using default settings (Auto features, convergence threshold=0.00001, the maximum number of background points=10,000, regularization multiplier=1). A logistic probability map was generated showing the relative probability of the presence of filoviruses survival on a scale ranging between 0 and 148. The occurrence data was subdivided into k-folds where 25% was set aside for testing the accuracy of the model, whereas 75% was used for training the model. However, there were few presence records (10) for the Marburg cases therefore, all the records were used in training the model. The Receiver Operating Curve (ROC) was used to assess the overall model predictive performance, a measure of the ability of the model to distinguish presence from absence of a species with a value of 1 indicating a perfect prediction while 0.5 is as good as a random prediction49,50. A jackknife test was used to evaluate individual covariate importance in the model developments (Supporting Information S3 File; see Appendix). To improve model robustness, 100 replicates were averaged for the final model outputs. MaxEnt outputs were imported into ArcGIS 10.3 mapping software to develop final maps.
Results
The bat occurrence and filovirus outbreak locations
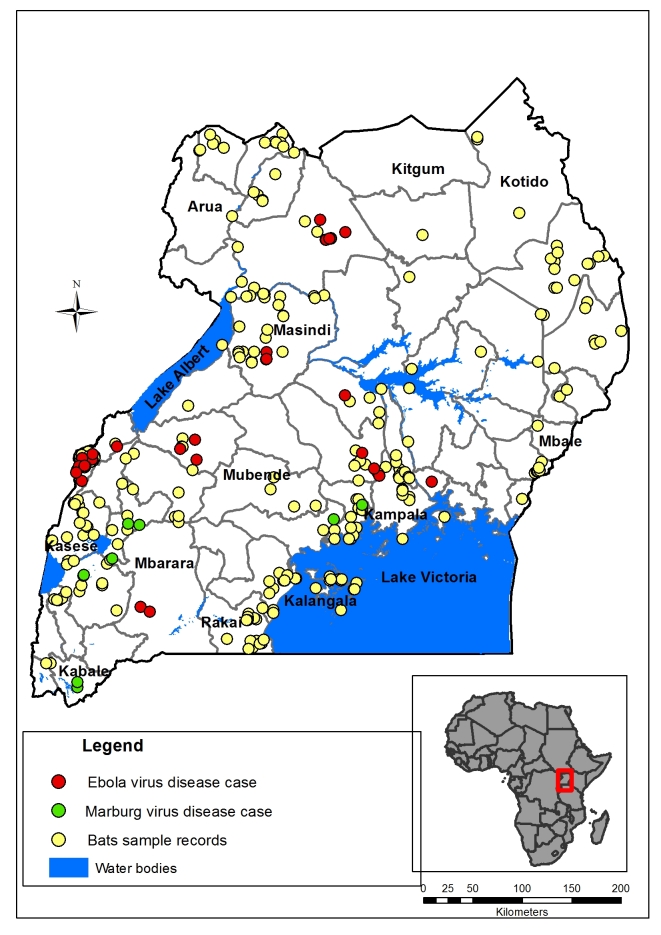
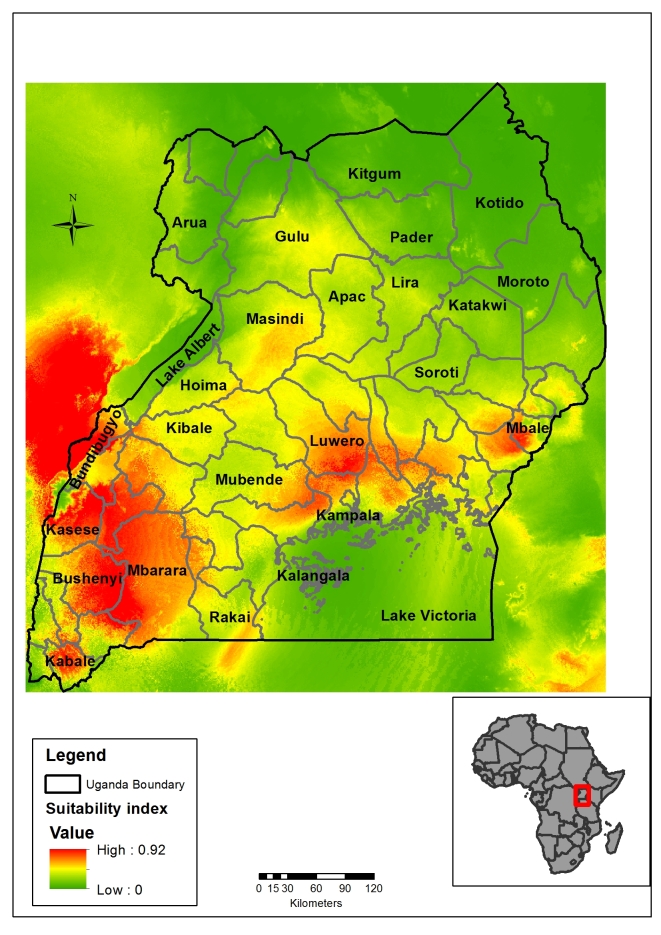
As shown in Figure 1, bats are distributed all over Uganda, with a high distribution around water bodies which is a core need for survival. Areas around Lake Victoria, River Nile, and Western Rift Valley have high numbers of bats. Their locality is in line with regions that have reported filovirus outbreaks in Uganda.
Map of Uganda showing outbreak locations of Ebola and Marburg virus diseases and bat locations included in the Maxent modeling Environment.
Bat distribution in Uganda
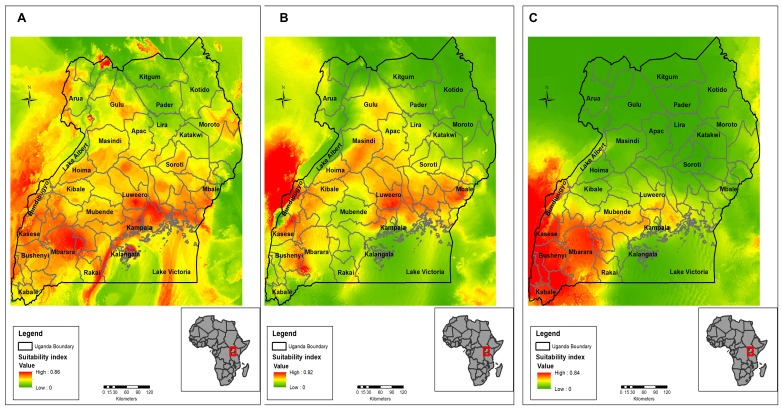
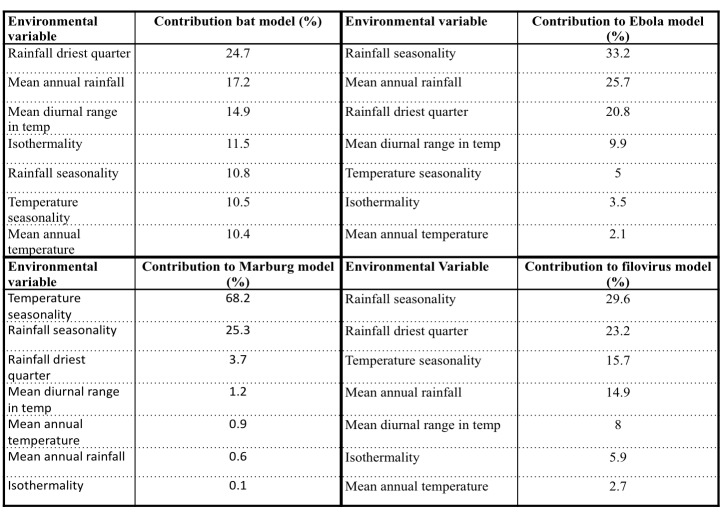
From 100 bootstrap replicates, a bat distribution map was generated (mean AUC=0.80; SD=0.012). Compared to a random prediction of AUC 0.5, our model was able to distinguish presence from the absence of bats within the geographic space with a high accuracy51. The relative probability of presence (RPP) ranged from highly suitable areas represented by red to orange colors to unsuitable areas represented by the green color in Figure 2A. The map shows that most areas in Uganda are suitable habitats for bats (both insect and fruit bats) with high RPP occurring in the following districts; Mbarara, Bushenyi, Bundibugyo and Kabale located in the western part of Uganda, around Lake Victoria (Kampala and Luweero districts) and in eastern region of Mbale and Soroti districts. Moderately suitable regions largely cover most parts of Uganda. The RPP of bats were mainly influenced by rainfall driest quarter with 24.7%, mean annual rainfall with 17.2%, mean diurnal range in temperature with 14.5%, and isothermality with 11.5% (Table 2).
Maps showing bats, EVD and MVD distribution in Uganda with high Relative Probability Presence represented in red while low in green.
A: Relative probability of presence of bats, hypothesized as reservoirs of filoviruses (AUC=0.80), B: Relative probability of presence of Ebola Virus disease outbreak (AUC=0.90), C: Relative probability of presence of Marburg Virus disease outbreak (AUC=0.92.
Table 2: Environmental variable contribution in the MaxEnt prediction models
Ebola virus distribution
High RPP for EVD outbreak was predicted in more than half of the country with hotspots in Western Rift valley districts of Bundibugyo, Masindi, Kibale and Hoima, Kasese, Kabarole, Kamwenge, Bushenyi and Ibanda as shown in Figure 2B (mean AUC=0.90; SD=0.024). In Central Uganda, Luweero, Kayunga, Mpigi, Kampala, Mityana and Nakasongola districts are predicted as potential areas for EVD outbreaks. In the eastern part of the country, it is mainly the Busoga region along River Nile and Mbale district around Mt. Elgon that are potential EVD hot spots. Other places that have not recorded outbreaks before but are predicted as potential probable areas for the spread of EVD include areas surrounding Lake Victoria and around Mount Elgon. A low RPP for EVD outbreak was predicted in North Eastern Uganda (Karamoja region) and Northern Uganda in the districts of Kitgum and Pader. Rainfall seasonality (33.2%), Mean annual rainfall (22.7%), rainfall of the driest quarter(20.8%) and mean diurnal range in Temperature (9.9%) had the highest relative contribution in predicting Ebola virus ecological suitability (Table 2).
Marburg virus disribution
The map in Figure 2C shows that Western, Southwestern and Central Uganda are potential areas for outbreaks of Marburg cases(AUC=0.92). Unlike predicted potential areas for EVD, predicted areas for MVD are mainly in the western sub-regions of Ankole, Tooro, Bunyoro, and Rwenzori region extending into DRC. Areas in the North and Eastern part of Uganda have a low or no relative probability of presence for MVD outbreaks as shown by the green color in Figure 2C. Temperature seasonality (68.2%) and rainfall seasonality (25.3%) contributed heavily to the model prediction (Table 2). Notably, temperature seasonality had the highest influence in MVD model compared to other variable contributions in all the models. However, the occurrence points were few in number to give us an accurate prediction.
Filovirus distribution
Combining Marburg and Ebola virus occurrence points (Figure 3), we see the range of the possible distribution of filovirus, mainly in western, southwestern Uganda, Victoria basin districts and eastern Uganda (mean AUC=0.90; SD =0.023). Predictor variables that contributed more than 75% in the model include; rainfall seasonality (29.6%), rainfall of the driest quarter (26.3%), Temperature seasonality and mean annual rainfall (14.9%) (Table 2).
Map showing areas of the relative probability of the presence of filovirus (Ebola and Marburg virus) outbreak in Uganda.
(AUC=0.9)
Variable Contribution to the prediction models
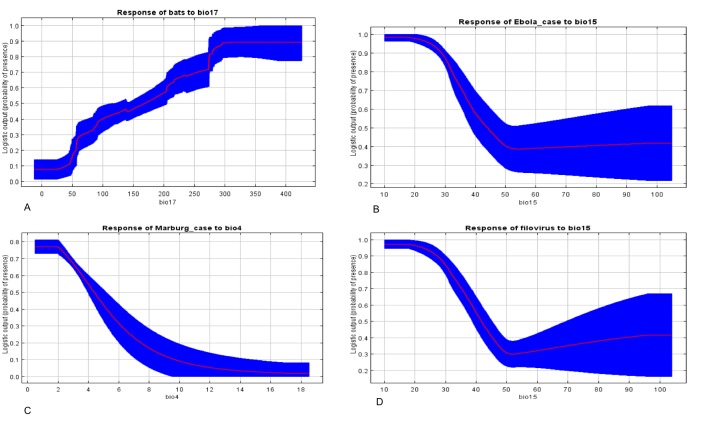
Figure 4, shows the response curve of the most important variable for each of the models (The response curves of all the predictor variables in all the four models are in Supporting Information S4 File; see Appendix). The response curves show the mean response of the 100 replicate MaxEnt runs (red) and the mean +/- one standard deviation. Figure 4Asuggests that probability of bats occurrence are optimal at 30 – 40 degree Celcius during the driest quarter(Bio17). MVD occurs in areas where temperature variability (Bio4) is minimal (Figure 4C) whereas EVD (Figure 4B) and both the filovirus (Figure 4D) occurs in areas with minimal rainfall variability (Bio15).Bio4 and Bio15 show how temperature and rainfall vary over a given year based on standard deviation. The response curves, show that MVD occurs in areas with low variability of temperature and EVD / Filoviruses occur in areas with low variability of rainfall. Bio4 contributes 68% to the relative probability of occurrence of MVD, which indicates that MVD is limited when there is high variability in temperature across the year. Rainfall variables contributed about 75% to the to the relative probability of occurrence of EVD. The results indicate that EVD is limited by the amount of rainfall received in an area. Higher rainfall increases the relative probability of occurrence of EVD.
Response curves of environmental variables that contribute highest to each of the prediction models.
A: Rainfall driest quarter(BIO17) vs Relative probability of bat presence. B: Rainfall seasonality(BIO15) vs. Relative probability of presence of Ebola virus outbreak; C: Temperature seasonality(BIO4) vs. Relative probability of presence of Marburg virus outbreak; D: Rainfall seasonality(BIO15) vs Relative probability of presence of Ebola or Marburg virus disease outbreak
Discussion
We used seven environmental variables in this model prediction. This was after assessing for collinearity in the model and removing all the collinear variables. Variable contribution assessment as shown inTable 2 showed that rainfall variables were the most important predictors. The importance of rainfall or precipitation and moderate to high temperature was highlighted by Peterson et al (2004) when they modeled filovirus distribution in Africa using GARP model29,31. Rainfall is important for the obvious reason that it provides water which is very important for bats survival52,53. Rainfall also provides for the development of fruiting trees that provide roosting areas for bats as well as food for fruit bats. Uganda is endowed with many water bodies and several rainforests, and hence bat distribution tends to be all over the country as seen in Figure 2A. Bats are hypothesized to be reservoirs for filoviruses; their distribution tends to correlate with that of filovirus predicted niches (Figure 3). Although we have some progress with Marburg virus in trying to describe bats as a source of infection for humans7,8,10,54, more research needs to be done especially on the reservoir for Ebola virus as these models can only give a clue as to the possible surveillance sites and possible areas to focus the research and to identify other potential reservoirs for filovirus. Temperature and rainfall seasonality were the most important environmental variables contributing to spatial prediction model for the Ebola and Marburg viruses. Seasonality has been found to be key in outbreaks of filoviruses, especially MVD as was reported in an ecological study by Amman et al. 20128. In this study, outbreaks of MVD are associated with the birthing seasons of adult juvenile bats when the virus circulation was high. This is further validated by a high percentage contribution (68.2%) of temperature seasonality into the MVD outbreak prediction model (Table 2). The relative probability of the presence of a Marburg outbreak is higher (80%) and at very low-temperature seasonality, which is a standard deviation (SD) over monthly values (Figure 4C). Therefore, areas with fewer variations in monthly temperature and rainfall are more likely to experience MVD and EVD outbreaks and this has been predicted by the models in Figures 2 & 3. The areas shown on the risk maps with a high relative probability of the presence of an outbreak are mainly in the South, the West and Central Uganda that have minimal temperature and rainfall variations compared to North Eastern Uganda that is not predicted for filovirus outbreaks except for bat presence. Bat presence model is mainly influenced by the variable rainfall driest quarter (24.7%) and mean annual rainfall (17.2%) (Table 2). As these variables increase, the relative probability of the presence of bats tends to increase. Areas of high rainfall are more likely to be forested or with many fruiting trees that provide a suitable habitat for bats, and this is true for three-quarters (75%) of Uganda.
Whereas Pigott et al (2015) used environmental covariates with a spatial resolution of 5km in their models55,56 , we used Africlim data with 1km spatial resolution. High-resolution data increases the accuracy of the models, and this was observed in our study by a high AUC greater than 0.8 recorded in all models.
The predictions show that a big part of Uganda, a country of 34 million people is at risk of a filovirus outbreak. This is more so in the Lake Victoria basin districts and in the Albertine Rift region districts and the areas that occur in between (Figure 2 & 3). The Albertine Rift region provides a variety of habitats characteristic of the East African savannahs and the West African rain forests that are suitable for reservoirs of filoviruses. According to Uganda National Meteorological department, these are the areas that receive near or above normal seasonal rainfall, and seasonal temperature variations are minimal57. Moreover, we see from variable contribution (Table 2), response curves (Figure 4x) and Jackknife test (Supporting Information S3 File; see Appendix) that rainfall and temperature seasonality were the most important variables in predicting outbreaks. The lower the variability in rainfall and temperature, the higher the relative probability of presence and vice versa and an increase in mean rainfall variables increases relative probability of having a filovirus outbreak (Figure 4). Indeed, six filovirus outbreaks have happened in this region, one caused by Bundibugyo ebolavirus in Bundibugyo district in the plains of Rwenzori mountains2, Sudan Ebolavirus in Kibale district5 and four outbreaks of Marburg virus all linked to Python cave and Kitaka gold mines in Kamwenge, Ibanda, and Rubirizi districts6,9,58,59. This remains a high-risk area with cross-border movement between Uganda and DRC where another EVD outbreak happened in 2012 in the neighboring Isiro region60 The Albertine Rift of East Africa needs to remain under heightened surveillance especially now that oil exploration will be taking place bringing an invasion of virgin lands by humans and interaction of wildlife and humans. Important to note also in this region has six national parks of Uganda (Queen Elizabeth National Park, Murchison Falls National Park, Kibale Forest National Park, Semiliki National Park, Bwindi Impenetrable National Park and Mgahinga National Park) on Uganda side and several other national parks on the DRC and Rwanda side as well as several forest reserves all of which harbor various species of bats and other possible reservoirs of filoviruses. All outbreaks of Marburg virus disease in Uganda have been investigated, and all originate from the old gold mines found in Ibanda and Kamwenge district6,9 in the Western Rift Valley which validates MVD distribution model in Figure 2C as it shows these as high-risk areas for filovirus outbreaks. A similar finding was obtained by Peterson and Samy 2016 in a recent model using MaxEnt as they predicted Sudan Ebola virus to occur in North Western Uganda between Lake Albert and Lake Vitoria61. We also see areas that have not had EVD outbreaks before such as West Nile region being predicted potential areas for EVD outbreak. These include areas along River Nile and areas bordering South Sudan and DRC Figure 2B). From Table 2, we see that rainfall variable contribute a higher percentage of the relative probability of presence for filovirus habitants. These areas receive average annual rainfall between 100-120mm and are endowed with high vegetation cover and water bodies all of which make the region conducive for reservoirs of filoviruses
Another area of high concern predicted by this model is Lake Victoria basin and districts in Nile River basin in Central districts of Uganda. Uganda has reported three outbreaks of filoviruses previously detected in these regions in the districts of Luweero4,5 and Mpigi11,62. This also can be attributed to the variety of habitats provided by water bodies, forests, swamps and high presence of fruit bats and other wildlife in this region. For example, the Kasokero cave that is the habitat of many Egyptian fruit bats that are known to harbor Marburg virus is found just on the banks of Lake Victoria in Masaka district, and several pathogens have been isolated from this cave 63. This is at the same time a highly-populated region with Uganda’s capital in the middle and needs to be heightened surveillance. We also predicted other regions that have not heard outbreaks of filoviruses in the past such as the Eastern region of Mbale, Busia and Tororo districts near the Mt. Elgon regions bordering with Kenya. This also still attributed to by the presence of suitable conditions for survival of putative reservoirs of Ebola and Marburg viruses. An outbreak happened in neighboring Kenya in Kitum cave64,65. These newly detected hotspots need to be kept under surveillance for early outbreak detection and response.
Limitations
We build on filovirus risk mapping efforts by Pigott et al32,33,56 and Peterson et al29,31,61 all of which have been done at the continental level of Africa. Their work was more ecologically oriented and more focused on identifying the ecological niche of species, they lacked country specific details that we bring in this publication with a bias in public health surveillance and outbreak detection rather that ecological niche identification. For public health surveillance of a country like Uganda, all filovirus species (Marburg virus, 5 Ebola virus species) are of public health importance. This makes our models more sensitive as opposed to specific risk map and hence more useful tools to the surveillance activities. There is already enough evidence of filovirus outbreaks in Uganda, especially areas predicted by our models. Focused surveillance needs to be done in these areas and bring additional surveillance in other new predicted areas where we have not heard outbreaks before. So we think modeling the map at a genus level (filovirus) level as opposed to species level is more informative for surveillance but may not be the best for ecological studies for which is not the purpose of this study. We know that disease outbreak is a combination of very many factors, not only suitable environmental covariates. However, we were not able to include as many factors as possible in this model because of lack of or poor quality data for Uganda specifically. We did not use bats as a predictor in our model because of their widespread distribution all over Uganda, otherwise doing this would lead to misleading interpretation and bias of potential outbreak hotspots as being the whole country. Another point would have been good to include in the prediction model are socio-economic factors since they play a big role in the outbreak of filoviruses.
Conclusion
Ecological niche modeling techniques have been widely used in predicting where disease outbreaks are likely to occur, more specifically where species have suitable living conditions depending on their environmental factors. The MaxEnt modeling algorithm uses presence only occurrence data and has been useful to estimate species’ niche in environmental space where absence records for a species are not available as it is the case with filoviruses. Given the public and global importance of filoviruses, developing models that predict where they are likely to occur is very important, and efforts in this direction have been done focusing on the African continent. In this paper, however, we focus on Uganda as one of the affected countries; and develop a country-specific prediction map. We show which places in Uganda that are hot spots for filovirus disease outbreaks and hence a focus on surveillance for early detection. Until now, no verified true reservoir for Ebola virus has been identified, and studies in this direction are still ongoing. In the absence of a known reservoir, these risk maps will help in early focused surveillance and early detection to avoid a global catastrophe like it happened in West Africa in 2014. Minimal seasonal variations in temperature and rainfall were important predictors of a filovirus outbreak. We believe these risk maps will be important in targeted surveillance, research and epidemic preparedness for Uganda. The results from this study also confirm previous findings that suggest that Filoviruses are mainly limited by the amount of rainfall received in an area.
Appendix
Supporting Information
S1 File: https://doi.org/10.6084/m9.figshare.5306875
S2 File: https://doi.org/10.6084/m9.figshare.5306908
Corresponding Author
Dr. Luke Nyakarahuka: n3luke@covab.mak.ac.ug; nyakarahuka@gmail.com
Competing Interests
The authors have declared that no competing interests exist.
Data Availability
All data is available in the paper and supporting files which can be found on figshare as follows: S1 File: Occurrence dataset used (Filovirus and Bats Occurrence coordinates) (10.6084/m9.figshare.5306875 <https://doi.org/10.6084/m9.figshare.5306875>); S2 File: Results of the quantitative comparisons of environmental variables to test for multicollinearity (10.6084/m9.figshare.5306908 <https://doi.org/10.6084/m9.figshare.5306908>); S3 File: A jackknife test result to evaluate individual covariate importance in the model developments (10.6084/m9.figshare.5306914 <https://doi.org/10.6084/m9.figshare.5306914>); S4 File: The response curves of all the predictor variables in all the four models (10.6084/m9.figshare.5306932 <https://doi.org/10.6084/m9.figshare.5306932>).
Acknowledgments
We thank Dr. Robert Kityo of Makerere University for bat occurrence data he provided us. We thank the Viral hemorrhagic fevers program and staff of Uganda virus Research Institute in collaboration with United States Centers for Disease Control and Prevention (CDC) for the surveillance activities and outbreak Investigations which generate some of the data we were using such as filovirus outbreak occurrence points.
Biographies
Luke Nyakarahuka is an Epidemiologist with a background in Veterinary Medicine and Masters in Public Health(MPH) from Makerere University Kampala Uganda. He is a zoonotic disease Epidemiologist at Uganda Virus Research Institute especially focussing on surveillance and outbreak investigation of viral hemorrhagic fevers including Ebola and Marburg viruses. He is a lecturer of Epidemiology and Veterinary Public Health at Makerere University.
I am an ecologist with strong interests in biogeography and climate change.
Gladys Mosomtai was born in 1988 in Mogotio, Kenya. She received her bachelor degree in Environmental Planning and Management in Kenyatta University, Kenya, in 2012 and her master degree in Geospatial Information Systems and Remote Sensing in Deadan Kimathi University of Technology, Kenya, in 2017. Recently she received ARPHIS-DAAD scholarship to pursue her doctoral degree and she will be conducting her research on modelling the impact of agro-ecological factors for coffee pest and disease dynamic prediction at International Centre for Insect Physiology and Ecology, Kenya. She is a passionate user of earth observation technology in pursuing her research interests in landscape ecology, landscape genetics, landscape epidemiology, movement ecology, species distribution modelling, food security, disaster risk management and climate change. She is competent in R programming language, various GIS and remote sensing softwares and has a good command in statistical analysis and data mining techniques. She has the ability to perform tasks with a good technical understanding and soundness whilst being able to come up with solutions in regards to complex scientific issues.
I have good interpersonal relationship and the ability to work with other people as well as independently with minimal supervision. I am a committed, reliable, a clear-headed person. I am a hard working person with a sound and good thinking attitude and good communication skills with the potential to carry out resource mobilization from different stakeholders. Besides, I have a very good command of English language. I have strong attributes, knowledge and skills in designing, planning, management and implementation of Projects and programs at a strategic level. I hold academic capabilities and competences acquired from the following disciplines: • Bachelor’s degree in Veterinary Medicine (BVM) which provides a foundation for the overall responsibility for managing animal health aimed at improved livestock production which in turn contributes to alleviation of poverty and ensuring food security. • Masters of Preventive Veterinary Medicine (MPVM) that compliments the first degree with the provisions for modeling, implementing, monitoring and evaluation of the animal diseases prevention and control strategies. • Postgraduate Diploma in Integrated Rural planning and development ( PGDIRPD) PhD in Veterinary Epidemiology and Public Health at the Norwegian University of Life Sciences with a focus on Zoonotic diseases • Masters of Arts degree in Rural Development (MARD) The Postgraduate Diploma and MARD provides multi-disciplinary knowledge and skills to effectively, design, plan and implementation of rural community development interventions so as to sustainably manage community resources. This therefore will ultimately contribute to socioeconomic transformation of the communities especially the poorest of the poor. These and many other interventions are aimed at improving livelihoods and contribute to poverty reduction and sustainable development. SELECTED PUBLICATIONS 1. Kankya C, Muwonge A, Olet S, Munyeme M, Biffa D, Opuda- Asibo J, Skjerve, E, Oloya J. Factors associated with pastoral community knowledge and occurrence of mycobacterial infections in Human – Animal interface areas of Nakasongola and Mubende District, Uganda. BMC Public Health 2010 Aug 10: 10: 471 2. Muwonge A, Kankya C, Godfroid J, Djønne B, Munyeme M, Biffa D, Opuda- Asibo J, Skjerve, E, Prevalence and associated risk factors of mycobacterial infection in slaughter pigs from Mubende district in Uganda. Tropical Animal health and production 2010 June 42(5) 905-13 3. Kankya C, Muwonge A, Djønne B, Munyeme M, Opuda-Asibo J, Skjerve E, Oloya J, Edvardsen V, Johansen TB: Isolation of non-tuberculous mycobacteria (NTM) from pastoral ecosystems of Uganda: Public Health significance. BMC Public Health 2011, 11:320 4. Kankya C, Mugisha A, Muwonge A, Skjerve E, Kyomugisha E, Oloya J: Myths, perceptions,knowledge,attitudes, and practices(KAP)linked to mycobacterial infection management among Ugandan pastoralists. Advance Tropical Medicine and Public health international 2011; 1(4) 111-124 5. Muwonge,A. Ashemeire A. Kankya C, Biffa D, Oloya J, Skjerve E. Book Chapter No: 2 Nontuberculous mycobacteria in Uganda: a problem or not? In Global view in HIV Infection. Published 2011. 6. Muwonge A, Kankya C, Johansen T.B, Djønne B, Godfroid J, Biffa D, Edvardsen V, Skjerve E : Non-tuberculous mycobacteria isolated from slaughtered pigs in Mubende district, Uganda. Accepted for publication in BMC Veterinary Research, 2012 7. Adrian Muwonge, Hetron M Munang'andu, Clovice Kankya, Demelash Biffa, Chris Oura, Eystein Skjerve and James Oloya: African swine fever among slaughter pigs in Mubende district, Uganda. Trop Anim Health Prod (2012) PMID 22367736
Am interested in veterinary virus research works. Currently am the PI Foot-and-Mouth Disease at COVAB, Mak. Uganda. I hold a Bachelors Degree of Veterinary Medicine, a Masters of Veterinary Preventive Medicine and a Ph.D.
Educational and professional background I was born on August 17 1957 in Steinkjer, Norway. After high school I went on to study veterinary medicine at the Norwegian School of Veterinary Science (NVH), to graduate in 1983. After 4 years as a research assistant at the NVH I got my PhD in 1987 on a work on Trichinella and joined the staff at the Department of Food Hygiene, later to merge into the Department of Food Safety and Infectious Biology. Besides one year in the Netherlands (1989-90), one year at the University of California, Davis (1997-98) and one year at Cornell University (2013-2014) I have stayed at the Norwegian School of Veterinary Science, the present Veterinary Faculty of the Norwegian University NMBU. Starting as a scientific assistant (1983-87), I worked as assistant professor (1987-2001). In 2001 I was appointed full professor in the epidemiology of food-borne diseases, in 2014 changed to Professor of Veterinary Public Health. I am also a diplomat of the European College of veterinary Public Health. In the years 2006-2015 I was leading the Centre for Epidemiology and Biostatistics at NMBU 2006-2015. I am now responsible for the scientific group Veterinary Public Health at the Faculty. Research A lead in my research has been topics related to Veterinary Public health and One Health. A starting focus was studies in the epidemiology of meat-borne zoonoses, including parasites as Trichinella, Toxoplasma and Taenia, and bacteria as Campylobacter, Salmonella, Yersinia, Listeria and STEC. The last year I have turned more to theoretical modelling of zoonotic infections, including risk assessment, also maintaining a work on Brucella and Mycobacterium bovis epidemiology in African countries. I have been involved in a range of collaborative projetcs with African partners, and is the Norwegian project leader of the current NORHED project Capacity Building in Zoonotic diseases Management using integrated approach to Ecosystems health (CAPAZOMANINTECO) at the human-livestock–wildlife interface in Eastern and Southern Africa. Adding to this a major focus on control of diseases in salmonids has been a main focus over recent years. Skjerve has authored or co-authored some 165 international papers, written a number of international book chapters, and a considerable number of papers, reports and books in Norwegian. I have supervised and co-supervised more than 40 PhD students in Norway and abroad.
Funding Statement
We are grateful for funding from Norwegian Agency for Development Cooperation (NORAD) through the Norwegian Program for Capacity Building in Higher Education and Research for Development (NORHED) project of Capacity Building in Zoonotic diseases Management using integrated approach to Ecosystems health at the human-livestock–wildlife interface in Eastern and Southern Africa. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Contributor Information
Luke Nyakarahuka, 1) Department of Food Safety and Infection Biology, Norwegian University of Life Sciences, Oslo, Norway; 2) Department of Biosecurity, Ecosystems and Veterinary Public Health, Makerere University, Kampala Uganda; 3) Department of Arbovirology, Emerging and Re-Emerging disease, Uganda Virus Research Institute, Entebbe, Uganda.
Samuel Ayebare, Climate Change and Biodiversity Unit, Wildlife Conservation Society, Bronx, New York, United States of America.
Gladys Mosomtai, Earth Observation Unit, International Centre for Insect Physiology and Ecology, Nairobi, Kenya.
Clovice Kankya, Department of Biosecurity, Ecosystems and Veterinary Public Health, Makerere University, Kampala, Uganda.
Julius Lutwama, Department of Arbovirology, Emerging and Re-Emerging diseases, Uganda Virus Research Institute, Entebbe, Uganda.
Frank Norbert Mwiine, Department of Biomolecular Resources and Biolab Sciences, Makerere University, Kampala, Uganda.
Eystein Skjerve, Department of Food Safety and Infection Biology, Norwegian University of Life Sciences, Oslo, Norway.
References
- 1.Okware SI, Omaswa FG, Zaramba S, Opio A, Lutwama JJ, Kamugisha J, et al. An outbreak of Ebola in Uganda. Trop Med Int Health. 2002;7(12):1068-75. PubMed PMID: 12460399. [DOI] [PubMed]
- 2.Wamala JF, Lukwago L, Malimbo M, Nguku P, Yoti Z, Musenero M, et al. Ebola hemorrhagic fever associated with novel virus strain, Uganda, 2007-2008. Emerg Infect Dis. 2010;16(7):1087-92. doi: 10.3201/eid1607.091525. PubMed PMID: 20587179; PubMed Central PMCID: PMCPMC3321896 [DOI] [PMC free article] [PubMed]
- 3.Nyakarahuka L, Kankya C, Krontveit R, Mayer B, Mwiine FN, Lutwama J, et al. How severe and prevalent are Ebola and Marburg viruses? A systematic review and meta-analysis of the case fatality rates and seroprevalence. BMC Infect Dis. 2016;16(1):708. doi: 10.1186/s12879-016-2045-6. PubMed PMID: 27887599 [DOI] [PMC free article] [PubMed]
- 4.Shoemaker T, MacNeil A, Balinandi S, Campbell S, Wamala JF, McMullan LK, et al. Reemerging Sudan Ebola virus disease in Uganda, 2011. Emerg Infect Dis. 2012;18(9):1480-3. doi: 10.3201/eid1809.111536. PubMed PMID: 22931687; PubMed Central PMCID: PMCPMC3437705. [DOI] [PMC free article] [PubMed]
- 5.Albarino CG, Shoemaker T, Khristova ML, Wamala JF, Muyembe JJ, Balinandi S, et al. Genomic analysis of filoviruses associated with four viral hemorrhagic fever outbreaks in Uganda and the Democratic Republic of the Congo in 2012. Virology. 2013;442(2):97-100. doi: 10.1016/j.virol.2013.04.014. PubMed PMID: 23711383. [DOI] [PMC free article] [PubMed]
- 6.Adjemian J, Farnon EC, Tschioko F, Wamala JF, Byaruhanga E, Bwire GS, et al. Outbreak of Marburg hemorrhagic fever among miners in Kamwenge and Ibanda Districts, Uganda, 2007. J Infect Dis. 2011;204 Suppl 3:S796-9. doi: 10.1093/infdis/jir312. PubMed PMID: 21987753; PubMed Central PMCID: PMCPMC3203392. [DOI] [PMC free article] [PubMed]
- 7.Towner JS, Amman BR, Sealy TK, Carroll SA, Comer JA, Kemp A, et al. Isolation of genetically diverse Marburg viruses from Egyptian fruit bats. PLoS Pathog. 2009;5(7):e1000536. doi: 10.1371/journal.ppat.1000536. PubMed PMID: 19649327; PubMed Central PMCID: PMCPMC2713404. [DOI] [PMC free article] [PubMed]
- 8.Amman BR, Carroll SA, Reed ZD, Sealy TK, Balinandi S, Swanepoel R, et al. Seasonal pulses of Marburg virus circulation in juvenile Rousettus aegyptiacus bats coincide with periods of increased risk of human infection. PLoS Pathog. 2012;8(10):e1002877. doi: 10.1371/journal.ppat.1002877. PubMed PMID: 23055920; PubMed Central PMCID: PMCPMC3464226. [DOI] [PMC free article] [PubMed]
- 9.Knust B, Schafer IJ, Wamala J, Nyakarahuka L, Okot C, Shoemaker T, et al. Multidistrict Outbreak of Marburg Virus Disease-Uganda, 2012. J Infect Dis. 2015;212 Suppl 2:S119-28. doi: 10.1093/infdis/jiv351. PubMed PMID: 26209681. [DOI] [PMC free article] [PubMed]
- 10.Amman BR, Nyakarahuka L, McElroy AK, Dodd KA, Sealy TK, Schuh AJ, et al. Marburgvirus resurgence in Kitaka Mine bat population after extermination attempts, Uganda. Emerg Infect Dis. 2014;20(10):1761-4. doi: 10.3201/eid2010.140696. PubMed PMID: 25272104; PubMed Central PMCID: PMCPMC4193183. [DOI] [PMC free article] [PubMed]
- 11.Nyakarahuka L, Ojwang J, Tumusiime A, Balinandi S, Whitmer S, Kyazze S, et al. Isolated Case of Marburg Virus Disease, Kampala, Uganda, 2014. Emerg Infect Dis. 2017;23(6):1001-4. doi: 10.3201/eid2306.170047. PubMed PMID: 28518032. [DOI] [PMC free article] [PubMed]
- 12.Jahrling PB, Geisbert TW, Dalgard DW, Johnson ED, Ksiazek TG, Hall WC, et al. Preliminary-Report - Isolation of Ebola Virus from Monkeys Imported to USA. Lancet. 1990;335(8688):502-5. doi: Doi 10.1016/0140-6736(90)90737-P. PubMed PMID: WOS:A1990CT01500005. [DOI] [PubMed]
- 13.Pooley S, Fa JE, Nasi R. Ebola and bushmeat. New Sci. 2014;224(2989):31-. PubMed PMID: WOS:000342656200017
- 14.Leroy EM, Rouquet P, Formenty P, Souquiere S, Kilbourne A, Froment JM, et al. Multiple Ebola virus transmission events and rapid decline of central African wildlife. Science. 2004;303(5656):387-90. doi: 10.1126/science.1092528. PubMed PMID: 14726594. [DOI] [PubMed]
- 15.Leroy EM, Kumulungui B, Pourrut X, Rouquet P, Hassanin A, Yaba P, et al. Fruit bats as reservoirs of Ebola virus. Nature. 2005;438(7068):575-6. doi: 10.1038/438575a. PubMed PMID: 16319873. [DOI] [PubMed]
- 16.Jaax NK, Davis KJ, Geisbert TJ, Vogel P, Jaax GP, Topper M, et al. Lethal experimental infection of rhesus monkeys with Ebola-Zaire (Mayinga) virus by the oral and conjunctival route of exposure. Archives of Pathology & Laboratory Medicine. 1996;120(2):140-55. PubMed PMID: WOS:A1996TV90500005. [PubMed]
- 17.Geisbert TW, Pushko P, Anderson K, Smith J, Davis KJ, Jahrling PB. Evaluation in nonhuman primates of vaccines against Ebola virus. Emerging Infectious Diseases. 2002;8(5):503-7. PubMed PMID: WOS:000175415300010. 18. Formenty P, Boesch C, Wyers M, Steiner C, Donati F, Dind F, et al. Ebola [DOI] [PMC free article] [PubMed]
- 18.Formenty P, Boesch C, Wyers M, Steiner C, Donati F, Dind F, et al. Ebola virus outbreak among wild chimpanzees living in a rain forest of Cote d'Ivoire. Journal of Infectious Diseases. 1999;179:S120-S6. doi: Doi 10.1086/514296. PubMed PMID: WOS:000078400300022. [DOI] [PubMed]
- 19.Rouquet P, Froment JM, Bermejo M, Kilbourn A, Karesh W, Reed P, et al. Wild animal mortality monitoring and human Ebola outbreaks, Gabon and Republic of Congo, 2001-2003. Emerg Infect Dis. 2005;11(2):283-90. doi: 10.3201/eid1102.040533. PubMed PMID: 15752448; PubMed Central PMCID: PMCPMC3320460. [DOI] [PMC free article] [PubMed]
- 20.Allela L, Boury O, Pouillot R, Delicat A, Yaba P, Kumulungui B, et al. Ebola virus antibody prevalence in dogs and human risk. Emerg Infect Dis. 2005;11(3):385-90. doi: 10.3201/eid1103.040981. PubMed PMID: 15757552; PubMed Central PMCID: PMCPMC3298261. [DOI] [PMC free article] [PubMed]
- 21.Weingartl HM, Embury-Hyatt C, Nfon C, Leung A, Smith G, Kobinger G. Transmission of Ebola virus from pigs to non-human primates. Sci Rep. 2012;2:811. doi: 10.1038/srep00811. PubMed PMID: 23155478; PubMed Central PMCID: PMCPMC3498927. [DOI] [PMC free article] [PubMed]
- 22.Team E. Ebola Reston Virus Detected Pigs in the Philippines. Eurosurveillance. 2009;14(4):11-. PubMed PMID: WOS:000267921400004. [PubMed]
- 23.Marsh GA, Haining J, Robinson R, Foord A, Yamada M, Barr JA, et al. Ebola Reston Virus Infection of Pigs: Clinical Significance and Transmission Potential. Journal of Infectious Diseases. 2011;204:S804-S9. doi: 10.1093/infdis/jir300. PubMed PMID: WOS:000295991400009. [DOI] [PubMed]
- 24.Towner JS, Amman BR, Sealy TK, Carroll SAR, Comer JA, Kemp A, et al. Isolation of Genetically Diverse Marburg Viruses from Egyptian Fruit Bats. Plos Pathogens. 2009;5(7). doi: ARTN e1000536 10.1371/journal.ppat.1000536. PubMed PMID: WOS:000269224500012. [DOI] [PMC free article] [PubMed]
- 25.Peterson AT. Ecological niche modelling and understanding the geography of disease transmission. Vet Ital. 2007;43(3):393-400. PubMed PMID: 20422515. [PubMed]
- 26.Batista TA, Gurgel-Goncalves R. Ecological niche modelling and differentiation between Rhodnius neglectus Lent, 1954 and Rhodnius nasutus Stal, 1859 (Hemiptera: Reduviidae: Triatominae) in Brazil. Mem Inst Oswaldo Cruz. 2009;104(8):1165-70. PubMed PMID: 20140378. [DOI] [PubMed]
- 27.Hawlitschek O, Porch N, Hendrich L, Balke M. Ecological niche modelling and nDNA sequencing support a new, morphologically cryptic beetle species unveiled by DNA barcoding. PLoS One. 2011;6(2):e16662. doi: 10.1371/journal.pone.0016662. PubMed PMID: 21347370; PubMed Central PMCID: PMCPMC3036709. [DOI] [PMC free article] [PubMed]
- 28.Mudenda NB, Malone JB, Kearney MT, Mischler PD, Nieto Pdel M, McCarroll JC, et al. Modelling the ecological niche of hookworm in Brazil based on climate. Geospat Health. 2012;6(3):S111-23. doi: 10.4081/gh.2012.129. PubMed PMID: 23032277. [DOI] [PubMed]
- 29.Peterson AT, Bauer JT, Mills JN. Ecologic and geographic distribution of filovirus disease. Emerg Infect Dis. 2004;10(1):40-7. doi: 10.3201/eid1001.030125. PubMed PMID: 15078595; PubMed Central PMCID: PMCPMC3322747. [DOI] [PMC free article] [PubMed]
- 30.WorldClim. WorldClim - Global Climate Data, Free climate data for ecological modeling and GIS 2016 [cited 2017 16.January 2017]. Available from: http://www.worldclim.org/bioclim.
- 31.Peterson AT, Lash RR, Carroll DS, Johnson KM. Geographic potential for outbreaks of Marburg hemorrhagic fever. Am J Trop Med Hyg. 2006 Jul;75(1):9-15. PubMed PMID:16837700. [DOI] [PubMed]
- 32.Pigott DM, Golding N, Mylne A, Huang Z, Henry AJ, Weiss DJ, et al. Mapping the zoonotic niche of Ebola virus disease in Africa. Elife. 2014;3:e04395. doi: 10.7554/eLife.04395. PubMed PMID: 25201877; PubMed Central PMCID: PMCPMC4166725. [DOI] [PMC free article] [PubMed]
- 33.IPigott DM, Golding N, Mylne A, Huang Z, Weiss DJ, Brady OJ, et al. Mapping the zoonotic niche of Marburg virus disease in Africa. Trans R Soc Trop Med Hyg. 2015;109(6):366-78. doi: 10.1093/trstmh/trv024. PubMed PMID: 25820266; PubMed Central PMCID: PMCPMC4447827. [DOI] [PMC free article] [PubMed]
- 34.Phillips SJ, Dudík M. Modeling of species distributions with Maxent: new extensions and a comprehensive evaluation. Ecography. 2008;31(2):161-75.
- 35.P Anderson R, Dudík M, Ferrier S, Guisan A, J Hijmans R, Huettmann F, et al. Novel methods improve prediction of species’ distributions from occurrence data. Ecography. 2006;29(2):129-51.
- 36.Chamaille L, Tran A, Meunier A, Bourdoiseau G, Ready P, Dedet JP. Environmental risk mapping of canine leishmaniasis in France. Parasit Vectors. 2010;3:31. doi: 10.1186/1756-3305-3-31. PubMed PMID: 20377867; PubMed Central PMCID: PMCPMC2857865. [DOI] [PMC free article] [PubMed]
- 37.Kantzoura V, Kouam MK, Feidas H, Teofanova D, Theodoropoulos G. Geographic distribution modelling for ruminant liver flukes (Fasciola hepatica) in south-eastern Europe. Int J Parasitol. 2011;41(7):747-53. doi: 10.1016/j.ijpara.2011.01.006. PubMed PMID: 21329694. [DOI] [PubMed]
- 38.Mischler P, Kearney M, McCarroll JC, Scholte RG, Vounatsou P, Malone JB. Environmental and socio-economic risk modelling for Chagas disease in Bolivia. Geospat Health. 2012;6(3):S59-66. doi: 10.4081/gh.2012.123. PubMed PMID: 23032284. [DOI] [PubMed]
- 39.Zeimes CB, Olsson GE, Ahlm C, Vanwambeke SO. Modelling zoonotic diseases in humans: comparison of methods for hantavirus in Sweden. Int J Health Geogr. 2012;11:39. doi: 10.1186/1476-072X-11-39. PubMed PMID: 22984887; PubMed Central PMCID: PMCPMC3517350. [DOI] [PMC free article] [PubMed]
- 40.Chikerema SM, Murwira A, Matope G, Pfukenyi DM. Spatial modelling of Bacillus anthracis ecological niche in Zimbabwe. Prev Vet Med. 2013;111(1-2):25-30. doi: 10.1016/j.prevetmed.2013.04.006. PubMed PMID: 23726015. [DOI] [PubMed]
- 41.Mweya CN, Kimera SI, Kija JB, Mboera LE. Predicting distribution of Aedes aegypti and Culex pipiens complex, potential vectors of Rift Valley fever virus in relation to disease epidemics in East Africa. Infect Ecol Epidemiol. 2013;3. doi: 10.3402/iee.v3i0.21748. PubMed PMID: 24137533; PubMed Central PMCID: PMCPMC3797365. [DOI] [PMC free article] [PubMed]
- 42.Nogareda C, Jubert A, Kantzoura V, Kouam MK, Feidas H, Theodoropoulos G. Geographical distribution modelling for Neospora caninum and Coxiella burnetii infections in dairy cattle farms in northeastern Spain. Epidemiol Infect. 2013;141(1):81-90. doi: 10.1017/S0950268812000271. PubMed PMID: 22370223. [DOI] [PMC free article] [PubMed]
- 43.Mughini-Gras L, Mulatti P, Severini F, Boccolini D, Romi R, Bongiorno G, et al. Ecological niche modelling of potential West Nile virus vector mosquito species and their geographical association with equine epizootics in Italy. Ecohealth. 2014;11(1):120-32. doi: 10.1007/s10393-013-0878-7. PubMed PMID: 24121802. [DOI] [PubMed]
- 44.Mylne A, Brady OJ, Huang Z, Pigott DM, Golding N, Kraemer MU, et al. A comprehensive database of the geographic spread of past human Ebola outbreaks. Sci Data. 2014;1:140042. doi: 10.1038/sdata.2014.42. PubMed PMID: 25984346; PubMed Central PMCID: PMCPMC4432636. [DOI] [PMC free article] [PubMed]
- 45.Kityo R, Howell K, Nakibuka M, Ngalason W, Tushabe H, Webala P. East African Bat Atlas. Makerere University and University of Dar es Salaam, Kampala, Uganda. 2009.
- 46.Platts PJ, Omeny PA, Marchant R. AFRICLIM: high‐resolution climate projections for ecological applications in Africa. African Journal of Ecology. 2015;53(1):103-8.
- 47.Warren DL, Glor RE, Turelli M. ENMTools: a toolbox for comparative studies of environmental niche models. Ecography. 2010;33(3):607-11.
- 48.Young N, Carter L, Evangelista P. A MaxEnt model v3. 3.3 e tutorial (ArcGIS v10). Fort Collins, Colorado. 2011.
- 49.Townsend Peterson A, Papeş M, Eaton M. Transferability and model evaluation in ecological niche modeling: a comparison of GARP and Maxent. Ecography. 2007;30(4):550-60.
- 50.Anderson RP, Lew D, Peterson AT. Evaluating predictive models of species’ distributions: criteria for selecting optimal models. Ecological modelling. 2003;162(3):211-32.
- 51.Swets JA. Measuring the accuracy of diagnostic systems. Science. 1988;240(4857):1285-93. [DOI] [PubMed]
- 52.Russo D, Cistrone L, Jones G. Sensory ecology of water detection by bats: a field experiment. PLoS One. 2012;7(10):e48144. doi: 10.1371/journal.pone.0048144. PubMed PMID: 23133558; PubMed Central PMCID: PMCPMC3483877. [DOI] [PMC free article] [PubMed]
- 53.Adams RA, Hayes MA. Water availability and successful lactation by bats as related to climate change in arid regions of western North America. J Anim Ecol. 2008;77(6):1115-21. doi: 10.1111/j.1365-2656.2008.01447.x. PubMed PMID: 18684132. [DOI] [PubMed]
- 54.Amman BR, Jones ME, Sealy TK, Uebelhoer LS, Schuh AJ, Bird BH, et al. Oral shedding of Marburg virus in experimentally infected Egyptian fruit bats (Rousettus aegyptiacus). J Wildl Dis. 2015;51(1):113-24. doi: 10.7589/2014-08-198. PubMed PMID: 25375951; PubMed Central PMCID: PMCPMC5022530. [DOI] [PMC free article] [PubMed]
- 55.Pigott DM, Golding N, Mylne A, Huang Z, Weiss DJ, Brady OJ, et al. Mapping the zoonotic niche of Marburg virus disease in Africa. Trans R Soc Trop Med Hyg. 2015;109(6):366-78. doi: 10.1093/trstmh/trv024. PubMed PMID: 25820266; PubMed Central PMCID: PMCPMC4447827. [DOI] [PMC free article] [PubMed]
- 56.Pigott DM, Millear AI, Earl L, Morozoff C, Han BA, Shearer FM, et al. Updates to the zoonotic niche map of Ebola virus disease in Africa. Elife. 2016;5. doi: 10.7554/eLife.16412. PubMed PMID: 27414263; PubMed Central PMCID: PMCPMC4945152. [DOI] [PMC free article] [PubMed]
- 57.UNMA. seasonal forecasts 2016 [cited 2017 18th May 2017]. Available from: https://www.unma.go.ug/index.php/climate/seasonal-performance.
- 58.Centers for Disease C, Prevention. Imported case of Marburg hemorrhagic fever - Colorado, 2008. MMWR Morb Mortal Wkly Rep. 2009;58(49):1377-81. PubMed PMID: 20019654. [PubMed]
- 59.Timen A, Koopmans MP, Vossen AC, van Doornum GJ, Gunther S, van den Berkmortel F, et al. Response to imported case of Marburg hemorrhagic fever, the Netherland. Emerg Infect Dis. 2009;15(8):1171-5. doi: 10.3201/eid1508.090015. PubMed PMID: 19751577; PubMed Central PMCID: PMCPMC2815969. [DOI] [PMC free article] [PubMed]
- 60.Kratz T, Roddy P, Tshomba Oloma A, Jeffs B, Pou Ciruelo D, de la Rosa O, et al. Ebola Virus Disease Outbreak in Isiro, Democratic Republic of the Congo, 2012: Signs and Symptoms, Management and Outcomes. PLoS One. 2015;10(6):e0129333. doi: 10.1371/journal.pone.0129333. PubMed PMID: 26107529; PubMed Central PMCID: PMCPMC4479598. [DOI] [PMC free article] [PubMed]
- 61.Peterson AT, Samy AM. Geographic potential of disease caused by Ebola and Marburg viruses in Africa. Acta Trop. 2016;162:114-24. doi: 10.1016/j.actatropica.2016.06.012. PubMed PMID: 27311387. [DOI] [PubMed]
- 62.WHO. Marburg virus disease - Uganda: WHO; 2014 [cited 2016 5-09-2016]. Available from: http://www.who.int/csr/don/10-october-2014-marburg/en/.
- 63.Kalunda M, Mukwaya LG, Mukuye A, Lule M, Sekyalo E, Wright J, et al. Kasokero virus: a new human pathogen from bats (Rousettus aegyptiacus) in Uganda. Am J Trop Med Hyg. 1986;35(2):387-92. PubMed PMID: 3082234. [DOI] [PubMed]
- 64.Johnson BK, Ocheng D, Gitau LG, Gichogo A, Tukei PM, Ngindu A, et al. Viral haemorrhagic fever surveillance in Kenya, 1980-1981. Trop Geogr Med. 1983;35(1):43-7. PubMed PMID: 6684336. [PubMed]
- 65.Smith DH, Johnson BK, Isaacson M, Swanapoel R, Johnson KM, Killey M, et al. Marburg-virus disease in Kenya. Lancet. 1982;1(8276):816-20. PubMed PMID: 6122054. [DOI] [PubMed]
- 66.Leendertz SA, Gogarten JF, Dux A, Calvignac-Spencer S, Leendertz FH. Assessing the Evidence Supporting Fruit Bats as the Primary Reservoirs for Ebola Viruses. Ecohealth. 2016;13(1):18-25. doi: 10.1007/s10393-015-1053-0. PubMed PMID: 26268210. [DOI] [PMC free article] [PubMed]
- 67.Mari Saez A, Weiss S, Nowak K, Lapeyre V, Zimmermann F, Dux A, et al. Investigating the zoonotic origin of the West African Ebola epidemic. EMBO Mol Med. 2015;7(1):17-23. doi: 10.15252/emmm.201404792. PubMed PMID: 25550396; PubMed Central PMCID: PMCPMC4309665. [DOI] [PMC free article] [PubMed]
- 68.Omilabu SA, Salu OB, Oke BO, James AB. The West African ebola virus disease epidemic 2014-2015: A commissioned review. Niger Postgrad Med J. 2016;23(2):49-56. doi: 10.4103/1117-1936.186299. PubMed PMID: 27424613. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data is available in the paper and supporting files which can be found on figshare as follows: S1 File: Occurrence dataset used (Filovirus and Bats Occurrence coordinates) (10.6084/m9.figshare.5306875 <https://doi.org/10.6084/m9.figshare.5306875>); S2 File: Results of the quantitative comparisons of environmental variables to test for multicollinearity (10.6084/m9.figshare.5306908 <https://doi.org/10.6084/m9.figshare.5306908>); S3 File: A jackknife test result to evaluate individual covariate importance in the model developments (10.6084/m9.figshare.5306914 <https://doi.org/10.6084/m9.figshare.5306914>); S4 File: The response curves of all the predictor variables in all the four models (10.6084/m9.figshare.5306932 <https://doi.org/10.6084/m9.figshare.5306932>).