Abstract
ActRIIB (activin receptor type-2B) is an activin receptor subtype constitutively expressed in the whole body, playing a role in cellular proliferation, differentiation, and metabolism. For its various physiological activities, ActRIIB interacts with activin and multiple other ligands including myostatin (MSTN), growth differentiation factor 11 (GDF11), and bone morphogenetic protein 9 (BMP9). Notably, the protein-protein interaction (PPI) between ActRIIB and MSTN negatively controls muscular development. Therefore, this PPI has been targeted for effective treatment of muscle degenerative diseases such as muscular dystrophy and sarcopenia. Here, we report the identification of ligand-selective peptidic ActRIIB-antagonists by phage display technology. Our peptides bound to the extracellular domain of ActRIIB, inhibited PPIs between ActRIIB expressed on the cell surface and its ligands, and subsequently suppressed activation of Smad that serves as the downstream signal of the ActRIIB pathway. Interestingly, these peptidic antagonists displayed different ligand selectivities; the AR2mini peptide inhibited multiple ligands (activin A, MSTN, GDF11, and BMP9), AR9 inhibited MSTN and GDF11, while AR8 selectively inhibited MSTN. This is the first report of artificial peptidic ActRIIB-antagonists possessing ligand-selectivity.
Abbreviation: ActRIIB, activin receptor type-2B; MSTN, myostatin; GDF11, growth differentiation factor 11; BMP9, bone morphogenetic protein 9; PPI, protein-protein interaction; ALK, activin receptor type-1B
Keywords: ActRIIB, Antagonist, Ligand-selectivity, Peptide, Phage display
Highlights
-
•
Artificial ActRIIB-antagonist peptides were discovered by phage display.
-
•
These peptides selectively bound to the extracellular domain of ActRIIB.
-
•
They antagonized ActRIIB expressed on the cell surface.
-
•
They presented multiple ligand selectivities.
1. Introduction
ActRIIB (activin receptor type-2B) is a receptor serine/threonine kinase that interacts with multiple ligands of the TGFβ superfamily, such as activin, MSTN (myostatin), GDF11 (growth differentiation factor 11), and BMP9 (bone morphogenetic protein 9) [1], [2]. Upon interaction, ligand/ActRIIB complex recruits activin receptor type-1 (ALK) forming a ligand/ActRIIB/ALK (2:2:2) hexamer complex to activate intracellular Smad signaling (Smad is the downstream signal of the ActRIIB pathway) [3]. Though ActRIIB is constitutively expressed in the whole body, its various physiological functions are tissue-selective, depending on the distribution and timing of ligand/ALK expression. Among ActRIIB ligands, MSTN plays a dominant role in both developing and adult human skeletal muscle [4]. MSTN-knockout mice exhibit skeletal muscle hypertrophy, indicating that MSTN acts as a negative regulator of muscle development. Thus, inhibitors of the ActRIIB-MSTN protein-protein interaction (PPI) provide a novel strategy to treat muscle degenerative disorders like muscular dystrophy and sarcopenia.
Some approaches have been explored to reduce MSTN function by either blocking the interaction with its receptor or through MSTN natural inhibitor overexpression [5]. Among them, monoclonal antibodies neutralizing MSTN have been well studied (e.g., MYO-029, PF-06252616, LY-2495655, and REGN-1033) [6]. The injection of a soluble form of ActRIIB (ACE-536) has been reported to improve the amyotrophic phenotype in animal models [7]. In addition to blockers that mask MSTN, the ActRIIB antagonist antibody BYM-338 has been studied [8]. However, treatment with antibodies block multiple ligands due to the high molecular weight of antibodies, leading to undesirable side effects. For example, Suragani et al. reported that injection of GDF11 to mice causes anemia, suggesting that inhibition of the PPI between ActRIIB and GDF11 would have the side effect of increasing the population of red blood cells to abnormal levels [9]. Since, as described above, ActRIIB interacts with multiple ligands and exerts various physiological functions, ligand-selective antagonists would be advantageous.
Recently, peptides have attracted much attention as an alternative to antibodies for PPI modulation because they exhibit high affinity and selectivity for target proteins compared to antibodies, in spite of their approximately 100 times smaller molecular weight [10]. Phage display is a powerful technology for the generation of PPI-modulating peptides [11], [12], [13], as it allows the construction of large and diverse peptide libraries (containing >10 billion distinct peptides), and the identification of target-binding peptide sequences through an affinity selection technique called biopanning.
To generate peptidic ActRIIB antagonists, we panned random peptide libraries displayed on T7 phage against the extracellular domain of ActRIIB, which was fused to human IgG-Fc (ActRIIB-Fc). We successfully identified ActRIIB-Fc binding peptides that we classified into three types. Importantly, these peptides possess the desirable ligand-selective antagonist activity.
2. Materials and methods
2.1. Preparation of recombinant ActRIIB-Fc and MSTN
The mammalian pcDNA3.4 vector was used to create the construct pcDNA3.4/hACVR2B(1-135)-Fc, expressing a fusion protein comprising human ACVR2B-ECD (residues Met1–Leu135, NCBI Reference Sequence: NNM_001106) and the Fc region of human immunoglobulin G at the C-terminus. The construct was transfected into Expi293F cells (at a final density of 2.5 × 106 cells/mL) using the ExpiFectamine 293 Transfection Kit and Opti-MEM Reduced Serum Medium (Life Technologies, Carlsbad, CA, USA) in Expi293 Expression Medium. After 18 h, Enhancer 1 and Enhancer 2 were added and the cultures were expanded for 5 days. After incubation, the supernatant containing recombinant protein was separated from cells by centrifugation. To purify recombinant proteins, the supernatant was loaded onto an SP-Sepharose Fast-Flow column (GE Healthcare, Piscataway, NJ, USA) equilibrated in buffer A (50 mM Sodium Acetate, pH 4.0). The column was washed with buffer A and the protein was eluted with a linear gradient from 0 to 1 M NaCl in the same buffer. The eluate was loaded onto a HiLoad 26/60 Superdex 200 pg column (GE Healthcare) using PBS (phosphate-buffered saline: 10 mM phosphate, 2.7 mM KCl, and 137 mM NaCl). Purified proteins were concentrated with AmiconUltra15 centrifugal filter units (MWCO = 30 kDa; Merck-Millipore, Billerica, MA, USA). Protein concentration was determined using the BCA Protein Assay Kit (Thermo Fisher Scientific, Waltham, MA, USA) using BSA (bovine serum albumin) as a standard.
The mammalian pcDNA3.4 vector was also used to create the pcDNA3.3/hMSTN-His construct that expressed human MSTN (NCBI Reference Sequence: NM_005259) as a fusion protein with a His-tag at its C-terminus. To generate an FS293F stable cell line secreting MSTN pro-peptide, FS293F cells were transfected with the construct using Neofection (ASTEC, Shime, Japan) according to the manufacturer's instructions. Transfected cells were selected by G418 treatment (500 μg/mL) for 14 days. The expression of recombinant MSTN pro-peptide in the stable cells was detected using western blotting analysis with anti-His6-peroxidase. The selected cell clones were cultured in FreeStyle expression medium (Life Technologies) containing 250 μg/mL G418 for 3 days. The supernatant containing recombinant protein was separated from the cultured cells by centrifugation. For protein purification, the supernatant was loaded onto a 5 mL Ni-NTA Superflow Cartridge (QIAGEN, Hilden, Germany). The Ni-NTA column was washed with 20 mM imidazole and then the protein was eluted with 250 mM imidazole in affinity buffer [50 mM Tris (pH 8.0) and 300 mM NaCl]. The eluate was loaded onto a HiLoad 26/60 Superdex 200 pg using SDX buffer [20 mM Tris (pH 7.5) and 150 mM NaCl]. To generate mature MSTN, the eluate was treated with the protease furin (produced internally) and the endoproteinase AspN at 37 °C for 16 h. The treated protein was bound to a reverse-phase PLRP-S column (Agilent Technologies, Santa Clara, CA, USA) and then eluted with an acetonitrile gradient in 0.1% trifluoroacetic acid (TFA). The eluted mature MSTN was freeze-dried and then dissolved in 10 mM HCl. Protein concentration was determined using the BCA Protein Assay Kit with BSA as a standard.
2.2. Chemical synthesis of peptides
AR2mini, AR2mut, AR8, and AR9 were synthesized internally by standard 9-fluorenylmethyloxycarbonyl (F-moc)-based solid-phase peptide synthesis (SPPS) using the Syro Wave microwave-assisted peptide synthesizer (Biotage, Uppsala, Sweden) with disulfide bond formation after cleavage from the solid support, followed by purification by reversed-phase high-performance liquid chromatography (RP-HPLC). Peptide purity was ascertained by analytical RP-HPLC, and the identity of the peptides was confirmed by matrix-assisted laser desorption ionization-time of flight mass spectrometry (MALDI-TOF MS).
For disulfide bond formation in AR2mini, AR2mut, and AR8, a linear crude peptide containing two acetamidomethyl-Cys residues (AR2mini, AR2mut: 0.03 mmol; AR8: 0.02 mmol) was dissolved in 90% acetic acid (0.2–0.3 mM peptide). Iodine was added to the solution (10–13.3 eq.) and the mixture was stirred overnight at room temperature. After removal of excess iodine by activated charcoal, the solution was filtered. For disulfide bond formation in AR9, a linear crude peptide with two Cys residues (SH-free form, 0.02 mmol) was dissolved in 50% acetonitrile, then CLEAR-OXTM resin (1.5 eq.) was added to the solution. After stirring for 7 h at room temperature, the resulting solution was filtered. The obtained peptide solutions were applied to preparative RP-HPLC using appropriate columns [(AR2mini, AR8: Daisopak SP-100-5-ODS-P column, 20 × 250 mm; DaiSO, Osaka, Japan)(AR2mut and AR9: Kinetex 5 µm XB-C18 100 A AX column, 21.1 × 250 mm; Phenomenex. Torrance, CA, USA)]. Linear density gradient elution (60 min) was performed with 0.1% TFA in water (eluent A) and 0.1% TFA in acetonitrile (eluent B) at a flow rate of 8.0 mL/min, at the following ratios (A/B): AR2mini, 71/29–61/39; AR2mut: 79/21–69/31; AR8: 79/21–69/31; AR9: 76/24–66/34. The fractions containing the product peptides were collected and lyophilized. The obtained peptide amounts were 5.9 mg AR2mini, 5.8 mg AR2mut, 6.7 mg AR8, and 2.5 mg AR9.
2.3. Peptide screening by phage display
The T7 phage-displayed random peptide libraries; X12, X16, X20, CX7–10C, X3CX7–10CX3 (X is a mixture of twenty natural amino acids), were internally constructed using mixed oligonucleotides as the DNA template and the T7Select 10-3 vector (Merck Millipore). Insert DNA were purified by QIAquick PCR Purification kit (QIAGEN, Hilden, Germany) and ligated into the T7Select 10-3 vector, according to the manufacture's T7Select System Manual. The total library diversity was estimated to be 1.4 × 1010 by plaque forming unit (pfu). For screening of T7 phage libraries, 20 μg of ActRIIB-Fc was immobilized on 200 μL of Protein A Dynabeads (Invitrogen, Carlsbad, CA, USA) in PBS (Cat. No. 045-29795; Wako, Osaka, Japan) containing 0.5% BSA. After washing beads with PBS containing 0.1% Tween20 (PBST), they were incubated with the mixture of phage libraries for 1 h at room temperature and washed with PBST (3 times for the first round, 5 times for the second round, and 10 times for third–fifth rounds). Bound phages were eluted with 200 μL of 0.5% SDS and used to infect 80 mL of E. coli BLT5615 cells (Merck Millipore) in log-phase growth for phage amplification. After bacteriolysis, phages were recovered from the culture supernatant by centrifugation and PEG-precipitation, dissolved in 1 mL of PBS, and 0.5 mL of the phages were used for the next round of biopanning.
2.4. Plate ELISA for binding evaluation
For phage binding evaluation, the wells of a Corning polystyrene microplate (Cat. No. 3700; Corning, Corning, NY, USA) were coated with goat anti-human Fc polyclonal antibody (10 μg/mL) (Jackson ImmunoResearch, West Grove, PA, USA) at 4 °C for overnight and blocked with 0.5% BSA in PBS at room temperature for 2 h. Fc-fused proteins were captured by the antibody and then phage solution was added to the wells. After 3 times washing with PBST, bound phages were detected using horseradish peroxidase (HRP)-conjugated anti-T7 antibody (5000-fold dilution by 0.5% BSA in PBS) (Merck Millipore). The amount of HRP in each well was measured by the detection reagent tetramethylbenzidine (Wako, Osaka, Japan).
For ActRIIA-Fc (Cat. No. 340-R2-100/CF; R&D Systems, McKinley Place, MN, USA) or ActRIIB-Fc binding, the wells of a Nunc MaxiSorp microplate (Cat. No. 460–518; Thermo Fisher Scientific) were directly coated with each ligand at 4 °C for overnight and blocked with 0.5% BSA in PBS at room temperature for 2 h. Fc-fused proteins were added to the wells. After 3 times washing with PBST, bound Fc-fused proteins were detected using HRP-conjugated goat anti-human IgG Fc antibody (5000-fold dilution by 0.5% BSA in PBS) (Cat. No. ab97225; Abcam, Cambridge, UK). The amount of HRP in each well was quantitated by measuring the produced signal after addition of a chemical luminescence reagent obtained from Wako.
For peptide competition experiments, Corning polystyrene microplates were coated with a goat anti-human Fc polyclonal antibody at 4 °C for overnight and blocked with 0.5% BSA in PBS at room temperature for 2 h. Fc-fused proteins were captured by the antibody, and biotinylated peptide solution (2 μM) with/without non-labeled peptide (100 μM) was added to the wells. After 3 times washing with PBST, the bound biotinylated peptide was detected using HRP-conjugated streptavidin (1000-fold dilution by 0.5% BSA in PBS) (Vector Laboratories Inc., Burlingame, CA, USA). The amount of HRP in each well was measured using the detection reagent tetramethylbenzidine. Percent inhibition was calculated using the values of wells without non-labeled peptide as “0% inhibition” controls and the values from wells without biotinylated peptide as “100% inhibition” controls.
2.5. SPR binding assay
SPR biosensor assays were performed on Biacore 3000 and Biacore S200 instruments equipped with the CM5 sensor chip (GE healthcare). For the immobilization of ActRIIA-Fc and ActRIIB-Fc, PBS supplied with 0.01% Surfactant P20 (GE healthcare) was used as the running buffer with the instrument temperature set at 22 °C. ActRIIA-Fc and ActRIIB-Fc were immobilized onto the sensor chip following the standard amine-coupling procedure according to the manufacturer's instructions. The final immobilization levels of ActRIIA and ActRIIB were around 9000 RU and 3000 RU, respectively.
For interaction experiments, PBS with 0.01% Surfactant P20 and 1% DMSO was used as the running buffer, with instrument temperature at 22 °C. Sample peptide solutions of different concentrations were injected sequentially onto the sensor chip surface at a flow rate of 75 μL/min for 60 s, after which the dissociation was monitored for up to 60 s.
Data processing and analysis were performed using Scrubber v2.0 (BioLogic Software, Campbell, Australia) and the Biacore S200 evaluation software (GE healthcare). Sensorgrams were double-referenced and global fitting of the concentration series to 1:1 binding models was performed for the determination of the binding rate constants, namely kon and koff. The dissociation constant (KD) was calculated by the following equation: KD = koff/kon. For samples showing fast off-rates, KD was calculated with steady state affinity evaluation, i.e., by fitting a plot of steady state binding level vs. concentration to a binding model with an equilibrium of 1:1.
2.6. Gene expression analysis
Total RNA was isolated using the RNeasy Mini Kit (QIAGEN) according to the manufacturer's instructions. Complementary DNA synthesis from total RNA was performed using the PrimeScript RT Reagent Kit (TaKaRa, Shiga, Japan). Quantitative polymerase chain reaction (QPCR) was performed on an ABI PRISM 7900 Sequence Detection System using the TaqMan Fast Advanced Master Mix (Thermo Fisher Scientific) and commercially available primers, as well as the FAM-labeled TaqMan probes (TaqMan Gene Expression Assays; Life Technologies) shown below. Gene expression levels were normalized to 36B4, which was used as an internal control.
| Gene symbol | Assay ID (TaqMan gene expression assays) |
|---|---|
| ACVR2A | Hs00176148_m1 |
| ACVR2B | Hs00609603_m1 |
| BMPR2 | Hs00155658_m1 |
| TGFBR2 | Hs00234253_m1 |
| ACVR1B(ALK4) | Hs00244715_m1 |
| TGFBR1(ALK5) | Hs00610324_m1 |
| RPLP0 (36B4) (internal control) | RPLP0-F: 5′-AAACGAGTCCTGGCCTTGTCT-3′ |
| RPLP0-R: 5′-GCAGATGGATCAGCCAAGAAG-3′ | |
| RPLP0-P: 5′-AGACGGATTACACCTTCCCACTTGCTGA-3′ |
2.7. Cell-based Smad reporter assays
Activin A (Cat. No. 338-AC), GDF11 (Cat. No. 1958-GD), and BMP9 (Cat. No. 3209-BP) were purchased from R&D Systems. Assays were performed as previously described [14]. We constructed vectors pGL4.28 containing a Smad2/3-specific response element with Firefly luciferase as the reference gene and pGL4.26 containing a Smad1/5/8-response element with NanoLuc. The reporter construct was transfected into HEK293T cells using the Fugene HD reagent (Promega, Fitchburg, WI, USA). Transfected cells were seeded on Corning 384-well plates (15 μL containing 1 × 104 cells in each well) in reaction medium, i.e., DMEM (Dulbecco's modified Eagle's medium) containing 0.1% BSA. After 2 h, 15 μL of reaction medium containing ligand (0.33 ng/mL of Activin A, or; 1 ng/mL of TGFβ, MSTN, or GDF11, or; 1.5 ng/mL of BMP9) and/or the indicated concentration of peptide was added to each well After additional incubation (4 h for Smad2/3 and 20 h for Smad1/5/8), cells were lysed to measure luciferase activity using the Bright-Glo Luciferase Assay System (Promega). Percent inhibition was calculated using the luminescence values from wells without peptide as 0% inhibition controls and values from wells without ligands as 100% inhibition controls. IC50 values were calculated by Prism 5 (GraphPad Software, La Jolla, CA, USA).
3. Results
3.1. Identification of ActRIIB-binding peptide sequences by phage display screening
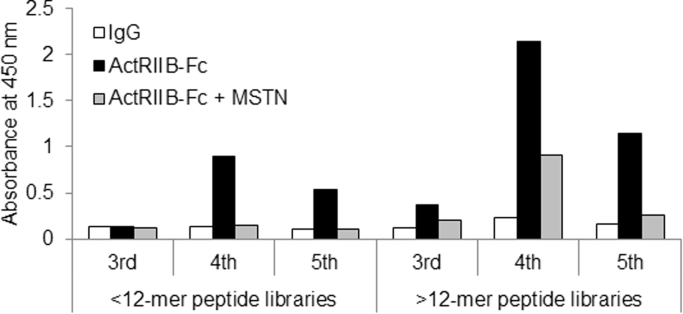
To identify peptides able to bind to the extracellular domain of ActRIIB, we panned T7 phage-displayed random peptide libraries against ActRIIB-Fc. ActRIIB-binding polyclonal phages were successfully collected (Fig. 1). Furthermore, their ability to bind ActRIIB decreased in the presence of MSTN, suggesting that these polyclonal phages included clones that were potential ActRIIB-antagonists. After screening by binding activities of monoclonal phages, DNA sequence analysis was performed on positive phages. We discovered one major cluster (AR2 family), one minor cluster (AR9 family), and one singleton (AR8). Among them, AR2mini (VCFGTSVRRICV-OH), AR9 (HYKCWPKTLFKCWNH-OH), and AR8 (PFQCTRARDGWYYCTKS-OH) were chemically synthesized.
Fig. 1.
Binding activity of polyclonal phages to ActRIIB-Fc. Binding activity of polyclonal phages to ActRIIB-Fc in the absence or presence of MSTN was evaluated by plate ELISA. Phage binding was detected by applying an HRP-labeled anti-T7 phage antibody measuring absorbance at 450 nm. The binding activities of phages displaying random peptides shorter than 12 residues (left column) or longer than 12 residues (right column) are displayed.
3.2. Evaluation by SPR analysis of the binding activities of synthetic peptides to ActRIIB-Fc and ActRIIA-Fc
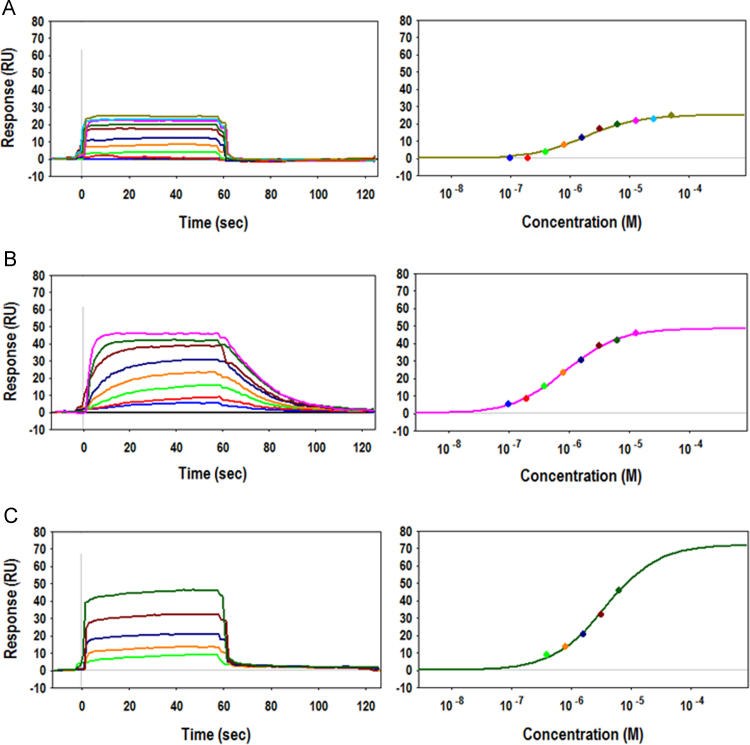
Binding activities of synthetic peptides to ActRIIB-Fc were evaluated by SPR binding assays. AR2mini, AR9, and AR8 were found to bind to ActRIIB-Fc in a peptide concentration-dependent manner, with respective KD values of 1.8 µM (koff > 1 × 10−1 s−1), 0.86 µM (kon = 5.0 × 104 M−1 s−1, koff = 4.9 × 10−2 s−1), and 3.6 µM (koff > 1 × 10−1 s−1) (Fig. 2 and Table 1). Interestingly, while the binding affinities of these peptides were similar, AR9 dissociated more slowly than AR2mini or AR8. We also evaluated the binding activity of the peptides to another activin receptor subtype, namely ActRIIA. As shown in Table 1, they exhibited cross-binding activity to ActRIIA-Fc; however, the respective affinities were significantly weaker than those for ActRIIB-Fc. AR9 was the most selective of the three peptides. The SPR data clearly indicated that our peptides bound directly and selectively to the extracellular domain of ActRIIB.
Fig. 2.
Binding activities and ligand selectivities of peptides as determined by SPR. Sensorgrams (left) and equilibrium plots (right) for the interaction of (A) AR2mini, (B) AR9, and (C) AR8 with immobilized ActRIIB-Fc. The top concentration was 100 μM. Serial 2-fold dilutions were performed.
Table 1.
IC50 and KD values of peptides.
|
KD (μM) |
Antagonist activity (IC50, μM) |
|||||||
|---|---|---|---|---|---|---|---|---|
| Name | Sequence | ActRIIA | ActRIIB | TGFβ | Activin | MSTN | GDF11 | BMP9 |
| AR2mini | VCFGTSVRRICV-OH | 19 | 1.8 | > 100 | 5 | 6 | 18 | 12 |
| AR9 | HYKCWPKTLFKCWNH-OH | > 100 | 0.86 | > 100 | > 100 | 14 | 22 | > 100 |
| AR8 | PFQCTRARDGWYYCTKS-OH | 18 | 3.4 | > 100 | > 100 | 40 | > 100 | > 100 |
| AR2mut | VCDGTSVPRICV-OH | N.T. | > 100 | > 100 | > 100 | > 100 | > 100 | > 100 |
N.T. = not tested.
3.3. Antagonist activities of synthetic peptides in Smad reporter assays
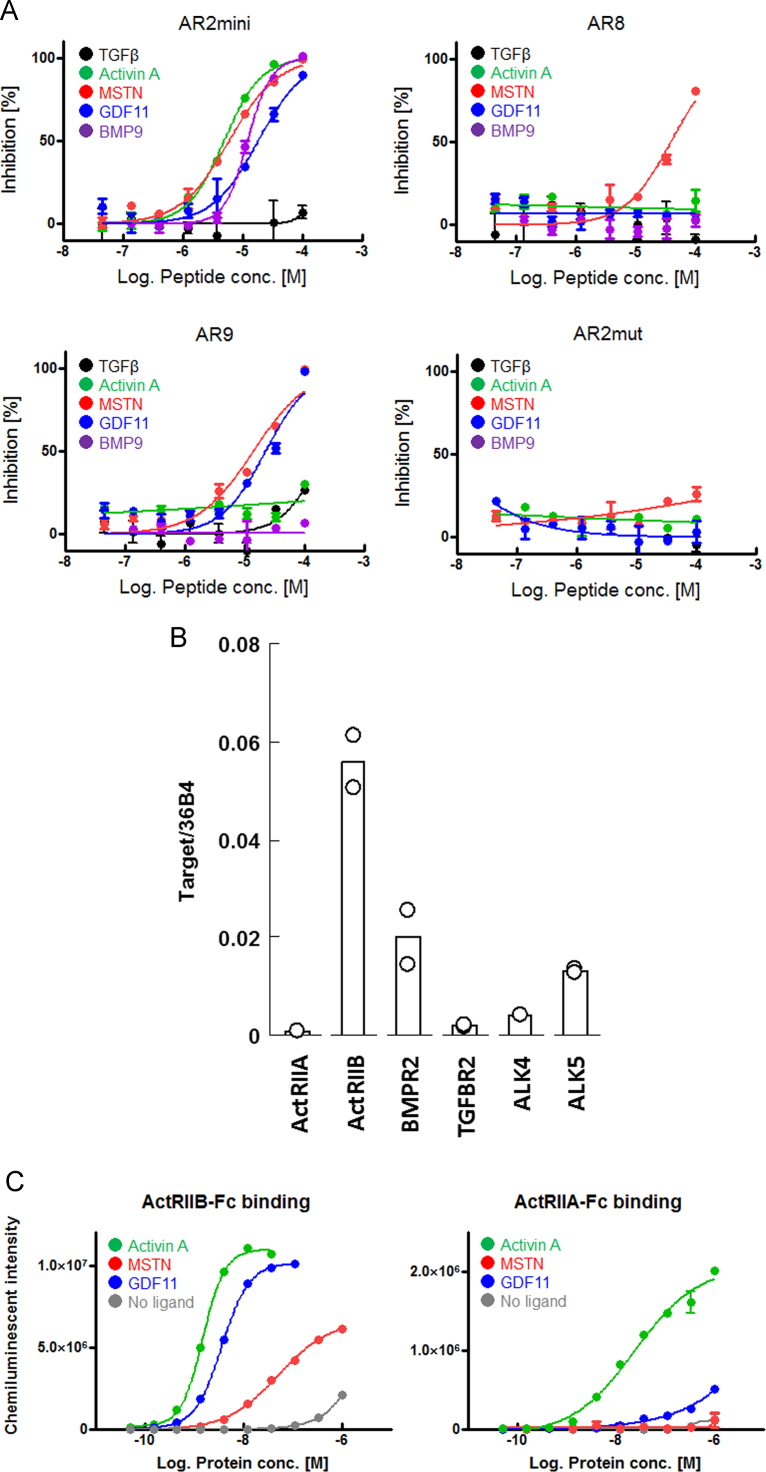
We evaluated the antagonist activity of four peptides, namely AR2mini, AR9, AR8, and AR2mut (negative control) against various ligands (TGFβ, activin A, MSTN, GDF11, and BMP9) using Smad2/3 or Smad1/5/8 reporter assays as previously described [14]. Results are shown in Fig. 3A and Table 1. AR2mini inhibited the signaling of all ligands with the exception of TGFβ. AR9 inhibited the signaling of MSTN and GDF11. Notably, AR8 selectively inhibited MSTN signaling. These antagonist activities were not a result of peptide cytotoxicity (data not shown). The negative control (AR2mut) did not display antagonist activity against any of the tested ligands.
Fig. 3.
Antagonist activities of peptides in Smad reporter assays. (A) Receptor activation by TGFβ, activin A, MSTN, or GDF11 was detected by Smad2/3 reporter assays. Receptor activation by BMP9 was detected by Smad1/5/8 reporter assays. Percent inhibition was calculated as described in the “Materials and methods” section. (B) Gene expression analysis of activin receptors in the HEK293T cells that were used in the Smad reporter assays. Target mRNA expressions were normalized to the expression of the internal control gene 36B4. (C) Ligand-binding activities of ActRIIB-Fc and ActRIIA-Fc as determined by plate ELISA.
To assess whether these antagonist activities are related to ActRIIB or other receptors, we evaluated the mRNA levels of ActRIIB and ActRIIA in the HEK293T cells that were used in this Smad reporter assays. Results showed that the cells dominantly expressed ActRIIB rather than ActRIIA mRNA (Fig. 3B). Moreover, plate ELISA revealed that ActRIIB-Fc binds to activin A, MSTN, and GDF11 in a concentration-dependent manner, whereas ActRIIA-Fc binds to activin A and GDF11 but not MSTN (Fig. 3C). PPIs between ActRIIB-Fc/ActRIIA-Fc and BMP9 have been previously reported [3]. These data supported that our peptides bound to ActRIIB expressed on HEK293T cells, inhibited PPI between the extracellular domain of ActRIIB and the ligands, and subsequently suppressed the activation of Smad, i.e. the downstream signal of the ActRIIB pathway.
3.4. Peptide binding competition assays using plate ELISA
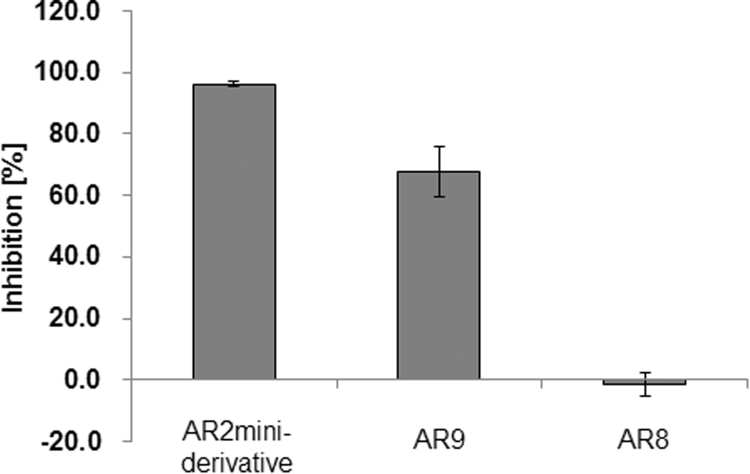
To further characterize the binding of each peptide, competition assays were performed by plate ELISA. The binding activity of biotinylated AR2mini-derived peptide to ActRIIB-Fc was evaluated in the presence of non-tagged peptides. Results showed that the AR2mini-derived peptide (positive control) and AR9 competitively inhibited the binding of the biotinylated AR2mini-derived peptide. On the other hand, AR8 did not compete with the biotinylated peptide. These data suggest that AR2mini and AR9 recognize the same binding site or partially overlapping binding sites on the extracellular domain of ActRIIB, while AR8 recognizes an independent binding site on the ActRIIB extracellular domain (Fig. 4).
Fig. 4.
Binding competition assay. The binding activity of a biotinylated AR2mini-derivative peptide (2 µM) to ActRIIB-Fc was evaluated in the presence of non-labeled peptides (AR2mini-derivative, AR9, or AR8) at a concentration of 100 µM. Percent inhibition was calculated as described in the “Materials and methods” section.
4. Discussion
ActRIIB exerts various physiological functions by interacting with multiple ligands. Structural information on the ActRIIB/activin complex (1S4Y, 1NYS, and 1NYU) [15], [16], the follistatin/MSTN complex (3SEK, 3HH2) [17], [18], GDF11 (5E4G) [19], and BMP9 (1ZKZ) [20] has been reported. These data show that the three-dimensional structures of the ligands are similar. MSTN and GDF11, in particular, also have high sequence homology (89%) [3]. It may be concluded that ligands bind to the same site on ActRIIB. Therefore, peptides displaying ligand-selective ActRIIB antagonist activity would be advantageous compared to neutralizing or antagonist antibodies. In this study, we successfully discovered three ligand-selective peptidic antagonists: AR2mini, AR8, and AR9.
At the present stage, we have not elucidated the mechanism underlying their ligand-selectivity. Our binding competition assays revealed that the extracellular domain of ActRIIB has at least two binding sites for our peptides: one for AR2min and AR9, and the other for AR8. Since our peptides have no sequence similarity with the natural ligands (activin A, MSTN, etc.), binding site prediction is difficult. We have considered X-ray crystal structure analysis to elucidate the mechanism of action of the peptides, but it has not been successful yet. In addition to X-ray crystal structure analysis, other biophysics strategies such as cross-link MS and hydrogen-deuterium exchange (HDX)-MS would be effective in elucidating the binding sites and modes of our peptides.
The ligand selectivity of our peptides is an attractive feature, but both the binding- and the inhibition-activity values were in the micromolar range, i.e., too high for in vivo evaluation. Since the binding affinities between ActRIIB and ligands are in the picomolar range [3], enhancement of our peptides’ affinity by at least 100–1000 times will be required for them to be considered lead compounds for use as ActRIIB antagonists. However, the available crystal structures indicate that there are no druggable pockets for peptides or conventional small molecule compounds on the surface of the ActRIIB extracellular domain, suggesting that the required binding affinity improvement is unlikely to be achieved by using monomeric peptides. There are indications that the use of dimeric peptides may prove to be effective. First, ActRIIB is a dimeric receptor and previous studies have demonstrated that dimeric peptides possess higher activity than monomeric peptides against several dimeric receptors, e.g., FGFR and TrkB [21], [22]. Moreover, preliminary results showed that the pseudo-dimeric GST-fused AR2mini showed increased antagonist activity compared with the AR2mini monomer peptide (data not shown).
In conclusion, we are confident that the current study will serve as the starting point for the generation of ligand-selective ActRIIB antagonists, though further optimization, as described above, will be required for our peptides to be used as tool/lead molecules for ActRIIB targeting.
Author contributions
K. Sakamoto discovered the peptide sequences by phage display screening, performed the Smad reporter assays and plate ELISA, and wrote most of this paper. Y. Kanematsu-Yamaki conceived the chemical synthesis of the peptides. Y. Kamada performed SPR analysis. M. Oka conceived the gene expression analysis. T. Ohnishi started up Smad reporter assays. M. Miwa prepared the recombinant proteins. T. Asami and H. Inooka supervised and supported this work.
Conflict of interest
No potential conflicts of interest are disclosed.
Contributor Information
Kotaro Sakamoto, Email: sakamoto-kotaro@ichimaru.co.jp.
Taiji Asami, Email: taiji.asami@takeda.com.
References
- 1.McPherron A.C., Lawler A.M., Lee S.J. Regulation of skeletal muscle mass in mice by a new TGF-beta superfamily member. Nature. 1997;387:83–90. doi: 10.1038/387083a0. [DOI] [PubMed] [Google Scholar]
- 2.Wakefield L.M., Hill C.S. Beyond TGFβ: roles of other TGFβ superfamily members in cancer. Nat. Rev. Cancer. 2013;13:328–341. doi: 10.1038/nrc3500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Townson S.A., Martinez-Hackert E., Greppi C. Specificity and structure of a high affinity activin receptor-like kinase 1 (ALK1) signaling complex. J. Biol. Chem. 2012;287:27313–27325. doi: 10.1074/jbc.M112.377960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Trendelenburg A.U., Meyer A., Rohner D. Myostatin reduces Akt/TORC1/p70S6K signaling, inhibiting myoblast differentiation and myotube size. Am. J. Physiol. Cell. Physiol. 2009;296:C1258–C1270. doi: 10.1152/ajpcell.00105.2009. [DOI] [PubMed] [Google Scholar]
- 5.Smith R.C., Lin B.K. Myostatin inhibitors as therapies for muscle wasting associated with cancer and other disorders. Curr. Opin. Support. Palliat. Care. 2013;7:352–360. doi: 10.1097/SPC.0000000000000013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wagner K.R., Fleckenstein J.L., Amato A.A. A phase I/IItrial of MYO-029 in adult subjects with muscular dystrophy. Ann. Neurol. 2008;63:561–571. doi: 10.1002/ana.21338. [DOI] [PubMed] [Google Scholar]
- 7.Carlson C.G., Bruemmer K., Sesti J. Soluble activin receptor type IIB increases forward pulling tension in the mdx mouse. Muscle Nerve. 2011;43:694–699. doi: 10.1002/mus.21944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lach-Trifilieff E., Minetti G.C., Sheppard K. An antibody blocking activin type II receptors induces strong skeletal muscle hypertrophy and protects from atrophy. Mol. Cell. Biol. 2014;34:606–618. doi: 10.1128/MCB.01307-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Suragani R.N., Cadena S.M., Cawley S.M. Transforming growth factor-β superfamily ligand trap ACE-536 corrects anemia by promoting late-stage erythropoiesis. Nat. Med. 2014;20:408–414. doi: 10.1038/nm.3512. [DOI] [PubMed] [Google Scholar]
- 10.Nevola L., Giralt E. Modulating protein-protein interactions: the potential of peptides. Chem. Commun. (Camb.) 2015;51:3302–3315. doi: 10.1039/c4cc08565e. [DOI] [PubMed] [Google Scholar]
- 11.Sakamoto K., Sogabe S., Kamada Y. Discovery of GPX4 inhibitory peptides from random peptide T7 phage display and subsequent structural analysis. Biochem. Biophys. Res. Commun. 2017;482:195–201. doi: 10.1016/j.bbrc.2016.11.035. [DOI] [PubMed] [Google Scholar]
- 12.Sakamoto K., Sogabe S., Kamada Y. Discovery of high-affinity BCL6-binding peptide and its structure-activity relationship. Biochem. Biophys. Res. Commun. 2017;482:310–316. doi: 10.1016/j.bbrc.2016.11.060. [DOI] [PubMed] [Google Scholar]
- 13.Sakamoto K., Adachi Y., Komoike Y. Novel DOCK2-selective inhibitory peptide that suppresses B-cell line migration. Biochem. Biophys. Res. Commun. 2017;483:183–190. doi: 10.1016/j.bbrc.2016.12.170. [DOI] [PubMed] [Google Scholar]
- 14.Korchynskyi O., ten Dijke P. Identification and functional characterization of distinct critically important bone morphogenetic protein-specific response elements in the Id1 promoter. J. Biol. Chem. 2002;277:4883–4891. doi: 10.1074/jbc.M111023200. [DOI] [PubMed] [Google Scholar]
- 15.Greenwald J., Vega M.E., Allendorph G.P. A flexible activin explains the membrane-dependent cooperative assembly of TGF-beta family receptors. Mol. Cell. 2004;15:485–489. doi: 10.1016/j.molcel.2004.07.011. [DOI] [PubMed] [Google Scholar]
- 16.Thompson T.B., Woodruff T.K., Jardetzky T.S. Structures of an ActRIIB: activin A complex reveal a novel binding mode for TGF-beta ligand: receptor interactions. EMBO J. 2003;22:1555–1556. doi: 10.1093/emboj/cdg156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cash J.N., Angerman E.B., Kattamuri C. Structure of myostatin·follistatin-like 3: n-terminal domains of follistatin-type molecules exhibit alternate modes of binding. J. Biol. Chem. 2012;287:1043–1053. doi: 10.1074/jbc.M111.270801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cash J.N., Rejon C.A., McPherron A.C. The structure of myostatin: follistatin 288: insights into receptor utilization and heparin binding. EMBO J. 2009;28:2662–2676. doi: 10.1038/emboj.2009.205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Padyana A.K., Vaidialingam B., Hayes D.B. Crystal structure of human GDF11. Acta Crystallogr. F: Struct. Biol. Commun. 2016;72:160–164. doi: 10.1107/S2053230X16001588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Brown M.A., Zhao Q., Baker K.A. Crystal structure of BMP-9 and functional interactions with pro-region and receptors. J. Biol. Chem. 2005;280:25111–25118. doi: 10.1074/jbc.M503328200. [DOI] [PubMed] [Google Scholar]
- 21.Sakamoto K., Kawata Y., Masuda Y. Discovery of an artificial peptide agonist to the fibroblast growth factor receptor 1c/βKlotho complex from random peptide T7 phage display. Biochem. Biophys. Res. Commun. 2016;480:55–60. doi: 10.1016/j.bbrc.2016.10.009. [DOI] [PubMed] [Google Scholar]
- 22.Ohnishi T., Sakamoto K., Asami-Odaka A. Generation of a novel artificial TrkB agonist, BM17d99, using T7 phage-displayed random peptide libraries. Biochem. Biophys. Res. Commun. 2017;483:101–106. doi: 10.1016/j.bbrc.2016.12.186. [DOI] [PubMed] [Google Scholar]