Abstract
Circulating fetal nucleated cells (CFNCs) in maternal blood offer an ideal source of fetal genomic DNA for noninvasive prenatal diagnostics (NIPD). We developed a class of nanoVelcro microchips to effectively enrich a subcategory of CFNCs, i.e., circulating trophoblasts (cTBs) from maternal blood, which can then be isolated with single-cell resolution by a laser capture microdissection (LCM) technique for downstream genetic testing. We first established a nanoimprinting fabrication process to prepare the LCM-compatible nanoVelcro substrates. Using an optimized cTB-capture condition and an immunocytochemistry protocol, we were able to identify and isolate single cTBs (Hoechst+/CK7+/HLA-G+/CD45−, 20 μm > sizes > 12 μm) on the imprinted nanoVelcro microchips. Three cTBs were polled to ensure reproducible whole genome amplification on the cTB-analysis. Using maternal blood samples collected from expectant mothers carrying a single fetus, the cTB-derived aCGH data were able to detect fetal genders and chromosomal aberrations, which had been confirmed by standard clinical practice. Our results support the use of nanoVelcro microchips for cTB-based noninvasive prenatal genetic testing, which holds potential for further development toward future NIPD solution.
Keywords: noninvasive prenatal testing (NIPT), nanoVelcro assays, circulating trophoblasts, single-cell analysis, array comparative genomic hybridization (aCGH)
Graphical abstract
The current gold standard for diagnosing fetal genetic abnormalities involves invasive procedures1,2 such as amniocentesis (AC, >16 weeks of gestational age, GA) and chorionic villus sampling (CVS, 10–12 weeks of GA), by which fetal cells are harvested for karyotyping and genetic testing. These procedures provide accurate information for clinical decision making. However, concerns have been raised regarding their invasiveness and increased risk of miscarriage (0.6–2%).3 Therefore, significant research endeavors have been devoted to developing noninvasive prenatal diagnostics4 (NIPD) to address this unmet need. Among the noninvasive approaches for prenatal genetic testing, cell-free fetal DNA5–7 (cffDNA)-based tests have demonstrated their sensitivity in detecting fetal aneuploidy (e.g., trisomies 21, 18, and 13) in large-scale clinical trials.8 However, cffDNA is highly fragmented and compounded by a huge background of maternal DNA, limiting its diagnostic utility for other genetic abnormalities, such as microdeletions and duplications.
In contrast to cffDNA, circulating fetal nucleated cells (CFNCs)4,9 in maternal blood are well-preserved and possess pure genomic DNA, making it an ideal candidate for NIPD. Feto-maternal cellular trafficking10 is the bidirectional passage of cells that results in the presence of CFNCs in maternal circulation. However, the detection, isolation, and characterization of CFNCs in maternal blood samples have been technically challenging due to their fragile nature and extremely low abundance11 (<6 cells mL−1 of maternal blood in normal pregnancy, compared to maternal hematologic cells of 109 cells mL−1). Throughout the past three decades,4 several different working mechanisms of CFNC enrichment, such as gradient centrifuge,12,13 magnetic-activated cell sorting (MACS),14,15 fluorescence-activated cell sorting (FACS),16,17 microchip technologies,18,19 filtration,20 and high-throughput microscopy,11,21 have been developed. In order to access the full potential of CFNCs for NIPD, the following challenges have to be addressed: (i) creating a robust rare-cell assay capable of isolating pure and intact CFNCs with well-preserved genomic DNA and (ii) coupling with downstream genetic testing beyond the detection of aneuploidy.
Among CFNCs4 that have been identified in maternal circulation, a circulating trophoblast21 (cTB) is one of the ideal targets considering its (i) short lifespan (a few days), which excludes the possibility of isolating cTBs from prior pregnancies, (ii) representation of fetal karyotype and genotype (except for the rare circumstances where discrepancy can result from confined placenta mosaicism), and (iii) expression of a collection of biomarkers22 that can be used for both enrichment and identification. Recent studies led by Baylor researchers demonstrated that cTBs could be recovered by either a single-cell picking technique21 or an optimized MACS approach15 from maternal blood (as early as 10 weeks of GA) and subjected to molecular characterizations by array comparative genomic hybridization (aCGH) and next-generation sequencing.
In contrast to the existing rare-cell sorting approaches, we pioneered the concept of the “nanoVelcro” rare-cell assay,23,24 in which a capture antibody-coated nanosubstrate substantially enhances the performance of rare-cell enrichment from blood. By integrating the nanoVelcro substrate with an overlaid microfluidic chaotic mixer, a nanoVelcro microchip was then created. In our pilot studies for circulating tumor cells (CTCs), nanoVelcro microchips demonstrated high sensitivity and specificity for enumerating CTCs in different solid tumors, including prostate cancer,25–27 lung cancer,28–30 pancreatic cancer,31 kidney cancer,32 and melanoma.33 Beyond enumeration, we were able to isolate pure CTCs individually by coupling a commercial laser capture microdissection (LCM) technique with nanoVelcro microchips coated by electrospun poly(lactic-co-glycolic acid) (PLGA) nanofibers. The CTCs isolated via nanoVelcro-LCM technology contain well-preserved genomic DNA compatible for downstream molecular characterizations, specifically, targeted mutational analysis,31,33 whole exome sequencing,34 and whole genome sequencing.35 In the interest of broadening the general applicability of nanoVelcro microchips, we continue to improve upon its performance and to explore its utility for the detection and characterization of cTBs, demonstrating the feasibility of a CFNC-based noninvasive prenatal testing (NIPT).
Herein we introduce a class of nanoVelcro microchips to effectively enrich cTBs from maternal blood, which can then be isolated from the nanoVelcro substrates by the LCM technique for downstream genetic testing. We first developed a nanoimprinting fabrication process36 (Figure 1) to prepare the LCM-compatible nanoVelcro substrates to avoid any of the technical challenges (i.e., long production time and limited reproducibility33,34) encountered previously from using the electrospinning nanofabrication process. A chip holder was adopted to assemble a microfluidic chaotic mixer37 onto the imprinted nanoVelcro substrate, yielding nanoVelcro microchips (Figure 2a). In order to enrich cTBs, biotinylated anti-EpCAM (a trophoblast surface marker) was grafted onto the imprinted nanoVelcro substrates via biotin–streptavidin-mediated conjugation. To optimize the cTB-capture performance (Figure 2b–d) of the imprinted nanoVelcro microchips, the devices were tested with artificial blood samples, prepared by spiking DIO-stained trophoblast cell lines (i.e., JEG-3/JAR/BeWo cells) from choriocarcinoma into freshly isolated human white blood cells (WBCs). Simultaneously, an immunocytochemistry (ICC) protocol was established to identify cTBs (Hoechst+/CK7+/HLA-G+/CD45−, 20 μm > sizes > 12 μm) from nonspecifically captured WBCs (Hoechst+/CK7−/HLA-G−/CD45+, 15 μm > sizes > 8 μm) and cellular debris on the imprinted nanoVelcro substrates (Figure 3b). To demonstrate the clinical utility of the imprinted nanoVelcro microchips for CFNC-based NIPT, we conducted cTB capture and characterization using maternal blood samples collected from two groups: (i) 6 expectant mothers (GA = 8–14 weeks) with clinically confirmed normal male fetuses (Table 1) and (ii) 9 expectant mothers (GA = 15–23 weeks, including two unknown) with clinically confirmed genetic aberrations in their fetuses (Table 2). A general workflow (Figure 3a) was developed for our cTB-based NIPT. On average, we detected 3 to 15 cTBs per 2 mL of blood in these maternal blood samples. For the cTBs subjected to aCGH analysis, we demonstrated an accurate and high-quality detection of fetal genders and fetal chromosomal aberrations in a total of 15 cases. Meanwhile, the feto-parental relationship of the isolated cTBs and their matching maternal/paternal cells and/or umbilical cord tissues were validated by short tandem repeats (STR) fingerprints. Overall, we introduced an imprinted nanoVelcro microchip and a protocol for cTB-based NIPT that can be further developed toward a NIPD.
Figure 1.
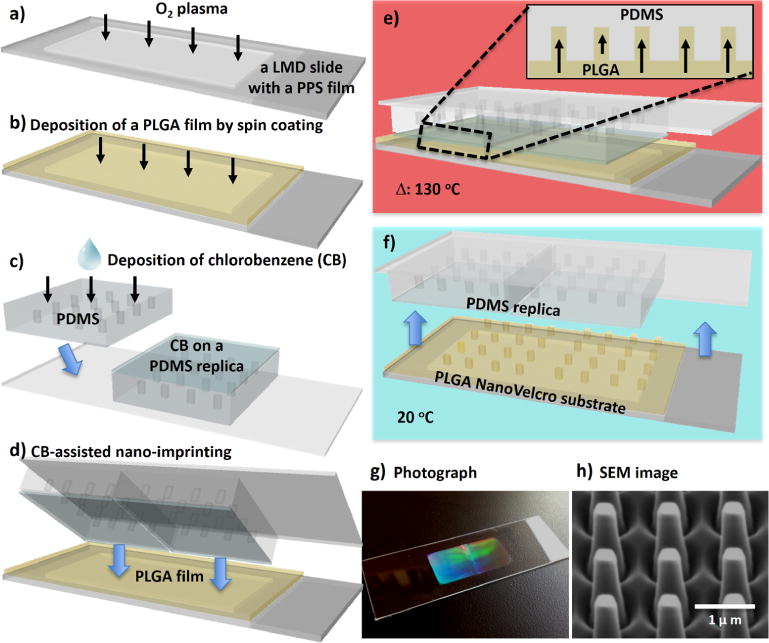
Workflow developed for the fabrication of imprinted PLGA nanoVelcro substrate. (a) A laser microdissection (LMD) slide was first cleaned by O2 plasma; (b) a 1.0-μm-thick PLGA film was spin-coated onto the plasma-treated LMD slide; (c) chlorobenzene (CB) was sprayed onto PDMS replicates; (d) immediately after, the CB-coated replicates were pressed onto the spin-coated PLGA film; (e) the assembled layers were then heated to 130 °C. PLGA nanopillars were pulled into the nanofeatures on the bottom of the PDMS replicates, as a result of the absorbance of CB into PDMS matrix; (f) after the assembled layers were cooled to 20 °C, the PDMS replicates were peeled off from the substrate to give (g) an imprinted PLGA nanoVelcro substrate on an LMD slide; (h) SEM image shows the topography of the imprinted PLGA-nanopillar features.
Figure 2.
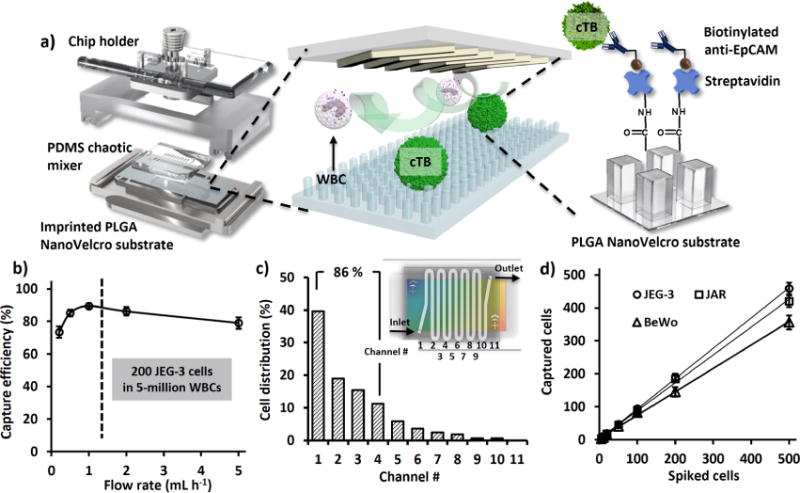
(a) A nanoVelcro microchip is assembled using a chip holder to hold together a chaotic mixer on top of an imprinted PLGA nanoVelcro substrate. The PLGA nanoVelcro substrate will be functionalized with capture agent (i.e., anti-EpCAM) for affinity capture of cTBs. (b) In the presence of anti-EpCAM, artificial cTB samples (200 JEG-3 cells in 5 million WBCs) were used to test the device performances at flow rates of 0.2, 0.5, 1.0, 2.0, and 5.0 mL h−1. (c) Spatial distribution of nanosubstrate-immobilized cTBs along the serpentine microchannel. 85% of cTBs were captured in the first 4 microchannels (8 cm). (d) Under the optimal flow rate of 1.0 mL h−1, the cTB-capture efficiencies were determine at different cTB numbers ranging from 5 to 500 cells mL−1. Here three trophoblast cell lines (JEG-3, JAR, and BeWo) were used.
Figure 3.
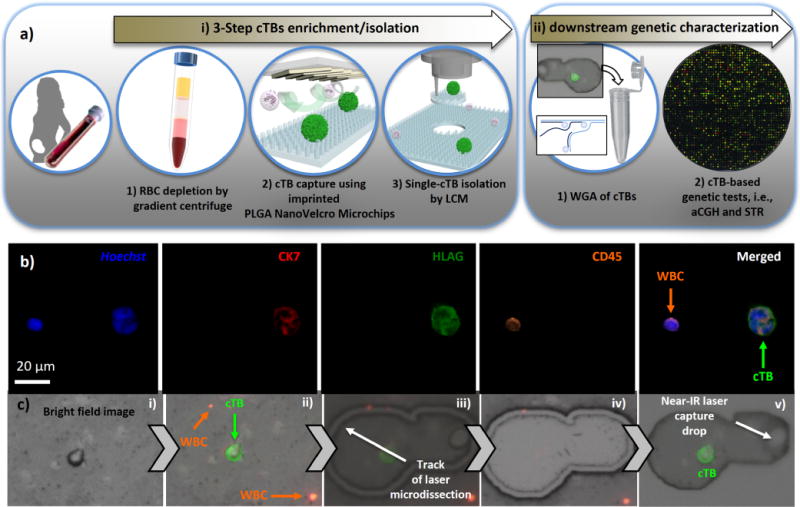
(a) A general workflow of cTB-based NIPT approach can be divided into two major sections, including (i) three-step cTB enrichment/isolation and (ii) downstream genetic characterization by aCGH and STR. Three-step cTB enrichment and isolation start from (i) RBC depletion by gradient centrifuge, (ii) affinity capture of cTBs onto nanoVelcro microchips in the presence of anti-EpCAM capture agent, and end with (iii) cTB isolation by LCM. For downstream genetic characterization, three individually isolated cTBs were pooled together in a 0.5 mL PCR tube for WGA. The resulting amplified DNA was then subjected to aCGH and STR assay. (b) Representative micrographs of cTBs obtained from maternal blood samples. (c) Micrograph images recording stepwise operation of LCM-based single cTB isolation on an imprinted PLGA nanoVelcro substrate, including (i/ii) microscopy imaging/identification of cTB and (iii–v) LCM of a single cTB.
Table 1.
Detailed Clinical Characteristics of the Six Expectant Mothers Who Participated in Our Studies
| molecular analyses
|
||||||
|---|---|---|---|---|---|---|
| patient | age (y/o) | gestational age (week) | cTBs # per 2 mL blood | PGD/ultrasound | array CGH | STR |
| #1 | 34 | 10 | 14 | XY | XY | N/A |
| #2 | 23 | 14 | 11 | XY | XY | matched |
| #3 | 31 | 13 | 22 | XY | XY | N/A |
| #4 | 20 | 9 | 26 | XY | XY | N/A |
| #5 | 33 | 12 | 20 | XY | XY | matched |
| #6 | 38 | 8 | 31 | XY | XY | matched |
Table 2.
Clinical Characteristics of the Seven Expectant Mothers and Results of the Molecular Analysis for cTBs and Cord Tissue
| molecular analyses
|
|||||||
|---|---|---|---|---|---|---|---|
| patient | age (y/o) | gestational age (week) | invasive diagnostic method/diagnosis/gender | cTBs # per 2 mL blood | PGD/ultrasound | cord tissue microarray | STR |
| #7 | 35 | 21 | AC/trisomy 21/F | 12 | trisomy 21/F | trisomy 21/F | N/A |
| #8 | 42 | 23 | AC/trisomy 21/F | 21 | trisomy 21/F | trisomy 21/F | N/A |
| #9 | 45 | 15 | CVS/trisomy 21/M | 25 | trisomy 21/M | trisomy 21/M | N/A |
| #10 | 44 | 21 | AC/trisomy 21/M | 16 | trisomy 21/M | trisomy 21/M | N/A |
| #11 | 35 | 20 | AC/trisomy 18/M | 12 | trisomy 18/M | trisomy 18/M | N/A |
| #12 | 38 | 17 | AC/trisomy 13/M | 14 | trisomy 13/M | trisomy 13/M | N/A |
| #13 | NA | NA | AC/XXXXY/M | 10 | XXXXY/M | XXXXY/M | N/A |
| #14 | NA | NA | AC/trisomy 18, XYY/M | 16 | trisomy 18, XYY/M | trisomy 18, XYY/M | N/A |
| #15 | 37 | 19 | AC/Del(9p) Dup(14q)/M | 18 | Del(9p) Dup(14q)/M | Del(9p) Dup(14q)/M | N/A |
RESULTS AND DISCUSSION
The procedure developed for the fabrication of imprinted PLGA nanoVelcro microchips is summarized in Figure 1. Prior to the fabrication of imprinted PLGA nanoVelcro microchips, a set of parental PMMA (poly(methyl methacrylate)) nanopillar features (200 nm in diameter, 1.5 μm in length, 0.8 μm in spacing) was introduced on a silicon wafer via e-beam lithography (Supporting Information, Section 1). Subsequently, a polydimethylsiloxane (PDMS) replicate was prepared by curing the prepolymers on the PMMA-nanopillar features. Figure 1a–f depicts a workflow developed for the fabrication of imprinted PLGA nanoVelcro substrates. First, a 5% PLGA polymer (MW = 43–100 K) solution in acetonitrile was spin-coated onto an O2 plasma-treated laser microdissection (LMD) slide (with a 1.2-μm-thick suspended poly(phenylene) sulfide (PPS) membrane). Second, through the use of an organic solvent (chlorobenzene)-assisted nanoimprinting process,38 the embedded nanofeatures were effectively transferred from PDMS replicates onto the PLGA film. Here, a metal sandwich holder was used to apply pressure (150 ± 20 g cm−2) onto the assembled layers, while the imprinting process was carried out at 130 °C, a temperature above the glass transition temperature of PLGA. Once the assembled layer cooled to 20 °C, the PDMS replicates were peeled off to give a PLGA nanoVelcro substrate on an LMD slide. The resulting imprinted nanoVelcro substrate (Figure 1g) exhibits rainbow diffraction patterns that signify the presence of regular nanopillar features on the surface. This observation is supported by the surface topographies of PLGA-nanopillar features examined by a scanning electron microscope (SEM, Figure 1h). In addition to the advantage of an optical transparency that makes the integration with the LCM technique possible, further benefits introduced by the imprinted PLGA nanosubstrates include (i) a dramatically improved production efficiency and reproducibility as compared to the earlier electrospinning processes33,34 and (ii) effective reduction of nonspecific absorption of cell-free DNA in blood once the substrate surface was treated with pH-8.4 phosphate-buffered saline (PBS). A custom-designed chip holder was produced to assemble an overlaid PDMS chaotic mixer25 (with a 22 cm microchannel) onto the imprinted nanoVelcro substrate (Figure 2a) to give the imprinted PLGA nanoVelcro microchip. Prior to conducting affinity capture of cTBs, NHS chemistry33,34 was employed to covalently attach streptavidin onto PLGA nanosubstrates, followed by conjugation of biotinylated anti-EpCAM. Similarly, we utilized syringe pumps and syringes to inject cell suspensions and agents through the imprinted nanoVelcro microchips at an optimized flow rate.
In search of an optimal cTB-capture condition for nanoVelcro microchips, we prepared an artificial cTB sample (containing prestained 200 JEG-3 cells and 5 million WBCs in 0.5 mL of RPMI medium) as a model system. We first examined the capture performance at different flow rates (i.e., 0.2 to 5.0 mL h−1) in the presence of trophoblast capture agent, anti-EpCAM. The results (Figure 2b) illustrate optimal cell capture performance at 1.0 mL h−1. Meanwhile, we analyzed the spatial distribution (Figure 2c) of the immobilized cTBs along the 22 cm serpentine microchannel (inset in Figure 2c). About 85% of the cTBs were captured in the first four microchannels (8 cm), suggesting that the 22 cm channel length was sufficient to achieve an appropriate capture performance. Using the optimal capture conditions (anti-EpCAM capture agent, flow rate = 1.0 mL h−1), we validated that the capture efficiency (Figure 2d) remained consistent over a range of spiked cTB numbers (i.e., 5 to 500 cTB mL−1) across three different types of TB cell lines (i.e., JEG-3, JAR, and BeWo).
We then adopted the optimal cTB capture conditions to examine the feasibility of the cTB-based NIPT approach. The general workflow (Figure 3a) can be divided into two major sections: (i) three-step cTB enrichment/isolation and (ii) downstream genetic characterization by aCGH and STR. The three-step cTB enrichment/isolation (Figure 3a) commenced with a gradient centrifuge method that removed red blood cells (RBCs) from the maternal blood. The resulting peripheral blood mononuclear cell (PBMC) suspension (containing cTBs) was then introduced onto the imprinted nanoVelcro microchips (with anti-EpCAM capture agent) for affinity capture. After capturing cTBs onto nanoVelcro substrates, a four-color ICC protocol was implemented for parallel staining of Hoechst+, CK7, HLA-G, and CD45. In conjunction with the use of fluorescence microscopy, quantitative image cytometry39 data covering Hoechst+ nuclear staining, CK7/HLA-G/CD45 expressions, and cell sizes may be used to identify cTBs (Hoechst+/CK7+/HLA-G+/CD45−, 20 μm > sizes > 12 μm) from nonspecifically captured WBCs (Hoechst+/CK7−/HLA-G−/CD45+, 15 μm > sizes > 8 μm) and cellular debris on the imprinted nanoVelcro substrates (Figure 3b). Subsequently, the LCM technique (ArcturusXT, Thermo-Fisher) was employed to isolate the identified cTBs onto LCM caps. To ensure sufficient cTB DNA for downstream molecular analysis, three cTBs were captured onto an LCM cap. The detailed workflow and timeline for single-cTB isolation are summarized in Figure S2 of the Supporting Information.
We recruited two cohorts of expectant mothers for feasibility studies using the cTB-based NIPT approach. The first cohort (Table 1) included 6 pregnancies (GA = 8–14 weeks) from in vitro fertilization clinics. Through their preimplantation genetic diagnostic (PGD)40 tests and ultrasound examinations, each expectant mother was confirmed to carry a single male fetus with no detectable genetic abnormalities. The second cohort (Table 2) included 9 pregnancies (GA = 15–23 weeks) with fetal genetic aberrations, confirmed by either CVS or AC as part of the standard care. For each cTB-based NIPT study, 2 mL of maternal blood was used. Duplicated tests were conducted for all of the patients. Following the three-step workflow (Figure 3a), cTBs in these maternal blood samples were captured onto the imprinted nanoVelcro substrates and identified according to ICC/size criteria (Hoechst+/CK7+/HLA-G+/CD45−, 20 μm > sizes > 12 μm). Figure 3b illustrates fluorescence micrographs recorded for a cTB immobilized on the imprinted nanoVelcro substrate. For the healthy cohort (Table 1), we detected 3 to 6 cTBs on the imprinted nanoVelcro substrates from 2 mL of these maternal blood samples. Interestingly, a significantly higher number of cTBs (5 to 15 cTBs per 2 mL of maternal blood) were observed for the diseased cohort (Table 2). As the third step of cTB isolation, LCM was employed to precisely harvest (Figure 3c) individual cTBs. To ensure the robustness of downstream genetic characterization, 3 cTBs were pooled on each LCM cap and transferred to a 0.5 mL PCR tube, in which whole-genome amplification (WGA) using an REPLI-g single-cell WGA kit (Qiagen) was conducted. After DNA quantification by a Qubit 3.0 fluorometer (Thermo-Fisher, >150 ng of amplified DNA), the cTB-derived WGA DNA was subjected to aCGH (Agilent, SurePrint G3 Human Catalog 8×60K) and STR assay (GenePrint 10 system).
For the healthy cohort (Table 1), the male fetal gender, determined by PGD and ultrasound, was used to validate cTB-derived aCGH data. To further confirm the feto-parental relationship, maternal and/or paternal STR fingerprints were generated using maternal and/or paternal blood samples and compared with cTB-derived fetal STR fingerprints. Figure 4a shows a representative aCGH datum obtained from a male fetus carried by an expectant mother (#1, Table 1). As shown in Figure 4b, the STR analysis results indicated that at least half of the cTB-derived STR signatures could be tracked back to either maternal or paternal WBC-derived STR fingerprints, confirming fetal–parental relationships. After demonstrating the cTB-based NIPT for a normal pregnancy, we further explored the use of our NIPT approach to detect fetal chromosomal aberrations using blood samples collected from the diseased cohort (Table 2). In this cohort, expectant mothers #7–10 carried fetuses with trisomy 21, and expectant mothers #11 and #12 carried fetuses with trisomy 18 and trisomy 13, respectively. Expectant mother #13 carried a fetus with tetrasomy X (i.e., four copies of X chromosomes). Expectant mother #14 carried a fetus with dual chromosomal aberrations combining trisomy 18 and disomy Y. Finally, rare dual chromosomal aberrations (9p deletion and 14q duplication) were detected in the fetus carried by expectant mother #15. We noted that all of the aCGH data (Figures 5a and 6) from the 9 study subjects correctly detected fetal genders and clinically confirmed chromosomal aberrations. As a result of termination of pregnancy, umbilical tissues were collected to verify the origin of the matching cTBs. Figure 5b shows STR signatures obtained from the cTBs, maternal blood, and the matching umbilical tissue, revealing (i) accurate fetomaternal relationship between the cTBs and maternal cells and (ii) superb consistency between the cTBs and matching umbilical tissue. In addition to the detection of trisomies 13, 18, and 21, we also performed fluorescence in situ hybridization (FISH) in the matching umbilical cord tissues for the chromosomal aberrations involving chromosomes X and Y for expectant mothers #13 and #14. The inset in Figure 6b shows a fluorescence micrograph signifying the four copies of chromosome X (red dots) and single chromosome Y (green dots) for expectant mother #13. The inset in Figure 6c shows the duplicated Y chromosome (green dots) and triplicated chromosome 18 (blue dots) for expectant mother #14. Finally, Figure 6d shows the cTB-derived aCGH data observed for expectant mother #15, where 17.2 Mb deletion in the 9p area [del(9)(p24.3p22.2)] and 11.7 Mb duplication in the 14q area [dup(14)(q32.13q32.33)] were detected, under the resolution of 1.8 Mb.
Figure 4.
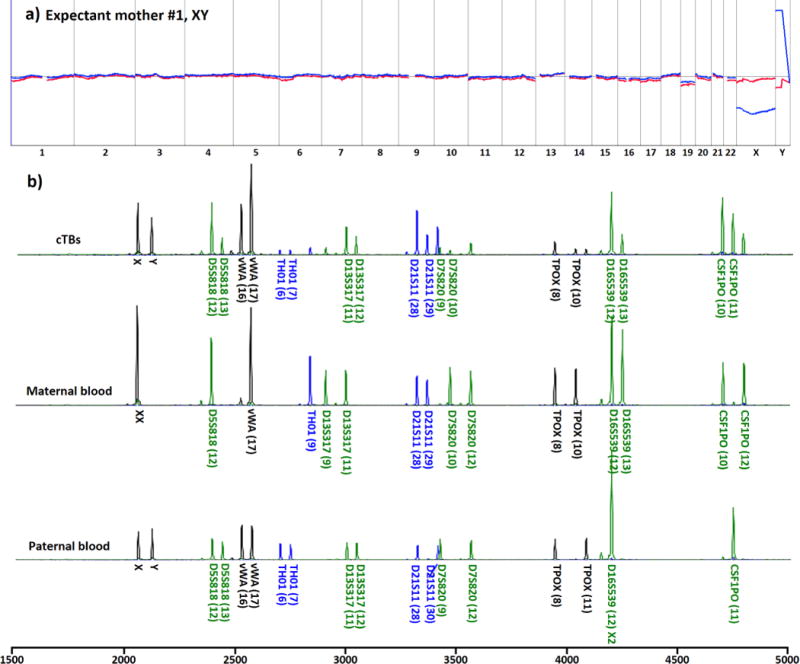
aCGH and STR data from cTBs. (a) cTB-derived aCGH data showed no genomic abnormalities in the fetus. The data were derived from three cTBs from a male fetus carried by an expectant mother with a healthy fetus (#1 in Table 1). (b) STR genomic fingerprinting confirmed the fetal–parental relationship between cTBs and their matching maternal and paternal WBCs.
Figure 5.
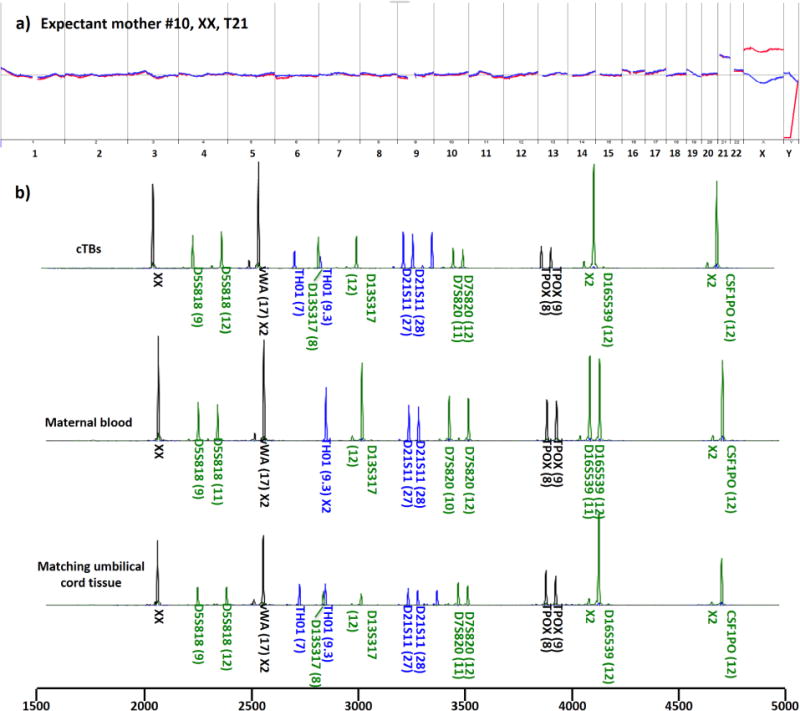
aCGH and STR data from cTBs. (a) cTB-derived aCGH data showed trisomy 21 in the fetus. The data were derived from three cTBs from a male fetus carried by an expectant mother (#10 in Table 2) with a diseased fetus with confirmed fetal trisomy 21. (b) STR analysis confirmed fetal–parental relationship between cTBs and maternal cells and showed consistency between cTBs and the matching umbilical tissue.
Figure 6.
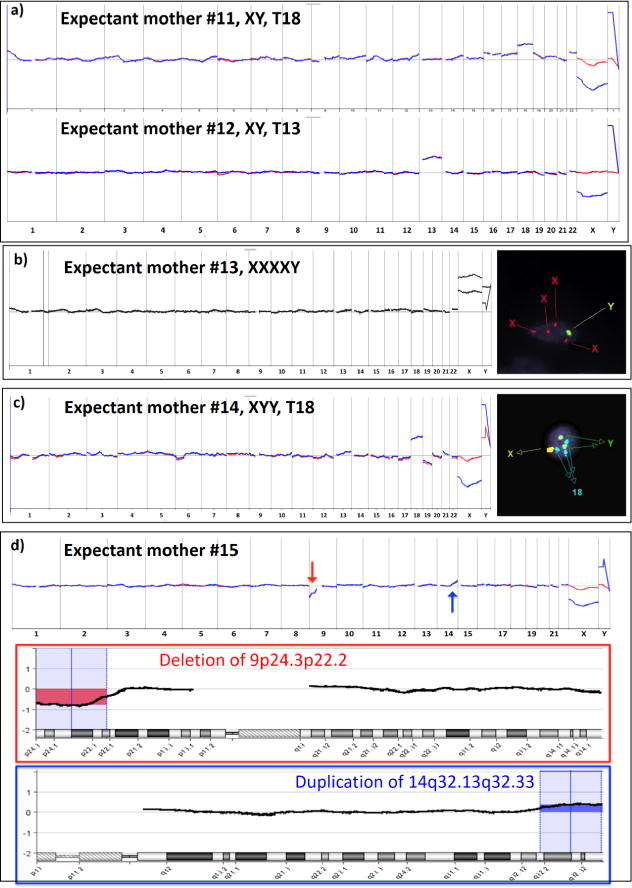
Genetic characterization of cTBs in maternal blood samples collected from expectant mothers (#11–15, Table 2) with confirmed chromosomal aberrations in the fetuses. (a) cTB-derived aCGH data obtained from expectant mothers #11 and 12 showed trisomy 18 and trisomy 13, respectively. (b) The finding of fetal tetrasomy X in expectant mothers #13 by aCGH was confirmed by FISH on matching umbilical cord tissue. (c) The finding of fetal dual chromosomal aberrations (trisomy 18 and disomy Y) in aCGH was confirmed by FISH on matching umbilical cord tissue. (d) Detecting fetal chromosomal aberrations from the expectant mother #15: A red arrow indicates copy number loss from 9p24.3 to 9p22.2 (17.2 Mb), and a blue arrow indicates copy number gain from 14q32.13 to 14q32.33 (11.7 Mb), under the resolution of 1.8 Mb.
CONCLUSIONS
We successfully developed a nanoimprinting fabrication process to prepare the LCM-compatible nanoVelcro substrates to overcome the technical challenges encountered previously from using the electrospinning nanofabrication process. We then explored the use of these imprinted nanoVelcro microchips to develop an NIPT approach through a streamlined workflow to isolate and characterize cTBs in maternal blood. Initial optimization studies using artificial maternal blood samples containing cTB cells concluded that in the presence of anti-EpCAM nanoVelcro microchips exhibited consistent cTB-capture performance (>70%). Furthermore, an ICC protocol was established to identify cTBs from nonspecifically captured WBCs and cellular debris on the imprinted nanoVelcro substrates. In conjunction with the use of the LCM technique, individual cTBs can be specifically isolated with single-cell precision. Subsequently, we determined that pooling 3 cTBs on an LCM cap promises reproducible WGA on the cTB DNA, thus paving the way for obtaining cTB-derived aCGH data and STR fingerprints of sufficient quality. Using maternal blood samples collected from expectant mothers who carried a single fetus, our NIPT approach generated cTB-derived aCGH and STR fingerprints under the optimal conditions. The cTB-derived aCGH data correctly showed fetal genders and chromosomal aberrations, which had been confirmed by standard clinical practice (e.g., AC, CVS, and/or ultrasound). In parallel, the STR fingerprints from cTB and maternal/paternal blood confirmed feto-parental relationships. These pilot studies support the feasibility4 of conducting cTB-based NIPT. With appropriate validation in large-scale clinical studies, this cTB-based NIPT holds the potential to evolve into a noninvasive prenatal diagnostics solution.
MATERIALS AND METHODS
Reagents
Poly(lactic-co-glycolic acid) used in the present study was purchased from DURECT Corporation (Cupertino, CA, USA; lactide:glycolide = 50:50, inherent viscosity range 0.55–0.75 dL g−1). Hexafluoroisopropanol (≥99%) employed for dissolving PLGA was supplied by Sigma-Aldrich and used as received. To covalently conjugate streptavidin onto the imprinted PLGA nanosubstrates through NHS chemistry, activation agents, i.e., 1-ethyl-3-[3-(dimethylamino)propyl] carbodiimide (EDC) and N-hydroxylsulfo-succinimide (sulfo-NHS), were used and obtained from Sigma-Aldrich. Laser capture microdissection slides with predeposited 1.2-μm-thick PPS membrane supplied by Leica were employed as the substrates for fabrication of the PLGA nanoVelcro substrates by using a nanoimprinting approach. All other reagents and solvents were purchased from Sigma-Aldrich and used as received unless otherwise noted.
Trophoblast Cell Lines
Three trophoblast cell lines (i.e., JEG-3, JAR, and BeWo) from choriocarcinoma were purchased from American Type Culture Collection. Under the recommended conditions, the three cell lines were cultured for the preparation of artificial samples in the following validation and optimization studies. Before spiking into the healthy donor’s blood as artificial samples, the cells were collected, counted, and prestained with DIO for further use.
Blood Sample Collection
Human blood samples, including healthy donors’ blood and maternal blood samples from expectant mothers, were collected for the presented studies. Blood samples were drawn prior to any procedure following a patient-informed consent process covered by UCLA Institutional Review Board approval (IRB #13-001264). The maternal blood sample collection in PacGenomics laboratory (Agoura Hills, CA, USA) was covered under Western IRB (IRB #1140744). Approximately 10–20 mL was collected from volunteers into anticoagulant ACD Vacutainer tubes (Becton-Dickinson). The artificial blood samples for optimization and validation studies were prepared by spiking 5–500 trophoblast cells into the WBC suspensions derived from healthy donors’ blood.
cTB Enrichment and Staining
Maternal peripheral venous blood samples were obtained and collected in heparin Vacutainer tubes (10 mL) from the expectant mothers. Before gradient centrifugation, the blood samples were first diluted 1:1 with PBS. The gradient separation solution was prepared by adding 2 mL of separation solution (Histopaque-1119) in 15 mL centrifuge tubes. Then, 6 mL of diluted blood sample was carefully placed on top of the above-described gradient separation solutions. After being tightly capped, the tube was then centrifuged at 800g for 25 min at room temperature. The PBMC layer containing cTBs should be located above the Histopaque-1119 separation solution layer. After aspirating and discarding the top layer (plasma), the PBMC layer containing cTB (approximate 2 mL) could be easily collected and transferred to another 15 mL centrifuge tube. Then, 9 mL of PBS was added and mixed with the collected cell suspension. The resulting cell suspension was then centrifuged at 300g for 10 min at room temperature. After careful removal of the supernatant, the cell pellet containing cTBs was finally isolated and collected from whole blood for the following cTB capture using the imprinted PLGA nanoVelcro microchips.
The centrifugally enriched maternal blood samples with a cell pellet containing cTBs were first resuspended into 200 μL with RPMI with 5% fetal bovine serum and then injected into the imprinted PLGA nanoVelcro microchip at a flow rate of 1.0 mL h−1. After the suspension sample solution containing cTBs was fully flowed through the imprinted PLGA nanoVelcro microchip, the on-chip enriched cTBs were first fixed with ethanol (95%) at a flow rate of 1.0 mL h−1 for 10 min. The imprinted PLGA nanoVelcro microchip was briefly washed with washing buffer for 10 min at a flow rate of 1.0 mL h−1. Then, the captured cTBs were identified by a four-color ICC protocol for parallel staining of Hoechst, anti-CK7 (PE), anti-HLA-G (FITC), and anti-CD45 (TRITC) to distinguish cTBs from nonspecifically captured WBCs on the nanoVelcro substrates. (Detailed information is provided in the Supporting Information, Section 3.)
Fabrication of Imprinted PLGA nanoVelcro Substrates
The LMD slide (LCM slides: PPS-membrane slides 1.2 μm from Leica) was first oxygen plasma treated for 1 min. A PLGA solution (5 wt % in acetonitrile) was then spin-coated (1500 rpm for 30 s) onto the oxygen plasma treated LCM slide. After being baked on a hot plate at a temperature of 120 °C for 10–20 s, the LMD slide coated with PLGA film was ready for nanoimprinting.
The PDMS replicate was first spin-coated with chlorobenzene (1500 rpm for 30 s) and then stuck onto the PLGA-coated LMD slide. Afterward, a metal sandwich holder was used to apply pressure (150 ± 20 g cm−2) onto the assembled layers, while the imprinting process was carried out at 130 °C, a temperature above the glass transition temperature of PLGA. Once the assembled layer cooled to 20 °C, the PDMS replicates were peeled off to give a PLGA nanoVelcro substrate on an LMD slide. Then, the imprinted PLGA nanoVelco substrate was ready for the subsequent chemical conjugation33,34 (NHS chemistry) to covalently attach streptavidin on the surface.
Fabrication of PDMS Chaotic Mixers
The chaotic mixer component was fabricated using a soft lithography method. The patterned silicon master mold is produced by a standard two-step photolithographic process. In the first step, a thin layer (110 μm) of negative photoresist (SU8-2100, MicroChem Corp., Newton, MA, USA) was spin-coated onto a 3 in. silicon wafer (Silicon Quest, San Jose, CA, USA). After UV exposure and development, a serpentine fluidic channel with rectangular shape in the cross section was obtained (length 22 cm and width 1.0 mm). In the second step, another layer (50 μm) of negative photoresist (SU8-2025, MicroChem Corp.) was spin-coated on the same wafer. Prior to the UV exposure the second mask was aligned (Karl Suss America Inc., Waterbury, VT, USA) to the previous pattern with the new pattern to be fabricated. The new pattern created the ceiling “ridges” that induce chaotic mixing within the fluid channel. A well-mixed PDMS prepolymer (GE Silicones, Waterford, NY, USA; RTV 615 A and B in 10:1 ratio) was poured onto the surface of the mold to replicate the pattern, producing a PDMS chaotic mixer (approximately 6 mm thick) after curing in an oven at 80 °C for 48 h. Before assembly with the imprinted PLGA nanoVelcro substrate, two through-holes were punched at both ends of the fluidic channel on the PDMS chaotic mixer to connect the tubing.
Assembly and Surface Modification of Imprinted PLGA NanoVelcro Microchips
The imprinted PLGA nanoVelcro substrate was allowed to instantly assemble with an overlaid PDMS chaotic mixer through a slide-in and click-on approach by using a custom-designed metal sandwich holder as shown in Figure 2a. To obtain the stable streptavidin-coated imprinted PLGA nanoVelcro substrate used in our studies, the EDC/NHS chemistry was applied as reported2,3 to create a covalent bond between the carboxylic acid end groups and free amines on the streptavidin molecules. The imprinted PLGA nanoVelcro substrate was first reacted in 0.5 mL of 1× PBS with EDC (8.0 mg mL−1) and sulfo-NHS (2.0 mg mL−1) to convert the terminal carboxyl group to an amine-reactive sulfo-NHS ester. A concentration of 250 μg mL−1 of streptavidin was then reacted with the NHS-functionalized carboxyl group in 1.0 mL of 1× PBS at room temperature. After careful washing with PBS three times to remove excess reactants, the streptavidin-modified imprinted PLGA nanoVelcro substrate was treated with 50 μg mL−1 biotinylated anti-EpCAM for 60 min at room temperature. After the brief wash with PBS three times, the fully biological functionalized imprinted PLGA nanoVelcro microchip was ready for the capture of cTBs in the blood samples.
Supplementary Material
Acknowledgments
This work was supported by National Institutes of Health (R33-CA174562).
Footnotes
Supporting Information
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acsnano.7b03073.
Additional information including details of PLGA chip preparation, cTB capture optimization, cTB enrichment and staining, LCM isolation method, and genetic characterization (PDF)
ORCID
Yu-Sheng Hsiao: 0000-0003-2072-1844
Hsian-Rong Tseng: 0000-0001-9028-8527
Notes
The authors declare the following competing financial interest(s): Following the guideline of UCLA Conflict of Interest Review Committee (CIRC), the authors would like to disclose: (i) The intellectual property that arose from this study has been licensed to FetoLumina Technologies Corp.; and (ii) Tom Lee and Hsian-Rong Tseng have their financial interests in FetoLumina Technologies Corp. given their roles as the founders of the company.
References
- 1.Dimaio MS, Fox JE, Mahoney MJ. Prenatal Diagnosis: Cases and Clinical Challenges. 1st. Wiley-Blackwell; 2010. [Google Scholar]
- 2.Beckmann CRB, Herbert W, Laube D, Ling F, Smith R. Obstetrics and Gynecology. Lippincott Williams & Wilkins; Philadelphia: 2013. [Google Scholar]
- 3.Mujezinovic F, Alfirevic Z. Procedure-Related Complications of Amniocentesis and Chorionic Villous Sampling: a Systematic Review. Obstet Gynecol. 2007;110:687–694. doi: 10.1097/01.AOG.0000278820.54029.e3. [DOI] [PubMed] [Google Scholar]
- 4.Beaudet AL. Using Fetal Cells for Prenatal Diagnosis: History and Recent Progress. Am J Med Genet, Part C. 2016;172:123–127. doi: 10.1002/ajmg.c.31487. [DOI] [PubMed] [Google Scholar]
- 5.Lo YM, Corbetta N, Chamberlain PF, Rai V, Sargent IL, Redman CW, Wainscoat JS. Presence of Fetal DNA in Maternal Plasma and Serum. Lancet. 1997;350:485–487. doi: 10.1016/S0140-6736(97)02174-0. [DOI] [PubMed] [Google Scholar]
- 6.Chiu RW, Chan KC, Gao Y, Lau VY, Zheng W, Leung TY, Foo CH, Xie B, Tsui NB, Lun FM, Zee BC, Lau TK, Cantor CR, Lo YM. Noninvasive Prenatal Diagnosis of Fetal Chromosomal Aneuploidy by Massively Parallel Genomic Sequencing of DNA in Maternal Plasma. Proc Natl Acad Sci USA. 2008;105:20458–20463. doi: 10.1073/pnas.0810641105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fan HC, Blumenfeld YJ, Chitkara U, Hudgins L, Quake SR. Noninvasive Diagnosis of Fetal Aneuploidy by Shotgun Sequencing DNA from Maternal Blood. Proc Natl Acad Sci USA. 2008;105:16266–16271. doi: 10.1073/pnas.0808319105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Norton ME, Jacobsson B, Swamy GK, Laurent LC, Ranzini AC, Brar H, Tomlinson MW, Pereira L, Spitz JL, Hollemon D, Cuckle H, Musci TJ, Wapner RJ. Cell-Free DNA Analysis for Noninvasive Examination of Trisomy. N Engl J Med. 2015;372:1589–1597. doi: 10.1056/NEJMoa1407349. [DOI] [PubMed] [Google Scholar]
- 9.Walknowska J, Conte FA, Grumbach MM. Practical and Theoretical Implications of Fetal-Maternal Lymphocyte Transfer. Lancet. 1969;1:1119–1122. doi: 10.1016/s0140-6736(69)91642-0. [DOI] [PubMed] [Google Scholar]
- 10.Bianchi DW, Robert E. Gross Lecture. Fetomaternal Cell Trafficking: a Story that Begins with Prenatal Diagnosis and may End with Stem Cell Therapy. J Pediatr Surg. 2007;42:12–18. doi: 10.1016/j.jpedsurg.2006.09.047. [DOI] [PubMed] [Google Scholar]
- 11.Emad A, Bouchard EF, Lamoureux J, Ouellet A, Dutta A, Klingbeil U, Drouin R. Validation of Automatic Scanning of Microscope Slides in Recovering Rare Cellular Events: Application for Detection of Fetal Cells in Maternal Blood. Prenatal Diagn. 2014;34:538–546. doi: 10.1002/pd.4345. [DOI] [PubMed] [Google Scholar]
- 12.Bhat NM, Bieber MM, Teng NN. One-step enrichment of nucleated red blood cells. A Potential Application in Perinatal Diagnosis. J Immunol Methods. 1993;158:277–280. doi: 10.1016/0022-1759(93)90224-u. [DOI] [PubMed] [Google Scholar]
- 13.Kwon KH, Jeon YJ, Hwang HS, Lee KA, Kim YJ, Chung HW, Pang MG. A High Yield of Fetal Nucleated Red Blood Cells Isolated Using Optimal Osmolality and a Double-Density Gradient System. Prenatal Diagn. 2007;27:1245–1250. doi: 10.1002/pd.1888. [DOI] [PubMed] [Google Scholar]
- 14.Mavrou A, Kouvidi E, Antsaklis A, Souka A, Kitsiou Tzeli S, Kolialexi A. Identification of Nucleated Red Blood Cells in Maternal Circulation: a Second Step in Screening for Fetal Aneuploidies and Pregnancy Complications. Prenatal Diagn. 2007;27:150–153. doi: 10.1002/pd.1640. [DOI] [PubMed] [Google Scholar]
- 15.Kolvraa S, Singh R, Normand EA, Qdaisat S, van den Veyver IB, Jackson L, Hatt L, Schelde P, Uldbjerg N, Vestergaard EM, Zhao L, Chen R, Shaw CA, Breman AM, Beaudet AL. Genome-Wide Copy Number Analysis on DNA from Fetal Cells Isolated from the Blood of Pregnant Women. Prenatal Diagn. 2016;36:1127–1134. doi: 10.1002/pd.4948. [DOI] [PubMed] [Google Scholar]
- 16.Herzenberg LA, Bianchi DW, Schroder J, Cann HM, Iverson GM. Fetal Cells in the Blood of Pregnant Women: Detection and Enrichment by Fluorescence-Activated Cell Sorting. Proc Natl Acad Sci USA. 1979;76:1453–1455. doi: 10.1073/pnas.76.3.1453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.de Wit H, Nabbe KC, Kooren JA, Adriaansen HJ, Roelandse-Koop EA, Schuitemaker JH, Hoffmann JJ. Reference Values of Fetal Erythrocytes in Maternal Blood During Pregnancy Established Using Flow Cytometry. Am J Clin Pathol. 2011;136:631–636. doi: 10.1309/AJCPHL3VXY0VMLXL. [DOI] [PubMed] [Google Scholar]
- 18.Kantak C, Chang CP, Wong CC, Mahyuddin A, Choolani M, Rahman A. Lab-on-a-Chip Technology: Impacting Non-Invasive Prenatal Diagnostics (NIPD) Through Miniaturisation. Lab Chip. 2014;14:841–854. doi: 10.1039/c3lc50980j. [DOI] [PubMed] [Google Scholar]
- 19.He ZB, Guo F, Feng C, Cai B, Lata JP, He RX, Huang QQ, Yu XL, Rao L, Liu HQ, Guo SS, Liu W, Zhang YZ, Huang TJ, Zhao XZ. Fetal Nucleated Red Blood Cell Analysis for Non-Invasive Prenatal Diagnostics Using a Nanostructure Microchip. J Mater Chem B. 2017;5:226–235. doi: 10.1039/c6tb02558g. [DOI] [PubMed] [Google Scholar]
- 20.Mouawia H, Saker A, Jais JP, Benachi A, Bussieres L, Lacour B, Bonnefont JP, Frydman R, Simpson JL, Paterlini-Brechot P. Circulating Trophoblastic Cells Provide Genetic Diagnosis in 63 Fetuses at Risk for Cystic Fibrosis or Spinal Muscular Atrophy. Reprod BioMed Online. 2012;25:508–520. doi: 10.1016/j.rbmo.2012.08.002. [DOI] [PubMed] [Google Scholar]
- 21.Breman AM, Chow JC, U’Ren L, Normand EA, Qdaisat S, Zhao L, Henke DM, Chen R, Shaw CA, Jackson L, Yang Y, Vossaert L, Needham RH, Chang EJ, Campton D, Werbin JL, Seubert RC, Van den Veyver IB, Stilwell JL, Kaldjian EP, Beaudet AL. Evidence for Feasibility of Fetal Trophoblastic Cell-Based Noninvasive Prenatal Testing. Prenatal Diagn. 2016;36:1009–1019. doi: 10.1002/pd.4924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jain CV, Kadam L, van Dijk M, Kohan-Ghadr HR, Kilburn BA, Hartman C, Mazzorana V, Visser A, Hertz M, Bolnick AD, Fritz R, Armant DR, Drewlo S. Fetal Genome Profiling at 5 Weeks of Gestation after Noninvasive Isolation of Trophoblast Cells from the Endocervical Canal. Sci Transl Med. 2016;8:363re4. doi: 10.1126/scitranslmed.aah4661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lin M, Chen JF, Lu YT, Zhang Y, Song J, Hou S, Ke Z, Tseng HR. Nanostructure Embedded Microchips for Detection, Isolation, and Characterization of Circulating Tumor Cells. Acc Chem Res. 2014;47:2941–2950. doi: 10.1021/ar5001617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen JF, Zhu Y, Lu YT, Hodara E, Hou S, Agopian VG, Tomlinson JS, Posada EM, Tseng HR. Clinical Applications of NanoVelcro Rare-Cell Assays for Detection and Characterization of Circulating Tumor Cells. Theranostics. 2016;6:1425–1439. doi: 10.7150/thno.15359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang S, Liu K, Liu J, Yu ZT, Xu X, Zhao L, Lee T, Lee EK, Reiss J, Lee YK, Chung LW, Huang J, Rettig M, Seligson D, Duraiswamy KN, Shen CK, Tseng HR. Highly Efficient Capture of Circulating Tumor Cells by Using Nanostructured Silicon Substrates with Integrated Chaotic Micromixers. Angew Chem, Int Ed. 2011;50:3084–3088. doi: 10.1002/anie.201005853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lu YT, Zhao L, Shen Q, Garcia MA, Wu D, Hou S, Song M, Xu X, Ouyang WH, Ouyang WW, Lichterman J, Luo Z, Xuan X, Huang J, Chung LW, Rettig M, Tseng HR, Shao C, Posadas EM. NanoVelcro Chip for CTC Enumeration in Prostate Cancer Patients. Methods. 2013;64:144–152. doi: 10.1016/j.ymeth.2013.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen JF, Ho H, Lichterman J, Lu YT, Zhang Y, Garcia MA, Chen SF, Liang AJ, Hodara E, Zhau HE, Hou S, Ahmed RS, Luthringer DJ, Huang J, Li KC, Chung LW, Ke Z, Tseng HR, Posadas EM. Subclassification of Prostate Cancer Circulating Tumor Cells by Nuclear Size Reveals Very Small Nuclear Circulating Tumor Cells in Patients with Visceral Metastases. Cancer. 2015;121:3240–3251. doi: 10.1002/cncr.29455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ke Z, Lin M, Chen JF, Choi JS, Zhang Y, Fong A, Liang AJ, Chen SF, Li Q, Fang W, Zhang P, Garcia MA, Lee T, Song M, Lin HA, Zhao H, Luo SC, Hou S, Yu HH, Tseng HR. Programming Thermoresponsiveness of NanoVelcro Substrates Enables Effective Purification of Circulating Tumor Cells in Lung Cancer Patients. ACS Nano. 2015;9:62–70. doi: 10.1021/nn5056282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhao L, Tang C, Xu L, Zhang Z, Li X, Hu H, Cheng S, Zhou W, Huang M, Fong A, Liu B, Tseng HR, Gao H, Liu Y, Fang X. Enhanced and Differential Capture of Circulating Tumor Cells from Lung Cancer Patients by Microfluidic Assays Using Aptamer Cocktail. Small. 2016;12:1072–1081. doi: 10.1002/smll.201503188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.He W, Xu D, Wang Z, Xiang X, Tang B, Li S, Hou M, Zhang Y, Chen JF, Lin M, Wang L, Hou S, Tseng HR, Kuang M, Ke ZF. Detecting ALK-Rearrangement of CTC Enriched by Nanovelcro Chip in Advanced NSCLC Patients. Oncotarget. 2016 doi: 10.18632/oncotarget.8305. [DOI] [Google Scholar]
- 31.Ankeny JS, Court CM, Hou S, Li Q, Song M, Wu D, Chen JF, Lee T, Lin M, Sho S, Rochefort MM, Girgis MD, Yao J, Wainberg ZA, Muthusamy VR, Watson RR, Donahue TR, Hines OJ, Reber HA, Graeber TG, Tseng HR, Tomlinson JS. Circulating Tumour Cells as a Biomarker for Diagnosis and Staging in Pancreatic Cancer. Br J Cancer. 2016;114:1367–1375. doi: 10.1038/bjc.2016.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liu S, Tian Z, Zhang L, Hou S, Hu S, Wu J, Jing Y, Sun H, Yu F, Zhao L, Wang R, Tseng HR, Zhau HE, Chung LW, Wu K, Wang H, Wu JB, Nie Y, Shao C. Combined Cell Surface Carbonic Anhydrase 9 and CD147 Antigens Enable High-Efficiency Capture of Circulating Tumor Cells in Clear Cell Renal Cell Carcinoma Patients. Oncotarget. 2016;7:59877–59891. doi: 10.18632/oncotarget.10979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hou S, Zhao L, Shen Q, Yu J, Ng C, Kong X, Wu D, Song M, Shi X, Xu X, OuYang WH, He R, Zhao XZ, Lee T, Brunicardi FC, Garcia MA, Ribas A, Lo RS, Tseng HR. Polymer Nanofiber-Embedded Microchips for Detection, Isolation, and Molecular Analysis of Single Circulating Melanoma Cells. Angew Chem, Int Ed. 2013;52:3379–3383. doi: 10.1002/anie.201208452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhao L, Lu YT, Li F, Wu K, Hou S, Yu J, Shen Q, Wu D, Song M, OuYang WH, Luo Z, Lee T, Fang X, Shao C, Xu X, Garcia MA, Chung LW, Rettig M, Tseng HR, Posadas EM. High-Purity Prostate Circulating Tumor Cell Isolation by a Polymer Nanofiber-Embedded Microchip for Whole Exome Sequencing. Adv Mater. 2013;25:2897–2902. doi: 10.1002/adma.201205237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jiang R, Lu YT, Ho H, Li B, Chen JF, Lin M, Li F, Wu K, Wu H, Lichterman J, Wan H, Lu CL, OuYang W, Ni M, Wang L, Li G, Lee T, Zhang X, Yang J, Rettig M, et al. A Comparison of Isolated Circulating Tumor Cells and Tissue Biopsies using Whole-Genome Sequencing in Prostate Cancer. Oncotarget. 2015;6:44781. doi: 10.18632/oncotarget.6330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hsiao YS, Luo SC, Hou S, Zhu B, Sekine J, Kuo CW, Chueh DY, Yu HH, Tseng HR, Chen P. 3D Bioelectronic Interface: Capturing Circulating Tumor Cells onto Conducting Polymer-Based Micro/Nanorod Arrays with Chemical and Topographical Control. Small. 2014;10:3012–3017. doi: 10.1002/smll.201400429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Stroock AD, Dertinger SK, Ajdari A, Mezic I, Stone HA, Whitesides GM. Chaotic Mixer for Microchannels. Science. 2002;295:647–651. doi: 10.1126/science.1066238. [DOI] [PubMed] [Google Scholar]
- 38.Park JY, Hendricks NR, Carter KR. Solvent-Assisted Soft Nanoimprint Lithography for Structured Bilayer Heterojunction Organic Solar Cells. Langmuir. 2011;27:11251–11258. doi: 10.1021/la201809g. [DOI] [PubMed] [Google Scholar]
- 39.Sun J, Masterman-Smith MD, Graham NA, Jiao J, Mottahedeh J, Laks DR, Ohashi M, DeJesus J, Kamei K, Lee KB, Wang H, Yu ZT, Lu YT, Hou S, Li K, Liu M, Zhang N, Wang S, Angenieux B, Panosyan E. A Microfluidic Platform for Systems Pathology: Multiparameter Single-Cell Signaling Measurements of Clinical Brain Tumor Specimens. Cancer Res. 2010;70:6128–6138. doi: 10.1158/0008-5472.CAN-10-0076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sermon K, Van Steirteghem A, Liebaers I. Preimplantation Genetic Diagnosis. Lancet. 2004;363:1633–1641. doi: 10.1016/S0140-6736(04)16209-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


