Abstract
Background
Understanding the validity of data from electronic data research networks is critical to national research initiatives and learning healthcare systems for cardiovascular care. Our goal was to evaluate the degree of agreement of electronic data research networks compared with data collected by standardized research approaches in a cohort study.
Methods
We linked individual-level data from The Multi-Ethnic Study of Atherosclerosis (MESA), a community-based cohort, with HealthLNK, a 2006–2012 database of electronic health records (EHRs) from six, Chicago health systems. To evaluate the correlation and agreement of blood pressure (BP) in HealthLNK as compared with in-person MESA examinations, and BMI in HealthLNK compared with MESA, we used Pearson Correlation Coefficients and Bland-Altman plots. Using diagnoses in MESA as the criterion standard, we calculated the performance of HealthLNK for hypertension (HTN), obesity, and diabetes diagnosis using ICD-9 codes and clinical data. We also identified potential myocardial infarctions (MIs), strokes, and heart failure events in HealthLNK and compared them with adjudicated events in MESA.
Results
Of the 1,164 MESA participants enrolled at the Chicago Field Center, 802 (68.9%) participants had data in HealthLNK. The correlation was low for systolic BP (0.39; P<0.0001). Compared with MESA, HealthLNK overestimated systolic BP by 6.5 mmHg (95%CI: 4.2, 7.8). There was a high correlation between BMI in MESA and HealthLNK (0.94; P<0.0001). HealthLNK underestimated BMI by 0.3 kg/m2 (95%CI: −0.4, −0.1). Using ICD-9 codes and clinical data, the sensitivity and specificity of HealthLNK queries for HTN were 82.4% and 59.4%, for obesity were 73.0% and 89.8%, and for diabetes were 79.8% and 93.3%. Compared with adjudicated CVD events in MESA, the concordance rates for MI, stroke, and heart failure were, respectively, 41.7% (5/12), 61.5% (8/13), and 62.5% (10/16).
Conclusions
These findings illustrate the limitations and strengths of electronic data repositories compared with information collected by traditional standardized epidemiologic approaches for the ascertainment of CVD risk factors and events.
Keywords: Cardiovascular disease risk factors, Cardiovascular events, Cardiovascular research, Epidemiology
Background
Since the passage of the Health Information Technology for Economic and Clinical Health (HITECH) Act in 2009, the use of electronic health records (EHRs) has become nearly ubiquitous with almost 100% adoption among hospitals and 80% among ambulatory offices.1,2 Leading scientific organizations and experts view leveraging electronic data as essential to the future of cardiology research, public health surveillance, and quality improvement initiatives.3–9
For example, in April 2017, the American Heart Association published a scientific statement that highlighted the central role of EHR data in learning healthcare systems—care systems in which scientific evidence is applied at the point of clinical care while also generating data for improving health care delivery and scientific discovery—for cardiovascular (CVD) care.7 In 2016, the National Institutes of Health (NIH) and the White House launched the largest national cohort study ever undertaken in the United States.10 Electronic health record data will comprise a large portion of phenotyping and outcome ascertainment for the one million-plus person Precision Medicine Initiative (PMI) “All of Us” Research Program. However, few studies have confirmed or addressed whether electronic data repositories can accurately characterize an individual’s health over time.
To better understand the strengths and weaknesses of electronic data research networks and traditional cohorts for the ascertainment of demographics, CVD risk factors, and CVD events, we directly compared individual-level data from HealthLNK11—a research database of approximately 2.95 million Chicago area residents with extracted electronic data merged from six, large health systems—to data from the Multi-Ethnic Study of Atherosclerosis (MESA), a community-based, cardiovascular disease (CVD) cohort. We hypothesized there would be a large amount of discordance for demographics, blood pressure (BP), body mass index (BMI), CVD risk factors, and CVD events between HealthLNK and MESA.
Methods
Sample
HealthLNK Data Repository is a database spanning 2006 to 2012 with data for approximately 2.95 million unique patients from 5 academic health systems and a large, county health system in Chicago.11 These systems comprise approximately 42% of the total inpatient beds in Chicago. Analysts at each institution extracted data from their data warehouse based on a specified data model. Data types included demographics, diagnostic codes from inpatient, outpatient, and emergency department encounters, procedure codes, medications, laboratory measurements recorded as structured data, and vital signs data. HealthLNK investigators then, using a secure matching algorithm,12 aggregated, de-duplicated, and de-identified the data.
Investigators for MESA, an ongoing community-based, cardiovascular cohort, enrolled 6,814 men and women ages 45 to 84 years from six, US communities from 2000–2002.13 The Chicago Field Center, based at Northwestern University, enrolled 1,164 participants. Data from the MESA baseline examination (July 2000–August 2002), Examination 4 (September 2005–May 2007), and Examination 5 (April 2010–December 2011) and adjudicated CVD event data through 2012 were used.
Using the same secure, matching algorithm used to link the HealthLNK institutions,12 we linked individuals who were included both in the HealthLNK database and enrolled at the MESA Chicago Field Center. Northwestern University IRB approved this study and granted a waiver of consent.
Data Collection
Demographics
In HealthLNK, gender and race/ethnicity were collected as part of routine clinical care without any standard method of collection. In MESA, study researchers collected race/ethnicity and gender via participant questionnaire as part of the baseline examination. In MESA, race and ethnicity were characterized on the basis of participants’ responses to the race/ethnicity questions modeled on the year 2000 US Census, whereas in HealthLNK, race and ethnicity (referring to non-Hispanic vs Hispanic) were reported in two, separate questions. In HealthLNK, only the year of birth was available. In MESA, we only had access to age on the date of Exam 4 or 5.
Medication, laboratory, blood pressure and anthropometric data
In HealthLNK, medication, laboratory, BP, and anthropometric data were measured as part of routine care. In MESA, these data were collected during standardized, in-person examinations. Participants were asked to bring in all of their medications for in-person examinations. For BP, MESA participants sat in a chair for five minutes in a quiet room. Then a MESA trained clinic staff member obtained three measurements using an automatic, oscillometric BP cuff. The average of the last 2 of 3 measurements were used as reported BP value. Height and weight were measured with participants wearing light clothing and no shoes.
Cardiovascular risk factors
In HealthLNK, we defined hypertension (HTN) by ICD-9 codes (401.XX, 402.XX, 403.XX, 404.XX, 405.XX, 437.2), 14,15 a systolic BP ≥ 140 mm Hg or diastolic BP ≥ 90 mm Hg on two measurements in different months or use of anti-hypertensive medications. In MESA, HTN was defined as systolic BP ≥ 140 mm Hg or diastolic BP ≥ 90 mm Hg or reported use of any anti-hypertensive medication. We defined obesity in HealthLNK by ICD-9 codes (278.0, 278.00, 278.01, 278.03) or a BMI ≥ 30 kg/m2, whereas in MESA, investigators defined obesity as BMI ≥ 30 kg/m2 measured during in-person examination. In HealthLNK, we defined diabetes with ICD-9 codes (249.XX, 250.XX, 357.2, 362.0X), 14,16 hemoglobin A1C ≥ 6.5%, or diabetes medication use. In MESA, diabetes was defined as either use of diabetes medications or a fasting glucose ≥ 126 mg/dL.
Additional details for MESA collection and ascertainment of demographics, laboratory, anthropometric, and CVD risk factors have been previously described.13 Data from the most recent MESA examination (either Exam 4 or 5) were used for ascertainment of risk factor status.
From the matched sample, we applied the following limitations. In MESA, participants had to attend either Exam 4 or Exam 5 and have data on the risk factor of interest. In HealthLNK, participants were required to have demographics data and at least one other data type required for risk factor ascertainment (encounters, medications, laboratory testing, vital signs).
Cardiovascular Events
The CVD events of interest were myocardial infarction (MI), stroke, and heart failure (HF). In Supplemental Table 1, we summarize the methods for detecting potential CVD events in HealthLNK and adjudicated CVD events in MESA. For HealthLNK, we chose sensitive methods to identify as many potential events as possible. We included diagnostic codes for relevant events in either primary or secondary position and in inpatient or outpatient encounters, recognizing that outpatient encounters likely represented episodes of follow-up for a prior event. These algorithms were adapted from multiple sources.17–20 Duplicate potential events within a given month were removed.
Details for event ascertainment and adjudication in MESA have been previously published.21 Briefly, study staff contacted participants every 9 to 12 months, either via phone or during in-person examinations. Trained staff asked participants about all interim outpatient cardiovascular diagnoses, cardiovascular treatments, and hospitalizations. Study staff obtained medical records for all potential events. Two physicians reviewed the medical records of each event independently, assigned event dates, and adjudicated any differences among themselves, or with consultation from remainder of the events committee when needed.
To determine reasons for discordance between MESA events and HealthLNK potential events, we performed a manual review of selected MESA event files. We reviewed the files for all MESA adjudicated events from 2006 to 2012 not found in HealthLNK. We next applied criteria with higher specificity to each set of potential HealthLNK CVD events (Supplemental Table 1). We then reviewed the MESA Chicago Field Center case files to ascertain reasons for discordance. The charts were reviewed by a MESA Chicago Field Center staff member with oversight from two study investigators (FSA, NBA).
Data visualizations and data quality checks
We first characterized the HealthLNK dataset through a series of analyses and visualizations that enabled us to perform data quality checks. We assessed for data breadth (the types of data for each individual), data plausibility (whether any of the data appeared grossly inconsistent with general medical knowledge about possible values), data missingness (the absence of data for a variable in an observation), and data density (the frequency of observations for an individual).22,23
Statistical analysis
Demographics
For gender, we first calculated the percentage of data categorized as “other” or missing in HealthLNK. Excluding those records, we then calculated concordance rate as the number of participants with the same labeled gender in HealthLNK and MESA over the total number of individuals in HealthLNK labeled as male or female. As with gender, for race/ethnicity we first calculated the percentage of participants in HealthLNK with “other” or missing values and then excluded those records. We then calculated the concordance rate for race/ethnicity for those with race/ethnicity reported as Hispanic, Asian, white, and black, in HealthLNK and MESA. In HealthLNK, we calculated the estimated age during the year of MESA Exam 4 or Exam 5. Because we did not have the exact date of birth in HealthLNK, we considered a HealthLNK age concordant with the age in MESA if it were the same value or within one year of the MESA age at time of examination.
Blood pressure and body mass index
We first identified participants in HealthLNK with at least one measurement of BP. We then identified participants in MESA who attended Exam 4 or Exam 5. We compared the BP value from MESA with the HealthLNK BP value in the nearest month and year to the MESA examination date. If multiple BP measurements were present within nearest month and year in HealthLNK, then the median value was used. If an individual attended both Exam 4 and Exam 5, we used the data from Exam 5 only.
We performed an individual-level analysis by calculating agreement using Bland-Altman plots24,25 and correlation using the Pearson’s correlation coefficient. We used the same methodology for the analysis of the BMI data. We also created fluctuation diagrams26 as another method to visualize disagreement between HealthLNK and MESA blood pressure and BMI measurements.
CVD Risk Factors
For each CVD risk factor (HTN, obesity, diabetes), using MESA as the criterion standard, we calculated sensitivity, specificity, and the positive and negative predictive value for the identification of participants in HealthLNK, based on ICD-9 codes only and then based on ICD-9 codes and clinical data. We evaluated the data types in HealthLNK that led to the identification of individuals who met criteria for risk factor diagnosis in both MESA and HealthLNK.
Cardiovascular Events
For each CVD event—MI, stroke, and HF—we generated diagrams for concordant and discordant events. An event was classified as concordant if it was adjudicated by the MESA committee, met criteria in HealthLNK, and shared the same month and year in MESA and HealthLNK. We calculated the concordance rate as the number of concordant events over the total number adjudicated events identified in MESA from 2006 to 2012. For selected, discordant adjudicated MESA events and potential HealthLNK events, we reviewed Chicago Field Center MESA event files and categorized reasons for discordance.
Sensitivity Analyses
We performed a sensitivity analysis by examining data from individuals who had a BP or BMI measurement within a one-year window and three-month window of the date of the participant’s selected MESA examination. To evaluate the performance of HealthLNK on a subset of well-phenotyped participants who received regular care at HealthLNK institutions, we re-analyzed data on CVD risk factors for individuals who were seen at least twice within a given year at HealthLNK institutions and had at least one record of all available data types used in the HealthLNK CVD risk factor algorithms (encounters, medications, laboratory testing, vital signs).
Results
Sample, Visualizations, and Demographics
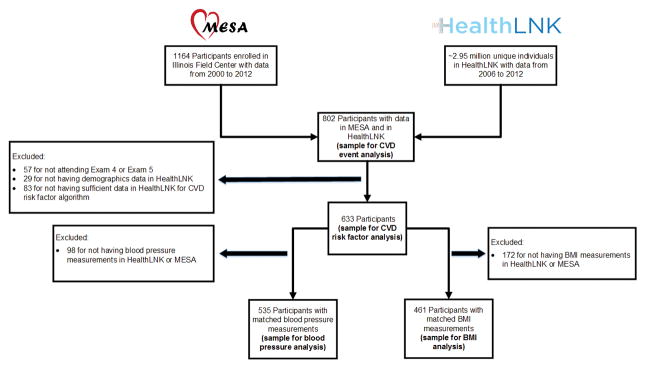
We linked data from 802 individuals in MESA and HealthLNK (Table 1, Figure 1). In terms of data breadth (the types of available data), demographic data were available in nearly all participants (96.1%), whereas encounter diagnostic codes, medication data, and laboratory data were available for between 70% and 80% of the sample (Supplemental Table 2). Only 55.2% of the sample had all types of data available. In terms of data density (frequency of observations over time), 615 participants (76.7%) received care for any type of visit at least two times within a given year at HealthLNK institutions.
Table 1.
Baseline characteristics of participants based on MESA Examination 1 data
| Characteristics | Overall (n=802) |
|---|---|
| Age, mean (SD), years | 61.4 (1.8) |
| Male, no. (%) | 364 (45.4) |
| Race/Ethnicity, no. (%) | |
| White | 439 (54.7) |
| Black | 229 (28.6) |
| Asian | 134 (16.7) |
| Education, no. (%) | |
| Less than high school or less | 45 (5.6) |
| High school graduate or equivalent | 52 (6.5) |
| More than high school | 705 (87.9) |
Figure 1. Flow diagram for main analyses.
MESA = The Multi-Ethnic Study of Atherosclerosis. CVD = Cardiovascular. BMI = body mass index.
Demographics
In MESA, gender and race/ethnicity were available for all 802 individuals. In HealthLNK, gender was either labeled as “other” or was missing for 46 (5.7%) individuals (Supplemental Table 3 and 4). Of those with a recorded gender in HealthLNK, the concordance rate with MESA was 99.3% (751/756). In HealthLNK, race and ethnicity were labeled as “other” or “missing” for 202 (25.2%) individuals. Of those with a recorded race or ethnicity in HealthLNK, the concordance rate with MESA was 98.8% (593/600). For the 715 individuals for whom we had MESA age at time of MESA exam 4 or 5 and an estimated age in HealthLNK at time of MESA exam, 99.3% (710/715) were concordant.
Blood pressure, body mass index, risk factors, and events
Blood pressure and body mass index
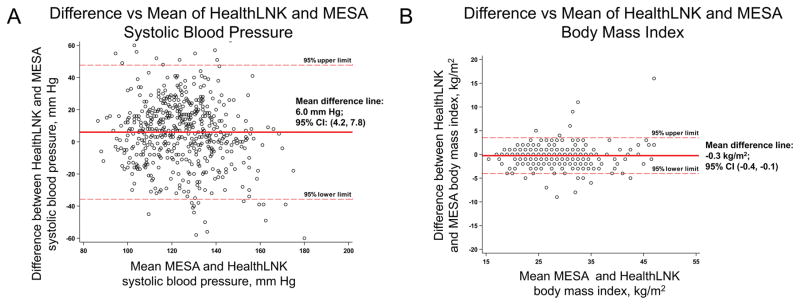
We identified 535 individuals who participated in MESA Exam 4 or Exam 5 and had matched HealthLNK BP data. The median (IQR) months between the MESA Exam date and the nearest HealthLNK date (either before or after the MESA Exam date) and HealthLNK dates were 6.3 (1.8, 17.1) months. Figure 2A shows the agreement in systolic BP between measurements in MESA and HealthLNK on an individual level using Bland-Altman plots. The mean difference for systolic BP for the population is 6.0 mm Hg (95%CI: 4.2, 7.8). There is large amount of variability in the difference between measurements in HealthLNK and MESA across all BP values. Compared to systolic BP, diastolic BP had lower mean difference (3.5 mm Hg; 95%CI: 2.5, 4.4) and similar variability (Supplemental Figure 1). The correlation for systolic BP and diastolic BP between HealthLNK and MESA were, respectively, 0.39 and 0.40 (P<0.0001 for both).
Figure 2. Bland-Altman plots for MESA and HealthLNK systolic blood pressure and body mass index.
A) Panel A shows the agreement between systolic blood pressure measurements from MESA in-person examinations and HealthLNK. B) Panel B shows agreement between body mass index measurements from MESA in-person examinations and HealthLNK.
We identified 461 individuals with matched BMI data in MESA and HealthLNK. The median (IQR) months between the MESA Exam and HealthLNK dates were 9.4 (2.3, 20.6) months. The mean difference in the population was −0.3 kg/m2 (95%CI: −0.4, −0.1), and, compared to BP, there was less variability in the difference between individual measurements of BMI between HealthLNK and MESA (Figure 2B). The correlation for BMI between HealthLNK and MESA was 0.94 (P<0.0001).
Fluctuations diagrams for blood pressure and BMI are included in the Supplemental Figures 2, 3, and 4.
Cardiovascular risk factors
Table 2 shows the sensitivity and specificity of HealthLNK algorithms for CVD risk factors compared to MESA, the criterion standard in this analysis. Data are shown for ICD-9 codes alone .and for ICD-9 codes with clinical data. Using ICD-9 diagnostic codes alone, the sensitivity and specificity of HealthLNK for the diagnosis of HTN were 71.2% (242/340) and 73.0% (214/293). The addition of clinical data (BP and medications) to ICD-9 codes increased sensitivity and decreased specificity for HTN to 82.4% (280/340) and 59.4% (174/293). Compared to using ICD-9 codes alone, the addition of BMI increased the sensitivity of HealthLNK for obesity from 30.9% (47/152) to 73.0% (111/152) and decreased the specificity from 97.5% (469/481) to 89.8% (432/481). Using ICD-9 diagnostic codes alone, the sensitivity and specificity of HealthLNK for the diagnosis of diabetes were 77.5% (69/89) and 95.6% (517/541). The addition of clinical data (medications and laboratory values) to ICD-9 codes increased sensitivity and decreased specificity for diabetes diagnosis to 79.8% (71/89) and 93.3% (505/541). Additional details for HealthLNK and MESA CVD risk factor diagnosis comparisons are included in the Supplemental Tables 5–10.
Table 2.
Comparison of cardiovascular risk factor prevalence in HealthLNK compared to MESA (criterion standard) with ICD-9 codes alone and with ICD-9 codes and clinical data
| ICD-9 Codes Alone | ICD-9 Codes and Clinical Data* | Change with Addition of Clinical Data | ||
|---|---|---|---|---|
| Hypertension | Sensitivity | 71.2% | 82.4% | +11.2 |
| Specificity | 73.0% | 59.4% | −13.6 | |
| Obesity | Sensitivity | 30.9% | 73.0% | +42.1 |
| Specificity | 97.5% | 89.8% | −7.7 | |
| Diabetes | Sensitivity | 77.5% | 79.8% | +2.3 |
| Specificity | 95.6% | 93.3% | −2.3 |
For hypertension, the clinical data comprised medication use and blood pressure measurements (two measurements ≥ 140/90 in two different months). For obesity, clinical data comprised body mass index ≥30 kg/m2. For diabetes, clinical data comprised of medication use and hemoglobin A1C ≥ 6.
Analysis of the data types in HealthLNK that led to the identification of true positive participants with HTN (n=280), obesity (n=111), and diabetes (n=71) revealed that for HTN, obesity, and diabetes, respectively, clinical data in the absence of ICD-9 codes led to the identification of 13.6% (38/280), 55.9% (62/111), and 2.8% (2/71) of true positive diagnoses (Supplemental Tables 11–13).
Cardiovascular Events
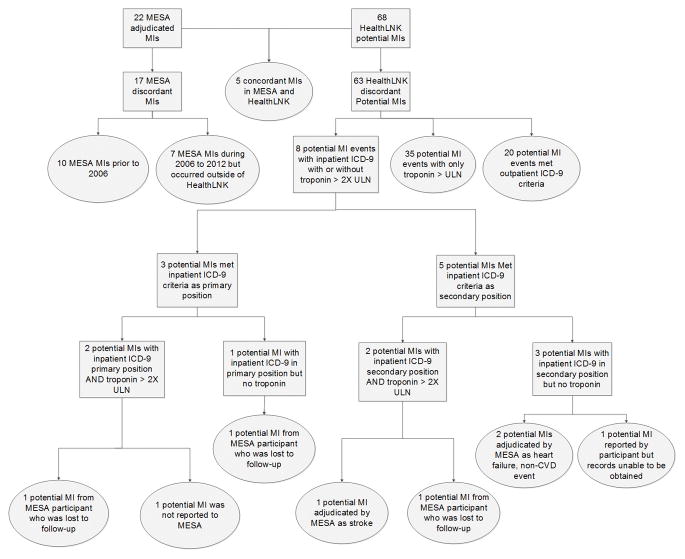
Figure 3 summarizes concordance and discordance between adjudicated MIs in MESA and potential MIs in HealthLNK. We identified in MESA a total of 22 adjudicated MIs, of which 12 occurred from 2006 to 2012. We identified 68 potential MIs in HealthLNK using the initial, sensitive criteria. Of the 12 MESA events that occurred from 2006 to 2012, 5/12 (41.7%) were concordant in HealthLNK.
Figure 3. Diagram of Concordant and Discordant Myocardial Infarctions in MESA and Potential Myocardial Infarctions in HealthLNK.
ULN = upper limit of normal. MI = myocardial infarction. CVD = cardiovascular disease.
Seven MESA-adjudicated MIs occurred during 2006 to 2012 but these events were not captured in HealthLNK because they occurred at hospitals not included in HealthLNK. Two of these occurred in different states. In HealthLNK, we initially identified 63 individuals with potential MIs that were not identified in MESA. Eight of these potential MIs in HealthLNK met the stricter MI definition of inpatient diagnosis codes with or without troponins greater than twice the upper limit of normal. Of these 8 potential events, 3 occurred among MESA participants after they were lost to follow-up, 1 event was not reported, 1 event was reported but records could not be obtained, and an additional 3 events were reported but determined by the adjudication committee as not being a MI and instead as stroke, HF, or non-CVD event. Supplemental Figures 5 and 6 summarize the findings for stroke and HF. The concordance rates for stroke and HF were, respectively, 61.5% (8/13), and 62.5% (10/16). Reasons for discordance were overall similar.
Sensitivity Analysis
In the sensitivity analysis for BP and BMI measurements, we found that using a more closely matched time window led to a similar amount of agreement (Supplemental Figures 7 and 8). In the sensitivity analysis for a limited subset of patients with at least two encounters in a given year in HealthLNK and all available data types, compared to the main sample in the main CVD risk factor analysis, overall the performance of the CVD risk factor algorithms resulted in higher sensitivity and slightly lower specificity. The sensitivity and specificity for HTN were 95.3% (201/211) and 51.0% (107/210). For obesity, the sensitivity was 85.1% (80/94) and specificity was 86.9% (284/327). For diabetes, the sensitivity and specificity were 94.4% (51/54) and 92.3% (337/365).
Discussion
In this analysis of data from individuals in MESA, a community-based cohort, and HealthLNK, an electronic research database from six health systems, we found differences in individual-level BP, the prevalence of CVD risk factors, and CVD events, and good agreement between BMI measurement, gender, race/ethnicity, and age. To our knowledge, this is the first study to directly compare data from an electronic data research network to a large, CVD cohort.
By linking to a traditional, epidemiological cohort, this study highlights the potential of using data from electronic research networks but also the need for a better understanding of the validity and data quality of those data. This need is particularly relevant to the success of studies such as the PMI All of Us Research Program,10 other national research initiatives,27,28 the future of CVD epidemiology,3–5 and the development of learning healthcare systems.7 Although several studies have evaluated the different dimensions of EHR data quality and developed EHR phenotyping algorithms,16,29–32 they frequently are from a single healthcare institution and use data from clinical care as a gold standard reference, including paper charts, patient and physician interviews, standardized patient encounters, registry data, and claims data.23
Fort et al. linked EHR data from a single institution to a local community population health survey in New York and found good agreement at the population-level between EHR and a community-based survey for gender, race, blood pressure, and BMI.33 In our study at the individual-level, for gender, race/ethnicity, and age, we found good agreement between HealthLNK and MESA after excluding the large amount of missing data, a frequent problem in electronic data repositories.34 We also found good agreement for BMI between electronic and research-grade data, but a significant mean difference in blood pressure measurements. In light of the large amount of intra-individual variability in blood pressure over time, the variability in measurement techniques, and medical reasons for measurement in health care settings, the difference in blood pressure measurement between two time points in MESA and HealthLNK is consistent with prior studies. Findings from a single site in the Systolic Blood Pressure Intervention Trial (SPRINT) trial also found similar variability between in-clinic and research-grade measurements within the same day among a smaller, predominantly male sample with chronic kidney disease.35 These results underscore the challenges of developing risk models using blood pressure measurements from the EHR in contrast to research-grade measurements, which at least remove most of the variability from measurement and setting. The results may also inform 1) the design of next-generation epidemiology cohorts and pragmatic clinical trials that include EHR measurements of blood pressure as part of the study design and 2) the translation of cohort-based risk scores to clinical practice.
Numerous studies have evaluated the validity of administrative data, primarily limited to ICD-9 data, for risk factors and event ascertainment.36–42 However, administrative data lack the richness of EHR data, which include anthropometric measurements and diagnostic testing results. In recent years, national research consortiums, such as eMERGE (electronic medical records and genomics) Network, the NIH Collaboratory, and other research groups, have developed, tested, and validated EHR detection algorithms for various cardiovascular disease risk factors and events, including diabetes, HTN, heart failure, and coronary heart disease.16,31,32 Many of these algorithms are developed at a single center, tested at other centers, and adapted if needed.30 Consistent with prior studies, our results underscore the additive value of clinical data to ICD codes in case detection, especially for obesity, which has previously been reported as under-detected in administrative databases.43,44 Of the three, HealthLNK risk factor algorithms, diabetes had the best performance. For diabetes, in contrast to HTN and obesity, clinical data had little additive value over ICD-9 codes alone. The sensitivity analysis of CVD risk factors illustrates that the addition a filter requiring two encounters within a year time period and a minimum amount of data type availability increases the sensitivity of EHR risk factor detection algorithms. This finding underscores the importance of defining minimal data requirements over time (data density) or across data types (data breadth) for EHR detection algorithms.
Our findings suggest that accurate event detection in cohort studies and pragmatic clinical trials using clinical data research networks alone may be challenging. Electronic data networks may need to be supplemented with patient-reported data and claims data for better event ascertainment. For example, in ADAPTABLE, a pragmatic clinical trial, investigators are using this three pronged strategy to monitor for events.45 These results also highlight the limitations of event ascertainment in traditional cohort studies, which include participant loss to follow-up, participant underreporting of events, and challenges in obtaining records from healthcare institutions.
Moreover, development of better EHR algorithms to identify and classify CVD events and subtypes is needed. Our algorithm for MI detected three potential MI events in HealthLNK that had a secondary ICD diagnosis code for MI, but they were adjudicated in MESA as stroke, heart failure, and a non-CVD event. Even if for each of these hospitalizations, the treating clinicians or the person responsible for coding considered the patient to have had a MI (such as a Type 2 MI), such events may not have met the MESA adjudication criteria for MI, which require a specific combination of symptoms, ECG changes, and biomarkers. Few prior studies on EHR detection algorithms for MI go beyond diagnosis codes to use data such as ECGs, biomarkers, and text extracted by natural language processing.46 Using diverse data types may lead to EHR algorithms with better detection performance and the ability to classify events based on criteria established in consensus statements, including differentiation between Type 1 and Type 2 MIs based on the Third Universal Definition of Myocardial Infarction.47
These results also have important implications for learning healthcare systems for cardiovascular care. In an AHA statement, the writing committee outlined 44 action steps across 3 domains to facilitate the creation of learning healthcare systems.7 This study highlights the need for further research on the quality of data from electronic sources, the development of better detection and classification algorithms for CVD events using diverse data types, and the incremental value of linking EHR data to other data sources. Moreover, redesigning systems with better usability may facilitate the collection of higher quality and more complete data from patients by clinicians at the point of care.
There are limitations to this study. First, this analysis reflects the experience of a single MESA field center and of the six HealthLNK institutions with a sample of 802 individuals; however, HealthLNK may reflect the experience and coverage of many electronic data research networks, especially in urban centers. Second, because HealthLNK was first created in 2010, the data model differs from the current ones. Extraction methods, standards, and data models have evolved, which limits the generalizability of these results. For example, vital sign data were linked by month and year and not by encounter, making it difficult to differentiate outpatient and inpatient measurements. However, we used the median values within a given month and year if multiple were present to address the variability inherent BP measurements. Third, in light of the numerous, published algorithms for administrative and EHR data, we could not identify a criterion standard algorithm for each CVD risk factor and event. The definitions were chosen based on a literature review and study team expertise. Lastly, the impact of our findings regarding CVD events remains unclear given the limited number of CVD events that occurred in this sample from 2006 to 2012.
In conclusion, we found areas of agreement and disagreement between BP and BMI measurements, CVD risk factor diagnosis, and CVD events between a community-based cohort and an electronic data research network. These findings help elucidate the strengths and limitations of using electronic data networks for research compared with traditional epidemiological studies and may inform the design of the next-generation of cardiovascular epidemiological cohorts and pragmatic clinical trials and the creation of learning healthcare systems for cardiovascular care. Future work is needed to explore how these differences in data sources may impact future studies examining the relationship between exposures and outcomes.
Supplementary Material
Clinical Perspective.
What is new?
Data from electronic health records will be essential to the success of national research initiatives and the development of learning healthcare systems for cardiovascular care.
Little is known about how the quality of data from electronic data sources directly compares with data collected by standardized research approaches.
This study evaluated the degree of agreement of cardiovascular risk factors and events from electronic data research networks compared with data collected by standardized research approaches in a traditional, cardiovascular cohort study.
What are the clinical implications?
We identified areas of agreement and disagreement between blood pressure, cardiovascular risk factor diagnosis, and cardiovascular events between an electronic data repository and a traditional cardiovascular cohort with standardized measurements.
These findings illustrate the limitations and strengths of using electronic data repositories compared with information collected by standardized epidemiologic approaches for cardiovascular research, learning healthcare systems, and public health surveillance.
Acknowledgments
The authors thank the other investigators, the staff, and the participants of the MESA study for their valuable contributions. A full list of participating MESA investigators and institutions can be found at http://www.mesa-nhlbi.org. The authors would like to acknowledge support from the HealthLNK Data Repository, including Bala Hota, MD, Rush University Medical Center, Bill Galanter, MD, University of Illinois at Chicago Hospital and Health Sciences System, and David Meltzer, MD, PhD, University of Chicago Medical Center.
Funding Souces: This research was supported by contracts HHSN268201500003I, N01-HC-95159, N01-HC-95160, N01-HC-95161, N01-HC-95162, N01-HC-95163, N01-HC-95164, N01-HC-95165, N01-HC-95166, N01-HC-95167, N01-HC-95168 and N01-HC-95169 from the National Heart, Lung, and Blood Institute and by grants UL1-TR-000040 and UL1-TR-001079 from National Center for Research Resources. Research reported in this publication was supported, in part, by the National Institutes of Health’s National Center for Advancing Translational Sciences, Grant Number UL1TR001422. Dr. Ahmad is supported by the National Heart, Lung, and Blood Institute of the National Institutes of Health under Award number T32HL069771 and by a 2015 Research Fellowship Award from the Heart Failure Society of America. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health, MESA Investigators, or HealthLNK Investigators.
Footnotes
Conflict of Interest Disclosures: Dr. Kho reports that he is a co-founder and equity holder of Health DataLink, LLC, with $0 current value. The other authors report conflicts.
References
- 1.Office of the National Coordinator for Health Information Technology. Non-federal Acute Care Hospital Electronic Health Record Adoption. [Accessed April 4, 2017];Health IT Quick-Stat #47. 2016 May; dashboard.healthit.gov/quickstats/pages/FIG-Hospital-EHR-Adoption.php.
- 2.Office of the National Coordinator for Health Information Technology. [Accessed April 4, 2017];Office-based Physician Electronic Health Record Adoption. 2016 Dec; https://dashboard.healthit.gov/quickstats/pages/physician-ehr-adoption-trends.php.
- 3.Lauer MS, Kiley JP, Mockrin SC, Mensah GA, Hoots WK, Patel Y, Cook NL, Patterson AP, Gibbons GH. National Heart, Lung, and Blood Institute (NHLBI) Strategic Visioning. Circulation. 2015;131:1106–1109. doi: 10.1161/CIRCULATIONAHA.115.015712. [DOI] [PubMed] [Google Scholar]
- 4.Roger VL, Boerwinkle E, Crapo JD, Douglas PS, Epstein JA, Granger CB, Greenland P, Kohane I, Psaty BM. Strategic transformation of population studies: recommendations of the working group on epidemiology and population sciences from the National Heart, Lung, and Blood Advisory Council and Board of External Experts. Am J Epidemiol. 2015;181:363–368. doi: 10.1093/aje/kwv011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hlatky MA, Douglas PS, Cook NL, Wells B, Benjamin EJ, Dickersin K, Goff DC, Hirsch AT, Hylek EM, Peterson ED, Roger VL, Selby JV, Udelson JE, Lauer MS. Future Directions for Cardiovascular Disease Comparative Effectiveness Research. J Am Coll Cardiol. 2012;60:569–580. doi: 10.1016/j.jacc.2011.12.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dzau VJ, McClellan MB, McGinnis JM, Burke SP, Coye MJ, Diaz A, Daschle TA, Frist WH, Gaines M, Hamburg MA, Henney JE, Kumanyika S, Leavitt MO, Parker RM, Sandy LG, Schaeffer LD, Steele GD, Thompson P, Zerhouni E. Vital Directions for Health and Health Care: Priorities From a National Academy of Medicine Initiative. JAMA. 2017;317:1461–1470. doi: 10.1001/jama.2017.1964. [DOI] [PubMed] [Google Scholar]
- 7.Maddox TM, Albert NM, Borden WB, Curtis LH, Ferguson TB, Kao DP, Marcus GM, Peterson ED, Redberg R, Rumsfeld JS, Shah ND, Tcheng JE American Heart Association Council on Quality of Care and Outcomes Research; Council on Cardiovascular Disease in the Young; Council on Clinical Cardiology; Council on Functional Genomics and Translational Biology; and Stroke Council. The Learning Healthcare System and Cardiovascular Care: A Scientific Statement From the American Heart Association. Circulation. 2017;135:e826–e857. doi: 10.1161/CIR.0000000000000480. [DOI] [PubMed] [Google Scholar]
- 8.Vasan RS, Benjamin EJ. The Future of Cardiovascular Epidemiology. Circulation. 2016;133:2626–2633. doi: 10.1161/CIRCULATIONAHA.116.023528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Solomon SD, Pfeffer MA. The Future of Clinical Trials in Cardiovascular Medicine. Circulation. 2016;133:2662–2670. doi: 10.1161/CIRCULATIONAHA.115.020723. [DOI] [PubMed] [Google Scholar]
- 10.Collins FS, Varmus H. A new initiative on precision medicine. N Engl J Med. 2015;372:793–795. doi: 10.1056/NEJMp1500523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Galanter WL, Applebaum A, Boddipalli V, Kho A, Lin M, Meltzer D, Roberts A, Trick B, Walton SM, Lambert BL. Migration of patients between five urban teaching hospitals in Chicago. J Med Syst. 2013;37:9930. doi: 10.1007/s10916-013-9930-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kho AN, Cashy JP, Jackson KL, Pah AR, Goel S, Boehnke J, Humphries JE, Kominers SD, Hota BN, Sims SA, Malin BA, French DD, Walunas TL, Meltzer DO, Kaleba EO, Jones RC, Galanter WL. Design and implementation of a privacy preserving electronic health record linkage tool in Chicago. J Am Med Inform Assoc. 2015;22:ocv038–ocv1080. doi: 10.1093/jamia/ocv038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bild DE, Bluemke DA, Burke GL, Detrano R, Diez Roux AV, Folsom AR, Greenland P, Jacobs DR, Jr, Kronmal R, Kiang L, Nelson JC, O’Leary D, Saad MF, Shea S, Szklo M, Tracy RP. Multi-Ethnic Study of Atherosclerosis: Objectives and Design. Am J Epidemiol. 2002;156:871–881. doi: 10.1093/aje/kwf113. [DOI] [PubMed] [Google Scholar]
- 14.CMS Virtual Research Data Center. [Accessed April 4, 2017];CMS Chronic Conditions Data Warehouse (CCW) CCW Condition Algorithms. https://www.ccwdata.org/cs/groups/public/documents/document/ccw_chronic_cond_algos.pdf. Published September 2015.
- 15.Goyal A, Norton CR, Thomas TN, Davis RL, Butler J, Ashok V, Zhao L, Vaccarino V, Wilson PWF. Predictors of Incident Heart Failure in a Large Insured Population: A One Million Person-Year Follow-Up Study. Circ Heart Fail. 2010;3:698–705. doi: 10.1161/CIRCHEARTFAILURE.110.938175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Spratt SE, Pereira K, Granger BB, Batch BC, Phelan M, Pencina M, Miranda ML, Boulware E, Lucas JE, Nelson CL, Neely B, Goldstein BA, Barth P, Richesson RL, Riley IL, Corsino L, Hinz ERM, Rusincovitch S, Green J, Barton AB, Group TDP, Kelley C, Hyland K, Tang M, Elliott A, Ruel E, Clark A, Mabrey M, Morrissey KL, Rao J, Hong B, Pierre-Louis M, Kelly K, Jelesoff N. Assessing electronic health record phenotypes against gold-standard diagnostic criteria for diabetes mellitus. J Am Med Inform Assoc. 2017;24:e121–e126. doi: 10.1093/jamia/ocw123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mentz RJ, Newby LK, Neely B, Lucas JE, Pokorney SD, Rao MP, Jackson LR, Grau-Sepulveda MV, Smerek MM, Barth P, Nelson CL, Pencina MJ, Shah BR. Assessment of Administrative Data to Identify Acute Myocardial Infarction in Electronic Health Records. J Am Coll Cardiol. 2016;67:2441–2442. doi: 10.1016/j.jacc.2016.03.511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.American Heart Association. [Accessed April 4, 2017];GWTG-Stroke ICD-9 Diagnosis Code Definitions. http://www.heart.org/idc/groups/heart-public/@wcm/@hcm/@gwtg/documents/downloadable/ucm_310115.pdf.
- 19.American Heart Association. [Accessed April 4, 2017];Get with the Guidelines -- Heart Failure ICD-9 Codes. https://www.heart.org/idc/groups/heart-public/@wcm/@hcm/@gwtg/documents/downloadable/ucm_309111.pdf.
- 20.Kokotailo RA, Hill MD. Coding of stroke and stroke risk factors using international classification of diseases, revisions 9 and 10. Stroke. 2005;36:1776–1781. doi: 10.1161/01.STR.0000174293.17959.a1. [DOI] [PubMed] [Google Scholar]
- 21. [Accessed April 4, 2017];MESA Manual of Operations. https://www.mesa-nhlbi.org/PublicDocs/MesaMOO/Appendix11_MESA_ClinicalEvents_MOP.pdf. Revised March 12, 2004.
- 22.Weiskopf NG, Hripcsak G, Swaminathan S. Defining and measuring completeness of electronic health records for secondary use. J Biomed Inform. 2013;46:830–836. doi: 10.1016/j.jbi.2013.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Weiskopf NG, Weng C. Methods and dimensions of electronic health record data quality assessment: enabling reuse for clinical research. J Am Med Inform Assoc. 2013;20:144–151. doi: 10.1136/amiajnl-2011-000681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bland JM, Altman DG. Applying the right statistics: analyses of measurement studies. Ultrasound Obstet Gynecol. 2003;22:85–93. doi: 10.1002/uog.122. [DOI] [PubMed] [Google Scholar]
- 25.Bland JM, Altman DG. Statistical methods for assessing agreement between two methods of clinical measurement. Lancet. 1986;327:307–310. doi: 10.1016/s0140-6736(86)90837-8. [DOI] [PubMed] [Google Scholar]
- 26.Rice K, Lumley T. Graphics and statistics for cardiology: comparing categorical and continuous variables. Heart. 2016;102:349–355. doi: 10.1136/heartjnl-2015-308104. [DOI] [PubMed] [Google Scholar]
- 27.Richesson RL, Hammond WE, Nahm M, Wixted D, Simon GE, Robinson JG, Bauck AE, Cifelli D, Smerek MM, Dickerson J, Laws RL, Madigan RA, Rusincovitch SA, Kluchar C, Califf RM. Electronic health records based phenotyping in next-generation clinical trials: a perspective from the NIH Health Care Systems Collaboratory. J Am Med Inform Assoc. 2013;20:e226–e231. doi: 10.1136/amiajnl-2013-001926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Curtis LH, Brown J, Platt R. Four health data networks illustrate the potential for a shared national multipurpose big-data network. Health Aff (Millwood) 2014;33:1178–1186. doi: 10.1377/hlthaff.2014.0121. [DOI] [PubMed] [Google Scholar]
- 29.Wei W-Q, Teixeira PL, Mo H, Cronin RM, Warner JL, Denny JC. Combining billing codes, clinical notes, and medications from electronic health records provides superior phenotyping performance. J Am Med Inform Assoc. 2016;23:e20–e27. doi: 10.1093/jamia/ocv130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Newton KM, Peissig PL, Kho AN, Bielinski SJ, Berg RL, Choudhary V, Basford M, Chute CG, Kullo IJ, Li R, Pacheco JA, Rasmussen LV, Spangler L, Denny JC. Validation of electronic medical record-based phenotyping algorithms: results and lessons learned from the eMERGE network. J Am Med Inform Assoc. 2013;20:e147–e154. doi: 10.1136/amiajnl-2012-000896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kirby JC, Speltz P, Rasmussen LV, Basford M, Gottesman O, Peissig PL, Pacheco JA, Tromp G, Pathak J, Carrell DS, Ellis SB, Lingren T, Thompson WK, Savova G, Haines J, Roden DM, Harris PA, Denny JC. PheKB: a catalog and workflow for creating electronic phenotype algorithms for transportability. J Am Med Inform Assoc. 2016;23:1046–1052. doi: 10.1093/jamia/ocv202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bielinski SJ, Pathak J, Carrell DS, Takahashi PY, Olson JE, Larson NB, Liu H, Sohn S, Wells QS, Denny JC, Rasmussen-Torvik LJ, Pacheco JA, Jackson KL, Lesnick TG, Gullerud RE, Decker PA, Pereira NL, Ryu E, Dart RA, Peissig P, Linneman JG, Jarvik GP, Larson EB, Bock JA, Tromp GC, de Andrade M, Roger VL. A Robust e-Epidemiology Tool in Phenotyping Heart Failure with Differentiation for Preserved and Reduced Ejection Fraction: the Electronic Medical Records and Genomics (eMERGE) Network. J of Cardiovasc Trans Res. 2015;8:475–483. doi: 10.1007/s12265-015-9644-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fort D, Weng C, Bakken S, Wilcox AB. Considerations for using research data to verify clinical data accuracy. AMIA Jt Summits Transl Sci Proc. 2014;2014:211–217. [PMC free article] [PubMed] [Google Scholar]
- 34.Hersh WR, Weiner MG, Embi PJ, Logan JR, Payne PRO, Bernstam EV, Lehmann HP, Hripcsak G, Hartzog TH, Cimino JJ, Saltz JH. Caveats for the use of operational electronic health record data in comparative effectiveness research. Med Care. 2013;51:S30–S37. doi: 10.1097/MLR.0b013e31829b1dbd. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Agarwal R. Implications of Blood Pressure Measurement Technique for Implementation of Systolic Blood Pressure Intervention Trial (SPRINT) J Am Heart Assoc. 2017;6:e004536. doi: 10.1161/JAHA.116.004536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hlatky MA, Ray RM, Burwen DR, Margolis KL, Johnson KC, Kucharska-Newton A, Manson JE, Robinson JG, Safford MM, Allison M, Assimes TL, Bavry AA, Berger J, Cooper-DeHoff RM, Heckbert SR, Li W, Liu S, Martin LW, Perez MV, Tindle HA, Winkelmayer WC, Stefanick ML. Use of Medicare data to identify coronary heart disease outcomes in the Women’s Health Initiative. Circ Cardiovas Qual Outcomes. 2014;7:157–162. doi: 10.1161/CIRCOUTCOMES.113.000373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Psaty BM, Delaney JA, Arnold AM, Curtis LH, Fitzpatrick AL, Heckbert SR, McKnight B, Ives D, Gottdiener JS, Kuller LH, Longstreth WTJ. Study of Cardiovascular Health Outcomes in the Era of Claims Data The Cardiovascular Health Study. Circulation. 2016;133:156–164. doi: 10.1161/CIRCULATIONAHA.115.018610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kucharska-Newton AM, Heiss G, Ni H, Stearns SC, Puccinelli-Ortega N, Wruck LM, Chambless L. Identification of Heart Failure Events in Medicare Claims: The Atherosclerosis Risk in Communities (ARIC) Study. J of Card Failure. 2016;22:48–55. doi: 10.1016/j.cardfail.2015.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kumamaru H, Judd SE, Curtis JR, Ramachandran R, Hardy C, Rhodes JD, Safford MM, Kissela BM, Howard G, Jalbert JJ, Brott TG, Setoguchi S. Validity of Claims-Based Stroke Algorithms in Contemporary Medicare Data Reasons for Geographic and Racial Differences in Stroke (REGARDS) Study. Circ Cardiovas Qual Outcomes. 2014;7:611–619. doi: 10.1161/CIRCOUTCOMES.113.000743/-/DC1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Muggah E, Graves E, Bennett C, Manuel DG. Ascertainment of chronic diseases using population health data: a comparison of health administrative data and patient self-report. BMC Public Health. 2013;13:c4226. doi: 10.1186/1471-2458-13-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Saczynski JS, Andrade SE, Harrold LR, Tjia J, Cutrona SL, Dodd KS, Goldberg RJ, Gurwitz JH. A systematic review of validated methods for identifying heart failure using administrative data. Pharmacoepidemiol Drug Saf. 2012;21:129–140. doi: 10.1002/pds.2313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Quach S, Blais C, Quan H. Administrative data have high variation in validity for recording heart failure. Can J Cardiol. 2010;26:306–312. doi: 10.1016/s0828-282x(10)70438-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Martin B-J, Chen G, Graham M, Quan H. Coding of obesity in administrative hospital discharge abstract data: accuracy and impact for future research studies. BMC Health Serv Res. 2014;14:70. doi: 10.1186/1472-6963-14-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Al Kazzi ES, Lau B, Li T, Schneider EB, Makary MA, Hutfless S. Differences in the Prevalence of Obesity, Smoking and Alcohol in the United States Nationwide Inpatient Sample and the Behavioral Risk Factor Surveillance System. PLoS ONE. 2015;10:e0140165–11. doi: 10.1371/journal.pone.0140165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.ADAPTABLE Investigators. [Accessed April 4, 2017];Aspirin Dosing: A Patient-Centric Trial Assessing Benefits and Long-term Effectiveness (ADAPTABLE) Study Protocol. http://pcornet.org/wp-content/uploads/2015/06/ADAPTABLE-Protocol-Final-Draft-6-4-15_for-post_06-26-.pdf. Published June 5, 2015.
- 46.Rubbo B, Fitzpatrick NK, Denaxas S, Daskalopoulou M, Yu N2, Patel RS, Hemingway H UK Biobank Follow-up and Outcomes Working Group. Use of electronic health records to ascertain, validate and phenotype acute myocardial infarction: A systematic review and recommendations. Int J Cardiol. 2015;187:705–11. doi: 10.1016/j.ijcard.2015.03.075. [DOI] [PubMed] [Google Scholar]
- 47.Thygesen K, Alpert JS, Jaffe AS, Simoons ML, Chaitman BR, White HD Joint ESC/ACCF/AHA/WHF Task Force for the Universal Definition of Myocardial Infarction. Third Universal Definition of Myocardial Infarction. Circulation. 2012;126:2020–2035. doi: 10.1161/CIR.0b013e31826e1058. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.