Abstract
Due to the dynamic, condition-dependent nature of brain activity, interest in estimating rapid functional connectivity (FC) changes that occur during resting-state functional magnetic resonance imaging (rs-fMRI) has recently soared. However, studying dynamic FC is methodologically challenging, due to the low signal-to-noise ratio of the blood oxygen level dependent (BOLD) signal in fMRI and the massive number of data points generated during the analysis. Thus, it is important to establish methods and summary measures that maximize reliability and the utility of dynamic FC to provide insight into brain function. In this study, we investigated the reliability of dynamic FC summary measures derived using three commonly used estimation methods - sliding window (SW), tapered sliding window (TSW), and dynamic conditional correlations (DCC) methods. We applied each of these techniques to two publicly available rs-fMRI test-retest data sets - the Multi-Modal MRI Reproducibility Resource (Kirby Data) and the Human Connectome Project (HCP Data). The reliability of two categories of dynamic FC summary measures were assessed, specifically basic summary statistics of the dynamic correlations and summary measures derived from recurring whole-brain patterns of FC (“brain states”). The results provide evidence that dynamic correlations are reliably detected in both test-retest data sets, and the DCC method outperforms SW methods in terms of the reliability of summary statistics. However, across all estimation methods, reliability of the brain state-derived measures was low. Notably, the results also show that the DCC-derived dynamic correlation variances are significantly more reliable than those derived using the non-parametric estimation methods. This is important, as the fluctuations of dynamic FC (i.e., its variance) has a strong potential to provide summary measures that can be used to find meaningful individual differences in dynamic FC. We therefore conclude that utilizing the variance of the dynamic connectivity is an important component in any dynamic FC-derived summary measure.
1 Introduction
The functional organization of the brain has a rich spatio-temporal structure that can be probed using functional connectivity (FC) measures. Defined as the undirected association between functional magnetic resonance imaging (fMRI) time series from two or more brain regions, FC has been shown to change with age (Betzel et al., 2014; Gu et al., 2015), training (Bassett et al., 2015, 2011), levels of consciousness (Hudson et al., 2014), and across various stages of sleep (Tagliazucchi and Laufs, 2014). Traditionally, FC has been assumed to be constant across a given experimental run. However, recent studies have begun to probe the temporal dynamics of FC on shorter timescales (i.e., seconds instead of entire runs lasting many minutes) (Hutchison et al., 2013a; Preti et al., 2016). Such rapid alterations in FC are thought to allow the brain to continuously sample various configurations of its functional repertoire (Sadaghiani et al., 2015; Preti et al., 2016). These studies of dynamic FC have also enabled the classification of whole-brain dynamic FC profiles into distinct “brain states”, defined as recurring whole-brain connectivity profiles that are reliably observed across subjects throughout the course of a resting state run (Calhoun et al., 2014). A common approach to determining the presence of such coherent brain states across subjects is to perform k-means clustering on the correlation matrices across time. Brain states can then be summarized as the patterns of connectivity at each centroid, and additional summary metrics such as the amount of time each subject spends in a given state can be computed. Using this definition of brain state, it has been shown that the patterns of connectivity describing each state are reliably observed across groups and individuals (Yang et al., 2014), while other characteristics such as the amount of time spent in specific states and the number of transitions between states vary with meaningful individual differences such as age (Hutchison and Morton, 2015; Marusak et al., 2017) or disease status (Damaraju et al., 2014; Rashid et al., 2014). However, this approach towards understanding what has recently been termed the “chronnectome” is still in its infancy (Calhoun et al., 2014).
A number of methodological issues have limited the interpretability of existing studies using dynamic connectivity. For instance, detecting reliable and neurally-relevant dynamics in FC is challenging when there are no external stimuli to model. Dynamic FC research generally relies upon the use of resting state fMRI (rs-fMRI) data and therefore, it is unclear whether the states that are identified accurately reflect underlying cognitive states. Another issue is that dynamic FC methods substantially increase the number of data points to consider initially (e.g., a T × d (time-by-region) input matrix becomes a d×d×T array). This is in contrast to statistical methods that reduce the dimensionality of the data. Also, the signal-to-noise ratio of the blood oxygen level dependent (BOLD) signal in rs-fMRI is low, and it is often unclear whether observed fluctuations in the temporal correlation between brain regions should be attributed to dynamic neural activity, non-neural biological signals (such as respiration or cardiac pulsation), or noise (Handwerker et al., 2012; Hlinka and Hadrava, 2015). Due to these methodological challenges, metrics of dynamic FC are sensitive to the method used to estimate them (Lindquist et al., 2014; Hlinka and Hadrava, 2015; Leonardi and Van De Ville, 2015), and uncertainty remains regarding the appropriate estimation method to use. An important concern moving forward is to establish methods that maximize the reliability of dynamic FC metrics, which in turn will enhance our ability to use individual variability in dynamic FC metrics to understand individual variability in behavior and cognitive function.
The most widely used method for detecting dynamic FC is the sliding window (SW) method, in which correlation matrices are computed over fixed-length, windowed segments of the fMRI time series. These time segments can be derived from individual voxels (Handwerker et al., 2012; Hutchison et al., 2013b; Leonardi and Van De Ville, 2015), averaged over pre-specified regions of interest (Chang and Glover, 2010), or estimated using data-driven methods such as independent component analysis (Allen et al., 2012a; Yaesoubi et al., 2015). Observations within the fixed-length window can be given equal weight as in the conventional SW method, or allowed to gradually enter and exit the window as it is shifted across time, a strategy that is used by the tapered sliding window (TSW) method (Allen et al., 2012a). Potential pitfalls of the family of SW methods include the use of arbitrarily chosen fixed-length windows, disregard of values outside of the windows, and an inability to handle abrupt changes in connectivity patterns.
Model-based multivariate volatility methods attempt to address these shortcomings through flexible modeling of dynamic correlations and variances. Widely used to forecast time-varying conditional correlations in financial time series, model-based multivariate volatility methods have consistently been shown to outperform SW methods (Hansen and Lunde, 2005). The dynamic conditional correlations (DCC) method is an example of a model-based multivariate volatility method that has recently been introduced to the neuroimaging field (Lindquist et al., 2014). Considered as one of the best multivariate generalized auto-regressive conditional heteroscedastic (GARCH) models (Engle, 2002), the DCC method effectively estimates all model parameters through quasi-maximum likelihood methods. Additionally, the asymptotic theory of the DCC model provides a mechanism for statistical inference that is not readily available when using other techniques for estimating dynamic correlations, though such mechanisms are currently under development (Kudela et al., 2017). In a previous study, simulations and analyses of experimental rs-fMRI data suggested that the DCC method achieved the best overall balance between sensitivity and specificity in detecting temporal changes in FC (Lindquist et al., 2014). Specifically, it was shown that the DCC method was less susceptible to noise-induced temporal variability in correlations compared to the SW method and other multivariate volatility methods.
The goal of this study was to identify estimation methods that provide accurate and reliable measures of various dynamic FC metrics. In particular, we compared the reliability of summary measures estimated using a family of SW methods (that represent the most commonly used dynamic FC estimation methods) and those estimated using the DCC method (that represents a more advanced model-based multivariate volatility method). We assessed the reliability of two types of dynamic FC summary measures: 1) basic summary statistics, specifically the mean and variance of dynamic FC across time, and 2) statistics derived from brain states, specifically the dwell time and number of change points between states. We compared the reliability of these methods using two publicly available rs-fMRI test-retest data sets: 1) the Multi-Modal MRI Reproducibility Resource (Kirby) data set (Landman et al., 2011), which used a well-established echo planar imaging (EPI) sequence with a repetition time (TR) of 2000 ms, and 2) the Human Connectome Project 500 Subjects Data Release (HCP) data set (Van Essen et al., 2013), which used a simultaneous multi-slice EPI sequence with a TR of 720 ms. These two data sets differ in terms of the acquisition parameters used and in the preprocessing steps performed to clean the data, with acquisition and processing parameters for the former representing well-established procedures used by many rs-fMRI researchers, and those for the latter representing cutting-edge procedures designed to optimize data quality. We hypothesized that the DCC-estimated dynamic FC summary measures would be more reliable than those estimated using the conventional SW and TSW methods, and that dynamic FC summary measures obtained using the HCP data would be more reliable than those obtained using the Kirby data.
2 Methods
2.1 Image Acquisition
2.1.1 Kirby Data
We used the Multi-Modal MRI Reproducibility Resource (Kirby) from the F.M. Kirby Research Center to evaluate the reliability of dynamic FC summary measures obtained using a typical-length, standard EPI sequence, which were cleaned using established preprocessing procedures. This resource is publicly available at http://www.nitrc.org/projects/multimodal. Please see Landman et al. (2011) for a detailed explanation of the entire acquisition protocol. Briefly, this resource includes data from 21 healthy adult participants who were scanned on a 3T Philips Achieva scanner. The scanner is designed to achieve 80 mT/m maximum gradient strength with body coil excitation and an eight channel phased array SENSitivity Encoding (SENSE) (Pruessmann et al., 1999) head-coil for reception. Participants completed two scanning sessions on the same day, between which participants briefly exited the scan room and a full repositioning of the participant, coils, blankets, and pads occurred prior to the second session. A T1-weighted (T1w) Magnetization-Prepared Rapid Acquisition Gradient Echo (MPRAGE) structural run was acquired during both sessions (acquisition time = 6 min, TR/TE/TI = 6.7/3.1/842 ms, resolution = 1×1×1.2 mm3, SENSE factor = 2, flip angle = 8°). A multi-slice SENSE-EPI pulse sequence (Stehling et al., 1991; Pruessmann et al., 1999) was used to acquire one rs-fMRI run during each session, where each run consisted of 210 volumes sampled every 2 s at 3-mm isotropic spatial resolution (acquisition time: 7 min, TE = 30 ms, SENSE acceleration factor = 2, flip angle = 75°, 37 axial slices collected sequentially with a 1-mm gap). Participants were instructed to rest comfortably while remaining as still as possible, and no other instruction was provided. We will refer to the first rs-fMRI run collected as session 1 and the second as session 2. One participant was excluded from data analyses due to excessive motion.
2.1.2 HCP Data
We used the 2014 Human Connectome Project 500 Parcellation+Timeseries+Netmats (HCP500-PTN) data release to evaluate the reliability of dynamic FC summary measures obtained using a larger data set of 523 healthy adults, sampled at a higher temporal frequency for a longer duration, and cleaned using cutting-edge preprocessing procedures. This resource is publicly available at http://humanconnectome.org. Please see Van Essen et al. (2013) for a detailed explanation of the entire acquisition protocol. Briefly, all HCP MRI data were acquired on a customized 3T Siemens connectome-Skyra 3T scanner, designed to achieve 100 mT/m gradient strength. Participants completed two scanning sessions on two separate days. A T1w MPRAGE structural run was acquired during each session (acquisition time = 7.6 min, TR/TE/TI = 2400/2.14/1000 ms, resolution = 0.7×0.7×0.7 mm3, SENSE factor = 2, flip angle = 8°). A simultaneous multi-slice pulse sequence with an acceleration factor of eight (Uğurbil et al., 2013) was used to acquire two rs-fMRI runs during each session, which consisted of 1200 volumes sampled every 0.72 seconds, at 2-mm isotropic spatial resolution (acquisition time: 14 min 24 sec, TE = 33.1 ms, flip angle = 52°, 72 axial slices). Participants were instructed to keep their eyes open and fixated on a cross hair on the screen, while remaining as still as possible. Within sessions, phase encoding directions for the two runs were alternated between right-to-left (RL) and left-to-right (LR) directions. Counterbalancing the order of the different phase-encoding acquisitions for the rs-fMRI runs across days was adopted on October 1, 2012 (RL followed by LR on Day 1; LR followed by RL on Day 2). Prior to that, rs-fMRI runs were acquired using the RL followed by LR order on both days. We limited our analyses to data from the 461 participants included in the HCP500-PTN release who completed the full rs-fMRI protocol. We will refer to the two runs collected during the first visit as sessions 1A and 1B and the two collected during the second visit as sessions 2A and 2B. Note that subjects did not exit the scanner between runs collected on the same day.
2.2 Image Processing
2.2.1 Kirby Data
SPM8 (Wellcome Trust Centre for Neuroimaging, London, United Kingdom) (Friston et al., 1994) and MATLAB (The Mathworks, Inc., Natick, MA) were used to preprocess the Kirby data. In order to allow the stabilization of magnetization, four volumes were discarded at acquisition, and an additional volume was discarded prior to preprocessing. Slice timing correction was performed using the slice acquired at the middle of the TR as a reference, and rigid body realignment parameters were estimated to adjust for head motion. Structural runs were registered to the first functional frame and spatially normalized to Montreal Neurological Institute (MNI) space using SPM8’s unified segmentation-normalization algorithm (Ashburner and Friston, 2005). The estimated rigid body and nonlinear spatial transformations were applied to the rs-fMRI data, which were then high pass filtered using a cutoff frequency of 0.01 Hz. Rs-fMRI data were then spatially smoothed using a 6-mm full-width-at-half-maximum Gaussian kernel (i.e., twice the nominal size of the rs-fMRI acquisition voxel).
The Group ICA of fMRI toolbox (GIFT) (http://mialab.mrn.org/software/gift; Medical Image Analysis Lab, Albuquerque, New Mexico) was used to estimate the number of independent components (ICs) in the data, to perform data reduction via principal component analysis (PCA) prior to independent component analysis (ICA), and then to perform group independent component analysis (GICA) (Calhoun et al., 2001) on the PCA-reduced data. Estimation of the number of ICs was guided by order selection using the minimum description length (MDL) criterion (Li et al., 2007). Across subjects and sessions, 56 was the maximum estimated number of ICs and 39 was the median. Prior to GICA, the image mean was removed from each time point for each session, and three steps of PCA were performed. Individual session data were first reduced to 112 principal components, and the reduced session data were then concatenated within subjects in the temporal direction and further reduced to 56 principal components. Finally, the data were concatenated across subjects and reduced to 39 principal components. The dimensionality of individual session PCA (i.e., 112) was chosen by doubling the estimated maximum IC number (i.e., 56), to ensure robust back-reconstruction (Allen et al., 2011, 2012b) of subject- and session-specific spatial maps and time courses from the group-level independent components. Using the ICASSO toolbox (Himberg et al., 2004), ICA was repeated on these 39 group-level principal components 10 times, utilizing the Infomax algorithm with random initial conditions (Bell and Sejnowski, 1995). ICASSO clustered the resulting 390 ICs across iterations using a group average-link hierarchical strategy, and 39 aggregate spatial maps were defined as the modes of the clusters. Subject-and session-specific spatial maps and time courses were generated from these aggregate ICs using the GICA3 algorithm, which is a method based on PCA compression and projection (Erhardt et al., 2011).
We compared the spatial distribution of each of the group-level, aggregate ICs to a publicly available set of 100 unthresholded t-maps of ICs estimated using rs-fMRI data collected from 405 healthy participants (Allen et al., 2012a). These t-maps have already been classified as resting state networks (RSNs) or noise by a group of experts, and the 50 components classified as RSNs have been organized into seven large functional groups: visual (Vis), auditory (Aud), somatomotor (SM), default mode (DMN), cognitive-control (CC), sub-cortical (SC) and cerebellar (Cb) networks. Henceforth, we refer to these as the Allen components (all 100) and the Allen RSNs (50 signal components). For each of the group-level spatial maps, we calculated the percent variance explained by the seven sets of Allen RSNs. The functional assignment of each Kirby component was determined by the set of Allen components that explained the most variance, and if the top two sets of Allen RSNs explained less than 50% of the variance in a Kirby component, the Kirby component was labeled as noise. Subject- and run-specific time series from the components then served as input for the dynamic FC analyses described below.
2.2.2 HCP Data
We used the preprocessed and artifact-removed rs-fMRI data as provided by the HCP500-PTN data release. The preprocessing and the artifact-removing procedures performed on the data are explained in detail elsewhere (Glasser et al., 2013; Smith et al., 2013; Griffanti et al., 2014; Salimi-Khorshidi et al., 2014), and briefly described below. Each run was minimally preprocessed (Glasser et al., 2013; Smith et al., 2013), and artifacts were removed using the Oxford Center for Functional MRI of the Brain’s (FMRIB) ICA-based X-noiseifier (ICA + FIX) procedure (Griffanti et al., 2014; Salimi-Khorshidi et al., 2014). At this point in the processing pipeline, rs-fMRI data from each run were represented as a time series of grayordinates, a combination of cortical surface vertices and subcortical standard-space voxels (Glasser et al., 2013). Each run was temporally demeaned and variance normalized (Beckmann and Smith, 2004). All four runs for 461 subjects were fed into MELODIC’s Incremental Group-Principal Component Analysis (MIGP) algorithm, which estimated the top 4500 weighted spatial eigenvectors. GICA was applied to the output of MIGP using FSL’s MELODIC tool (Beckmann and Smith, 2004) using five different dimensions (i.e., number of independent components: 25, 50, 100, 200, 300). In this study, we used the data corresponding to dimension d = 50 to perform further dynamic FC analysis, which was closest to the dimension used for the Kirby data (i.e., 39). Dual-regression was then used to map group-level spatial maps of the components onto each subject’s time series data (Filippini et al., 2009). For dual-regression, the time series of each of the runs were first concatenated within subjects in the following order: Day 1 LR, Day 1 RL, Day 2 LR, Day 2 RL (http://www.mail-archive.com/hcp-users@humanconnectome.org/msg02054.html; S. M. Smith, personal communication, 24 October 2015). Then the full set of group-level maps were used as spatial regressors against each subject’s full time series (4800 volumes) to obtain a single representative time series per IC. The functional assignment of each component was determined as described above (refer to section 2.2.1) using the Allen RSNs. Subject- and run-specific time series from the components then served as input for the dynamic FC analyses described below.
2.3 Computing Dynamic Functional Connectivity
Dynamic FC between multiple regions of the brain are often represented using either a covariance or correlation matrix, that represent the relationship between different brain regions or components. In this study, the elements of the correlation matrix were estimated using the SW, TSW and DCC methods.
2.3.1 Sliding Window Methods
Perhaps the simplest approach for estimating the elements of the covariance/correlation matrix is to use the SW method. Here, a time window of fixed length w is selected, and data points within that window are used to calculate the correlation coefficients. The window is thereafter shifted across time and a new correlation coefficient is computed for each time point. The general form of the estimate of the SW correlation is given by
| (1) |
where , i = 1, 2 represents the estimated time-varying mean.
The SW method gives equal weight to all observations within w time points in the past and 0 weight to all others. Hence, the removal of a highly influential outlying data point will cause a sudden change in the dynamic correlation that may be mistaken for an important aspect of brain connectivity. To circumvent this issue, Allen and colleagues (Allen et al., 2012a) suggested the use of a TSW method. Here, the sliding window (assumed to have width = 22 TRs) is convolved with a Gaussian kernel (σ = 3 TRs). This allows points to gradually enter and exit the window as it moves across time. It should be noted that t is defined to be the middle of the subsequent window, thus giving equal weight to future and past values.
Thus, both SW- and TSW-derived correlations can be seen as special cases of the following formula:
| (2) |
where wts is the weight when considering the contribution of point s in calculating the dynamic correlation for index t (at clock time t × TR from the start of the run). For the SW method, wts = 1 for t−w−1 ≤ s ≤ t−1 and wts = 0 otherwise. In general, however, the weights could be determined by any kernel distribution (Wand and Jones, 1994).
The window-length parameter needs to be carefully chosen to avoid introducing spurious fluctuations (Shakil et al., 2016). For the Kirby data, we used a window length of 30 TRs, which is the suggested optimal window-length for rs-fMRI data collected using a standard EPI sequence with a sampling frequency of 2 seconds (Leonardi and Van De Ville, 2015). For the HCP data, we investigated sliding window lengths of 15, 30, 45, 60, 75, 90, 105, and 120 TRs because it was unclear how the increased sampling frequency used to collect the HCP data would influence what is considered the optimal window length. However, we only show results for 30, 60, and 120 TRs (hereon referred to as SW30, SW60 and SW120, respectively) due to the consistency of the results. These three window lengths allowed us to compare the reliability of dynamic FC methods using a window consisting of a similar number of volumes as the Kirby data (SW30; ~ 22 s), a window covering a similar amount of time (SW120; ~ 86 s), and an intermediate window length (SW60; ~ 43 s).
2.3.2 DCC Method
The DCC model (Engle, 2002) for estimating conditional variances and correlations has become increasingly popular in the finance literature over the past decade. Before introducing DCC, we must first discuss generalized autoregressive conditional heteroscedastic (GARCH) processes (Engle, 1982; Bollerslev, 1986), which are often used to model volatility in univariate time series. They provide flexible models for the variance in much the same manner that commonly used time series models, such as Autoregressive (AR) and Autoregressive Moving Average (ARMA), model the mean. GARCH models express the conditional variance of a single time series at time t as a linear combination of past values of the conditional variance and of the squared process itself. To illustrate, let us assume that we are observing a univariate process
| (3) |
where εt is a N(0, 1) random variable and σt represents the time-varying conditional variance term we seek to model. In a GARCH(1,1) process the conditional variance is expressed as
| (4) |
where ω > 0, α, β ≥ 0 and α + β < 1. Here the term α controls the impact of past values of the time series on the variance and β controls the impact of past values of the conditional variance on its present value.
While many multivariate GARCH models can be used to estimate dynamic correlations, it has been shown that the DCC model outperforms the rest (Engle, 2002). To illustrate the DCC method, assume yt is a bivariate mean zero time series with conditional covariance matrix Σt. The first order form of DCC can be expressed as follows:
| (5) |
| (6) |
| (7) |
| (8) |
| (9) |
| (10) |
The DCC algorithm consists of two steps. In the first step (Eqs. 5–7), univariate GARCH(1,1) models are fit (Eq. 5) to each of the two univariate time series that make up yt, and used to compute standardized residuals (Eq. 7). In the second step (Eqs. 8–10), an exponentially weighted moving average (EWMA) window is applied to the standardized residuals to compute a non-normalized version of the time-varying correlation matrix Rt (Eq. 8). Here represents the unconditional covariance matrix of εt, which is estimated as:
| (11) |
and (θ1, θ2) are non-negative scalars satisfying 0 < θ1 + θ2 < 1. Eq. 9 is simply a rescaling step to ensure a proper correlation matrix is created, while Eq. 10 computes the time-varying covariance matrix.
In contrast to the standard implementation of SW and TSW methods, where observations n-steps forward in time are given the same weight as observations n-steps backwards in time, the DCC estimates the conditional correlation. Specifically, the current estimate of conditional correlation is updated using a linear combination of past estimates of the conditional correlation and current observations. In this respect, the model shares similarities with the time series models commonly used to describe fMRI noise, such as the AR and ARMA models (Purdon and Weisskoff, 1998), where the current noise estimate is influenced by its past values, not its future values. In resting state experiments the exact timing of the dynamic correlation is typically not meaningful in itself. Therefore in this setting, the manner in which the window is defined is unimportant and will simply result in a time shift of half the window length size. However, we still believe that the conditional correlation should be used because it provides a more suitable estimate of the correlation at a specific time point, which can be critical particularly if it is important to link the dynamic correlation to the timing of a specific task or emotion.
The model parameters (ω1, α1, β1, ω2, α2, β2, θ1, θ2) can be estimated using a two-stage approach. In the first stage, time-varying variances are estimated for each time series. In the second stage, the standardized residuals are used to estimate the dynamic correlations {Rt}. This two-stage approach has been shown to provide estimates that are consistent and asymptotically normal with a variance that can be computed using the generalized method of moments approach (Engle and Sheppard, 2001; Engle, 2002).
The description above assumes a bivariate time series. However, in practice yt will often be N-variate with N > 2. There are two ways to deal with such data. First, it is possible to fit an N-variate version of DCC. A second option is to perform a “massive bi-variate analysis” where the bivariate connection between each pair of time courses is fit separately. We opted for the latter approach, as it provides increased flexibility (i.e., more variable parameters) at the cost of increased computation time. Also note that like AR-processes, DCC can be defined to incorporate longer lags. However, in this work we limit ourselves to the first order variant.
2.4 Summarizing Dynamic Functional Connectivity
As previously mentioned, estimating dynamic FC initially increases the number of observations to consider. To illustrate, suppose we have data from d regions measured at T time points, for a total of d × T data points. After computing dynamic correlations, the initial output consists of T separate d × d correlation matrices that together represent time varying correlations. However, as each matrix is symmetric, there are unique observations only in the lower triangular portion of the matrix, and our final output consists of a total of d(d − 1)/2 × T data points. Nonetheless, rather than providing data reduction, the analysis increases the total number of available data points. For this reason, there is a need to identify ways to meaningfully and reliably summarize this information.
2.4.1 Mean and Variance of Dynamic Functional Connectivity
We explored two basic summary statistics of pairwise dynamic FC: the dynamic FC mean and variance. The mean is presumed to give roughly equivalent information as the standard sample correlation coefficient, while the variance can be used to more directly assess the dynamics of FC. If an edge is involved in frequent state-changes (i.e., exhibits greater FC dynamics), it should exhibit consistently higher variation in correlation strength across time when compared to edges whose FC remains more static throughout an experimental run. Hence, we propose that the degree of variability should reasonably be included in any summary of dynamic FC.
2.4.2 Brain States
Another emerging method for summarizing dynamic FC is the classification of brain states, or recurring whole-brain patterns of FC that appear repeatedly across time and subjects (Calhoun et al., 2014). Following the method of Allen and colleagues (Allen et al., 2012a), we used k-means clustering to estimate recurring brain states across subjects, separately for each run within a session. We then compared the results across runs and sessions to assess reliability. First, we reorganized the lower triangular portion of each subject’s d × d × T dynamic correlation data into a matrix with dimensions (d(d − 1)/2) × T, where d is the number of nodes and T is the number of time points. Then we concatenated the data from all subjects into a matrix with dimensions (d(d − 1)/2) and (T × N), where N is the number of subjects. Finally, we applied k-means clustering, where each of the resulting cluster centroids represented a recurring brain state.
The number of clusters was chosen based on computing the within-group sum of squares for each candidate number of clusters k = 1,….10 and picking the ‘elbow’ in the plot (the point at which the slope of the curve leveled off) (Everitt et al., 2001). K-means clustering was repeated 50 times, using random initialization of centroid positions, in order to increase the chance of escaping local minima. Additional summary measures, such as the amount of time each subject spends in each state (i.e., dwell time) and the number of transitions from one brain state to another (i.e., number of change points), were computed and assessed for reliability across runs and sessions.
2.5 Evaluating the Reliability of Dynamic Functional Connectivity Methods
2.5.1 Mean and Variance of Dynamic Functional Connectivity
The primary goal of this work was to investigate the test-retest reliability of dynamic FC summary statistics computed using three different estimation methods: SW, TSW, and DCC. We assessed the reliability of basic summary statistics (dynamic FC mean and variance) using both the intra-class correlation coefficient (ICC) (Shrout and Fleiss, 1979) and the image intra-class correlation (I2C2) (Shou et al., 2013), which is a generalization of the ICC to images. Specifically, we used ICC to assess the reliability of individual elements (i.e., edges) of mean and variance matrices, and I2C2 to assess the omnibus reliability of the mean and variance of dynamic FC across the brain. The omnibus measure of reliability was computed to provide a single value that indicates the degree of reliability across the entire brain.
The ICC is defined as follows:
| (12) |
where denotes the between-subject variance, the within-subject variance, and the variance of the observed data. To interpret the results, we use the conventions from Cicchetti (1994), where an ICC-score less than 0.40 is poor; between 0.40 and 0.59 is fair; between 0.60 and 0.74 is good; and between 0.75 and 1.00 is excellent.
Based on the classical image measurement error (CIME) (Carroll et al., 2006), the I2C2 coefficient can be defined as follows:
| (13) |
where KX is the within-subject covariance, KU is the covariance of the replication error, and KW = KX+KU is the covariance of the observed data. Using method of moments estimators, calculating I2C2 is both quick and scalable. In theory, both the ICC and I2C2 produce values between 0 and 1, where 0 indicates exact independence of the measurements and 1 indicates perfect reliability. However, due to the manner in which they are estimated, both of these measures could potentially take negative values, which are interpreted as indicating low reliability. We calculated I2C2 and ICC for the summary measures described above across Kirby sessions 1 and 2 and across HCP sessions 1A, 1B, 2A, and 2B.
2.5.2 Brain States
Brain states were matched across runs, sessions, and dynamic FC methods by maximizing their spatial correlation. After matching, we evaluated the reliability of brain states derived from each estimation method by calculating the spatial correlation of corresponding brain states across runs and sessions for each method. We also used the ICC to quantify the inter-run reliability of the estimated dwell time and number of change points for each estimation method.
3 Results
3.1 Mean and Variance of Dynamic Functional Connectivity
3.1.1 Kirby Data
We first assessed the reliability of the mean and variance of estimated dynamic correlations by applying SW, TSW and DCC methods to the Kirby data.
The reliability of dynamic correlation means was highly consistent across all estimation methods
We computed the I2C2 score for the mean of dynamic correlations across all pairs of components, which produced an omnibus reliability measure across the brain. As can be seen by the overlapping confidence intervals presented in the left panel of Figure 1A, the I2C2 of dynamic correlation means was similar across all estimation methods (95% confidence intervals (CIs) for SW, TSW and DCC methods were [0.51, 0.65], [0.50, 0.64], and [0.51, 0.62] respectively). For comparison, the 95% CI for the static correlation was [0.52, 0.66].
Figure 1. Reliability of dynamic correlation means and variances from the Kirby data.
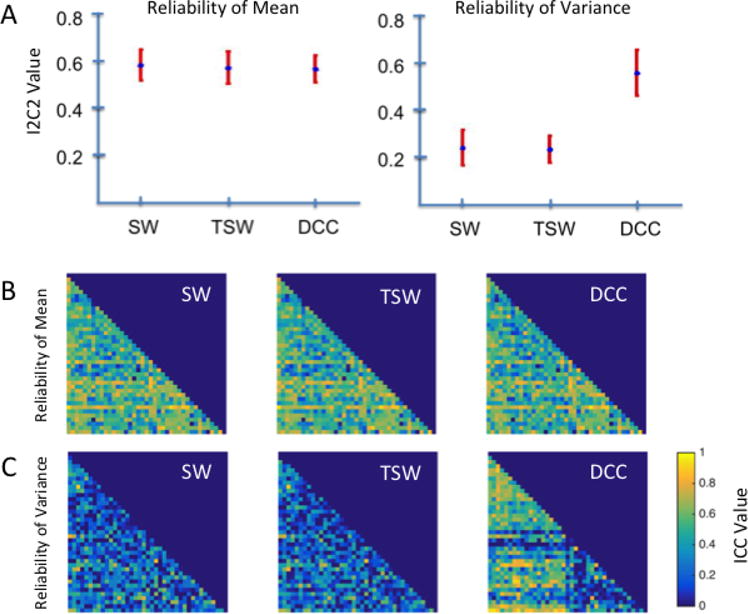
A) Omnibus reliability of dynamic correlation means and variances across all component pairs, or edges, for sliding windows (SW), tapered sliding windows (TSW) and dynamic conditional correlations (DCC) methods, as measured by the image intra-class correlation (I2C2). The mean I2C2 values across components are represented by blue dots, and the 95% confidence interval (CI) is represented by red bars. B) Edge-wise reliability of dynamic correlation means as measured using the intra-class correlation (ICC). C) Edge-wise reliability of dynamic correlation variances as measured using the ICC. Dynamic correlation means were similarly reliable across estimation methods using both omnibus and edge-wise reliability measures. In contrast, DCC-derived variances were more reliable than SW- and TSW-derived variances.
To investigate how the reliability of dynamic correlation means varied across the brain, we additionally computed the ICC for the mean of dynamic correlations between each pair of components (i.e., each edge) across participants. Figure 1B illustrates the ICC matrices for the correlation means between component pairs for all three estimation methods. Consistent with the omnibus I2C2 findings, ICC matrices for the correlation means between component pairs show similar ICC values and patterns across all estimation methods. For all of the methods, the majority of edges fall in the fair-to-good range; see Table 1.
Table 1.
Summary of the ICC results for the Kirby data. For each method (SW. TSW, DCC, and static correlation), and statistic (mean or variance) we show the proportion of edges whose reliability falls in the poor, fair, good, and excellent range. We repeat this for all edges, as well as for only edges between two signal nodes. Note the static correlation consists of a single metric that is comparable to the mean of the other methods
| Method | Statistic | Edges | Poor | Fair | Good | Excellent |
|---|---|---|---|---|---|---|
|
| ||||||
| SW | Mean | All | 0.2200 | 0.3900 | 0.3495 | 0.0405 |
| Mean | S-S | 0.2476 | 0.4952 | 0.2429 | 0.0143 | |
| Var | All | 0.8084 | 0.1417 | 0.0445 | 0.0054 | |
| Var | S-S | 0.8714 | 0.1048 | 0.0143 | 0.0095 | |
|
| ||||||
| TSW | Mean | All | 0.2389 | 0.3981 | 0.3293 | 0.0337 |
| Mean | S-S | 0.2762 | 0.4952 | 0.2238 | 0.0048 | |
| Var | All | 0.7949 | 0.1619 | 0.0391 | 0.0040 | |
| Var | S-S | 0.8476 | 0.1333 | 0.0143 | 0.0048 | |
|
| ||||||
| DCC | Mean | All | 0.2645 | 0.3914 | 0.3104 | 0.0337 |
| Mean | S-S | 0.3476 | 0.4381 | 0.2095 | 0.0048 | |
| Var | All | 0.4345 | 0.2942 | 0.2159 | 0.0553 | |
| Var | S-S | 0.2190 | 0.4190 | 0.3381 | 0.0238 | |
|
| ||||||
| Static | All | 0.2024 | 0.4116 | 0.3401 | 0.0459 | |
| S-S | 0.2095 | 0.5286 | 0.2524 | 0.0095 | ||
Variances of dynamic correlations estimated using the DCC method displayed the highest reliability
We computed the I2C2 score for the variance of dynamic correlations across all pairs of components, which produced an omnibus reliability measure across the brain. In contrast to that of the dynamic correlation means, the I2C2 of dynamic correlation variances differed substantially across the estimation methods. As illustrated in the right panel of Figure 1A, the variance of DCC-estimated dynamic correlations was significantly more reliable across the brain (95% CI: [0.43, 0.63]) than the variance of dynamic correlations estimated using the SW (95% CI: [0.17, 0.32]) and the TSW methods (95% CI: [0.18, 0.29]).
Similarly, as shown in Figure 1C, the ICC matrix for the variance of DCC-estimated dynamic correlations is distinct from the those for the correlation variances estimated using the SW and TSW methods. Specifically, as seen in Table 1, a majority of the DCC-estimated edge variances fall in the fair-to-excellent range. In contrast, roughly 80% of the SW- and TSW-estimated edges fall in the poor range.
Reliability of edges connecting signal components were higher for DCC-estimated FC measures
While the variances of dynamic correlations estimated using the DCC method displayed the highest overall reliability, it was also observed that certain edges still displayed low ICC values (Figure 1C, right panel). Such observed variability in the reliability across edges for dynamic correlation variances led us to further investigate the relationship between dynamic correlation variances and the degree of reliability as measured using ICC. We therefore plotted the dynamic correlation variances for each edge for session 1 against the ICC of the dynamic correlation variance for that edge estimated using the SW, TSW and DCC methods (Figure 2A). In this figure, each point represents a single edge. Specifically, the red dots indicate edges between two signal components (identified through matching with the Allen RSNs), while blue dots represent edges between either two noise components or between a noise and a signal component. Interestingly, Figure 2A shows that for all three estimation methods, the edges between two signal components display higher variance as compared to edges involving at least one noise component. Strikingly, the degree of this separation between the signal-signal edges and the signal-noise/noise-noise edges was greatest for the DCC method, mainly because the dynamic correlation variances for edges involving a non-signal component appeared to shrink toward zero using the DCC method. We also related the dynamic correlation means with the correlation variances of each edge. We found that the correlation variances between the signal-signal edges increased as the absolute value of the correlation means decreased for all estimation methods (Figure 2B). Similar to our findings describing the relationship between dynamic correlation variances and reliability of each edge (ICC), the degree of separation between signal-signal edges and edges involving at least one noise component was greatest for the DCC method. When focusing solely on signal-signal edges, according to Table 1, roughly 80% of DCC edges fall in fair-to-excellent range. In contrast, for SW and TSW roughly 85% of edges fall in the poor range.
Figure 2. Comparison of dynamic functional connectivity involving signal and noise components from the Kirby data.
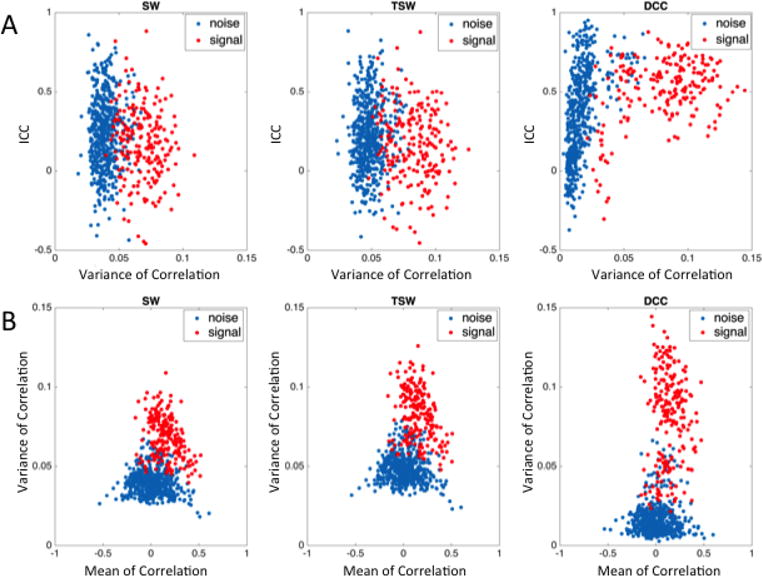
A) The relationship between variance of dynamic functional connectivity (FC) of each edge and reliability of that variance estimated using SW, TSW, and DCC methods. B) The relationship between dynamic correlation means and variances for each edge. For both A and B, each point represents a single edge, where red dots indicate edges composed of two signal components and blue dots indicate edges that contain at least one noise component. Compared to dynamic correlation variances derived using the SW and TSW methods, DCC-derived correlation variances for edges involving a noise component appears to shrink more towards zero, thus creating greater separation between signal-signal edges and all other edges. Additionally, for all estimation methods, the variances of dynamic correlations between signal components increased as the absolute value of the dynamic correlation means between signal components decreased.
We further investigated which functional edges were reliably more variable by visualizing edge variances that were averaged across Kirby subjects for each session. Figures 3A–C show the SW-, TSW-, and DCC-estimated correlation variances of each edge and session, with components sorted according to their classification of signal or noise. Again, we see that the separation between signal-signal edges and edges including at least one noise component were enhanced using the DCC method, as compared to SW and TSW methods. Figure 3D focuses on DCC-estimated correlation variances for signal-signal edges only, with components sorted according to their assignment to one of seven functional domains based on the Allen labels. Within both sessions, time-dependent edges between visual components (color-coded as pink) and both cognitive control (olive green) and default mode (grey) components appeared to be particularly variable, with variance values above 0.12. In contrast, edges involving the cerebellum (blue) and subcortical structures (light green) showed very little volatility, with variance values below 0.08.
Figure 3. Edge variances averaged across subjects for each Kirby session and method.
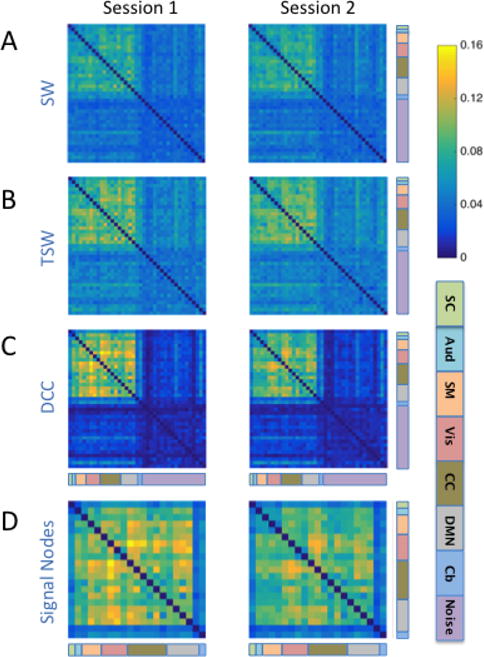
The variances of A) SW-, B) TSW-, and C) DCC-derived dynamic correlations for each edge averaged over all 20 subjects for each session. Note that dynamic FC variances are higher for signal-signal edges than for edges involving at least one noise component for all methods. D) DCC-derived dynamic FC variance of signal-signal edges. The functional label assigned to each signal node is indicated using the color code at the bottom right of the figure. [SC: subcortical (mint green); Aud: auditory (aqua); SM: somatomotor (orange); Vis: visual (pink); CC: cognitive control (olive green); DMN: default mode network (grey); Cb: cerebellum (blue); Noise: light purple]. Within both sessions, time-dependent edges between Vis components and both CC and DMN components appeared to be particularly variable (variance values above 0.12). In contrast, edges involving the cerebellum (blue) and sub-cortical structures (light green) showed very little volatility (variance values below 0.08).
3.1.2 HCP Data
The results for the Kirby and HCP data sets were highly consistent. One important difference between the Kirby and HCP data sets is the higher temporal sampling frequency with which the HCP data were collected. Due to the cutting-edge nature of HCP data acquisition and processing approaches, little information exists on how such high sampling frequencies impact the optimal window length for SW methods. Thus, we compared the reliability of dynamic correlation means and variances estimated using the SW method with varying window lengths (30, 60, and 120 TRs are presented here, though we fit lengths ranging from 15 to 120 TRs), as well as those estimated using the DCC method. Due to the almost identical reliability results observed between the SW and TSW methods, the results for the TSW-method are not presented.
The reliability of dynamic correlation means was highly consistent across all estimation methods
Our omnibus reliability findings for the HCP data were highly consistent with those observed for the Kirby data. As can be seen in the left panel of Figure 4A, the I2C2 for dynamic correlation means was similar across all methods, where 95% CIs for the SW30, SW60, SW120, and DCC methods were [0.45, 0.48], [0.45, 0.48], [0.44, 047], and [0.44, 0.47] respectively. For comparison purposes, the 95% CI for the static correlation was [0.454, 0.483].
Figure 4. Reliability of dynamic correlation means and variances from the HCP data.
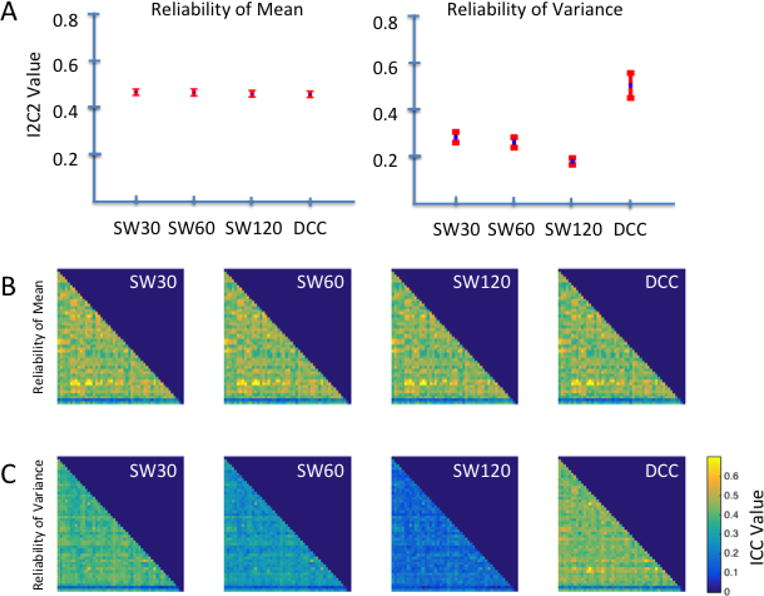
A) Omnibus reliability of dynamic correlation means and variances across all components pairs obtained using SW methods with varying window lengths of 30, 60, and 120 TRs (SW30, SW 60, and SW120 respectively) and the DCC method. Omnibus reliability is measured using I2C2; the mean I2C2 values across components for each method are represented by blue dots, and the 95% CIs are represented by red bars. B) Edge-wise reliability of dynamic correlation means as measured using the ICC. C) Edge-wise reliability of dynamic correlation variances as measured using the ICC. Dynamic correlation means were similarly reliable across estimation methods using both omnibus and edge-wise reliability measures. In contrast, DCC-derived variances were more reliable than those derived using the SW methods.
To investigate how the reliability of dynamic correlation means varied across the brain, we additionally computed the ICC for the mean of dynamic correlations between each pair of components across participants. Consistent with the omnibus I2C2 findings in Figure 4A, we observed that ICC matrices for the mean correlation between component pairs show similar patterns of reliability across edges, regardless of the method used to estimate dynamic correlations (Figure 4B). Additionally, most edges show a similar level of reliability, with the exception of edges involving components 42 and 49. The mean dynamic correlations for edges involving component 42 were more reliable than most edges, while those for edges involving component 49 were less reliable than most. Based on the Allen labels these components were classified as cerebellum and noise, respectively. The average ICC for SW30, SW60, SW120, and DCC derived edge means were 0.45 (sd: 0.09), 0.44 (sd: 0.09), 0.44 (sd: 0.09), and 0.43 (sd:0.09) respectively. According to Table 2, for all methods roughly 70 – 75% of all edges fall in the fair-to-good range.
Table 2.
Summary of the ICC results for the HCP data. For each method (SW. TSW, DCC, and static correlation), and statistic (mean or variance) we show the proportion of edges whose reliability falls in the poor, fair, good, and excellent range. We repeat this for all edges, as well as for only edges between two signal nodes. Note the static correlation consists of a single metric that is comparable to the mean of the other methods.
| Method | Statistic | Edges | Poor | Fair | Good | Excellent |
|---|---|---|---|---|---|---|
|
| ||||||
| SW30 | Mean | All | 0.2286 | 0.7388 | 0.0327 | 0 |
| Mean | S-S | 0.1744 | 0.7987 | 0.0269 | 0 | |
| Var | All | 0.6857 | 0.3135 | 0.0008 | 0 | |
| Var | S-S | 0.6641 | 0.3346 | 0.0013 | 0 | |
|
| ||||||
| SW60 | Mean | All | 0.2465 | 0.7249 | 0.0286 | 0 |
| Mean | S-S | 0.1987 | 0.7782 | 0.0231 | 0 | |
| Var | All | 0.9935 | 0.0065 | 0 | 0 | |
| Var | S-S | 0.9910 | 0.0090 | 0 | 0 | |
|
| ||||||
| SW120 | Mean | All | 0.2784 | 0.6939 | 0.0278 | 0 |
| Mean | S-S | 0.2308 | 0.7462 | 0.0231 | 0 | |
| Var | All | 1 | 0 | 0 | 0 | |
| Var | S-S | 1 | 0 | 0 | 0 | |
|
| ||||||
| DCC | Mean | All | 0.2776 | 0.6971 | 0.0253 | 0 |
| Mean | S-S | 0.2231 | 0.7564 | 0.0205 | 0 | |
| Var | All | 0.2416 | 0.7510 | 0.0073 | 0 | |
| Var | S-S | 0.1756 | 0.8167 | 0.0077 | 0 | |
|
| ||||||
| Static | All | 0.2767 | 0.6955 | 0.0278 | 0 | |
| S-S | 0.2282 | 0.7487 | 0.0231 | 0 | ||
Variances of dynamic correlations estimated using the DCC method displayed the highest reliability
In contrast to that of the dynamic correlation means, the I2C2 of dynamic correlation variances differed substantially among estimation methods. As illustrated in the right panel of Figure 4A, the variance of dynamic correlations estimated using the DCC method was significantly more reliable (95% CI: [.44, .55]) than the SW-derived dynamic correlation variances, which decayed as the window length increased (95% CIs: [0.25, 0.30], [0.23, 0.27], and [0.16, 0.19] for SW30, SW60, and SW120, respectively).
Similarly, as shown in Figure 4C, the ICC matrix for DCC-derived edge variances were visually distinct from the ICC matrices for SW-derived edge variances. Overall, for DCC-estimated edge variances 75% of all ICC values fall in the fair range. In contrast, for SW30 70% were in the poor range, while for SW60 and SW120 almost all values were in the poor range. The average ICC for DCC-estimated edge variance was 0.43 (sd:0.07), compared to 0.37 (sd: 0.07) for SW30, 0.27 (sd: 0.06) for SW60, and 0.17 (sd: 0.04) for SW120. Additionally, the inverse relationship between sliding window length and reliability of dynamic correlation variance appears to be fairly consistent across the brain, as is apparent from the gradual darkening of the three ICC matrices for SW-estimated variances moving from left to right in Figure 4C.
Reliability of edges connecting signal components were higher for DCC-estimated FC measures
To further probe edge variance reliability patterns in the HCP data, we plotted the dynamic correlation variance for each edge averaged across all sessions against the ICC of the dynamic correlation variance for that edge using the SW30, SW60, SW120, and DCC methods (Figure 5A). In this figure, each point represents a single edge. Red dots indicate edges between two signal components (identified through matching with the Allen RSNs), while blue dots represent edges between either two noise components or between a noise and a signal component. Similar to the Kirby data findings, dynamic correlation variances for signal-noise/noise-noise edges appear to shrink more towards zero when using the DCC method compared to the SW methods. Notably, compared to the Kirby data there is a higher presence of blue dots embedded in the cluster of red signal-signal edges. This may indicate that we were overly aggressive in labeling components as noise. In general, the percent variance explained by the Allen RSNs was lower for HCP components compared to Kirby components, which may be due to spatial discrepancies between the HCP greyordinate data mapped back into volume space and the Allen components.
Figure 5. Comparison of the dynamic correlation means and variances of each edge from the HCP data.
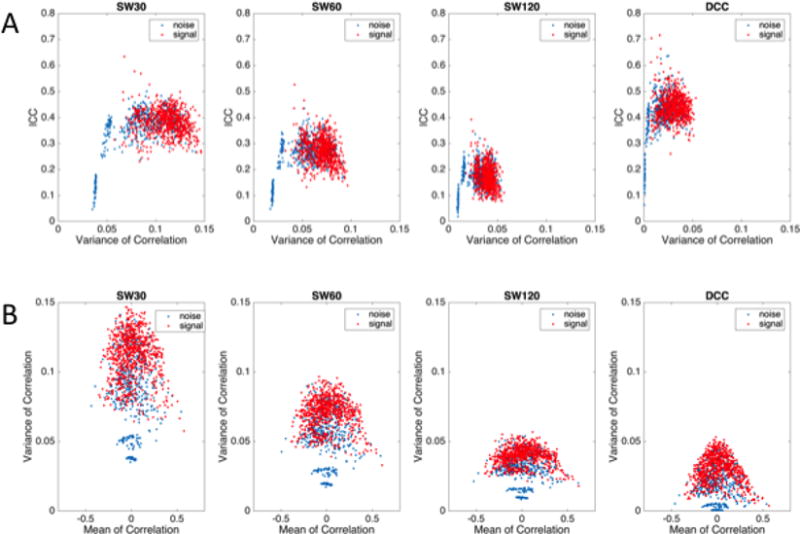
A) The relationship between variance of dynamic functional connectivity (FC) of each edge and reliability of that variance estimated using SW30, SW60, SW120, and DCC methods. B) The relationship between dynamic correlation means and variances for each edge. For both A and B, each point represents a single edge, where red dots indicate edges composed of two signal components and blue dots indicate edges that contain at least one noise component.Compared to dynamic correlation variances derived using the SW methods, DCC-derived correlation variances for edges involving a noise component appears to shrink more towards zero. In addition, for all estimation methods, the variances of dynamic correlations between signal components increased as the absolute value of the dynamic correlation means between signal components decreased.
Figure 5B shows the mean dynamic correlation plotted against the variance of each edge. In these plots, each point represents one edge and different colors are used to discriminate between signal-signal and signal-noise/noise-noise edges. Similar to the Kirby data, across all estimation methods, the dynamic correlation variances decrease as the absolute value of the dynamic correlation means increase.
We further investigated which functional edges were reliably more variable by visualizing DCC-derived edge variances averaged across subjects for Session 1A, 1B, 2A, and 2B separately, with components sorted by their classification based on the Allen RSNs (Figure 6). Again, the results show a remarkable consistency in edge variances across runs. In particular, note the heightened variation between visual, cognitive control, and DMN components, which is consistent with the edge variance patterns observed in the Kirby data. Despite the FIX de-noising correction performed on the HCP data that intended to remove nuisance signals, our Allen RSN matching procedure labeled some of the HCP components as noise (purple components in Figure 6). However, note our discussion above regarding the fact that some of these components are potentially mis-labeled. The average variances for edges involving an HCP component labeled as noise was .01 (sd: .01), while the average variance for signal edges was .03 (sd: .01).
Figure 6. DCC-derived edge variances averaged across all subjects in each of the four runs from HCP data.
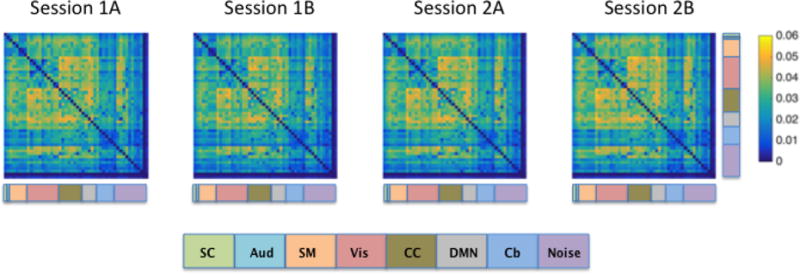
HCP data was collected over two visits that occurred on separate days, with two runs collected during each visit. Across sessions, phase encoding directions for the two runs were alternated between right-to-left (RL) and left-to-right (LR) directions. Sessions 1A and 2B indicate runs collected using the RL phase encoding direction, while sessions 1B and 2A indicate runs collected using the LR direction. The functional label assigned to each signal node is indicated using the color code at the bottom of the figure.
3.2 Brain States
3.2.1 Kirby Data
Next, we examined the reliability of brain state-derived measures using Kirby signal components.
Two recurring whole-brain patterns were identified as brain states in the Kirby data
Brain state clustering was performed separately for each rs-fMRI session and estimation method, resulting in six independent analyses (3 methods x 2 sessions). The optimal number of brain states was estimated to be two. Figure 7 illustrates the two brain states for each session derived from the SW, TSW, and DCC methods. Using the between-session spatial correlation as a measure of reliability, we found that brain states 1 and 2 were highly reliable across sessions regardless of the dynamic connectivity method used (Table 3). Across sessions and estimation methods, State 2 was characterized by stronger correlations (both positive and negative) relative to State 1. Moderate to strong negative correlations between sensory systems, namely auditory (aqua), somatomotor (orange), and visual (pink) components, were present in State 2 but were reduced in State 1. Similarly, negative correlations within the DMN (grey) components were present in State 2 and were reduced in State 1.
Figure 7. Brain states from the Kirby data.
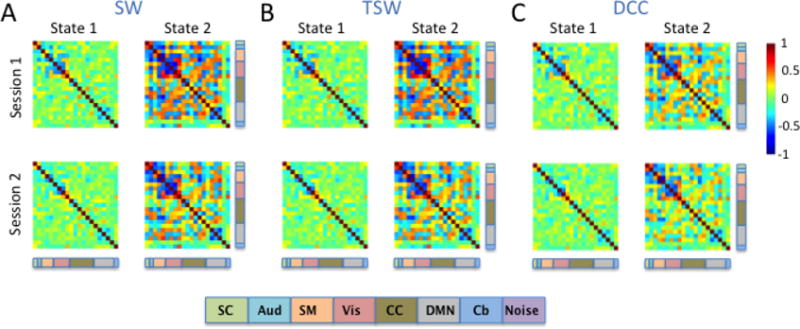
Two brain states were identified by k-means clustering the A) SW, B) TSW, and C) DCC output of signal nodes for sessions 1 and 2 separately. Brain states were highly consistent across all estimation methods. The functional label assigned to each signal node is indicated using the color code at the bottom of the figure. [SC: subcortical (mint green); Aud: auditory (aqua); SM: somatomotor (orange); Vis: visual (pink); CC: cognitive control (olive green); DMN: default mode network (grey); Cb: cerebellum (blue)].
Table 3.
Between-session Pearson correlations of the brain states estimated from the Kirby data
| DCC | SW | TSW | |
|---|---|---|---|
| State 1 | 0.95 | 0.94 | 0.95 |
| State 2 | 0.97 | 0.89 | 0.90 |
Across all estimation methods, reliability of the brain state-derived measures was low
Figure 8 shows box plots of the average time spent in each brain state (dwell time; left column) and the number of transitions (change points; right column) across subjects for each estimation method and session. In both sessions, regardless of the estimation method used to derive dynamic correlations, Kirby subjects on average spent the most time in State 1. As can be seen from the box plots, there was a great deal of between-subject variability with regards to the amount of time spent in each state; however, state dwell time for each subject was correlated across runs. Table 4 lists the reliability of estimated dwell times and the number of change points derived from each estimation method as measured by the ICC. DCC-derived dwell times were the most reliable, and fall in the good range. Similarly, there was a great deal of between-subject variability in the estimated number of change points for all estimation methods (Figure 8; right column). On average across subjects, state changes occurred more frequently when estimated from DCC-derived FC than when estimated from SW- and TSW-derived FC for both sessions. Generally, the reliability of state-change frequency estimates was fair for SW and TSW and poor for DCC.
Figure 8. Brain-state-derived summary measures for each session and method from the Kirby data.
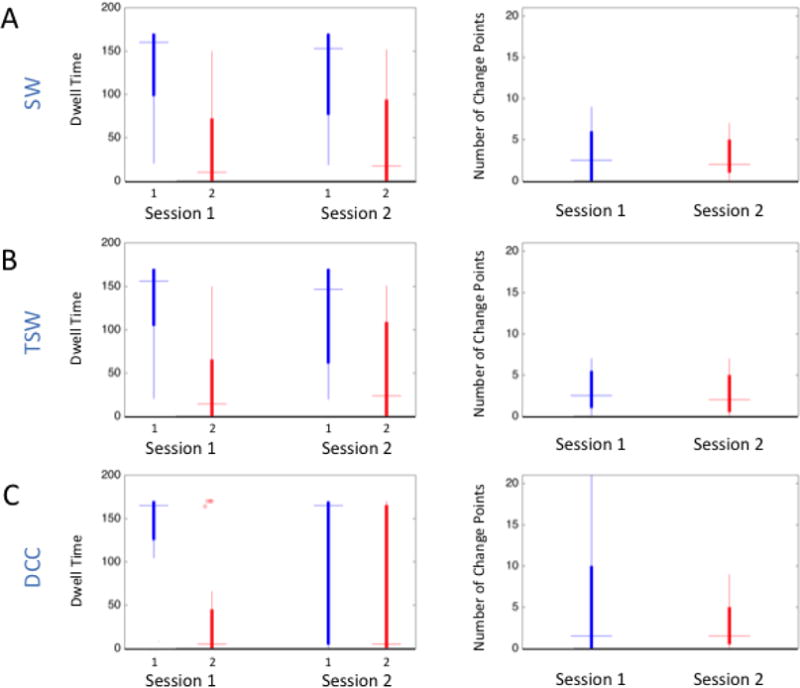
The left column contains box plots of the average time spent in each brain state (dwell time) in TRs for each session estimated using the A) SW, B) TSW, and C) DCC methods. The right column contains box plots of the number of transitions (change points) across subjects. On average, subjects spent more time in State 1 than State 2 across sessions and methods.
Table 4.
Reliability of brain state dwell times and of the number of change points estimated from the Kirby data
| DCC ICC | SW ICC | TSW ICC | |
|---|---|---|---|
| State 1 Dwell Time | 0.61 | 0.56 | 0.53 |
| State 2 Dwell Time | 0.61 | 0.56 | 0.53 |
| Number of Change Points | 0.04 | 0.41 | 0.39 |
3.2.2 HCP Data
The results for the Kirby and HCP data sets were somewhat consistent.
Three recurring whole-brain patterns were identified as brain states in the HCP data
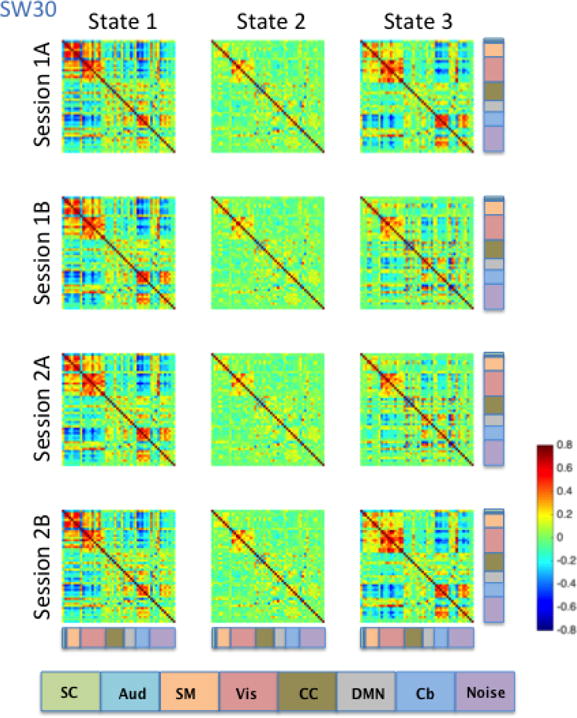
Brain state clustering was performed separately for each dynamic correlation estimation method (SW30, SW60, SW120, and DCC) and rs-fMRI run, resulting in 16 independent analyses (4 estimation methods x 4 runs). The optimal number of brain states was estimated to be three. Brain states were matched across runs and dynamic FC methods by maximizing their spatial correlation. Figures 9–12 illustrate the three brain states determined by applying k-means clustering to the results derived from the SW30, SW60, SW120, and DCC methods respectively.
Figure 9. SW30-derived brain states averaged across subjects for each of the four HCP sessions.
Brain states were identified using the cluster centers from k-means clustering. The functional label assigned to each signal node is indicated using the color code located at the bottom of the figure.
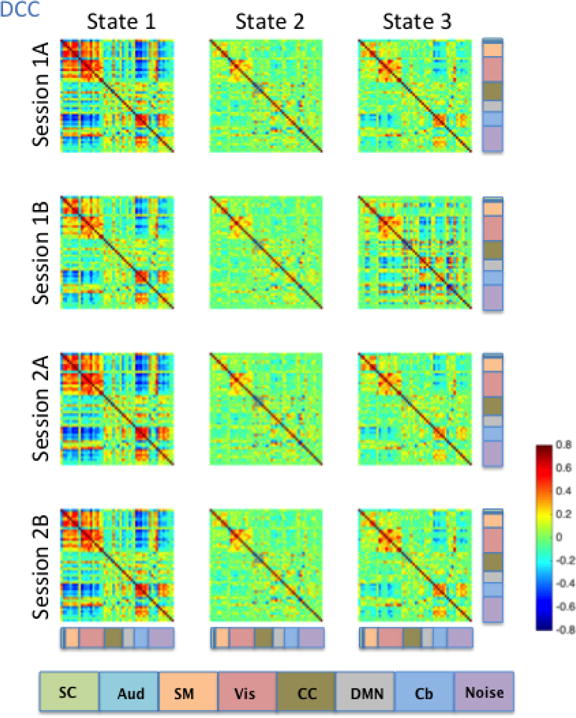
Figure 12. DCC-derived brain states averaged across subjects for each of the four HCP runs.
Brain states were determined using the cluster centers from k-means clustering. The functional label assigned to each signal node is indicated using the color code located at the bottom of the figure.
Consistent with the Kirby brain states, there was a great deal of similarity in brain states across the four HCP sessions. States 1, 2 and 3 all showed moderate to high correlations among signal components representing sensory systems: visual (Vis), somatomotor (SM), and auditory (Aud) components (Figures 9–12). In states 1 and 3, a set of components in the cerebellum (Cb, light blue) showed negative correlations with visual, somatomotor, and auditory components. These negative correlations were not observed in State 2. The HCP states were similar to those obtained from the Kirby data, particularly with regard to the second state in both cases, though it is important to note that the number and placement of the components in each HCP RSN do not map directly onto one another exactly. Using the between-run spatial correlation for each brain state as a measure of its reliability, we found that brain states 1 and 2 were similarly reliable regardless of the dynamic connectivity method used (Table 5). The only clear difference in inter-run similarity was with regard to State 3. The third brain state was in general slightly less reliable across runs, with the exception of SW120 (0.66, 0.71, and 0.95 for SW30, SW60, and SW120, and r = 0.80 for DCC, respectively).
Table 5.
Average between-session Pearson correlations of the brain states from the HCP data
| DCC | SW30 | SW60 | SW120 | |
|---|---|---|---|---|
| State 1 | 0.98 | 0.95 | 0.93 | 0.97 |
| State 2 | 0.98 | 0.98 | 0.98 | 0.98 |
| State 3 | 0.80 | 0.66 | 0.71 | 0.95 |
Across all approaches, reliability of the brain state-derived measures was low
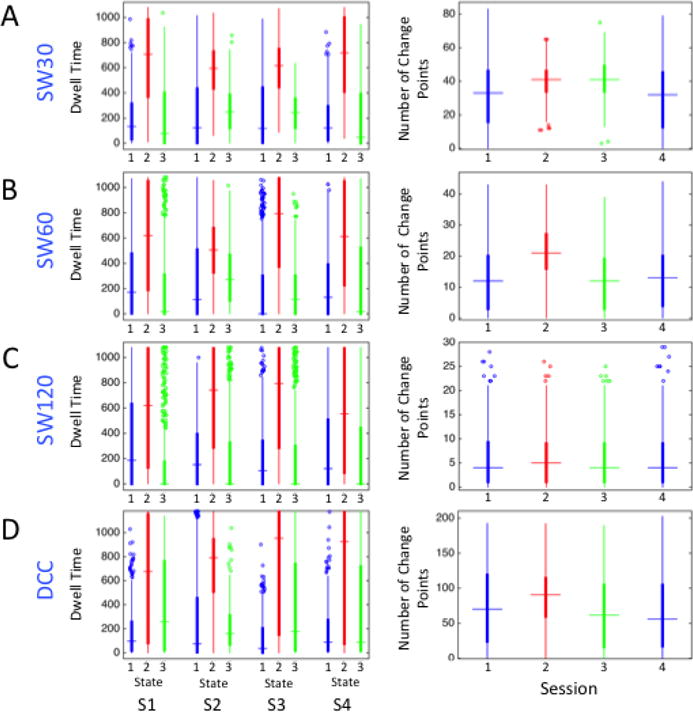
Figure 13 shows box plots of the dwell time (left) and number of change points (right) for each estimation method and session. In all four sessions, regardless of the estimation method used to derive dynamic correlations, HCP subjects on average spent the most time in State 2, while the relative dwell time ranking of States 1 and 3 varied with the estimation method used. There was also a great deal of between-subject variability with regard to the amount of time spent in each state, and the reliability of dwell time estimates varied across states and methods used to derive them (Table 6). For States 1 and 2, DCC- and SW120-derived dwell times were more reliable than SW30- and SW60-derived dwell times; however, DCC-derived State 3 dwell times were less reliable than SW120-derived dwell times.
Figure 13. Brain-state-derived summary measures for each session and method, from HCP data.
Box plots of the dwell time in TRs and the number of change points estimated using the A) SW30, B) SW60, C) SW120, and D) DCC methods.
Table 6.
Reliability of dwell times and number of change points for brain states estimated from the HCP data
| DCC ICC | SW30 ICC | SW60 ICC | SW120 ICC | |
|---|---|---|---|---|
| State 1 Dwell Time | 0.31 | 0.27 | 0.25 | 0.34 |
| State 2 Dwell Time | 0.51 | 0.44 | 0.46 | 0.58 |
| State 3 Dwell Time | 0.26 | −0.06 | 0.01 | 0.52 |
| Number of Change Points | 0.26 | 0.24 | 0.21 | 0.26 |
As was the case when comparing dynamic FC methods applied to the Kirby data, more frequent state changes were indicated by DCC-derived brain states than by the SW methods across all four HCP runs (Figure 13). On average, subjects switched states every 136 s (averaging across the four runs) using SW120-derived brain states, every 52 s using SW60-derived brain states, every 22 s using SW30-brain states, and every 12 s using DCC-derived brain states, as shown in Figure 13. In other words, brain state derived measures obtained using the DCC and SW30 methods displayed more frequent state changes than those obtained using the SW60 and SW120 methods. The relatively high rate of state changes for the DCC-derived measures can be attributed almost entirely to the existence of more frequent transitions when in State 3. Generally, the reliability of state-change frequency estimates was quite low for all of the methods, as shown in Table 6.
4 Discussion
Identification of dynamic FC estimation methods and summary measures that maximize reliability is important to provide accurate insight into brain function. Here, we compared the reliability of summary statistics and brain states derived from commonly used non-parametric estimation methods (SW and TSW) to a model-based method (DCC). Given the previously demonstrated susceptibility of SW methods to noise-induced temporal variability in correlations (Lindquist et al., 2014), we set out to compare the reliability of these methods when applied to two rs-fMRI test-retest data sets with potentially varying levels of noise: 1) the Kirby data set, which was collected using a typical-length, standard EPI sequence and then cleaned using established standard preprocessing procedures; and 2) the HCP data set, which was collected using a cutting-edge multiband EPI sequence optimized to produce higher temporal resolution images and was cleaned using more aggressive preprocessing procedures. Consistent with our hypothesis, we found that the model-based DCC method consistently outperformed the non-parametric SW and TSW methods, which is in line with findings from our previous work (Lindquist et al., 2014). Specifically, the DCC method demonstrated the highest reliability of dynamic FC summary statistics in both data sets, and was best able to differentiate between signal components and noise components based on the variance of the dynamic connectivity values. Reliability of the brain state-derived measures, however, were low across all estimation methods, with no method clearly outperforming the others.
Reliability of the Mean and Variance of Dynamic Functional Correlations
We found that the mean of dynamic correlations derived from all estimation methods was equivalently reliable (Figures 1A–B and 4A–B). This observation was not surprising, as all methods should result in average dynamic correlations that roughly correspond to the sample correlation and thus should be similarly reliable. For all methods, we observed that the dynamic correlation variances decreased as the absolute value of the dynamic correlation means increased, which is consistent with recent work by Thompson and colleagues (Thompson and Fransson, 2015) (Figures 2 and 5). While more prominent in the HCP data (Figure 5), this pattern was also observed for signal-signal edges in the Kirby data (red data points in Figure 2). However, the reliability of dynamic correlation variance was significantly higher when derived using the DCC method than when derived from SW and TSW methods. This observation held true for both the Kirby data (Figures 1A and C) and for the HCP data (Figures 4A and C). For SW methods, as the window lengths increased, the reliability of the dynamic correlation variance (Figures 4A and C), as well as the estimated dynamic correlation variance (Figure 5), decreased - i.e., SW30 resulted in the largest correlation variance values that were most reliable, while SW120 resulted in the smallest correlation variance values that were least reliable. This increase in the observed dynamic correlation variance values with decreasing window length is expected, as the SW methods are more susceptible to noise when smaller window sizes are used. Similarly, the decrease in reliability with increasing window length is expected; assuming a constant time series length, the total of number of samples used to calculate the mean and variance of dynamic FC decreases as the window length increases. Identifying dynamic FC estimation methods that maximize the reliability of correlation variance estimates are particularly important, given that edges between brain regions involved in larger or frequent state changes should exhibit consistently higher correlation variation than edges involving brain regions whose functional connectivity and network membership remain more stable throughout an experimental session.
Previous studies have found that SW methods are susceptible to noise and suboptimal at estimating dynamic FC, both when connectivity changes are gradual (Lindquist et al., 2014) and when they are abrupt (Shakil et al., 2016). Performance of the SW method is especially poor when small window sizes are applied to standard EPI data, as dynamic changes in connectivity produce spurious correlations when only a small number of time points are taken into account (Shakil et al., 2016). For all three estimation methods we explored, the variance of the dynamic correlations of edges involving two signal components was higher as compared to the variance of the dynamic correlations of edges involving at least one noise component (Figures 2 and 5). This is important, as it may indicate that neuronally relevant signal fluctuation demonstrate higher variance. This observation was true for both the Kirby (Figure 2) and the HCP (Figure 5) data sets and is consistent with previous literature (Allen et al., 2012a). Moreover, the degree of separation between the variability of signal-signal edges and edges that include at least one noise component in the Kirby data was larger for the DCC method than for the SW or TSW methods; DCC-derived variance of edges involving noise components appeared to shrink more toward zero (Figure 2A). In the HCP data, similar decreased variability (i.e., the shrinking of dynamic correlation variance toward zero) was observed in clusters of edges across all three estimation methods (Figure 5A), and DCC-derived variance of all edges were shifted more toward zero compared to the SW methods. Unlike in the Kirby data, however, the degree of separation between the variability of signal-signal edges and edges that include at least one noise component was not enhanced by the DCC method. This may be due to the fact that most of the HCP noise-related components were actively removed using the FIX algorithm (Griffanti et al., 2014; Salimi-Khorshidi et al., 2014).
In terms of maximizing reliability when comparing different window lengths, the shortest window length (SW30) performed the best out of the three windows lengths tested for the HCP data and was the most similar to the DCC (Figures 4A and C). However, in terms of minimizing the bias in dynamic correlation variance (shrinking the variance of edges involving noise components toward zero), the longest window length (SW120) performed the best and was the most similar to the DCC method (Figure 5). These findings provide further evidence that the DCC method is less susceptible to the temporal variability in correlations induced by noise (Lindquist et al., 2014), even when applied to data that has been aggressively cleaned.
Focusing on the variability of dynamic correlations between signal components, we observed a group of edges that consistently displayed higher variance than others edges in both the Kirby data (Figure 3D) and the HCP data (Figure 6); namely the edges involving visual, cognitive control, and default mode regions. Notably, these regions are consistently identified as functional hubs (Buckner et al., 2009), and are recognized as some of the most globally connected regions in the brain (Cole et al., 2010). Connectivity between these brain regions has also been previously described as highly variable using TSW methods (Allen et al., 2012a), suggesting that this finding is robust. Interestingly, the DCC-derived variance estimates for these edges were more reliable than SW- or TSW-derived variance estimates (Figure 2), suggesting that the more reliable DCC-derived estimates may in turn increase the likelihood of detecting nuances in dynamic connectivity that might otherwise be missed by methods more susceptible to noise. Exploring whether the use of more reliable dynamic FC outcome measures also improves the reliability of related brain-behavior relationships is an important area of future work.
Finally, there is the issue of long-term test-retest reliability. All of the Kirby data were acquired within the same day, while different parts of the HCP data were acquired on different days (two sessions acquired on each of two days). Although a thorough investigation is outside the scope of the current manuscript, we do note a decrease in similarity in the results across days for the HCP data set. For example, the I2C2 score for the mean of the dynamic correlations across all pairs of components measured using DCC, which produces an omnibus reliability measure across the brain, is on average 0.55 when comparing two sessions from the same day, while it is on average 0.44 when comparing two sessions from different days. Similarly, the I2C2 score for the variance of dynamic correlations across all pairs of components is on average 0.515 when comparing two sessions from the same day, while it is on average 0.415 when comparing two sessions from different days. Future work is needed to determine how reliable the dynamic correlation is between sessions that are further apart in time.
Reliability of Brain States
Our results suggest that the DCC method provides the best estimate for the correlation variance (Figures 1 and 4). However, whether that is also true for the estimation and characterization of brain states is less clear. Interestingly, the three brain states identified from the HCP data (Figures 9–12) follow similar patterns to the most common occurring states for healthy individuals found in an earlier study probing recurring brain states conducted by Damaraju and colleagues (Damaraju et al., 2014). The brain states estimated using the Kirby data (Figure 7) also follow a similar pattern, but the negative correlations observed between the sensory networks (Aud, SM, and Vis networks) are more pronounced, perhaps due to the smaller number of networks estimated. The observed similarity of brain states across methods and data sets suggests that the most robust features of dynamic connectivity will emerge regardless of the method used to estimate them. It is critical that future research design studies to probe the functional relevance of these brain states. This can be achieved by relating these patterns of brain network organization both to neuronal measurements and to behavioral and cognitive outcomes.
In the Kirby data, DCC-derived brain states were equally or more reliable than SW- and TSW-derived brain states (Table 3). However, the two methods that produced the most reliable connection variances in the HCP data, the DCC and SW30 methods, produced the least reliable State 3, as exhibited by the reduced spatial correlation for State 3 across the four HCP sessions (Table 5). One possible explanation is that this may be due to increased sensitivity of these methods to features of the HCP data acquisition, which varied between runs. The DCC and SW30 methods had high inter-run reliability for State 3 between sessions collected using the same phase encoding direction (between Sessions 1A and 2B, which were collected using a left-to-right phase encoding direction, and between Sessions 1B and 2A, which were collected using a right-to-left phase encoding direction), but lower reliability across pairs of runs collected using opposite phase encoding directions. It may be that the DCC and SW30 methods, which were more reliable in terms of correlation variances, are able to detect subtle, systematic data acquisition biases that were introduced to brain states-derived measures that SW60 and SW120 methods cannot.
Regardless of the method used to estimate dynamic connectivity, metrics derived from the brain states (dwell time and number of change points) were generally less reliable than the mean and variance of dynamic functional connectivity; this was true for both Kirby brain states (ICC values in Table 4 compared with Figure 1) and HCP brain states (ICC values in Table 6 compared with Figure 4). An important limitation is that they may be affected by aging and other uncontrolled factors, so additional research is needed. An important methodological issue that may have impacted the reliability of brain state metrics is the difficulty associated with determining the actual number of latent brain states present in the data. Prior to applying the k-means clustering algorithm to the dynamic correlation matrices, we had to specify the number of brain states into which the algorithm should divide the data. Many approaches have been developed to find an approximate optimal cluster number k. Here we adopted the popular ‘Elbow Method’ to identify the appropriate number of states (Tibshirani et al., 2001). This approach is ad hoc, and there are several more sophisticated methods that build statistical models to formalize the ‘elbow’ heuristic, including the ‘gap statistic’ (Tibshirani et al., 2001). However, in practice the ‘Elbow Method’ usually achieves better performance and strikes a balance between computational efficiency and accuracy. Nevertheless, we cannot rule out the possibility that the overall lower reliability of brain state metrics was, in part, due to our choice regarding the number of latent brain states present in the data.
An important alternative possibility is that average brain states, which are detected reliably with the DCC method and display similar patterns across both DCC and SW approaches, reflect participant traits that are relatively stable. Summary metrics such as dwell time and number of change points, however, may reflect participant states that meaningfully change both across days and even within a session. These states could be due to factors such as arousal and attention that may themselves be unreliable in interesting ways. Future research designed to probe this possibility is needed.
Finally, it is important to note that throughout we assumed subjects are in a single state at a given time period. However, recent research (Leonardi et al., 2014; Miller et al., 2016) has suggested the possibility that a subject may simultaneously be in multiple overlapping states. In such a setting, the choice of appropriate summary measure would change compared to the single-state setting.
Kirby vs. HCP Data Sets
We are encouraged by the level of agreement between our results for these two very different data sets. In general, the confidence intervals for the reliability estimates of the dynamic correlation means and variances were smaller for the HCP data set (Figure 4) compared to the confidence intervals for the Kirby data set (Figure 1). This increased confidence in our estimates of reliability is expected given that the HCP data set includes significantly more observations than the Kirby data set. At the same time, however, we are intrigued that despite almost a sixfold increase in the amount of data collected for each HCP participant compared to the amount of data collected for each Kirby participant, the reliability of dynamic correlation means and variances were relatively similar (Figures 1 and 4) - though the HCP data set did show a significantly lower I2C2 value for the mean dynamic correlation than the Kirby data set. The observed similarity in reliability is consistent with the results from a previous study that showed that truncating multi-band data sets (up to half the original amount of data) did not significantly change the stability of RSNs, as long as moderate acceleration factors were used (Chen et al., 2015). It is possible that the amount of data acquired for each Kirby subject is sufficient to achieve maximum reliability. Alternately, another possible explanation for the observed similarity in the reliability of the dynamic FC derived measures between the two data sets is that the introduction of structured noise by the multi-band image acquisition scheme reduced the reliability of the dynamic FC measures obtained using the HCP data. Specifically, while the simultaneous acquisition of multiple slices significantly increases the temporal resolution of the data, the multi-band acquisition also introduces strong noise amplification to the images during the subsequent un-folding of the simultaneously acquired slices during the image reconstruction process (Xu et al., 2013). The amount of noise amplification increases as the acceleration factor (i.e., number of slices simultaneously acquired at each TR) increases, and to minimize the introduction of undesired structured noise and other image artifacts, use of an acceleration factor larger than eight is generally not recommended (Smith et al., 2013; Chen et al., 2015; De Martino et al., 2015). This is important, as the HCP data uses an acceleration factor of eight, which is at the higher end of the recommended acceleration factor. At this time, however, a thorough analysis of the many differences between the two data sets and how they interact to impact the reliability of FC dynamics is beyond the scope of this paper.
Comparison to Reliability Studies of Static Functional Connectivity
A number of studies have previously evaluated the reliability of static FC in resting-state fMRI data. A particular focus has been on determining the necessary scan length needed to obtain reliable estimates (Van Dijk et al., 2010; Anderson et al., 2011; Birn et al., 2013). While increased scan length has consistently been shown to improve reliability, different studies have reached widely varying conclusions about the necessary length required, with recommendations ranging from 5 minutes (Whitlow et al., 2011; Liao et al., 2013) to 90 minutes (Laumann et al., 2015). Studies have similarly indicated that increasing temporal resolution (Birn et al., 2013; Zuo et al., 2013; Liao et al., 2013) results in improved reliability. In our study, we examined data with varying scan duration and temporal resolution. As mentioned above, these settings had less clear effects on the reliability of dynamic FC than previously observed for static FC.
In terms of preprocessing, several studies have found that global signal regression tends to worsen the reliability of static FC (Zuo et al., 2013; Liao et al., 2013), while nuisance regression tends to improve it (Zuo et al., 2013). In addition, the use of functional versus anatomical regions of interest (ROIs) (Anderson et al., 2011) have been shown to improve reliability. In our study we didn’t explicitly study the effects of these different preprocessing choices, however we did let them guide the choices we made throughout.
Finally, it is interesting to note that for all methods, the mean of the dynamic correlation and the static correlation were similarly reliable.
Considerations for Dynamic Functional Connectivity Estimation Methods
The results of our study provide evidence that dynamic features of functional connectivity can be reliably estimated, and that the model-based DCC method outperforms its non-parametric counterparts (i.e., SW and TSW methods). However, several aspects of the dynamic FC estimation process exist of which one should be aware when interpreting results from dynamic FC data. First, it is important to note that the dynamic FC estimation methods discussed in this work are descriptive in nature, and do not involve specific inferential tests or methods. While this reflects the majority of current dynamic connectivity studies, such tests are now being introduced into the field. For example, Zalesky et al. (2014) have developed a univariate test statistic to measure the extent of time-varying fluctuations in the time-resolved correlation coefficients between pairs of brain regions. We anticipate significant developments in this area in coming years. Secondly, it is important to recognize that model selection procedures were not employed when applying DCC. These include plots of the auto-correlation or partial correlation functions to determine model order, and diagnostic test to validate the model assumptions. In addition, although we have shown that the dynamic FC measures obtained using the model-based DCC method are more reliable than SW and TSW methods, we recognize that I2C2 and ICC scores in the range of .6 indicate that room for improvement remains in the reliability of DCC-derived dynamic FC measures. It is thus worth exploring variations or alternatives to the DCC method that might improve upon its accuracy and reliability.
Finally, it should be noted that there exist alternative approaches in the literature that may also improve upon sliding-window approaches. For example, wavelet decompositions effectively use an adaptive windowing approach, like DCC, and may thus also provide improved results (e.g., Yaesoubi et al., 2017). Likewise, there exist alternative methods for detecting brain states that use techniques from change point analysis (e.g., Cribben et al., 2012, 2013; Xu and Lindquist, 2015) that may similarly improve upon results obtained using k-means clustering. However, we leave further comparisons for future work.
5 Conclusions
The primary aim of this project was to identify dynamic FC estimation methods and summary measures that maximize reliability and the utility of dynamic FC, by comparing the reliability of dynamic FC summary measures computed using the SW, TSW, and DCC methods. Additionally, comparison of the estimation methods’ reliability between a data set acquired using a conventional data acquisition/processing approach and that acquired using a more cutting-edge approach was enabled by utilizing the Kirby and HCP data sets. The results of our study provide evidence that dynamic features of FC can be reliably estimated in both data sets when the model-based DCC method is used. This is significant, as there is a clear need in the field for summary measures that can be used to find meaningful individual differences in dynamic FC, and it is the degree and patterns of the fluctuations of dynamic FC (i.e., the variance) that may provide the most interesting information. We therefore believe that utilizing the variance of the dynamic connectivity is a crucial component in any dynamic FC-derived summary measure. The study also showed that across all estimation methods, reliability of the brain state-derived measures are low, indicating that caution should be taken when analyzing and interpreting dynamic FC summary measures derived from brain states and that further efforts to develop more reliable approaches to calculating brain states are necessary.
Figure 10. SW60-derived brain states averaged across subjects for each of the four HCP sessions.
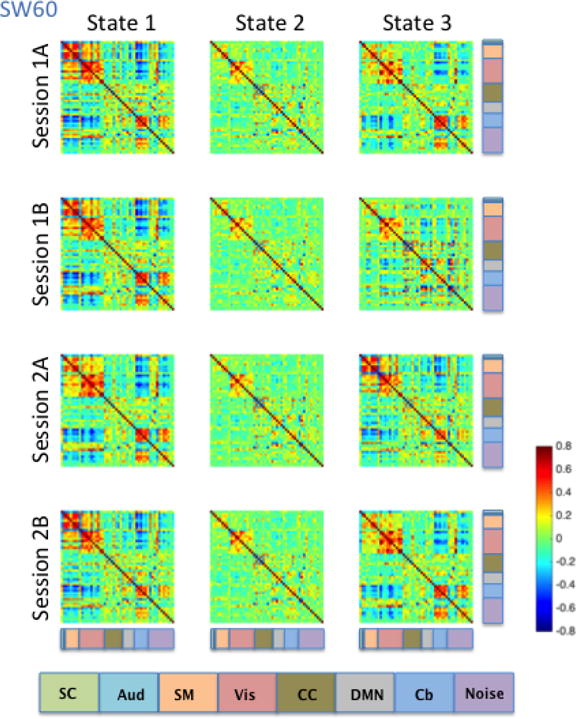
Brain states were determined using the cluster centers from k-means clustering. The functional label assigned to each signal node is indicated using the color code located at the bottom of the figure.
Figure 11. SW120-derived brain states averaged across subjects for each of the four HCP sessions.
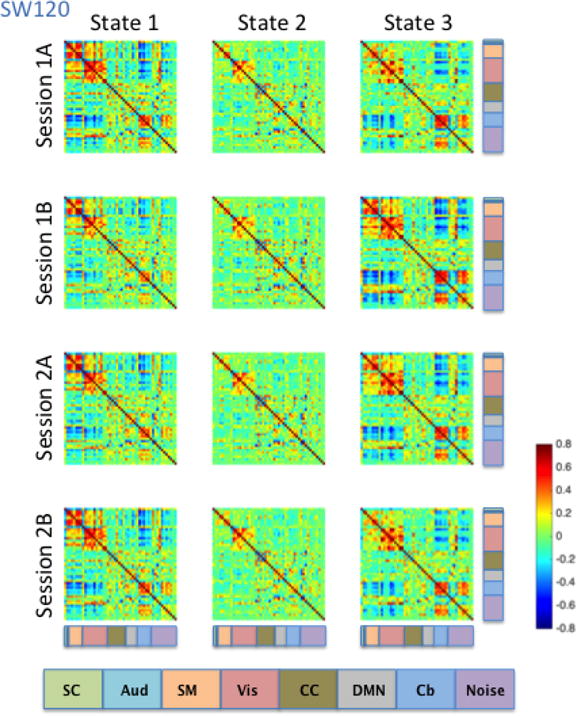
Brain states were determined using the cluster centers from k-means clustering. The functional label assigned to each signal node is indicated using the color code located at the bottom of the figure.
Acknowledgments
Data were provided by the Human Connectome Project, WU-Minn Consortium (Principal Investigators: David Van Essen and Kamil Ugurbil; 1U54MH091657), which was funded by the McDonnell Center for Systems Neuroscience at Washington University and the 16 NIH Institutes and Centers that support the NIH Blueprint for Neuroscience Research. The work presented in this paper was supported in part by NIH grants R01 EB016061, R01 EB012547, and P41 EB015909 from the National Institute of Biomedical Imaging and Bioengineering, R01 MH095836 and K01 MH109766 from the National Institute of Mental Health, and the Craig H. Neilsen Foundation (Project Number 338419).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Allen EA, Damaraju E, Plis SM, Erhardt EB, Eichele T, Calhoun VD. Tracking whole-brain connectivity dynamics in the resting state. Cerebral Cortex. 2012a doi: 10.1093/cercor/bhs352. page bhs352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen EA, Erhardt EB, Damaraju E, Gruner W, Segall JM, Silva RF, Havlicek M, Rachakonda S, Fries J, Kalyanam R, et al. A baseline for the multivariate comparison of resting-state networks. Frontiers in systems neuroscience. 2011;5 doi: 10.3389/fnsys.2011.00002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen EA, Erhardt EB, Wei Y, Eichele T, Calhoun VD. Capturing inter-subject variability with group independent component analysis of fmri data: a simulation study. NeuroImage. 2012b;59(4):4141–4159. doi: 10.1016/j.neuroimage.2011.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson JS, Ferguson MA, Lopez-Larson M, Yurgelun-Todd D. Reproducibility of single-subject functional connectivity measurements. American journal of neuroradiology. 2011;32(3):548–555. doi: 10.3174/ajnr.A2330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashburner J, Friston KJ. Unified segmentation. NeuroImage. 2005;26(3):839–851. doi: 10.1016/j.neuroimage.2005.02.018. [DOI] [PubMed] [Google Scholar]
- Bassett DS, Wymbs NF, Porter MA, Mucha PJ, Carlson JM, Grafton ST. Dynamic reconfiguration of human brain networks during learning. Proceedings of the National Academy of Sciences. 2011;108(18):7641–7646. doi: 10.1073/pnas.1018985108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassett DS, Yang M, Wymbs NF, Grafton ST. Learning-induced autonomy of sensorimotor systems. Nature Neuroscience. 2015;18(5):744–751. doi: 10.1038/nn.3993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beckmann CF, Smith SM. Probabilistic independent component analysis for functional magnetic resonance imaging. Medical Imaging, IEEE Transactions on. 2004;23(2):137–152. doi: 10.1109/TMI.2003.822821. [DOI] [PubMed] [Google Scholar]
- Bell AJ, Sejnowski TJ. An information-maximization approach to blind separation and blind deconvolution. Neural computation. 1995;7(6):1129–1159. doi: 10.1162/neco.1995.7.6.1129. [DOI] [PubMed] [Google Scholar]
- Betzel RF, Byrge L, He Y, Goñi J, Zuo XN, Sporns O. Changes in structural and functional connectivity among resting-state networks across the human lifespan. NeuroImage. 2014;102:345–357. doi: 10.1016/j.neuroimage.2014.07.067. [DOI] [PubMed] [Google Scholar]
- Birn RM, Molloy EK, Patriat R, Parker T, Meier TB, Kirk GR, Nair VA, Meyerand ME, Prabhakaran V. The effect of scan length on the reliability of resting-state fmri connectivity estimates. Neuroimage. 2013;83:550–558. doi: 10.1016/j.neuroimage.2013.05.099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bollerslev T. Generalized autoregressive conditional heteroskedasticity. Journal of econometrics. 1986;31(3):307–327. [Google Scholar]
- Buckner RL, Sepulcre J, Talukdar T, Krienen FM, Liu H, Hedden T, Andrews-Hanna JR, Sperling RA, Johnson KA. Cortical hubs revealed by intrinsic functional connectivity: mapping, assessment of stability, and relation to alzheimer’s disease. The Journal of Neuroscience. 2009;29(6):1860–1873. doi: 10.1523/JNEUROSCI.5062-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calhoun V, Adali T, Pearlson G, Pekar J. A method for making group inferences from functional mri data using independent component analysis. Human brain mapping. 2001;14(3):140–151. doi: 10.1002/hbm.1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calhoun VD, Miller R, Pearlson G, Adalı T. The chronnectome: time-varying connectivity networks as the next frontier in fMRI data discovery. Neuron. 2014;84(2):262–274. doi: 10.1016/j.neuron.2014.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll RJ, Ruppert D, Stefanski LA, Crainiceanu CM. Measurement error in nonlinear models: a modern perspective. Chapman and Hall/CRC; 2006. [Google Scholar]
- Chang C, Glover GH. Time–frequency dynamics of resting-state brain connectivity measured with fmri. NeuroImage. 2010;50(1):81–98. doi: 10.1016/j.neuroimage.2009.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Vu A, Xu J, Moeller S, Ugurbil K, Yacoub E, Feinberg D. Evaluation of highly accelerated simultaneous multi-slice epi for fmri. NeuroImage. 2015;104:452–459. doi: 10.1016/j.neuroimage.2014.10.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cicchetti DV. Guidelines, criteria, and rules of thumb for evaluating normed and standardized assessment instruments in psychology. Psychological assessment. 1994;6(4):284. [Google Scholar]
- Cole MW, Pathak S, Schneider W. Identifying the brain’s most globally connected regions. NeuroImage. 2010;49(4):3132–3148. doi: 10.1016/j.neuroimage.2009.11.001. [DOI] [PubMed] [Google Scholar]
- Cribben I, Haraldsdottir R, Atlas LY, Wager TD, Lindquist MA. Dynamic connectivity regression: determining state-related changes in brain connectivity. NeuroImage. 2012;61(4):907–920. doi: 10.1016/j.neuroimage.2012.03.070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cribben I, Wager TD, Lindquist MA. Detecting functional connectivity change points for single-subject fmri data. Frontiers in Computational Neuroscience. 2013;7 doi: 10.3389/fncom.2013.00143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damaraju E, Allen EA, Belger A, Ford JM, McEwen S, Mathalon DH, Mueller BA, Pearlson GD, Potkin SG, Preda A, Turner JA, Vaidya JG, van Erp TG, Calhoun VD. Dynamic functional connectivity analysis reveals transient states of dysconnectivity in schizophrenia. Neuroimage: Clinical. 2014;5:298–308. doi: 10.1016/j.nicl.2014.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Martino F, Moerel M, Ugurbil K, Formisano E, Yacoub E. Less noise, more activation: multiband acquisition schemes for auditory functional mri. Magnetic Resonance in Medicine. 2015;74(2):462–467. doi: 10.1002/mrm.25408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engle R. Dynamic conditional correlation: A simple class of multivariate generalized autoregressive conditional heteroskedasticity models. Journal of Business & Economic Statistics. 2002;20(3):339–350. [Google Scholar]
- Engle RF. Autoregressive conditional heteroscedasticity with estimates of the variance of united kingdom inflation. Econometrica: Journal of the Econometric Society. 1982:987–1007. [Google Scholar]
- Engle RF, Sheppard K. Technical report. National Bureau of Economic Research; 2001. Theoretical and empirical properties of dynamic conditional correlation multivariate garch. [Google Scholar]
- Erhardt EB, Rachakonda S, Bedrick EJ, Allen EA, Adali T, Calhoun VD. Comparison of multi-subject ica methods for analysis of fmri data. Human brain mapping. 2011;32(12):2075–2095. doi: 10.1002/hbm.21170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Everitt BS, Landau S, Leese M. Cluster analysis. A member of the Hodder Headline Group, London 2001 [Google Scholar]
- Filippini N, MacIntosh BJ, Hough MG, Goodwin GM, Frisoni GB, Smith SM, Matthews PM, Beckmann CF, Mackay CE. Distinct patterns of brain activity in young carriers of the apoe-ε4 allele. Proceedings of the National Academy of Sciences. 2009;106(17):7209–7214. doi: 10.1073/pnas.0811879106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friston KJ, Holmes AP, Worsley KJ, Poline J, Frith CD, Frackowiak RS, et al. Statistical parametric maps in functional imaging: a general linear approach. Human brain mapping. 1994;2(4):189–210. [Google Scholar]
- Glasser MF, Sotiropoulos SN, Wilson JA, Coalson TS, Fischl B, Andersson JL, Xu J, Jbabdi S, Webster M, Polimeni JR, et al. The minimal preprocessing pipelines for the human connectome project. NeuroImage. 2013;80:105–124. doi: 10.1016/j.neuroimage.2013.04.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffanti L, Salimi-Khorshidi G, Beckmann CF, Auerbach EJ, Douaud G, Sexton CE, Zsoldos E, Ebmeier KP, Filippini N, Mackay CE, et al. Ica-based artefact removal and accelerated fmri acquisition for improved resting state network imaging. NeuroImage. 2014;95:232–247. doi: 10.1016/j.neuroimage.2014.03.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu S, Satterthwaite TD, Medaglia JD, Yang M, Gur RE, Gur RC, Bassett DS. Emergence of system roles in normative neurodevelopment. Proceedings of the National Academy of Sciences. 2015;112(44):13681–13686. doi: 10.1073/pnas.1502829112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Handwerker DA, Roopchansingh V, Gonzalez-Castillo J, Bandettini PA. Periodic changes in fmri connectivity. NeuroImage. 2012;63(3):1712–1719. doi: 10.1016/j.neuroimage.2012.06.078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen PR, Lunde A. A forecast comparison of volatility models: does anything beat a garch (1, 1)? Journal of applied econometrics. 2005;20(7):873–889. [Google Scholar]
- Himberg J, Hyvärinen A, Esposito F. Validating the independent components of neuroimaging time series via clustering and visualization. NeuroImage. 2004;22(3):1214–1222. doi: 10.1016/j.neuroimage.2004.03.027. [DOI] [PubMed] [Google Scholar]
- Hlinka J, Hadrava M. On the danger of detecting network states in white noise. Frontiers in Computational Neuroscience. 2015;9 doi: 10.3389/fncom.2015.00011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson AE, Calderon DP, Pfaff DW, Proekt A. Recovery of consciousness is mediated by a network of discrete metastable activity states. Proceedings of the National Academy of Sciences of the United States of America. 2014;111(25):9283–9288. doi: 10.1073/pnas.1408296111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutchison RM, Morton JB. Tracking the Brain’s Functional Coupling Dynamics over Development. The Journal of Neuroscience. 2015;35(17):6849–6859. doi: 10.1523/JNEUROSCI.4638-14.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutchison RM, Womelsdorf T, Allen EA, Bandettini PA, Calhoun VD, Corbetta M, Della Penna S, Duyn JH, Glover GH, Gonzalez-Castillo J, et al. Dynamic functional connectivity: promise, issues, and interpretations. NeuroImage. 2013a;80:360–378. doi: 10.1016/j.neuroimage.2013.05.079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutchison RM, Womelsdorf T, Gati JS, Everling S, Menon RS. Resting-state networks show dynamic functional connectivity in awake humans and anesthetized macaques. Human brain mapping. 2013b;34(9):2154–2177. doi: 10.1002/hbm.22058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kudela M, Harezlak J, Lindquist MA. Assessing uncertainty in dynamic functional connectivity. NeuroImage. 2017;149:165–177. doi: 10.1016/j.neuroimage.2017.01.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landman BA, Huang AJ, Gifford A, Vikram DS, Lim IAL, Farrell JA, Bogovic JA, Hua J, Chen M, Jarso S, et al. Multi-parametric neuroimaging reproducibility: a 3-t resource study. NeuroImage. 2011;54(4):2854–2866. doi: 10.1016/j.neuroimage.2010.11.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laumann TO, Gordon EM, Adeyemo B, Snyder AZ, Joo SJ, Chen MY, Gilmore AW, McDer-mott KB, Nelson SM, Dosenbach NU, et al. Functional system and areal organization of a highly sampled individual human brain. Neuron. 2015;87(3):657–670. doi: 10.1016/j.neuron.2015.06.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leonardi N, Shirer WR, Greicius MD, Van De Ville D. Disentangling dynamic networks: Separated and joint expressions of functional connectivity patterns in time. Human brain mapping. 2014;35(12):5984–5995. doi: 10.1002/hbm.22599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leonardi N, Van De Ville D. On spurious and real fluctuations of dynamic functional connectivity during rest. Neuroimage. 2015;104:430–436. doi: 10.1016/j.neuroimage.2014.09.007. [DOI] [PubMed] [Google Scholar]
- Li YO, Adalı T, Calhoun VD. Estimating the number of independent components for functional magnetic resonance imaging data. Human brain mapping. 2007;28(11):1251–1266. doi: 10.1002/hbm.20359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao XH, Xia MR, Xu T, Dai ZJ, Cao XY, Niu HJ, Zuo XN, Zang YF, He Y. Functional brain hubs and their test–retest reliability: a multiband resting-state functional mri study. Neuroimage. 2013;83:969–982. doi: 10.1016/j.neuroimage.2013.07.058. [DOI] [PubMed] [Google Scholar]
- Lindquist MA, Xu Y, Nebel MB, Caffo BS. Evaluating dynamic bivariate correlations in resting-state fmri: A comparison study and a new approach. NeuroImage. 2014;101:531–546. doi: 10.1016/j.neuroimage.2014.06.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marusak HA, Calhoun VD, Brown S, Crespo LM, Sala-Hamrick K, Gotlib IH, Thomason ME. Dynamic functional connectivity of neurocognitive networks in children. Human Brain Mapping. 2017;38(1):97–108. doi: 10.1002/hbm.23346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller RL, Yaesoubi M, Turner JA, Mathalon D, Preda A, Pearlson G, Adali T, Calhoun VD. Higher dimensional meta-state analysis reveals reduced resting fmri connectivity dynamism in schizophrenia patients. PloS one. 2016;11(3):e0149849. doi: 10.1371/journal.pone.0149849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preti MG, Bolton TA, Van De Ville D. The dynamic functional connectome: State-of-the-art and perspectives. NeuroImage. 2016 doi: 10.1016/j.neuroimage.2016.12.061. [DOI] [PubMed] [Google Scholar]
- Pruessmann KP, Weiger M, Scheidegger MB, Boesiger P, et al. Sense: sensitivity encoding for fast mri. Magnetic resonance in medicine. 1999;42(5):952–962. [PubMed] [Google Scholar]
- Purdon PL, Weisskoff RM. Effect of temporal autocorrelation due to physiological noise and stimulus paradigm on voxel-level false-positive rates in fmri. Human brain mapping. 1998;6(4):239–249. doi: 10.1002/(SICI)1097-0193(1998)6:4<239::AID-HBM4>3.0.CO;2-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rashid B, Damaraju E, Pearlson GD, Calhoun VD. Dynamic connectivity states estimated from resting fmri identify differences among schizophrenia, bipolar disorder, and healthy control subjects. Frontiers in Human Neuroscience. 2014;8 doi: 10.3389/fnhum.2014.00897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadaghiani S, Poline JB, Kleinschmidt A, D’Esposito M. Ongoing dynamics in large-scale functional connectivity predict perception. Proceedings of the National Academy of Sciences. 2015;112(27):8463–8468. doi: 10.1073/pnas.1420687112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salimi-Khorshidi G, Douaud G, Beckmann CF, Glasser MF, Griffanti L, Smith SM. Automatic denoising of functional mri data: combining independent component analysis and hierarchical fusion of classifiers. NeuroImage. 2014;90:449–468. doi: 10.1016/j.neuroimage.2013.11.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shakil S, Lee CH, Keilholz SD. Evaluation of sliding window correlation performance for characterizing dynamic functional connectivity and brain states. NeuroImage. 2016 doi: 10.1016/j.neuroimage.2016.02.074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shou H, Eloyan A, Lee S, Zipunnikov V, Crainiceanu A, Nebel M, Caffo B, Lindquist M, Crainiceanu C. Quantifying the reliability of image replication studies: The image intraclass correlation coefficient (i2c2) Cognitive, Affective, & Behavioral Neuroscience. 2013;13(4):714–724. doi: 10.3758/s13415-013-0196-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shrout P, Fleiss J. Intraclass correlations: Uses in assessing rater reliability. Psychological Bulletin. 1979;86(2):420–428. doi: 10.1037//0033-2909.86.2.420. [DOI] [PubMed] [Google Scholar]
- Smith SM, Beckmann CF, Andersson J, Auerbach EJ, Bijsterbosch J, Douaud G, Duff E, Feinberg DA, Griffanti L, Harms MP, et al. Resting-state fmri in the human connectome project. NeuroImage. 2013;80:144–168. doi: 10.1016/j.neuroimage.2013.05.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stehling MK, Turner R, Mansfield P. Echo-planar imaging: magnetic resonance imaging in a fraction of a second. Science. 1991;254(5028):43–50. doi: 10.1126/science.1925560. [DOI] [PubMed] [Google Scholar]
- Tagliazucchi E, Laufs H. Decoding wakefulness levels from typical fMRI resting-state data reveals reliable drifts between wakefulness and sleep. Neuron. 2014;82(3):695–708. doi: 10.1016/j.neuron.2014.03.020. [DOI] [PubMed] [Google Scholar]
- Thompson WH, Fransson P. The mean–variance relationship reveals two possible strategies for dynamic brain connectivity analysis in fmri. Frontiers in human neuroscience. 2015;9 doi: 10.3389/fnhum.2015.00398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tibshirani R, Walther G, Hastie T. Estimating the number of clusters in a data set via the gap statistic. Journal of the Royal Statistical Society: Series B (Statistical Methodology) 2001;63(2):411–423. [Google Scholar]
- Uğurbil K, Xu J, Auerbach EJ, Moeller S, Vu AT, Duarte-Carvajalino JM, Lenglet C, Wu X, Schmitter S, Van de Moortele PF, et al. Pushing spatial and temporal resolution for functional and diffusion mri in the human connectome project. Neuroimage. 2013;80:80–104. doi: 10.1016/j.neuroimage.2013.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Dijk KR, Hedden T, Venkataraman A, Evans KC, Lazar SW, Buckner RL. Intrinsic functional connectivity as a tool for human connectomics: theory, properties, and optimization. Journal of neurophysiology. 2010;103(1):297–321. doi: 10.1152/jn.00783.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Essen DC, Smith SM, Barch DM, Behrens TE, Yacoub E, Ugurbil K, Consortium WMH et al. The wu-minn human connectome project: an overview. NeuroImage. 2013;80:62–79. doi: 10.1016/j.neuroimage.2013.05.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wand MP, Jones MC. Kernel smoothing. Crc Press; 1994. [Google Scholar]
- Whitlow CT, Casanova R, Maldjian JA. Effect of resting-state functional mr imaging duration on stability of graph theory metrics of brain network connectivity. Radiology. 2011;259(2):516–524. doi: 10.1148/radiol.11101708. [DOI] [PubMed] [Google Scholar]
- Xu J, Moeller S, Auerbach EJ, Strupp J, Smith SM, Feinberg DA, Yacoub E, UȈgurbil K. Evaluation of slice accelerations using multiband echo planar imaging at 3t. NeuroImage. 2013;83:991–1001. doi: 10.1016/j.neuroimage.2013.07.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y, Lindquist MA. Dynamic connectivity detection: an algorithm for determining functional connectivity change points in fmri data. Frontiers in Neuroscience. 2015;9 doi: 10.3389/fnins.2015.00285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yaesoubi M, Miller RL, Calhoun VD. Mutually temporally independent connectivity patterns: A new framework to study the dynamics of brain connectivity at rest with application to explain group difference based on gender. NeuroImage. 2015;107:85–94. doi: 10.1016/j.neuroimage.2014.11.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yaesoubi M, Miller RL, Calhoun VD. Time-varying spectral power of resting-state fmri networks reveal cross-frequency dependence in dynamic connectivity. PloS one. 2017;12(2):e0171647. doi: 10.1371/journal.pone.0171647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z, Craddock RC, Margulies DS, Yan CG, Milham MP. Common intrinsic connectivity states among posteromedial cortex subdivisions: Insights from analysis of temporal dynamics. Neuroimage. 2014;93:124–137. doi: 10.1016/j.neuroimage.2014.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zalesky A, Fornito A, Cocchi L, Gollo LL, Breakspear M. Time-resolved resting-state brain networks. Proceedings of the National Academy of Sciences. 2014;111(28):10341–10346. doi: 10.1073/pnas.1400181111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuo XN, Xu T, Jiang L, Yang Z, Cao XY, He Y, Zang YF, Castellanos FX, Milham MP. Toward reliable characterization of functional homogeneity in the human brain: preprocessing, scan duration, imaging resolution and computational space. Neuroimage. 2013;65:374–386. doi: 10.1016/j.neuroimage.2012.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]


