Abstract
Adoptive transfer of anti-inflammatory FOXP3+ Tregs has gained attention as a new therapeutic strategy for auto-inflammatory disorders such as Inflammatory Bowel Disease. The isolated cells are conditioned in vitro to obtain a sufficient number of anti-inflammatory FOXP3+ Tregs that can be reintroduced into the patient to potentially reduce the pathologic inflammatory response. Previous evidence suggests that microbiota metabolites can potentially condition cells during the in vitro expansion/differentiation step. However, the number of combinations of cytokines and metabolites that can be varied is large, preventing a purely experimental investigation which would determine optimal cell therapeutic outcomes. To address this problem, a combined experimental and modeling approached is investigated here: an artificial neural network model was trained to predict the steady-state T cell population phenotype after differentiation with a variety of host cytokines and the microbial metabolite indole. This artificial neural network model was able to both reliably predict the phenotype of these T cell populations and also uncover unexpected conditions for optimal Treg differentiation that were subsequently verified experimentally.
Keywords: T cell, indole, gut microbiome, artificial neural network
Subject Category: Systems Biotechnology
INTRODUCTION
The human gastrointestinal (GI) tract is inhabited by approximately 1014 microorganisms of numerous species, presenting a diverse range of pro- and anti-inflammatory signals to the mucosal immune system in the GI tract. In order to maintain homeostasis in the GI tract, the mucosal immune system, also known as gut associated lymphoid tissue (GALT), must simultaneously be able to clear pathogenic bacteria from the system while preserving the non-pathogenic commensal bacteria in the GI tract (Kamada et al, 2013; Hill & Artis, 2010). Of the many immune cell types that compose the GALT, CD4+ T helper (Th)-cells have a unique role as orchestrators of the overall immune response (Koboziev et al, 2010). Activation of naïve T-cells in the unique cytokine/metabolite environment of the GALT promotes immune tolerance in physiologic, homeostatic conditions (Coombes & Powrie, 2008). On the other hand, Th cell activation can also result in inflammation in the GI tract (Shale et al, 2013), illustrating the delicate balance that the immune response of the GI tract must maintain: tolerance of abundant commensal antigens and clearance of pathogenic antigens. Genetic predisposition and/or environmental stressors such as infection or antibiotic use can disrupt the delicate homeostasis in the GI tract, giving rise to chronic intestinal inflammation that leads to inflammatory bowel diseases (IBD) (Gkouskou et al, 2014).
Treg and Th17 cells are two major Th cell lineages found at mucosal sites, such as the GI tract (Shale et al, 2013). Tregs comprise a heterogeneous lineage of T lymphocytes that induce immune tolerance by cytokine secretion and modulation of either APC or effector lymphocyte function (Sakaguchi et al, 2010; Vignali et al, 2008); thus, Tregs are an essential cellular determinant of immune homeostasis in most tissues. Tregs are classified based on FOXP3 expression, CD25HI expression, and a number of other surface or functional markers, and are functionally identified by their suppression of effector T-cells in vitro and in vivo (Sakaguchi et al, 2010). In the thymus, “natural” Tregs (nTregs) are produced when FOXP3 is induced in developing T lymphocytes that bind tightly with thymic stromal cell self peptide-MHC complex; in the periphery, “induced” Tregs (iTregs) are produced when Th cells are activated by antigen presenting cells in the presence of IL-2 and TGF-β (Sakaguchi et al, 2008). Perturbation of the Treg population in circulation and affected tissue is linked to autoimmune disorders (Baecher-Allan & Hafler, 2006) and IBD (Eastaff-Leung et al, 2009).
Th17 cells play an important role in autoimmune and aberrant inflammatory responses of mucosal surfaces in the GI tract (Weaver et al, 2013). Th17 cells comprise a class of activated Th cells that express high levels of RORγT, IL-17, IL-17F, IL-21, and IL-22; in addition, Th17 cells are reported to readily differentiate to IFN-γ+, Tbet+ Th1-like cells in vivo (Zúñiga et al, 2013). In the periphery, Th17 cells are produced when Th cell activation occurs in the presence of TGF-β and inflammatory cytokines such as IL-1β, IL-6, and IL-23 (Shale et al, 2013). Th17 cells and their secreted cytokines are increased in IBD (Eastaff-Leung et al, 2009), and genome-wide association studies have linked IBD and polymorphisms in the receptor for IL-23, a main driver of pro-inflammatory Th17 differentiation (Weaver et al, 2013). Mouse models of IBD have shown a similar importance for Th17 cells in pathogenesis (Ahern et al, 2008). Promotion of a Th17 response and subsequent inhibition of Tregs is thought to be important in the pathogenesis of aberrant inflammation in the GI tract (Ahern et al, 2008, 2010).
A healthy microbiota is an essential factor in full development of the Treg population and homeostasis in the GI tract (Atarashi et al, 2011; Ishikawa et al, 2008). Th17 cells are similarly dependent on microbiota (i.e., segmented filamentous bacteria, SFB) presence for proliferation in the GI tract, but the role of Th17 cells is reported to be clearance, rather than tolerance, of antigen (Shale et al, 2013; Basu et al, 2013). In addition to responding to foreign antigen, Th17 cells may also contribute to epithelial barrier integrity (Rutz et al, 2013). The balance between Treg and Th17 responses in the GI tract has multiple regulatory nodes that, when dysregulated, can lead to pathologic inflammation, that includes epithelial insult, microbiota dysbiosis, and development of genetic or epigenetic abnormalities in immune signaling pathways.
One hypothesis that has gained attention recently is that production of metabolites from the microbiota alters T cell differentiation (Rooks & Garrett, 2016). A number of groups have reported that short-chain fatty acids, a microbiota-derived product of dietary fiber breakdown, can promote both Treg and Th17 differentiation (Furusawa et al, 2013; Arpaia et al, 2013; Park et al, 2015). The microbiota-derived metabolite indole has recently been shown to be a potent augmenter of Treg differentiation and inhibitor of Th17 differentiation (Katepalli et al, 2013), suggesting that indole has a unique ability among the known microbiota-derived metabolites to consistently promote tolerance and inhibit inflammation. In support of the role of indole specifically as a tolerogenic signal from the microbiota, both IBD patients and colitic mice have a lower level of indole in stool samples (Hisamatsu et al, 2012; Schicho et al, 2010). Indole has also been linked to epithelial barrier integrity through aryl hydrocarbon receptor (AhR)-mediated responses (Bansal et al, 2010; Jin et al, 2014), further corroborating indole’s role as a cross-kingdom signaling molecule in vivo.
A promising new therapy for IBD involves culturing naïve T cells from a patient, differentiating the isolated naïve T cells to anti-inflammatory Treg cells, and transplanting them back into the patient in a technique known as Treg adoptive transfer (Fischbach et al, 2013; Fantini et al, 2006; Haribhai et al, 2009). By introducing Treg cells via adoptive transfer, the ultimate goal is to shift the balance back to a homeostatic level of Treg cells to counteract the pro-inflammatory Th17 cells. Despite the relative success of Treg adoptive transfer moving into clinical trials, questions remain about the best way to induce Tregs during the in vitro culture step (Himmel et al, 2012). The physiologic conditions that promote Treg differentiation in the GALT are not fully understood, but microbiota metabolites are emerging as key factors that shape T cell differentiation in the GALT. The ability of indole to promote anti-inflammatory Treg and inhibit pro-inflammatory Th17 differentiation, as well as the decrease of indole in the stool of IBD patients, suggests that indole from the microbiota could be an important point of regulation missing in vivo that can be reintroduced during the in vitro culture step of Treg adoptive transfer.
Although the signaling pathways underlying the influence of host cytokines on T cell differentiation are beginning to be uncovered (Carbo et al, 2013), little is known about the effect of bacterial metabolites on these signaling cascades. Unfortunately, the number of host cytokines and bacterial metabolites present in this system poses a challenge for a purely experimental approach aimed at elucidating the effects on T cell differentiation. Even if the effects of host cytokines on the differentiation process are assumed to be known, adding a bacterial metabolite with unknown effect forces the experimenter to investigate the interactions between the bacterial metabolite and host cytokines over a wide range, with control experiments that leave out the bacterial metabolite. Therefore, understanding these interactions at a wide range of concentrations quickly becomes experimentally cumbersome. One solution to this challenge is to use a combined experimental/modeling approach which reduces the amount of data needed for making predictions. Empirical models, such as artificial neural networks (ANNs), are particularly well suited for the modeling task as they are very flexible in their structure and the type of behavior that they can predict. Specifically, multilayer perceptrons with sigmoid transfer functions have been proven to be able to represent any general function arbitrarily well (provided a sufficient number of neurons are used) (Cybenko, 1989), enabling these metabolite effects to be sufficiently modeled in the absence of knowledge on the particular interconnections and nonlinear effects in these complex differentiation pathways. This work employs ANNs as a tool for uncovering the complex relationship between microbial metabolites and T cell differentiation into Th17 and Treg progeny with the ultimate goal to derive host cytokine and bacterial metabolite conditions which will result in high levels of anti-inflammatory Treg and low levels of pro-inflammatory Th17 after differentiation.
MATERIALS AND METHODS
Chemicals
Sodium Propionate, sodium acetate, sodium butyrate and sodium chloride (all from Sigma-Aldrich) were solubilized in media and added to culture. Indole was solubilized in DMF (both from Sigma-Aldrich) at 2M and brought to final concentration in media. MGCD0103 (Selleck Chemicals) was solubilized in DMF at 2 mM and brought to final concentration in media. A 0.05% DMF solvent control was used for all experiments.
In vitro T Cell Differentiation
CD4+ CD25- T-cells were isolated to high purity (>98%) from pooled spleen and mesenteric lymph nodes of wild type C57BL/6 mice (Jackson labs) with a BD FACS Aria II flow cytometer. Fc Block (BD), αCD4-efluor450 (eBioscience clone GK1.5), and αCD25-PECy7 (eBioscience clone PC61.5) antibodies were used for staining before sorting.
Isolated murine CD4+ CD25− T cells were cultured at an initial concentration of 1x105 cells/well in RPMI 1640 supplemented with 2-mercaptoethanol, gentamicin, penicillin, streptomycin, and 10% FCS (all from Life Technologies) in a 96-well round bottom plate (Falcon) coated with 5 μg/mL αCD3 (BioXcell clone 145-2C11) and 2 μg/mL αCD28 (BioXcell clone 37.51) for 72 hours. For conditioning media, 0–20 ng/mL IL-6 (Peprotech), 0–10 ng/mL IL-23 (R&D), 0–10 ng/mL TGF-β (Peprotech), 0–500 U/mL IL-2 (Roche), 10 μg/mL αIL-4 (BioXCell clone 11B11), and 10 μg/mL αIFN-γ (BioXCell clone R4-6A2) were added before culture.
Intracellular Cytokine and Transcription Factor Staining
For the last four hours of culture, cells were treated with golgi plug (BD), PMA and ionomycin (both from Sigma-Aldrich). Cells were fixed with the FOXP3 fixation buffer (eBioscience) or 4% paraformaldehyde (Sigma-Aldrich), treated with permeabilization buffer (eBioscience) and stained with appropriate antibodies. The antibodies used were: αIL-17a-PE or –APC (eBioscience clone eBio17B7 or BD Biosciences clone N49-653) and αFOXP3-APC or –FITC (eBioscience clone FJK-16s or BD Biosciences clone 259D/C7).
Experimental Design
Initial combinatorial experimental studies were generated using concentrations of IL-6, IL-23, IL-2, TGF-β, and indole that included a null concentration, a functional concentration, and a concentration that results in maximal effect on promoting Treg or Th17 differentiation (Supplemental Table I). Functional concentrations of host cytokines have been reported previously (Sekiya & Yoshimura, 2016) and saturating conditions were evaluated from dose-response curves (data not shown). Physiologically-relevant concentrations of indole were determined from previous reports of human fecal concentrations in the low millimolar range (Karlin et al, 1985; Zuccato et al, 1993). These selected concentrations were investigated in a full factorial experimental design and are represented in Table I. Each combination was evaluated thrice (technical replicates) on cultures arising from two different animals (biological replicates). Means were computed for the technical replicates and are shown in Supplemental Tables I and II; furthermore, these mean values were used for model training (S. Table I) and validation (S. Table II).
Table I.
Stimulation conditions used in the initial fully factorial experimental studies.
| Differentiation Signal | Training Concentrations | Units |
|---|---|---|
| IL-6 | {0, 10, 20} | ng/mL |
| IL-23 | {0, 5, 10} | ng/mL |
| IL-2 | {0, 100, 500} | U/mL |
| TGF-β | {0, 0.5, 2, 10} | ng/mL |
| Indole | {0, 0.5, 1} | mM |
Artificial Neural Network Modeling
The ANN models considered contained between two and five neurons with sigmoid transfer functions in a single hidden layer. The percentages of Treg (FOXP3+) and Th17 (IL17+) cells (outputs) were predicted from the concentrations of cytokines and indole in the stimulation media (inputs). The inputs were log-transformed prior to training. The data were divided into training, testing, and validation sets in a 70:15:15 ratio with all replicates in the same set and all extreme points in the training set. Models were trained by the Levenberg-Marquardt algorithm in the MATLAB Neural Network Toolbox.
Model Selection
Retaining the extreme points in the training set, the data were divided 10,000 times by randomly assigning the remaining sets of experimental conditions to the training, testing, or validation sets while maintaining the 70:15:15 data division. ANNs with sigmoid transfer functions in a single hidden layer and a linear output layer (i.e. multilayer perceptrons) of differing sizes were trained on each data partition. Then, a normalized version of the popular Akaike Information Criterion (AIC) (Akaike, 1974) was used to select a model of the appropriate size. The interested reader is referred to the wide body of literature on information criteria for model selection, e.g. (Kuha, 2004; Ljung, 1999).
Briefly, for a model with np parameters, the normalized AIC (nAIC ) can be written as
where θ̂N is the vector of estimated parameters given the first N measurements and ei(θ̂N) represents the i -th prediction error. The nAIC was computed for all ANNs trained on different data partitions and the hidden layer size with a minimum expected nAIC (E[nAIC] ) was used to select the appropriate size of the ANN.
RESULTS
Initial Experimental Studies
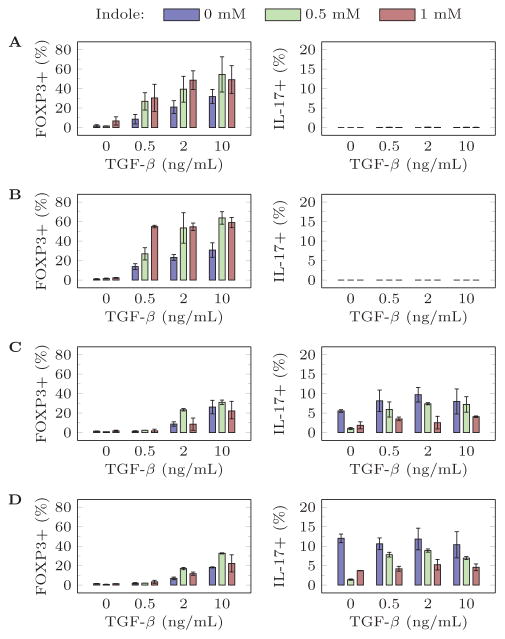
Since the ANN is data-driven, limited initial experimental studies based on a full factorial design of experiments was used to collect data for training. Selected two-way relationships from these initial screens are presented in Figure 1. Note that some concentrations did not produce any IL17+ cells (Figure 1a and Figure 1b). A positive correlation between Th 17 induction and the pro-inflammatory cytokines IL-6 and IL-23 is observed, whereas a low concentration of TGF-β optimally induces Th17 (Figure 1c and Figure 1d). TGF-β positively correlates with Treg differentiation (Figure 1). All of these relationships are supported by the existing experimental literature (Carbo et al, 2013). Interestingly, indole promotes Treg and inhibits Th17 differentiation at all cytokine concentrations with varying levels of dose response. These results support the hypothesis that microbial metabolites influence Th differentiation in the GALT and that indole can provide an important signal for stabilizing Treg populations prior to adoptive transfer.
Figure 1.
Selected relationships in the initial training data set with IL2 = 100 U/mL and (A) IL6 = 0 ng/mL, IL23 = 0 ng/mL (B) IL6 = 0 ng/mL, IL23 = 5 ng/mL (C) IL6 = 10 ng/mL, IL23 = 0 ng/mL (D) IL6 = 10 ng/mL, IL23 = 5 ng/mL.
Model Selection and Training Performance
ANNs of differing size were evaluated to determine a model that best represents the complex interconnections between the differentiation signals and resulting differentiated T cell distributions while avoiding overfitting. Overfitting a model to data enables a model to perform exceedingly well on the existing data, but poorly when new data are considered; however, it is the ability to predict, rather than the goodness of the fit, that is of interest for a model. The normalized Akaike Information Criterion (nAIC ) was used to determine the model size that best represents the data while minimizing overfitting. The expected nAIC (E[nAIC] ) over all data partitions for differing neural network sizes is presented in Table II. These results indicate that an ANN with four neurons in a single hidden layer is sufficient for describing these data. Then, the neural networks with four neurons were ranked by the Euclidean distance of the testing R2 to the point (1,1); the network with the minimum distance was selected for all subsequent analysis. The performance of the final ANN on the training, testing, and validation sets is presented in Table III.
Table II.
The E[nAIC] for different neural network sizes.
| Number of Hidden Layer Neurons | E[nAIC] |
|---|---|
| 2 | 10.98 |
| 3 | 10.65 |
| 4 | 10.54 |
| 5 | 10.56 |
Table III.
Performance of the final ANN.
| R2 | |||
|---|---|---|---|
| Training | Validation | Testing | |
| % FOXP3+ | 0.8969 | 0.9046 | 0.9006 |
| % IL-17+ | 0.7321 | 0.7619 | 0.6427 |
Model Predictions
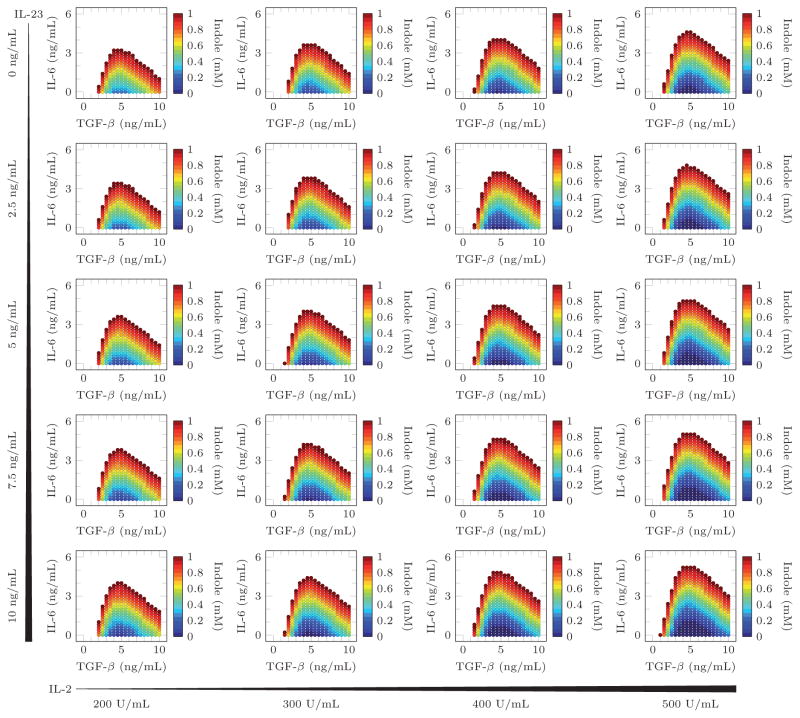
The ANN was then used to predict cytokine and indole concentrations that gave maximal (>50%) Treg induction minimal (<5%) Th17 induction. These predictions are visualized in Figure 2. Only the minimum concentration of indole necessary to achieve the desired phenotype is shown since higher concentrations always preserved or improved the resulting phenotype. As expected from the training data, in the absence of IL-6 and at a low TGF-β concentration, a high concentration of indole is necessary to sufficiently polarize the differentiated T cell distribution to favor Treg differentiation and suppress Th17 differentiation. However, the model predicts that at intermediate TGF-β concentrations, only a small amount of indole is required to obtain the desired differentiated T cell distribution. Furthermore, as the TGF-β concentration continues to increase, higher doses of indole are again needed to obtain high Treg and low Th17 induction.
Figure 2.
Input concentrations predicted to produce more than 50% FoxP3+ and less than 5% Th17 cells. The indole concentrations represent the minimum amount of indole needed to satisfy the prediction criteria.
These model predictions were verified by testing a higher resolution of indole concentrations with a low, intermediate, or high concentration of TGF-β (Supplemental Table II). The results of these validation experiments are presented in Figure 3 and Figure 4. The validation experiments with IL6 = 0 ng/mL failed to produce any IL17+ cells. Interestingly, in the presence of low concentration pro-inflammatory IL-6 (3 ng/mL), the model predicts that a high indole concentration should still be able to maximize Treg and minimize Th17 differentiation but only at an intermediate concentration of TGF-β. This result was experimentally verified and, indeed, indole has maximal anti-inflammatory effects on T cell differentiation only at an intermediate TGF-β concentration.
Figure 3.
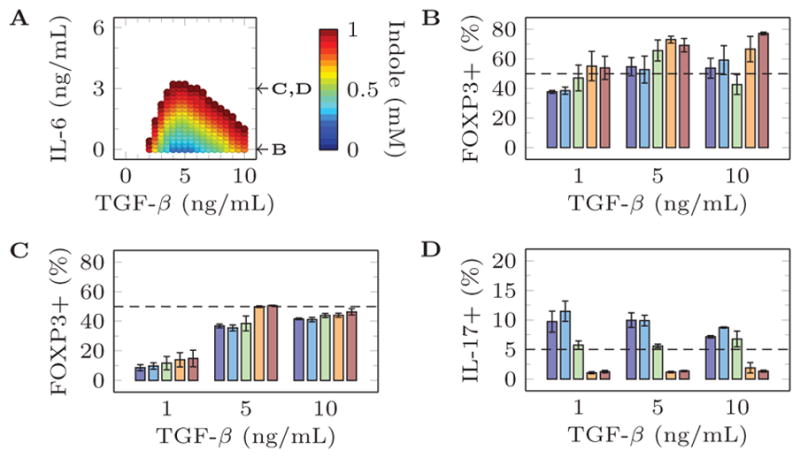
Comparison of model predictions and experimental validation for 0 ng/mL Il-23, 200 U/mL IL-2, and selected concentrations of other metabolites. (a) Model predictions producing more than 50% FoxP3+ and less than 5% Th17 cells. (b) Experimental validation of percentage of FOXP3+ cells after stimulation with 0 ng/mL Il-23, 200 U/mL IL-2, 0 ng/mL IL-6, {1,5,10} ng/mL TGF-β, and {0, 0.25, 0.5, 0.75, 1} mM Indole. (c) Experimental validation of percentage of FOXP3+ cells after stimulation with 0 ng/mL Il-23, 200 U/mL IL-2, 3 ng/mL IL-6, {1,5,10} ng/mL TGF-β, and {0, 0.25, 0.5, 0.75, 1} mM Indole. (d) Experimental validation of percentage of IL-17+ cells after stimulation with 0 ng/mL Il-23, 200 U/mL IL-2, 3 ng/mL IL-6, {1,5,10} ng/mL TGF-β, and {0, 0.25, 0.5, 0.75, 1} mM Indole.
Figure 4.
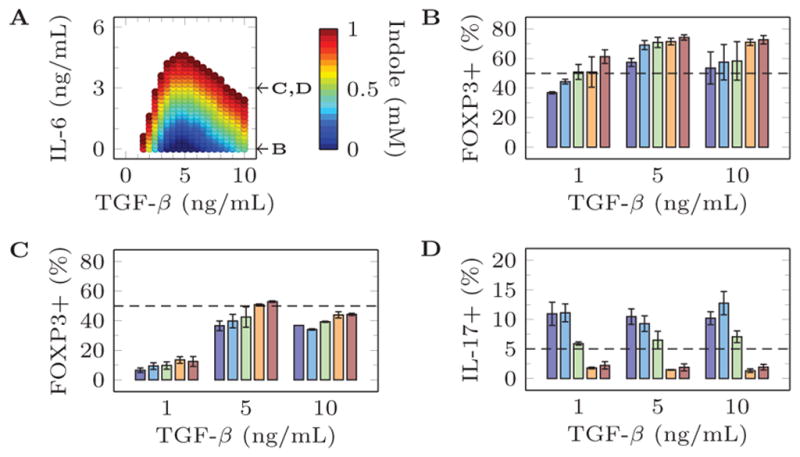
Comparison of model predictions and experimental validation for 0 ng/mL Il-23, 500 U/mL IL-2, and selected concentrations of other metabolites. (a) Model predictions producing more than 50% FoxP3+ and less than 5% Th17 cells. (b) Experimental validation of percentage of FOXP3+ cells after stimulation with 0 ng/mL Il-23, 500 U/mL IL-2, 0 ng/mL IL-6, {1,5,10} ng/mL TGF-β, and {0, 0.25, 0.5, 0.75, 1} mM Indole. (c) Experimental validation of percentage of FOXP3+ cells after stimulation with 0 ng/mL Il-23, 500 U/mL IL-2, 3 ng/mL IL-6, {1,5,10} ng/mL TGF-β, and {0, 0.25, 0.5, 0.75, 1} mM Indole. (d) Experimental validation of percentage of IL-17+ cells after stimulation with 0 ng/mL Il-23, 500 U/mL IL-2, 3 ng/mL IL-6, {1,5,10} ng/mL TGF-β, and {0, 0.25, 0.5, 0.75, 1} mM Indole.
DISCUSSION
The use of T cells as a therapeutic poses a unique challenge due to their intricate nature and an incomplete understanding of mechanisms that underlie clinical efficacy. Despite these obstacles, Treg adoptive transfer has significant clinical potential, suggesting that cellular therapeutics will move forward based on their end-stage effect without a complete understanding of the mechanisms leading to alleviation of auto-immune and inflammatory diseases.
A T cell activated towards the Treg phenotype in vivo likely receives a diverse array of significant signals that are not fully known, let alone understood at a mechanistic level. Essential host cytokines have been identified that are necessary for Treg differentiation, but microbiota-derived metabolites are actively being identified that have a specialized role to promote Tregs in the GI tract. This additional knowledge of physiologic Treg conditioning holds promise for further refining cellular therapeutics, because they offer additional points of modulation of the cellular therapeutic before reintroducing the cells into a patient. As these diverse physiological cues are better understood, they can be incorporated into cellular therapeutic approaches and personalized treatment protocols can be established.
A fully experimental approach to investigate the effects of indole on the T cell differentiation process was both cost- and time-prohibitive. If five experimental conditions for each cytokine/metabolite were investigated, a full factorial experimental design results in 3125 different experimental conditions. However, building an empirical model from data obtained from a coarser input grid results in accurate predictions of the T cell population phenotype, while reducing experimental burden. Furthermore, since empirical models are data-agnostic, many model types, estimation procedures, model analysis techniques, etc. can be found in widely-available commercial and open-source programming environments. Using an ANN to build an empirical model of T cell differentiation proved computationally feasible and experimentally manageable. The ANN supplemented a traditional experimental approach by allowing conclusions to be drawn from a continuous set of input conditions rather than only at the discrete input conditions that were actually implemented experimentally. Furthermore, the ANN trained on a limited set of training data produced unexpected predictions about T cell activation in the presence of microbiota metabolites at concentrations that were not included in the training set (e.g. at 5 ng/mL TGF-β, Figure 1); these experimental conditions were subsequently verified experimentally (Figures 3 and 4). These results support the use of the neural network model as a tool to predict the complex interplay between the multiple signals a T cell receives during activation and ultimately guide cellular therapeutics toward a tailored approach, providing therapeutic applicability across multiple auto-immune and auto-inflammatory diseases currently not being considered as strong candidates for Treg adoptive transfer therapy.
Optimal T cell conditions heavily favored the use of indole in differentiating toward a high Treg/low Th17 phenotype acceptable for adoptive transfer applications. Naïve T cells were able to tolerate much higher amounts of IL-6 in the presence of indole while still adopting a phenotype with low numbers of Th17 cells, supporting the hypothesis that indole preserves homeostasis in the GI tract. Furthermore, the range of host cytokine inputs that produce acceptable T cell population phenotypes is greatly increased by the addition of indole, suggesting that adding indole to T cell stimulation conditions may help stabilize the Treg population and more reliably produce the desired high Treg/low Th17 T cell population in vitro.
The results described herein contribute to indole’s role in immunomodulation. Indole is a major extracellular metabolite produced from gut microbiota (Bansal et al, 2010) and low millimolar concentrations have been reported in human feces (Karlin et al, 1985; Zuccato et al, 1993) and mice cecum and feces (Jin et al, 2014). Indole has been shown to be an AhR antagonist in Caco-2 cells (Jin et al, 2014). Although some evidence suggests that agonist/antagonist AhR activity is cell/tissue-dependent (Jordan, 2007), AhR agonists have been shown to enhance Th17 differentiation in vitro (Duarte et al, 2013), suggesting that indole acts as an AhR antagonist in the in vitro T cell differentiation reported herein. Furthermore, since indole has been shown to be a weak AhR agonist/partial AhR antagonist in complex colon crypts (Jin et al, 2014) and traditional AhR agonists have shown differential effects in vivo, biasing T cells toward the Treg phenotype via indole and other AhR antagonists in an adoptive transfer scheme is potentially more advantageous than directly affecting in vivo T cell populations via microbial metabolite-derived therapies.
The extreme complexity inherent to cells is both the reason cellular therapy is promising and also difficult to clinically implement. The necessity to analyze a patient’s individual inflammatory status and cellular response to cytokine, metabolite, and other as yet to be determined factors could be a roadblock to the development of cellular therapy as a broadly utilized, personalized treatment. This work starts to address this problem by generating a highly adaptable protocol that uses a limited set of empirical data representing T cell response to conditioning factors in order to successfully predict the response of the cells to a wide range of combinations of conditioning factors that would require a prohibitive amount of supplies and labor to explore experimentally.
Supplementary Material
Acknowledgments
The authors appreciate partial financial support from NIH via grant R01AI110642.
Footnotes
AUTHOR CONTRIBUTIONS
A.J., J.H., and R.C.A. conceptualized the project. S.S. performed the experiments. D.P.H. performed the modeling. A.J. and J.H. supervised successful completion of the project. D.P.H. and S.S. wrote the first draft of the manuscript. J.H., A.J., and R.C.A., D.P.H., and S.S. edited the manuscript.
CONFLICT OF INTEREST
The authors declare no conflict of interest.
References
- Ahern PP, Izcue A, Maloy KJ, Powrie F. The interleukin-23 axis in intestinal inflammation. Immunol Rev. 2008;226:147–159. doi: 10.1111/j.1600-065X.2008.00705.x. [DOI] [PubMed] [Google Scholar]
- Ahern PP, Schiering C, Buonocore S, McGeachy MJ, Cua DJ, Maloy KJ, Powrie F. Interleukin-23 Drives Intestinal Inflammation through Direct Activity on T Cells. Immunity. 2010;33:279–288. doi: 10.1016/j.immuni.2010.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akaike H. A new look at the statistical model identification. Autom Control IEEE Trans On. 1974;19:716–723. [Google Scholar]
- Arpaia N, Campbell C, Fan X, Dikiy S, van der Veeken J, deRoos P, Liu H, Cross JR, Pfeffer K, Coffer PJ, Rudensky AY. Metabolites produced by commensal bacteria promote peripheral regulatory T-cell generation. Nature. 2013;504:451–455. doi: 10.1038/nature12726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atarashi K, Tanoue T, Shima T, Imaoka A, Kuwahara T, Momose Y, Cheng G, Yamasaki S, Saito T, Ohba Y, Taniguchi T, Takeda K, Hori S, Ivanov II, Umesaki Y, Itoh K, Honda K. Induction of Colonic Regulatory T Cells by Indigenous Clostridium Species. Science. 2011;331:337–341. doi: 10.1126/science.1198469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baecher-Allan C, Hafler DA. Human regulatory T cells and their role in autoimmune disease. Immunol Rev. 2006;212:203–216. doi: 10.1111/j.0105-2896.2006.00417.x. [DOI] [PubMed] [Google Scholar]
- Bansal T, Alaniz RC, Wood TK, Jayaraman A. The bacterial signal indole increases epithelial-cell tight-junction resistance and attenuates indicators of inflammation. Proc Natl Acad Sci. 2010;107:228–233. doi: 10.1073/pnas.0906112107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basu R, Hatton RD, Weaver CT. The Th17 family: flexibility follows function. Immunol Rev. 2013;252:89–103. doi: 10.1111/imr.12035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carbo A, Hontecillas R, Kronsteiner B, Viladomiu M, Pedragosa M, Lu P, Philipson CW, Hoops S, Marathe M, Eubank S, Bisset K, Wendelsdorf K, Jarrah A, Mei Y, Bassaganya-Riera J. Systems modeling of molecular mechanisms controlling cytokine-driven CD4+ T cell differentiation and phenotype plasticity. PLoS Comput Biol. 2013;9:e1003027. doi: 10.1371/journal.pcbi.1003027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coombes JL, Powrie F. Dendritic cells in intestinal immune regulation. Nat Rev Immunol. 2008;8:435–446. doi: 10.1038/nri2335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cybenko G. Approximation by superpositions of a sigmoidal function. Math Control Signals Syst. 1989;2:303–314. [Google Scholar]
- Duarte JH, Di Meglio P, Hirota K, Ahlfors H, Stockinger B. Differential Influences of the Aryl Hydrocarbon Receptor on Th17 Mediated Responses in vitro and in vivo. [Accessed December 14, 2016];PLoS ONE. 2013 :8. doi: 10.1371/journal.pone.0079819. Available at: http://www.ncbi.nlm.nih.gov/pmc/articles/PMC3828240/ [DOI] [PMC free article] [PubMed]
- Eastaff-Leung N, Mabarrack N, Barbour A, Cummins A, Barry S. Foxp3+ Regulatory T Cells, Th17 Effector Cells, and Cytokine Environment in Inflammatory Bowel Disease. J Clin Immunol. 2009;30:80–89. doi: 10.1007/s10875-009-9345-1. [DOI] [PubMed] [Google Scholar]
- Fantini MC, Becker C, Tubbe I, Nikolaev A, Lehr HA, Galle P, Neurath MF. Transforming growth factor β induced FoxP3+ regulatory T cells suppress Th1 mediated experimental colitis. Gut. 2006;55:671–680. doi: 10.1136/gut.2005.072801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischbach MA, Bluestone JA, Lim WA. Cell-Based Therapeutics: The Next Pillar of Medicine. Sci Transl Med. 2013;5:179ps7–179ps7. doi: 10.1126/scitranslmed.3005568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furusawa Y, Obata Y, Fukuda S, Endo TA, Nakato G, Takahashi D, Nakanishi Y, Uetake C, Kato K, Kato T, Takahashi M, Fukuda NN, Murakami S, Miyauchi E, Hino S, Atarashi K, Onawa S, Fujimura Y, Lockett T, Clarke JM, et al. Commensal microbe-derived butyrate induces the differentiation of colonic regulatory T cells. Nature. 2013;504:446–450. doi: 10.1038/nature12721. [DOI] [PubMed] [Google Scholar]
- Gkouskou KK, Deligianni C, Tsatsanis C, Eliopoulos AG. The gut microbiota in mouse models of inflammatory bowel disease. [Accessed May 12, 2016];Front Cell Infect Microbiol. 2014 :4. doi: 10.3389/fcimb.2014.00028. Available at: http://www.ncbi.nlm.nih.gov/pmc/articles/PMC3937555/ [DOI] [PMC free article] [PubMed]
- Haribhai D, Lin W, Edwards B, Ziegelbauer J, Salzman NH, Carlson MR, Li S-H, Simpson PM, Chatila TA, Williams CB. A central role for induced regulatory T cells in tolerance induction in experimental colitis. J Immunol Baltim Md 1950. 2009;182:3461–3468. doi: 10.4049/jimmunol.0802535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill DA, Artis D. Intestinal Bacteria and the Regulation of Immune Cell Homeostasis. Annu Rev Immunol. 2010;28:623–667. doi: 10.1146/annurev-immunol-030409-101330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Himmel ME, Yao Y, Orban PC, Steiner TS, Levings MK. Regulatory T-cell therapy for inflammatory bowel disease: more questions than answers. Immunology. 2012;136:115–122. doi: 10.1111/j.1365-2567.2012.03572.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hisamatsu T, Okamoto S, Hashimoto M, Muramatsu T, Andou A, Uo M, Kitazume MT, Matsuoka K, Yajima T, Inoue N, Kanai T, Ogata H, Iwao Y, Yamakado M, Sakai R, Ono N, Ando T, Suzuki M, Hibi T. Novel, Objective, Multivariate Biomarkers Composed of Plasma Amino Acid Profiles for the Diagnosis and Assessment of Inflammatory Bowel Disease. PLOS ONE. 2012;7:e31131. doi: 10.1371/journal.pone.0031131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishikawa H, Tanaka K, Maeda Y, Aiba Y, Hata A, Tsuji NM, Koga Y, Matsumoto T. Effect of intestinal microbiota on the induction of regulatory CD25+ CD4+ T cells. Clin Exp Immunol. 2008;153:127–135. doi: 10.1111/j.1365-2249.2008.03668.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin U-H, Lee S-O, Sridharan G, Lee K, Davidson LA, Jayaraman A, Chapkin RS, Alaniz R, Safe S. Microbiome-Derived Tryptophan Metabolites and Their Aryl Hydrocarbon Receptor-Dependent Agonist and Antagonist Activities. Mol Pharmacol. 2014;85:777–788. doi: 10.1124/mol.113.091165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jordan VC. SERMs: Meeting the Promise of Multifunctional Medicines. J Natl Cancer Inst. 2007;99:350–356. doi: 10.1093/jnci/djk062. [DOI] [PubMed] [Google Scholar]
- Kamada N, Seo S-U, Chen GY, Núñez G. Role of the gut microbiota in immunity and inflammatory disease. Nat Rev Immunol. 2013;13:321–335. doi: 10.1038/nri3430. [DOI] [PubMed] [Google Scholar]
- Karlin DA, Mastromarino AJ, Jones RD, Stroehlein JR, Lorentz O. Fecal skatole and indole and breath methane and hydrogen in patients with large bowel polyps or cancer. J Cancer Res Clin Oncol. 1985;109:135–141. doi: 10.1007/BF00391888. [DOI] [PubMed] [Google Scholar]
- Katepalli M, Mueller C, Steinmeyer S, Jayaraman A, Alaniz R. The microbiota-derived signal indole regulates T-cell fate by reciprocal control of Th17 and Treg development (P3178) J Immunol. 2013;190:61.14–61.14. [Google Scholar]
- Koboziev I, Karlsson F, Grisham MB. Gut-associated lymphoid tissue, T cell trafficking, and chronic intestinal inflammation. Ann N Y Acad Sci. 2010;1207:E86–E93. doi: 10.1111/j.1749-6632.2010.05711.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuha J. AIC and BIC: Comparisons of Assumptions and Performance. Sociol Methods Res. 2004;33:188–229. [Google Scholar]
- Ljung L. System Identification: Theory for the User. Upper Saddle River, NJ: Prentice Hall PTR; 1999. [Google Scholar]
- Park J, Kim M, Kang SG, Jannasch AH, Cooper B, Patterson J, Kim CH. Short-chain fatty acids induce both effector and regulatory T cells by suppression of histone deacetylases and regulation of the mTOR-S6K pathway. Mucosal Immunol. 2015;8:80–93. doi: 10.1038/mi.2014.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rooks MG, Garrett WS. Gut microbiota, metabolites and host immunity. Nat Rev Immunol. 2016;16:341–352. doi: 10.1038/nri.2016.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutz S, Eidenschenk C, Ouyang W. IL-22, not simply a Th17 cytokine. Immunol Rev. 2013;252:116–132. doi: 10.1111/imr.12027. [DOI] [PubMed] [Google Scholar]
- Sakaguchi S, Miyara M, Costantino CM, Hafler DA. FOXP3+ regulatory T cells in the human immune system. Nat Rev Immunol. 2010;10:490–500. doi: 10.1038/nri2785. [DOI] [PubMed] [Google Scholar]
- Sakaguchi S, Yamaguchi T, Nomura T, Ono M. Regulatory T Cells and Immune Tolerance. Cell. 2008;133:775–787. doi: 10.1016/j.cell.2008.05.009. [DOI] [PubMed] [Google Scholar]
- Schicho R, Nazyrova A, Shaykhutdinov R, Duggan G, Vogel HJ, Storr M. Quantitative Metabolomic Profiling of Serum and Urine in DSS-Induced Ulcerative Colitis of Mice by 1H NMR Spectroscopy. J Proteome Res. 2010;9:6265–6273. doi: 10.1021/pr100547y. [DOI] [PubMed] [Google Scholar]
- Sekiya T, Yoshimura A. In Vitro Th Differentiation Protocol. In: Feng X-H, Xu P, Lin X, editors. TGF-β Signaling. Springer; New York: 2016. [Accessed November 2, 2016]. pp. 183–191. Available at: http://dx.doi.org/10.1007/978-1-4939-2966-5_10. [Google Scholar]
- Shale M, Schiering C, Powrie F. CD4+ T-cell subsets in intestinal inflammation. Immunol Rev. 2013;252:164–182. doi: 10.1111/imr.12039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vignali DAA, Collison LW, Workman CJ. How regulatory T cells work. Nat Rev Immunol. 2008;8:523–532. doi: 10.1038/nri2343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weaver CT, Elson CO, Fouser LA, Kolls JK. The Th17 Pathway and Inflammatory Diseases of the Intestines, Lungs and Skin. Annu Rev Pathol. 2013;8:477–512. doi: 10.1146/annurev-pathol-011110-130318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuccato E, Venturi M, Di Leo G, Colombo L, Bertolo C, Doldi SB, Mussini E. Role of bile acids and metabolic activity of colonic bacteria in increased risk of colon cancer after cholecystectomy. Dig Dis Sci. 1993;38:514–519. doi: 10.1007/BF01316508. [DOI] [PubMed] [Google Scholar]
- Zúñiga LA, Jain R, Haines C, Cua DJ. Th17 cell development: from the cradle to the grave. Immunol Rev. 2013;252:78–88. doi: 10.1111/imr.12036. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.