Abstract
Recent structural characterizations of the p51 and p66 monomers has established an important starting point for understanding the maturation pathway of the HIV-1 reverse transcriptase p66/p51 heterodimer. This process requires a metamorphic transition of the polymerase domain leading to formation of a p66/p66′ homodimer that exists as a structural heterodimer. In order to better understand the drivers for this metamorphic transition, we have performed NMR studies of 15N-labeled RT216 - a construct that includes the fingers and most of the palm domains. These studies are consistent with the conclusion that the p66 monomer exists as a spring-loaded complex. Initial dissociation of the fingers/palm:connection complex allows the fingers/palm to adopt an alternate, more stable structure, reducing the rate of re-association and facilitating subsequent maturation steps. One of the drivers for an initial extension of the fingers/palm domains is identified as a straightening of helix E relative to its conformation in the monomer by eliminating a bend of ~ 50° near residue Phe160. NMR and CD data also are consistent with the conclusion that a hydrophobic surface of palm domain that becomes exposed after the initial dissociation, as well as the intrinsic conformational preferences of the palm domain C-terminal segment, facilitate formation of the β-sheet structure that is unique to the active polymerase subunit. Spectral comparisons based on 15N-labeled constructs are all consistent with previous structural conclusions based on studies of 13C-methyl-labeled constructs.
Keywords: HIV-1 reverse transcriptase, NMR, nuclear magnetic resonance, metamorphic, structural maturation
INTRODUCTION
Reverse transcription of the single-stranded viral RNA genome into double-stranded DNA is an essential step in the human immunodeficiency virus (HIV) life-cycle that is dependent on the enzymatic activities of the retroviral enzyme reverse transcriptase (RT). Mature HIV-1 RT is a p66/p51 heterodimer composed of a p66 subunit containing the active polymerase and RNase H sites, and a second p51 subunit that is formed from the same polymerase domain that has adopted an alternate fold in which the polymerase active site is buried and nonfunctional. This unusual structure provides a mechanism for responding to the strong selective pressure to minimize genome size of RNA viruses [1]; the HIV viral genome is only 9.2 kB [2], a value typical for RNA viruses [3, 4]. Reliance on a single protein that adopts two alternate folding patterns provides a means of increasing the information density of the HIV genome. In the case of RT, this economy of coding comes at the cost of a complex maturation process, such that a pair of p66 precursors develop along different pathways, one forming the polymerase and RNase H active sites, while the second adopts an inactive fold with a buried polymerase active site, and loses most of its RH domain. Shortly after the structure of the RT heterodimer was first determined [5, 6], a structural analysis led Steitz and coworkers to conclude that the inactive structure of the polymerase domain that is present in the p51 subunit is more stable than the active structure observed in the p66 subunit [7]. Based on this analysis, they proposed that homodimers formed from either p51 or p66 would be structurally asymmetric, formed by combining a pair of polymerase domains that have adopted the two alternate structures.
Recent NMR, crystallographic, and molecular dynamics studies have provided structural information about the p66 monomer precursor and elucidated the three major steps involved in formation of the heterodimer [1, 8–11]: 1) Unimolecular rearrangement of the monomer subdomains to a structure approximating the mature p66 subunit; 2) initial homodimer formation by combining the structurally isomerized p66 with the predominant monomer; 3) very slow conformational changes that result in subunit-specific RH-domain unfolding, exposing the buried proteolysis site within the RH domain. These studies have elucidated many additional details about the factors that underlie these complex conformational transitions. Further, they provide basic insights into the mechanisms by which the metamorphic polymerase domain is able to adopt alternate structures.
As more structural information for complex biological molecules has become available, metamorphic proteins, i.e. proteins for which there is not a unique relationship between sequence and structure, have been increasingly identified [12–16]. It thus becomes of increasing interest to identify the factors that influence and are involved in the corresponding metamorphic structural transition. For HIV-1 reverse transcriptase, the metamorphic transition involves tension that exists between the p66 monomer structure that corresponds to a global energy minimum, and the intrinsic conformational preferences of its component domains [10]. For example, recent studies indicate that subsequent to dissociation of the fingers/palm from the connection domain, the domains undergo a conformational expansion that alters the relative orientation of the fingers and palm. However, understanding of the structural basis for this transition remains limited.
Despite the substantial progress that has been made in the development of RT-targeted drugs, RT remains an important target due to the ability of the enzyme to develop drug-resistant mutations [17, 18], and to the toxicity of nucleoside RT inhibitors (NRTIs) [19], that must be administered chronically since there is as yet no cure for this illness. This studies described here have two objectives: 1) Utilization of 15N-labeling approach to further evaluate earlier conclusions based largely on 13C-methyl labeling studies [8, 10, 11], and to further investigate the basis for the metamorphic transition that is an essential step of RT maturation, and 2) to compare results with recent NMR studies of 15N-labeled p66 reported by Sharaf et al. [20] that reached a different conclusion about the nature of the p66/p66 homodimer.
Materials and Methods
Cloning, expression, and purification of RT216
Truncated RT (named RT216) was constructed from wild type p51 expression vector by adding a stop codon at 217 using the QuickChange XL site directed mutagenesis kit (Stratagene), and transformed into BL21(DE)3 codon plus RIL. The RT216 expression was induced by the addition of 0.5 mM IPTG at A600 ~0.9 followed by growth of cells at 22°C for 18 hours. The cell pellets were lysed in 50 mM Tris-HCl, 5% Glycerol, and 1 mM EDTA pH 8.0 buffer (Buffer A) by sonication. The cell lysate was centrifuged at 30000 g for 30 min, and the supernatant was loaded on the Q Sepharose Fast Flow column and HiTrap SP HP column connected in tandem. When the OD280 of flow through was observed to be less than 0.01, the HiTrap SP HP column was eluted with a 0 to 1 M NaCl gradient of buffer A. The fractions containing RT216 were pooled based on SDS PAGE analysis. The pooled RT216 samples were concentrated to small volume (< 10ml), and further purified by gel filtration chromatography on a HiLoad 26/60 Superdex-200 column using a buffer of 50 mM Tris-HCl 200 mM NaCl 1mM EDTA pH8.0.
The uniformly U-[2H,13C,15N]-labeled RT216 samples were expressed in E. coli BL21(DE)3 codon plus RIL using M9 deuterated (99% D2O) medium containing U-[2H, 13C] glucose (2 g/L) and 15NH4Cl (1 g/L) as the sole carbon and nitrogen source. The NMR samples were concentrated to 0.6 mM and exchanged into 25 mM Tris-HCl-d11, 8% D2O, 0.02% NaN3 pH 6.8. The U-[15N]RH domain of HIV-1 RT and the U-[15N]thumb domain constructs, were prepared as described previously [8], using M9 minimal media containing 15NH4Cl (1 g/L). As noted previously the RH domain construct includes a four residue Met-Asn-Glu-Leu sequence prior to Tyr427. Preparation of the p66ΔPL also followed the procedure previously described, however the U-[2H,15N] labeling pattern used the media described above for RT216 with U-[2H] glycerol (2 g/L) replacing the doubly labeled glucose.
NMR experiments
NMR data in the present study were collected on an Agilent DD2 800 MHz and Varian INOVA 600 MHz NMR spectrometers, each equipped with a 5-mm Varian 1H[13C,15N] triple-resonance cryogenically cooled probe at 25 °C. Chemical shift assignments of RT216, the isolated thumb domain, and the isolated RH domain were assigned as described previously [8, 21, 22]. The sequential backbone and Cβ resonance assignments were established by the combined analysis of TROSY HNCA, HN(CO)CA, HN(CA)CB, and HN(COCA)CB spectra [23] using triply labeled U-[2H,13C,15N]RT216. Resonance assignments were challenging as a result of spectral congestion, the presence of 18 proline residues in the RT216 construct, and additional broadening of some resonances due to exchange processes. We thus utilized triply labeled U-[2H,13C,15N]RT216, and also made use of selectively labeled samples containing [15N]-alanine, [15N]-leucine, or [15N]-valine (Supplementary Figure S1). A total of 161 resonances of the expected 197 (=215-18) non-proline amide resonances were assigned, corresponding to 81 % coverage of the protein, and deposited in the BMRB (accession No.: 25292). Of particular note were the shifts for Gly155, for which δ(1H,15N) = (3.78, 99.62), well beyond the typical range of values. Examination of RT crystal structures indicates that these extreme upfield shifts result from the close proximity of the Gly155 NH to the indole ring of Trp153.
The 1H-15N TROSY HSQC spectra of U-[2H,15N]p66ΔPL [24] were obtained using Agilent’s gNfhsqc experiment in Biopack (Agilent, Santa Clara, CA). In the 1H dimension, 1024 complex points were acquired with a sweep width of 14 ppm using a relaxation delay of 1 s. In the indirect 15N dimension, 128 complex points were acquired with a spectral width of 29.6 ppm, and the 15N offset was set to 118.886 ppm. The residual water peak was suppressed using the 3919 WATERGATE sequence [25]. The 1H-13C HMQC spectrum of [U-2H,13CH3-Ile]p51 was obtained as described previously [10]. All NMR data were processed by NMRPipe [26] and analyzed with NMRViewJ [27].
Molecular dynamics simulations
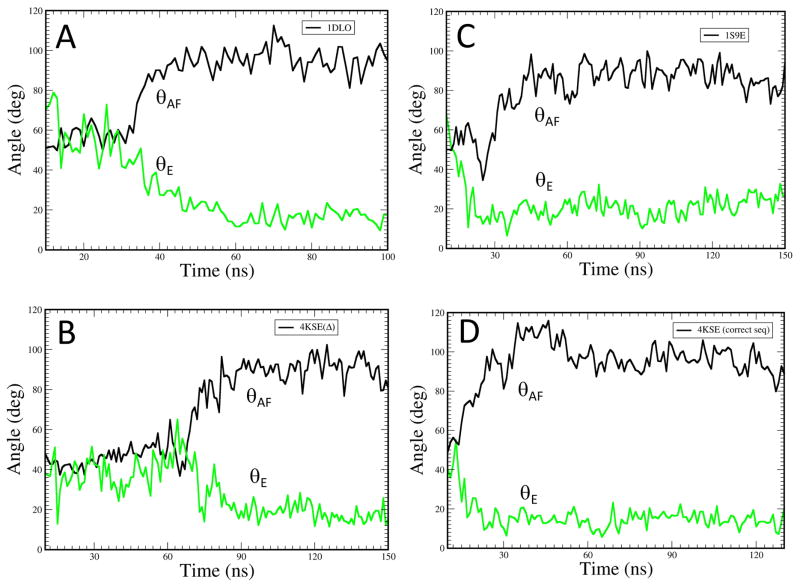
The molecular dynamics (MD) simulations treated the segment from 1-237 following the protocol described previously [10]. Four systems were considered: (i) residues 1-236 of the p51 subunit from the X-ray crystal structure (pdb ID 1DLO) with residues 219 to 230 introduced using initial coordinates for the p66 subunit (pdb ID 1DLO), (ii) residues 1-256 of the p51ΔPL monomer from the X-ray crystal structure (pdb ID 4KSE(Δ)), (iii) residues 1-236 of the p51 subunit from the X-ray crystal structure (pdb ID: 1S9E) which includes residues 219-230; and (iv) residues 1-236 of the p51subunit of the p51ΔPL monomer (pdb ID: 4KSE) with residues 219 to 230 introduced using initial coordinates for the p66 subunit (pdb ID 1DLO). Simulations used the standard Amber FF10 force field for amino acid residues; 100–150 ns production MD runs of the solvated peptides were carried out to evaluate various dynamic properties. The angle θAF between helices A and F was determined as described previously, and the angle θE in these constructs was determined based on helical segments: 156-160 and 162-166.
Circular dichroism (CD) spectroscopy
Secondary structure of two peptides corresponding to the palm domain C-terminal segment (PFL - RT(226-241) –PFLWMGYELHPDKWTV), or to a more water soluble analog containing three substitutions, (PYK - RT(226-241)(F227Y,L228K,V241K) –PYKWMGYELHPDKWTK) were determined by circular dichroism. The CD signal from 190 nm to 260 nm was measured on a Jasco J-810 CD spectrophotometer using a 1 mm quartz cell at 20 °C containing 10–20 μM peptide concentrations in 10 mM sodium phosphate, pH 8.0, with fractional ethanol volume ranging from 0 (for the water-soluble analog) to 40% under the following settings: 1 nm bandwidth, 0.2 nm data pitch, 2 sec response, and 200 nm/min scanning speed. CD spectra from three individual measurements were averaged and processed using Spectral Manager software version 1.5 (Jasco Corporation, Japan) with a curve-smoothing function. The secondary structure content of the peptide was estimated from the DichroWeb on-line server using the CDSSTR method (Whitmore and Wallace, 2004), and data visualization was performed with Excel software (Microsoft corporation, USA). Predicted CD spectra for residues 226-241 corresponding to each subunit of RT were calculated from the PDB2CD on-line web server (http://pdb2cd.cryst.bbk.ac.uk) [28].
RESULTS
Structural Background
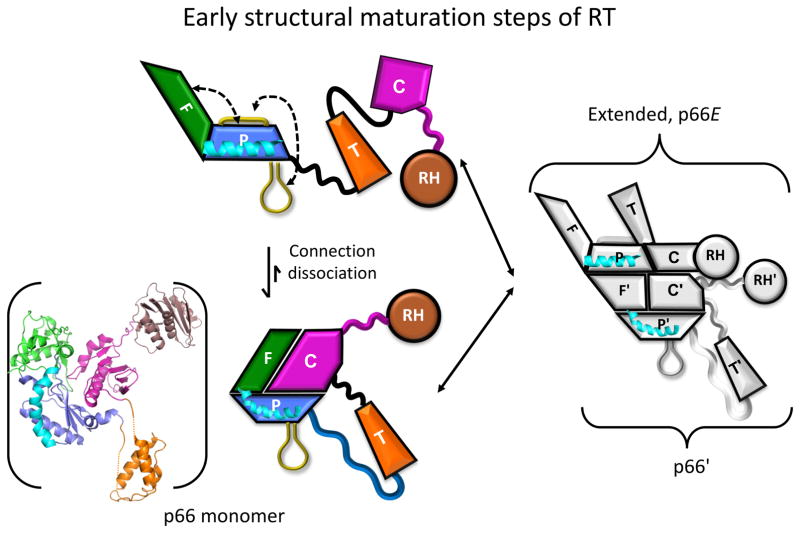
The RT dimer represents an extremely challenging target for direct structural analysis by NMR. Consequently, it has proven useful to analyze the results of NMR studies in the context of available structural information. A schematic illustration of the early maturation steps based on previous studies is shown in Figure 1 [8, 10, 11]. The p66 monomer, shown as both a ribbon diagram and schematic representation, consists of a globular structure corresponding to a complex of the connection and fingers/palm domains, while the thumb and RH domains are loosely connected by flexible linking segments derived from unravelling of the palm, thumb, and connection domains [8]. The fingers and palm domain sequences are discontinuous and so cannot be separated [5, 29]. The structure of the fingers/palm:connection complex is similar to that of the p51 subunit of the RT heterodimer (e.g. [30]). Support for this structure was derived from crystallographic and NMR studies of constructs in which a segment of the C-terminal palm loop corresponding to residues 219-230 was deleted, as well as studies demonstrating that this deletion does not significantly alter the monomer structure [8]. Dissociation of the connection from the fingers/palm allows further domain rearrangements, exposing surfaces involved in dimer formation. The re-arranged monomer is then able to interact with the more stable (not rearranged) monomer, forming an initial p66/p66′ homodimer that exists as a structural heterodimer. Subsequently, a series of conformational changes occur primarily in the p66′ subunit that culminate with the subunit-specific destabilization and unfolding of the p66′ RH′ domain [1](not shown). RH domain unfolding allows the protease access to the monomer-inaccessible cleavage site.
Figure 1. Early steps of RT structural maturation.
The p66 monomer is shown in the lower left using both a ribbon diagram and a schematic representation. Domains are color coded as: fingers (green); palm (blue); thumb (orange); connection (magenta), and RH (brown), while helix E is colored cyan. Initial dissociation of the connection from the fingers/palm is rapidly followed by expansion of angle between the fingers and palm domains (see Figure 2), and recruitment of the C-terminal palm loop to the exposed hydrophobic surface of the palm domain. The re-arranged p66 domains form an initial complex with the monomer, followed by slower maturation steps (not shown).
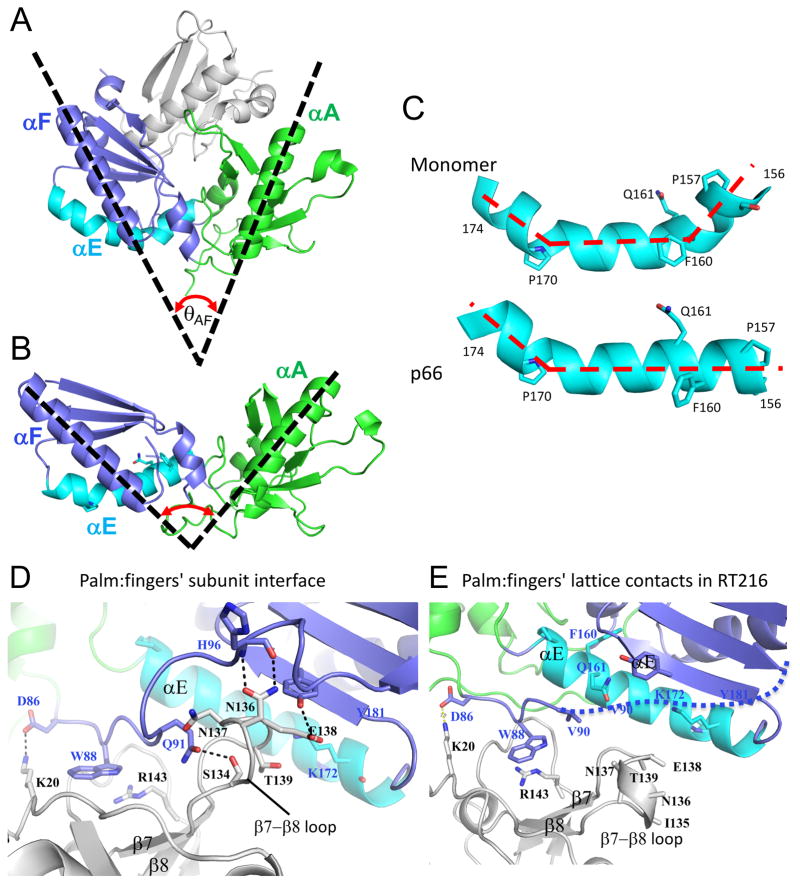
Previous studies have suggested that the p66 monomer is spring-loaded, so that after dissociation of the connection domain, the angle between the fingers and palm domains expands significantly (Figure 2A–C; [10]). Figure 2A illustrates the more bent conformation of the fingers/palm observed in the monomer construct with disordered palm loop residues 219-230 deleted (pdb: 4KSE, [8]), where the angle θAF between helix αA in the fingers and αF in the palm is about 45°. A similar, acute angle characterizes the p51 subunit of RT. In contrast, the crystal structure of a construct corresponding to the isolated fingers and most of the palm, RT216, adopts a more expanded structure in which θAF ~ 100° (Figure 2B), similar to the fingers/palm angle in the p66 subunit of RT. A further comparison reveals that in the monomer helix αE exhibits a sharp bend near residue Phe160, but straightens out in the isolated RT216 construct (Figure 2C). Helix E is located mostly in the palm but extends to the fingers-palm interface. The enhanced stability of the straighter helix geometry is presumably one of the factors that favors the extended structure of the isolated fingers/palm construct. It is thus of interest to determine whether this bend is also present in solution.
Figure 2. Structural comparisons of the palm/loop domains.
A) Structure of the complex formed by the fingers (green)/palm (blue) and connection (gray) domains in the p51ΔPL monomer (pdb: 4KSE), and helix αE is indicated in cyan. The angle θAF formed by helices A and F is ~ 50°. B) Ribbon diagram of the isolated RT216 (pdb: 1HAR, [42] construct that includes the fingers and most of the palm domains color coded as in A. For the isolated construct, θAF expands to ~100°. C) Structural comparison of palm domain helix E (residues 156-174) in the structures shown in A and B, illustrating elimination of the bend near Phe160, Gln161. D) Ribbon diagram showing the region of the p66/p51 subunit interface formed from the p51 fingers (gray) and the p66 palm (blue) domains (pdb: 1DLO). The p66 fingers domain (green) does not contribute to the interface. E) Ribbon diagram showing the lattice contacts in the RT216 crystal structure that correspond to the region shown in panel D. The region of the p66/p51 interface formed near palm residue W88 also forms a lattice contact, however the region of the interface involving β7–β8 does not.
The fingers and palm domains form part of the inter-subunit interface in RT, leading to the possibility that similar interactions in the isolated RT216 construct may lead to aggregation. The interface between the p51 fingers and p66 palm domains in the RT heterodimer is shown in Figure 2D. This interface involves two components: 1) the p66 (palm domain) region near Trp88, and 2) the fingers domain (p51) region involving the β7- β8 loop. Interestingly, a similar but less complete interface exists at the lattice contracts of the reported RT216 crystal structure (Figure 2E, pdb: 1HAR). In this case, palm domain residue Trp88 forms a similar set of interactions with fingers domain residues on an adjacent molecule, however the β7- β8 loop is poorly defined and does not interact with palm domain. Further, the palm domain segment from residues 90-104 is disordered (Figure 2E). These results are consistent with the possibility of intermolecular interactions of the RT216 construct, but suggest that the interface may be less extensive than observed in the RT heterodimer.
Solution behavior of the isolated fingers/palm domains
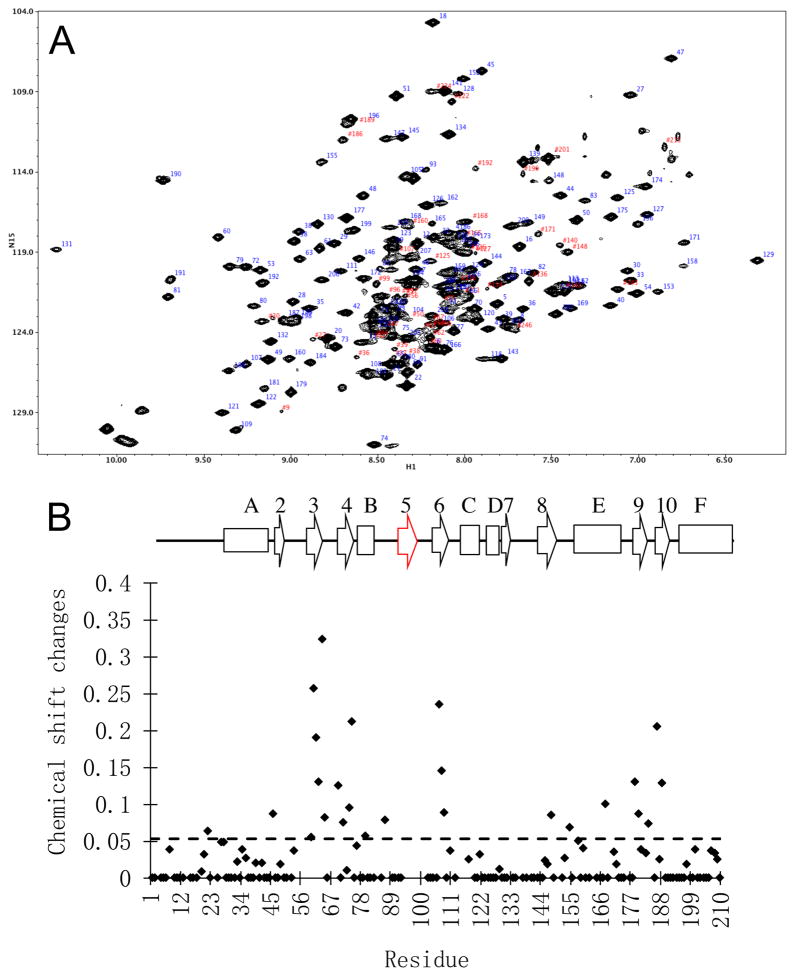
In order to evaluate the solution behavior of the RT216 construct discussed above, isotopically-labeled RT216 was expressed and characterized by NMR. The 1H-15N HSQC spectrum of doubly labeled U-[13C,15N]-RT216 showed many well dispersed resonances, but was also characterized by some regions with a high degree of congestion and uneven resonance intensities in the random coil region near δ1H = 8.5 ppm. One characteristic of RT216 that tends to be unfavorable for NMR analysis is the presence of multiple, solvent-exposed aromatic sidechains. A solvent exposure analysis [31] of the RT216 crystal structure (pdb: 1HAR) indicates that seven aromatic residues: Trp24, Phe61, Trp71, Trp88, Tyr181, Tyr183, and Tyr188 have a fractional solvent exposure ranging from 24 % to 88%. Exposed aromatic sidechains can mediate aggregation as well as broadening resonances of nearby residues due to internal motion. Significant improvement in spectral resolution and homogeneity was obtained using triple U-[2H,13C,15N] labeling (Figure 3a), although several congested areas remained. Missing assignments include several short segments bounded by proline residues, a central region of helix E (164-170), as well as several segments that are also disordered in the RT216 crystal structure (Supplementary Figure S2). A significant number of the problematic residues in the RT216 construct are readily observed in crystal structures of the full RT molecule, most probably due to additional stabilizing interactions at the domain boundaries.
Figure 3. Solution behavior of RT216.
A) 1H-15N TROSY spectrum of U-[2H,13C,15N]RT216. B) Concentration-dependent shift behavior of RT216 calculated as using shift differences between 650 μM and 45 μM RT216. The secondary structure diagram at the top is based on the identifications given by Wang et al. [7] indicating similar secondary structure organization for p66 and p51 (Table 1). Strand β5 is not identified in either the isolated RT216 construct (pdb: 1HAR) or the p51ΔPL monomer (pdb: 4KSE), and the monomer also contains short helical segments involving residues 98-103 and 134-138.
As summarized above, the fingers and palm domains form an intersubunit interface (Figure 2D), a portion of which is also present as a lattice contact surface in the crystal structure of RT216 (Figure 2E). Based on these structures, it was anticipated that dimerization or formation of higher aggregates could be occurring for the RT216 construct. In order to evaluate the effects of intermolecular complex formation, concentration-dependent NMR studies of the labeled RT216 were obtained over the range from 45 to 650 μM (Figure 3B). Secondary structure identification is given in Table 1. The two most concentration sensitive regions of the protein identified are two β-sheets: β3–β4 on the fingers domain and β6–β9–β10 on the palm (Supplementary Figure S2). Both of these sheets contain solvent-exposed hydrophobic residues that presumably mediate the aggregation. Truncation of the palm domain in RT216 removes a region of the structure that would normally interact with the hydrophobic surface of the NNRTI pocket, exposing NNRTI-binding residues Leu100, Val106, Val108, Tyr181, Tyr183, and Tyr188 to the solvent. The hydrophobic region of the fingers domain includes a region that interacts with the RNA or DNA substrate. Although few concentration-dependent shifts were observed for residues located in regions that form the fingers:palm interface in the RT heterodimer (Figure 2D), fingers domain amide resonances for Lys20, Val21, Lys22, Asn57, and Tyr56, that would be positioned near palm domain residue Trp88 if aggregation were occurring were unassigned, possibly as a result of monomer-dimer exchange broadening.
Table 1.
Fingers/palm secondary structure
| Secondary structure | p66a | p51a | RT216b | p51ΔPL monomer |
|---|---|---|---|---|
| β1 | 19–23 | 20–24 | - | |
| αA | 27–44 | 28–44 | 28–43 | 27–44 |
| β2 | 47–51 | 49–52 | 47–49 | 47–49 |
| β3 | 57–63 | 58–64 | 60–63 | 60–63 |
| β4 | 71–76 | 71–76 | 72–75 | 72–75 |
| αB | 78–85 | 78–85 | 78–83 | 77–84 |
| β5 | 93–97 | 93–97 | - | - |
| α-helix | 98–103 | |||
| β6 | 104–112 | 105–110 | 105–110 | 105–110 |
| αC | 113–118 | 112–116 | 112–117 | 112–118 |
| αD | 124–128 | 123–128 | 125–128 | 124–129 |
| β7 | 128–133 | 129–132 | 129–132 | 130–132 |
| α-helix | 134–138 | |||
| β8 | 140–146 | 141–146 | 142–146 | 142–146 |
| αE | 154–174 | 154–175 | 156–174 | 154–175 |
| β9 | 178–183 | 178–183 | 178–183 | 179–183 |
| β10 | 186–191 | 186–191 | 186–191 | 186–191 |
| αF | 195–212 | 198–212 | 195–212 | 194–212 |
| β11 | 214–218 | 215–217 |
Secondary structure identification from [7].
Secondary structure from PyMol analysis of pdb entry 1HAR for RT216.
Secondary structure from PyMol analysis of pdb entry 4KSE for p51ΔPL.
Since the major difference between the fingers/palm structure in the monomer and in the isolated fingers/palm construct involves a change in relative domain orientation, we attempted to obtain RDC data using multiple alignment media. None of these approaches was successful, perhaps as a consequence of the solvent-exposed aromatic residues noted above. We subsequently turned to a more detailed analysis of helix αE. This helix, which has been considered to extend from residue 154-174 [7] is unusual, containing Pro residues at positions 157 and 170. Residues 164-170 in the center of this helix are positioned near milder bends in the helix and correspond to regions of increased local instability that limited assignments. However, the major kink in the helix is not located at these positions, but near Phe160, and assignments were made for several residues immediately before and after Phe160 (Supplementary Figure S3).
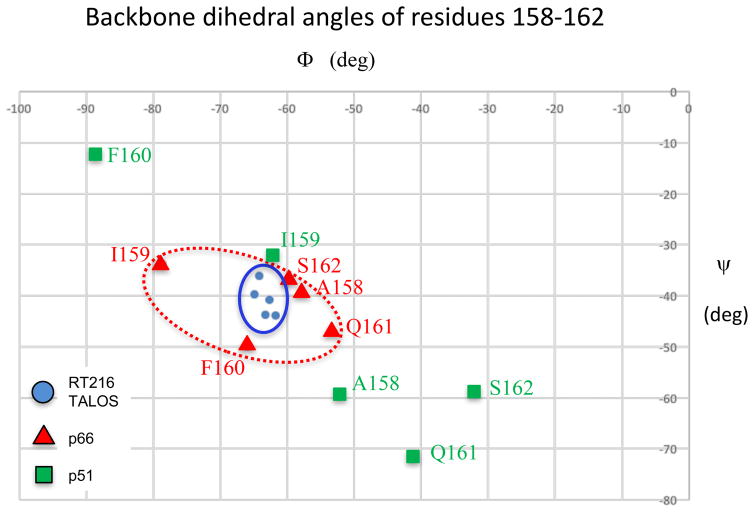
A TALOS+ analysis of the shifts [32] provided the most useful approach for assessing the solution conformation of helix αE near the bend. As shown in Figure 4, TALOS analysis of the shift data for residues 158-162 yielded a set of tightly clustered phi and psi values consistent with α-helical structure. Comparisons with the phi/psi values for the same residues in the isolated RT216 construct (pdb: 1HAR) or the p66 subunit of the RT heterodimer (pdb: 1DLO) are also consistent with α-helix geometry, while the corresponding values for the RT p51 subunit or the p51ΔPL monomer (pdb: 4KSE) show much larger variations. Thus, the TALOS analysis supports the conclusion that in solution, RT216 adopts a helix E geometry that lacks the sharp bend that is present in the monomer, and is thus consistent with the conformation observed in the extended conformation present in the p66 subunit of the RT heterodimer.
Figure 4. Conformational analysis of helix E.
Plots compare the phi/psi angles for residues 158-162 located on either side of the bend in helix E. Color coding is: RT p66 subunit (Δ red, pdb: 1DLO); RT p51 subunit (□, green, pdb: 1DLO), TALOS+ analysis of NMR shifts for the corresponding residues of RT216 (○, blue).
Molecular dynamics simulations of the fingers/palm
We previously reported molecular dynamics simulations for the isolated fingers/palm domains indicating that over a time frame of about 100 ns, the more expanded conformation observed in the p66 subunit or the isolated fingers/palm construct persists (θAF ~ 100°), while the more bent conformation (θAF ~ 50°) present in the monomer is not stable once the fingers/palm has been separated from the connection domain [10]. It appears that the bent conformation of αE in the monomer is stabilized by the extensive interface between the fingers/palm and the connection domains. These as well as additional simulations were again used to evaluate the bend in helix E (θE), based on a comparison of the helix direction in segments 156-160 and 162-164. Initial bends of ~ 40° – 60° are reduced to ~20° on the same time scale as the increase in θAF (Figure 5). Each simulation starts from a slightly different structure at time zero: 1DLO, 1S9E, 4KSE and 4KSEΔ (see Methods). Straightening of helix E appears to be concerted with the change in θAF in simulations A and B, and to slightly precede the change in θAF in simulations C and D. These results suggest that the more stable helical conformation helps select the extended conformation and perhaps may precede the overall conformational expansion.
Figure 5. Molecular dynamics simulations of helix E.
Time-dependent behavior of θAF (black curve) corresponding to the angle between helices A and F, and θE (green curve) describing the bend in helix E, for the fingers/palm domains from molecular dynamics simulations starting with the following constructs: A) residues 1-236 of the p51 subunit of 1DLO with the segment from 219-230 modeled into the structure as described in Methods; B) residues 1-256 of the p51subunit of the p51ΔPL monomer (pdb: 4KSE); C) residues 1-236 of the p51 subunit of structure 1S9E that includes residues 219-230; D) residues 1-236 of the p51ΔPL monomer with the deleted 219-230 residue segment added to the initial model as described in Methods.
Intrinsic conformational preferences of the palm C-terminal residues
In addition to changes in the fingers/palm orientation, the metamorphic transition involves formation of a short, 4-stranded β-sheet that is not present in the monomer. This sheet is formed from residues 226-241 corresponding to the C-terminus of the palm domain (Supplementary Figure S4), and residues 314-318 at the beginning of the connection domain (using recently proposed domain boundaries [1]). We previously suggested that, as in the case of the expanded fingers/palm angle, this conformational transformation arises after connection domain dissociation removes constraints of the monomer, allowing the intrinsic conformational preferences of the peptide segments to be expressed. The conformational preferences of the palm domain C-terminal peptide, residues 226-241 (peptide PFL: PFLWMGYELHPDKWTV), were evaluated using circular dichroism. Unsurprisingly, the extremely hydrophobic peptide PFL was predicted to have a high tendency toward aggregation based on analysis by the web-server TANGO [33]. We therefore also evaluated a more hydrophilic analog: RT(226-241) containing three residue substitutions: F227Y, L228K, and V241K (peptide PYK: PYKWMGYELHPDKWTK) that appeared likely to increase water solubility without significantly perturbing the expected residue interactions. Consistent with this expectation, the TANGO analysis indicated that the hydrophilic analog has no significant tendency to aggregate, and both isolated peptides are predicted to have β-sheet structure, although with low probability.
CD spectra of the hydrophilic analog were obtained in water, while due to solubility limitations, the hydrophobic peptide spectra were obtained in 10 – 40% ethanol at peptide concentrations from 10 – 20 μM. Analysis of the spectra using the web-based program DICHROWEB [34] showed that both peptides exist in solution as mixtures of β-turns and β-strands (consistent with β-sheet structure) and disordered structures (Supplementary Figure S5 and Supplementary Table S1). The fractional β-sheet ranged from 28 – 62 % for PFL, with the highest values obtained in 40% ethanol, consistent with the interpretation that aggregation is reduced in this solvent. For the more hydrophilic analog, the β-sheet probabilities determined in water ranged from 41–56 %. CD spectra of the PFL peptide obtained in 40 % ethanol were compared with theoretical spectra for residues 226-241 in the each subunit of RT generated using web server PDB2CD [28] (Supplementary Figure S5). The spectra for the palm domain segment in the p66 subunit that contains the β-sheet structure are much more similar to the experimental CD spectra, than the spectra generated for residues 226-241 in the p51 subunit (Supplementary Figure S5). These results support the intrinsic tendency of the peptide to adopt the β-sheet structure.
NMR spectra of the U-[2H,15N]p66ΔPL monomer
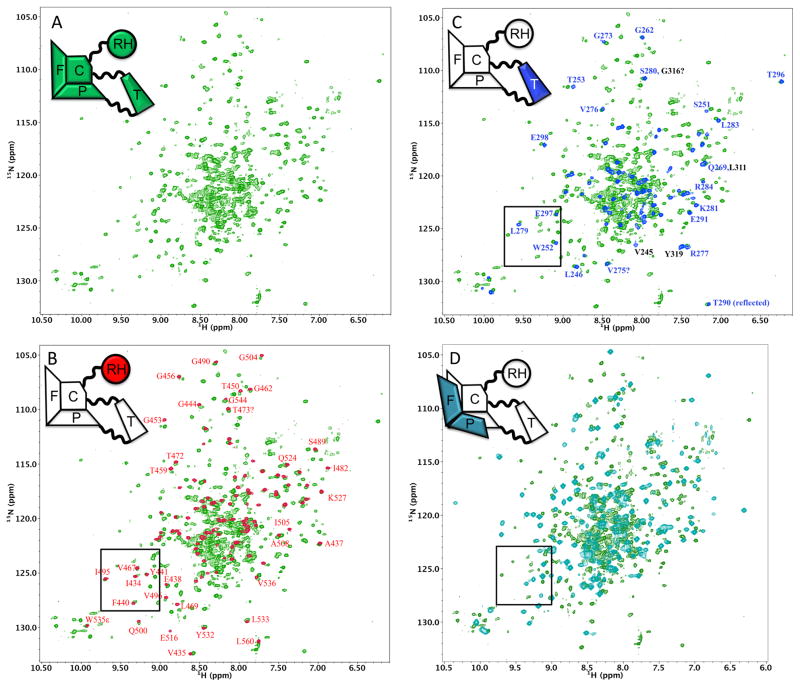
In addition to the more specific questions about the conformational changes observed for the fingers/palm, perhaps the most significant question about the metamorphic transition of the RT polymerase domain is whether it is primarily a unimolecular rearrangement that occurs prior to dimer formation, or whether it occurs subsequently. Kinetic studies have led to inconsistent conclusions [35, 36], and more recently NMR studies using 15N-labeling [20] also have led to conclusions that are inconsistent with those based on earlier methyl-labeling studies [8, 10, 11]. Since the structure shown in Figure 1 is based on both crystallographic and NMR studies of methyl labeled constructs, it was of interest to determine to what extent these earlier conclusions are also consistent with NMR analysis of 15N-labeled constructs. Following the NMR approaches previously utilized by both groups, we compared the TROSY spectra of U-[2H,15N]p66ΔPL with spectra obtained for three constructs of the 15N-labeled individual domains: the thumb, RH, and RT216 corresponding to the fingers and most of the palm domains (Figure 6). We note that the structure shown in Figure 1 (see also Figure 1C of reference [8]), would imply very close agreement of the monomer spectra with those of the RH and thumb domains, but much poorer agreement with the spectrum of RT216.
Figure 6. NMR spectra of the p66ΔPL monomer.
A) 1H-15N TROSY spectrum of U[2H,15N]p66ΔPL. B) Overlay of the 1H-15N HSQC spectrum of U-[15N]RH domain (red) with the spectrum in panel A (green). C) Overlay of the 1H-15N HSQC spectrum of U-[15N]thumb domain (blue) with the spectrum in panel A (green). D) Overlay of the 1H-15N TROSY spectrum of U-[2H,13C,15N]RT216 (cyan) with the spectrum in panel A (green). Since the data were obtained using somewhat different experimental conditions, labeling, and NMR experiments, the offsets in panels B and C were selected to optimize spectrum consistency. This was generally not feasible for the overlay in D, since optimization of some regions resulted in much poorer agreement in others. The box shown in the spectra corresponds to the spectral region highlighted by Sharaf et al. [20].
TROSY spectra of U-[2H,15N]p66ΔPL overlaid with amide spectra for U-[15N]RH, U[15N]thumb, and U-[2H,13C,15N]RT216 are shown in Figure 6A–D. Since these comparisons involved TROSY spectra of the labeled RT216 and p66ΔPL but 1H-15N HSQC spectra for the RH and thumb domains, and since the samples were studied under similar but not identical conditions, the overlays were optimized to facilitate resonance comparisons rather than using a constant shift reference. Assignments for some of the more well dispersed resonances are indicated in the figures. As is apparent from Figures 6B and 6C, the overlays with the isolated RH and thumb domains show excellent agreement.
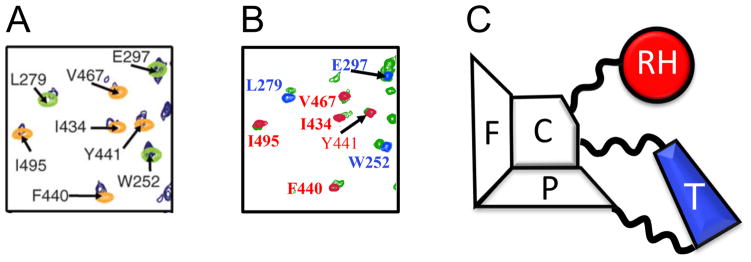
An expanded region of the spectra illustrating these comparisons is shown in Figure 7. Panel A, derived from Figure 3A of Sharaf et al. [20] shows a spectral region of the putative p66/p66 homodimer overlaid with color-coded resonances for the isolated RH and thumb domains. These spectral results were used as the basis for a model of a symmetric p66/p66 homodimer. Figure 7B shows the same overlay derived from spectra in Figure 6 that correspond to the p66ΔPL monomer (green), RH (red), and thumb (blue) domains. A color-coded schematic of the monomer is shown in Figure 7C. Comparison of Figures 7A and 7B shows that although attributed to very different species, the spectral overlays are extremely similar, indicating that the corresponding structures are also very similar. In view of the spectral similarity of Figures 7A and 7B and the consistency of the spectral data in Figure 7B with the monomer structure in Figure 7C, the data in Figure 7A are also consistent with a structure similar to that of the monomer (Figure 7C).
Figure 7. Comparison of an expanded spectral region of the p66/p66 dimer and the p66ΔPL monomer.
A) region of the 1H-15N spectrum presented by [20]: (Δ1H,Δ15N) ~ (9.1–9.8 ppm, 122–128 ppm). Color coding is: p66, dark blue; thumb, green; RH, yellow. B) Corresponding region of the 1H-15N spectrum of U-[2H,15N]p66ΔPL (green) overlaid with spectra for the RH (red) and thumb (blue) domains. C) Schematic illustration of the p66 monomer structure based on Figure 1, color coded to illustrate the domain origin of the corresponding resonances.
In contrast with the above comparisons, the agreement with the RT216 spectrum is substantially poorer (Figure 6D) - a result that is anticipated on the basis of the structural data in Figures 1 and 2. Specifically, the isolated RT216 construct has a different conformation than the corresponding segment of the p51ΔPL monomer (Figures 2A,2B). More importantly, there are extensive interface contacts between the connection domain and the fingers/palm in the monomer that are not present in the extended p66 subunit of the heterodimer or the RT216 construct. This interface contains several aromatic residues that will produce significant shift perturbations for residues in the fingers/palm domains. The much poorer consistency between the chemical shift pattern in p66ΔPL monomer and in the RT216 construct is completely consistent with expectations based on the available structural information and Figures 1 and 2. Conversely, the NMR spectral comparisons presented above provide strong support for the p66 monomer structure shown in Figure 1 that was previously deduced from structural data for p51ΔPL combined with NMR studies of methyl labeled p66ΔPL and the component domains. Specifically, in the p66 monomer structure, the RH and thumb domains are structurally isolated and internally mobile, consistent with amide spectra comparisons summarized above. Alternatively, the RT216 does not show good agreement, again consistent with the structure shown in Figure 1. In summary, the NMR studies of the 15N-labeled constructs presented here and in the previous study by Sharaf et al. [20] are consistent with the monomer structure of Figure 1.
Spectral comparison of the U-[2H,15N]p66ΔPL monomer with spectra of the reported U[2H,15N]p66 homodimer
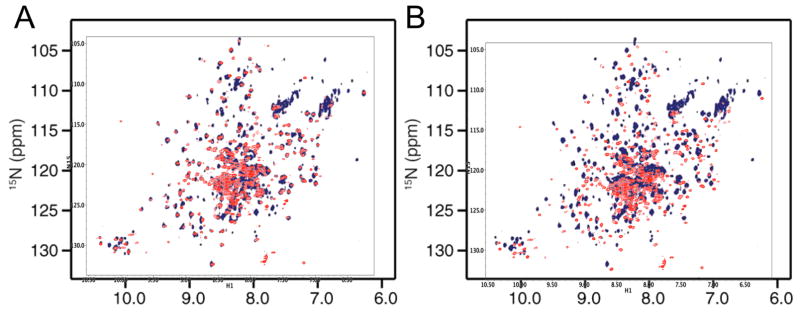
In the previous section we observed that the amide spectrum of the U-[2H,15N]p66ΔPL monomer agrees closely with spectra for the isolated thumb and RH domain, but much less closely with spectra for RT216. The spectral comparisons presented in the previous section are identical with comparisons made by Sharaf et al. [20] for the similarly labeled p66/p66 homodimer. Hence, we can conclude that the amide spectra for the p66ΔPL monomer must be very similar with the spectrum attributed to the p66/p66 homodimer. Two spectral overlays shown in Figure 8 compare the amide resonances of the p66ΔPL monomer with the published spectra for the homodimer. The comparisons in Figure 8 optimized the resonance overlap (Figure 8A) or utilized the chemical shift axes from the two studies (Fig. 8B). The spectra are very similar but not identical. In addition to differences in experimental conditions, one important basis for a difference is the absence of the palm loop segment from 219-230 corresponding to residues KKHQKEPPFLWM in the spectrum of the p66ΔPL monomer. In addition to the loss of the corresponding amide resonances, this segment contains three aromatic sidechains that, depending on the motional characteristics, may produce shift and broadening effects of nearby residues. Thus, at least one of the missing TrpNHε resonances in Figure 8A (bottom, lower left) can be attributed to the loop deletion. The spectral offsets in Figure 8B approximate those expected to arise from a comparison in which only one spectrum utilized a TROSY sequence and intensity differences in the amide sidechain regions are also suggestive of this difference. Overall, the comparison is consistent with very similar, if not identical structures.
Figure 8. Comparison of monomer and dimer spectra.
A) Overlaid NMR spectrum attributed to the p66/p66 homodimer (dark blue, [20]), with the 1H-15N TROSY spectrum obtained for U-[2H,15N]p66ΔPL (red). The offsets were selected to optimize resonance consistency. B) Overlaid spectra as in A using the reported referencing.
Effects of exchange
The symmetric p66/p66 homodimer model presented by Sharaf and coworkers is based largely on the conclusion that only a single set of resonances is observed for both species. This analysis is limited by the presence of both monomers and dimers in the sample and by the absence of information on chemical exchange rates. In the event that monomer and dimer exchange is fast on the chemical shift difference time scale, the monomer becomes a rapidly exchanging intermediate that will equilibrate the shifts of the two subunits of the dimer:
| (1) |
Sharaf et al. [37] conclude that for p51, the high KD value of ~ 0.3 mM is consistent with fast exchange, if dimer formation is assumed to be diffusion limited. This would presumably preclude conclusions about the symmetry of the p51/p51 homodimer. The estimated diffusion-controlled association rate constant of 106 M−1s−1 is nearly 6 orders of magnitude faster than the heterodimer association rate constant of 1.7 x M−1s−1 reported for RT heterodimer formation [36]. Sharaf et al. suggested that for the p66 sample, the lower Kd of ~ 4 μM would be sufficient to overwhelm contributions from the p66 monomer, however this does not necessarily eliminate the effects described by Eq. 1 above. Thus, subunit shift differences are still subject to equilibration if the monomer-dimer exchange rates are sufficiently fast.
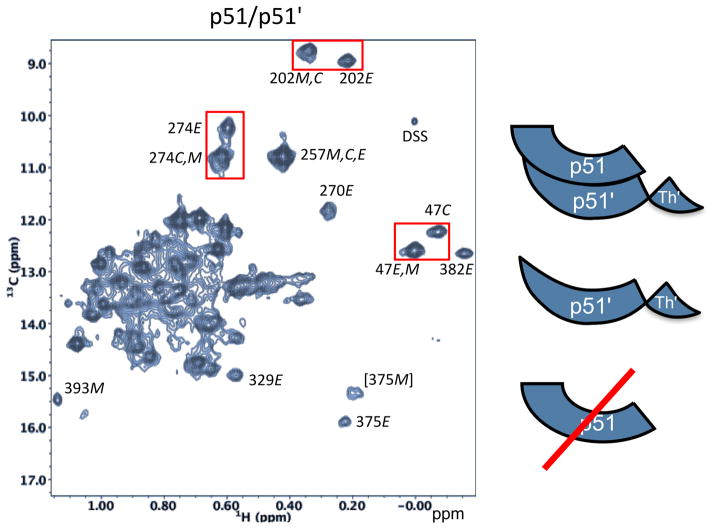
In contrast with the analysis of Sharaf et al., NMR studies of [13CH3-Met]- and [13CH3-Ile]-labeled p51 and p66 homodimers showed multiple chemical shift differences for the two subunits, consistent with slow chemical exchange for both the p66 and p51 homodimers [8, 10, 11]. For the purpose of comparison, the 1H-13C HMQC spectrum of a sample containing [13CH3-Ile]p51 obtained under very high salt conditions that enhance the dimer/monomer ratio is shown in Figure 9. Even under high salt conditions a significant monomer fraction remains, as indicated most directly by the observation of an Ile393 resonance attributed specifically to the monomer species. Based on comparison with spectra for the [13CH3-Ile]-labeled p66/p51 heterodimer [8], several of the more highly resolved resonances are readily attributed to either the compact or extended structures of the polymerase domain. The high salt conditions used to increase the dimer/monomer ratio in this study is apparently not essential for achieving the slow exchange conditions, since separate methyl resonances in the p51 dimer have been observed at lower salt concentrations (200 mM KCl) [11]. We note also that none of the resonances are attributed to the extended (active) polymerase conformation of the monomer (p51E), which is considerably less stable and present at a very low concentration.
Figure 9. 1H-13C HMQC spectrum of [U-2H,13CH3-Ile]p51.
Species contributing to the spectrum, illustrated in the schematic on the right, include the p51/p51′ homodimer and the p51 monomer. Resonance intensities suggest that the sample is ~ 80% dimeric. Resonances are attributed to the extended conformation (labeled E), the compact (monomer) structure (labeled C), and to the monomer (labeled M) as indicated. Resolved resonances for Ile47 (fingers domain), Ile202 (palm domain), and Ile274 (thumb domain). The spectrum was obtained under conditions that favor dimerization: 800 mM KCl, 20 mM MgCl2. Despite using these conditions, a significant Ile393 resonance attributed to the monomer species is also present.
As discussed previously, the monomer – dimer equilibrium is slow for many resonances in p51 because a substantial structural rearrangement – the metamorphic transition – must occur prior to dimer formation. For the isolated polymerase domain, the structural isomerization-dimerization process is described by the relations [10, 11]:
| (2) |
The first step above will be slow since it entails major structural changes that involve domain rearrangements as well as the formation of new secondary structural elements discussed above [10]. In general, each subunit corresponds to a separate set of resonances. For some resonances, e.g. Ile47, Ile202, and Ile274, the shift differences are small but easily resolved (Figure 9); in some cases, e.g. Ile257, the very small shift difference limits resolution of the two forms, while in others, e.g. Ile270 and Ile382, the shift difference is so large that only one of the two corresponding peaks is observed, while the resonance arising from the other subunit is broadened and/or shifted into a more congested region of the spectrum. Thus, it is seen that even in the example of the weakly associated p51/p51′ homodimer, monomer – dimer exchange is slow, consistent with the earlier analysis that this requires a structural, metamorphic transition prior to dimer formation (Eq. 2).
Discussion
Although conformational heterogeneity is a nearly universal characteristic of proteins, the significant structural variations characterizing metamorphic proteins have been less extensively documented. Nevertheless, the increasing number of examples becoming available leads to interesting new problems of protein folding and structural inter-conversion. The polymerase domain of HIV-1 reverse transcriptase adopts either of two alternate structures that fulfill two different functions. As a comparison, the interconversion of open and closed positions of the thumb domain are achieved by bond rotations and occur on a much faster time scale that is typical for conformational transitions [38]. Atypically, the extended and compact structures of p66 must both be present simultaneously in order to form the p66/p66′ homodimer precursor and the RT heterodimer. As first proposed by Wang et al., [7] and subsequently substantiated by Zheng et al. [8, 11], the compact fold characterizing the inactive domain is considerably more stable than the extended fold of the active domain, and the homodimers exist as structural heterodimers. Much of the energy required to stabilize the extended, active fold of the polymerase domain is provided by the energy of dimerization. As a consequence, the net energy available for dimer stabilization is limited. Thus, despite a very large dimer interface area of ~ 4500 Å2 [7], the apparent dimer Kd values are in the micromolar range [39, 40]. This contrasts with many examples of considerably smaller interfaces characterized by nM stability [41]. This effect is even more dramatic for the p51/p51′ homodimer: a 2500 Å2 interface [7] corresponds to a Kd value of ~230 μM [40] or higher [20].
As noted above, recent analysis of NMR data by Sharaf et al. supporting the existence of a long-lived, symmetric p66/p66 homodimer was based in part on the assumption of diffusion-controlled dimerization, with an association rate constant ~ 106 M−1s−1 [37]. Although earlier kinetic measurements were interpreted as supporting rapid initial dimer formation followed by slower conformational adjustments [35], a much slower association rate constant, ka = 1.7 M−1s−1, was subsequently determined [36]. This study attributed the earlier kinetic results to an artefactual consequence of the acetonitrile added to stabilize the monomer. However, since both the p51 and p66 monomers adopt compact structures that resemble the p51 subunit of RT [8], all p51 and p66 dimerization reactions – whether they involve homo- or hetero-dimer formation – are rate limited by the requirement for an initial metamorphic structural transition. The slow exchange behavior expected on the basis of the slower rate constant has been observed in previous NMR studies of both p51 and p66 homodimers [8, 10, 11] and is consistent with the observation of multiple p51 Ile methyl resonances in Figure 9.
As concluded in previous studies [8–10], the metamorphic transition is triggered by dissociation of a spring-loaded fingers/palm:connection complex. Subsequent to dissociation, the fingers/palm and connection domains each adopt alternate conformations that are not constrained by the interdomain interactions present in the monomer. Previous analysis based on crystallographic and molecular modeling studies indicated that upon dissociation, the fingers/palm adopts a more extended conformation, similar to that observed in the p66 subunit of RT [10, 42], however, the basis for this transition has been unclear. In the present study, we have shown that one of the drivers for this structural change is likely the preference of helix αE to eliminate the sharp kink near Phe160 that is present in the monomer (pdb: 4KSE; [8] extending the standard helical geometry (Figure 2C). Although the solution behavior of the RT216 construct, containing the fingers and most of the palm domains, is not ideal, TALOS+ analysis supports the conclusion that in solution, as in the crystal, this pronounced bend is not present.
The most typical feature of metamorphic transitions is a change in secondary structure. For p66, the most significant secondary structure change involves formation of a short β-sheet from residues 226-241 at the C-terminus of the palm domain and residues 314-318 at the N-terminus of the connection domain. This β-sheet is not present in the monomer structure or in the p51 subunit of RT. As with the fingers/palm expansion, this transition also appears to be spring-loaded: dissociation of the fingers/palm:connection complex in the monomer removes the structural constraint on the palm domain exposing the hydrophobic surface of the palm and allowing the intrinsic conformational preferences of palm residues 226-241 to be expressed. The interactive nature of the exposed hydrophobic surface of the β-sheet of the palm domain (β6–β9–β10) is demonstrated by concentration-dependent shifts that reveal the availability of this surface for interactions with hydrophobic residues. A CD analysis of the palm domain C-terminal peptide as well as a hydrophilic analog support the propensity of this sequence for β-sheet formation. The β-sheet formed from the palm C-terminus, corresponding to β12–β13–β14 in the p66 RT subunit, is amphiphilic, having both hydrophilic and hydrophobic surfaces, so that β-sheet formation and stabilization is further supported by the interaction of the hydrophobic surfaces of the two β-sheets (Supplementary Figure S4). Thus, the results are consistent with the conclusion that both factors – interaction with the exposed β-sheet and intrinsic conformational preferences of the palm C-terminal segment, contribute to the metamorphic transition.
As shown above, spectral comparisons of 15N-labeled p66ΔPL monomer with 15N-labeled RH, thumb, and RT216 (fingers/palm) domains exhibit the patterns predicted on the basis of the monomer structure shown in Figure 1. Thus, the RH and thumb domain spectra are in very good agreement with the p66 monomer spectrum, while the RT216 spectrum is not. These same comparisons recently were reported for the 15N-labeled p66/p66 homodimer, but interpreted to support a stable, symmetric homodimer structure [37]. Thus, it must be concluded that either a long-lived symmetric homodimer is formed from a pair of monomers, each of which closely resembles the isolated monomer, or that the spectra attributed to the homodimer were dominated by contributions from the smaller and more flexible monomer species. Although the existence of a short-lived symmetric homodimer formed from a pair of monomer-like structures is difficult to completely rule out, the stable, long-lived symmetric homodimer proposed by Sharaf and coworkers is inconsistent with many reported homodimer characteristics, including the activity of the p66/p66 homodimer, which requires the presence of at least one domain with an active fold and exposed catalytic site [39, 43, 44].
The second explanation – that the spectra attributed to the p66/p66 homodimer were instead dominated by the smaller and more flexible monomer species, is further supported by the spectral comparisons shown in Figures 6–8. The structure of the p66 monomer corresponds to a group of domains connected by flexible linking segments. The largest of these is the fingers/palm:connection complex with MW ~ 40 kD, while the RH and thumb domains are ~ 15 kD and 7 kD, respectively. In contrast, the p51/p51 homodimer forms a globular structure of ~ 100 kD, and the p66/p66 homodimer is even larger. The bias toward observation of the monomer vs. dimer forms is thus larger than that suggested by the molecular weight difference. Several groups have observed that RT preparations are heterogeneous [39, 45]. Our previous studies of dimer formation have generally exhibited greater variability than is typically encountered for simpler proteins, and in some instances, evidence of monomer species persists well beyond that predicted by a simple kinetic dimerization model. Among potentially complicating factors, intermediate species formed during the metamorphic transition may become trapped as domain-swapped species (e.g., [9, 46]), and the presence of unstructured connecting loops make the protein susceptible to adventitious proteases.
The NMR studies summarized above provide further insight into the mechanism of the metamorphic transition. Comparison with recently reported studies of 15N-labeled p51 and p66 further supports our previous determinations of monomer structure and supports the conclusion that the long-lived p51 and p66 homodimers exist as structural heterodimers. This conclusion is consistent with the analysis originally put forth by Steitz and coworkers [7] and inconsistent with the recent analysis of Sharaf et al. [20].
Supplementary Material
Acknowledgments
The authors are grateful for a critical reading of this manuscript by Dr. Peter M. Thompson and Dr. Jason Williams, NIEHS. This research was performed as part of the Intramural Research Program of the NIH and National Institute of Environmental Health Sciences (NIEHS), Research Project Number Z01-ES050147 to REL. EFD is supported by NIH and NIEHS under delivery order HHSN273200700046U.
List of Abbreviations
- CD
circular dichroism
- domain
for clarity, both the domains and polymerase subdomains are simply referred to as domains
- HSQC
Heteronuclear Single-Quantum Coherence spectroscopy
- p66
includes residues 1-560 of HIV-1 reverse transcriptase (RT)
- p66ΔPL
RT residues 1-560 with palm domain residues 219-230 deleted
- p51
corresponds to the polymerase domain, and generally includes constructs that terminate at residue Trp426 corresponding to the polymerase-RH domain boundary [1] used in this study, up to constructs that terminate at Phe440, corresponding to the protease cleavage site within the RNase H domain
- RT
HIV-1 reverse transcriptase
- RT216
residues 1-216 of the p66 chain – includes the fingers and most of the palm subdomains
- TROSY
transverse relaxation-optimized spectroscopy
References
- 1.London RE. Structural Maturation of HIV-1 Reverse Transcriptase-A Metamorphic Solution to Genomic Instability. Viruses. 2016;8 doi: 10.3390/v8100260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sierra S, Kupfer B, Kaiser R. Basics of the virology of HIV-1 and its replication. J Clin Virol. 2005;34:233–244. doi: 10.1016/j.jcv.2005.09.004. [DOI] [PubMed] [Google Scholar]
- 3.Belshaw R, Pybus OG, Rambaut A. The evolution of genome compression and genomic novelty in RNA viruses. Genome research. 2007;17:1496–1504. doi: 10.1101/gr.6305707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Holmes EC. Error thresholds and the constraints to RNA virus evolution. Trends Microbiol. 2003;11:543–546. doi: 10.1016/j.tim.2003.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kohlstaedt LA, Wang J, Friedman JM, Rice PA, Steitz TA. Crystal-Structure at 3.5 Angstrom Resolution of Hiv-1 Reverse-Transcriptase Complexed with an Inhibitor. Science. 1992;256:1783–1790. doi: 10.1126/science.1377403. [DOI] [PubMed] [Google Scholar]
- 6.Jacobo-Molina A, Ding JP, Nanni RG, Clark AD, Lu XD, Tantillo C, Williams RL, Kamer G, Ferris AL, Clark P, Hizi A, Hughes SH, Arnold E. Crystal-Structure of Human-Immunodeficiency-Virus Type-1 Reverse-Transcriptase Complexed with Double-Stranded DNA at 3.0 Angstrom Resolution Shows Bent DNA. Proceedings of the National Academy of Sciences of the United States of America. 1993;90:6320–6324. doi: 10.1073/pnas.90.13.6320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang J, Smerdon SJ, Jager J, Kohlstaedt LA, Rice PA, Friedman JM, Steitz TA. Structural Basis of Asymmetry in the Human-Immunodeficiency-Virus Type-1 Reverse-Transcriptase Heterodimer. Proceedings of the National Academy of Sciences of the United States of America. 1994;91:7242–7246. doi: 10.1073/pnas.91.15.7242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zheng X, Pedersen LC, Gabel SA, Mueller GA, Cuneo MJ, DeRose EF, Krahn JM, London RE. Selective unfolding of one Ribonuclease H domain of HIV reverse transcriptase is linked to homodimer formation. Nucleic acids research. 2014;42:5361–5377. doi: 10.1093/nar/gku143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zheng X, Pedersen LC, Gabel SA, Mueller GA, DeRose EF, London RE. Unfolding the HIV-1 reverse transcriptase RNase H domain - how to lose a molecular tug-of-war. Nucleic acids research. 2016;44:1776–1788. doi: 10.1093/nar/gkv1538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zheng X, Perera L, Mueller GA, DeRose EF, London RE. Asymmetric conformational maturation of HIV-1 reverse transcriptase. Elife. 2015;4:e06359. doi: 10.7554/eLife.06359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zheng XH, Mueller GA, Cuneo MJ, DeRose EF, London RE. Homodimerization of the p51 Subunit of HIV-1 Reverse Transcriptase. Biochemistry. 2010;49:2821–2833. doi: 10.1021/bi902116z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Murzin AG. Biochemistry - Metamorphic proteins. Science. 2008;320:1725–1726. doi: 10.1126/science.1158868. [DOI] [PubMed] [Google Scholar]
- 13.Bryan PN, Orban J. Proteins that switch folds. Curr Opin Struc Biol. 2010;20:482–488. doi: 10.1016/j.sbi.2010.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Markley JL, Kim JH, Dai ZQ, Bothe JR, Cai K, Frederick RO, Tonelli M. Metamorphic protein IscU alternates conformations in the course of its role as the scaffold protein for iron-sulfur cluster biosynthesis and delivery. Febs Lett. 2013;587:1172–1179. doi: 10.1016/j.febslet.2013.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tuinstra RL, Peterson FC, Kutlesa S, Elgin ES, Kron MA, Volkman BF. Interconversion between two unrelated protein folds in the lymphotactin native state. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:5057–5062. doi: 10.1073/pnas.0709518105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tseng R, Goularte NF, Chavan A, Luu J, Cohen SE, Chang YG, Heisler J, Li S, Michael AK, Tripathi S, Golden SS, LiWang A, Partch CL. Structural basis of the day-night transition in a bacterial circadian clock. Science. 2017:355. doi: 10.1126/science.aag2516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Waheed AA, Tachedjian G. Why Do We Need New Drug Classes for HIV Treatment and Prevention? Curr Top Med Chem. 2016;16:1343–1349. doi: 10.2174/1568026616999151013124606. [DOI] [PubMed] [Google Scholar]
- 18.Iyidogan P, Anderson KS. Current perspectives on HIV-1 antiretroviral drug resistance. Viruses. 2014;6:4095–4139. doi: 10.3390/v6104095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lewis W, Dalakas MC. Mitochondrial Toxicity of Antiviral Drugs. Nature Medicine. 1995;1:417–422. doi: 10.1038/nm0595-417. [DOI] [PubMed] [Google Scholar]
- 20.Sharaf NG, Poliner E, Slack RL, Christen MT, Byeon IJ, Parniak MA, Gronenborn AM, Ishima R. The p66 immature precursor of HIV-1 reverse transcriptase. Proteins. 2014 doi: 10.1002/prot.24594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zheng X, Mueller GA, Derose EF, London RE. Metal and ligand binding to the HIV-RNase H active site are remotely monitored by Ile556. Nucleic acids research. 2012;40:10543–10553. doi: 10.1093/nar/gks791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pari K, Mueller GA, DeRose EF, Kirby TW, London RE. Solution structure of the RNase H domain of the HIV-1 reverse transcriptase in the presence of magnesium. Biochemistry. 2003;42:639–650. doi: 10.1021/bi0204894. [DOI] [PubMed] [Google Scholar]
- 23.Yang DW, Kay LE. Improved (HN)-H-1-detected triple resonance TROSY-based experiments. J Biomol Nmr. 1999;13:3–10. doi: 10.1023/A:1008329230975. [DOI] [PubMed] [Google Scholar]
- 24.Pervushin KV, Wider G, Wuthrich K. Single transition-to-single transition polarization transfer (ST2-PT) in [N-15,H-1]-TROSY. J Biomol Nmr. 1998;12:345–348. doi: 10.1023/A:1008268930690. [DOI] [PubMed] [Google Scholar]
- 25.Sklenar V, Piotto M, Leppik R, Saudek V. Gradient-Tailored Water Suppression for H-1-N-15 Hsqc Experiments Optimized to Retain Full Sensitivity. J Magn Reson Ser A. 1993;102:241–245. [Google Scholar]
- 26.Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A. Nmrpipe - a Multidimensional Spectral Processing System Based on Unix Pipes. J Biomol Nmr. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 27.Johnson BA, Blevins RA. Nmr View - a Computer-Program for the Visualization and Analysis of Nmr Data. J Biomol Nmr. 1994;4:603–614. doi: 10.1007/BF00404272. [DOI] [PubMed] [Google Scholar]
- 28.Mavridis L, Janes RW. PDB2CD: a web-based application for the generation of circular dichroism spectra from protein atomic coordinates. Bioinformatics (Oxford, England) 2017;33:56–63. doi: 10.1093/bioinformatics/btw554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ding J, Jacobo-Molina A, Tantillo C, Lu X, Nanni RG, Arnold E. Buried surface analysis of HIV-1 reverse transcriptase p66/p51 heterodimer and its interaction with dsDNA template/primer. J Mol Recognit. 1994;7:157–161. doi: 10.1002/jmr.300070212. [DOI] [PubMed] [Google Scholar]
- 30.Hsiou Y, Ding J, Das K, Clark AD, Hughes SH, Arnold E. Structure of unliganded HIV-1 reverse transcriptase at 2.7 angstrom resolution: Implications of conformational changes for polymerization and inhibition mechanisms. Structure. 1996;4:853–860. doi: 10.1016/s0969-2126(96)00091-3. [DOI] [PubMed] [Google Scholar]
- 31.Willard L, Ranjan A, Zhang H, Monzavi H, Boyko RF, Sykes BD, Wishart DS. VADAR: a web server for quantitative evaluation of protein structure quality. Nucleic acids research. 2003;31:3316–3319. doi: 10.1093/nar/gkg565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shen Y, Delaglio F, Cornilescu G, Bax A. TALOS plus: a hybrid method for predicting protein backbone torsion angles from NMR chemical shifts. J Biomol Nmr. 2009;44:213–223. doi: 10.1007/s10858-009-9333-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fernandez-Escamilla AM, Rousseau F, Schymkowitz J, Serrano L. Prediction of sequence-dependent and mutational effects on the aggregation of peptides and proteins. Nat Biotechnol. 2004;22:1302–1306. doi: 10.1038/nbt1012. [DOI] [PubMed] [Google Scholar]
- 34.Whitmore L, Wallace BA. DICHROWEB, an online server for protein secondary structure analyses from circular dichroism spectroscopic data. Nucleic acids research. 2004;32:W668–W673. doi: 10.1093/nar/gkh371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Divita G, Rittinger K, Geourjon C, Deleage G, Goody RS. Dimerization Kinetics of Hiv-1 and Hiv-2 Reverse-Transcriptase - a 2-Step Process. Journal of molecular biology. 1995;245:508–521. doi: 10.1006/jmbi.1994.0042. [DOI] [PubMed] [Google Scholar]
- 36.Venezia CF, Meany BJ, Braz VA, Barkley MD. Kinetics of Association and Dissociation of HIV-1 Reverse Transcriptase Subunits. Biochemistry. 2009;48:9084–9093. doi: 10.1021/bi9010495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sharaf NG, Poliner E, Slack RL, Christen MT, Byeon IJL, Parniak MA, Gronenborn AM, Ishima R. The p66 immature precursor of HIV-1 reverse transcriptase. Proteins. 2014;82:2343–2352. doi: 10.1002/prot.24594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ivetac A, McCammon JA. Elucidating the Inhibition Mechanism of HIV-1 Non-Nucleoside Reverse Transcriptase Inhibitors through Multicopy Molecular Dynamics Simulations. Journal of molecular biology. 2009;388:644–658. doi: 10.1016/j.jmb.2009.03.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cabodevilla JF, Odriozola L, Santiago E, Martinez-Irujo JJ. Factors affecting the dimerization of the p66 form of HIV-1 reverse transcriptase. Eur J Biochem. 2001;268:1163–1172. doi: 10.1046/j.1432-1327.2001.01939.x. [DOI] [PubMed] [Google Scholar]
- 40.Venezia CF, Howard KJ, Ignatov ME, Holladay LA, Barkley MD. Effects of efavirenz binding on the subunit equilibria of HIV-1 reverse transcriptase. Biochemistry. 2006;45:2779–2789. doi: 10.1021/bi051915z. [DOI] [PubMed] [Google Scholar]
- 41.Chen JM, Sawyer N, Regan L. Proteinprotein interactions: General trends in the relationship between binding affinity and interfacial buried surface area. Protein Science. 2013;22:510–515. doi: 10.1002/pro.2230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Unge T, Knight S, Bhikhabhai R, Lovgren S, Dauter Z, Wilson K, Strandberg B. 2.2 A resolution structure of the amino-terminal half of HIV-1 reverse transcriptase (fingers and palm subdomains) Structure. 1994;2:953–961. doi: 10.1016/s0969-2126(94)00097-2. [DOI] [PubMed] [Google Scholar]
- 43.Dufour E, El Dirani-Diab R, Boulme F, Fournier M, Nevinsky G, Tarrago-Litvak L, Litvak S, Andreola ML. p66/p51 and p51/p51 recombinant forms of reverse transcriptase from human immunodeficiency virus type 1 - Interactions with primer tRNA(Lys3), initiation of cDNA synthesis, and effect of inhibitors. Eur J Biochem. 1998;251:487–495. doi: 10.1046/j.1432-1327.1998.2510487.x. [DOI] [PubMed] [Google Scholar]
- 44.Bavand MR, Wagner R, Richmond TJ. Hiv-1 Reverse-Transcriptase - Polymerization Properties of the P51 Homodimer Compared to the P66/P51 Heterodimer. Biochemistry. 1993;32:10543–10552. doi: 10.1021/bi00091a003. [DOI] [PubMed] [Google Scholar]
- 45.Wilson JE, Wright LL, Martin JL, Haire SE, Ray PH, Painter GR, Furman PA. Recombinant human immunodeficiency virus type 1 reverse transcriptase is heterogeneous. J Acq Immun Def Synd. 1996;11:20–30. doi: 10.1097/00042560-199601010-00003. [DOI] [PubMed] [Google Scholar]
- 46.Christen MT, Menon L, Myshakina NS, Ahn J, Parniak MA, Ishima R. Structural basis of the allosteric inhibitor interaction on the HIV-1 reverse transcriptase RNase H domain. Chem Biol Drug Des. 2012;80:706–716. doi: 10.1111/cbdd.12010. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.