Abstract
FOXA2, a member of the forkhead family of transcription factors, plays essential roles in liver development and bile acid homeostasis. In this study, we report a 2.8 Å co-crystal structure of the FOXA2 DNA-binding domain (FOXA2-DBD) bound to a DNA duplex containing a forkhead consensus binding site (GTAAACA). FOXA2-DBD adopts the canonical winged-helix fold, with helix H3 and wing 1 regions mainly mediating the DNA recognition. Although the wing 2 region was not defined in the structure, isothermal titration calorimetry (ITC) assays suggested that this region was required for optimal DNA binding. Structure comparison with the FOXA3-DBD bound to DNA revealed more major groove contacts and less minor groove contacts in the FOXA2 structure compared to the FOXA3 structure. Structure comparison with the FOXO1-DBD bound to DNA showed that different forkhead proteins could induce different DNA conformations upon binding to identical DNA sequences. Our findings provide the structural basis for FOXA2 protein binding to a consensus forkhead site and elucidate how members of the forkhead protein family bind different DNA sites.
Keywords: Forkhead transcription factor, crystal structure, protein-DNA binding, isothermal titration calorimetry, DNA shape readout
Introduction
Forkhead box (FOX) proteins comprise a large family of transcription factors (TFs), members of which display functional diversity and participate in cellular processes ranging from development to immunity and metabolism1–2. More than 170 FOX family members have been identified from different species and classified into 19 subfamilies (from FOXA to FOXS)3–4. FOXA, also known as hepatocyte nuclear factor 3 (HNF3), was initially discovered as a key transcriptional regulator in the liver and many endoderm-derived tissues5. Members of the FOXA subfamily can remodel nucleosomes and, as pioneer factors, facilitate DNA binding of other TFs6. In mammals, the FOXA subfamily consists of FOXA1 (HNF3α), FOXA2 (HNF3β), and FOXA3 (HNF3γ). FOXA2 is a master regulator of gene expression in the liver, participating in liver-specific gene transcription and related physiological activities7. This protein is essential for hepatic bile acid homeostasis and prevention of cholestatic liver injury7–9. Recent evidence suggests that FOXA2 can affect the proliferation and invasiveness of pancreatic cancer cells and act as a tumor suppressor gene in pancreatic cancer10.
FOX family proteins contain a relatively conserved DNA binding domain (DBD), known as the winged-helix or forkhead domain. The DBD of FOX is typically composed of three parts: three α-helices at the N-terminus, a three-stranded β-sheet, and two less-conserved winged loops at the C-terminus (wings 1 and 2). The main DNA recognition region is located on the third helix (H3), which binds DNA by inserting into the DNA major groove. The amino acid sequence of H3 exhibits high homology among all FOX family members. Most paralogous FOX proteins bind to the canonical DNA response element 5′-RYAAAYA-3′ (R = A or G, Y = C or T)11–13. Sequence divergence in the wing regions contributes to differences in DNA binding14–15. The wing regions carry basic amino acids, which potentially recognize DNA structural features in the flanking regions of FOX binding sites, as previously observed for other transcription factor families16–18. For forkhead proteins, a recent analysis of HT-SELEX data demonstrated that binding specificity predictions improve for models that augment nucleotide sequence with DNA shape features19. In protein binding microarray experiments, forkhead proteins were shown to have the ability to specifically bind to alternate distinct DNA motifs20. These findings merit structural studies that compare binding mechanisms of paralogous FOX transcription factors.
The human full-length FOXA2 protein contains two transcription activation domains and a forkhead domain (Supplementary Figure S1A)21. Its DBD displays high sequence homology (95%) to FOXA1 and FOXA3 (Supplementary Figure S1B)22. The co-crystal structure of a FOXA3/DNA complex was previously reported23, but the DNA used for crystallization contained a nonconsensus binding site. Genome-wide analysis of FOXA2-binding sites by ChIP-seq in human and mouse adult liver tissues suggested that FOXA2 binds to the consensus sequence (5′-GTAAACA-3′) of the FOX family24–25. In this study, we determined the co-crystal structure of FOXA2-DBD bound to a 16-bp DNA containing a consensus site (5′-GTAAACA-3′). We compared our structure with previously solved FOXA3 and FOXO1 co-crystal structures, and employed biochemical analyses to study FOXA2 binding to different DNA sites.
Materials and Methods
Protein expression and purification
The coding region of human FOXA2-DBD (residues 157–258) was cloned in the pGEX-6P1 vector. All constructs were confirmed by DNA sequencing (GenScript, NanJing, China). Protein was expressed in the Escherichia coli Rosetta BL21 (DE3) cells and induced by 0.5 mM IPTG for 6 h at 297 K. Protein was purified by using glutathione-Sepharose (GE Healthcare) according to a standard protocol. The GST affinity tag was removed by PresScission cleavage overnight at 277 K. Protein was further purified by cation-exchange (Mono S 5/50GL, GE Healthcare) and size-exclusion chromatography (Superdex 75 10/300 GL, GE Healthcare). Peak fractions were collected and concentrated to ~26 mg/mL. The final protein was stored in storage buffer (10 mM Hepes, 300 mM NaCl, 10 mM MgCl2, 0.5 mM TCEP, pH = 7.5) at 193 K.
Duplex DNA preparation
All DNAs were purchased from Genscript (NanJing, China) and purified as described previously26. The DBE2 (Daf-16 family binding element 2) sequence contains strands 5′-TGCAAAATGTAAACAAGACT-3′ and 5′-AGTCTTGTTTACATTTTGCA-3′27. The TTR (transthyretin) sequence contains strands 5′-TTGACTAAGTCAATCA-3′ and 5′-TGATTGACTTAGTCAA-3′23.
Crystallization, data collection, and structure determination
The FOXA2-DNA complex was prepared by mixing purified FOXA2 protein and DNA at 1:1.2 molar ratio with the final protein concentration of 10 mg/mL. Crystals were initially screened with the Hampton Natrix Kit by sitting drop vapor diffusion at 291 K. Crystals were optimized by the hanging drop method at 291 K, by using a well solution of 80 mM Mg(OAc)2, 50 mM MES buffer, and 16–20% PEG4K (pH 6.5). Crystals were stabilized in harvest/cryoprotectant buffer containing 80 mM Mg(OAc)2, 50 mM MES buffer, 30% PEG4K, and 20% glycerol (pH 6.5) and flash-frozen with liquid nitrogen for cryo-crystallography. Data were collected at the Shanghai Synchrotron Radiation Facility (SSRF) and BL17U1 beamline.
The co-crystal structure was solved as described previously28. Molecular replacement was carried out using Phaser from the PHENIX package29. A previously solved FOXA3 structure (PDB ID 1VTN, Chain C) was used as initial search model in the molecular replacement. The structure was further refined by using PHENIX. Statistical results of the crystallographic analysis are presented in Table 1. Figures were generated by PyMol30. DNAproDB was used to generate schematic diagrams of protein-nucleic acid interactions31.
Table 1.
Data collection and refinement statistics.
| FOXA2-DBD/DNA complex | |
|---|---|
| Resolution range [Å] | 33.64 – 2.796 (2.896 – 2.796) |
| Space group | C 1 2 1 |
| Unit cell (a, b, c, α, β, γ) | 195.126, 71.932, 72.155, 90.0, 103.14, 90.0 |
| Total reflections | 178752 (17848) |
| Unique reflections | 23535 (2364) |
| Multiplicity | 7.5 (7.5) |
| Completeness [%] | 96.80 (86.05) |
| Mean I/sigma(I) | 18.52 (2.71) |
| Wilson B-factor | 68.73 |
| R-merge | 0.1666 (3.753) |
| R-meas | 0.1789 |
| R-work | 0.2298 (0.3499) |
| R-free | 0.2811 (0.3964) |
| Number of non-hydrogen atoms | 5425 |
| Macromolecules | 5384 |
| Water | 41 |
| Protein residues | 460 |
| RMS (bonds) | 0.005 |
| RMS (angles) | 1.05 |
| Ramachandran favored [%] | 91 |
| Ramachandran outliers [%] | 3.4 |
| Clashscore | 10.38 |
| Average B-factor | 69.40 |
| Macromolecules | 69.50 |
| Solvent | 58.70 |
ITC assays
ITC measurements were obtained at 298 K on a NANO ITC instrument (TA Instruments). Duplex DNA and purified protein were dialyzed overnight in storage buffer (25 mM Hepes, 300 mM NaCl, 10 mM MgCl2, 1 mM TCEP, pH 7.0). Next, 50 μL of DNA at 100–200 μM was injected into 15–20 μM FOXA2 protein solution (300 μL per sample cell). A 240s interval between injections was used, to permit the signal to return to baseline before the next injection. Each injection volume was 2 μL. All data were analyzed by NANO Analysis software (TA Instruments). Heat generated by DNA dilution was used as a control. Standard free energies of binding and stoichiometry were obtained from the Kd and ΔH values derived from ITC curve fitting, respectively. To ensure data reliability, all experiments were repeated at least three times.
DNA structure and protein-DNA interaction analysis
Structures of DNA bound to FOXA2 (this study), FOXA3 (PDB ID 1VTN)23, and FOXO1 (PDB ID 3CO7)27 were analyzed with CURVES32. For comparison of these structures, we renamed the FOXA3 chains to align the structures and perform binding site analysis in the same strand orientation. DNA structural features were compared between different binding sites. For visualization of protein-DNA interactions, contact maps were generated with the DNAproDB tool31. Labels were customized for simplicity. In certain cases, the recognition helix was renamed to “H3” for agreement with the standard forkhead nomenclature. To calculate the electrostatic potential in the DNA minor groove, DelPhi33 was used to solve the nonlinear Poisson-Boltzmann equation as previously described34. To analyze the structure of unbound DNA targets, all-atom Monte Carlo (MC) simulations were performed by using a previously published method and protocol35, 36. All possible hydrogen atom (H) positions were calculated for donor atoms (D) that satisfy specified geometrical criteria with acceptor atoms (A) in the vicinity. The criteria used are: the H-A distance is < 2.7Å, the D-A distance is < 3.35Å, the D-H-A angle is > 90° and the H-A-AA angle is > 90°, where AA is the atom attached to the acceptor. All possible hydrogen bonds were identified by finding all the prospective atoms which satisfy given geometric criteria between the hydrogen bond donors (D) and acceptors (A), and the D-A distance is less than 3.35Å37.
PDB accession numbers
Atomic coordinates and structural factors of the FOXA2-DBD/DNA complex were deposited at the RCSB Protein Data Bank under accession code 5X07.
Results
Overall structure of the FOXA2-DBD/DNA complex
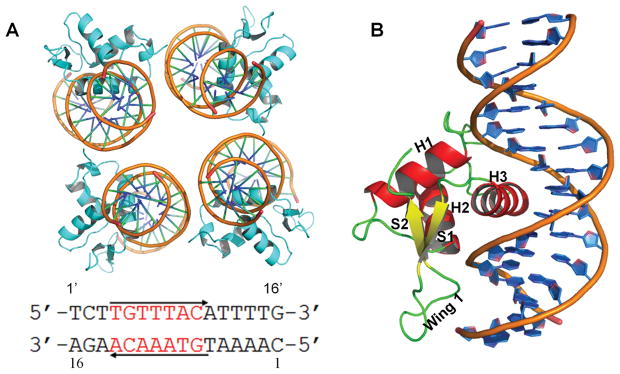
We determined the co-crystal structure of FOXA2-DBD (residues 157–257) bound to a 16-bp double-stranded DNA containing a consensus binding element (5′-GTAAACA-3;) (Figure 1A). Co-crystals diffracted to a resolution of 2.8 Å and belonged to space group C2 (Table 1). The asymmetric unit contained four FOXA2 forkhead domains and four double-stranded DNA segments. Each FOXA2 monomer adopted the canonical winged-helix fold23, 27 and comprised three α-helixes (H1, H2, and H3), two antiparallel β strands (S1 and S2), and a loop (wing 1) between S1 and S2 (Figure 1B). Electron density was not observed for wing 2 of the FOXA2 forkhead domain, suggesting that this region might be flexible and disordered in the crystal. This finding was surprising because this region of FOXA2 is highly homologous to that of FOXA3, for which a previously published structure revealed the C-terminal region forming an extended loop (wing 2).
Figure 1.
Co-crystal structures of the FOXA2-DBD/DBE2 complex. (A) Overall structure of the FOXA2 forkhead domain bound to the DBE2 element in an asymmetric unit. FOXA2 molecules are shown in cyan, and DNA targets in orange. Sequence of the 16-bp DNA duplex (DBE2) used in the crystallization is listed below. (B) Structural representation of the FOXA2-DBD (chain C) bound to the DBE2 DNA (chains A and B). Secondary structure elements are labeled.
DNA recognition by the FOXA2-DBD
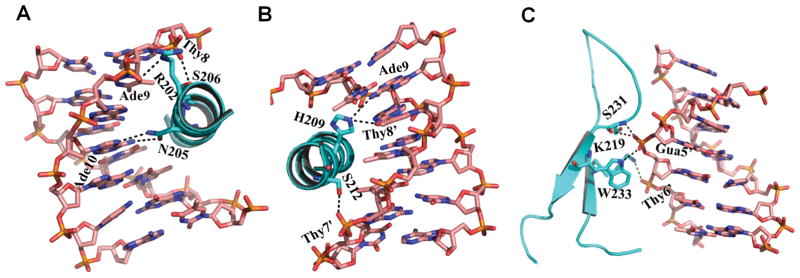
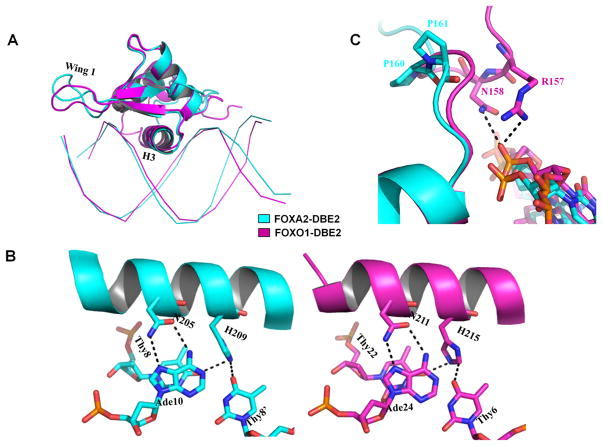
DNA recognition by FOXA2 was dominated by H3, which inserted deeply into the major groove (Figure 2). Asn205 formed bidentate hydrogen bonds with Ade10, whereas His209 recognized Thy8′ and Ade9 with base-specific hydrogen bonds (Figures 2A & 2B). Arg202, Ser206, and Ser212 formed hydrogen bonds with the phosphate groups of Ade9, Thy8, and Thy7′, respectively (Figures 2A & 2B). Numerous van der Waals contacts also formed between FOXA2 and its corresponding DNA targets. In addition to helix H3, wing 1 of FOXA2 contributed to DNA binding. A pair of hydrogen bonds formed between the Gua5′ phosphate group and Ser231/Trp233. Lys219 interacted with the phosphate group of Thy6′ (Figure 2C).
Figure 2.
DNA recognition by the FOXA2 DBD domain. (A, B) Hydrogen bonds between helix 3 of FOXA2 and DNA. (C) Interactions between wing 1 and DNA.
Comparison of DNA binding affinity using isothermal titration calorimetry (ITC)
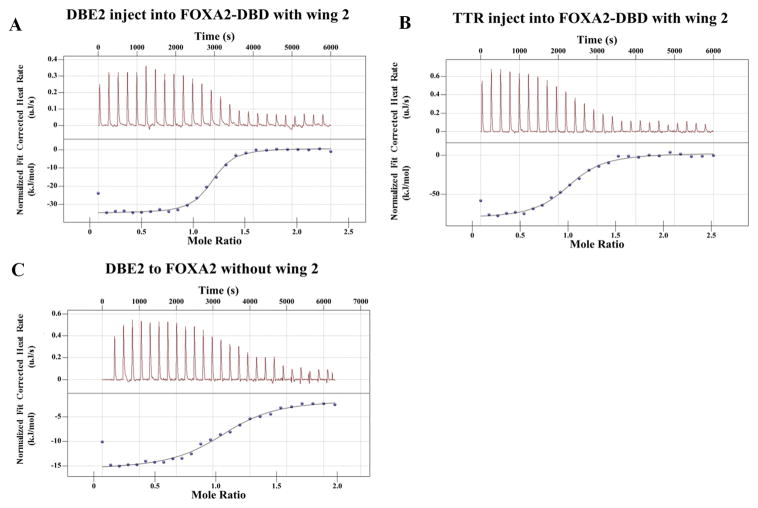
In the previously solved FOXA3-DBD/DNA structure, the binding site of FOXA3 was 5′-GACTAAGTCAACC-3, which is quite different from the consensus FOX binding site (5′-GTAAACA-3′). We used ITC assays to compare the binding affinities of FOXA2-DBD (same fragment as in the crystal structure determination) to different DNA target sites. Figure 3 shows representative binding isotherms and a detailed analysis of thermodynamic binding parameters. According to these results, the FOXA2 DBD bound to DNA with a 1:1 stoichiometric ratio. Kd of the FOXA2 forkhead domain binding to the transthyretin (TTR) site was estimated to be ~1.5 μM. Kd of FOXA2 binding to the Daf-16 family binding element 2 (DBE2) was estimated to be ~105 nM (Table 2). Thus, FOXA2 bound to the consensus site with a higher binding affinity.
Figure 3.
Binding affinity measurement of protein-DNA interactions by quantitative ITC. (A–C) Quantification of the interaction between FOXA2 DBD and DNA derived from different DNA by ITC. Representative power-response curves (top) and heats of reaction normalized to the moles of protein injected (bottom).
Table 2.
Thermodynamic parameters of the interaction. All data were measured at 298 K in 25 mM Hepes (pH 7.5), 300 mM NaCl, 10 mM MgCl2, and 1 mM TCEP buffer. Values represent at least three independent experiments corresponding to the averaged deviation.
| Protein | DNA | Kd (nM) | N | ΔH (KJ/mol) | ΔS (J/mol×K) |
|---|---|---|---|---|---|
| FOXA2 (157–257) | TTR | 1500±200 | 1.0±0.1 | −12.26±0.5 | 58.2±0.8 |
| FOXA2 (157–257) | DBE2 | 105±20 | 1.2±0.1 | −35.54±2 | 14±5 |
| FOXA2 (157–239) | DBE2 | 3500±300 | 1.1±0.1 | −14.38±0.6 | 55.8±0.5 |
In contrast to the previously reported FOXA3-DBD/TTR structure, the electron density of wing 2 (residues 240–258) in the FOXA2-DBD/DBE2 structure could not be observed. This result suggested that wing 2 was flexible and disordered in the crystal. To test whether wing 2 contributes to FOXA2-DBD/DNA binding, we constructed a truncated fragment of FOXA2 without wing 2 and measured the DNA binding ability using ITC. The binding affinity of FOXA2 was reduced by ~30-fold when the C-terminal wing 2 region was removed (Table 2). These results suggested that wing 2 of FOXA2 was important for optimal DNA binding.
Structural comparison between FOXA2 and FOXA3 DBDs bound to dissimilar DNA sequences
We sought to compare the properties of protein-DNA binding among FOXA subfamily members by superimposing the FOXA3-DBD/DBE2 structure with the previously reported structure of the FOXA3-DBD/TTR complex (PDB ID 1VTN)23. Despite the high sequence homology between FOXA2 and FOXA3 (Supplementary Figure S1B), their structures differed when bound to different DNA targets (Figure 4A). In the previously reported structure, wing 2 was ordered, whereas it was flexible in the FOXA2-DBD/DBE2 structure (Figure 4A).
Figure 4.
Comparison with previous FOXA3-DBD/TTR structure (PDB ID 1VTN)23. (A) Overall structure comparison between FOXA2 (cyan) and previous FOXA3 (orange). (B) Bidentate hydrogen bonds between Asn and Ade were conserved in both structures. (C) A hydrogen bond formed between His209 and Thy8′ in the FOXA2-DBD/DBE2 structure but not in the FOXA3-DBD/TTR structure. (D) FOXA3 Arg210 formed hydrogen bonds with DNA in the FOXA3-DBD/TTR structure.
In addition to analyzing the overall structure, we compared the structures in terms of their protein-DNA interactions. Bidentate hydrogen bonds between Asn and Ade were conserved in both structures (Figure 4B). His209 in the FOXA2-DBD/DBE2 structure formed a hydrogen bond with Thy8′, whereas this hydrogen bond did not exist in the FOXA3-DBD/TTR structure because the corresponding nucleic acid was replaced with guanine (Figure 4C). Arg210 of FOXA3 (corresponding to Arg250 in FOXA2) formed hydrogen bonds with Thy4 and Gua31, whereas these interactions were not observed in the FOXA2-DBD/DBE2 structure because this region was flexible in that structure (Figure 4D).
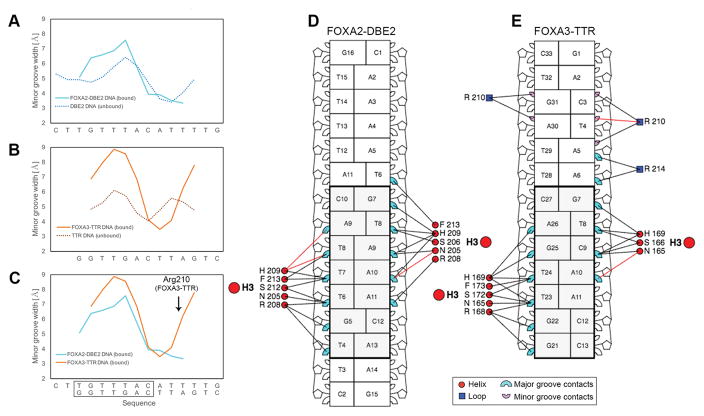
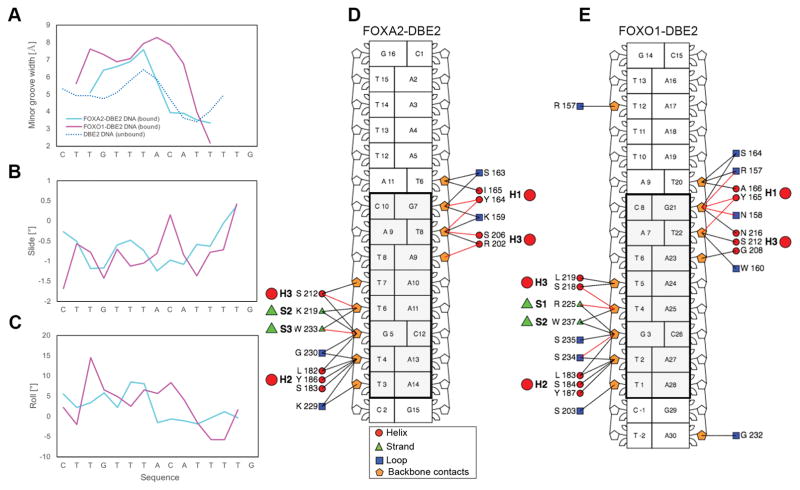
We also analyzed DNA structural features for the DBE2 DNA bound by FOXA2-DBD. This analysis revealed a widening of the minor groove within the center of the core binding site, with a maximum located at the TpA step, followed by an adjacent narrowing (Figure 5A, cyan line) around the AT-rich flanking region. To differentiate between an intrinsic or induced shape, we performed all–atom MC simulations of the free DNA. This analysis revealed that the deformation pattern was intrinsic and already apparent in the shape of the unbound DNA target (Figure 5A, dotted blue line).
Figure 5.
Structural analysis of FOXA2 and FOXA3 DNA binding sites. (A–C) Minor groove width of FOXA-bound and unbound DNAs, showing comparisons between (A) FOXA2 bound DBE site vs. unbound DBE2 site, (B) FOXA3 bound TTR site vs. unbound TTR site, and (C) FOXA2 bound DBE2 vs. FOXA3 bound TTR. (D–E) Schematic diagrams show interactions between the forkhead domain and the DNA major and minor groove in a nucleotide-residue interaction map for (D) FOXA2-DBD/DBE2 and (E) FOXA3-DBD/TTR. Nucleotide-residue red and black lines represent interactions which involve hydrogen bonds or other contacts, respectively. Images were generated with DNAproDB 31. Black rectangles (panels C, D and E) highlight the core binding element recognized by these FOXA proteins.
Next, we investigated whether the DNA structural features of FOXA binding sites could be important for DNA shape readout38. In the case of FOX proteins, besides the major groove recognition, wings 1 and 2 could a play role in shape readout at the edges and flanking regions of the core binding site. In addition to our current FOXA2-DBD/DBE2 complex, we also analyzed the DNA structure of the previously published FOXA3-DBD/TTR complex due to its protein similarity and well-defined electron density of wing 223.
The DNA target site in the FOXA3-DBD/DNA complex (TTR) adjusted to protein binding in a similar manner as the target bound by FOXA2 (DBE2) (Figure 5A–B), despite differences in the sequences of their core DNA binding sites. Comparing the FOXA3-bound to the FOXA2-bound target, we found that the minor groove width of FOXA3-DBD/TTR followed the same pattern as that of FOXA2-DBD/DBE2, with an increased maximum (by ~1.3 Å) for the FOXA3-bound DNA and a similar minimum (Figure 5C). For the free DNA, maximum and minimum minor groove width were comparable for the two sequences, although the maximum was shifted by one nucleotide position and the minimum by two positions (Figure 5A–B). For the FOXA3-DBD/TTR target site, the region of minor groove narrowing coincided with the location where Arg210 of wing 2 inserted into the minor groove. This is also a region of increased negative electrostatic potential (Figure S3), which would favor Arg210 interaction with the DNA. Arg210 is not exactly located at the minor groove minimum; instead, it is in closer proximity to the A30/T4 base pair (Figure 5E), where it forms a hydrogen bond with Thy4. This observation is consistent with an electrostatic potential-dependent recognition of narrow minor groove regions39.
DNAproDB analysis of the protein-DNA contacts of the complexes highlighted an intricate pattern of interactions (Figure 5D–5E). The analysis indicated a greater number of contacts for the FOXA2-DBD in the major groove than for the FOXA3-DBD, whereas the latter showed contacts in the minor groove that were not observed in the FOXA2-DBD/DBE2 complex. Therefore, the well-defined electron density of FOXA3-DBD/TTR wing 2 might be a result of enhanced interactions between wing 2 and the minor groove, as a way to stabilize the complex due to the decreased number of contacts in the major groove.
Overall, we found that highly homologous FOXA2 and FOXA3 DBDs induce a similar deformation to their DNA target sites upon binding, in spite of their different DNA sequences. This is observed in a scenario where the FOXA2-DBD displays an increased number of hydrogen bonds in the major groove when compared to the FOXA3-DBD, whereas the wing 2 region is disordered. These differences may contribute to binding affinity differences of the FOXA2 protein when binding to different DNA target sites.
Structure comparison between FOXA2 and FOXO1 DBDs bound to similar DNA sequences
In order to study the structural variations induced upon binding of different FOX proteins to the same DNA sequence, we compared the DNA conformation of our FOXA2 complex to that of the previously published FOXO131, both bound to DNA sequence DBE2 (Figure 6). The proteins shared similar overall structures, with slight differences in the wing 1 region and the N-terminal tail (Figure 6A). The major groove in the FOXA2-DBD/DBE2 structure was wider than in the FOXO1-DBD/DBE2 structure (Figure 6A). We also examined interactions between the proteins and their DNA target sites. In the structures, asparagine and histidine in the FOXA2 and FOXO1 proteins, respectively, formed the same interaction network with the corresponding bases (Figure 6B). A major difference in DNA binding lies at the N-terminal tail. The N-terminal tail of FOXO1 forms a closer contact to DNA, and Arg157 and Asn158 of FOXO1 interact with DNA backbone directly (Figure 6C). The sequence differences in this region between two proteins may contribute to this difference. The N-terminal tail of FOXA2 has two prolines (Pro160 and Pro161), in contrast to the same region in FOXO1.
Figure 6.
Comparison with the previous FOXO1-DBD/DBE2 structure (PDB ID: 3CO7). (A) Overall structure comparison between FOXA2-DBD/DBE2 (cyan) and FOXO1-DBD/DBE2 (magenta). (B) Most hydrogen bonding interactions were conserved between the structures. (C) Structure comparison shows that the N-terminal tail of FOXO1-DBD is located closer to the minor groove although the proteins is bound to the same DNA. Arg157 and Asn158 of FOXO1 (magenta) interact with the DNA backbone. The presence of Pro160 and Pro161 of FOXA2 (cyan) orients the N-terminal tail of FOXA2-DBD away from the minor groove.
Despite the DNA binding sequence being the same in both the FOXA2 and FOXO1 complexes, the minor groove width changes observed upon protein binding are strikingly different. In particular, the minor groove width changes on the DBE2 target DNA induced upon protein binding in both complexes (Figure 7A) deviated more from each other than the changes observed upon the binding of more similar FOXA proteins bound to different DNA binding sites (FOXA2-DBD/DBE2 and FOXA3-DBD/TTR, Figure 5C). Compared to the FOXA2-bound DNA, the DNA bound by FOXO1 has its maximum minor groove width shifted by one nucleotide position and was more distorted at the edges of the core binding site (Figure 7A, magenta line). Furthermore, the minor groove width maximum was increased by ~1.8 Å and the minimum was decreased by ~1.2 Å compared to the values of free DNA. These findings indicate a more distorted DNA when compared to the same sequence bound by FOXA2-DBD (Figure 7A, cyan line). Other DNA shape parameters also varied as a result of accommodating these two different proteins (Figure 7C–D), particularly at the CpA step.
Figure 7.
Structural analysis of DNA in complex with FOXA2 and FOXO1 DBDs. (A–C) Comparison of DNA shape features between FOXA2 and FOXO1 complexes, showing (A) minor groove widths of bound and unbound DNAs, (B–C) slide and roll angles for FOXA2 and FOXO1 DBE2 DNAs. Bound DBE2 is shown in cyan (FOXA2) and magenta (FOXO1), while unbound DBE2 is shown as a dotted blue line. (D–E) Schematic diagram shows interactions between the forkhead domain and the DNA backbone as a nucleotide-residue interaction map for (D) FOXA2-DBD/DBE2 and (E) FOXO1-DBD/DBE2. Backbone contacts display slight differences between FOXA2-DBD/DBE2 and FOXO1-DBD/DBE2 complexes. Nucleotide-residue contacts (red and black lines) represent interactions that involve hydrogen bonds or other contacts, respectively. Images were obtained using DNAproDB31. Black rectangles (panels D and E) highlight the core binding element in the DBE2 DNA site.
Although neither protein showed base-specific contacts with the minor groove, the N-terminal tail of FOXO1 was directed towards the minor groove, allowing for an increased number of closer contacts with the backbone compared to FOXA2 (Figure 6C; Figure 7D–E, loop residues contacts with the backbone). This positioning could explain the widening of the minor groove observed in this region (Figure 6A & Figure 7A). Furthermore, the wing 1 region differed between these two proteins, leading to slightly different contacts to the backbone of the flanking regions (Figure 7 and Figure S2).
Discussion
FOXA2 plays important roles in the initiation of liver development and normal bile acid homeostasis 8. In human and mouse adult liver tissues, ChIP-seq analysis showed that FOXA2 preferentially bound to the FOX consensus sequence 5′-RTAAAYA-3′ (R=A/G, Y=C/T) 24–25. The FOXA3 protein could also bind to a nonconsensus DNA site23. It has, however, remained unclear how FOXA proteins recognize different DNA binding sites. Hydroxyl radical footprinting and site-directed mutagenesis indicated that the wing regions, especially wing 1, are crucial for optimal DNA binding 14. In this study, we determined the co-crystal structure of FOXA2 bound to a DNA consensus site. ITC data showed that FOXA2 bound to DBE2 with a higher binding affinity than to TTR. These observations suggest that the highly similar FOX proteins FOXA2 and FOXA3 may bind to different DNA targets through adaptions of their protein-DNA interactions.
Most forkhead proteins recognize a common binding site (RTAAAYA, R = A/G, Y = C/T)11. We observed that FOXA2 bound to a DAF-16 family binding element (5′-GTAAACAA-3′), which was also the recognition element of FOXO1. Although FOX proteins recognized the DNA binding element with remarkable similarity, their DNA binding specificities displayed marked variability. Each forkhead protein has its own nucleotide preference at the R and Y positions in the core motif and flanking regions11. For example, although DBE1 and DBE2 share a common core motif, FOXO1 exhibited 5-fold higher binding affinity to DBE2 than to DBE1 because of the thymine-rich 3′ flanking sequence 27. On the other hand, different FOX proteins could induce different DNA deformations (e.g., strong bending of the DNA helix11). Our study suggests that, upon binding of different FOX proteins, DNA can adapt through changes in shape parameters.
Wing regions might play important roles in the DNA binding of FOX proteins40–41. Nuclear magnetic resonance data revealed that wing 1 is disordered in the absence of DNA42–43. This region undergoes rearrangement upon DNA binding, binding to the minor groove through direct hydrogen bonds and van der Waals contacts42. Amino acid composition and length variations might contribute to DNA binding differences. Wing 1 in the FOXP subfamily is much shorter than wing 1 in the FOXA subfamily and makes limited DNA contacts. Therefore, the FOXP forkhead domain binds DNA with lower binding affinity than that of FOXA44. Wing 1 might act as a cofactor recognition target. In the crystal structure of the FOXP2/NFAT/DNA ternary complex, wing 1 served as a primary interaction site of the two TFs45. Wing 2 is well ordered upon DNA binding in some co-crystal structures but disordered in others. In the FOXA3-DBD/DNA structure, wing 2 formed an extended loop that bound DNA extensively; in the FOXP2-DBD/DNA and FOXM-DBD/DNA structures, wing 2 adopted an α-helical structure (H5)44, 46. Although wing 2 cannot be defined in both the FOXO1-DBD/DNA and FOXA2-DBD/DNA structures27, biochemical studies suggest that this region is important for optimal DNA binding.
The N-terminal tail of the forkhead domain might also contribute to DNA binding. Interestingly, for the DBE2 DNA binding site, this region was shown to interact with the DNA backbone in the FOXO1 complex, but not in the FOXA2 complex. By investigating structural features of other forkhead structures, we found that the positioning of the N-terminal tail varies across other members of this family, showing interactions with the DNA backbone in FOXK2 and FOXM1 complexes, but not with that of FOXP347. Sequence differences in the region among these proteins might contribute to the differences in DNA binding since this region is not conserved among forkhead transcription factors (Figure S1). In addition, DNA sequence also contributes to the binding differences. In the case of FOXO1, the N-terminal tail binds to DBE2 DNA27, but not to G6PC1 DNA48.
In summary, the structure of the FOXA2-DBD/DNA complex revealed the mechanism by which the FOXA2 forkhead domain binds to the consensus FOX binding sequence (5′-GTAAACA-3′). ITC experiments showed that FOXA2-DBD binds to the DBE2 consensus site with higher binding affinity than to the TTR site. This result is consistent with our structural observations that FOXA2-DBD formed more interactions with the DBE2 site than with the TTR site. In addition, structural comparison with the FOXO1-DBD/DBE2 complex showed that different forkhead proteins could induce different DNA deformations when binding to the same DNA target site.
Supplementary Material
Acknowledgments
The authors thank Minjiong Li from the staff of the Shanghai Synchrotron Radiation facility (SSRF) for help with data collection. This work was supported by the National Natural Science Foundation of China (grants 81372904 and 81570537 to Y.C.; grant 81272971 to Z.C.) and the National Institutes of Health (grants U01GM103804 and R01GM106056 to R.R.; grants R01AI113009 and R01GM064642 to L.C.). R.R. is an Alfred P. Sloan Research Fellow.
Footnotes
Supplemental Information includes three figures which can be found with this article online.
Competing financial interests
The authors declare no competing financial interests.
References
- 1.Hannenhalli S, Kaestner KH. The evolution of Fox genes and their role in development and disease. Nature reviews Genetics. 2009;10(4):233–40. doi: 10.1038/nrg2523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lam EW, Brosens JJ, Gomes AR, Koo CY. Forkhead box proteins: tuning forks for transcriptional harmony. Nature reviews Cancer. 2013;13(7):482–95. doi: 10.1038/nrc3539. [DOI] [PubMed] [Google Scholar]
- 3.Tuteja G, Kaestner KH. SnapShot: forkhead transcription factors I. Cell. 2007;130(6):1160. doi: 10.1016/j.cell.2007.09.005. [DOI] [PubMed] [Google Scholar]
- 4.Tuteja G, Kaestner KH. Forkhead transcription factors II. Cell. 2007;131(1):192. doi: 10.1016/j.cell.2007.09.016. [DOI] [PubMed] [Google Scholar]
- 5.Friedman JR, Kaestner KH. The Foxa family of transcription factors in development and metabolism. Cellular and molecular life sciences : CMLS. 2006;63(19–20):2317–28. doi: 10.1007/s00018-006-6095-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Iwafuchi-Doi M, Donahue G, Kakumanu A, Watts JA, Mahony S, Pugh BF, Lee D, Kaestner KH, Zaret KS. The Pioneer Transcription Factor FoxA Maintains an Accessible Nucleosome Configuration at Enhancers for Tissue-Specific Gene Activation. Molecular cell. 2016;62(1):79–91. doi: 10.1016/j.molcel.2016.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bochkis IM, Schug J, Rubins NE, Chopra AR, O’Malley BW, Kaestner KH. Foxa2-dependent hepatic gene regulatory networks depend on physiological state. Physiological genomics. 2009;38(2):186–95. doi: 10.1152/physiolgenomics.90376.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bochkis IM, Rubins NE, White P, Furth EE, Friedman JR, Kaestner KH. Hepatocyte-specific ablation of Foxa2 alters bile acid homeostasis and results in endoplasmic reticulum stress. Nature medicine. 2008;14(8):828–36. doi: 10.1038/nm.1853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gosalia N, Yang R, Kerschner JL, Harris A. FOXA2 regulates a network of genes involved in critical functions of human intestinal epithelial cells. Physiological genomics. 2015;47(7):290–7. doi: 10.1152/physiolgenomics.00024.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vorvis C, Hatziapostolou M, Mahurkar-Joshi S, Koutsioumpa M, Williams J, Donahue TR, Poultsides GA, Eibl G, ID Transcriptomic and CRISPR/Cas9 technologies reveal FOXA2 as a tumor suppressor gene in pancreatic cancer. Am J Physiol Gastrointest Liver Physiol. 2016;310(11):G1124–1137. doi: 10.1152/ajpgi.00035.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pierrou S, Hellqvist M, Samuelsson L, Enerbäck S, Carlsson P. Cloning and characterization of seven human forkhead proteins: binding site specificity and DNA bending. EMBO J. 1994;13(20):5002–5012. doi: 10.1002/j.1460-2075.1994.tb06827.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen X, Ji Z, Webber A, Sharrocks AD. Genome-wide binding studies reveal DNA binding specificity mechanisms and functional interplay amongst Forkhead transcription factors. Nucleic acids research. 2016;44(4):1566–78. doi: 10.1093/nar/gkv1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Unutmaz D, Koh KP, Sundrud MS, Rao A. Domain Requirements and Sequence Specificity of DNA Binding for the Forkhead Transcription Factor FOXP3. PLoS ONE. 2009;4(12):e8109. doi: 10.1371/journal.pone.0008109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cirillo LA, Zaret KS. Specific interactions of the wing domains of FOXA1 transcription factor with DNA. J Mol Biol. 2007;366(3):720–724. doi: 10.1016/j.jmb.2006.11.087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Boura E, Silhan J, Herman P, Vecer J, Sulc M, Teisinger J, Obsilova V, Obsil T. Both the N-terminal loop and wing W2 of the forkhead domain of transcription factor Foxo4 are important for DNA binding. The Journal of biological chemistry. 2007;282(11):8265–75. doi: 10.1074/jbc.M605682200. [DOI] [PubMed] [Google Scholar]
- 16.Schöne S, Jurk M, Helabad MB, Dror I, Lebars I, Kieffer B, Imhof P, Rohs R, Vingron M, Thomas-Chollier M, Meijsing SH. Sequences flanking the core-binding site modulate glucocorticoid receptor structure and activity. Nature Communications. 2016;7:12621. doi: 10.1038/ncomms12621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Levo M, Zalckvar E, Sharon E, Dantas Machado AC, Kalma Y, Lotam-Pompan M, Weinberger A, Yakhini Z, Rohs R, Segal E. Unraveling determinants of transcription factor binding outside the core binding site. Genome Research. 2015;25(7):1018–1029. doi: 10.1101/gr.185033.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dror I, Zhou T, Mandel-Gutfreund Y, Rohs R. Covariation between homeodomain transcription factors and the shape of their DNA binding sites. Nucleic acids research. 2014;42(1):430–441. doi: 10.1093/nar/gkt862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yang L, Orenstein Y, Jolma A, Yin Y, Taipale J, Shamir R, Rohs R. Transcription factor family-specific DNA shape readout revealed by quantitative specificity models. Molecular Systems Biology. 2017;13(2) doi: 10.15252/msb.20167238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nakagawa S, Gisselbrecht SS, Rogers JM, Hartl DL, Bulyk ML. DNA-binding specificity changes in the evolution of forkhead transcription factors. Proceedings of the National Academy of Sciences. 2013;110(30):12349–12354. doi: 10.1073/pnas.1310430110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pani L, OD, Porcella A, Qian X, Lai E, Costa RH. Hepatocyte nuclear factor 3 beta contains two transcriptional activation domains, one of which is novel and conserved with the Drosophila fork head protein. Molecule Cell Biology. 1992;12(9):3723–3732. doi: 10.1128/mcb.12.9.3723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lalmansingh AS, Karmakar S, Jin Y, Nagaich AK. Multiple modes of chromatin remodeling by Forkhead box proteins. Biochimica et biophysica acta. 2012;1819(7):707–15. doi: 10.1016/j.bbagrm.2012.02.018. [DOI] [PubMed] [Google Scholar]
- 23.Clark KL, HE, Lai E, Burley SK. Co-crystal structure of the HNF-3/fork head DNA-recognition motif resembles histone H5. nature. 1993;364(6436):412–420. doi: 10.1038/364412a0. [DOI] [PubMed] [Google Scholar]
- 24.Wederell ED, Bilenky M, Cullum R, Thiessen N, Dagpinar M, Delaney A, Varhol R, Zhao Y, Zeng T, Bernier B, Ingham M, Hirst M, Robertson G, Marra MA, Jones S, Hoodless PA. Global analysis of in vivo Foxa2-binding sites in mouse adult liver using massively parallel sequencing. Nucleic acids research. 2008;36(14):4549–64. doi: 10.1093/nar/gkn382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Levitsky VG, Kulakovskiy IV, Ershov NI, Oshchepkov DY, Makeev VJ, Hodgman TC, Merkulova TI. Application of experimentally verified transcription factor binding sites models for computational analysis of ChIP-Seq data. BMC Genomics. 2014:15–80. doi: 10.1186/1471-2164-15-80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen Y, Bates DL, Dey R, Chen PH, Machado AC, Laird-Offringa IA, Rohs R, Chen L. DNA binding by GATA transcription factor suggests mechanisms of DNA looping and long-range gene regulation. Cell reports. 2012;2(5):1197–206. doi: 10.1016/j.celrep.2012.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Brent MM, Anand R, Marmorstein R. Structural basis for DNA recognition by FoxO1 and its regulation by posttranslational modification. Structure. 2008;16(9):1407–16. doi: 10.1016/j.str.2008.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wu D, Guo M, Philips MA, Qu L, Jiang L, Li J, Chen X, Chen Z, Chen L, Chen Y. Crystal Structure of the FGFR4/LY2874455 Complex Reveals Insights into the Pan-FGFR Selectivity of LY2874455. PLOS ONE. 2016;11(9):e0162491. doi: 10.1371/journal.pone.0162491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Adams PD, Afonine PV, Bunkóczi G, Chen VB, Echols N, Headd JJ, Hung LW, Jain S, Kapral GJ, Grosse Kunstleve RW, McCoy AJ, Moriarty NW, Oeffner RD, Read RJ, Richardson DC, Richardson JS, Terwilliger TC, Zwart PH. The Phenix software for automated determination of macromolecular structures. Methods. 2011;55(1):94–106. doi: 10.1016/j.ymeth.2011.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Warren LD. PyMOL User’s Guide. Delano Scientific; San Carlos, CA: 2004. [Google Scholar]
- 31.Sagendorf JM, Berman HM, Rohs R. DNAproDB: an interactive tool for structural analysis of DNA-protein complexes. Nucleic Acids Research. 2017;45:W89–W97. doi: 10.1093/nar/gkx272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lavery R, Sklenar H. Defining the structure of irregular nucleic acids: conventions and principles. J Biomol Struct Dyn. 1989;6(4):655–67. doi: 10.1080/07391102.1989.10507728. [DOI] [PubMed] [Google Scholar]
- 33.Honig B, Nicholls A. Classical electrostatics in biology and chemistry. Science. 1995;268(5214):1144–1149. doi: 10.1126/science.7761829. [DOI] [PubMed] [Google Scholar]
- 34.Rohs R, West SM, Sosinsky A, Liu P, Mann RS, Honig B. The role of DNA shape in protein-DNA recognition. Nature. 2009;461(7268):1248–1253. doi: 10.1038/nature08473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rohs R, Sklenar H, Shakked Z. Structural and energetic origins of sequence-specific DNA bending: Monte Carlo simulations of papillomavirus E2-DNA binding sites. Structure. 2005;13(10):1499–509. doi: 10.1016/j.str.2005.07.005. [DOI] [PubMed] [Google Scholar]
- 36.Zhang X, Dantas Machado AC, Ding Y, Chen Y, Lu Y, Duan Y, Tham KW, Chen L, Rohs R, Qin PZ. Conformations of p53 response elements in solution deduced using site-directed spin labeling and Monte Carlo sampling. Nucleic acids research. 2014;42(4):2789–2797. doi: 10.1093/nar/gkt1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.McDonald IK, Thornton JM. Satisfying Hydrogen Bonding Potential in Proteins. Journal of Molecular Biology. 1994;238(5):777–793. doi: 10.1006/jmbi.1994.1334. [DOI] [PubMed] [Google Scholar]
- 38.Rohs R, Jin X, West SM, Joshi R, Honig B, Mann RS. Origins of specificity in protein-DNA recognition. Annual review of biochemistry. 2010;79:233–69. doi: 10.1146/annurev-biochem-060408-091030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rohs R, West SM, Sosinsky A, Liu P, Mann RS, Honig B. The role of DNA shape in protein-DNA recognition. nature. 2009;461(7268):1248–53. doi: 10.1038/nature08473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Overdier DG, Porcella A, Costa RH. The DNA-binding specificity of the hepatocyte nuclear factor 3/forkhead domain is influenced by amino-acid residues adjacent to the recognition helix. Mol Cell Biol. 1994;14(4):2755–2766. doi: 10.1128/mcb.14.4.2755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Marsden I, Jin C, Liao X. Structural changes in the region directly adjacent to the DNA-binding helix highlight a possible mechanism to explain the observed changes in the sequence-specific binding of winged helix proteins. J Mol Biol. 1998;278(2):293–9. doi: 10.1006/jmbi.1998.1703. [DOI] [PubMed] [Google Scholar]
- 42.Liu PP, Chen YC, Li C, Hsieh YH, Chen SW, Chen SH, Jeng WY, Chuang WJ. Solution structure of the DNA-binding domain of interleukin enhancer binding factor 1 (FOXK1a) Proteins. 2002;49(4):543–553. doi: 10.1002/prot.10227. [DOI] [PubMed] [Google Scholar]
- 43.Jin C, Marsden I, Chen X, Liao X. Dynamic DNA contacts observed in the NMR structure of winged helix protein-DNA complex. J Mol Biol. 1999;289(4):683–690. doi: 10.1006/jmbi.1999.2819. [DOI] [PubMed] [Google Scholar]
- 44.Stroud JC, Wu Y, Bates DL, Han A, Nowick K, Paabo S, Tong H, Chen L. Structure of the forkhead domain of FOXP2 bound to DNA. Structure. 2006;14(1):159–66. doi: 10.1016/j.str.2005.10.005. [DOI] [PubMed] [Google Scholar]
- 45.Wu Y, Borde M, Heissmeyer V, Feuerer M, Lapan AD, Stroud JC, Bates DL, Guo L, Han A, Ziegler SF, Mathis D, Benoist C, Chen L, Rao A. FOXP3 controls regulatory T cell function through cooperation with NFAT. Cell. 2006;126(2):375–87. doi: 10.1016/j.cell.2006.05.042. [DOI] [PubMed] [Google Scholar]
- 46.Littler DR, Alvarez-Fernandez M, Stein A, Hibbert RG, Heidebrecht T, Aloy P, Medema RH, Perrakis A. Structure of the FoxM1 DNA-recognition domain bound to a promoter sequence. Nucleic acids research. 2010;38(13):4527–38. doi: 10.1093/nar/gkq194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chen Y, Chen C, Zhang Z, Liu CC, Johnson ME, Espinoza CA, Edsall LE, Ren B, Zhou XJ, Grant SFA, Wells AD, Chen L. DNA binding by FOXP3 domain-swapped dimer suggests mechanisms of long-range chromosomal interactions. Nucleic acids research. 2015;43(2):1268–1282. doi: 10.1093/nar/gku1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Singh P, Han EH, Endrizzi JA, O’Brien RM, Chi YI. Crystal structures reveal a new and novel FoxO1 binding site within the human glucose-6-phosphatase catalytic subunit 1 gene promoter. Journal of Structural Biology. 2017;198(1):54–64. doi: 10.1016/j.jsb.2017.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.