Abstract
In response to growing needs for quantitative biochemical and cellular assays that address whether the extracellular matrix (ECM) acts as a mechanochemical signal converter to co-regulate cellular mechanotransduction processes, a new assay is presented where plasma fibronectin fibers are manually deposited onto elastic sheets, while force-induced changes in protein conformation are monitored by fluorescence resonance energy transfer (FRET). Fully relaxed assay fibers can be stretched at least 5–6 fold, which involves Fn domain unfolding, before the fibers break. In native fibroblast ECM, this full range of stretch-regulated conformations coexists in every field of view confirming that the assay fibers are physiologically relevant model systems. Since alterations of protein function will directly correlate with their extension in response to force, the FRET vs. strain curves presented herein enable the mapping of fibronectin strain distributions in 2D and 3D cell cultures with high spatial resolution. Finally, cryptic sites for fibronectin’s N-terminal 70-kD fragment were found to be exposed at relatively low strain, demonstrating the assay’s potential to analyze stretch-regulated protein-rotein interactions.
Keywords: Fibronectin, Protein unfolding, Mechanotransduction, Mechanochemical signal conversion, Fluorescence resonance energy transfer, FRET
1. Introduction
In addition to soluble biochemical cues such as growth factors and cytokines, cells are also known to sense and respond to physical factors, including the mechanical properties of their environment (as reviewed in Discher et al., 2005; Ingber, 2006; Vogel and Sheetz, 2006) and the local topography of their substrates (Dalby et al., 2004; Dalby et al., 2005). Differences in substrate rigidity (Engler et al., 2006; Galbraith et al., 2002; Giannone and Sheetz, 2006; Kostic and Sheetz, 2006; Yeung et al., 2005) as well as micro- and nanoscale features (Dalby et al., 2005) have been correlated with profound effects on cell adhesion, morphology, proliferation, and gene regulation. The rigidity of a substrate, for example, determines whether or not mammary epithelial cells up-regulate integrin expression and differentiate into a malignant phenotype (Paszek et al., 2005), and also dictates whether mesenchymal stem cells differentiate into bone, muscle, or neuronic tissue (Engler et al., 2006). Yet, the underlying molecular mechanisms by which physical factors, including force and the mechanical properties of matrices, are converted into biochemical signals that ultimately regulate protein expression are only beginning to be explored (Kostic and Sheetz, 2006). One focus is to identify proteins whose structure/function relationship can be altered by mechanical forces, causing for example the opening of ion channels, a change in the spatial presentation of binding sites, or the exposure of otherwise cryptic binding sites (for reviews see Bustamante et al., 2004; Kung, 2005; Vogel, 2006; Vogel and Sheetz, 2006; Arcangeli and Becchetti, 2006).
On the intracellular side, mechanochemical signal conversion through protein unfolding has been demonstrated. The phosphorylation sites of p130Cas become exposed by tensile force (Sawada et al., 2006) which leads to the downstream activation of Rap1 upon stretch (Tamada et al., 2004). Also, two isoforms of the cytoskeletal protein spectrin within red blood cells showed enhanced exposure of otherwise cryptic cysteines as a function of shear stress and time, as did nonmuscle myosin IIA and vimentin within mesenchymal stem cells (Johnson et al., 2007).
On the extracellular side, fibronectin (Fn) can be unfolded by cell generated tension which causes at least a partial loss of secondary structure of its domains (Baneyx et al., 2002; Smith et al., 2007). Fn exists in a broad range of conformations, once it is harvested by cells from solution and incorporated into their own matrix as revealed by fluorescence resonance energy transfer (FRET) (Baneyx et al., 2002; Barker et al., 2005; Smith et al., 2007).
Deciphering the physiological significance of force-induced unfolding events of Fn and whether any of Fn’s versatile functions are up- or down-regulated in a mechanoresponsive manner has been hampered due to the lack of well suited assays. Fn plays a critical role in a wide variety of processes including embryogenesis and wound healing (Hynes, 1990; Midwood et al., 2006; Pankov and Yamada, 2002; Sechler and Schwarzbauer, 1998), and its fibrillogenesis into matrix fibers is tightly regulated (for reviews see Mao and Schwarzbauer, 2005; Wierzbicka-Patynowski and Schwarzbauer, 2003). This large 440 kD multimodular dimeric protein contains multiple surface-exposed molecular recognition sites and cryptic binding sites buried in the fully folded module that regulate fibrillogenesis and mediate cell adhesion, as well as interactions with other ECM proteins, and proteolytic activity (Magnusson and Mosher, 1998; Mao and Schwarzbauer, 2005; Pankov and Yamada, 2002; Romberger, 1997; Vogel, 2006). First indications also exist that downstream cell signaling is impacted by the conformation of Fn (Keselowsky et al., 2005; Lan et al., 2005). When adsorbed from solution to biomaterial surfaces, its conformation is for example impacted by the underlying surface chemistry (Altroff et al., 2003; Antia et al., 2006; Baugh and Vogel, 2004; Garcia et al., 1999; Halter et al., 2005) which can then alter integrin binding (Keselowsky et al., 2005). The conformations Fn assumes on synthetic surfaces, however, are rather distinct from those of native Fn in soluble and fibrillar forms (reviewed in Antia et al., 2006; Baugh and Vogel, 2004; Halter et al., 2005). Fn polymerization into fibers is thought to proceed as cryptic self-assembly sites are exposed by cell contractility (Baneyx and Vogel, 1999; Mao and Schwarzbauer, 2005; Ohashi et al., 1999; Zhong et al., 1998).
Single-molecule force-spectroscopy experiments provided the first indications that Fn might serve as a mechanochemical signal converter. Major insights into force-induced unfolding pathways are available (Oberhauser et al., 2002; Samori et al., 2005; Wang et al., 2001), as well as high-resolution structural models relating unfolding of FnIII modules to altered protein functions as derived from steered molecular dynamics simulations (Craig et al., 2004a; Craig et al., 2004b; Craig et al., 2001; Vogel, 2006).
Native, cell-derived Fn matrices, however, are not ideal systems for investigating force-induced potentially altered structure–function relationships. A key complication is the complex and interwoven nature of ECM fibers, where the conformation of Fn can vary from fiber to fiber and even within fibers as they are not freely suspended (Baneyx et al., 2001; Baneyx et al., 2002; Chen et al., 1997; Peters et al., 1998).
Our aim was thus to develop a mechanical strain assay where the conformation of Fn can be adjusted externally on demand and force-induced Fn extensions can be optically measured. To probe how the alterations of the structure of stretched Fn impact the displayed biochemistry and cellular behavior, such a strain assay needs to be amenable to cell culture environments. To tune the conformation of Fn, Fn fibers were drawn from a concentrated Fn solution (Ahmed and Brown, 1999; Ejim et al., 1993; Wojciak-Stothard et al., 1997) and were deposited onto a stretchable substrate which was mounted onto a custom-built, one-dimensional strain device.
2. Results and discussion
2.1. Manually deposited Fn fibers bundle into fiber cables similar to cell-derived fibers and promote cell adhesion and guidance
While it has been reported previously that manually deposited fibers pulled from concentrated solutions of soluble Fn resemble in vivo Fn fibers in diameter and composition (Wojciak-Stothard et al., 1997), the latter was difficult to confirm because the exact conformation and the nature of the lateral interactions between adjacent Fn molecules in fibers is unknown (Mao and Schwarzbauer, 2005). To briefly characterize the morphology of our deposited fibers, it should be noted that the average diameter (3.7± 1.0 µm) of fibers deposited from a 0.76 mg/mL Fn solution, as observed via fluorescence confocal microscopy, is similar to that of the thickest fibers and branching areas found in cell culture matrices (Chen et al., 1978). The average fiber diameter can be moderately adjusted via the pulling procedure. Our fibers were typically between 2 and 5 µm in diameter when the fibers were deposited out of 0.2 mg/mL or 2.6 mg/mL Fn solutions, respectively. Also the length of a fiber can be controlled by drawing it out of a solution to a desired length before bringing it in contact with the PDMS substrate. Fibers used in this study were generally 1 to 2 cm long. Fibronectin fibers in 24-h Fn matrix produced by NIH-3T3 fibroblasts in cell culture form a mesh-like network where smaller fibrils merge to form bundles and later branched again (Chen et al., 1978).
Similarly, fluorescence images of manually deposited fibers (Fig. 1) show that submicron fibers emerge from the surface of the drop and bundle together to form larger cables of fibrils (Fig. 1B,C), in agreement with previously published images of such artificial Fn fibers (Ejim et al., 1993; Wojciak-Stothard et al., 1997). Indeed, cryo-scanning electron microscopic images suggest that Fn fibers exist as ‘cables’ comprised of individual fibrous strands of ~5–15 nm in diameter and larger (Chen et al., 1978; Dzamba and Peters, 1991; Peters et al., 1998; Singer, 1979) which are proposed to be held together by hydrogen bonds, intermolecular beta-strand swapping (Briknarova et al., 2003; Litvinovich et al., 1998), disulfide bonds which are potentially formed by cryptic disulfide isomerase activity (Langenbach and Sottile, 1999), and other weak electrostatic interactions (Chen and Mosher, 1996; Morla et al., 1994). Although the exact location and properties of these bonds are unknown, it has been observed that they are strong enough to render cell-derived Fn fibers irreversibly insoluble in 1% de-oxycholate (McKeown-Longo and Mosher, 1983), which is a phenomenon we observed with manually deposited Fn fibers as well (data not shown).
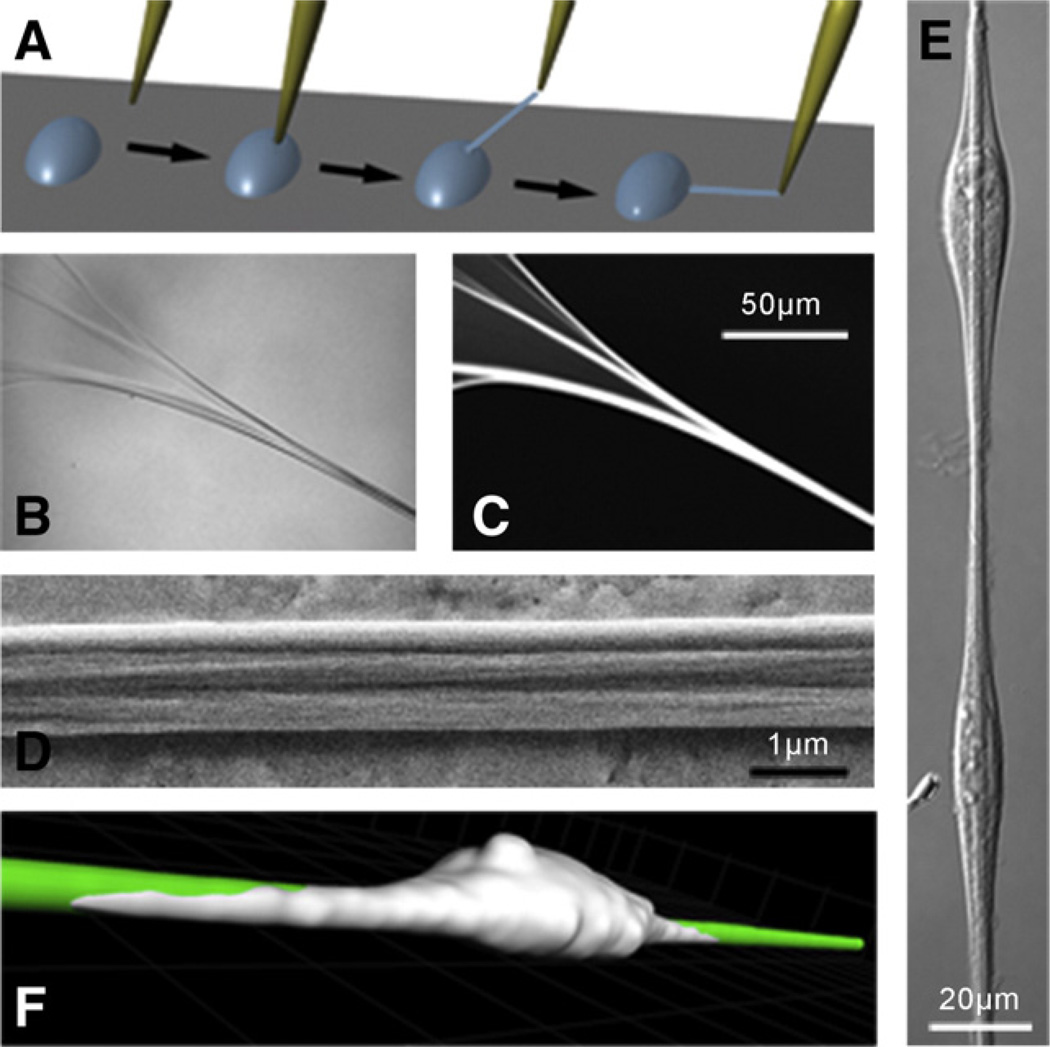
Fig. 1.
Fabrication and characterization of manually deposited fibronectin fibers. (A) A pipette tip is submersed slowly into a concentrated solution (0.76 mg/mL) of fibronectin (Fn) and removed to generate polymerized Fn fibers that were deposited onto stretchable silicone sheets. (B) Differential Interference Contrast (DIC) and (C) fluorescence images of a fiber as it extends away from the droplet (upper-left corner) reveal a bundling behavior that is confirmed via (D) a Scanning Electron Micrograph taken along the length of the fiber. (E) DIC image of Human Foreskin Fibroblasts (HFFs) adhered to and oriented along the length of a fiber. (F) 3D-confocal microscopy reconstruction of a Human Umbilical Vein Endothelial Cell (HUVEC) spread on a manually deposited fiber: the cell is fluorescently labeled with a CellTrace™ Far Red dye (Invitrogen) and cultured for 1 h after seeding onto an Alexa Flour® 488-labeled fiber. A dual-channel z-stack and Imaris™ software (Bitplane) were used to render the image shown. Grid marks represent 10 µm. The image is representative of cells on manually deposited fibers and confirms that cell adhesion occurs on the top and sides of a fiber but not onto the underlying substrate.
Furthermore, Human Foreskin Fibroblasts (HFFs) and Human Umbilical Vein Endothelial Cells (HUVECs) were observed to adhere and polarize along the axis of the manually deposited fibers (Fig. 1E,F), a phenomenon shown previously by others (Ahmed and Brown, 1999; Ahmed et al., 2000; Wojciak-Stothard et al., 1997), confirming that cell adhesion sites are exposed on the surface of the fibers.
2.2. Establishing a quantitative FRET vs. strain curve to read out Fn extensions in 2D and 3D matrices
To conduct a conformational FRET analysis of manually deposited Fn fibers, plasma Fn dimers were chemically denatured and site-specifically labeled with acceptors on the four native and free cysteines (buried within type III7 and III15 domains of the Fn dimer (Lai et al., 1984; Mosher and Johnson, 1983; Smith et al., 1982)) and randomly labeled with donors on free lysines (Fn-d/a, see Materials and methods). Circular dichroism spectroscopy of FRET-labeled Fn confirmed that refolding occurs in physiological buffer (Smith et al., 2007).
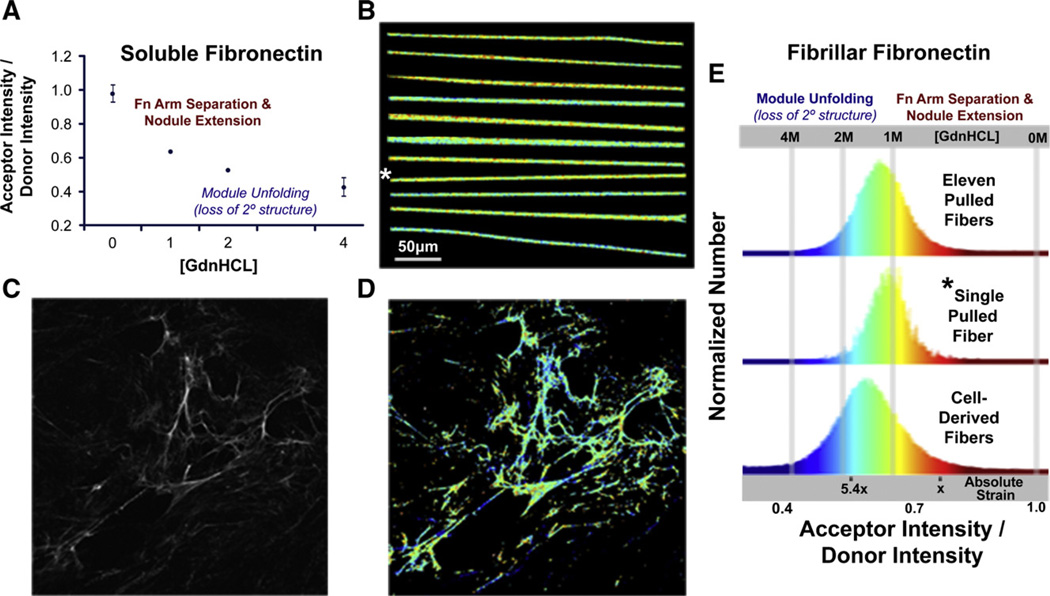
Ratiometric FRET images of manually deposited fibers show little color change along a single fiber (visualized as false colors in Fig. 2B), whereas fibers from a 24-h cell-derived matrix (Fig. 2C) show more locally heterogeneous conformations (Fig. 2D). The frequency at which a given ratio between the intensity of the acceptor channel and the donor channel (IA/ID ratio) is found within a field of view containing a total number of pixels (640 pixel×640 pixel) is displayed in the FRET histograms (Fig. 2E). The histogram for the manually deposited fibers peaks at an IA/ID of 0.62, which corresponds to the mean value recorded from Fn in solution at ~1 M GdnHCl. Histograms for each of 11 different pulled fibers are provided in Supplementary Fig. S1B. Variations of the pulling rate by which the fibers were drawn out of solution, from 1.0 mm/s to 0.2 mm/s, had no major impact on the conformational distribution (data not shown). The width of the histogram originates from the pixel-to-pixel variations of the local averaged Fn conformation and from instrument noise (as further discussed in Smith et al., 2007). The widths of the histograms taken from Fn-d/a in solution (1 M GdnHCl; data not shown) and in manually deposited fibers (Fig. 2E) were similar.
Fig. 2.
Spatially resolved conformational mapping by Fluorescence Resonance Energy Transfer (FRET) and comparison of FRET histograms of manually deposited fibronectin and extracellular matrix fibers of living fibroblasts. (A) Plasma fibronectin, dual-labeled with donors and acceptors (Fn-d/a—see Materials and methods), is subjected to increasing concentrations of guanidine hydrochloride (GdnHCL) to calibrate a ratio of the acceptor intensity divided by the donor intensity (IA/ID) to known conformational states of fibronectin in solution. In concentrations greater than 1.5 M GdnHCL, circular dicroism indicates that fibronectin undergoes a rapid and significant loss of secondary structure causing the Fn modules to unfold (Alexander et al., 1979; Baugh and Vogel, 2004). (B) Reproducibility of conformational distribution: 11 manually deposited Fn fibers containing 5% Fn-d/a were laid onto a transparent sheet of silicone and the pixel-resolved IA/ID values were mapped to a color gradient representing module-unfolded (blue) or dimer-arm separated (red) Fn conformations. Images of individually deposited fibers here were mounted next to each other for presentation purposes (though it is possible manually, see Fig. 5). (C) A fluorescence image of a 24-h cell-derived Fn matrix derived from NIH-3T3 fibroblasts (Baneyx et al., 2001) is shown beside the (D) corresponding false color FRET map. (E) The top histogram represents IA/ID data collected from every pixel analyzed from the eleven fibers shown in (B). The middle histogram shows the IA/ID data collected from the single fiber in (B) as denoted by the *. The bottom histogram shows the IA/ID data collected from the cell-derived Fn matrix image in (D). The absolute strain scale represents the average IA/ID values of Fn fibers, from fully relaxed (nodules of quatenary structure are present), to extended, to unfolded, to the point where they begin to break (5.4×), as described in the text and in Fig. 3. Scale bar in (B) is the same scale as the images in (C) and (D).
To stretch Fn fibers, a one-dimensional strain device was constructed to hold a thin rectangular silicone sheet and stretch it over four times its length (Fig. S2A–E, see Materials and methods). Fibers are first deposited onto the silicone sheet (Fig. S2A), rinsed and incubated with a PBS or BSA solution, and then stretched to a desired length (Fig. S2B). A holding chamber is then used to sandwich and capture the strained substrate (Fig. S2D) before it is removed from the strain device. Finally, the bottom half of the chamber is removed after the ends of the substrate are fastened to the side (Fig. S2E) in order to allow a microscope objective to reach the substrate for imaging. Fibers can also be first deposited on pre-strained substrates such as in Fig. S2B and relaxed to a position such as in Fig. S2A to remove any residual tension that may exist as they are originally deposited from solution. Shown in Fig. 3A is the resulting IA/ID histograms from relaxed, strained, and control fibers as they compare to cell-derived 24-h Fn matrices, and Fig. 3B shows the corresponding color and relative length-changes that are possible to achieve with the strain device.
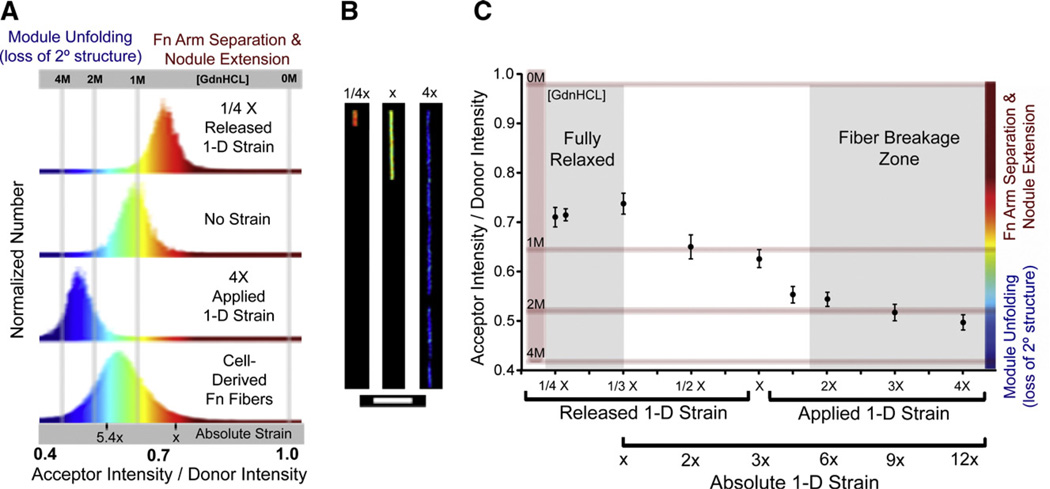
Fig. 3.
FRET vs. strain dependency of manually deposited fibronectin fibers probed by externally stretching a supporting silicone sheet. (A) Histograms of pixel-resolved acceptor channel / donor channel ratios (IA/ID) from representative fibers shown in (B) and cell-derived fibers from Fig. 2D,E containing 5% dual-labeled fibronectin. The absolute strain scale represents the average IA/ID values of Fn fibers that are fully relaxed or extended to the point where they begin to break (5.4×), as described in the text and shown in (C). The fibers are subjected to 1/4× release of strain (orange), no strain (green), or 4× applied strain (blue) and show that distinct and uniform conformational changes can be induced via mechanical strain and each mean IA/ID can be found within the histogram of cell-derived fibers. For presentation purposes, the fibers in (B) have been cropped to scale in order to demonstrate the length reductions and extensions achievable via the strain device. (C) A plot of the average IA/ID±standard deviation versus applied to or released strain from manually deposited Fn fibers (see Materials and methods). Average values were obtained from at least 10 fibers to compute a single data point. Released strain is acquired by pre-straining the silicone substrate before pulling fibers and then relaxing the strain device. IA/ID values are calibrated to chemical denaturing data (Fig. 2A). The absolute scale of strain is shown as well, assuming that 1/3× is the point of full relaxation. Scale bar in (B)=50 µm.
Thus, in order to begin establishing an absolute IA/ID vs. strain curve, we first deposited Fn fibers on silicone sheets that were pre-strained to 4×. Subsequent relaxation of the fibers led to a uniform increase of their IA/ID ratios which indicated progressively more refolding of fibronectin until the sheet has been relaxed to about 1/3 of its initial length (Fig. 3C). With further sheet relaxation, the fibers began to bulge which likely explains the slightly lower IA/ID values observed at 1/4 the substrate length (Fig. 3C).
The 1/3× relaxation point shown in Fig. 3C represented the amount of strain release leading to maximal FRET recovery.
After having determined that the drawing process by which the fibers are pulled out of the droplet causes considerable pre-straining of fibronectin, for which we have to compensate by relaxing the substrate, we can now establish an absolute extension versus FRET curve for fibronectin fibrils (Fig. 3E). This is significant because it allows us to read out the strain and thus the conformation of fibronectin by optical means, whether the cells are cultured in two or three dimensions.
2.3. Fully relaxed fibers do not contain the globular Fn conformation found in solution
When the Fn fibers are fully relaxed, the average IA/ID ratio is 0.74±0.02, which lies between the 0 M (0.98) and 1 M (0.64) GdnHCL calibration points, indicating that fully relaxed fibrillar Fn does not return to its globular solution conformation where its two dimer arms are folded upon themselves. These results confirm previous data obtained with fully relaxed cell-derived fibronectin matrices which also do not contain the tightly folded quaternary structure otherwise seen in solution (Smith et al., 2007). These findings thus invalidate a recent model postulating that relaxed fibronectin fibers are composed of arrays of compactly folded fibronectins in which the opposing dimeric arms cross each other (Abu-Lail et al., 2005; Erickson, 2002).
2.4. Fn fibers show signs of breakage only when stretched over 5–6 times their resting length
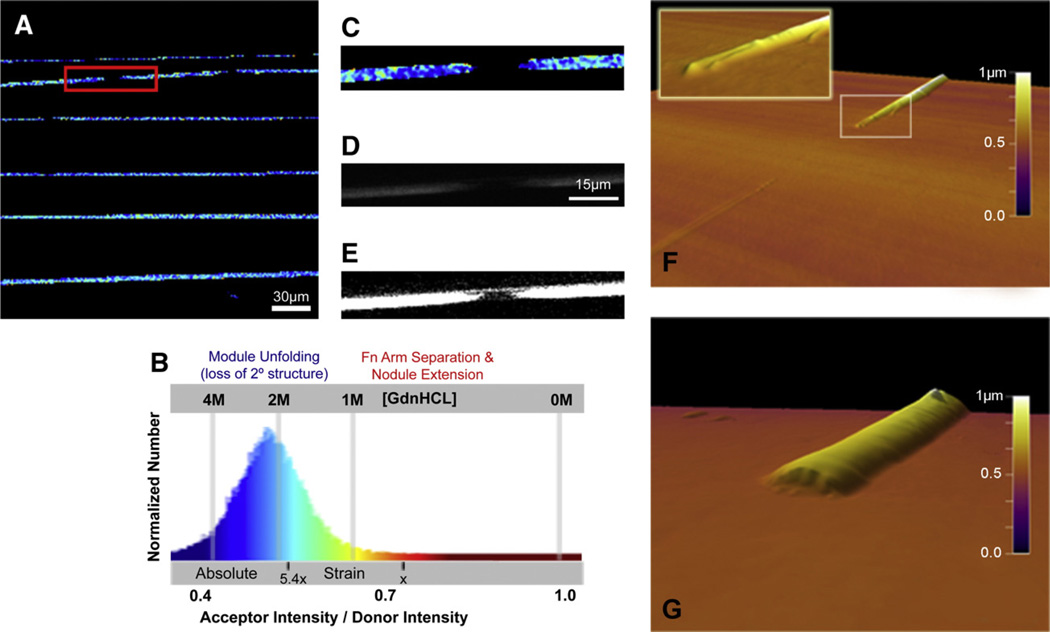
To establish the extension and the associated optical signature at which the fibers begin to rupture, Fn fibers were deposited onto unstrained silicone sheets (Fig. S2A). After applying uniaxial strain, the observed IA/ID ratios decreased as expected (Fig. 3C), and beginning at a 1.8× length increase, the fibers began to fracture at a few sites into randomly distributed large segments, each hundreds of micrometers long. In the 2× strain field of the view shown in Fig. 4A (as calibrated using data shown in Fig. 3C and Fig. 4B), a magnified image of one of the breaks (Fig. 4C,D) and a contrast-enhanced fluorescence image (Fig. 4E), shows that a small amount of Fn is left attached to the substrate. Since the IA/ID values continued to decrease along the length of the fiber segments when further strained until the limit of the strain device was reached at 4× (Fig. 3C), this may indicate that the adhesion strength of the fiber bundle to the silicone substrate is considerable and does not allow for major slippage between the strained fiber and the silicone sheet once the first breakages have occurred. Atomic force microscopy (AFM) images of fiber breakage areas reveal two kinds of breaks; one where a small layer of fibronectin remains attached to the underlying silicone sheet and the broken fiber ends appear tapered (Fig. 4F), and another where the break results in relatively blunt fibers and the fibronectin layer left behind exists closer to the fractured ends (Fig. 4G). Finally, the IA/ID values continued to decrease homogenously along the length of the fibrous segments upon further strain until the limit of the strain device was reached at 4× (Fig. 3C).
Fig. 4.
Fn fiber breakage for fibers that are adherent to an elastic silicone substrate. (A) A spatially resolved FRET map of manually deposited Fn fibers subjected to a 2× increase in substrate length shows areas of fiber breakage and large areas of mechanically induced unfolded Fn Type III modules, as confirmed via (B) a histogram of every pixel’s acceptor intensity / donor intensity (IA/ID, see Materials and methods) and calibrated to a guanidinium hydrochloride (GdnHCL) denaturing curve. The absolute strain scale represents the average IA/ID values of Fn fibers that are fully relaxed or extended to the point where they begin to break (5–6×), as described in the text and in Fig. 3. (C) Zooming in on an example breakage location in a fiber as noted by the red box in (A). (D) A fluorescence image of the break, especially a (E) contrast-enhanced imaged, reveals that a small number of individual strands are being pulled apart and left behind in the breakage gap that are not detected in our FRET measurements due to the necessary analysis thresholding (see Materials and methods). (F) AFM image confirming the presence of a small amount of fibrillar Fn left on the substrate and a tapered end to the broken fiber. Inset zoomed-in image of a fractured fiber end shows the receding strands. (G) Some breaks also show only a small amount of Fn left behind on the substrate and show more blunted fibers ends. Scale bar in (A)=30 µm, Scale bar in (D)=15 µm and is the same scale as the images in (C) and (E).
Bringing the fiber strain and relaxation data together, mechanically stretched Fn-fibers began to show signs of breakage at a relative length increase of 1.8× compared to fibers deposited onto unstrained silicone sheets, and can thus be strained at least 5–6 times with respect to their resting state (5.4=1.8 / 0.333). It is currently believed that Fn fibers can be stretched three- to four-fold beyond their resting length (Ohashi et al., 1999). This conclusion was derived from various cell culture measurements of the lengths of chimeric GFP or YFP-Fn fibers before and after breakage or detachment of fibers under cell-dependent strain, from fiber rupture from laser damage, or after inhibition of cell contractility with cyto-chalasin B. Since physisorption to a substrate might have an impact on strain at which the fibers break, future work is needed to determine the extension at which freely suspended fibers rupture. The FRET ratios seen at the onset of fiber fracture, however, are also seen in native cell-made matrix (see Fig. 2D).
2.5. High extensions of Fn fibers involve module unfolding
These data on the total range of extensibility of manually deposited Fn fibers further support recent conclusions that native, cell-derived Fn matrices are unfolded by cell traction forces (Baneyx et al., 2001; Baneyx et al., 2002; Smith et al., 2007). FRET measurements of dual-labeled Fn matrices supported a model whereby Fn fiber elasticity involves unfolding of typeIII modules (Baneyx et al., 2001; Baneyx et al., 2002). The possibility of force-induced unfolding of Fn modules was initially suggested based on purely theoretical calculations (Erickson, 1994). The model assumed that Fn exists with a compactly folded quaternary structure in resting fibers, and that fibril elasticity originates from a separation of the overlapping dimer arms (quaternary structure) followed by an extension into a straight string of modules (loss of quaternary but not of secondary structure) (Abu-Lail et al., 2005). If one assumes that module unfolding does not occur prior to fiber breakage, this quaternary structural model only allows for a maximum 4-fold extension from compactly folded (~40 nm) to extended (~130 nm; Erickson, 2002).We show here, however, that manually deposited Fn fibers can be stretched 5- to 6-fold prior to the onset of fiber breakage. Furthermore, fully relaxed manually deposited Fn fibers do not contain a measurable population of Fn in the fully compact conformation (Fig. 3C and Smith et al., 2007). Therefore, Fn module unfolding must contribute to the extraordinary extensibility of Fn fibers. While our measurements cannot directly determine the IA/ID ratio below which mechanically stretched fibers unfold, CD measurements from Fn in solution indicate that Fn unfolding occurs at concentrations of GdnHCl well below 2 M. Khan et al. 1990 reported a 26% loss in secondary structure for the central cell binding fragment of Fn (modules FnIII1–11) from a baseline of 5 (0 to 1.2 M GdnHCl) to ~3.7×102 deg×cm2×dmol−1 (2 M GdnHCl). A 26% loss of secondary structure within the FnIII1–11 fragment would roughly correspond to complete unfolding of three FnIII modules. If a FnIII module is unfolded mechanically (not chemically), it lengthens from ~3.2 nm to a contour length of ~28.5 nm (Oberhauser et al., 2002). If three modules are unfolded mechanically, an extended Fn molecule which is estimated to be 130 nm in length prior unfolding (Engel et al., 1981), would thus increase in length to 206 nm (130+3 * (28.5–3.2)), or 1.6-fold. Interestingly, Fig. 3 shows that the average end-to-end distance of Fn increases roughly by a factor of 2 due to mechanical stretching at IA/ID ratios that correspond to those measured at 1.5 (0.58) and 2 M GdnHCl in solution (0.52), and are thus in agreement with our results derived from this strain. Since publishing 360 our first paper on using FRET to probe conformational alterations of Fn (Baneyx et al., 2001), the FRET signature of each batch of progressively denatured Fn in solution was determined. This allows for a comparison between batch to batch variations of labelled Fn, and for other groups to see whether their labelling protocols lead to comparable results. Certainly, Fn has different conformations in solution and in the fibrillar states. However, if the two dimeric Fn arms are separated by breaking stabilizing electrostatic interactions, the labelled domains do not have to move far to completely eliminate inter-arm FRET. Similarly, if two FRET probes are on adjacent domains, domain unfolding whether induced by force or chemically, rapidly leads to a distance increase that exceeds the Förster radius. Experiments are underway to identify the exact point at which fibrillar Fn starts to lose secondary structure by force-induced unfolding.
Data from these assay fibers also indicate that the fully relaxed state of fibrils is not composed of Fn dimers whose arms are already extended into a straight string of folded modules. Instead, some arms partially fold back upon themselves (intramonomer) as indicated by the finding that the fully relaxed fibers have IA/ID ratios which lie roughly half-way between the average IA/ID value of compact Fn found in solution at 0 M GdnHCl (0.98) and the IA/ID value of extended Fn found at 1.5 M GdnHCl (0.58) (Figs. 2A, 3C). This conclusion agrees with other spectroscopic observations (Smith et al., 2007) and high-resolution Cryo-scanning electron microscopy images showing that nodules decorate the otherwise smooth surface of the fibers (Chen et al., 1997; Peters et al., 1998). From the observed nodule sizes, itwas suggested that each nodule can contain ~4 FnIII modules. These nodules are shown in the Cryo-SEM studies to essentially disappear within fibers extended between cells, presumably because such fibers are under mechanical tension (Chen et al., 1997; Peters et al., 1998). Therefore, Fn fibrils upon stretch transition from a nodular to a smooth fibril structure where the nodules gradually disappear and their content integrates into the smooth fibers as the end-to-end distance of the fiber is increased. Future research must address whether the nodules completely disappear before the first Fn modules unfold as the fibers are strained, or if nodules and module unfolded can coexist at intermediate strains. If straightening of the nodules takes less energy than breaking secondary structure, fiber extensibility might have two regimes: one associated with the loss of quaternary structure resulting from the backfolding of a few Fn modules into nodules and a second from loss of secondary structure at higher tensions.
2.6. The range of conformations observed from relaxed to highly strained manually deposited Fn fibers simultaneously exists within one field of view within ECM of living fibroblasts
In addition to the microscopic structural similarities and similar average IA/ID ratios of cell-derived and manually deposited Fn fibers (0.59±0.09 vs. 0.62±0.06, respectively), further evidence that the assay fibers can serve as physiologically relevant and useful model systems lies in the observation that they can be strained or relaxed to yield IA/ ID ratios that span the range of IA/ID ratios found in single fields of view of ECM (Fig. 2E). When strained or relaxed, manually deposited fibers maintain a relatively narrow conformational distribution (Fig. 3C); the data points in the resulting FRET vs. strain curve for both relaxed and strained fibers had an average IA/ID standard deviation of only 0.018 (Fig. 3C), compared to 0.089 found within cell-derived fibers (Fig. 2E). On absolute scales, cellular fibers had IA/ID ratios between 0.42 and 0.81 (from 4 M to 0.5 M GdnHCL equivalents) and a few particularly long and thin fibrils (Fig. 2D) showed IA/ID ratios between 0.42 and 0.58 (similar to Fn in 1.5–4 M GdnHCL).
2.7. Fn fibers can be manually deposited in various orientations and curves to generate a variety of molecular conformations in one field of view after simultaneous one-dimensional strain and relaxation
An advantageous property of a 1D stretch assay is that when a rectangular sheet is stretched 3× their length in one dimension the width compresses ~0.59× in the orthogonal direction. Therefore, depositing “grids” of fibers onto 3× pre-strained sheet followed by a full relaxation of the sheet results in horizontal fibers relaxed 1/3× and vertical fibers stretched 1.7× their end-to-end length, corresponding to the fully relaxed conformation and near-breakage point on the IA/ID vs. strain calibration curve, respectively (Fig. 3C). Therefore, this method of pre-straining the sheet before depositing fibers in various angles and curves and subsequently relaxing it renders the highest possible range in homogeneous fibrillar Fn conformations existing in a single field of view without fiber breakage.
2.8. Mechanically strained Fn fibers expose cryptic binding sites for the N-terminal 70 kD Fn fragment at early extensions
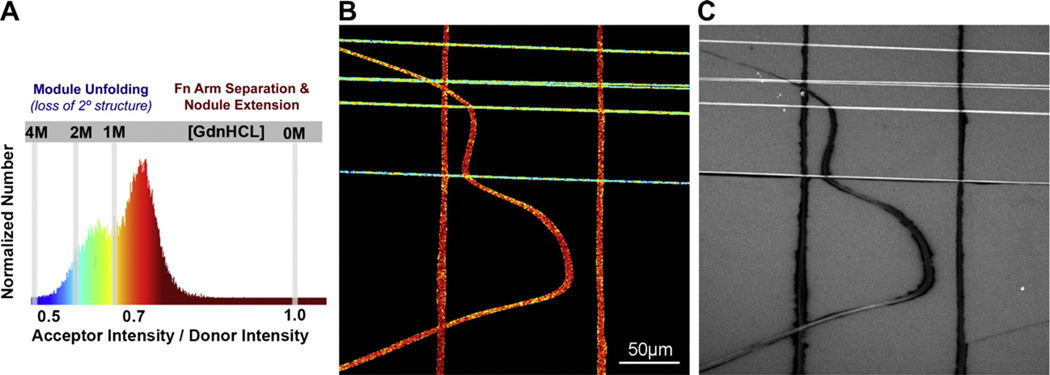
A number of studies have demonstrated previously that either proteolytic cleavage or heat-, chemical-, or substrate-induced unfolding of soluble Fn reveals a variety of cryptic protein binding sites (for reviews see Pankov and Yamada, 2002; Vogel, 2006). However, to our knowledge only one study has investigated how protein binding relates to mechanically induced changes in fibrillar Fn conformation (Zhong et al., 1998) showing that the binding of Fn, the N-terminal 70 kD Fn fragment, and the L8 monoclonal antibody to cell-made Fn fibers was decreased when cells were treated with Rho inhibitors. Since Rho stimulates cell contractility, the authors concluded that cell-generated tension is needed to expose Fn’s binding sites for these proteins. Fig. 5C now shows that the binding of the 70 kD fragment to Fn is indeed highly dependent on the Fn conformation. While the Rho inhibition studies only relaxed a native cell matrix which is, as we now know, composed of a broad range of conformations, we therefore used the assay presented above to generate fibers of defined conformations to examine how the binding of the 70-kd fragment relates to the conformation of Fn fibers as detected via FRET. Fig. 5C shows a representative field of view of the far-red channel (645 nm–745 nm) after Fn-d/a labeled-fibers were coated with Alexa Flour® 633-labeled 70-kd fragment (See Materials and methods). In agreement with the results by Zhong et al. (1998), horizontal fibers strained 1.7× clearly bound more of the 70-kd fragment than the 1/ 3×-relaxed vertical fibers. Interestingly, the curved fiber in the image contains both vertical and more horizontal areas along its length as well as the transitional conformations between them. The 70 kDa Fn fragment appeared to begin binding to the curved fiber as the conformation of the Fn molecules within transitioned early in their extensibility range from a red to a yellow color, corresponding to IA/ID values calibrated to the ~1 M GdnHCL point, which as discussed above likely represents the transition between the resting to the extended conformation prior to the loss of major secondary structure. In the broader context, the 70 kD binding studies show that the assay is well suited for the study of strain-dependent protein–protein interactions.
Fig. 5.
Stretch-dependent adsorption of the 70-kd N-terminal Fn fragment to a wide range of fibrillar Fn conformations generated in a single field of view. (A) Fn fibers were manually deposited onto a 3× pre-strained silicone sheet in curves and anti-parallel orientations and then fully relaxed to simultaneously strain horizontally oriented fibers in (B) 1.7× and relax vertically oriented fibers in (B) 1/3×. A histogram of the resulting IA/ID for each pixel analyzed (see Materials and methods) is shown and color-coded so that the corresponding image map (B) can be used to easily identify highly strained fibers (blue) and fully relaxed fibers (red) and conformations in between. (C) A solution of Alexa Flour® 633-labeled 70-kd Fn fragment was adsorbed to the substrate for 10 min, rinsed 4× with PBS, then imaged in a far-red channel (645 nm–745 nm). Stretched fibers (horizontal orientation) clearly bind more of the protein fragment than relaxed fibers (vertical orientation), and the curved fiber shows that the transition between relatively low and high binding occurs where the region of the fiber transitions between a vertical and horizontal alignment.
3. Conclusions
A new strain assay is presented which is well suited for biochemical and cellular assays aimed at investigating if Fn within the extracellular matrix can act as a mechanotransducer. Essential features of the assay are the ability of tuning the conformational distribution of Fn by the application of external strain, and of probing the strain-induced protein conformational changes within a large number of parallel fibers or fiber grids. This assay has the capability to detect strain-dependent protein–protein interactions.
Future studies need to examine whether integrins or other proteins involved in outside-in cell signaling might bind to ECM fibers in a stretch-dependent manner which could then result in a stretch-dependent regulation of signaling pathways. Since it has been shown so far only that most cell types are capable of probing the rigidity of their environment (Discher et al., 2005; Engler et al., 2006; Galbraith et al., 2002; Giannone and Sheetz, 2006; Ingber, 2006; Kostic and Sheetz, 2006; Vogel and Sheetz, 2006; Yeung et al., 2005), the assay introduced here could play a key role in the design of experiments that might distinguish between the force-induced functional changes of ECM molecules and of fiber rigidity on cell function.
4. Materials and methods
4.1. Isolation of Fn
Human plasma Fn was isolated from human plasma (Swiss Red Cross) via a previously described procedure involving the use of gelatin-sepharose chromatography (Engvall and Ruoslahti, 1977). Briefly, the serum was first centrifuged at 2000 RPM for 10 min to ensure the removal of all blood cells. The supernatant was then supplied with 10 mM ethylenediami-netetraacetic acid (EDTA) and 2 mM phenylmethylsulfonyl fluoride (PMSF) and centrifuged at 10,000 RPM for 15 min. The resulting supernatant was then passed through a sepharose 4B column (Sigma) and then through a gelatin-sepharose 4B column until the eluant contained no detectable proteins (absorbance at 280 nm)—washing buffer was PBS with 10 mM EDTA and 2 mM PMSF. The column was then washed with 1 M NaCl followed by 1 M urea. Finally, the Fn was eluted using 6 M urea and its purity was confirmed using western blotting and silver staining (not shown).
4.2. Cell culture and fluorescent cell labeling
NIH-3T3 mouse fibroblasts were obtained from ATCC and cultured in Dulbecco’s Modified Eagle’s Medium (DMEM) with 10% Fetal Calf Serum (FCS) (Gibco). Human Umbilical Vein Endothelial Cells (HUVECs) were obtained from PromoCell and cultured in endothelial cell growth medium plus supplement (PromoCell). Both cell types were incubated in a humidified 37 °C chamber with 5% CO2. Only cells below passage 15 were used for the experiments described in this report. For Fig. 1E, HUVECs were fluorescently labeled with CellTrace™ Far Red DDAO-SE (Invitrogen) via the manufacturer’s protocol.
4.3. Fluorophore conjugation of Fn
Fn was dual-labeled with Alexa Fluor® 488 succinimidyl ester and Alexa Fluor® 546 maleimide for FRET experiments via a modified procedure previously described (Baneyx et al., 2001). Plasma Fn contains two free sulfhydryls per subunit (Smith et al., 1982) which are buried within the tertiary structure of the Fn III7 and III15 modules (Mosher and Johnson, 1983), which requires that they be exposed by denaturant for fluorophore conjugation. Briefly, Fn in 6 M urea taken from the final step of the isolation procedure was further denatured by the addition of guanidine hydrochloride (GdnHCL, 4 M final concentration) and incubated at room temperature for 5 min. A 25-fold molar excess of Alexa Fluor® 546 maleimide was then added to the denatured Fn solution, mixed by gentle pipetting, and incubated at room temperature for 2 h with gentle rocking. Free dye from the solution was removed via size-exclusion chromatography with phosphate buffered saline (PBS, pH 7.4) through a PD10 column and the concentration of remaining Fn was determined via a spectrophotometer (MBA2000, PerkinElmer Instruments). At this point the free cysteine residues located within modules FnIII7 and FnIII15 (4 total per Fn dimer) are specifically labeled with dye.
Next, sodium bicarbonate was added to the solution to a final concentration of 0.1 M at pH 8.7 for amine labeling. A 110-fold molar excess of Alexa Fluor® 488 succinimidyl ester was then added to the solution and allowed to incubate for 1 h at room temperature with gentle rocking to randomly label amine residues. Free dye was again removed from the solution via size-exclusion chromatography and the eluant solution was determined via spectrophotography to have Fn labeled on average with 6.7 donors (Alexa Fluor® 488) and 3.7 acceptors (Alexa Fluor® 546). The point at which fragments contaminated the eluant was determined via Western blotting (not shown) and only the volume before this point was collected for experiments. The Fn solution was stored in frozen 10 µl aliquots and only used when thawed for less than 5 days. This batch of dual-labeled Fn (Fn-d/a) was used to collect all the FRET data in Figs. 2–4 presented in this report. The batch used for Fig. 5 was prepared by the same methods and had on average 6.0 donors and 3.6 acceptors.
4.4. Chemical denaturing curve, microscopy, and FRET analysis
To calibrate how FRET relates to the loss of quaternary (separation of the dimer arms) and tertiary structure, the IA/ID ratios were determined for Fn in denaturing solutions of 0 M, 1 M, 2 M, and 4 M GdnHCL. Bovine Serum Albumin (BSA)-coated glass coverslips and glass microscope slides were used to create 2 mm wide chambers to house the GdnHCL-Fn-d/a solutions and to prevent adsorption to the glass surfaces. 200 µm-thick silicone sheets (SMI, Saginaw, MI) were used for the chamber walls and spacer for the coverslip and slide. The Fn-d/a solution (~2 µL) was drawn into the chamber via capillary forces and each solution was imaged via scanning laser confocal microscopy (Olympus FV-1000). The samples were excited at 488 nm and the two emission detection windows were set at 514–526 nm (donor channel) and 566–578 nm (acceptor channel) to capture their peak emissions.
Spatially resolved FRET analysis (by pixel) was then performed using a custom script written in PHP (Php Hypertext Preprocessor, www.php.net) or Ruby (www.ruby-lang.org) together with image analysis software (GD graphics library, www.boutell.com/gd—ImageMagick Software Suite, www.imagemagick.org). The script first subtracted a dark current background from each channel (acquired from each experiment), then applied a 1-pixel radius Gaussian blur (with a 1-pixel standard deviation) to each channel to smooth errant intensity values. The intensity of the 12-bit acceptor channel (values from 0–4095) for each pixel was then divided by the intensity of the corresponding 12-bit donor channel (as long as each channel’s intensity value was above a threshold of at least 100 and not saturated at 4095) to yield IA/ID ratios. Decreasing IA/ID ratios indicate increased nodule extension and domain unfolding. To generate spatially resolved FRET maps, the IA/ID ratio for each pixel (0.33 µm×0.33 µm) was mapped to a false color, with red and blue representing compact and unfolded conformations, respectively.
4.5. Cell-derived Fn fibers
Fn-d/ -labeled cell-derived matrix was obtained by a modified procedure previously described (Baneyx et al., 2001). Briefly, a 10 µg/mL solution of unlabeled Fn (u-Fn) was first adsorbed onto a glass coverslip for 30 min at room temperature and before a near-confluent layer of NIH-3T3 cells was cultured. Thirty minutes later the cells were rinsed with media and then incubated in media containing 45 µg/mL of u-Fn and 5 µg/mL of Fn-d/a in order to avoid intermolecular FRET (Baneyx et al., 2001). After 24 h, the sample was rinsed with PBS at 37 °C and then filled with a 2 mM 6-Hydroxy-2,5,7,8-tetramethylchroman-2-carboxylic acid (Trolox) solution in PBS for live imaging of the matrix.
4.6. Manually deposited Fn fibers
Manually deposited Fn fibers were generated by a modified procedure previously described (Ejim et al., 1993). In all fibrillar experiments, soluble Fn-d/a was diluted into 95% unlabeled Fn to prevent intermolecular FRET. At this dilution factor, the acceptor intensity divided by donor intensity (IA/ID) ratio was independent of the Fn concentration, ensuring that the signals reported here originate from intramolecular FRET (Fig. S3). Briefly, a plastic pipette tip was cut into a sharp point and dipped into a 0.76 mg/mL 5% Fn-d/a / 95% u-Fn droplet on a 200 µm-thick silicone sheet (SMI, Saginaw, MI) that was pre-cleaned by immersion in ethanol for 10 min and gently drawn upward to induce Fn polymerization. The resulting fiber was then deposited onto the surface ~1–2 cm away from the droplet slightly pressing the tip into the elastic silicone sheet. Once the desired number and pattern of fibers were deposited, the droplet and sample would be allowed to dry for 5 min before rinsing four times with a 2% (w/v) bovine serum albumin (BSA) solution in PBS. Samples were left in this BSA solution at room temperature no longer than 2 h before use.
Once in contact with glass or silicone substrates, the fibers attach tightly along their full length, which was confirmed by using a microneedle to disrupt the fiber at various locations in both dry and aqueous environments. Instead of detaching and/or recoiling non-uniformly away from a break, the fiber stayed in place and only the immediate area around the microneedle was cut, independent of where the break occurred (not shown). This observation was also confirmed by the fact that the fibers did not detach from the silicone substrate when stretched and the spatially resolved IA/ID ratios were homogeneous when strain was applied or released (Fig. 3).
4.7. Fluorescent labeling and adsorption of the 70-kd Fn fragment
For adsorption studies, Alexa Fluor® 633 was used to fluorescently label the 70-kd N-terminal Fn fragment (Sigma) per the manufacturer’s protocol. After depositing Fn fibers on 3× strained silicone substrates as described above in various angles and curves, the substrate was fully relaxed resulting in a 1/3× relaxation of fibers in the direction of the one-dimensional strain and a 1.7× extension of perpendicular fibers. A ~0.1 g/L solution of the labeled 70-kd fragment in PBS was introduced to the fibers for 10 min at room temperature then rinsed 4× with PBS before imaging.
4.8. AFM of manually deposited fibronectin fibers
Surface scans of Fn fibers (Fig. 4I, J) were obtained by first depositing the fibers onto 4× pre-strained silicone substrates in an orientation perpendicular to the direction of 1D strain and rinsing 4× with PBS. After full relaxation of the substrate in PBS the fibers resulted in being strained 2× to induce fracturing. AFM of dried samples was used in AC-mode (Asylum Research, MFP-3D). The cantilever was an Olympus silicon cantilever AC160TS, operating slightly below the resonance frequency of 292 kHz at a scan rate of 0.3 Hz.
Supplementary Material
Acknowledgements
The authors gratefully acknowledge Jörg Albuschies for taking the AFM images shown in Fig. 4, Jean Schwarzbauer for providing us with the Fn isolation, Sheila Luna for the Fn purifications, Anne Wandrey for aid with SEM imaging, Kristopher Kubow, Delphine Gourdon, John Saeger, Gretchen Baneyx, and Meher Antia for their helpful discussions, and members of the ETH Materials department machine shop for constructing the custom strain device. This work was financially supported by the ETH Zurich (VV), the Human Frontier Science Program Organization (MLS), and from the Nanomedicine Development Center (NDC) “Nanotechnology Center for Mechanics in Regenerative Medicine” (NIH grant PN2 EY016586), that participates in the NIH Nanomedicine Development Center Network (NNDCN).
Abbreviations
- ECM
extracellular matrix
- Fn
Fibronectin
- FRET
fluorescence resonance energy transfer
- Fn-d/a
Fn labeled with donor and acceptor fluorophores
- GdnHCL
guanidine hydrochloride
- IA/ID
ratio of the acceptor intensity divided by the donor intensity
- u-Fn
unlabeled Fn
Footnotes
Appendix A. Supplementary data
Supplementary data associated with this article can be found, in the online version, at doi:10.1016/j.matbio.2008.02.003.
References
- Abu-Lail NI, Ohashi T, Clark RL, Erickson HP, Zauscher S. Understanding the elasticity of fibronectin fibrils: unfolding strengths of FN-III and GFP domains measured by single molecule force spectroscopy. Matrix Biol. 2005;25:175–184. doi: 10.1016/j.matbio.2005.10.007. [DOI] [PubMed] [Google Scholar]
- Ahmed Z, Brown RA. Adhesion, alignment, and migration of cultured Schwann cells on ultrathin fibronectin fibres. Cell Motil. Cytoskelet. 1999;42:331–343. doi: 10.1002/(SICI)1097-0169(1999)42:4<331::AID-CM6>3.0.CO;2-7. [DOI] [PubMed] [Google Scholar]
- Ahmed Z, Underwood S, Brown RA. Low concentrations of fibrinogen increase cell migration speed on fibronectin/fibrinogen composite cables. Cell Motil. Cytoskelet. 2000;46:6–16. doi: 10.1002/(SICI)1097-0169(200005)46:1<6::AID-CM2>3.0.CO;2-Z. [DOI] [PubMed] [Google Scholar]
- Alexander SS, Colonna G, Edelhoch H. The structure and stability of human plasma cold-insoluble globulin. J. Biol. Chem. 1979;254:1501–1505. [PubMed] [Google Scholar]
- Altroff H, Choulier L, Mardon HJ. Synergistic activity of the ninth and tenth FIII domains of human fibronectin depends upon structural stability. J. Biol. Chem. 2003;278:491–497. doi: 10.1074/jbc.M209992200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antia M, Islas LD, Boness DA, Baneyx G, Vogel V. Single molecule fluorescence studies of surface-adsorbed fibronectin. Biomaterials. 2006;27:679–690. doi: 10.1016/j.biomaterials.2005.06.014. [DOI] [PubMed] [Google Scholar]
- Arcangeli A, Becchetti A. Complex functional interaction between integrin receptors and ion channels. Trends Cell Biol. 2006;16:631–639. doi: 10.1016/j.tcb.2006.10.003. [DOI] [PubMed] [Google Scholar]
- Baneyx G, Vogel V. Self-assembly of fibronectin into fibrillar networks underneath dipalmitoyl phosphatidylcholine monolayers: role of lipid matrix and tensile forces. Proc. Natl. Acad. Sci. U. S. A. 1999;96:12518–12523. doi: 10.1073/pnas.96.22.12518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baneyx G, Baugh L, Vogel V. Coexisting conformations of fibronectin in cell culture imaged using fluorescence resonance energy transfer. Proc. Natl. Acad. Sci. U. S. A. 2001;98:14464–14468. doi: 10.1073/pnas.251422998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baneyx G, Baugh L, Vogel V. Fibronectin extension and unfolding within cell matrix fibrils controlled by cytoskeletal tension. Proc. Natl. Acad. Sci. U. S. A. 2002;99:5139–5143. doi: 10.1073/pnas.072650799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barker TH, Baneyx G, Cardo-Vila M, Workman GA, Weaver M, Menon PM, Dedhar S, Rempel SA, Arap W, Pasqualini R, Vogel V, Sage EH. SPARC regulates extracellular matrix organization through its modulation of integrin-linked kinase activity. J. Biol. Chem. 2005;280:36483–36493. doi: 10.1074/jbc.M504663200. [DOI] [PubMed] [Google Scholar]
- Baugh L, Vogel V. Structural changes of fibronectin adsorbed to model surfaces probed by fluorescence resonance energy transfer. J. Biomed. Mater. Res. 2004;69A:525–534. doi: 10.1002/jbm.a.30026. [DOI] [PubMed] [Google Scholar]
- Briknarova K, Akerman ME, Hoyt DW, Ruoslahti E, Ely KR. Anastellin, an FN3 fragment with fibronectin polymerization activity, resembles amyloid fibril precursors. J. Mol. Biol. 2003;332:205–215. doi: 10.1016/s0022-2836(03)00890-8. [DOI] [PubMed] [Google Scholar]
- Bustamante C, Chemla YR, Forde NR, Izhaky D. Mechanical processes in biochemistry. Annu. Rev. Biochem. 2004;73:705–748. doi: 10.1146/annurev.biochem.72.121801.161542. [DOI] [PubMed] [Google Scholar]
- Chen H, Mosher DF. Formation of sodium dodecyl sulfate-stable fibronectin multimers. Failure to detect products of thiol-disulfide exchange in cyanogen bromide or limited acid digests of stabilized matrix fibronectin. J. Biol. Chem. 1996;271:9084–9089. doi: 10.1074/jbc.271.15.9084. [DOI] [PubMed] [Google Scholar]
- Chen LB, Murray A, Segal RA, Bushnell A, Walsh ML. Studies on intercellular LETS glycoprotein matrices. Cell. 1978;14:377–391. doi: 10.1016/0092-8674(78)90123-x. [DOI] [PubMed] [Google Scholar]
- Chen Y, Zardi L, Peters DM. High-resolution cryo-scanning electron microscopy study of the macromolecular structure of fibronectin fibrils. Scanning. 1997;19:349–355. doi: 10.1002/sca.4950190505. [DOI] [PubMed] [Google Scholar]
- Craig D, Gao M, Schulten K, Vogel V. Structural insights into how the MIDAS ion stabilizes integrin binding to an RGD peptide under force. Structure. 2004a;12:2049–2058. doi: 10.1016/j.str.2004.09.009. [DOI] [PubMed] [Google Scholar]
- Craig D, Gao M, Schulten K, Vogel V. Tuning the mechanical stability of fibronectin type III modules through sequence variations. Structure. 2004b;12:21–30. doi: 10.1016/j.str.2003.11.024. [DOI] [PubMed] [Google Scholar]
- Craig D, Krammer A, Schulten K, Vogel V. Comparison of the early stages of forced unfolding for fibronectin type III modules. Proc. Natl. Acad. Sci. U. S. A. 2001;98:5590–5595. doi: 10.1073/pnas.101582198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalby MJ, Riehle MO, Sutherland DS, Agheli H, Curtis AS. Use of nanotopography to study mechanotransduction in fibroblasts—methods and perspectives. Eur. J. Cell Biol. 2004;83:159–169. doi: 10.1078/0171-9335-00369. [DOI] [PubMed] [Google Scholar]
- Dalby MJ, Riehle MO, Sutherland DS, Agheli H, Curtis AS. Morphological and microarray analysis of human fibroblasts cultured on nanocolumns produced by colloidal lithography. Eur Cell Mater. 2005;9:1–8. doi: 10.22203/ecm.v009a01. discussion 8. [DOI] [PubMed] [Google Scholar]
- Discher DE, Janmey P, Wang YL. Tissue cells feel and respond to the stiffness of their substrate. Science. 2005;310:1139–1143. doi: 10.1126/science.1116995. [DOI] [PubMed] [Google Scholar]
- Dzamba BJ, Peters DM. Arrangement of cellular fibronectin in noncollagenous fibrils in human fibroblast cultures. J. Cell Sci. 1991;100(Pt 3):605–612. doi: 10.1242/jcs.100.3.605. [DOI] [PubMed] [Google Scholar]
- Ejim OS, Blunn GW, Brown RA. Production of artificial-orientated mats and strands from plasma fibronectin: a morphological study. Biomaterials. 1993;14:743–748. doi: 10.1016/0142-9612(93)90038-4. [DOI] [PubMed] [Google Scholar]
- Engel J, Odermatt E, Engel A, Madri JA, Furthmayr H, Rohde H, Timpl R. Shapes, domain organizations and flexibility of laminin and fibronectin, two multifunctional proteins of the extracellular matrix. J. Mol. Biol. 1981;150:97–120. doi: 10.1016/0022-2836(81)90326-0. [DOI] [PubMed] [Google Scholar]
- Engler AJ, Sen S, Sweeney HL, Discher DE. Matrix elasticity directs stem cell lineage specification. Cell. 2006;126:677–689. doi: 10.1016/j.cell.2006.06.044. [DOI] [PubMed] [Google Scholar]
- Engvall E, Ruoslahti E. Binding of soluble form of fibroblast surface protein, fibronectin, to collagen. Int. J. Cancer. 1977;20:1–5. doi: 10.1002/ijc.2910200102. [DOI] [PubMed] [Google Scholar]
- Erickson HP. Reversible unfolding of fibronectin type III and immunoglobulin domains provides the structural basis for stretch and elasticity of titin and fibronectin. Proc. Natl. Acad. Sci. U. S. A. 1994;91:10114–10118. doi: 10.1073/pnas.91.21.10114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erickson HP. Stretching fibronectin. J. Muscle Res. Cell Motil. 2002;23:575–580. doi: 10.1023/a:1023427026818. [DOI] [PubMed] [Google Scholar]
- Galbraith CG, Yamada KM, Sheetz MP. The relationship between force and focal complex development. J. Cell Biol. 2002;159:695–705. doi: 10.1083/jcb.200204153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia AJ, Vega MD, Boettiger D. Modulation of cell proliferation and differentiation through substrate-dependent changes in fibronectin conformation. Mol. Biol. Cell. 1999;10:785–798. doi: 10.1091/mbc.10.3.785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giannone G, Sheetz MP. Substrate rigidity and force define form through tyrosine phosphatase and kinase pathways. Trends Cell. Biol. 2006;16:213–223. doi: 10.1016/j.tcb.2006.02.005. [DOI] [PubMed] [Google Scholar]
- Halter M, Antia M, Vogel V. Fibronectin conformational changes induced by adsorption to liposomes. J. Control. Release. 2005;101:209–222. doi: 10.1016/j.jconrel.2004.07.002. [DOI] [PubMed] [Google Scholar]
- Hynes RO. Fibronectins. New York: Springer-Verlag Inc; 1990. [Google Scholar]
- Ingber DE. Cellular mechanotransduction: putting all the pieces together again. Faseb J. 2006;20:811–827. doi: 10.1096/fj.05-5424rev. [DOI] [PubMed] [Google Scholar]
- Johnson CP, Tang HY, Carag C, Speicher DW, Discher DE. Forced unfolding of proteins within cells. Science. 2007;317:663–666. doi: 10.1126/science.1139857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keselowsky BG, Collard DM, Garcia AJ. Integrin binding specificity regulates biomaterial surface chemistry effects on cell differentiation. Proc. Natl. Acad. Sci. U. S. A. 2005;102:5953–5957. doi: 10.1073/pnas.0407356102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan MY, Medow MS, Newman SA. Unfolding transitions of fibronectin and its domains. Stabilization and structural alteration of the N-terminal domain by heparin. Biochem. J. 1990;270:33–38. doi: 10.1042/bj2700033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kostic A, Sheetz MP. Fibronectin rigidity response through Fyn and p130Cas recruitment to the leading edge. Mol. Biol. Cell. 2006;17:2684–2695. doi: 10.1091/mbc.E05-12-1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kung C. A possible unifying principle for mechanosensation. Nature. 2005;436:647–654. doi: 10.1038/nature03896. [DOI] [PubMed] [Google Scholar]
- Lai CS, Tooney NM, Ankel EG. Structure and flexibility of plasma fibronectin in solution: electron spin resonance spin-label, circular dichroism, and sedimentation studies. Biochemistry. 1984;23:6393–6397. doi: 10.1021/bi00321a017. [DOI] [PubMed] [Google Scholar]
- Lan MA, Gersbach CA, Michael KE, Keselowsky BG, Garcia AJ. Myoblast proliferation and differentiation on fibronectin-coated self assembled monolayers presenting different surface chemistries. Biomaterials. 2005;26:4523–4531. doi: 10.1016/j.biomaterials.2004.11.028. [DOI] [PubMed] [Google Scholar]
- Langenbach KJ, Sottile J. Identification of protein-disulfide isomerase activity in fibronectin. J. Biol. Chem. 1999;274:7032–7038. doi: 10.1074/jbc.274.11.7032. [DOI] [PubMed] [Google Scholar]
- Litvinovich SV, Brew SA, Aota S, Akiyama SK, Haudenschild C, Ingham KC. Formation of amyloid-like fibrils by self-association of a partially unfolded fibronectin type III module. J. Mol. Biol. 1998;280:245–258. doi: 10.1006/jmbi.1998.1863. [DOI] [PubMed] [Google Scholar]
- Magnusson MK, Mosher DF. Fibronectin: structure, assembly, and cardiovascular implications. Arterioscler. Thromb. Vasc. Biol. 1998;18:1363–1370. doi: 10.1161/01.atv.18.9.1363. [DOI] [PubMed] [Google Scholar]
- Mao Y, Schwarzbauer JE. Fibronectin fibrillogenesis, a cell-mediated matrix assembly process. Matrix Biol. 2005;24:389–399. doi: 10.1016/j.matbio.2005.06.008. [DOI] [PubMed] [Google Scholar]
- McKeown-Longo PJ, Mosher DF. Binding of plasma fibronectin to cell layers of human skin fibroblasts. J. Cell Biol. 1983;97:466–472. doi: 10.1083/jcb.97.2.466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Midwood KS, Mao Y, Hsia HC, Valenick LV, Schwarzbauer JE. Modulation of cell-fibronectin matrix interactions during tissue repair. J. Investig. Dermatol. Symp. Proc. 2006;11:73–78. doi: 10.1038/sj.jidsymp.5650005. [DOI] [PubMed] [Google Scholar]
- Morla A, Zhang Z, Ruoslahti E. Superfibronectin is a functionally distinct form of fibronectin. Nature. 1994;367:193–196. doi: 10.1038/367193a0. [DOI] [PubMed] [Google Scholar]
- Mosher DF, Johnson RB. In vitro formation of disulfide-bonded fibronectin multimers. J. Biol. Chem. 1983;258:6595–6601. [PubMed] [Google Scholar]
- Oberhauser AF, Badilla-Fernandez C, Carrion-Vazquez M, Fernandez JM. The mechanical hierarchies of fibronectin observed with single-molecule AFM. J. Mol. Biol. 2002;319:433–447. doi: 10.1016/S0022-2836(02)00306-6. [DOI] [PubMed] [Google Scholar]
- Ohashi T, Kiehart DP, Erickson HP. Dynamics and elasticity of the fibronectin matrix in living cell culture visualized by fibronectin-green fluorescent protein. Proc. Natl. Acad. Sci. U. S. A. 1999;96:2153–2158. doi: 10.1073/pnas.96.5.2153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pankov R, Yamada KM. Fibronectin at a glance. J. Cell. Sci. 2002;115:3861–3863. doi: 10.1242/jcs.00059. [DOI] [PubMed] [Google Scholar]
- Paszek MJ, Zahir N, Johnson KR, Lakins JN, Rozenberg GI, Gefen A, Reinhart-King CA, Margulies SS, Dembo M, Boettiger D, Hammer DA, Weaver VM. Tensional homeostasis and the malignant phenotype. Cancer Cell. 2005;8:241–254. doi: 10.1016/j.ccr.2005.08.010. [DOI] [PubMed] [Google Scholar]
- Peters DMP, Chen Y, Zardi L, Brummel S. Conformation of fibronectin fibrils varies: discrete globular domains of type III repeats detected. Microsc. Microanal. 1998;4:385–396. doi: 10.1017/s1431927698980369. [DOI] [PubMed] [Google Scholar]
- Romberger DJ. Fibronectin. Int. J. Biochem. Cell Biol. 1997;29:939–943. doi: 10.1016/s1357-2725(96)00172-0. [DOI] [PubMed] [Google Scholar]
- Samori B, Zuccheri G, Baschieri R. Protein unfolding and refolding under force: methodologies for nanomechanics. Chemphyschem. 2005;6:29–34. doi: 10.1002/cphc.200400343. [DOI] [PubMed] [Google Scholar]
- Sawada Y, Tamada M, Dubin-Thaler BJ, Cherniavskaya O, Sakai R, Tanaka S, Sheetz MP. Force sensing by mechanical extension of the Src family kinase substrate p130Cas. Cell. 2006;127:1015–1026. doi: 10.1016/j.cell.2006.09.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sechler JL, Schwarzbauer JE. Control of cell cycle progression by fibronectin matrix architecture. J. Biol. Chem. 1998;273:25533–25536. doi: 10.1074/jbc.273.40.25533. [DOI] [PubMed] [Google Scholar]
- Singer II. The fibronexus: a transmembrane association of fibronectin-containing fibers and bundles of 5 nm microfilaments in hamster and human fibroblasts. Cell. 1979;16:675–685. doi: 10.1016/0092-8674(79)90040-0. [DOI] [PubMed] [Google Scholar]
- Smith DE, Mosher DF, Johnson RB, Furcht LT. Immunological identification of two sulfhydryl-containing fragments of human plasma fibronectin. J. Biol. Chem. 1982;257:5831–5838. [PubMed] [Google Scholar]
- Smith ML, Gourdon D, Little WC, Kubow KE, Eguiluz RA, Luna-Morris S, Vogel V. Force-induced unfolding of fibronectin in the extracellular matrix of living cells. PLoS Biol. 2007;5:e268. doi: 10.1371/journal.pbio.0050268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamada M, Sheetz MP, Sawada Y. Activation of a signaling cascade by cytoskeleton stretch. Dev. Cell. 2004;7:709–718. doi: 10.1016/j.devcel.2004.08.021. [DOI] [PubMed] [Google Scholar]
- Vogel V. Mechanotransduction involving multimodular proteins: converting force into biochemical signals. Annu. Rev. Biophys. Biomol. Struct. 2006;35:459–488. doi: 10.1146/annurev.biophys.35.040405.102013. [DOI] [PubMed] [Google Scholar]
- Vogel V, Sheetz M. Local force and geometry sensing regulate cell functions. Nat. Rev. Mol. Cell Biol. 2006;7:265–275. doi: 10.1038/nrm1890. [DOI] [PubMed] [Google Scholar]
- Wang K, Forbes JG, Jin AJ. Single molecule measurements of titin elasticity. Prog. Biophys. Mol. Biol. 2001;77:1–44. doi: 10.1016/s0079-6107(01)00009-8. [DOI] [PubMed] [Google Scholar]
- Wierzbicka-Patynowski I, Schwarzbauer JE. The ins and outs of fibronectin matrix assembly. J. Cell Sci. 2003;116:3269–3276. doi: 10.1242/jcs.00670. [DOI] [PubMed] [Google Scholar]
- Wojciak-Stothard B, Denyer M, Mishra M, Brown RA. Adhesion, orientation, and movement of cells cultured on ultrathin fibronectin fibers. In Vitro Cell Dev. Biol. Anim. 1997;33:110–117. doi: 10.1007/s11626-997-0031-4. [DOI] [PubMed] [Google Scholar]
- Yeung T, Georges PC, Flanagan LA, Marg B, Ortiz M, Funaki M, Zahir N, Ming W, Weaver V, Janmey PA. Effects of substrate stiffness on cell morphology, cytoskeletal structure, and adhesion. Cell Motil. Cytoskelet. 2005;60:24–34. doi: 10.1002/cm.20041. [DOI] [PubMed] [Google Scholar]
- Zhong C, Chrzanowska-Wodnicka M, Brown J, Shaub A, Belkin AM, Burridge K. Rho-mediated contractility exposes a cryptic site in fibronectin and induces fibronectin matrix assembly. J. Cell Biol. 1998;141:539–551. doi: 10.1083/jcb.141.2.539. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.