ABSTRACT
In this study, we employed proteomics to identify mechanisms of posttranslational regulation on cell survival signaling proteins. We focused on Cu-Zn superoxide dismutase (SOD1), which protects cells from oxidative stress. We found that acylation of K122 on SOD1, while not impacting SOD1 catalytic activity, suppressed the ability of SOD1 to inhibit mitochondrial metabolism at respiratory complex I. We found that deacylase depletion increased K122 acylation on SOD1, which blocked the suppression of respiration in a K122-dependent manner. In addition, we found that acyl-mimicking mutations at K122 decreased SOD1 accumulation in mitochondria, initially hinting that SOD1 may inhibit respiration directly within the intermembrane space (IMS). However, surprisingly, we found that forcing the K122 acyl mutants into the mitochondria with an IMS-targeting tag did not recover their ability to suppress respiration. Moreover, we found that suppressing or boosting respiration levels toggled SOD1 in or out of the mitochondria, respectively. These findings place SOD1-mediated inhibition of respiration upstream of its mitochondrial localization. Lastly, deletion-rescue experiments show that a respiration-defective mutant of SOD1 is also impaired in its ability to rescue cells from toxicity caused by SOD1 deletion. Together, these data suggest a previously unknown interplay between SOD1 acylation, metabolic regulation, and SOD1-mediated cell survival.
KEYWORD: superoxide dismutase
INTRODUCTION
The regulation of cell signaling events through protein posttranslational modifications (PTMs), including phosphorylation, acylation, glycosylation, and ubiquitination, provides a means for cells to respond rapidly to environmental cues. Defects in the pathways that control PTMs underlie many human diseases and typically represent a fundamental breakdown in communication between the cell and its environment. Mass spectrometry (MS)-based approaches have dramatically expanded our view of PTMs in terms of both PTM variety and proteome coverage. More than 200 different PTMs have been described (www.uniprot.org/), and many, including phosphorylation and ubiquitination, regulate essentially every known biological process in the cell. With this understanding of the breadth of PTMs, the challenge is to “zoom in” so as to understand the functional consequences of specific PTMs. Thus, our goal is to understand how PTMs affect protein function, with particular focus on cell survival signaling nodes and pathways.
Here we used an affinity purification approach, coupled with liquid chromatography-tandem MS (LC–MS-MS), to identify PTMs in primary untreated mouse tissues. Using a recently developed PTM analysis tool (SAPH-ire [structural analysis of PTM hotspots]) to prioritize our functional studies, we evaluated PTMs from our experiment against all other experimentally observed PTMs curated for the Cu-Zn superoxide dismutase (SOD) domain family (1, 2). This analysis highlighted an acyl-modified lysine on SOD1, on which we focused in this study. SOD1 is a well conserved metalloenzyme best known for its catalytic role in the disproportionation of superoxide radicals (O2−) into molecular oxygen (O2) and hydrogen peroxide (H2O2) (3–11). Its expression is primarily cytosolic, although a fraction of SOD1 exists in the mitochondrial intermembrane space (IMS). Studies spanning multiple model organisms and cell lines have underscored the importance of SOD1 in oxidative-stress protection (reviewed in references 12 and 13). Accordingly, SOD1 is overexpressed in a variety of cancers, and SOD1 deletion in mice results in a higher incidence of liver cancer (6). Additionally, mutations in SOD1 that increase its propensity to aggregate cause toxicity in motor neurons, resulting in a familial form of amyotrophic lateral sclerosis (ALS) (reviewed in reference 13).
Several lines of evidence suggest that SOD1 biology may be more complex than initially thought. While SOD1 deletion is toxic in cultured cells due to an accumulation of oxidative damage (14, 15), titration of SOD1 expression against a SOD1-null background in Saccharomyces cerevisiae revealed that only a small fraction, representing about 1% of total SOD1, is required for protection against oxidative stress (16). This suggests that SOD1 may have additional functions beyond its traditional role in reactive oxygen species (ROS) scavenging. Indeed, studies suggest that SOD1 plays roles in zinc and copper buffering (11, 17) and in regulating gene transcription (18, 19). In addition, a recent study by Reddi and Culotta found that yeast SOD1 suppressed mitochondrial respiration (20). However, despite this emerging complexity in SOD1 biology and clear roles for SOD1 in human disease, we have a limited understanding of SOD1 regulation at a posttranslational level.
Here we uncover a novel regulatory mechanism by which a sirtuin-governed acylation within the electrostatic loop of SOD1, at K122, suppresses SOD1-mediated inhibition of mitochondrial metabolism in mammalian cells. This observation provided genetic tools to help us understand the relationship between SOD1 mitochondrial localization and metabolic regulation, as well as the potential contribution of this metabolic function of SOD1 to its role in promoting cell survival. Our data suggest a model in which sirtuin-mediated deacylation of SOD1 promotes its inhibition of respiration, which in turn, elevates levels of mitochondrial SOD1 and contributes to the prosurvival function of SOD1.
RESULTS
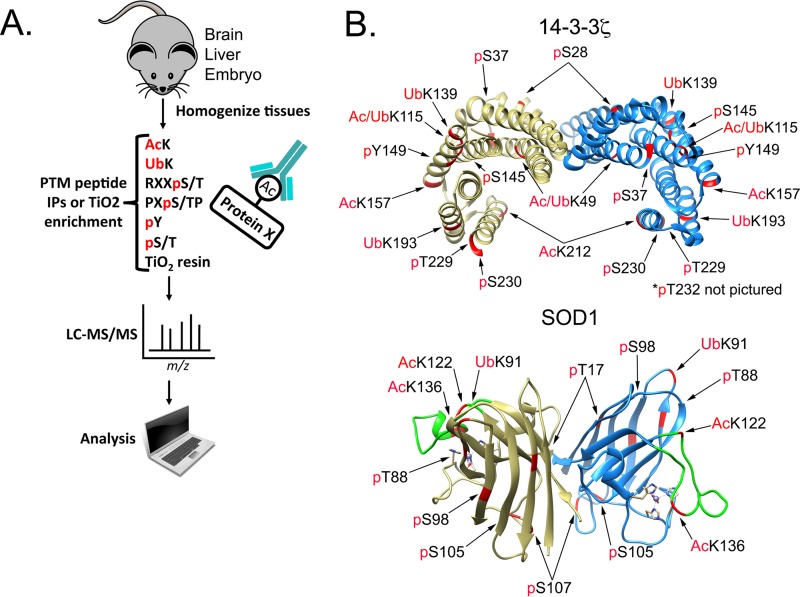
As a starting point, with the goal of identifying PTMs on cell survival signaling nodes, we used several PTM-specific antibody resins to compare in vivo PTMs across multiple mouse tissues (brain, liver, and embryo homogenates). The experimental layout, shown in Fig. 1A, included several phosphomotif, ubiquitin, and acetyl-lysine affinity resins. A complete set of database search results from this experiment is publicly available as a Scaffold file (Proteome Software Inc.) at https://discovery.genome.duke.edu/express/resources/3023/3023_PTMScanAll_withTiO2.sf3. In an effort to zoom in on PTMs on cell survival signaling nodes, we applied Gene Ontology analysis, as well as manual sorting by protein function. Two proteins of interest, 14-3-3ζ and SOD1, are shown in Fig. 1B. 14-3-3ζ is a phospho-serine/threonine binding protein that is overexpressed in a variety of cancers and promotes cell survival by directly modulating a network of phosphoproteins. In combining our PTM data sets, we identified PTMs of unknown function, including phosphorylation of Y149 (phospho-Y149) and ubiquitination of K139 (Ub-K139), on 14-3-3ζ, in addition to well-described PTMs, such as acetylation of K49 (Ac-K49) (21–23). In particular, acetylation of K49 is known to disrupt 14-3-3ζ interactions, and our previous work identified histone deacetylase 6 (HDAC6) as the K49-targeted lysine deacylase (KDAC) (23).
FIG 1.
Identification of PTMs on 14-3-3ζ and SOD1. (A) Brain, liver, and whole-embryo mouse tissues were homogenized and digested with trypsin. Peptides were subjected to affinity purification by the indicated antibody resin. Peptides were eluted and analyzed by LC–MS-MS. Proteomics data were analyzed with Scaffold software. IP, immunoprecipitation. (B) Crystal structures of human 14-3-3ζ (PDB accession no. 4IHL) and mouse SOD1 (PDB accession no. 3GTT) with PTMs identified in the proteomics data.
SAPH-ire identifies PTMs with high function potential in the SOD domain family.
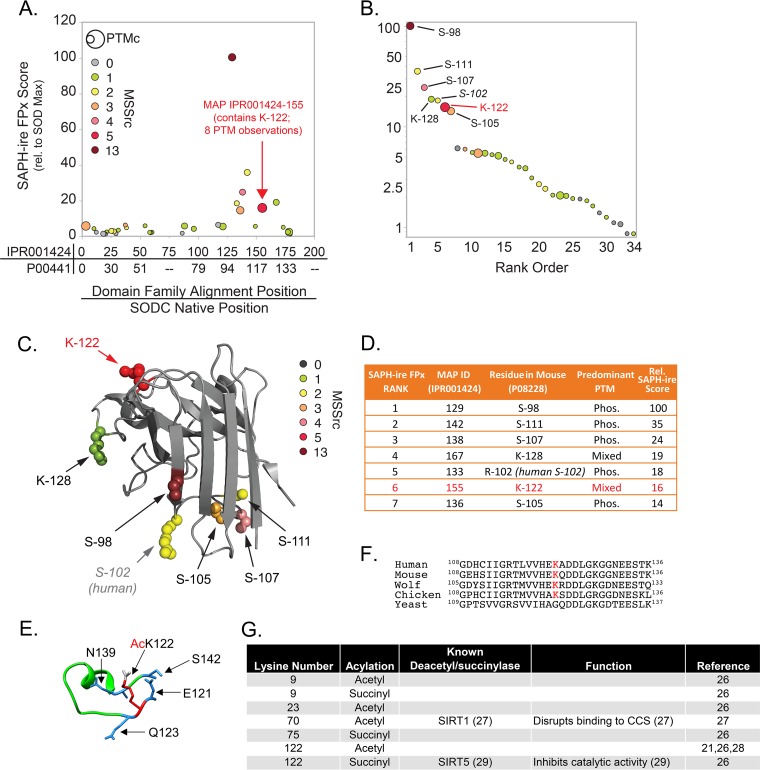
Our attention was also drawn to SOD1, which acts as one of the main modes of defense against oxidative stress by catalyzing the disproportionation of superoxide radicals (O2−) to molecular oxygen (O2) and hydrogen peroxide (H2O2). The lower panel of Fig. 1B shows the crystal structure of the SOD1 dimer and PTMs identified from our proteomics data. In an effort to prioritize PTMs on SOD1, we utilized SAPH-ire FPx, a machine learning-based PTM “hot spot” finder that examines experimentally identified PTMs and prioritizes them for the likelihood of biological function based on a number of parameters as described previously (1, 2, 24). By use of human SOD1 (UniProt ID P00441) as an anchor, PTMs were evaluated in the context of the entire eukaryotic SOD domain family (InterPro ID IPR001424). Interestingly, PTMs localized in the region between S98 and K128 exhibited the highest SAPH-ire FPx scores across the entire domain (Fig. 2A). In fact, the seven PTM sites with the highest FPx scores were all within this region (Fig. 2B to D). We became interested in an acylated lysine (K122) that not only was found in our mouse proteomics data set but also was identified by SAPH-ire in the region with the highest FPx scores and was one of the two highest-ranking modified lysine hot spots in the SOD domain family (Fig. 2A to D). Comparison of the two acyl-lysines (K122 and K128) within the top SAPH-ire cluster showed that K122 had the highest number (eight) of coaligned PTMs in the family (indicated by circle size in Fig. 2A to C) and the second-highest number (five) of independent MS-based observations (indicated by circle color in Fig. 2A to C), suggesting that K122 may be an important regulatory site. K122 sits within the electrostatic loop (Fig. 2E), which has nearby residues (H120 and D124) important for binding zinc and copper (25). K122 is conserved from humans to birds (Fig. 2F) and has been identified in previous studies, among other acyl-lysines, as acetylated and succinylated (21, 26–29) (Fig. 2G). A study by Lin et al. found that SIRT5 desuccinylates K122, and it was proposed to affect SOD1 antioxidant activity (29). In addition, a recent study using a site-specific antibody demonstrated that K122 is acetylated on endogenous SOD1 in multiple cell types throughout the murine nervous system (30), although the impact of the acetyl modification on SOD1 function was not demonstrated. With this background, we investigated the functional impact of K122 acylation on SOD1.
FIG 2.
SAPH-ire identifies PTMs with high function potential in the SOD domain family. (A) Plot of SAPH-ire function potential scores (FPx scores) for 34 modified-alignment positions (MAPs) (represented by circles) along the length of the SOD domain family. The domain family alignment position and corresponding native position of human SOD1 are plotted on the x axis for reference (from the N to the C terminus). The size of each circle is scaled to the number of distinct PTMs observed within a MAP (PTM count [PTMc]). The color of each circle indicates the number of independent literature sources documenting the observation of PTMs in that MAP (mass spectrometry source count [MSSrc]). (B) Rank-ordered plot of SAPH-ire function potential scores for the 34 SOD domain family MAPs. Residue positions correspond to mouse SOD1 residues found in each of the top seven domain family MAPs. (C) Projection of the top seven MAPs onto the crystal structure of mouse SOD1 (UniProt ID P08228; PDB ID 3GTT, chain A). (D) Table summarizing the identities and predominant PTMs observed for each of the top seven SOD MAPs. (E) Zoomed-in crystal structure of the mouse SOD1 (PDB ID 3GTT) electrostatic loop featuring K122 (red) modified with an acetyl group (gray) and surrounding amino acids (blue). (F) Sequence alignment of human, mouse, wolf, chicken, and yeast SOD1, with K122 highlighted in red. (G) Table indicating all known lysine acetyl or succinyl modifications of human SOD1, known deacetyl/succinylases, and phenotypes of the modifications.
Acyl-mimicking mutations at K122 have no effect on SOD1 dimerization or ROS-scavenging activity.
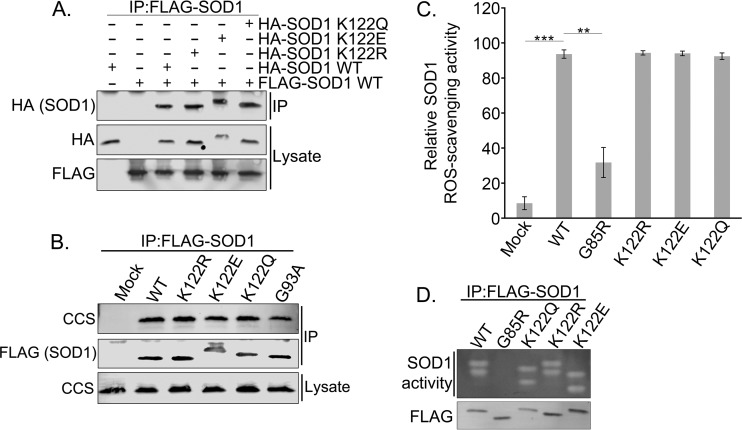
To examine the effect of acylation at K122 on various SOD1 functions, we generated acetyl-mimicking (K122Q) and succinyl-mimicking (K122E) mutations. Although there are structural differences between glutamine/glutamate and Nε-acyl-lysine, these mutations mimic the change in charge associated with the acyl modifications. One limitation of this approach is that the mutations abrogate any other lysine modification (e.g., ubiquitin) that may occur at the site; thus, comparison with the K-to-R control (which mimics a nonacylated lysine but also abrogates other lysine PTMs) is critical. To determine whether the acyl-mimicking mutants could dimerize with wild-type (WT) SOD1, we overexpressed Flag-tagged WT SOD1 together with either hemagglutinin (HA)-tagged WT SOD1 or HA-tagged mutant SOD1 in HEK 293 cells and saw no difference in dimerization between WT SOD1 and acyl-mimicking SOD1 mutants (Fig. 3A). We also saw no difference in the binding of these SOD1 mutants to the copper chaperone for superoxide dismutase (CCS) (Fig. 3B), which promotes the final stage of SOD1 folding and maturation. Furthermore, in contrast to the study by Lin et al. (29), we found no difference in ROS scavenging between WT SOD1 and the acyl-mimicking SOD1 mutants. This was verified in multiple ROS-scavenging assays with purified SOD1, and in a nitroblue tetrazolium (NBT) gel assay, including the ROS-scavenging-impaired G85R point mutant of SOD1 as a positive control (Fig. 3C and D).
FIG 3.
Acyl-mimicking mutations at K122 of SOD1 do not affect canonical SOD1 ROS-scavenging activity. (A) HEK 293 cells were cotransfected with Flag-tagged WT SOD1 and HA-tagged WT, K122R, K122E, or K122Q SOD1 expression plasmids. Flag-WT SOD1 was immunoprecipitated (IP) with Flag-agarose resin and was subjected to SDS-PAGE before being immunoblotted for HA-tagged binding partners. (B) HEK 293 cells were transfected with a Flag-tagged WT, K122R, K122E, K122Q, or G93A SOD1 expression plasmid. Flag-tagged SOD1 was immunoprecipitated with Flag-agarose resin and was subjected to SDS-PAGE before being immunoblotted for the CCS binding partner. (C) HEK 293 cells were transfected with Flag-tagged WT, G85R, K122R, K122E, or K122Q SOD1. SOD1 was immunoprecipitated with Flag-agarose resin and was then competitively eluted with Flag peptide. The eluted SOD1 activity was then measured colorimetrically. The experiment was carried out with three biological replicates, each with three technical replicates. Asterisks indicate significant differences (**, P < 0.01; ***, P < 0.001); error bars represent SEM. (D) Samples were prepared as for the assay for which results are shown in panel C. The eluted SOD1 was resolved through native PAGE, and SOD1 activity was assayed within the gel. White bands indicate active SOD1.
K122 acyl mimics inhibit SOD1-mediated control of mitochondrial respiration.
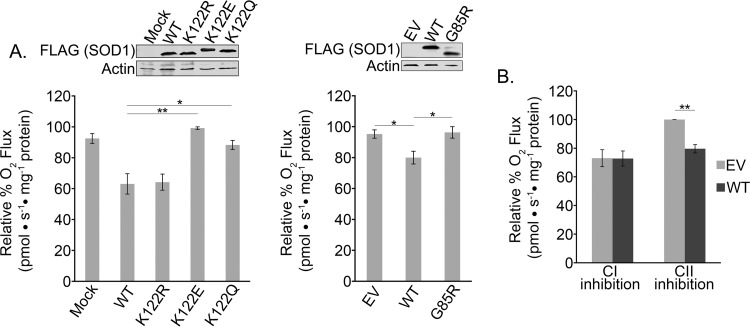
Although SOD1 is commonly thought of as an antioxidant, only 1% of SOD1 expressed in cells is necessary to keep ROS below cytotoxic levels (16). The rationale for such high levels of SOD1 has led others to propose that it may have important functions beyond the disproportionation of oxygen radicals (20). Previous studies have suggested that SOD1 plays roles in cellular zinc (17) and copper (11) buffering and as a nuclear transcription factor (18, 19). In addition, a more recent study by Reddi and Culotta demonstrated a novel function of yeast SOD1 in suppressing mitochondrial respiration through the modulation of casein kinase signaling (20). Given that lysine acylation is linked to metabolism (21, 31, 32), we decided to test the potential link between the K122 PTM and SOD1 antirespiratory activity. To test whether mammalian SOD1, like its yeast counterpart, suppresses respiration, we expressed WT human SOD1 in HEK 293 cells and saw a marked decrease in mitochondrial oxygen flux, suggesting conservation of this SOD1 metabolic function from yeast to humans (Fig. 4A). Importantly, in comparing this effect on respiration across the panel of acyl-mimicking SOD1 mutants, we found that the acetyl-mimicking (K122Q) and succinyl-mimicking (K122E) mutants completely lost their ability to suppress respiration. Like that of yeast SOD1, the inhibitory effect of mammalian SOD1 on respiration required its enzymatic activity, as evidenced by the fact that the G85R mutant, which shows ∼70% lower ROS-scavenging activity than the WT (Fig. 3C), failed to inhibit respiration (20). The K122R mutant, used as an additional control, was fully active in suppressing respiration, suggesting that the K122Q and K122E phenotypes were not due simply to mutation of the lysine, which could block other lysine PTMs, such as sumoylation or ubiquitination (Fig. 4A).
FIG 4.
Acyl-K122 mimics of SOD1 inhibit SOD1-mediated suppression of mitochondrial respiration. (A) (Left) HEK 293 cells were transfected with a Flag-tagged WT, K122R, K122E, or K122Q SOD1 expression plasmid, and oxygen flux was measured with an Oroboros O2K respirometer. Cell lysates were separated by SDS-PAGE and immunoblotted for Flag-SOD1. The experiment was carried out in quadruplicate. Asterisks indicate significant differences (*, P < 0.05; **, P < 0.01); error bars represent SEM. (Right) HEK 293 cells were transfected with an empty vector (EV) or a Flag-tagged WT or G85R SOD1 expression plasmid and were assayed as in the experiment for which results are shown on the left. The experiment was carried out in triplicate. (B) HEK 293 cells were transfected with an empty vector or a Flag-tagged WT SOD1 expression vector. Oxygen rates were measured with an Oroboros O2K respirometer after inhibition of complex I (CI) with rotenone (2 mM) or of CII with malonate (2.5 mM). Three technical replicates were carried out.
To determine which electron transport chain (ETC) subunit complex was affected by SOD1, we treated the SOD1-expressing cells with inhibitors of complexes I and II. While complex II does not transport protons across the intermembrane space, as does complex I, the two complexes work in parallel to transfer electrons to a pool of ubiquinone. Electrons from ubiquinone are then transferred stepwise to complex IV, where molecular oxygen is reduced to water. Because complex I and complex II work in parallel to transfer electrons, each can be inhibited separately and electrons can still be transferred through the uninhibited complex. When we inhibited complex II with malonate, SOD1 was still able to suppress respiration. However, when complex I was inhibited with rotenone, SOD1 had no additional effect on respiration (Fig. 4B), suggesting that SOD1 is inhibiting the ETC at complex I.
SOD1-mediated inhibition of respiration can be modulated via SIRT5.
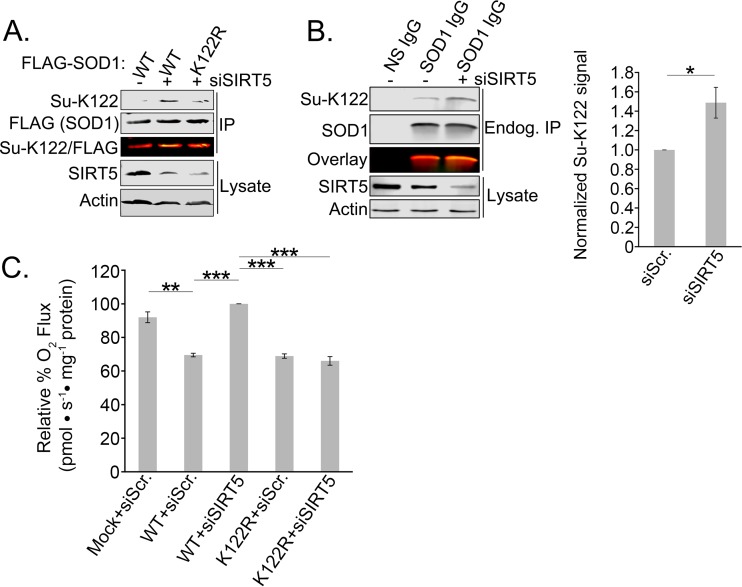
Lysine acylation is regulated by the activity of KDACs, which include the HDAC and sirtuin families. Unlike many KDACs, which prefer acetyl-lysine as their substrate, SIRT5 shows a preference for the longer-chain acylations of succinylation and malonylation (33). It has been shown previously that SIRT5 is the desuccinylase for K122 (29), and while SIRT5 is traditionally thought of as a mitochondrial sirtuin, recent studies have reported the presence of SIRT5 in the cytosol as well (34–36). We validated the role of SIRT5 in desuccinylating K122 by developing an antibody specific for succinylated K122 (Su-K122) (Fig. 5A). To confirm that succinylation occurs on endogenous SOD1, we depleted cells of SIRT5 and immunoprecipitated SOD1. Figure 5B shows the increase in endogenous SOD1 succinylation at K122 in SIRT5-depleted cells. Although acetylation of K122 clearly occurs on endogenous SOD1 (21, 26, 28, 30; also our proteomics data), our attempts to generate site-specific acetyl-K122 antibodies had only limited success: the antibodies showed marginal specificity to the acetylation. This complication, combined with the fact that K122E showed a slightly more robust effect in inhibiting the antirespiration function of SOD1 than K122Q, led us to focus primarily on K122 succinylation.
FIG 5.
Development of an antibody specific to acylated K122 of SOD1. SIRT5 inhibition results in succinylation of endogenous SOD1. (A) Flag-tagged WT or K122R SOD1 was overexpressed in HEK 293 cells with or without SIRT5 knocked down by siRNA. Flag-tagged SOD1 was immunoprecipitated with Flag-agarose resin, subjected to SDS-PAGE, and immunoblotted for succinyl-K122 (Su-K122) and total (Flag) SOD1. (B) Endogenous SOD1 was immunoprecipitated from HEK 293 cells with or without SIRT5 knocked down via siRNA. The immunoprecipitated SOD1 was then subjected to SDS-PAGE and immunoblotted with the Su-K122 antibody. The experiment was carried out in triplicate. The asterisk indicates a significant difference (*, P < 0.05); error bars represent SEM. (C) Flag-tagged WT or K122R SOD1 was overexpressed in HEK 293 cells with or without SIRT5 knocked down by siRNA. Oxygen rates were measured with an Oroboros O2K respirometer. The experiment was carried out in triplicate. Asterisks indicate significant differences (**, P < 0.01; ***, P < 0.001).
To determine whether we could block the antirespiratory activity of SOD1 by modulating the endogenous deacylation machinery, we depleted SIRT5 in cells expressing WT SOD1 or the nonsuccinylatable K122R SOD1 and measured respiration. Remarkably, we found that SIRT5 depletion recovered normal respiration levels in cells expressing WT SOD1 and that the recovery was overridden by the K122R mutant, which cannot be acylated (Fig. 5C). This suggests that SIRT5 modulates metabolism via desuccinylation of SOD1 at K122.
SOD1-mediated suppression of respiration is upstream of its mitochondrial localization.
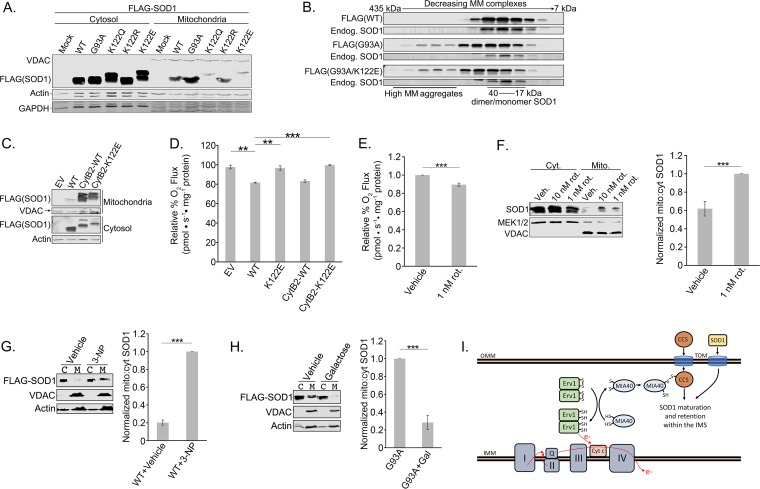
SOD1 is known to reside in the cytosol, the IMS of the mitochondria, and the nucleus (37). Our respiration data made us question whether the antirespiratory effect of SOD1 is exerted locally within the IMS. If so, we would predict that acylation of K122 would block SOD1 mitochondrial localization. As shown in Fig. 6A, we found decreased levels of the acyl-mimicking SOD1 mutants in the mitochondrial fraction. We included the ALS-linked G93A SOD1 mutant as a positive control because it is known to accumulate in the IMS (Fig. 6A) (38). Shaw and colleagues recently showed that neutralization of positively charged lysines by aspirin-induced lysine acetylation on SOD1 inhibited the propensity of SOD1 to aggregate (39). However, the acyl-mimicking mutant, in the context of the ALS-linked G93A mutation, did not decrease SOD1 aggregation (Fig. 6B). Together, these data suggest that acylation at K122 inhibits the mitochondrial accumulation of SOD1, and while only correlative, they raise the possibility that the mitochondrial pool of SOD1 directly inhibits respiration in the IMS.
FIG 6.
SOD1-mediated suppression of respiration is upstream of its mitochondrial localization. (A) HEK 293 cells were transfected with Flag-tagged WT, G93A, K122Q, K122R, or K122E SOD1. The cells were lysed, and mitochondria were fractionated from the cytosol. The two fractions were subjected to SDS-PAGE and immunoblotted for Flag-SOD1 present in the mitochondria versus the cytosol. (B) Flag-tagged WT, G93A, or G93A K122E SOD1 was induced with tetracycline for 48 h in T-REx 293 cells. The cells were lysed, and the lysates were then loaded onto a Superdex 200 10/300 GL size exclusion column and were eluted in fractions. The fractions were subjected to SDS-PAGE and immunoblotted for Flag-SOD1 and endogenous SOD1. MM, molecular mass. (C) Flag-tagged WT, CytB2-WT, or CytB2-K122E SOD1 was overexpressed in HEK 293 cells. The cells were lysed, and mitochondria were fractionated from the cytosol. The two fractions were subjected to SDS-PAGE and immunoblotted for Flag-SOD1 present in the mitochondria versus the cytosol. (D) Flag-tagged WT, K122E, CytB2-WT, or CytB2-K122E SOD1 was overexpressed in HEK 293 cells. Oxygen rates were measured with an Oroboros O2K respirometer. The experiment was carried out in triplicate. Asterisks indicate significant differences (**, P < 0.01; ***, P < 0.001); error bars represent SEM. (E) Rotenone (1 nM) or a vehicle was added to HEK 293 cells 30 min prior to measurement of oxygen consumption rates with an Oroboros O2K respirometer. The experiment was carried out in quadruplicate. (F) Rotenone at 1 or 10 nM or a vehicle was added to HEK 293 cells 30 min prior to lysis of the cells and fractionation of the mitochondria from the cytosol as in the experiment for which results are shown in panel A. The fractions were immunoblotted for the presence of endogenous SOD1 in the mitochondria and cytosol. In each of the six replicates, SOD1 levels were first normalized to mitochondrial and cytosolic loading controls and then normalized to the ratio of mitochondrial SOD1 in the 1 nM rotenone-treated sample. (G) Flag-tagged WT-SOD1 was overexpressed in HEK 293 cells. A 1 mM concentration of 3-NP was added for 16 to 18 h before the cells were lysed and mitochondria (M) were fractionated from the cytosol (C). The two fractions were subjected to SDS-PAGE and immunoblotted for Flag-SOD1 present in the mitochondria versus the cytosol. In each of the three replicates, Flag-SOD1 levels were normalized first to mitochondrial and cytosolic loading controls and then to the ratio of mitochondrial Flag-SOD1 in the 3-NP-treated sample. (H) Flag-tagged G93A SOD1 was overexpressed in HEK 293 cells. A 25 mM concentration of galactose in low-glucose (2 mM) medium was added 24 h before the cells were lysed and assayed as in the experiment for which results are shown in panel G. In each of the three replicates, Flag-SOD1 levels were normalized first to mitochondrial and cytosolic loading controls and then to the ratio of mitochondrial Flag-SOD1 in the untreated sample. (I) An Erv1/MIA40 disulfide relay promotes the IMS import of CCS, which in turn promotes the IMS retention of SOD1. The disulfide relay is reset when oxidized cytochrome c accepts an electron from Erv1. OMM, outer mitochondrial membrane; IMM, inner mitochondrial membrane. (Model based on that of Kawamata and Manfredi [41].)
Based on these data, our initial hypothesis was that acylation of K122 prevented SOD1 mitochondrial localization and thus blocked SOD1-mediated suppression of respiration. To test this further, we used an approach in which SOD1 is appended, in frame, to the mitochondrial import signal of Saccharomyces cerevisiae cytochrome b2 (CytB2) (40). Our prediction was twofold: first, that pushing WT SOD1 into the IMS would enhance SOD1-mediated suppression of respiration, and second, that forcing the K122E SOD1 mutant into the mitochondria would rescue its ability to suppress respiration. Figure 6C shows that the IMS-targeting tag resulted in robust accumulation of WT and K122E SOD1 in the mitochondrial fractions. However, to our surprise, the mitochondrial localization of SOD1 did not enhance its ability to suppress respiration (Fig. 6D), nor did it recover the ability of the K122E mutant to suppress respiration. This suggests that the suppression of respiration by SOD1 is independent of its mitochondrial localization, a possibility that agrees with data from yeast in which the respiration-suppressing signal from SOD1 originates in the cytosol (20).
An alternative explanation for the relationship between K122 acylation and SOD1 mitochondrial localization is that the effect of SOD1 on respiration is upstream of its mitochondrial localization. Indeed, there is evidence that the disulfide relay system that imports SOD1 into the IMS is dependent on electron flux through cytochrome c reductase (complex III) of the ETC (41, 42). Based on this model, up- or downregulation of respiratory activity would cause a corresponding decrease or increase in SOD1 mitochondrial localization. To test this, we first identified a dose of rotenone, a complex I inhibitor, that produced a 10 to 15% reduction in overall oxygen consumption (Fig. 6E), similar to what we had observed with WT SOD1 in our previous experiments. In support of the idea that inhibition of respiration drives the mitochondrial uptake of SOD1, treatment of cells with this dose of rotenone (1 nM) for 30 min significantly increased the accumulation of endogenous SOD1 in the mitochondrial pellet (Fig. 6F). Additionally, treatment of cells with 3-nitropropionic acid (3-NP), a complex II inhibitor, also triggered an increase in mitochondrial SOD1 levels (Fig. 6G). Conversely, boosting ETC activity by culturing cells in galactose nearly eliminated the IMS-localized G93A mutant SOD1 from the IMS (Fig. 6H). These results suggest that cellular respiration levels modulate SOD1 mitochondrial uptake, such that inhibition or stimulation of respiration can toggle SOD1 mitochondrial accumulation up or down, respectively (model in Fig. 6I). Furthermore, this suggests a potential feedback loop between SOD1 acylation, inhibition of respiration, and SOD1 mitochondrial import (see Discussion).
The SOD1 K122E acyl mimic is impaired in its ability to rescue the lethality of SOD1 deletion and reduce mitochondrial ROS levels.
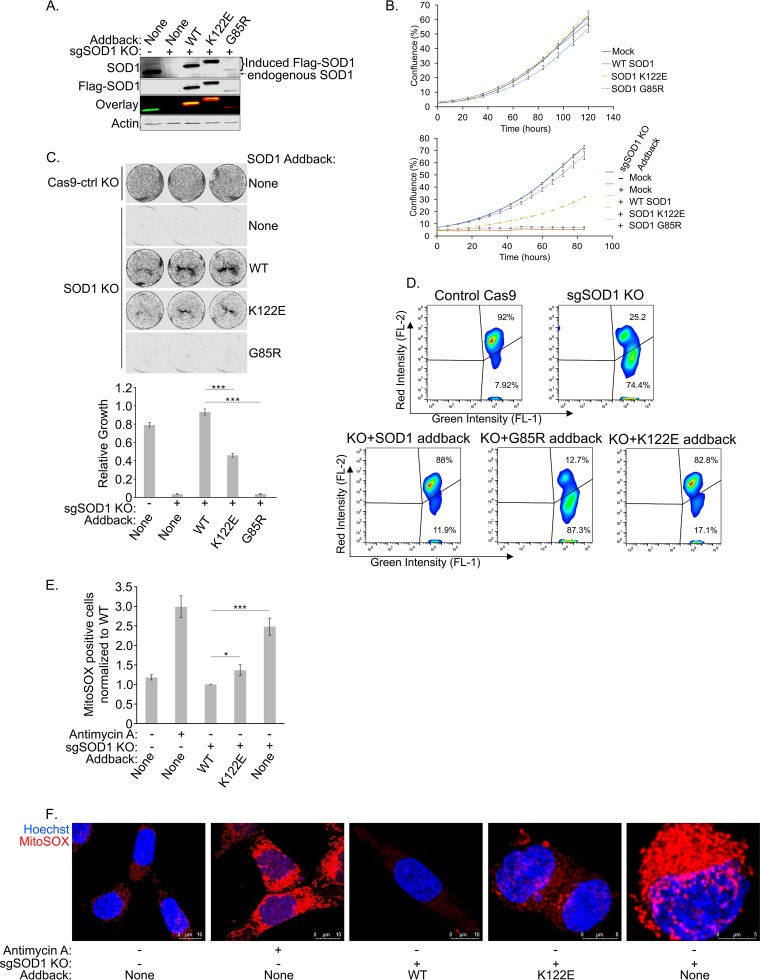
While the vital role of SOD1 in direct disproportionation of cytosolic ROS is well established, our observation that SOD1 suppresses respiration at complex I, and the link between respiration and SOD1 mitochondrial import, may indicate an added dimension to the role of SOD1 as an antioxidant. This is based on the decades-old observation that the ETC is a source of ROS (43, 44) and the possible role of the mitochondrial pool of SOD1 in scavenging ROS directly within the IMS (45, 46). The K122E mutant provided, to our knowledge, the first tool to address this question genetically, because this mutation inhibits the antirespiratory function of SOD1 while maintaining its enzymatic ROS-scavenging activity. We took advantage of an HCT-116 cell T-REx-based “flip-in” system wherein a gene of interest can be stably recombined with a specific locus under the control of a doxycycline (dox)-inducible promoter. This provides the advantage of controlling for copy number, which allows for a fair comparison between different derived cell lines. After generating a panel of stable doxycycline-inducible SOD1 variants with silent mutations at protospacer adjacent motif (PAM) sites, we used the clustered regularly interspaced short palindromic repeat (CRISPR)-Cas9 system to delete endogenous genomic SOD1 from each of the cell lines. Figure 7A shows that the inducible system gives near-endogenous levels of WT and K122E SOD1 expression and is completely refractory to the endogenous SOD1-targeted single-guide RNA (sgRNA). Of note, we had difficulty immunoblotting for the G85R mutant after the deletion of SOD1 due to the inability of that mutant to rescue cells from the acute toxicity of SOD1 knockout (KO).
FIG 7.
The SOD1 K122E acyl mimic is impaired in its ability to rescue the lethality of SOD1 deletion and reduce mitochondrial ROS levels. (A) Endogenous SOD1 was knocked out of HCT-116 T-REx cells using the CRISPR-Cas9 system. Expression of Flag-tagged SOD1 (with a mutated PAM site) was induced in the T-REx cells with doxycycline. The cells were lysed and were subjected to SDS-PAGE before being immunoblotted for endogenous SOD1 and overexpressed Flag-tagged SOD1. (B) (Top) SOD1 HCT-116 T-REx cells were seeded in 24-well plates and were grown in the absence of doxycycline. Cell confluence was measured by an IncuCyte Zoom system. Four technical replicates were carried out. Error bars represent SEM. (Bottom) Endogenous SOD1 was knocked out as described for panel A. Cells were plated and analyzed as described directly above but were grown in the presence of doxycycline. (C) After knockout of endogenous SOD1 and overexpression of Flag-tagged SOD1, the HCT-116 T-REx cells were seeded at 22,500 per well and were allowed to grow for 4 days before being fixed and stained with Giemsa stain. The wells were then imaged and quantified. Three technical replicates were carried out. Asterisks indicate significant differences (***, P < 0.001). (D) Cells prepared as for panel B (bottom) were stained with JC-1 to measure mitochondrial membrane potential. A live-cell population was gated from forward scatter and side scatter measurements, and FL-1 was plotted against FL-2. A higher FL-2/FL-1 ratio indicates a higher (healthier) mitochondrial membrane potential. (E) Cells prepared as for panel B (bottom) were stained with MitoSOX Red to measure mitochondrial superoxide content. A live-cell population was gated from forward scatter and side scatter measurements, from which a population of singlets was identified and selected. A threshold for positive MitoSOX staining was set based on the unstained control, and a ratio of BL-3-positive cells to singlets was calculated and normalized to WT addback cells. A higher ratio indicates a larger proportion of cells with superoxide present. Four replicates were carried out. Asterisks indicate significant differences (*, P < 0.05; ***, P < 0.001). (F) Cells prepared as for panel B (bottom) were stained with 5 μM MitoSOX Red for 10 min, and nuclei were counterstained. Prior to MitoSOX staining, positive-control cells were treated with 30 μM antimycin A for 40 min.
Previous studies have shown that SOD1 deletion in eukaryotes results in cell death as a result of ROS accumulation (14, 15). In agreement with this observation, the SOD1 KO lines showed a dramatic loss of cell growth as measured by an IncuCyte live-cell analysis system (Fig. 7B; the upper panel shows basal growth rates without doxycycline treatment) and infrared scanning of Giemsa-stained cultures after cell dilution and growth over a time course (Fig. 7C). The loss of growth caused by SOD1 KO was completely reversed in these cells by reconstituting WT SOD1 expression from the dox-inducible locus, indicating that growth defects were not due to off-target effects of the sgRNA. As a control, the G85R mutant, which shows reduced ROS scavenging and does not inhibit respiration, was unable to rescue the growth defect in SOD1 KO cells. In contrast, SOD1 K122E, which is active at ROS scavenging but defective at inhibition of respiration, showed an intermediate rescue of growth. JC-1 staining for mitochondrial membrane potential showed a similar trend, with a dramatic loss of membrane potential, a hallmark of oxidative stress, in the SOD1 KO and G85R mutant addback lines and a partial rescue of mitochondrial function in K122E-expressing cells (Fig. 7D). To measure ROS levels directly within the mitochondria for each treatment, we used the MitoSOX mitochondrial ROS probe. Flow cytometric quantitation of mitochondrial ROS mirrored the proliferation and JC-1 results: WT SOD1 rescued the increase in mitochondrial ROS levels caused by SOD1 KO, and the K122E mutant showed a partial rescue (Fig. 7E). Representative confocal images of this experiment are shown in Fig. 7F. Together, these data suggest that SOD1-mediated dampening of respiration contributes to SOD1-mediated cell survival. Possible mechanisms to explain this effect are discussed below.
DISCUSSION
We began this study with several affinity purification-based proteomics approaches to identify PTMs across multiple tissues with the primary goal of finding new mechanisms of regulation on cell survival signaling nodes. All proteomics data from this effort are freely available (see the link in Materials and Methods). By using the machine-learning approach of SAPH-ire (2), we prioritized several SOD1 PTMs for future study. Notably, a phosphorylation at S98 (or S99, depending on the numbering scheme) showed the highest SAPH-ire score. This phosphorylation sits within a solvent-exposed beta sheet of SOD1 and has been shown to regulate the role of SOD1 in transcription (18). It is also notable that seven SOD1 PTMs were grouped within a high–SAPH-ire–score cluster, and all sit within a relatively small region spanning S98 to K128, which includes part of the electrostatic loop.
We focused our initial studies on an acetylation within the electrostatic loop of SOD1 at K122. This lysine was intriguing for several reasons. First, of the acylated lysines within the top SAPH-ire cluster, it had the highest number of independent MS identifications (Fig. 2A). Second, it was conserved across multicellular eukaryotes and positioned within a known functional domain of SOD1. Third, we had previously identified this lysine as a sirtuin substrate by use of a biotin switch approach in Xenopus egg extracts (22), so its appearance in our proteomics data from mouse tissue was striking. Finally, during the course of this project, Lin and colleagues confirmed the site as succinylated (29), which corroborated the lysine as a potentially important site of regulation, since lysines that are acetylated are also commonly succinylated (26).
Two observations shed light on the potential mechanism by which acylation at K122 inhibits SOD1-mediated suppression of respiration. First, we found that the ROS-scavenging-impaired SOD1 G85R mutant fails to suppress respiration, which agrees with data from yeast that ROS-scavenging enzymatic activity is required for this new SOD1 function (20). Second, we found that the acyl-mimicking (K122E and K122Q) mutants of SOD1 fail to suppress respiration yet have fully functional ROS-scavenging enzymatic activity, which we confirmed through multiple assays. In aligning these two observations, we reasoned that acylation at K122 may disrupt a protein-protein interaction critical for suppressing respiration. Based on data from yeast, a clear candidate SOD1 interactor is casein kinase 1 gamma (CK1γ) (20). Unlike its yeast counterpart, human CK1γ has no clear analogous C-terminal degron, and we were unable to see the interaction by coimmunoprecipitation in human cells (due to poor specificity of antibodies). However, there are several isoforms of human casein kinase, and interestingly, a sequence roughly matching the yeast CK1γ degron exists in the alpha isoform. Ongoing investigations are exploring these possibilities. Furthermore, we attempted unbiased coimmunoprecipitation proteomics to identify proteins that interacted differentially with SOD1 WT versus the K122E mutant, but these provided strikingly few proteins (data not shown). Consultation with others in the SOD1 field revealed that this appears to be a fairly common observation and may indicate the need for cross-linking or other approaches to stabilize SOD1-interacting proteins for capture. Nevertheless, our data support the idea that K122 acylation disrupts a SOD1 interaction that is critical for the suppression of respiration, and our future efforts will focus on this mechanism.
In searching for a mechanism to explain the ability of K122 acylation to regulate the antirespiratory activity of SOD1, we found that acyl-mimicking SOD1 mutants differed from WT SOD1 in that they failed to accumulate in mitochondria. This result initially led us to think that the acyl-K122 mutants failed to suppress respiration because they were unable to accumulate within the IMS, where we presumed they were directly inhibiting the ETC. However, we found that forcing the K122E mutant into the IMS with an IMS-targeting tag failed to recover its ability to suppress respiration. In addition, the IMS-targeting tag did not confer additional respiration-suppressing activity on WT SOD1. Nevertheless, this initially surprising result actually agrees with data from Reddi and Culotta showing that yeast SOD1 suppresses mitochondrial respiration from the cytosol by interacting with and stabilizing CK1γ (20).
Our observation that forcing the K122E mutant back into the mitochondria failed to recover its ability to suppress respiration caused us to reconsider how SOD1 acylation and IMS accumulation could be linked. Previous work suggests that the import of SOD1 into the IMS depends on an Erv1/Mia40 disulfide relay system (reviewed in reference 41). The activity of this disulfide system is dependent on the oxidation of Erv1 by oxidized cytochrome c (47, 48). Therefore, under conditions in which respiration is highly active, the ratio of reduced to oxidized cytochrome c should increase, leading to a corresponding decrease in Erv1 oxidation and SOD1 mitochondrial import. Conversely, a drop in respiration (causing a buildup of oxidized cytochrome c) would increase SOD1 import into the IMS. In support of this model, we found that inhibiting respiration led to the accumulation of SOD1 in mitochondrial fractions, while boosting respiration with galactose caused a decrease in mitochondrial SOD1 levels. Similar results have been observed in astrocytes (49). Taking these findings together with the IMS phenotype of the acyl-SOD1 mutants, we favor the model that SOD1 acylation is upstream of SOD1 import into the IMS. Acylation of SOD1 blocks its antirespiratory function, in turn decreasing its import into the IMS. Conversely, we posit that the ability of SOD1 to suppress respiration feeds forward to drive its own IMS import and that acylation may serve as a brake on this mechanism.
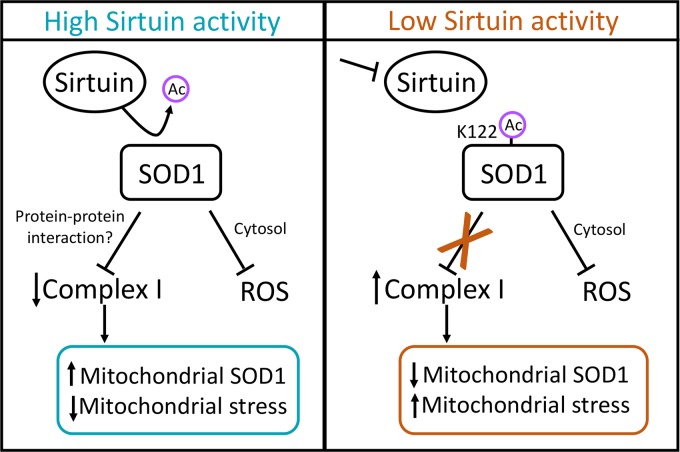
In the bigger picture, an intriguing implication of this model is a potentially expanded view of SOD1-mediated antioxidant defense: that the suppression of respiration mediated by SOD1 contributes to its antioxidant/prosurvival function (model in Fig. 8). This could occur through at least two possible mechanisms. (i) Inhibition of respiration drives SOD1 into the mitochondria, placing SOD1 directly in position to scavenge ROS as it leaks from complex III. In support of this idea, elevated mitochondrial SOD1 levels increase cell viability in a yeast system (46), and mitochondrially targeted SOD1 rescues the loss of mitochondrial density observed in SOD1-null mice (45). This model is intriguing, since a variety of stresses, such as ischemia/reperfusion and cell toxins, place strain on mitochondrial respiration and are known to cause spikes in mitochondrial ROS (50). (ii) Alternatively, the dampening of respiration at complex I by SOD1 may inhibit ROS production directly. Interestingly, genetic inhibition of respiration partially rescues the loss of viability caused by SOD1 deletion in yeast (51). Furthermore, metformin is known to suppress mitochondrial ROS via inhibition of complex I (52–54). Exactly how this may occur is more puzzling, since our understanding of ROS generation from the ETC, and complex I in particular, is limited. Prevailing dogma suggests that complex I only releases ROS into the matrix, but depending on the point at which complex I is inhibited, this inhibition could dampen the reduction of semiquinones or Fe-S clusters that may generate O2− elsewhere (55), or it may simply reduce the flow of electrons to complex III, a key producer of ROS within the IMS (50).
FIG 8.
Sirtuin activity model. High levels of sirtuin activity (SIRT5 for SOD1 succinylation) deacylate SOD1, which activates the respiration-suppressing activity of SOD1 at complex I. We posit that SOD1-mediated inhibition of respiration contributes to SOD1-mediated cell survival through two potential (non-mutually exclusive) mechanisms: (i) inhibition of respiration increases the mitochondrial pool of SOD1, placing SOD1 directly in position to scavenge ROS leaking into the IMS; (ii) dampening of respiration may directly reduce the production of ROS from the ETC.
We found that the impaired growth of SOD1-null cells, known to be caused by accumulation of oxidative damage (7, 8, 14, 51, 56), could be only partially rescued by the SOD1 K122E mutant, which is catalytically active against ROS but defective at inhibiting respiration. Taken together, our data suggest that SOD1-mediated inhibition of respiration, while clearly not the entire picture, may contribute to SOD1-mediated cell survival. Furthermore, the link between IMS import of SOD1 and respiratory activity suggests a feedback loop in which additional SOD1 may be recruited into mitochondria under conditions (e.g., mitochondrial damage leading to ROS production) in which reinforced ROS scavenging would be needed.
In summary, our data identify, to our knowledge, the first acylation on SOD1 that regulates its ability to suppress mitochondrial respiration. Given that sirtuin activity is linked to NAD+ levels, which, in turn, are linked to the overall metabolic state of the cell, SOD1 acylation may act as a sensor to link nutrient metabolism to SOD1-mediated suppression of respiration. As described above, this may provide an additional dimension to SOD1 antioxidant defense. These data raise additional questions. Does SOD1 acylation disrupt a protein-protein interaction that transmits the signal to complex I? Given that our data suggest that dampened respiration promotes SOD1 mitochondrial import, could the toxic mitochondrial accumulation of ALS-linked SOD1 mutants in neurons be caused by an underlying perturbation of mitochondrial metabolism in a feed-forward loop? Answers to these and related questions will help shed light on the increasingly complex picture of SOD1 biology.
MATERIALS AND METHODS
Mass spectrometry and PTM affinity purifications.
Trypsin-digested cell lysates from mouse embryo, brain, and liver were prepared by lysis in 8 M urea, normalized to approximately 2 mg/ml and 1.8 M urea, reduced with 10 mM dithiothreitol, alkylated with 20 mM iodoacetamide, and digested with tosylsulfonyl phenylalanyl chloromethyl ketone (TPCK) trypsin at 1:25 (wt/wt). Peptides were desalted with C18 solid-phase extraction (SPE) cartridges (Waters) and were lyophilized to dryness. By use of the PTMScan product line (Cell Signaling Technologies), aliquots (approximately 5 mg) of each lysate digest were enriched for posttranslational modifications including a series of phosphospecific motifs (AKT, mitogen-activated protein kinase [MAPK], phosphothreonine, or pY), as well as ubiquitin remnant (GG-K) or acetyl-lysine (Kac), as described previously (57–59). Titanium dioxide enrichment was performed as described previously (59) for comparison.
After immunoaffinity or titanium dioxide enrichment, peptides were analyzed using data-dependent acquisition (DDA) by a 3-fraction two-dimensional LC–MS-MS (LC-LC–MS-MS) method as described previously (60). Samples were resuspended in 12 μl of trifluoroacetic acid (TFA)-methyl cyanide (MeCN)-water (1/2/97, vol/vol/vol). Phosphopeptide samples additionally contained 20 mM citric acid. Approximately half of each sample was analyzed on a 2D nanoAcquity ultraperformance liquid chromatography (UPLC) system coupled to a Synapt G2 high-definition mass spectrometer (HDMS) (Waters Corporation). Searchable files (*.mgf) were generated from raw data in Mascot Distiller, and database searching for peptide identification (ID) was carried out with the Mascot search engine, v2.4 (Matrix Science, Inc.). The UniProt database (www.uniprot.org) with Mus musculus taxonomy and reviewed status was utilized, along with 10 ppm precursor and 0.04 Da product ions. A fixed modification of Cys with carbamidomethylation was required, and variable modifications included deamidation of NQ and oxidation of M, along with acetylated K, ubiquitin remnant, or phosphorylated S/T/Y. Full tryptic specificity was required. Data were curated in Scaffold, v4.7, using the PeptideProphet algorithm, to a 0.05% false discovery rate (FDR) for peptides and a 0.4% FDR for proteins. The Scaffold file is available for download at https://discovery.genome.duke.edu/express/resources/3023/3023_PTMScanAll_withTiO2.sf3. All mouse experiments were IACUC approved.
SAPH-ire.
SAPH-ire FPx was employed as described previously (2), with modifications for the domain-specific analysis of the superoxide dismutase (SOD) copper-zinc binding domain (InterPro ID IPR001424). FPx scores were calculated using the structure-free 8-factor neural network model as described previously (1). The SOD domain sequences for all eukaryotic proteins were obtained from Pfam (61). The multi-FASTA files generated for the domain family were subsequently aligned using MUSCLE with default parameters (62). The domain data set for SOD contained 79 unique sequences, within which 77 distinct PTMs could be retrieved from the public domain. PTMs coalesced into 34 modified-alignment positions (MAPs) in the family, the features of which were used for SAPH-ire FPx scoring.
Cell culture and reagents.
HEK293T cells were purchased from the ATCC. The Flp-In T-REx 293 host cell line was purchased from Thermo Fisher Scientific (catalog no. R78007). The Flp-In T-REx HCT-116 host cells were developed by Stephen Taylor (63). All cell lines were cultured in Dulbecco's modified Eagle's medium (DMEM; product no. 11965; Gibco) supplemented with 10% fetal bovine serum (FBS) at 37°C in a 5% CO2 incubator.
Antimycin A (catalog no. A8674), 3-nitropropionic acid (3-NP) (catalog no. N5636), galactose (catalog no. G5388), tetracycline (catalog no. T7660), and doxycycline (catalog no. D3447) were purchased from Sigma. Rotenone was purchased from Millipore (catalog no. 557368). Drugs used in cell culture were first dissolved in the recommended solvent and then diluted in cell culture medium to the final concentrations indicated elsewhere in Materials and Methods and in the figure legends.
Plasmid transfections and siRNA.
Cells were transfected with empty pcDNA3.1 vector or the same vector expressing affinity-tagged SOD1, with polyethylenimine (PEI; Polysciences Inc.) transfection reagent in a ratio of 1 μg DNA to 3 to 4 mg PEI. Site-directed mutants of FLAG-SOD1 were generated using the Agilent QuikChange II XL kit (200521) or the Q5 site-directed mutagenesis kit from New England BioLabs, Inc. (catalog no. E0554). Plasmids and selection reagents for Flp-In T-REx host cell lines were obtained from Thermo Fisher Scientific. Lipofectamine RNAiMAX reagent was purchased from Invitrogen (catalog no. 13778100). SMARTpool small interfering RNA (siRNA) against SIRT5 was obtained from GE Dharmacon (catalog no. M013448010005). The cytochrome b2 SOD1 cDNA was obtained from Giovanni Manfredi.
Generation of Flp-In T-REx cell lines.
The generation of T-REx cell lines with Flp recombinase-mediated gene insertion allows for tight control of the expression of a gene of interest at a single, actively transcribed genomic locus by the addition or removal of tetracycline or its derivatives. For further explanation, refer to the manufacturer's manual (catalog no. R78007; Thermo Fisher Scientific).
T-REx host cells were transiently transfected with pOG44 and pcDNA5/FRT/TO-FLAG-SOD1, including WT SOD1 and acyl-mimicking mutants, at a pOG44/pcDNA5 ratio of 9:1. Constructs were cotransfected into cells in 6-well plates when cells were approximately 60% confluent. After 48 h, cells were trypsinized, transferred to 15-cm cell culture dishes, and cultured in complete medium containing 150 μg/ml of hygromycin B selection reagent (catalog no. 10687010; Gibco). After 10 days, single colonies were transferred to individual wells of a 24-well plate and were cultured in medium without selection reagent. Plates were replicated, and expression was induced for 24 h with 1 μg/ml tetracycline in one set of the replicates, after which cells were harvested, lysed, and subjected to Western blot analysis as described below. Cells positive for the FLAG epitope were selected, and cells that had the lowest background expression when not induced, but still exhibited high inducible expression with tetracycline, were selected for use in experiments. For experiments, expression was induced with either tetracycline or doxycycline at a final concentration of 100 ng/ml.
Antibodies.
Primary mouse monoclonal anti-HA (catalog no. 7392), rabbit polyclonal anti-FLAG (catalog no. 807), and mouse monoclonal and rabbit polyclonal anti-beta-actin (catalog no. 8432 and 1616) antibodies were purchased from Santa Cruz Biotechnology. Primary mouse monoclonal anti-FLAG (catalog no. 8146), rabbit polyclonal anti-FLAG (catalog no. 2368), mouse monoclonal and rabbit polyclonal anti-human SOD1 (catalog no. 4266 and 2770), mouse monoclonal anti-HA (catalog no. 2367), rabbit monoclonal anti-SIRT5 (catalog no. 8779), and rabbit polyclonal anti-VDAC (anti-voltage-dependent ion channel) (catalog no. 4661) antibodies were purchased from Cell Signaling Technology, Inc. Primary mouse monoclonal anti-GAPDH (anti-glyceraldehyde-3-phosphate dehydrogenase) and rabbit monoclonal anti-MEK1/2 antibodies were purchased from Abcam (catalog no. 9484 and 178876). Anti-mouse and anti-rabbit secondary antibodies conjugated with IRDye 680RD and IRDye 800CW were purchased from Li-Cor (catalog no. 92668070, 92632210, 92668071, and 92632213).
For succinyl-Lys122 antibody purification, rabbits were immunized against the succinylated-SOD1 peptide (LVVHE-[succinyl-K]-ADDLGC), and serum was collected. The succinyl-Lys122 polyclonal antibody was immunopurified out of serum using a succinylated Lys122 SOD1 peptide as reported previously (64). In brief, the biotinylated (nonsuccinyl) Lys122 peptide was conjugated to streptavidin-agarose (catalog no. 20359; Thermo Scientific) and was gently rotated at 4°C overnight with rabbit serum diluted 1:1 in equilibration buffer (150 mM Tris [pH 7.5], 20 mM NaCl) to deplete nonsuccinyl Lys122-specific antibody from the serum. Following incubation, the serum was transferred to a column containing the succinylated Lys122 peptide conjugated to streptavidin resin and was again incubated overnight with gentle rotation. The resin was thereafter washed twice with equilibration buffer. The succinyl Lys122-specific antibody was then eluted from the resin with elution buffer (0.1 M glycine-HCl [pH 2.8]) by gravity elution. Fractions were then collected in microcentrifuge tubes and were neutralized with neutralization buffer (1 M Tris-HCl [pH 8.5]). The reactivity of each fraction was tested via Western blotting for the antibody titer.
The primary anti-human SOD1 succinyl-Lys122 antibody serum, biotinylated SOD1 succinyl-Lys122 peptide, and biotinylated SOD1 nonsuccinyl Lys122 peptide were produced by Eton Bioscience, Inc.
Western blotting and immunoprecipitation.
Cells were seeded in 10-cm or 6-well plates to 15% confluence. After 24 h, at about 30% confluence, T-REx cells were induced, or non-T-REx cells were either mock transfected or transfected with a SOD1 expression vector or an empty vector. After 8 h, cell culture medium was replaced with complete medium, and cells were incubated at 37°C in a cell culture incubator until 48 h posttransfection. Cells receiving drug treatment were treated within this incubation time. Cells treated with RNA interference were transfected with 100 nM SIRT5-directed or scrambled siRNA with Lipofectamine RNAiMAX. The knockdown was repeated to ensure efficient depletion of SIRT5. Then the cells were allowed to grow for 24 to 48 more hours. Following transfection and incubation, cells were washed twice and were harvested with ice-cold phosphate-buffered saline (PBS). Cells were then lysed in ice-cold coimmunoprecipitation buffer (10 mM HEPES [pH 7.5], 150 mM KCl, 0.1% NP-40) or in TNTE coimmunoprecipitation buffer (20 mM Tris [pH 7.8], 150 mM NaCl, 0.3% [wt/vol] Triton X-100, 5 mM EDTA), supplemented with protease inhibitors (catalog no. 88665; Pierce). Lysates were then cleared by centrifugation at 21,000 × g for 10 min at 4°C. The lysate protein concentration was determined by a DC protein determination assay (catalog no. 5000116; Bio-Rad). The lysate was either saved for analysis or incubated with 15 μl of anti-FLAG-agarose resin (catalog no. 2220; Sigma). Protein was eluted from the resin by incubation with modified Laemmli buffer for 5 min at 100°C. Lysates were likewise mixed and boiled in modified Laemmli buffer. Samples were then loaded onto SDS-PAGE gels. A BLUEstain protein ladder (catalog no. P007; Gold Bio) was used as a molecular weight reference. Gels were then transferred to a polyvinylidene difluoride (PVDF) membrane and were immunoblotted for proteins of interest. Proteins were visualized, and bands were quantified, using the Li-Cor Odyssey Classic or CLx imaging system and the Image Studio software package.
SOD1 activity assays.
For the in vitro colorimetric activity assay, HEK293T cells were either transfected with a FLAG-SOD1 plasmid or mock transfected. Cells were harvested and lysed, and FLAG was immunoprecipitated with FLAG epitope-conjugated agarose beads, after which the resin was washed three times in cold PBS. FLAG-SOD1 was then competitively eluted from the beads by incubation in a 200-ng/μl solution of purified FLAG peptide (ApexBio) with gentle shaking for 10 min at 4°C. Beads were then centrifuged at 8,200 × g for 30 s at 4°C. The FLAG-SOD1-containing supernatant was transferred to a new microcentrifuge tube; the elution was repeated; and the supernatant was added to the same tube. With the purified SOD1 or mock supernatants, SOD1 ROS scavenging activity was indirectly measured using a SOD assay kit purchased from Sigma (catalog no. 19160) according to the manufacturer's recommendations.
For the in situ SOD1 activity gel, purified FLAG-SOD1 was produced as in the colorimetric assay. SOD1 ROS scavenging activity was measured indirectly as a competitive reaction of superoxide with nitroblue tetrazolium (NBT) dye as described previously (65). Briefly, a portion of the purified SOD1 supernatants was mixed with 5× SOD loading dye (25 mM Tris-HCl [pH 6.8], 50% glycerol, 0.5% bromophenol blue) to a final concentration of 1× loading dye. Samples were then loaded onto an 8% native PAGE gel, and SOD1 was resolved at a constant 80 V until the loading dye reached the end of the gel. The gel apparatus was maintained at 4°C. The gel was then moved to a shallow container, immersed in 50 ml of staining solution (45 mM K2HPO4, 4.6 mM KH2PO4, 0.163 mM NBT, 0.266 mM riboflavin), and covered with foil. A 50-μl volume of N,N,N′,N′-tetramethylethylenediamine (TEMED) was then added, and the gel was incubated with gentle shaking for 1 h in the dark. Following incubation, the staining solution was removed, and the gel was first washed twice with distilled water and then immersed and incubated in distilled water with gentle shaking overnight, with exposure to ambient light. Clear bands on the blue-stained gel were interpreted as active SOD1. NBT (catalog no. N6639), K2HPO4 (catalog no. P3786), KH2PO4 (catalog no. P0662), and riboflavin (catalog no. R9504) were purchased from Sigma.
Cell survival assay.
Single-guide RNA (sgRNA) against SOD1 was cloned into the pSpCas9(BB)-2A-Puro (PX459) vector. PX459 was a gift from Feng Zhang (plasmid 48139; Addgene) (66). Human codon-optimized Cas9 from Streptococcus pyogenes and the sgRNA were expressed from the human cytomegalovirus (CMV) immediate early promoter and the human U6 promoter, respectively. The 20-bp SOD1-targeted sgRNA contained the sequence TTGCATCATTGGCCGCACAC with an NGG protospacer adjacent motif (PAM) site of TGG. Knockout efficiency was measured via Western blot analysis. The construct was transfected into T-REx HCT-116 cells. After 8 h, the transfection medium was removed and replaced with DMEM complete medium. Twenty-four hours posttransfection, the cell culture medium was replaced with DMEM complete medium containing 3 μg/ml puromycin selection reagent. After 48 h, the selection medium was removed and replaced with DMEM complete medium containing 100 ng/ml doxycycline. The doxycycline medium was replenished every 48 h, and cells were trypsinized as needed. Cells were allowed to grow until there were sufficient cells for the assays, after which cells were trypsinized and counted.
Growth assay with Giemsa stain.
For Giemsa staining, 22,500 cells per well were seeded into 6-well plates with 100 ng/ml doxycycline and were allowed to grow for 5 days. Cells were then washed twice with ice-cold PBS and were fixed for 30 min in ice-cold methanol at 4°C, after which the methanol was aspirated, and a 1:20 mixture of Giemsa stain (catalog no. 48900; Fluka Analytical) and PBS was added to cells, which were then incubated at room temperature for 45 min. Following staining, cells were washed three times with distilled water and were allowed to air dry. Wells were imaged and quantified using the Li-Cor Odyssey Classic imaging system and the Image Studio software package.
IncuCyte zoom analysis.
For IncuCyte analysis of control cells without SOD1 knockout, 12,500 cells per well were seeded into 12-well plates. For the SOD1 knockout/addback cells, 18,000 cells per well were seeded into 24-well plates. Plates were put into the IncuCyte system 1 day after seeding, and cell confluence was monitored via basic phase-contrast analysis for 80 to 120 h. Cell confluence was given as the mean for 4 wells per sample, with 16 images per well. Error bars indicate the standard errors of the means (SEM).
Cellular respiration.
HEK293T cells either were transiently transfected with 8 μg of a SOD1 plasmid or empty pcDNA3.1 or were mock transfected. Eight hours later, the cell culture medium was replenished, and cells were allowed to grow for a total of 48 h. For rotenone treatment, 1 nM rotenone or a vehicle was added to HEK 293 cells 30 min prior to measurement of oxygen consumption rates and was incubated at 37°C under a 5% CO2 atmosphere. Cells were then detached in culture dishes with 0.05% trypsin–EDTA (Gibco), and growth medium was added to the culture. Contents were transferred to a 15-ml conical tube and were centrifuged at room temperature. After removal of the supernatant, the cells were permeabilized and were prepared for the O2K analyzer (Oroboros Instruments, Innsbruck, Austria), and O2 flux was measured as described previously by Hodson et al. (67) except for a few changes outlined below. The data presented are taken from maximal O2 flux measurements, in which the full electron transport system capacity of oxidative phosphorylation was measured after the addition of carbonyl cyanide p-trifluoromethoxy-phenylhydrazone (FCCP). For the complex I and complex II inhibition assays, rotenone (2 mM) and malonate (2.5 mM) were added prior to O2 flux measurements. After respiration was measured, cellular protein concentrations from the same cells were determined as described in “Western blotting and immunoprecipitation” above. For the rotenone-treated cells, 2 million cells were counted and assayed in the Oroboros O2K analyzer.
Endogenous immunoprecipitation.
Endogenous immunoprecipitation of SOD1 was performed using the Dynabeads coimmunoprecipitation kit purchased from Thermo Fisher Scientific (catalog no. 14321D) according to the manufacturer's recommendations. To pull down SOD1, 7.5 mg of magnetic epoxy beads was conjugated to 37.5 μl of SOD1 antibody per sample. As an IgG control for SOD1 immunoprecipitation, primary anti-Chk1 was conjugated to magnetic beads.
Mitochondrial enrichment.
All mitochondrial isolations were carried out with the Mitochondria Isolation Kit for Cultured Cells, obtained from Thermo Scientific (catalog no. 89874). Mitochondrial pellets were isolated according to the manufacturer's protocol.
SOD1 aggregation assay.
T-REx HEK 293 cells with WT, G93A, or G93A K122E SOD1 were plated at 20% confluence in two 15-cm plates per sample. Tetracycline was added to the culture medium to induce expression of the SOD1 constructs, and cells were allowed to express SOD1 for 48 h. Tetracycline-containing medium was replenished after 24 h due to tetracycline instability. Cells were washed twice with ice-cold PBS and were removed from the plate with a cell scraper, and cells from the two 15-cm plates in each set were combined (e.g., cells from the two WT SOD1 plates were combined) to a total of three samples. Cells were lysed in 600 μl of TNTE coimmunoprecipitation buffer containing protease inhibitors by gentle rotation at 4°C for 25 min. Following incubation, cells were passaged 10 times through a 25-gauge needle. Lysates were cleared by centrifugation at 21,000 × g for 10 min at 4°C. Five hundred microliters of lysate was loaded onto a Superdex 200 10/300 GL size exclusion column, separated, and eluted with PBS on an ÄKTA pure fast protein liquid chromatography (FPLC) system (GE Healthcare). Lysates were fractionated at a rate of 0.5 ml/min into 48 fractions. Fractions were resolved via SDS-PAGE on a 15% gel and were immunostained for FLAG and endogenous SOD1.
Mitochondrial membrane potential assay.
Mitochondrial membrane potential in cells (the same cells as those used for the cell survival assay, described above) was assayed with the JC-1 mitochondrial membrane potential assay kit (catalog no. 113850; Abcam) based on an adaptation of the manufacturer's instructions. Specifically, for each sample, the medium was collected and was saved in a 12- by 75-mm polystyrene test tube. Cells were washed once with PBS, which was added to the corresponding test tube. Following the wash, cells were trypsinized and were collected into the same tube. Tubes were capped and were centrifuged at 1,000 × g for 5 min. The supernatant was discarded, and the cells were resuspended in 1 ml of warm complete medium. A 10-μl volume of 200 μM JC-1 in dimethyl sulfoxide (DMSO) was then added to each sample. No stain was added to a separate sample, as a control. After the addition of the stain, the samples were protected from light and were incubated at 37°C for 20 min, followed by a single wash with 2 ml of PBS. Cells were resuspended in 500 μl of PBS and were analyzed on an Accuri C6 flow cytometer. Emission spectra from excitation at 488 nm were detected with the FL-1 and FL-2 emission filters. Standard compensation was carried out based on control Cas9 cells.
Mitochondrial ROS determination.
The relative number of ROS-containing cells (the same cells as those used for the cell survival assay) was determined by a flow cytometry-based application of MitoSOX Red mitochondrial superoxide indicator (catalog no. M36008; Life Technologies). After cells were grown to 80% confluence in 12-well plates, 10 mM antimycin A was added to the positive-control cells to a final concentration of 30 μM, and cells were incubated for 40 min in a 5% CO2 incubator at 37°C. Following incubation, the cell medium was collected and was saved in a 12- by 75-mm polystyrene test tube. Cells were washed once with PBS, which was added to the corresponding test tube. Following the wash, cells were trypsinized and were collected into the same tube. Tubes were capped and were centrifuged at 600 × g for 3 min. The supernatant was discarded, and cells were washed once in 1 ml of warm complete medium. To stain cells, 5 mM MitoSOX Red was prepared in DMSO as recommended by the manufacturer and was then diluted to 5 μM in complete medium. All samples were resuspended in 1 ml of the 5 μM MitoSOX Red-containing medium, except for the negative control, which was resuspended in 1 ml of complete medium. After the addition of the stain, the samples were protected from light and were incubated for 10 min at 37°C under 5% CO2, followed by a 3-min centrifugation at 600 × g and a single wash with 1 ml PBS. Cells were resuspended in 1 ml of PBS, passed through a 70-μm cell strainer, diluted to approximately 500,000 cells/ml, and analyzed on either an Accuri C6 or an Attune acoustic focusing cytometer (Life Technologies). Emission spectra from excitation at 488 nm were detected with the BL-3 or FL-3 emission filter.
Live-cell confocal imaging.
Cells (the same as those used for the cell survival assay) were seeded at medium density into 35-mm glass-bottom microwell dishes (part no. P35GC-1.5-10-C; MatTek) and were allowed to adhere for 24 h. Prior to staining, positive-control cells were treated for 40 min with 30 μM antimycin A. To stain cells, MitoSOX Red was prepared as for the ROS determination assay, cells were washed once with warm complete medium, and 1 ml of MitoSOX medium was added to each plate, except for the negative control (1 ml complete medium). Cells were incubated protected from light for 10 min at 37°C under a 5% CO2 atmosphere, after which they were washed three times with warm complete medium. Nuclei were counterstained with 1 μg/ml Hoechst 33342 (catalog no. 62249; Pierce) according to the manufacturer's protocol, using a 10-min incubation time, followed by a PBS wash. Cells were then treated for at least 45 min with 1 ml of ProLong Live antifade reagent for live-cell imaging (catalog no. P36975; Molecular Probes) at 1:100 in complete DMEM. Finally, at the time of imaging, the plate was transferred to a humidified 5% CO2, 37°C minichamber adapted to the microscope stage.
Cells were imaged on a Leica (Buffalo Grove, IL, USA) TCS SP8 HyD microscope using the resonance scanning mode. Images were collected from a 63× oil objective. Data collection parameters were determined based on the fluorescence signal from the positive control. Images were oversampled and deconvolved with the Huygens Essential software package, v16.10 (Scientific Volume Imaging, The Netherlands), after which strong automatic dye separation was applied. All samples and images were treated identically.
Statistical analysis.
Statistical analysis was conducted either with one-way analysis of variance (ANOVA) with Tukey's post hoc test or with an unpaired, equal-variance, two-tailed t test. Where indicated, SEM were determined after the normalization of each replicate.
ACKNOWLEDGMENTS
We thank Giovanni Manfredi and Dennis Winge for guidance in SOD1 biology. We thank Jonathan Alder and J. C. Price for excellent technical help and key reagents. We thank Cell Signaling Technology, Inc., specifically Jeff Silva and Charles Farnsworth, for providing the PTM antibody resins for proteomics. We thank all members of the Andersen laboratory for constructive discussion and analysis of data.
We thank the Fritz B. Burns Foundation (gift to J.L.A.), the Simmons Center for Cancer Research (fellowships to C.J.B. and N.W.R.), and the National Institutes of Health (grants R15CA202619 to J.L.A. and R01GM117400 to M.P.T.) for funding.
We declare that we have no conflicts of interest with the contents of this article.
C.J.B. and N.W.R. conceived/performed experiments and cowrote the manuscript. K.R.G. performed experiments. J.B.M. and M.D.W. helped initiate the project and provided key laboratory support. R.R.P. and M.P.T. performed SAPH-ire analysis. B.T.B. and J.S.T. provided support with metabolic assays. E.J.S., J.W.T., and M.A.M. executed proteomics experiments. A.R.R. provided guidance throughout the course of the project. J.L.A. conceived experiments and cowrote the manuscript.
REFERENCES
- 1.Dewhurst HM, Torres MP. 2017. Systematic analysis of non-structural protein features for the prediction of PTM function potential by artificial neural networks. PLoS One 12:e0172572. doi: 10.1371/journal.pone.0172572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Torres MP, Dewhurst H, Sundararaman N. 2016. Proteome-wide structural analysis of PTM hotspots reveals regulatory elements predicted to impact biological function and disease. Mol Cell Proteomics 15:3513–3528. doi: 10.1074/mcp.M116.062331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.McCord JM, Fridovich I. 1969. Superoxide dismutase: an enzymic function for erythrocuprein (hemocuprein). J Biol Chem 244:6049–6055. [PubMed] [Google Scholar]
- 4.Reddi AR, Jensen LT, Naranuntarat A, Rosenfeld L, Leung E, Shah R, Culotta VC. 2009. The overlapping roles of manganese and Cu/Zn SOD in oxidative stress protection. Free Radic Biol Med 46:154–162. doi: 10.1016/j.freeradbiomed.2008.09.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sanchez RJ, Srinivasan C, Munroe WH, Wallace MA, Martins J, Kao TY, Le K, Gralla EB, Valentine JS. 2005. Exogenous manganous ion at millimolar levels rescues all known dioxygen-sensitive phenotypes of yeast lacking CuZnSOD. J Biol Inorg Chem 10:913–923. doi: 10.1007/s00775-005-0044-y. [DOI] [PubMed] [Google Scholar]
- 6.Elchuri S, Oberley TD, Qi W, Eisenstein RS, Jackson Roberts L, Van Remmen H, Epstein CJ, Huang TT. 2005. CuZnSOD deficiency leads to persistent and widespread oxidative damage and hepatocarcinogenesis later in life. Oncogene 24:367–380. doi: 10.1038/sj.onc.1208207. [DOI] [PubMed] [Google Scholar]
- 7.Blander G, de Oliveira RM, Conboy CM, Haigis M, Guarente L. 2003. Superoxide dismutase 1 knock-down induces senescence in human fibroblasts. J Biol Chem 278:38966–38969. doi: 10.1074/jbc.M307146200. [DOI] [PubMed] [Google Scholar]
- 8.Phillips J, Campbell S, Michard D, Charbonneau M, Hilliker AJ. 1989. Null mutation of copper/zinc superoxide in Drosophila confers hypersensitivity to paraquat and reduced longevity. Proc Natl Acad Sci U S A 86:2761–2765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Corson LB, Folmer J, Strain JS, Culotta VC, Cleveland DW. 1999. Oxidative stress and iron are implicated in fragmenting vacuoles of Saccharomyces cerevisiae lacking Cu, Zn superoxide dismutase. J Biol Chem 274:27590–27596. doi: 10.1074/jbc.274.39.27590. [DOI] [PubMed] [Google Scholar]
- 10.McNaughton RL, Reddi AR, Clement MH, Sharma A, Barnese K, Rosenfeld L, Gralla EB, Valentine JS, Culotta VC, Hoffman BM. 2010. Probing in vivo Mn2+ speciation and oxidative stress resistance in yeast cells with electron-nuclear double resonance spectroscopy. Proc Natl Acad Sci U S A 107:15335–15339. doi: 10.1073/pnas.1009648107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Culotta VC, Joh HD, Lin SJ, Slekar KH, Strain J. 1995. A physiological role for Saccharomyces cerevisiae copper/zinc superoxide dismutase in copper buffering. J Biol Chem 270:29991–29997. doi: 10.1074/jbc.270.50.29991. [DOI] [PubMed] [Google Scholar]
- 12.Papa L, Manfredi G, Germain D. 2014. SOD1, an unexpected novel target for cancer therapy. Genes Cancer 5:15–21. doi: 10.18632/genesandcancer.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Palomo GM, Manfredi G. 2015. Exploring new pathways of neurodegeneration in ALS: the role of mitochondria quality control. Brain Res 1607:36–46. doi: 10.1016/j.brainres.2014.09.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.De Freitas JM, Liba A, Meneghini R, Valentine JS, Gralla EB. 2000. Yeast lacking Cu-Zn superoxide dismutase show altered iron homeostasis. Role of oxidative stress in iron metabolism. J Biol Chem 275:11645–11649. [DOI] [PubMed] [Google Scholar]
- 15.Inoue E, Tano K, Yoshii H, Nakamura J, Tada S, Watanabe M, Seki M, Enomoto T. 2010. SOD1 is essential for the viability of DT40 cells and nuclear SOD1 functions as a guardian of genomic DNA. J Nucleic Acids 2010:795946. doi: 10.4061/2010/795946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Corson LB, Strain JJ, Culotta VC, Cleveland DW. 1998. Chaperone-facilitated copper binding is a property common to several classes of familial amyotrophic lateral sclerosis-linked superoxide dismutase mutants. Proc Natl Acad Sci U S A 95:6361–6366. doi: 10.1073/pnas.95.11.6361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wei JP, Srinivasan C, Han H, Valentine JS, Gralla EB. 2001. Evidence for a novel role of copper-zinc superoxide dismutase in zinc metabolism. J Biol Chem 276:44798–44803. doi: 10.1074/jbc.M104708200. [DOI] [PubMed] [Google Scholar]
- 18.Tsang CK, Liu Y, Thomas J, Zhang Y, Zheng XF. 2014. Superoxide dismutase 1 acts as a nuclear transcription factor to regulate oxidative stress resistance. Nat Commun 5:3446. doi: 10.1038/ncomms4446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wood LK, Thiele DJ. 2009. Transcriptional activation in yeast in response to copper deficiency involves copper-zinc superoxide dismutase. J Biol Chem 284:404–413. doi: 10.1074/jbc.M807027200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Reddi AR, Culotta VC. 2013. SOD1 integrates signals from oxygen and glucose to repress respiration. Cell 152:224–235. doi: 10.1016/j.cell.2012.11.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Choudhary C, Kumar C, Gnad F, Nielsen ML, Rehman M, Walther TC, Olsen JV, Mann M. 2009. Lysine acetylation targets protein complexes and co-regulates major cellular functions. Science 325:834–840. doi: 10.1126/science.1175371. [DOI] [PubMed] [Google Scholar]
- 22.Andersen JL, Thompson JW, Lindblom KR, Johnson ES, Yang CS, Lilley LR, Freel CD, Moseley MA, Kornbluth S. 2011. A biotin switch-based proteomics approach identifies 14-3-3ζ as a target of Sirt1 in the metabolic regulation of caspase-2. Mol Cell 43:834–842. doi: 10.1016/j.molcel.2011.07.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mortenson JB, Heppler LN, Banks CJ, Weerasekara VK, Whited MD, Piccolo SR, Johnson WE, Thompson JW, Andersen JL. 2015. Histone deacetylase 6 (HDAC6) promotes the pro-survival activity of 14-3-3ζ via deacetylation of lysines within the 14-3-3ζ binding pocket. J Biol Chem doi: 10.1074/jbc.M114.607580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dewhurst HM, Choudhury S, Torres MP. 2015. Structural analysis of PTM hotspots (SAPH-ire)—a quantitative informatics method enabling the discovery of novel regulatory elements in protein families. Mol Cell Proteomics 14:2285–2297. doi: 10.1074/mcp.M115.051177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rakhit R, Chakrabartty A. 2006. Structure, folding, and misfolding of Cu, Zn superoxide dismutase in amyotrophic lateral sclerosis. Biochim Biophys Acta 1762:1025–1037. doi: 10.1016/j.bbadis.2006.05.004. [DOI] [PubMed] [Google Scholar]
- 26.Weinert BT, Scholz C, Wagner SA, Iesmantavicius V, Su D, Daniel JA, Choudhary C. 2013. Lysine succinylation is a frequently occurring modification in prokaryotes and eukaryotes and extensively overlaps with acetylation. Cell Rep 4:842–851. doi: 10.1016/j.celrep.2013.07.024. [DOI] [PubMed] [Google Scholar]
- 27.Lin C, Zeng H, Lu J, Xie Z, Sun W, Luo C, Ding J, Yuan S, Geng M, Huang M. 2015. Acetylation at lysine 71 inactivates superoxide dismutase 1 and sensitizes cancer cells to genotoxic agents. Oncotarget 6:20578–20591. doi: 10.18632/oncotarget.3987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhao S, Xu W, Jiang W, Yu W, Lin Y, Zhang T, Yao J, Zhou L, Zeng Y, Li H, Li Y, Shi J, An W, Hancock SM, He F, Qin L, Chin J, Yang P, Chen X, Lei Q, Xiong Y, Guan KL. 2010. Regulation of cellular metabolism by protein lysine acetylation. Science 327:1000–1004. doi: 10.1126/science.1179689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lin Z, Xu H, Wang J, Lin Q, Ruan Z, Liu F, Jin W, Huang H, Chen X. 2013. SIRT5 desuccinylates and activates SOD1 to eliminate ROS. Biochem Biophys Res Commun 441:191–195. doi: 10.1016/j.bbrc.2013.10.033. [DOI] [PubMed] [Google Scholar]
- 30.Kaliszewski M, Kennedy AK, Blaes SL, Shaffer RS, Knott AB, Song W, Hauser HA, Bossy B, Huang TT, Bossy-Wetzel E. 2016. SOD1 lysine 123 acetylation in the adult central nervous system. Front Cell Neurosci 10:287. doi: 10.3389/fncel.2016.00287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Xiong Y, Guan KL. 2012. Mechanistic insights into the regulation of metabolic enzymes by acetylation. J Cell Biol 198:155–164. doi: 10.1083/jcb.201202056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hirschey MD, Zhao Y. 2015. Metabolic regulation by lysine malonylation, succinylation, and glutarylation. Mol Cell Proteomics 14:2308–2315. doi: 10.1074/mcp.R114.046664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Du J, Zhou Y, Su X, Yu JJ, Khan S, Jiang H, Kim J, Woo J, Kim JH, Choi BH, He B, Chen W, Zhang S, Cerione RA, Auwerx J, Hao Q, Lin H. 2011. Sirt5 is a NAD-dependent protein lysine demalonylase and desuccinylase. Science 334:806–809. doi: 10.1126/science.1207861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nishida Y, Rardin MJ, Carrico C, He W, Sahu AK, Gut P, Najjar R, Fitch M, Hellerstein M, Gibson BW, Verdin E. 2015. SIRT5 regulates both cytosolic and mitochondrial protein malonylation with glycolysis as a major target. Mol Cell 59:321–332. doi: 10.1016/j.molcel.2015.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Matsushita N, Yonashiro R, Ogata Y, Sugiura A, Nagashima S, Fukuda T, Inatome R, Yanagi S. 2011. Distinct regulation of mitochondrial localization and stability of two human Sirt5 isoforms. Genes Cells 16:190–202. doi: 10.1111/j.1365-2443.2010.01475.x. [DOI] [PubMed] [Google Scholar]
- 36.Rardin MJ, He W, Nishida Y, Newman JC, Carrico C, Danielson SR, Guo A, Gut P, Sahu AK, Li B, Uppala R, Fitch M, Riiff T, Zhu L, Zhou J, Mulhern D, Stevens RD, Ilkayeva OR, Newgard CB, Jacobson MP, Hellerstein M, Goetzman ES, Gibson BW, Verdin E. 2013. SIRT5 regulates the mitochondrial lysine succinylome and metabolic networks. Cell Metab 18:920–933. doi: 10.1016/j.cmet.2013.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Papa L, Hahn M, Marsh EL, Evans BS, Germain D. 2014. SOD2 to SOD1 switch in breast cancer. J Biol Chem 289:5412–5416. doi: 10.1074/jbc.C113.526475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vijayvergiya C, Beal MF, Buck J, Manfredi G. 2005. Mutant superoxide dismutase 1 forms aggregates in the brain mitochondrial matrix of amyotrophic lateral sclerosis mice. J Neurosci 25:2463–2470. doi: 10.1523/JNEUROSCI.4385-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Abdolvahabi A, Shi Y, Rhodes NR, Cook NP, Marti AA, Shaw BF. 2015. Arresting amyloid with Coulomb's law: acetylation of ALS-linked SOD1 by aspirin impedes aggregation. Biophys J 108:1199–1212. doi: 10.1016/j.bpj.2015.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Magrané J, Hervias I, Henning MS, Damiano M, Kawamata H, Manfredi G. 2009. Mutant SOD1 in neuronal mitochondria causes toxicity and mitochondrial dynamics abnormalities. Hum Mol Genet 18:4552–4564. doi: 10.1093/hmg/ddp421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kawamata H, Manfredi G. 2010. Import, maturation, and function of SOD1 and its copper chaperone CCS in the mitochondrial intermembrane space. Antioxid Redox Signal 13:1375–1384. doi: 10.1089/ars.2010.3212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bihlmaier K, Mesecke N, Terziyska N, Bien M, Hell K, Herrmann JM. 2007. The disulfide relay system of mitochondria is connected to the respiratory chain. J Cell Biol 179:389–395. doi: 10.1083/jcb.200707123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Takeshige K, Minakami S. 1979. NADH- and NADPH-dependent formation of superoxide anions by bovine heart submitochondrial particles and NADH-ubiquinone reductase preparation. Biochem J 180:129–135. doi: 10.1042/bj1800129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Raha S, Robinson BH. 2000. Mitochondria, oxygen free radicals, disease and ageing. Trends Biochem Sci 25:502–508. doi: 10.1016/S0968-0004(00)01674-1. [DOI] [PubMed] [Google Scholar]
- 45.Fischer LR, Igoudjil A, Magrané J, Li Y, Hansen JM, Manfredi G, Glass JD. 2011. SOD1 targeted to the mitochondrial intermembrane space prevents motor neuropathy in the Sod1 knockout mouse. Brain 134:196–209. doi: 10.1093/brain/awq314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sturtz LA, Diekert K, Jensen LT, Lill R, Culotta VC. 2001. A fraction of yeast Cu, Zn-superoxide dismutase and its metallochaperone, CCS, localize to the intermembrane space of mitochondria. A physiological role for SOD1 in guarding against mitochondrial oxidative damage. J Biol Chem 276:38084–38089. doi: 10.1074/jbc.M105296200. [DOI] [PubMed] [Google Scholar]
- 47.Kawamata H, Manfredi G. 2008. Different regulation of wild-type and mutant Cu, Zn superoxide dismutase localization in mammalian mitochondria. Hum Mol Genet 17:3303–3317. doi: 10.1093/hmg/ddn226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Reddehase S, Grumbt B, Neupert W, Hell K. 2009. The disulfide relay system of mitochondria is required for the biogenesis of mitochondrial Ccs1 and Sod1. J Mol Biol 385:331–338. doi: 10.1016/j.jmb.2008.10.088. [DOI] [PubMed] [Google Scholar]
- 49.Jacobson J, Duchen MR, Hothersall J, Clark JB, Heales SJ. 2005. Induction of mitochondrial oxidative stress in astrocytes by nitric oxide precedes disruption of energy metabolism. J Neurochem 95:388–395. doi: 10.1111/j.1471-4159.2005.03374.x. [DOI] [PubMed] [Google Scholar]
- 50.Zorov DB, Juhaszova M, Sollott SJ. 2014. Mitochondrial reactive oxygen species (ROS) and ROS-induced ROS release. Physiol Rev 94:909–950. doi: 10.1152/physrev.00026.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Longo VD, Gralla EB, Valentine JS. 1996. Superoxide dismutase activity is essential for stationary phase survival in Saccharomyces cerevisiae. Mitochondrial production of toxic oxygen species in vivo. J Biol Chem 271:12275–12280. [DOI] [PubMed] [Google Scholar]
- 52.Kelly B, Tannahill GM, Murphy MP, O'Neill LA. 2015. Metformin inhibits the production of reactive oxygen species from NADH:ubiquinone oxidoreductase to limit induction of interleukin-1β (IL-1β) and boosts interleukin-10 (IL-10) in lipopolysaccharide (LPS)-activated macrophages. J Biol Chem 290:20348–20359. doi: 10.1074/jbc.M115.662114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ouslimani N, Peynet J, Bonnefont-Rousselot D, Therond P, Legrand A, Beaudeux JL. 2005. Metformin decreases intracellular production of reactive oxygen species in aortic endothelial cells. Metabolism 54:829–834. doi: 10.1016/j.metabol.2005.01.029. [DOI] [PubMed] [Google Scholar]
- 54.Marycz K, Tomaszewski KA, Kornicka K, Henry BM, Wronski S, Tarasiuk J, Maredziak M. 2016. Metformin decreases reactive oxygen species, enhances osteogenic properties of adipose-derived multipotent mesenchymal stem cells in vitro, and increases bone density in vivo. Oxid Med Cell Longev 2016:9785890. doi: 10.1155/2016/9785890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lenaz G, Fato R, Genova ML, Bergamini C, Bianchi C, Biondi A. 2006. Mitochondrial complex I: structural and functional aspects. Biochim Biophys Acta 1757:1406–1420. doi: 10.1016/j.bbabio.2006.05.007. [DOI] [PubMed] [Google Scholar]
- 56.Glasauer A, Sena LA, Diebold LP, Mazar AP, Chandel NS. 2014. Targeting SOD1 reduces experimental non-small-cell lung cancer. J Clin Invest 124:117–128. doi: 10.1172/JCI71714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Davies MN, Kjalarsdottir L, Thompson JW, Dubois LG, Stevens RD, Ilkayeva OR, Brosnan MJ, Rolph TP, Grimsrud PA, Muoio DM. 2016. The acetyl group buffering action of carnitine acetyltransferase offsets macronutrient-induced lysine acetylation of mitochondrial proteins. Cell Rep 14:243–254. doi: 10.1016/j.celrep.2015.12.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kulej K, Avgousti DC, Sidoli S, Herrmann C, Della Fera AN, Kim ET, Garcia BA, Weitzman MD. 2017. Time-resolved global and chromatin proteomics during herpes simplex virus type 1 (HSV-1) infection. Mol Cell Proteomics doi: 10.1074/mcp.M116.065987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Soderblom EJ, Philipp M, Thompson JW, Caron MG, Moseley MA. 2011. Quantitative label-free phosphoproteomics strategy for multifaceted experimental designs. Anal Chem 83:3758–3764. doi: 10.1021/ac200213b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hoos MD, Richardson BM, Foster MW, Everhart A, Thompson JW, Moseley MA, Colton CA. 2013. Longitudinal study of differential protein expression in an Alzheimer's mouse model lacking inducible nitric oxide synthase. J Proteome Res 12:4462–4477. doi: 10.1021/pr4005103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Finn RD, Bateman A, Clements J, Coggill P, Eberhardt RY, Eddy SR, Heger A, Hetherington K, Holm L, Mistry J, Sonnhammer EL, Tate J, Punta M. 2014. Pfam: the protein families database. Nucleic Acids Res 42:D222–D230. doi: 10.1093/nar/gkt1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Edgar RC. 2004. MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics 5:113. doi: 10.1186/1471-2105-5-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Tighe A, Johnson VL, Taylor SS. 2004. Truncating APC mutations have dominant effects on proliferation, spindle checkpoint control, survival and chromosome stability. J Cell Sci 117:6339–6353. doi: 10.1242/jcs.01556. [DOI] [PubMed] [Google Scholar]
- 64.Goto H, Inagaki M. 2007. Production of a site- and phosphorylation state-specific antibody. Nat Protoc 2:2574–2581. doi: 10.1038/nprot.2007.374. [DOI] [PubMed] [Google Scholar]
- 65.Flohé L, Otting F. 1984. Superoxide dismutase assays. Methods Enzymol 105:93–104. doi: 10.1016/S0076-6879(84)05013-8. [DOI] [PubMed] [Google Scholar]
- 66.Ran FA, Hsu PD, Wright J, Agarwala V, Scott DA, Zhang F. 2013. Genome engineering using the CRISPR-Cas9 system. Nat Protoc 8:2281–2308. doi: 10.1038/nprot.2013.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Hodson AE, Tippetts TS, Bikman BT. 2015. Insulin treatment increases myocardial ceramide accumulation and disrupts cardiometabolic function. Cardiovasc Diabetol 14:153. doi: 10.1186/s12933-015-0316-y. [DOI] [PMC free article] [PubMed] [Google Scholar]