ABSTRACT
Loss-of-function mutations in 3′-to-5′ exoribonucleases have been implicated in hereditary human diseases. For example, PARN mutations cause a severe form of dyskeratosis congenita (DC), wherein PARN deficiency leads to human telomerase RNA instability. Since the DC phenotype in PARN patients is even more severe than that of loss-of-function alleles in telomerase components, we hypothesized that PARN would also be required for the stability of other RNAs. Here, we show that PARN depletion reduces the levels of abundant human Y RNAs, which might contribute to the severe phenotype of DC observed in patients. Depletion of PAPD5 or the cytoplasmic exonuclease DIS3L rescues the effect of PARN depletion on Y RNA levels, suggesting that PARN stabilizes Y RNAs by removing oligoadenylated tails added by PAPD5, which would otherwise recruit DIS3L for Y RNA degradation. Through deep sequencing of 3′ ends, we provide evidence that PARN can also deadenylate the U6 and RMRP RNAs without affecting their levels. Moreover, we observed widespread posttranscriptional oligoadenylation, uridylation, and guanylation of U6 and Y RNA 3′ ends, suggesting that in mammalian cells, the formation of a 3′ end for noncoding RNAs can be a complex process governed by the activities of various 3′-end polymerases and exonucleases.
KEYWORDS: 3′-end processing, DIS3L, dyskeratosis congenita, PARN, RNA degradation, RNA modification, Ro RNP, TOE1, U6 snRNA, Y RNA
INTRODUCTION
Proper 3′-end formation of RNAs plays an important role in regulating their function in the cell. Correct 3′-end formation is also important for the recruitment of a protective protein(s), which can serve to stabilize the RNA. For a number of stable noncoding RNAs (ncRNAs) in the cell, like snRNAs, snoRNAs, and human telomerase RNA (hTR), the 3′-end formation is regulated by coordinated activities of exonucleases that trim the precursor RNA to the mature form (1–4). These exonucleases compete with RNA binding proteins to regulate the levels and stability of the RNA, and perturbations in this equilibrium can lead to a disease state (5).
Mutations in 3′-to-5′ exonucleases have been shown to cause a number of different human diseases. For example, mutations in several core exosome components have been shown to cause diverse diseases. Mutations in EXOSC3 lead to pontocerebellar hypoplasia (6). Mutations in EXOSC8 have been shown to cause a form of spinal muscular atrophy (7). Finally, mutations in EXOSC2 have recently been shown to cause pleiotropic effects that include vision, hearing, and intellectual defects (8). More recently, it has been shown that mutations in the 3′-to-5′ exonuclease TOE1 also lead to pontocerebellar hypoplasia (9). How these specific mutations in exonucleases with overlapping functions lead to different human diseases remains unresolved.
Recently, mutations in PARN were identified that lead to familial pulmonary fibrosis, as well as a severe form of dyskeratosis congenita (DC) characterized by congenital defects and abnormally short telomere lengths (10, 11). We, along with others, have recently shown that PARN deadenylates hTR 3′ ends, and PARN deficiency leads to the oligoadenylation of hTR 3′ ends by PAPD5, leading to its degradation by EXOSC10 (12–15). The fact that the DC in PARN patients is even more severe than that from loss-of-function alleles in telomerase itself suggests that loss of PARN activity has other consequences that contribute to disease. One simple hypothesis is that PARN is also required for the stability of other RNAs.
In this work, we investigate how PARN regulates the processing and stability of several abundant ncRNAs. We found that PARN deadenylates both U6 and RMRP under normal conditions but has no significant effect on the steady-state levels of either of these RNAs. This suggests that many ncRNAs are substrates for PARN-mediated deadenylation, but without an effect on RNA stability. In contrast, the loss of PARN reduces the levels of human Y RNAs, which are abundant small RNA polymerase (Pol) III-transcribed RNAs that are posited to play a role in RNA quality control, initiation of DNA replication, DNA damage response, and histone mRNA processing (16–19). Low levels of Y RNAs could exacerbate the effect of PARN depletion on telomere maintenance, leading to the severe phenotype of DC observed in patients carrying PARN mutations. In addition, our deep sequencing of the 3′ ends of these noncoding RNAs uncovered widespread posttranscriptional modifications, as well as dynamic 3′-end formation, for the ncRNAs. These results suggest that the 3′ ends of ncRNAs are substrates of various exonucleases and noncanonical 3′-end polymerases, and these different 3′ ends could be a way to increase the diversity of RNPs formed by these ncRNAs in the cell.
RESULTS
PARN affects the levels of Y RNAs.
In order to identify novel substrates for PARN-mediated 3′-end deadenylation and processing in human cells, we first examined whether PARN depletion by small interfering RNAs (siRNAs) affects the levels of several ncRNAs. We examined the levels of U6 snRNA in PARN-depleted cells, since U6 has been shown to contain oligoadenylated ends in patients suffering from poikiloderma with neutropenia (20). We also examined the RMRP RNA, which is the catalytic component of the mitochondrial RNase P-like enzyme complex and was previously shown to contain posttranscriptionally added oligo(A) tails (21). We also examined the Y RNAs (also referred to as hY RNAs) Y1, Y3, Y4, and Y5 (in decreasing order of length), which are small Pol III-transcribed RNAs 83 to 112 nucleotides (nt) in length and are highly abundant in human cells (22, 23). The exact function of these abundant RNAs in the cell in unclear; however, they have been proposed to play a role in DNA damage response and DNA replication, among others (16, 19, 24, 25), and therefore, a deficiency in Y RNAs might have synergistic effects with defects in telomere elongation.
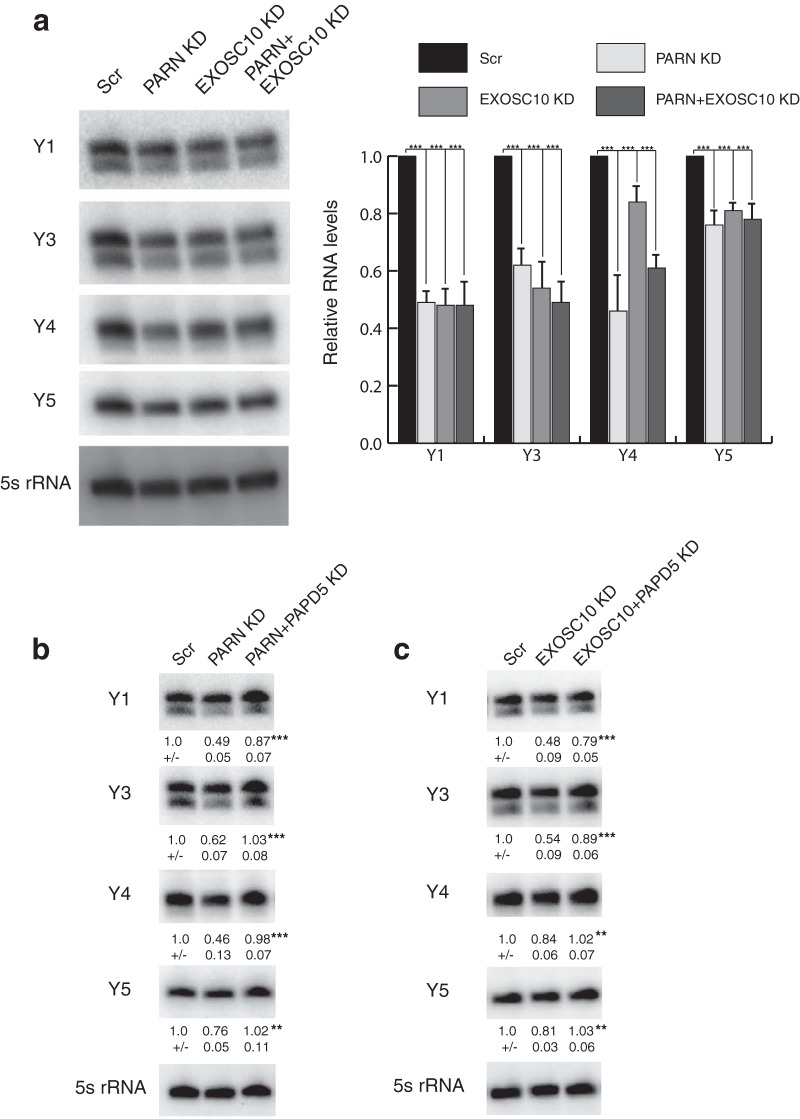
An important result was that PARN knockdown (KD) led to a nonuniform decrease in the levels of all four Y RNAs in HeLa cells (Fig. 1a). In contrast, PARN knockdown had essentially no effect on the levels of U6 snRNA or RMRP RNA, although PARN can affect the precise sequences of these RNAs' 3′ ends (see below).
FIG 1.
PARN and EXOSC10 depletion leads to a decrease in Y RNA levels. (a) Representative Northern blots depicting Y RNA levels under various conditions. RNA levels were normalized to a 5S rRNA loading control and are shown as a histogram (averages and standard deviations [SD] for three biological replicates). (b and c) Representative Northern blots depicting Y RNA levels under various conditions. RNA levels were normalized to a 5S rRNA loading control (averages and SD for four biological replicates). Scr, Scrambled control. **, P < 0.01; ***, P < 0.001. For panel a, each knockdown was compared to Scr. For panels b and c, double knockdowns were compared to individual knockdown.
Given the effect of PARN on Y RNA levels, we sought to understand the manner by which PARN maintains normal Y RNA levels. Earlier work has shown that for the telomerase RNA (hTR), PARN deficiency led to hTR degradation through EXOSC10, and coknockdown of PARN and EXOSC10 could recue the defect (12, 13). We therefore asked whether Y RNA levels could be rescued by EXOSC10 knockdown under low levels of PARN. Surprisingly, EXOSC10 knockdown led to a decrease in Y RNA levels as well, and coknockdown of PARN and EXOSC10 led to a slight decrease in Y RNA levels compared to PARN or EXOSC10 knockdown alone (Fig. 1a). This suggests that both PARN and EXOSC10 are required for the stability of Y RNAs in human cells, which is different from the specific mechanism by which PARN affects hTR levels (12).
EXOSC10 could be required for Y RNA stability, because the nuclear exosome could play a role in Y RNA 3′ processing or in trimming oligo(A) tails, similar to PARN, to prevent recruitment of 3′-to-5′ exonucleases. Consistent with the nuclear exosome having a role in Y RNA biogenesis, we also observed that knockdown of the core exosome subunit DIS3 also led to a reduction in Y RNA levels (see below).
By analogy to hTR, PARN and EXOSC10 might stabilize Y RNAs by removing oligo(A) tails added by PAPD5, which otherwise could recruit a 3′-to-5′ exonuclease that degrades the Y RNAs (26–29). Therefore, we asked whether coknockdown of PAPD5 with PARN or EXOSC10 could rescue Y RNA levels. We found that PARN and PAPD5 coknockdown was able to rescue all Y RNA levels (Fig. 1b). Similarly, coknockdown of PAPD5 and EXOSC10 led to a partial rescue of Y1 and Y3 and a complete rescue of Y4 and Y5 RNAs (Fig. 1c). These results suggest that Y RNAs are substrates for PAPD5-mediated oligoadenylation and that these oligo(A) tails are removed by PARN and EXOSC10 individually or in concert, leading to stabilization of the RNA. Note that these results imply that an additional 3′-to-5′ exonuclease can be recruited to oligoadenylated Y RNAs for their degradation.
DIS3L is involved in regulating the steady-state levels of Y RNAs.
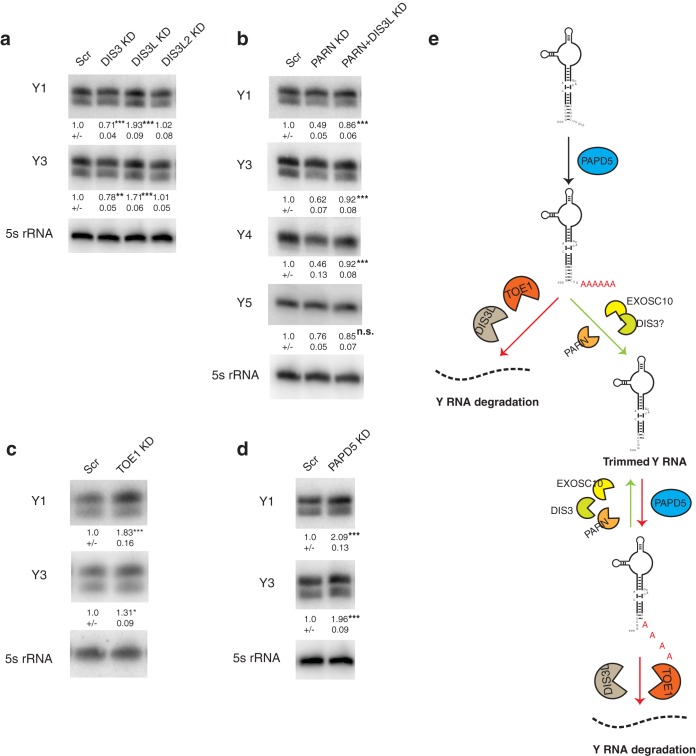
In order to identify the 3′-to-5′ exonuclease responsible for degradation of Y RNAs upon PARN or EXOSC10 knockdown, we performed a knockdown of various 3′-to-5′ exonucleases in HeLa cells. We knocked down DIS3, which is the catalytic component of the exosome possessing both 3′-to-5′ exo- and endonucleolytic activities (30); DIS3L, which is a DIS3-like exonuclease believed to interact with the exosome and degrades RNAs carrying oligoadenylated 3′ ends (31, 32); and DIS3L2, which is a second cytoplasmic 3′-to-5′ exonuclease that is thought to preferentially degrade oligouridylated RNAs independently of the core exosome (33–35).
An important result was that DIS3L knockdown led to an approximate 2-fold increase in the levels of Y1 and Y3 RNAs in HeLa cells (Fig. 2a). In contrast, DIS3 knockdown led to a decrease in Y RNA levels (∼30% for Y1 and ∼20% for Y3) and DIS3L2 knockdown had no significant effect on Y RNA levels. This is in contrast to recent reports that suggest that DIS3L2 targets 3′-uridylated Y RNAs for degradation (36, 37), although as discussed below, we found that only a small fraction of Y RNAs are uridylated in HeLa cells. Moreover, DIS3L coknockdown with PARN is sufficient to rescue Y RNA levels to control, suggesting that DIS3L is responsible for Y RNA degradation when PARN is limiting (Fig. 2b).
FIG 2.
DIS3L regulates Y RNA levels through oligoadenylation by PAPD5. (a) Representative Northern blots depicting Y RNA levels under various conditions. The RNA levels were normalized to a 5S rRNA loading control (averages ± SD for four biological replicates). (b) Representative Northern blots depicting Y RNA levels under DIS3L and PARN coknockdown compared to PARN knockdown alone. RNA levels were normalized to a 5S rRNA loading control (averages ± SD for three biological replicates). (c) Representative Northern blots depicting Y RNA levels under control or TOE1 KD. RNA levels were normalized to a 5S rRNA loading control (averages ± SD for three biological replicates). (d) Representative Northern blots depicting Y RNA levels under control or PAPD5 KD. RNA levels were normalized to a 5S rRNA loading control (averages ± SD for three biological replicates). (e) Model depicting the maturation and quality control of Y RNAs by various 3′-end modifiers and exonucleases. Depletion of PARN or EXOSC10 leads to oligoadenylation of Y RNAs by PAPD5 and degradation by DIS3L or TOE1. *, P < 0.05; ***, P < 0.001; n.s., not significant. For panels a, c, and d, individual knockdowns were compared to Scr. For panel b, double knockdowns were compared to PARN KD.
A previous study showed that DIS3L preferentially degrades oligoadenylated rRNA species in human cells (31). Another 3′-to-5′ exonuclease, TOE1, was recently shown to process U snRNAs by removing oligo(A) tails from their 3′ ends (9). We therefore asked if degradation through oligo(A) tail recognition is a general feature of Y RNA stability by knocking down TOE1 in cells. We found that TOE1 knockdown led to a 1.8-fold increase in Y1 RNA levels and a smaller but significant increase in Y3 levels (Fig. 2c). This suggests that oligoadenylation of Y RNAs could also lead to their degradation by TOE1.
Our rescue experiments with PARN and PAPD5 or EXOSC10 and PAPD5 coknockdown, along with an increase in Y RNA levels upon DIS3L (and TOE1) knockdown, suggest that PAPD5-mediated oligoadenylation leads to the degradation of Y RNAs. Therefore, a prediction would be that PAPD5 knockdown alone leads to an increase in Y RNA levels. Indeed, PAPD5 knockdown led to a 2-fold increase in the levels of Y1 and Y3 RNAs in HeLa cells, suggesting that PAPD5's activity is indeed required for the degradation of Y RNAs by DIS3L (Fig. 2d).
The above-mentioned knockdowns of PARN, EXOSC10, and PAPD5 were carried out using validated siRNA pools. However, to ensure that the results were due to the knockdown of the specific gene in each case, we repeated the experiments with different individual siRNAs against PARN, EXOSC10, and PAPD5. These individual siRNA experiments mimicked the siRNA pool knockdowns, and we again observed that PARN and EXOSC10 knockdown reduced Y RNA levels, while PAPD5 knockdown increased Y RNA levels (see Fig. S1 in the supplemental material).
Together, these data suggest a model where Y RNAs are trimmed by PARN and EXOSC10/DIS3 in conjunction with PAPD5 to a stable RNA that assembles into an RNP (Fig. 2e). In the absence of PARN or EXOSC10, the oligoadenylated Y RNA becomes a substrate for DIS3L or TOE1, which recognize the oligo(A) tails and degrade the RNA. DIS3L and TOE1 might also be responsible for the basal turnover of Y RNAs, as suggested by a 2-fold increase in Y RNA levels upon DIS3L or TOE1 knockdown (Fig. 2e).
Y RNAs are trimmed to a stable end and exhibit significant amounts of posttranscriptional modification at the 3′ end.
Our results with steady-state levels of Y RNAs in human cells suggest that Y RNAs are substrates for oligoadenylation and deadenylation. We next investigated whether these events are reflected in the 3′-end processing of Y RNAs under control or knockdown conditions. To do this, we performed RNA sequencing (RNA-Seq) on the 3′ ends of Y1 and Y3, the two Y RNAs most affected by PARN, EXOSC10, and PAPD5 knockdowns.
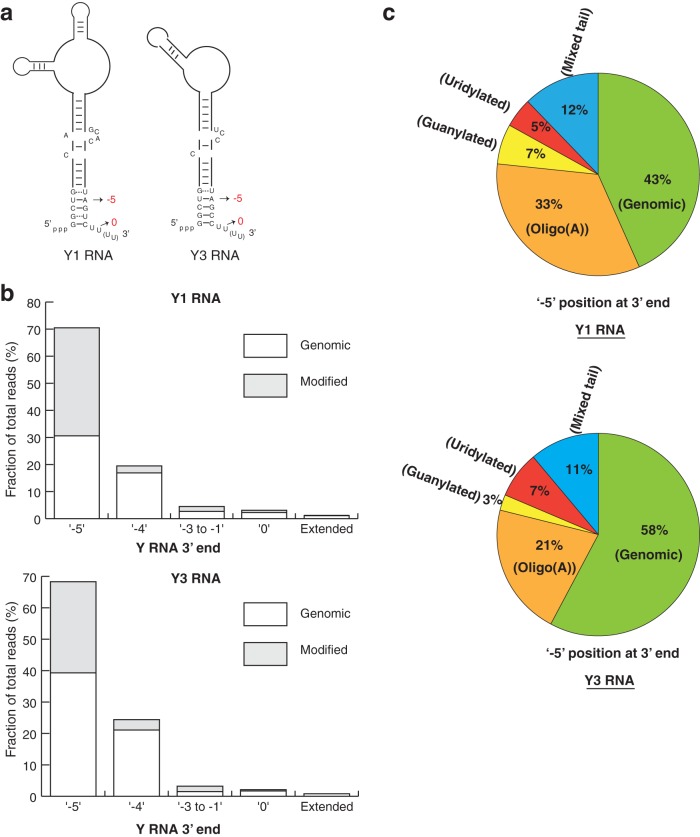
Sequencing Y1 and Y3 RNAs from HeLa cells revealed the following key points. First, we observed a distribution of 3′ ends with the most abundant RNA 3′ end corresponding to the −5 position, upstream of the canonical 3′ end identified previously (Fig. 3a and b). For Y1 and Y3 RNAs, ∼70% of total reads, including genomic (unmodified) reads and posttranscriptionally modified reads, mapped to the −5 genomic position (Fig. 3b). The next most abundant position is the −4 end (∼20% of total reads), while a very small fraction of reads mapped to the canonical 3′ end for Y1 and Y3 RNAs (∼3% and 2% for Y1 and Y3 RNAs, respectively) (Fig. 3b). While previous studies identified the sequence of Y RNAs based on biochemical purification and hybridization to genomic DNA, our deep sequencing of 3′ ends identified the precise location and relative abundance of each 3′ end in HeLa cells.
FIG 3.
Y RNAs are trimmed to an abundant end and contain a large proportion of posttranscriptional modifications. (a) Schematic depicting various 3′-end positions for Y1 and Y3 RNAs in human cells. (b) Clustered bar graph depicting the fraction of reads at each 3′-end position as a percentage of total sequencing reads in control cells. (c) Compositions of Y1 and Y3 RNAs at the abundant −5 position in control cells. Uridylated and guanylated reads represent reads that terminate in a U or a G, respectively. Mixed reads contain a combination of A's, U's, G's, and C's added posttranscriptionally.
A second important observation was that at every 3′ end on the Y RNAs, we observed a substantial number of 3′ ends that contained nonencoded A's, G's, or U's. For example, at the most abundant −5 3′ end, we observed that 57% and 42% of the reads for Y1 and Y3 were modified (Fig. 3c). The most abundant modification was oligo(A) tails, although we also observed RNAs modified with a tail ending in a G (guanylated) or a U (uridylated), as well as tails containing a mixture of these nucleotides (Fig. 3c). Similar results were observed at the −4 position and for the summations of RNAs trimmed into the body of the Y RNA (−3 to −1) (Fig. 4), although these reads were predominantly modified with oligo(A) tails. Reads corresponding to a mixture of these modifications suggest that the modifying enzymes target these RNAs as substrates in cis.
FIG 4.

Y RNAs are oligoadenylated at various 3′-end positions. Shown are the compositions of Y1 and Y3 RNAs at the abundant −5 position in control cells. Guanylated reads represent reads that terminate in a G. Mixed reads contain a combination of A's, U's, G's, and C's added posttranscriptionally.
These abundant posttranscriptional modifications suggest that Y RNAs potentially contain a variety of 3′ ends, with the most abundant species being −5 and −4 relative to the previously mapped 3′ ends. We found that the most abundant modified end contained a single A followed by either an A, a U, or a G nucleotide, in decreasing order of abundance, for both Y1 and Y3 RNAs (Fig. 5). A U or a G addition could potentially be stabilized through the formation of a wobble base pair or bulging of the stem, respectively, providing a rationale for the existence of stable species carrying these modifications (Fig. 5). Another interesting observation was that these posttranscriptionally added dinucleotides (AA, AU, or AG) were often followed by oligo(A) tails, which we classified as mixed tails for AU and AG dinucleotides in our analysis (Fig. 3c). This suggests that oligoadenylating enzymes have higher activity at the 3′ ends of Y1 and Y3 RNAs and can outcompete uridylating or guanylating enzymes for substrate modification.
FIG 5.
Various possible 3′-end structures for Y1 and Y3 RNAs based on the most common 3′-end modification at the −5 position. The red letters represent posttranscriptionally added nucleotides.
PARN, EXOSC10, and PAPD5 are involved in the processing of Y RNAs.
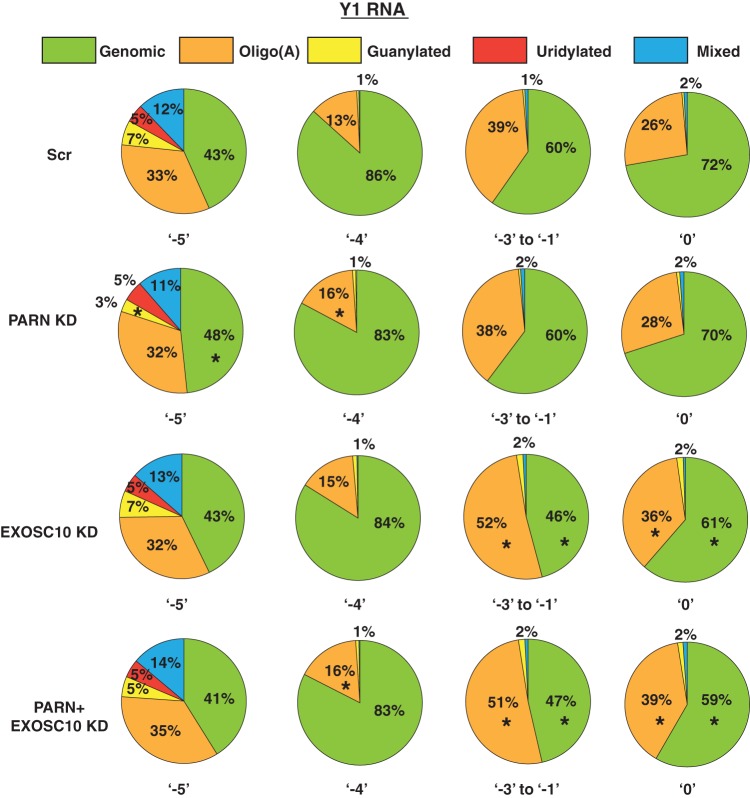
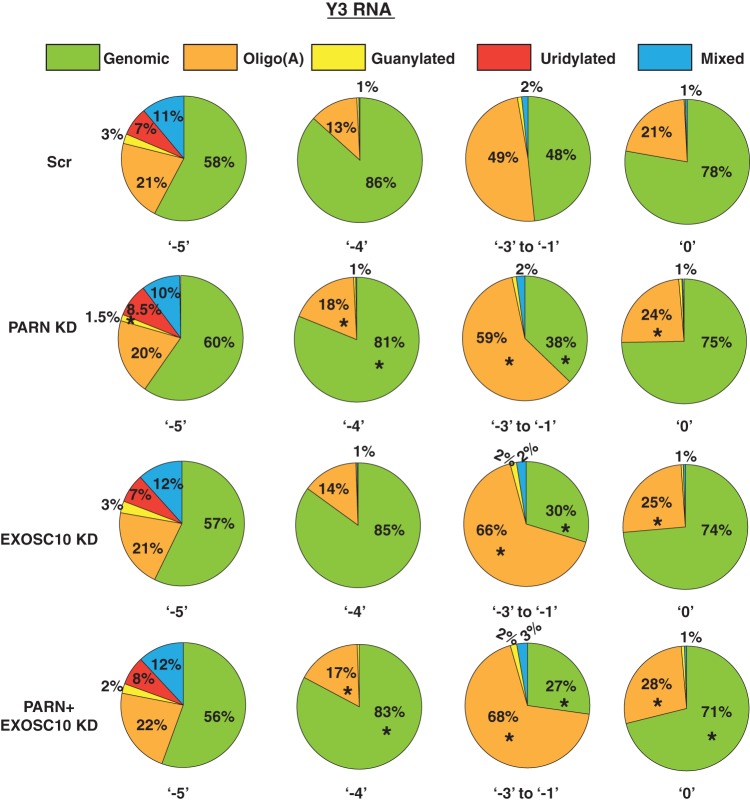
PARN and EXOSC10 knockdown leads to a reduction in the levels of Y RNAs in human cells. One possibility is that PARN and EXOSC10 protect Y RNAs from degradation by trimming the Y RNAs to the −5 position, which could be a boundary element formed by the binding of a protective protein, like the Ro protein (38). Therefore, we asked if the 3′ ends of Y1 and Y3 RNAs change upon the knockdown of PARN or EXOSC10 in human cells.
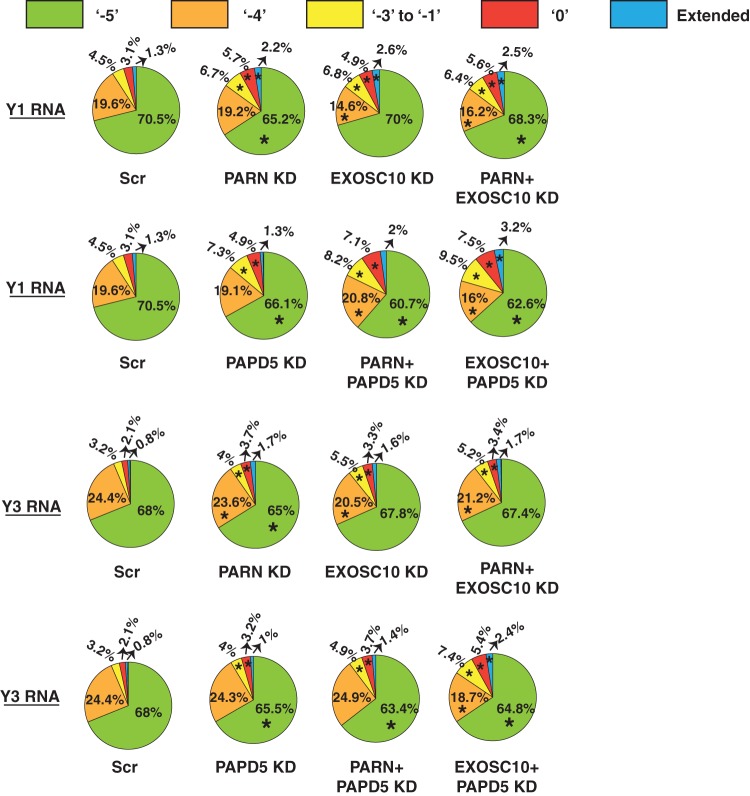
We found that PARN or EXOSC10 knockdown led to a small but clear increase in the percentage of reads corresponding to the internally chewed-up positions (−3 to −1), as well as the canonical 3′ end (0 position) and extended ends for both Y1 and Y3 RNAs compared to the control (Fig. 6). The increase in the fraction of reads at downstream ends was compensated for by a reduction in the fraction of reads terminating at either of the two abundant ends, −5 or −4 (Fig. 6). Interestingly, while PARN knockdown led to a decrease in the fraction of reads at the −5 position, EXOSC10 knockdown led to a decrease in the fraction of reads at the −4 position (Fig. 6). This suggests that these enzymes potentially trim Y RNAs at different positions.
FIG 6.
PARN and EXOSC10 are responsible for trimming Y RNAs to −5 and −4 positions in a PAPD5-dependent manner. The pie charts depict the fraction of reads at each 3′-end position under various knockdown conditions. *, P < 0.05. Each sector in the pie chart for knockdowns was compared to the corresponding sector in Scr.
Since Y RNAs are degraded through recognition of the oligo(A) tail, it is likely that the oligo(A) tail is also important for the 3′-end processing of Y RNAs to the trimmed end. Therefore, a prediction would be that PAPD5 knockdown leads to an increase in reads at the downstream ends and a reduction in reads at the trimmed end. Another prediction would be that PARN and PAPD5 coknockdown or EXOSC10 and PAPD5 coknockdown leads to an even greater effect on the fractions of reads at these different positions. Indeed, PAPD5 knockdown led to an increase in the fraction of reads corresponding to downstream ends and a decrease in the fraction of reads at the −5 end (Fig. 6). Second, PARN and PAPD5 or EXSOC10 and PAPD5 coknockdown led to an even greater increase in the fraction of reads at the downstream ends and a decrease in the fraction of reads at the −5 end. Together, these data suggest that PAPD5-mediated oligoadenylation and exonucleolytic processing by PARN and EXOSC10 play a role in Y RNA maturation. However, it is possible that other enzymes, such as DIS3, DIS3L, and TOE1, also play a role in this process due to the redundancy in Y RNA stability upon deficiency of any of these enzymes (Fig. 2).
PARN removes oligo(A) tails from Y RNA 3′ ends, which are added by PAPD5.
Since PARN is a deadenylase that removes oligo(A) tails from its substrates, we asked whether PARN depletion also affects the oligoadenylation of Y RNAs. As described above, Y1 and Y3 RNAs exhibit significant posttranscriptional modification at various 3′-end positions (Fig. 3c and 4). We found that PARN knockdown led to an increase in oligo(A) reads at ends from the −4 position to the 0 position but did not really affect total oligo(A) reads at the −5 position for Y1 and Y3 RNAs, respectively (Fig. 7 and 8). EXOSC10 knockdown also led to an increase in total oligo(A) reads at various positions from −4 to 0 for Y1 and Y3 RNAs; the effect was greater than that of PARN knockdown at some positions (e.g., positions −3 to −1 and 0 for Y1 RNA and −3 to −1 for Y3 RNA) (Fig. 7 and 8). This suggests that EXOSC10 also deadenylates Y1 and Y3 RNA 3′ ends, potentially protecting the RNA from degradation.
FIG 7.
PARN and EXOSC10 affect the 3′ modification status of Y1 RNA at different positions. The pie charts depict the fraction of genomic (unmodified) or posttranscriptionally modified reads at various 3′-end positions under the indicated knockdown condition. *, P < 0.05. Each sector in the pie chart for knockdowns was compared to the corresponding sector in Scr.
FIG 8.
PARN and EXOSC10 affect the 3′ modification status of Y3 RNA at different positions. The pie charts depict the fractions of genomic (unmodified) or posttranscriptionally modified reads at various 3′-end positions under the indicated knockdown conditions. *, P < 0.05. Each sector in the pie chart for knockdowns was compared to the corresponding sector in Scr.
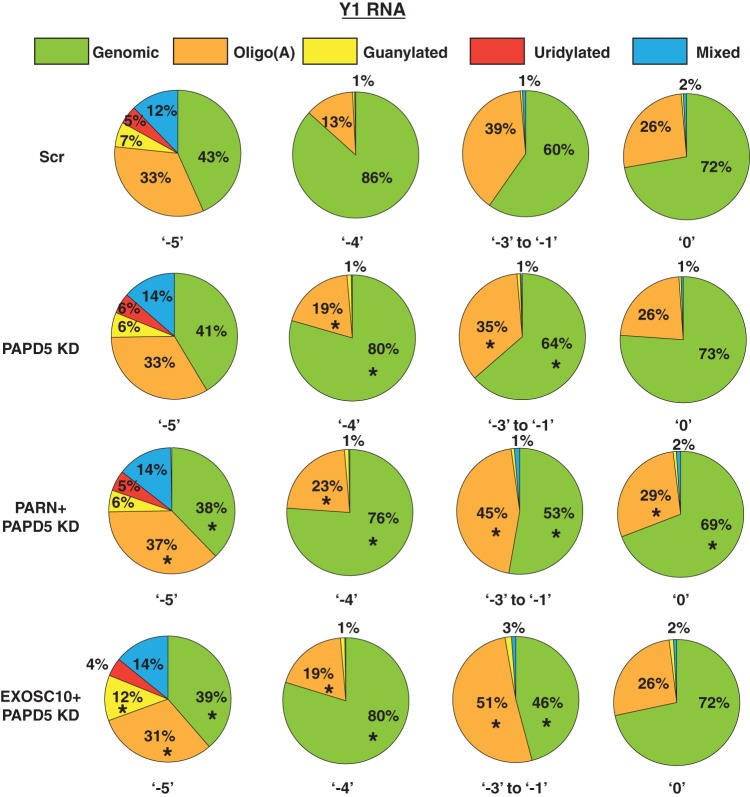
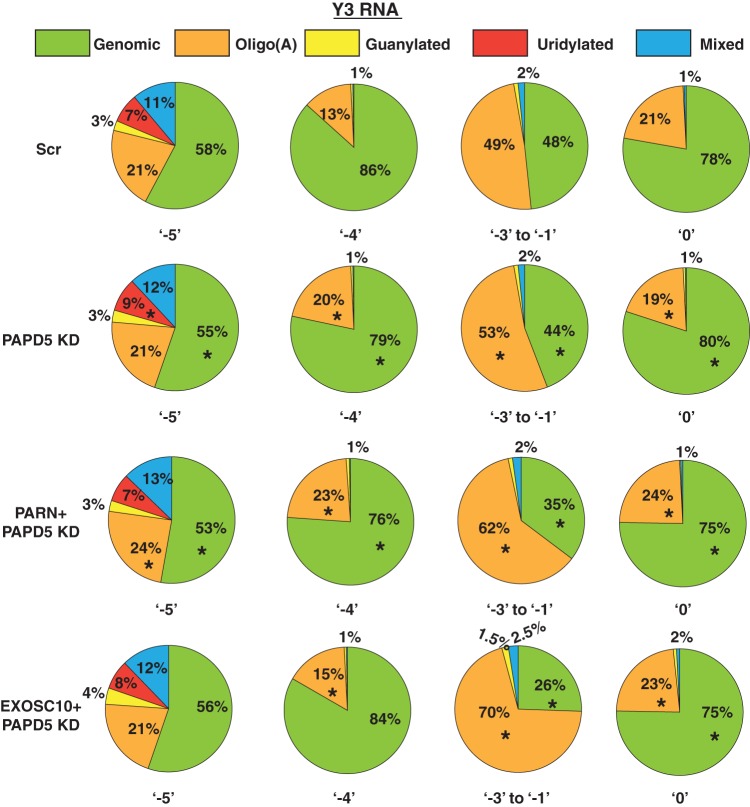
As discussed above, PAPD5-mediated oligoadenylation is involved in the trimming of Y RNAs to the −4 and −5 positions (Fig. 2e and 6). Therefore, a prediction would be that PAPD5 knockdown leads to a decrease in oligo(A) modification at the 3′ end and PARN and PAPD5 coknockdown or EXOSC10 and PAPD5 coknockdown should lead to an increase in oligo(A) modifications. Surprisingly, we found that PAPD5 knockdown did not lead to a large decrease in oligo(A) reads at various positions for both Y1 and Y3 RNAs (Fig. 9 and 10). In fact, at some positions, PAPD5 knockdown led to an increase in oligo(A) reads (discussed below). However, we did find that PARN and PAPD5 coknockdown or EXOSC10 and PAPD5 coknockdown led to an increase in oligo(A) reads at various positions for Y1 and Y3 RNAs compared to the control (Fig. 9 and 10). This suggests that PARN and EXOSC10 remove oligo(A) tails added by PAPD5 from the 3′ ends of Y RNAs.
FIG 9.
PAPD5 knockdown affects the level of oligoadenylation of Y1 RNA at different positions. The pie charts depict the fractions of genomic (unmodified) or posttranscriptionally modified reads at various 3′-end positions under the indicated knockdown conditions. *, P < 0.05. Each sector in the pie chart for knockdowns was compared to the corresponding sector in Scr.
FIG 10.
PAPD5 knockdown affects the level of oligoadenylation of Y3 RNA at different positions. The pie charts depict the fractions of genomic (unmodified) or posttranscriptionally modified reads at various 3′-end positions under the indicated knockdown conditions. *, P < 0.05. Each sector in the pie chart for knockdowns was compared to the corresponding sector in Scr.
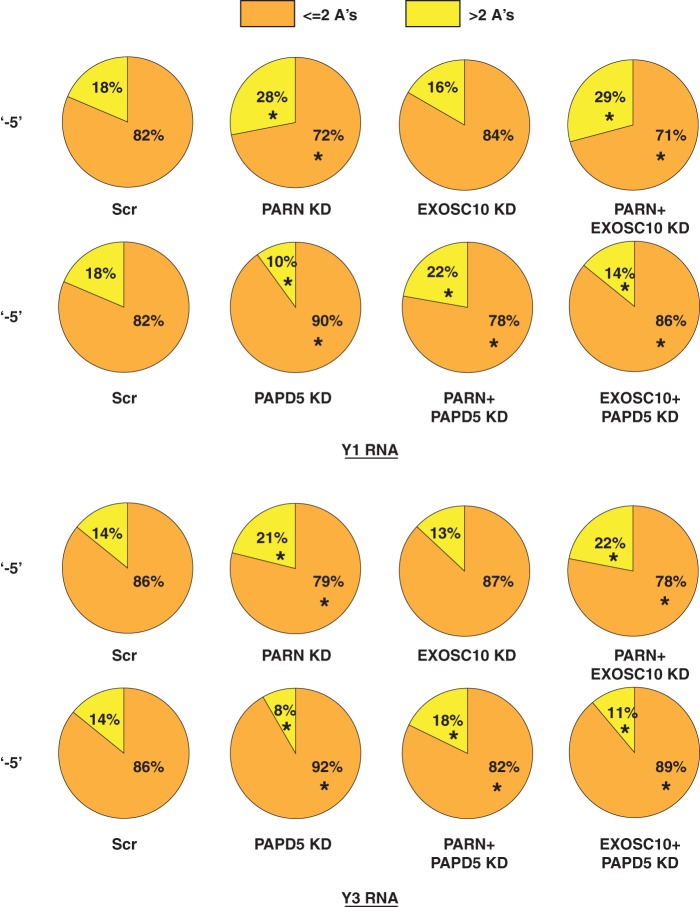
One of the predominant modifications for Y1 and Y3 RNAs is the addition of adenosine dinucleotides at the −5 position (Fig. 5). Therefore, it is possible that PARN and PAPD5 do affect the oligoadenylation of Y RNAs at this position but the effect on deadenylation, either due to PARN knockdown or due to PAPD5 knockdown, is masked by the overabundance of the adenosine dinucleotide modified species. Therefore, we separated the fraction of oligoadenylated reads that contained one or two A's at the −5 position from those that contained more than two A's and asked whether PAPD5 knockdown or PARN knockdown led to a change in the fraction of longer A tails at the −5 position. We found that PARN knockdown led to an increase in the fraction of longer oligo(A) reads, indirectly indicating the presence of a longer oligo(A) tail on average for both Y1 and Y3 RNAs (Fig. 11). Conversely, PAPD5 knockdown led to a decrease in the fraction of longer oligo(A) reads, indicating the presence of shorter oligo(A) tails on average at the −5 end. The shorter oligo(A) tails are probably due to deadenylation by PARN or EXOSC10 (Fig. 11). A similar trend was observed for oligo(A) modifications at the −4 position (data not shown).
FIG 11.
PARN knockdown and PAPD5 knockdown have antagonistic effects on oligoadenylation of Y1 and Y3 RNA 3′ ends. The pie charts depict the fractions of oligoadenylated reads containing less than two or more than two A's at various 3′ ends under the indicated knockdown conditions. *, P < 0.05. Each sector in the pie chart for knockdowns was compared to the corresponding sector in Scr.
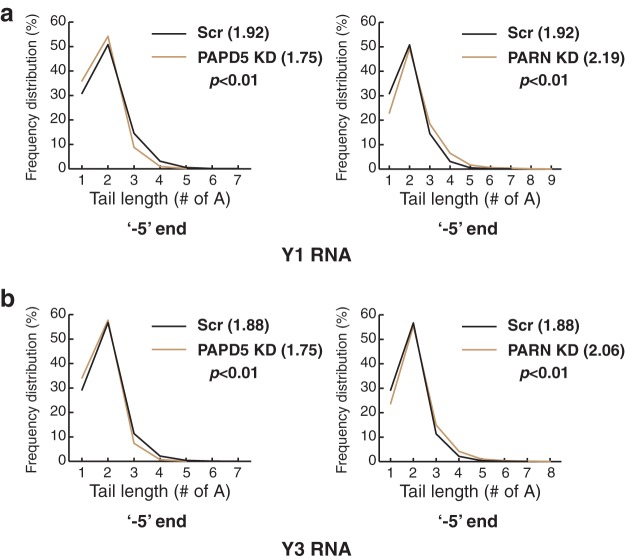
We further analyzed the A tail length at the −5 end upon PAPD5 or PARN knockdown to see whether the average tail length differed under these knockdown conditions. We found that for both Y1 and Y3 RNAs, the average number of A's decreased significantly upon PAPD5 knockdown and increased upon PARN knockdown (Fig. 12). This effect was also elicited in PAPD5 knockdown cells having shorter oligo(A) tails and PARN knockdown cells having longer oligo(A) tails (Fig. 12). However, as discussed above, the numbers and the distribution skew heavily toward the dinucleotide species (2 A's added to the −5 end), suggesting a possible function of the dinucleotide modification in stabilizing the stem through noncanonical G-A base pairing. Finally, it is also possible that the first two A's are added by a terminal adenylase other than PAPD5, which would obscure the analysis upon PAPD5 knockdown.
FIG 12.
PAPD5 knockdown leads to shorter oligo(A) tails for Y1 and Y3 RNAs. (a and b) Line plots depicting oligo(A) tail lengths at the canonical 3′ end in control, PAPD5 KD, and PARN KD cells for Y1 and Y3 RNA. The numbers in parentheses represent the average numbers of A's under each condition. Statistical differences were calculated using two-tailed unpaired Student t tests.
Altogether, two conclusions can be drawn from the analysis of oligo(A) modifications at the various 3′ ends of Y1 and Y3 RNAs. First, PAPD5 adds oligo(A) tails to the 3′ ends of Y RNAs and PARN removes oligo(A) tails from the 3′ ends of Y RNAs. Second, the adenosine dinucleotide modification is the predominant modified species at the −5 and −4 positions for Y RNAs. This could be due to the selective stabilization of the Y RNA stem when this modification is added at the 3′ end.
PARN deadenylates U6 snRNA and RMRP without affecting their stability.
U6 snRNA and RMRP are abundant ncRNAs that have previously been demonstrated to contain oligo(A) modifications at the 3′ end in human cells (20, 21). We therefore investigated whether PARN affects the 3′-end deadenylation and/or stability of these two RNAs.
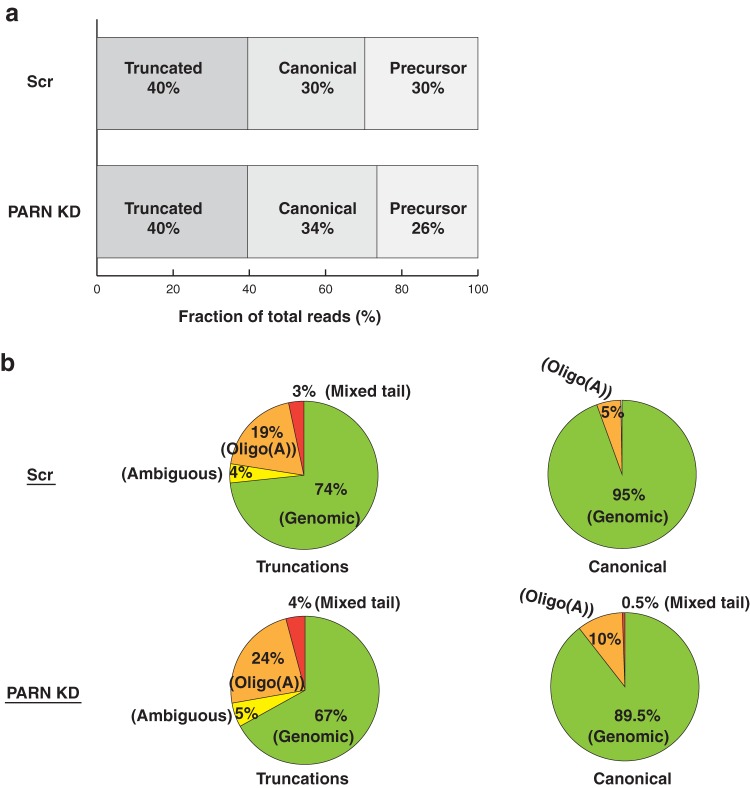
For U6 snRNA, a significant fraction of reads mapped to truncations and precursors, along with the canonical 3′ end (Fig. 13a). We classify truncations as reads shorter than the mature species, where the mature species are reads corresponding to canonical 3′-end positions (AUUUU[U]; Lsm 2-8 binding site plus 1 U, which could contain a 2′-3′ cyclic phosphate), and precursor species as reads that are uridylated and extend beyond the canonical 3′ end (AUUUUUU, AUUUUUUU, etc. [underlining indicates nontemplated additions]). In PARN knockdown cells, we found that the proportion of truncated reads did not change but the proportion of canonical end reads increased slightly, whereas the proportion of extended reads decreased in a compensatory manner (Fig. 13a).
FIG 13.
PARN depletion increases oligoadenylation of U6 snRNA. (a) Distribution of total U6 snRNA reads based on the 3′ end in control and PARN KD cells. Fractions were calculated by dividing the total reads for each category (including posttranscriptional modifications) by the total reads from sequencing. (b) Pie charts depicting genomic or modified reads for truncated and canonical 3′ ends of U6 snRNA. PARN knockdown leads to an increase in the fraction of oligoadenylated reads for both truncated and canonical species. Ambiguous reads are reads that end in a single A, which could be added co- or posttranscriptionally. Mixed reads are reads that contain both A's and U's.
The increase in the fraction of canonical reads in PARN knockdown could be due to more genomic reads or more posttranscriptionally modified reads. We therefore analyzed whether the 3′ end is modified posttranscriptionally for U6 snRNA. Analysis of the 3′-end modification status under control conditions suggested that U6 snRNA contains a significant number of oligo(A) modifications at both the truncated and canonical ends (Fig. 13b). This suggests that U6 snRNA is posttranscriptionally modified through the addition of oligo(A) tails at the 3′ end. We then asked whether PARN affects the 3′-end oligoadenylation of the U6 snRNAs. We found that for both truncated and canonical ends, PARN knockdown led to an increase in the fraction of oligo(A) reads (Fig. 13b). The net increase in the fraction of total reads at the canonical end, along with an increase in the fraction of oligoadenylated reads at this position, suggests that PARN deadenylates U6 snRNA 3′ ends, similar to what we have previously shown for hTR (12).
RMRP is the RNA component of the mitochondrial RNase P-like complex in human cells and was previously shown to contain posttranscriptional oligo(A) modifications at its 3′ end (21). We therefore sequenced the 3′ end of RMRP in control and PARN knockdown cells to investigate whether RMRP is a substrate for PARN-dependent deadenylation. A majority of the reads mapped to the canonical 3′ end (∼50%), and others mapped to precursor ends (∼36%) under control conditions. Upon PARN knockdown, the fraction of reads mapping to the canonical 3′ end decreased slightly (48.5% compared to 50%), and the fraction of reads corresponding to precursor ends increased slightly (37% compared to 36%).
Apart from genomic reads, the only posttranscriptional modification observed on RMRP was oligo(A) tails in both control and PARN knockdown cells. This suggests that RMRP is exclusively a substrate for oligoadenylating enzymes in human cells.
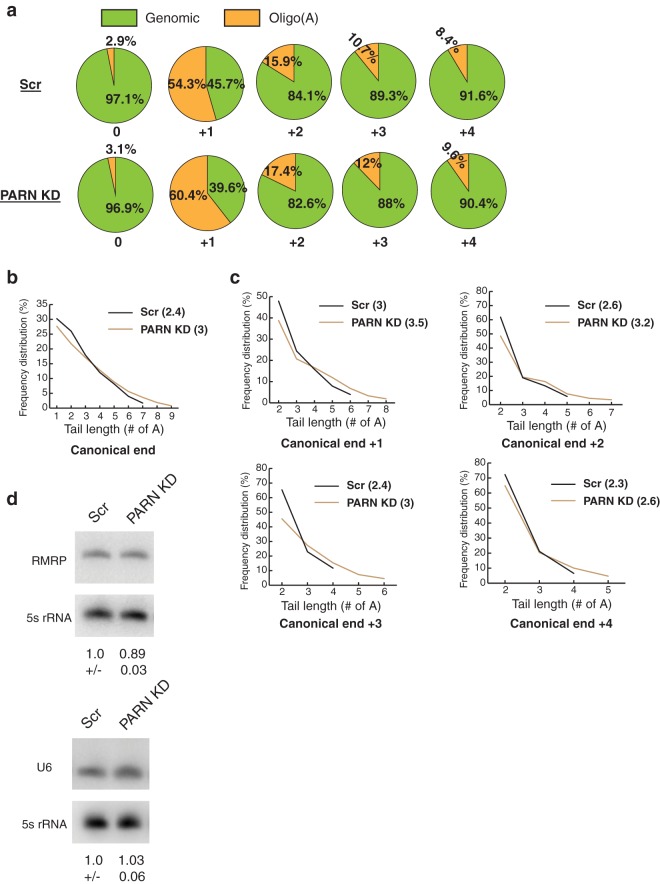
We then investigated how the fraction of oligo(A) tails changed in PARN knockdown cells compared to control cells. We found that, compared to control cells, the fraction of oligo(A) reads at canonical and precursor ends increased in PARN knockdown cells (Fig. 14a). The increase was small, but consistent, at all ends and was largest at the +1 end (54% in control cells versus 60% in PARN KD cells). To further investigate whether this increase in oligo(A) reads has an effect on the length of oligo(A) tails at RMRP ends, we plotted the oligo(A) tails at each end in control and PARN knockdown cells and calculated the mean number of A's present at each end under control or PARN knockdown. At the canonical 3′ end, oligo(A) tail length increased in PARN knockdown cells compared to control cells, along with an increase in the mean number of A's (2.4 in control versus 3.0 in PARN KD cells) (Fig. 14b). Similarly, at each precursor end, the length of the oligo(A) tails and the mean number of A's increased in PARN knockdown cells compared to control cells (Fig. 14c). Together, these results suggest that PARN removes oligo(A) tails from the 3′ end of RMRP.
FIG 14.
PARN KD leads to longer oligo(A) tails for RMRP. (a) Pie charts depicting the fractions of oligo(A) reads at different 3′ ends of RMRP in control and PARN KD cells. (b) Line plot depicting oligo(A) tail lengths at the canonical 3′ end in control and PARN KD cells. The numbers in parentheses represent the average number of A's under each condition. (c) Line plots depicting oligo(A) tail lengths at precursor positions in control and PARN KD cells. The numbers in parentheses represent the average number of A's under each condition. (d) Representative Northern blots depicting steady-state levels of U6 snRNA or RMRP in control and PARN KD cells. The RNA levels were normalized to a 5S rRNA loading control (averages ± SD for three biological replicates).
Since PARN deadenylates both U6 snRNA and RMRP, we asked whether PARN knockdown affects the steady-state levels of these RNAs. Unexpectedly, PARN knockdown had no effect on the steady-state levels of U6 and only a slight effect on the steady-state levels of RMRP (∼11% decrease compared to the control) (Fig. 14d). Together with the increase in the percentage of oligoadenylated reads for these RNAs upon PARN depletion, two possible conclusions can be reached. First, PARN acts to promote the stability of only a subpopulation of these RNAs, and therefore, the effect on steady-state levels is not significant. Second, PARN deadenylates the majority of these RNA populations, but the lack of PARN depletion is not sufficient to trigger the decay of these RNAs in a 3′-5′ manner, possibly due to the presence of protective protein complexes, like the Lsm 2-8 complex for U6 snRNA or the Rpp20-Rpp25 complex for RMRP (39, 40).
DISCUSSION
PARN′s possible role in regulating the stability of multiple ncRNAs.
PARN is mutated in a severe form of DC and familial pulmonary fibrosis (10, 11). We, along with others, have shown that PARN stabilizes hTR by removing oligo(A) tails from its 3′ end. However, it is possible that the severe phenotype of DC observed in patients with PARN mutations is due to the misregulation of other RNAs. We investigated a few stable noncoding RNAs to test this hypothesis and found that PARN knockdown leads to an increase in oligoadenylated ends for a number of ncRNAs, such as U6 and RMRP (Fig. 13 and 14). However, PARN knockdown does not affect the steady-state levels of either of these RNAs in HeLa cells (Fig. 14d). On the other hand, PARN knockdown affects both the processing and the stability of human Y RNAs (Fig. 1, 2, 6, 7, and 8).
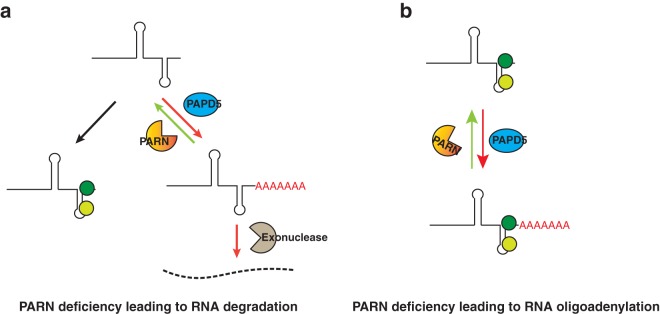
This suggests that PARN affects the processing of ncRNAs by two different mechanisms (Fig. 15). For some ncRNAs, PARN competes with PAPD5 to limit the addition of oligo(A) tails on the 3′ end. When PARN is limiting, oligo(A) tails are recognized by a 3′-to-5′ exonuclease, for example, EXOSC10 in the case of hTR or DIS3L for Y RNAs, which leads to the degradation of the RNA (Fig. 15a). For other ncRNAs, PARN competes with PAPD5 to limit the addition of oligo(A) tails on the 3′ end, but the 3′ end is well protected by a protein complex, which prevents RNA degradation even in the case of low PARN levels (Fig. 15b). This would hold true for RMRP and U6, as well as H/ACA snoRNAs described previously (41). One aspect of this process is that RNA turnover would simply require dissociation of the protective protein complex from the 3′ end and therefore could be used by the cell to regulate the levels of ncRNAs in a PARN-dependent manner (see below).
FIG 15.
Two modes of PARN-mediated regulation of ncRNAs. (a) PARN competes with PAPD5 to limit the addition of oligo(A) tails to the 3′ end of the RNA, and conditions where PARN is limiting lead to the degradation of the RNA through recognition of the oligo(A) tails, as is the case for hTR degradation by EXOSC10 or Y RNA degradation by DIS3L. (b) PARN removes oligo(A) tails from the 3′ end of the RNA, but the preassembled RNP is stabilized through the presence of protein partners at the end that block exonucleolytic degradation.
One interesting possibility is that PARN acts a general regulator of ncRNA stability in mammalian cells. PARN could exert its effect through two general mechanisms. First, programmed cellular pathways could downregulate or upregulate the expression of PARN in cells, depending on the response to stimuli at the cellular level or to cell stages during growth and development. Second, the proteins at the 3′ ends of ncRNAs, which presumably protect them from destabilization by PARN knockdown (as is the case for U6 or RMRP), could themselves be downregulated by the cell, which would sensitize the RNA to the activity of PARN. PARN-mediated RNA decay (PMD) could therefore be a widespread cellular phenomenon to establish homeostasis in cellular ncRNA (and mRNA) levels, and further research is warranted to test this hypothesis.
Y RNAs are processed by PARN and EXOSC10 in human cells.
Y RNAs are abundant RNA Pol III-transcribed ncRNAs in human cells, posited to play a role in DNA replication, histone mRNA maturation, and RNA quality control in the cell (42, 43). We found that Y RNA 3′ maturation is a complicated process governed by multiple 3′-to-5′ exonucleases. First, Y RNAs are trimmed to a position 5 nucleotides upstream of the canonical 3′ end in the steady state in HeLa cells (Fig. 3a and b). Second, this processing is promoted by PAPD5-mediated oligoadenylation of the 3′ end, since PAPD5 knockdown leads to a strong decrease in the percentage of reads terminating at the abundant end and an increase in the number of reads terminating at the mature end (Fig. 6). Third, both PARN and EXOSC10 are involved in the trimming of Y RNAs, since knockdown of the two enzymes causes an increase in the proportion of reads that terminate at the canonical end (Fig. 6).
We also found that Y RNA levels decrease upon both PARN and EXOSC10 knockdown in HeLa cells (Fig. 1a). Furthermore, Y RNA levels could be rescued partially or completely under each condition upon PAPD5 knockdown, providing direct evidence for 3′-end oligoadenylation being a destabilizing mechanism for Y RNAs (Fig. 1b and c). Interestingly, DIS3 knockdown also affects Y RNA levels in HeLa cells, although the effect is not as strong as that of EXOSC10 knockdown for Y1 and Y3 RNAs (Fig. 2a). DIS3 associates with the exosome in both the nucleus and the cytoplasm (31, 32), which suggests that EXOSC10 and DIS3 could have redundant functions in the trimming of Y RNAs to the abundant −5 end in conjunction with PARN.
Analysis of Y RNA levels in HeLa cells depleted of two other 3′-to-5′ exonucleases, DIS3L and DIS3L2, suggests that DIS3L might be responsible for degrading Y RNAs when PARN or EXOSC10/DIS3 enzyme is limiting in the cell. DIS3L knockdown leads to an ∼2-fold increase in the levels of Y1 and Y3 RNAs (Fig. 2a), and PAPD5 knockdown alone had a similar effect on Y RNA levels (Fig. 2d), suggesting that PAPD5-mediated oligoadenylation leads to the degradation of Y RNAs by DIS3L when PARN or EXOSC10 is limiting.
How do these different nucleases coregulate the maturation and stability of Y RNAs in the cell? Differential subcellular localization of Y RNAs, as well as of EXOSC10, DIS3, DIS3L, and PARN in human cells, provides some clues to this question. Human Y RNAs localize to both the nucleus and the cytoplasm, and the nuclear localization is observed at discrete puncta called perinucleolar compartments (PNCs) (43–45). DIS3 and PARN both shuttle between the nucleus and the cytoplasm, whereas EXOSC10 is predominantly localized to the nucleus in the cell (31, 32, 46, 47). DIS3L and DIS3L2 are both predominantly cytoplasmic enzymes, although DIS3L is also found at discrete sites in the nucleus (31–34). Therefore, Y RNA can be susceptible to the activities of these nucleases at different stages of its life cycle, based on its interaction with different protein partners. For example, Y RNAs could be trimmed to the abundant −5 end by PARN and EXOSC10 posttranscription in the nucleus, aided by oligoadenylation by PAPD5 (48–50). If PARN or EXOSC10/DIS3 is absent, the RNA could be recognized by the oligo(A) tails at its 3′ end, leading to degradation by DIS3L. Y RNAs could also be degraded in the cytoplasm by DIS3L after nuclear export, which could be a part of the normal recycling of the Y RNAs in human cells.
Y RNA deficiency could lead to the severe phenotype of DC observed in patients with PARN mutations.
The severe phenotype of DC observed due to PARN mutations is intriguing, not least because of severely short telomere lengths and earlier onset compared to mutations in the telomerase components, like DKC1 and TERC (11, 51, 52). Two previously published reports suggested that defects in Y RNA maturation could contribute to this severe phenotype. First, PARN patient cells have a defective response to UV irradiation stress compared to controls (52). Y RNAs, in complex with the Ro protein, accumulate in the nucleus upon UV stress as part of the cell's DNA damage response mechanism, suggesting an important function in adaptation to UV stress (53, 54). Therefore, PARN deficiency could negatively impact the cell's DNA damage response mechanism. Second, PARN patient cells arrest in the G0/G1 cell cycle phase (10). Y RNAs have been shown to be important for the initiation of DNA replication in mammalian cells, and Y RNA knockdown arrests cells in late G1 phase (16, 17, 24, 55). Interestingly, this function of Y RNAs is independent of its binding to the Ro protein, which suggests that newly transcribed Y RNAs in the nucleus could be important for this process (24). Therefore, lack of proper 3′-end processing of Y RNAs could impact their function upstream of interaction with Ro protein, which would be a likely scenario in PARN- or EXOSC10-deficient cells. Analysis of Y RNA levels and maturation in PARN patient cells would be important in establishing the causality between Y RNA function and the DC phenotype.
MATERIALS AND METHODS
Cell culture.
HeLa cells purchased from the ATCC were cultured in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum (FBS), 1× GlutaMax, penicillin-streptomycin, and normocin. Cells were regularly checked for mycoplasma contamination.
RNA interference in HeLa cells.
siRNAs targeting PAPD5, DIS3, DIS3L, TOE1, DIS3L2 (ON-TARGETplus SMARTpool formulation), and PARN (siGenome SMARTpool configuration) were purchased from Dharmacon. siRNAs targeting EXOSC10 were purchased from Qiagen (a pool of four siRNAs). Allstars negative-control siRNA from Qiagen was used as a negative control. Individual siRNAs targeting PARN, EXOSC10, or PAPD5 (used for Fig. S1 in the supplemental material) were purchased from Dharmacon in the ON-TARGETplus formulation. Approximately 100,000 cells were seeded per well in a six-well plate for transfections. Transfections were carried out 24 h after seeding with Interferin reagent (Polyplus) according to the manufacturer's instructions. siRNA concentrations were limited to 5 nM for each siRNA. Cells were harvested 48 to 60 h posttransfection.
Northern analysis of RNAs.
RNA extraction was carried out using the Quick RNA miniprep kit from Zymo Research. Approximately 5 μg of total RNA was separated on an 8% polyacrylamide gel and transferred to a nylon membrane (Nytran; GE Healthcare). Blots were probed with 32P-labeled probes and imaged on a Typhoon FLA 9500 phosphorimager. The probes used for specific RNAs were as follows: U6, ATATGGAACGCTTCACGAATTTGC; RMRP, AGCCGCGCTGAGAATGAGCCCCGTGT; Y1, GAACAAGGAGTTCGATCTGTAACTGACTG; Y3, GTGATCAATTAGTTGTAAACACCACTGCACTCGGACCAGCC; Y4, GATAACCCACTACCATCGGACCAGCC; Y5, GGGGAGACAATGTTAAATCAAC. The probes for 5S rRNA have been described previously (12). The Northern blots were quantified on Image Quant 5.2, and intensities were normalized to the loading control. Statistically significant differences were calculated using a two-tailed unpaired Student t test.
3′-end deep sequencing of RNAs.
Libraries for individual RNAs were prepared as previously described (12, 21). Briefly, 5 μg of total RNA was Ribozero treated to deplete ribosomal RNAs. The RNA was then dephosphorylated using rAPid alkaline phosphatase (Roche) and ligated to two different 3′ appendices. cDNA was prepared from the ligated RNA using Superscript III RT, and selected cDNA was amplified using barcoded reverse primers through 3′ rapid amplification of cDNA ends (RACE). Reverse primers were selected for unique matches across all the RNA sequences available in the NCBI database to rule out off-target amplification. RACE products corresponding to 100 to 500 nt in length were purified on a 1% agarose gel, and libraries were amplified using universal Illumina primers. The libraries were analyzed on a bioanalyzer and quantified on Qubit and were sequenced on an Illumina MiSeq desktop sequencer as previously described (21).
An excess of 200,000 reads was obtained from each library. Reads of interest were selected using barcodes in the reverse primer of 3′ RACE and the appendix, as well as the exact match for the RNA sequence downstream of the barcode. Any reads containing mismatches in these two sequences were not included in the analysis. The resultant reads were analyzed for information regarding the nature of the 3′ end and the number of reads under each condition. For determination of canonical ends, sequences were mapped to the RNA sequence deposited in the NCBI database. Differences were compared using Z scores of the two population proportions under different knockdown conditions and converted into P values for statistically significant differences.
Supplementary Material
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/MCB.00264-17.
REFERENCES
- 1.Allmang C, Kufel J, Chanfreau G, Mitchell P, Petfalski E, Tollervey D. 1999. Functions of the exosome in rRNA, snoRNA and snRNA synthesis. EMBO J 18:5399–5410. doi: 10.1093/emboj/18.19.5399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.van Hoof A, Lennertz P, Parker R. 2000. Three conserved members of the RNase D family have unique and overlapping functions in the processing of 5S, 5.8S, U4, U5, RNase MRP and RNase P RNAs in yeast. EMBO J 19:1357–1365. doi: 10.1093/emboj/19.6.1357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yong J, Kasim M, Bachorik JL, Wan L, Dreyfuss G. 2010. Gemin5 delivers snRNA precursors to the SMN complex for snRNP biogenesis. Mol Cell 38:551–562. doi: 10.1016/j.molcel.2010.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fu D, Collins K. 2003. Distinct biogenesis pathways for human telomerase RNA and H/ACA small nucleolar RNAs. Mol Cell 11:1361–1372. doi: 10.1016/S1097-2765(03)00196-5. [DOI] [PubMed] [Google Scholar]
- 5.Shukla S, Parker R. 2016. Hypo- and hyper-assembly diseases of RNA-protein complexes. Trends Mol Med 22:615–628. doi: 10.1016/j.molmed.2016.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wan J, Yourshaw M, Mamsa H, Rudnik-Schöneborn S, Menezes MP, Hong JE, Leong DW, Senderek J, Salman MS, Chitayat D, Seeman P, von Moers A, Graul-Neumann AL, Kornberg AJ, Castro-Gago M, Sobrido M-J, Sanefuji M, Shieh PB, Salamon N, Kim RC, Vinters HV, Chen Z, Zerres K, Ryan MM, Nelson SF, Jen JC. 2012. Mutations in the RNA exosome component gene EXOSC3 cause pontocerebellar hypoplasia and spinal motor neuron degeneration. Nat Genet 44:704–708. doi: 10.1038/ng.2254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Boczonadi V, Müller JS, Pyle A, Munkley J, Dor T, Quartararo J, Ferrero I, Karcagi V, Giunta M, Polvikoski T, Birchall D, Princzinger A, Cinnamon Y, Lützkendorf S, Piko H, Reza M, Florez L, Santibanez-Koref M, Griffin H, Schuelke M, Elpeleg O, Kalaydjieva L, Lochmüller H, Elliott DJ, Chinnery PF, Edvardson S, Horvath R. 2014. EXOSC8 mutations alter mRNA metabolism and cause hypomyelination with spinal muscular atrophy and cerebellar hypoplasia. Nat Commun 5:4287. doi: 10.1038/ncomms5287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Di Donato N, Neuhann T, Kahlert A-K, Klink B, Hackmann K, Neuhann I, Novotna B, Schallner J, Krause C, Glass IA, Parnell SE, Benet-Pages A, Nissen AM, Berger W, Altmüller J, Thiele H, Weber BHF, Schrock E, Dobyns WB, Bier A, Rump A. 2016. Mutations in EXOSC2 are associated with a novel syndrome characterised by retinitis pigmentosa, progressive hearing loss, premature ageing, short stature, mild intellectual disability and distinctive gestalt. J Med Genet 53:419–425. doi: 10.1136/jmedgenet-2015-103511. [DOI] [PubMed] [Google Scholar]
- 9.Lardelli RM, Schaffer AE, Eggens VRC, Zaki MS, Grainger S, Sathe S, Van Nostrand EL, Schlachetzki Z, Rosti B, Akizu N, Scott E, Silhavy JL, Heckman LD, Rosti RO, Dikoglu E, Gregor A, Guemez-Gamboa A, Musaev D, Mande R, Widjaja A, Shaw TL, Markmiller S, Marin-Valencia I, Davies JH, de Meirleir L, Kayserili H, Altunoglu U, Freckmann ML, Warwick L, Chitayat D, Blaser S, Ccedil;ağlayan AO, Bilguvar K, Per H, Fagerberg C, Christesen HT, Kibaek M, Aldinger KA, Manchester D, Matsumoto N, Muramatsu K, Saitsu H, Shiina M, Ogata K, Foulds N, Dobyns WB, Chi NC, Traver D, Spaccini L, Bova SM, Gabriel SB, Gunel M, Valente EM, Nassogne M-C, Bennett EJ, Yeo GW, Baas F, Lykke-Andersen J, Gleeson JG. 2017. Biallelic mutations in the 3′ exonuclease TOE1 cause pontocerebellar hypoplasia and uncover a role in snRNA processing. Nat Genet 49:457–464. doi: 10.1038/ng.3762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dhanraj S, Gunja SMR, Deveau AP, Nissbeck M, Boonyawat B, Coombs AJ, Renieri A, Mucciolo M, Marozza A, Buoni S, Turner L, Li H, Jarrar A, Sabanayagam M, Kirby M, Shago M, Pinto D, Berman JN, Scherer SW, Virtanen A, Dror Y. 2015. Bone marrow failure and developmental delay caused by mutations in poly(A)-specific ribonuclease (PARN). J Med Genet 52:738–748. doi: 10.1136/jmedgenet-2015-103292. [DOI] [PubMed] [Google Scholar]
- 11.Stuart BD, Choi J, Zaidi S, Xing C, Holohan B, Chen R, Choi M, Dharwadkar P, Torres F, Girod CE, Weissler J, Fitzgerald J, Kershaw C, Klesney-Tait J, Mageto Y, Shay JW, Ji W, Bilguvar K, Mane S, Lifton RP, Garcia CK. 2015. Exome sequencing links mutations in PARN and RTEL1 with familial pulmonary fibrosis and telomere shortening. Nat Genet 47:512–517. doi: 10.1038/ng.3278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shukla S, Schmidt JC, Goldfarb KC, Cech TR, Parker R. 2016. Inhibition of telomerase RNA decay rescues telomerase deficiency caused by dyskerin or PARN defects. Nat Struct Mol Biol 23:286–292. doi: 10.1038/nsmb.3184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tseng C-K, Wang H-F, Burns AM, Schroeder MR, Gaspari M, Baumann P. 2015. Human telomerase RNA processing and quality control. Cell Rep 13:2232–2243. doi: 10.1016/j.celrep.2015.10.075. [DOI] [PubMed] [Google Scholar]
- 14.Nguyen D, St-Sauveur VG, Bergeron D, Dupuis-Sandoval F, Scott MS, Bachand F. 2015. A polyadenylation-dependent 3′ end maturation pathway is required for the synthesis of the human telomerase RNA. Cell Rep 13:2244–2257. doi: 10.1016/j.celrep.2015.11.003. [DOI] [PubMed] [Google Scholar]
- 15.Moon DH, Segal M, Boyraz B, Guinan E, Hofmann I, Cahan P, Tai AK, Agarwal S. 2015. Poly(A)-specific ribonuclease (PARN) mediates 3′-end maturation of the telomerase RNA component. Nat Genet 47:1482–1488. doi: 10.1038/ng.3423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Christov CP, Gardiner TJ, Szuts D, Krude T. 2006. Functional requirement of noncoding Y RNAs for human chromosomal DNA replication. Mol Cell Biol 26:6993–7004. doi: 10.1128/MCB.01060-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Krude T, Christov CP, Hyrien O, Marheineke K. 2009. Y RNA functions at the initiation step of mammalian chromosomal DNA replication. J Cell Sci 122:2836–2845. doi: 10.1242/jcs.047563. [DOI] [PubMed] [Google Scholar]
- 18.Stein AJ, Fuchs G, Fu C, Wolin SL, Reinisch KM. 2005. Structural insights into RNA quality control: the Ro autoantigen binds misfolded RNAs via its central cavity. Cell 121:529–539. doi: 10.1016/j.cell.2005.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Köhn M, Ihling C, Sinz A, Krohn K, Hüttelmaier S. 2015. The Y3** ncRNA promotes the 3′ end processing of histone mRNAs. Genes Dev 29:1998–2003. doi: 10.1101/gad.266486.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hilcenko C, Simpson PJ, Finch AJ, Bowler FR, Churcher MJ, Jin L, Packman LC, Shlien A, Campbell P, Kirwan M, Dokal I, Warren AJ. 2013. Aberrant 3′ oligoadenylation of spliceosomal U6 small nuclear RNA in poikiloderma with neutropenia. Blood 121:1028–1038. doi: 10.1182/blood-2012-10-461491. [DOI] [PubMed] [Google Scholar]
- 21.Goldfarb KC, Cech TR. 2013. 3′ terminal diversity of MRP RNA and other human noncoding RNAs revealed by deep sequencing. BMC Mol Biol 14:23. doi: 10.1186/1471-2199-14-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kheir EGA, Krude T. 2017. Non-coding Y RNAs associate with early replicating euchromatin concordantly with the origin recognition complex (ORC). J Cell Sci 130:1239–1250. doi: 10.1242/jcs.197566. [DOI] [PubMed] [Google Scholar]
- 23.Hendrick JP, Wolin SL, Rinke J, Lerner MR, Steitz JA. 1981. Ro small cytoplasmic ribonucleoproteins are a subclass of La ribonucleoproteins: further characterization of the Ro and La small ribonucleoproteins from uninfected mammalian cells. Mol Cell Biol 1:1138–1149. doi: 10.1128/MCB.1.12.1138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gardiner TJ, Christov CP, Langley AR, Krude T. 2009. A conserved motif of vertebrate Y RNAs essential for chromosomal DNA replication. RNA 15:1375–1385. doi: 10.1261/rna.1472009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wolin SL, Belair C, Boccitto M, Chen X, Sim S, Taylor DW, Wang H-W. 2014. Non-coding Y RNAs as tethers and gates. RNA Biol 10:1602–1608. doi: 10.4161/rna.26166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.VaňáčovአWolf J, Martin G, Blank D, Dettwiler S, Friedlein A, Langen H, Keith G, Keller W. 2005. A new yeast poly(A) polymerase complex involved in RNA quality control. PLoS Biol 3:e189-12. doi: 10.1371/journal.pbio.0030189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kadaba S, Krueger A, Trice T, Krecic AM, Hinnebusch AG, Anderson J. 2004. Nuclear surveillance and degradation of hypomodified initiator tRNAMet in S. cerevisiae. Genes Dev 18:1227–1240. doi: 10.1101/gad.1183804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.LaCava J, Houseley J, Saveanu C, Petfalski E, Thompson E, Jacquier A, Tollervey D. 2005. RNA degradation by the exosome is promoted by a nuclear polyadenylation complex. Cell 121:713–724. doi: 10.1016/j.cell.2005.04.029. [DOI] [PubMed] [Google Scholar]
- 29.Rammelt C, Bilen B, Zavolan M, Keller W. 2011. PAPD5, a noncanonical poly(A) polymerase with an unusual RNA-binding motif. RNA 17:1737–1746. doi: 10.1261/rna.2787011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schmid M, Jensen TH. 2008. The exosome: a multipurpose RNA-decay machine. Trends Biochem Sci 33:501–510. doi: 10.1016/j.tibs.2008.07.003. [DOI] [PubMed] [Google Scholar]
- 31.Staals RHJ, Bronkhorst AW, Schilders G, Slomovic S, Schuster G, Heck AJR, Raijmakers R, Pruijn GJM. 2010. Dis3-like 1: a novel exoribonuclease associated with the human exosome. EMBO J 29:2358–2367. doi: 10.1038/emboj.2010.122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tomecki R, Kristiansen MS, Lykke-Andersen S, Chlebowski A, Larsen KM, Szczesny RJ, Drazkowska K, Pastula A, Andersen JS, Stepien PP, Dziembowski A, Jensen TH. 2010. The human core exosome interacts with differentially localized processive RNases: hDIS3 and hDIS3L. EMBO J 29:2342–2357. doi: 10.1038/emboj.2010.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lubas M, Damgaard CK, Tomecki R, Cysewski D, Jensen TH, Dziembowski A. 2013. Exonuclease hDIS3L2 specifies an exosome-independent 3“-5” degradation pathway of human cytoplasmic mRNA. EMBO J 32:1855–1868. doi: 10.1038/emboj.2013.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Malecki M, Viegas SC, Carneiro T, Golik P, Dressaire C, Ferreira MG, Arraiano CM. 2013. The exoribonuclease Dis3L2 defines a novel eukaryotic RNA degradation pathway. EMBO J 32:1842–1854. doi: 10.1038/emboj.2013.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Faehnle CR, Walleshauser J, Joshua-Tor L. 2014. Mechanism of Dis3l2 substrate recognition in the Lin28-let-7 pathway. Nature 514:252–256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pirouz M, Du P, Munafò M, Gregory RI. 2016. Dis3l2-mediated decay is a quality control pathway for noncoding RNAs. Cell Rep 16:1861–1873. doi: 10.1016/j.celrep.2016.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Labno A, Warkocki Z, Kuliński T, Krawczyk PS, Bijata K, Tomecki R, Dziembowski A. 2016. Perlman syndrome nuclease DIS3L2 controls cytoplasmic non-coding RNAs and provides surveillance pathway for maturing snRNAs. Nucleic Acids Res 44:10437–10453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Langley AR, Chambers H, Christov CP, Krude T. 2010. Ribonucleoprotein particles containing non-coding Y RNAs, Ro60, La and nucleolin are not required for Y RNA function in DNA replication. PLoS One 5:e13673-10. doi: 10.1371/journal.pone.0013673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mattijssen S, Welting TJM, Pruijn GJM. 2010. RNase MRP and disease. WIREs RNA 1:102–116. doi: 10.1002/wrna.9. [DOI] [PubMed] [Google Scholar]
- 40.Achsel T, Brahms H, Kastner B, Bachi A, Wilm M, Luhrmann R. 1999. A doughnut-shaped heteromer of human Sm-like proteins binds to the 3′-end of U6 snRNA, thereby facilitating U4/U6 duplex formation in vitro. EMBO J 18:5789–5802. doi: 10.1093/emboj/18.20.5789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Berndt H, Harnisch C, Rammelt C, Stohr N, Zirkel A, Dohm JC, Himmelbauer H, Tavanez JP, Huttelmaier S, Wahle E. 2012. Maturation of mammalian H/ACA box snoRNAs: PAPD5-dependent adenylation and PARN-dependent trimming. RNA 18:958–972. doi: 10.1261/rna.032292.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Köhn M, Pazaitis N, Hüttelmaier S. 2013. Why YRNAs? About versatile RNAs and their functions. Biomolecules 3:143–156. doi: 10.3390/biom3010143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kowalski MP, Krude T. 2015. Functional roles of non-coding Y RNAs. Int J Biochem Cell Biol 66:20–29. doi: 10.1016/j.biocel.2015.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Matera AG, Frey MR, Margelot K, Wolin SL. 1995. A perinucleolar compartment contains several RNA polymerase III transcripts as well as the polypyrimidine tract-binding protein, hnRNP I. J Cell Biol 129:1181–1193. doi: 10.1083/jcb.129.5.1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Farris AD, Puvion-Dutilleul F, Puvion E, Harley JB, Lee LA. 1997. The ultrastructural localization of 60-kDa Ro protein and human cytoplasmic RNAs: association with novel electron-dense bodies. Proc Natl Acad Sci U S A 94:3040–3045. doi: 10.1073/pnas.94.7.3040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Körner CG, Wahle E. 1997. Poly(A) tail shortening by a mammalian poly(A)-specific 3′-exoribonuclease. J Biol Chem 272:10448–10456. doi: 10.1074/jbc.272.16.10448. [DOI] [PubMed] [Google Scholar]
- 47.Gao M, Fritz DT, Ford LP, Wilusz J. 2000. Interaction between a poly(A)-specific ribonuclease and the 5′ cap influences mRNA deadenylation rates in vitro. Mol Cell 5:479–488. doi: 10.1016/S1097-2765(00)80442-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gendron M, Roberge D, Boire G. 2001. Heterogeneity of human Ro ribonucleoproteins (RNPS): nuclear retention of Ro RNPS containing the human hY5 RNA in human and mouse cells. Clin Exp Immunol 125:162–168. doi: 10.1046/j.1365-2249.2001.01566.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Fouraux MA, Bouvet P, Verkaart S, van Venrooij WJ, Pruijn GJM. 2002. Nucleolin associates with a subset of the human Ro ribonucleoprotein complexes. J Mol Biol 320:475–488. doi: 10.1016/S0022-2836(02)00518-1. [DOI] [PubMed] [Google Scholar]
- 50.Sim S, Weinberg DE, Fuchs G, Choi K, Chung J, Wolin SL. 2009. The subcellular distribution of an RNA quality control protein, the Ro autoantigen, is regulated by noncoding Y RNA binding. Mol Biol Cell 20:1555–1564. doi: 10.1091/mbc.E08-11-1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mason PJ, Bessler M. 2011. The genetics of dyskeratosis congenita. Cancer Genet 204:635–645. doi: 10.1016/j.cancergen.2011.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tummala H, Walne A, Collopy L, Cardoso S, de la Fuente J, Lawson S, Powell J, Cooper N, Foster A, Mohammed S, Plagnol V, Vulliamy T, Dokal I. 2015. Poly(A)-specific ribonuclease deficiency impacts telomere biology and causes dyskeratosis congenita. J Clin Invest 125:2151–2160. doi: 10.1172/JCI78963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chen X, Quinn AM, Wolin SL. 2000. Ro ribonucleoproteins contribute to the resistance of Deinococcus radiodurans to ultraviolet irradiation. Genes Dev 14:777–782. [PMC free article] [PubMed] [Google Scholar]
- 54.Chen X, Smith JD, Shi H, Yang DD, Flavell RA, Wolin SL. 2003. The Ro autoantigen binds misfolded U2 small nuclear RNAs and assists mammalian cell survival after UV irradiation. Curr Biol 13:2206–2211. doi: 10.1016/j.cub.2003.11.028. [DOI] [PubMed] [Google Scholar]
- 55.Wang I, Kowalski MP, Langley AR, Rodriguez R, Balasubramanian S, Hsu S-TD, Krude T. 2014. Nucleotide contributions to the structural integrity and DNA replication initiation activity of noncoding Y RNA. Biochemistry 53:5848–5863. doi: 10.1021/bi500470b. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.