ABSTRACT
Reductive genome evolution has purged many metabolic pathways from obligate intracellular Rickettsia (Alphaproteobacteria; Rickettsiaceae). While some aspects of host-dependent rickettsial metabolism have been characterized, the array of host-acquired metabolites and their cognate transporters remains unknown. This dearth of information has thwarted efforts to obtain an axenic Rickettsia culture, a major impediment to conventional genetic approaches. Using phylogenomics and computational pathway analysis, we reconstructed the Rickettsia metabolic and transport network, identifying 51 host-acquired metabolites (only 21 previously characterized) needed to compensate for degraded biosynthesis pathways. In the absence of glycolysis and the pentose phosphate pathway, cell envelope glycoconjugates are synthesized from three imported host sugars, with a range of additional host-acquired metabolites fueling the tricarboxylic acid cycle. Fatty acid and glycerophospholipid pathways also initiate from host precursors, and import of both isoprenes and terpenoids is required for the synthesis of ubiquinone and the lipid carrier of lipid I and O-antigen. Unlike metabolite-provisioning bacterial symbionts of arthropods, rickettsiae cannot synthesize B vitamins or most other cofactors, accentuating their parasitic nature. Six biosynthesis pathways contain holes (missing enzymes); similar patterns in taxonomically diverse bacteria suggest alternative enzymes that await discovery. A paucity of characterized and predicted transporters emphasizes the knowledge gap concerning how rickettsiae import host metabolites, some of which are large and not known to be transported by bacteria. Collectively, our reconstructed metabolic network offers clues to how rickettsiae hijack host metabolic pathways. This blueprint for growth determinants is an important step toward the design of axenic media to rescue rickettsiae from the eukaryotic cell.
KEYWORDS: Rickettsia, evolution, host-parasite relationship, host-pathogen interactions, intracellular parasites, metabolic modeling, phylogenetic analysis, phylogenomics
IMPORTANCE
A hallmark of obligate intracellular bacteria is the tradeoff of metabolic genes for the ability to acquire host metabolites. For species of Rickettsia, arthropod-borne parasites with the potential to cause serious human disease, the range of pilfered host metabolites is unknown. This information is critical for dissociating rickettsiae from eukaryotic cells to facilitate rickettsial genetic manipulation. In this study, we reconstructed the Rickettsia metabolic network and identified 51 host metabolites required to compensate patchwork Rickettsia biosynthesis pathways. Remarkably, some metabolites are not known to be transported by any bacteria, and overall, few cognate transporters were identified. Several pathways contain missing enzymes, yet similar pathways in unrelated bacteria indicate convergence and possible novel enzymes awaiting characterization. Our work illuminates the parasitic nature by which rickettsiae hijack host metabolism to counterbalance numerous disintegrated biosynthesis pathways that have arisen through evolution within the eukaryotic cell. This metabolic blueprint reveals what a Rickettsia axenic medium might entail.
INTRODUCTION
The members of the order Rickettsiales (Alphaproteobacteria) are obligate intracellular bacteria found in species across nearly every major lineage of Eukaryota (1). Robust phylogeny estimation places the families Rickettsiaceae, Anaplasmataceae, and Midichloriaceae as sisters to the mitochondrial progenitor (2), with the basal rickettsial lineage now recognized as a new order (Holosporales ord. nov.) (3). Rickettsial species of medical and agricultural significance are almost exclusively found in the families Rickettsiaceae and Anaplasmataceae (4), though we lack information regarding the impact on host fitness of many of the formally recognized species and most of the putative species. Despite this, all members of the order Rickettsiales can be considered metabolic parasites, as tremendous reductive genome evolution has resulted in a seemingly inextricable metabolic dependence on the eukaryotic cell (5).
Members of family Rickettsiaceae (e.g., Rickettsia and Orientia species) are unique among the members of the order Rickettsiales in lysing the host phagocytic vacuole and residing primarily in the host cytosol (6). This lifestyle affords access to a broad range of host metabolites, some of which may not be available to vacuole-enclosed bacteria. Indeed, members of the family Rickettsiaceae have a more diminished metabolic capability than other members of the order Rickettsiales (7–9). Unlike other notable cytosolic pathogens (e.g., Listeria monocytogenes, Shigella flexneri, Francisella tularensis), members of the family Rickettsiaceae require the host cell for replication and cannot persist extracellularly (10). For the genus Rickettsia, which contains dozens of formally recognized species ranging from nonpathogens to serious human pathogens (11), the doubling time is typically 8 to 12 h (12). It has been suggested that slow rickettsial growth correlates with a large array of transport systems and limiting metabolite availability in the host cytosol (13), a phenomenon better realized when considering the large numbers of rickettsiae capable of occupying a single cell.
The metabolic deficiency of Rickettsia spp. is well established, with studies from the pregenomics era illuminating a limited oxidative metabolism (14). Despite evidence of a functional pyruvate dehydrogenase complex (PDC) and tricarboxylic acid (TCA) cycle (15–20), glycolysis/gluconeogenesis enzymatic activities were undetectable (15, 17, 21). Import of host Glu (22, 23) and Gln (24) was determined, with a metabolic flux among Glu, Gln, and 2-oxaloglutarate indicating the importance of Glu as an energy source (15, 20, 23, 25, 26). Glu oxidation was shown to drive electron transport coupled to oxidative phosphorylation (22, 27–29), with generated ATP facilitating import of Pro (30, 31) and Lys (32, 33). Import of Ser and Gly was demonstrated to be critical for rickettsial growth (34, 35), with Ser-Gly interconversion via serine hydroxymethyltransferase (GlyA) observed (35). These findings, combined with characterized uptake of Met (36, 37), indicated that Rickettsia species are likely auxotrophic for the majority of proteogenic amino acids.
Despite generating ATP via Glu oxidation, Rickettsia species were shown to possess an ATP/ADP symporter, termed nucleotide translocase (Tlc1), which exchanges host ATP for bacterial ADP without a change in the total adenylate pool (38). Rickettsial membranes were also shown to be permeable to NAD+, another host energy source (39). Uptake of UDP-glucose was characterized and suggested to provide the main sugar source for the synthesis of the slime layer, lipopolysaccharide (LPS), and peptidoglycan (PGN) (21). Studies of nucleotide transport and metabolism revealed the import of AMP, GMP, and UMP, indicating that host-acquired ribonucleotides likely serve as building blocks for RNA synthesis and as precursors for deoxyribonucleotide production (40–42). Unlike ATP, the specific transport systems for NAD+, UDP-glucose, and nucleoside monophosphates were not identified; however, the emerging consensus was that rickettsiae contain an elaborate assemblage of transport systems for the acquisition of host metabolites (43).
The first reported genome sequence of a Rickettsia species, Rickettsia prowazekii, confirmed the limited metabolic capacity of rickettsiae, with genes underpinning glycolysis and biosynthesis of pentose phosphates, amino acids, and nucleotides largely absent (44). An abundance of pseudogenes pointed to “reductive genome evolution” (45–47), a characteristic shared by all subsequently sequenced Rickettsia genomes (1). Phylogenomics studies observed additional depleted metabolic pathways, particularly those for B vitamins, many cofactors, and pentose phosphates (48–50). Efforts in the postgenomics era identified rickettsial import of the glycerophospholipid precursors dihydroxyacetone phosphate (DHAP) and sn-glycerol 3-phosphate (G3P) (51, 52) and also the cosubstrate S-adenosylmethionine (SAM), for which a novel transporter (EamA) was characterized (53). Additionally, substrate ranges of four Tlc1 paralogs (Tlc2 to Tlc5) were investigated, revealing that none function in energy exchange, yet two import host ribonucleotides (Tlc4 [CTP, UTP, and GDP] and Tlc5 [GTP and GDP]) (54). Collectively, these genome-driven studies further defined rickettsial metabolic parasitism, illustrating the dependence of rickettsiae on host metabolites.
Although they possess limited metabolic activity, isolated rickettsiae are unable to grow extracellularly. Despite tremendous efforts in rickettsial genetic manipulation over the last 2 decades (55), the lack of an axenic medium continues to impede progress. Knowledge of the range of essential metabolites would greatly facilitate the establishment of an axenic culture, as has previously been shown for another obligate intracellular bacterium, Coxiella burnetii (56, 57). In the present study, we used 84 rickettsial genomes to reconstruct the Rickettsia metabolic and associated transport networks. Our results indicate that 51 host metabolites are required to compensate for the patchwork Rickettsia metabolic pathways and that the majority of cognate transporters for these metabolites are unknown. Our analysis also reveals several pathways that contain isolated holes (missing enzymes); similar patterns across a range of taxonomically diverse taxa imply alternative metabolic strategies shared by divergent bacteria. The comprehensive metabolic blueprint developed here illuminates what a successful Rickettsia axenic medium might entail. Furthermore, our work elucidates the parasitic nature by which rickettsiae pilfer host metabolites to counterbalance the many biosynthesis pathways that have disintegrated through evolution within the eukaryotic cell.
RESULTS AND DISCUSSION
Our metabolic network reconstruction entailed the analysis of 84 Rickettsia genomes, with 74 genomes comprising the three well-established rickettsial lineages (48, 58, 59): the transitional group (n = 10), the typhus group (n = 15), and the spotted fever group (n = 49). Ten additional genomes are from basal lineages that do not form a monophyletic group. When considering only closed genomes (n = 54), Rickettsia genomes range in size from 1.5 Mb (Rickettsia bellii strain RML369-C, with 1,429 coding sequences [CDS]) to 1.1 Mb (Rickettsia typhi strain Wilmington, with 838 CDS), with a core genome including 621 protein-encoding genes. Variation in accessory genomes is dominated by pseudogenization events, but for some species (e.g., Rickettsia buchneri [60] and Rickettsia peacockii [61]), it can also consist of extensive mobile genetic elements. The genes encoding components of the metabolic and transport networks described below make up 25% of the core genome, indicating a highly conserved strategy for parasitizing the eukaryotic cytoplasm.
Synthesis of cell envelope glycans requires host precursors.
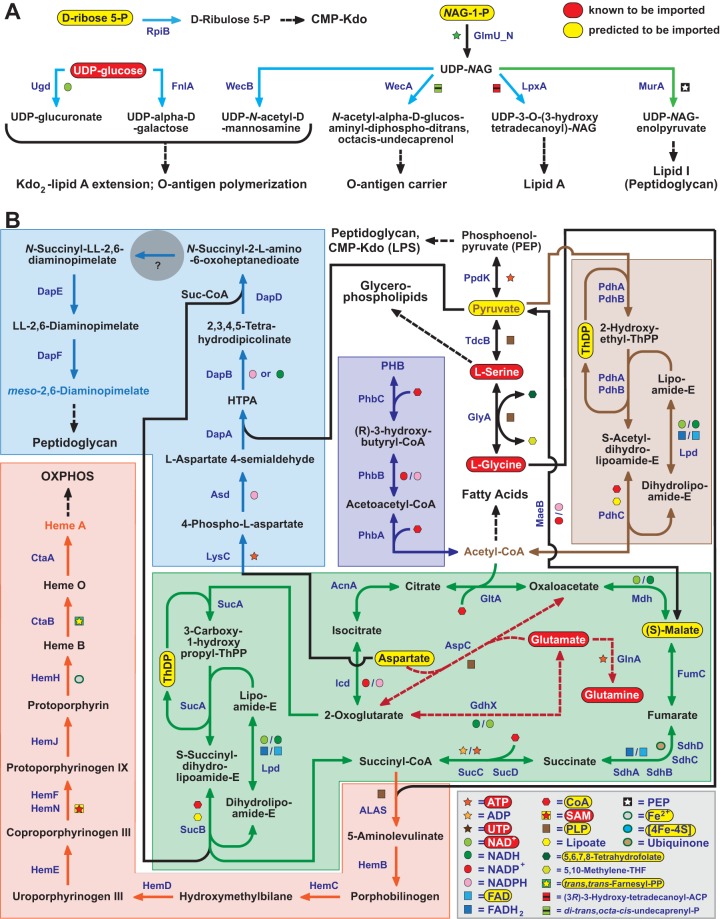
Without the products of glycolysis and the pentose phosphate pathway, rickettsiae must acquire host metabolites for the synthesis of PGN and LPS (Fig. 1A; see Fig. S1A in the supplemental material). The first amino sugar able to be synthesized in the stem pathway leading to PGN/LPS biosynthesis is UDP-N-acetyl-α-d-glucosamine (UDP-NAG), which is typically synthesized from glucosamine-1-P (GlcN-1-P) in bacteria by the bifunctional protein GlmU. The GlmU C-terminal acetyltransferase domain generates N-acetylglucosamine-1-P (NAG-1-P) from GlcN-1-P, while the N-terminal uridyltransferase domain converts NAG-1-P to UDP-NAG (62) (see Fig. S1B). However, the predominant eukaryotic pathway (including vertebrates and arthropods) generates NAG-1-P from NAG-6-P, not GlcN-1-P (see Fig. S1C), suggesting that rickettsiae acquire NAG-1-P from the host. Remarkably, the rickettsial GlmU proteins lack the entire C-terminal acetyltransferase domain (see Fig. S1D) and thus are streamlined for the conversion of host-acquired NAG-1-P to UDP-NAG. Generated UDP-NAG then enters pathways for PGN and LPS (both lipid A and O-antigen) biosynthesis (see Fig. S1E; Fig. S1F, respectively).
FIG 1 .
Rickettsia species synthesize cell envelope glycoconjugates from imported host sugars and fuel the TCA cycle with a range of host-acquired metabolites. (A) Previously shown to be imported, UDP-glucose is predicted to yield UDP-glucuronate and UDP-α-d-galactose, sugars likely to be used in LPS synthesis (light blue pathway lines). Synthesis of UDP-N-acetyl-d-mannosamine, another sugar likely incorporated into LPS, as well as pathways for O-antigen, lipid A, and lipid I of PGN (green pathway line), initiates with UDP-NAG. Without glycolysis enzymes, rickettsiae are predicted to import host NAG-1-P and convert this charged sugar to UDP-NAG via the uridyltransferase GlmU. d-Ribose 5-P, which is required to initiate CMP-Kdo synthesis, is also predicted to be imported from the host, provided that Rickettsia species lack enzymes of the pentose phosphate pathway. (B) Rickettsia species must acquire pyruvate for generation of PEP and acetyl-CoA. Pyruvate interconversions with Ser, Gly (via Ser), and malate are likely mediated by additional import of these molecules, ensuring that enough pyruvate enters the PDC to yield acetyl-CoA (brown). Aside from entering the TCA cycle (green), acetyl-CoA is also used in fatty acid biosynthesis and production of PHB (dark blue), a storage molecule that is metabolized when host energy sources are unavailable. Imported malate, glutamine (Gln), and glutamate (Glu) likely regulate the flow of acetyl-CoA into the TCA cycle, with Gln/Glu interconversions with 2-oxaloglutarate and oxaloacetate providing additional energy (dashed burgundy pathway lines). Generated aspartate is essential for initiation of the synthesis of DAP, which is used in PGN biosynthesis (light blue), a pathway nearly conserved except for a central hole (gray circle). DAP synthesis also requires generated succinyl-CoA, which is also used to synthesize porphyrins (orange). HTPA, (2S,4S)-4-hydroxy-2,3,4,5-tetrahydrodipicolinate.
Rickettsia species import six host metabolites that are required for the synthesis of cell envelope glycoconjugates and glycerophospholipids. Download FIG S1, PDF file, 2 MB (2MB, pdf) .
Copyright © 2017 Driscoll et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
UDP-NAG is also a source for the generation of UDP-N-acetyl-d-mannosamine, one of three sugars predicted to make up the precursors for extension of the Kdo2 (two 3-deoxy-d-manno-octulosonic acid residues)-lipid A acceptor and O-antigen polymerization (see Fig. S1F). The other two sugars, UDP-glucuronate and UDP-α-d-galactose, must derive from host-acquired UDP-glucose (21), which is a highly abundant amino sugar in vertebrate cells (63). The complete synthesis of lipid A also requires the import of host d-ribose 5-P, an essential precursor of CMP-Kdo synthesis. CMP-Kdo, which is added to lipid IV(A) in sequential steps (64), is typically synthesized from d-ribose 5-P using five enzymes (see Fig. S1F). The rickettsial CMP-Kdo biosynthesis pathway is complete, except for a single pathway hole (see Materials and Methods for a more complete definition of a pathway hole); it lacks the phosphatase KdsC (see Fig. S1G to I). Similarly, the rickettsial biosynthesis of diaminopimelate (DAP), a component of the PGN stem peptide, also contains a pathway hole; it lacks the N-succinyldiaminopimelate aminotransferase DapC or related enzymes (Fig. 1B; see Fig. S2). Otherwise, pathways for PGN and LPS biosynthesis are highly conserved in rickettsial genomes, indicating that acquisition of host NAG-1-P, UDP-glucose, and d-ribose 5-P suffices for initiation of the metabolism of cell envelope glycans (Fig. 1A).
The Rickettsia DAP biosynthesis pathway contains a hole for the conversion of N-succinyl-l-2-amino-6-oxopimelate to N-succinyl-ll-2,6-DAP. Download FIG S2, PDF file, 0.4 MB (454.3KB, pdf) .
Copyright © 2017 Driscoll et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
The Rickettsia glycerophospholipid biosynthesis pathway contains a hole for the conversion of G3P to LPA. Download FIG S3, PDF file, 0.4 MB (422.4KB, pdf) .
Copyright © 2017 Driscoll et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
A range of imported metabolites fuels the TCA cycle.
The TCA cycle is a key metabolic pathway that uses the oxidation of acetyl coenzyme A (acetyl-CoA) to generate energy, as well as important substrates for other biosynthetic processes. The principal source of acetyl-CoA is pyruvate, generated by the breakdown of sugars via glycolysis—a pathway that rickettsiae do not possess. Rickettsiae must obtain pyruvate for a variety of reasons, including use in DAP biosynthesis (leading to PGN) and for the generation of phosphoenolpyruvate (PEP), a cofactor in the biosynthesis of both PGN and LPS (Fig. 1B). Pyruvate interconversions with PEP, Ser, Gly (via Ser), and also malate from the TCA cycle indicate an intricate network for the regulation of pyruvate entry into the PDC, which generates acetyl-CoA. Rickettsiae have been shown previously to uptake Ser and Gly (34, 35), and our analysis suggests the need, and potentially the capacity, to import pyruvate and/or malate from host cells as well: all 84 rickettsial genomes encode Auxin Efflux Carrier (AEC) transporters (YfdV), which are known to transport small dicarboxylates including malate (65, 66), and almost all encode on an adjacent locus the gene for MaeB, which interconverts malate and pyruvate.
PDC-generated acetyl-CoA is used primarily for the TCA cycle and fatty acid biosynthesis (Fig. 1B). A pathway for the conversion of acetyl-CoA to polyhydroxybutyrate (PHB), a storage molecule that is metabolized when host energy sources are unavailable (67), is also found in most rickettsial genomes, though it is noticeably less conserved in spotted fever group species (see Fig. S10). This is consistent with the variability of observed intracytoplasmic vacuoles across different Rickettsia species (68), as these distinct structures are known to house accumulated polyhydroxyalkanoates such as PHB (69, 70).
Interestingly, a pathway for acetyl-CoA generation from acetate, employing acetate kinase and phosphate acetyltransferase, is highly conserved in typhus and transitional group rickettsiae but diminished in most other rickettsial lineages (data not shown). Like the PDC, this pathway requires CoA, a cofactor that rickettsiae must acquire from the host (discussed below), as well as acetate, which was previously shown to be imported by typhus group rickettsiae (71). Remarkably, only typhus group rickettsiae contain WecH, an enzyme used by Escherichia coli to acetylate O-antigen (72), indicating that these rickettsiae alone can incorporate acetate into LPS.
In addition to host-acquired malate and Rickettsia-generated acetyl-CoA, imported Gln and Glu also regulate the flow of acetyl-CoA into the TCA cycle (Fig. 1B). The presence in all rickettsiae of three enzymes underpinning these conversions (AspC, GdhX, and GlnA) is consistent with the essentiality of Glu/Gln uptake and the fact that Gln is the most abundant free amino acid in human blood and other tissues (73). Asp generated via these processes is essential for initiation of the synthesis of DAP, which was previously shown to be a component of the rickettsial PGN stem peptide (74). Succinyl-CoA produced by the TCA cycle is also required for DAP synthesis, as well as the synthesis of porphyrins important to electron transport. Like nonphotosynthetic eukaryotes and other members of the class Alphaproteobacteria, rickettsieae initiate porphyrin biosynthesis by forming δ-aminolevulinic acid from Gly and succinyl-CoA. Ten conserved enzymes, including the alternative protoporphyrinogen IX oxidase HemJ (75), are used to generate heme A, the prosthetic group of cytochromes associated with cytochrome c oxidase of the electron transport chain. Collectively, a wide array of characterized (Ser, Glyc, Glu, Gln) and predicted (pyruvate, malate) host metabolites, as well as numerous host cofactors (see “Rickettsieae cannot synthesize B vitamins and most other cofactors” below) are used by rickettsiae to drive the TCA cycle, which in turn feeds the pathways for the generation of DAP and heme A for PGN and oxidative phosphorylation, respectively.
Host metabolites and a noncanonical carboxylation complex are required to initiate fatty acid and glycerophospholipid biosynthesis.
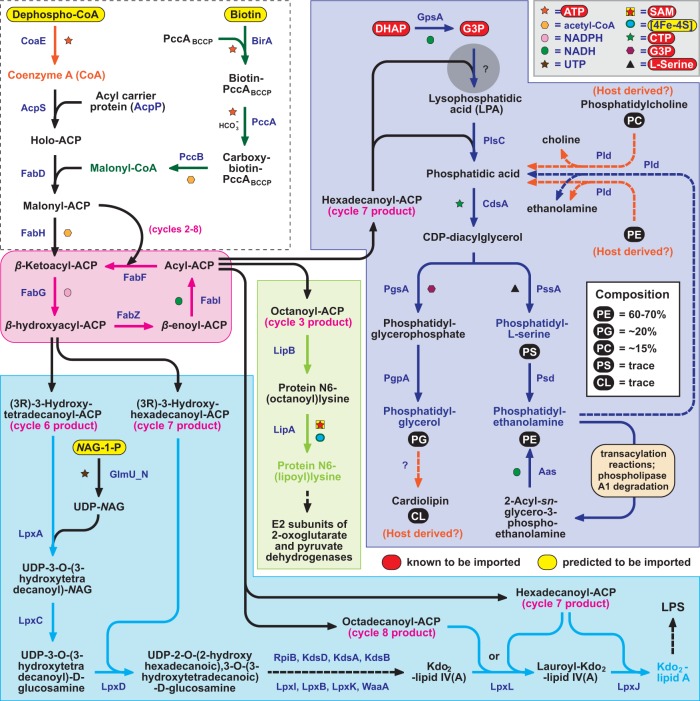
Bacterial fatty acid biosynthesis initiates with malonyl acyl carrier protein (malonyl-ACP), which is formed from holo-ACP and malonyl-CoA (Fig. 2). Rickettsial ACP synthesis requires CoA, which our analysis suggests is likely generated from host-acquired dephospho-CoA, since the CoA synthesis pathway contains only the terminal enzyme (dephospho-CoA kinase). Malonyl-CoA synthesis in bacteria occurs predominantly via the carboxylation of acetyl-CoA by acetyl-CoA carboxylase (ACC), a multisubunit complex consisting of a biotin carboxyl carrier protein (AccB), a biotin carboxylase (AccC), and a carboxyltransferase (AccA/AccD). Rickettsiae lack the genes that encode any of the ACC subunits; however, our analysis has identified all of the functional domains required for the carboxylation of acetyl-CoA in two conserved rickettsial proteins originally annotated as subunits of propionyl-CoA carboxylase (PCC). Rickettsiae have no other enzymes associated with propanoate metabolism; consequently, we propose that Rickettsia PCC substitutes for bacterial ACC in the generation of malonyl-CoA. Other lines of evidence support this proposition: (i) PCCs are included within a large family of diverse biotin-dependent carboxylases with highly variable substrate specificities (76); (ii) several archaeal biotin-dependent carboxylases are bifunctional, metabolizing both acetyl-CoA and propionyl-CoA (77, 78); and (iii) Wolbachia PCC can complement an E. coli ACC mutant (79). Finally, rickettsiae must still import host biotin to initiate acetyl-CoA carboxylation (60), as they are equipped with biotin ligase and the BioY transporter but lack the enzymes for de novo biotin synthesis (although see “In light of metabolic parasitism, what, exactly, is a Rickettsia endosymbiont?” below for a rare exception).
FIG 2 .
Rickettsia species synthesize fatty acids and glycerophospholipids from host precursors. (Dashed box) Predicted imported substrates dephospho-CoA and biotin are required for holo-ACP synthesis and loading of the biotin carboxyl carrier protein, respectively, which collectively lead to the formation of malonyl-ACP. As Rickettsia species lack ACC, a conserved PCC complex is predicted to generate malonyl-CoA (green). Type II fatty acid synthesis (pink) is utilized by Rickettsia species to generate octanoyl-ACP for lipoate synthesis (light green), β-hydroxyacyl-ACPs (14C and 16C) and acyl-ACPs (16C and 18C) for Kdo2-lipid A synthesis (light blue), and hexadecanoyl-ACP for glycerophospholipid synthesis (dark blue). Acyl chain incorporation into lipid A follows the structure deduced for R. typhi (171). While both DHAP and G3P are known to be imported from the host (51, 52), Rickettsia species lack enzymes to generate LPA via the first incorporation of hexadecanoyl-ACP (the gray circle represents this pathway hole). All enzymes subsequent to this step are highly conserved (see Fig. S10), generating the predominant glycerophospholipids characterized in Rickettsia membranes (inset at upper right) (81). Dashed lines illustrate possible Pld-mediated salvage pathways for bacterial PE, as well as host PE and PC (orange). If PC (82) and cardiolipin (81) are incorporated into Rickettsia membranes, both must be acquired from the host (orange).
Major offshoots of the fatty acid cycle used by rickettsiae include octanoyl-ACP for lipoate synthesis, β-hydroxyacyl-ACPs (14C and 16C) and acyl-ACPs (16C and 18C) for Kdo2-lipid A synthesis, and hexadecanoyl-ACP for glycerophospholipid synthesis (Fig. 2). For the latter pathway, hexadecanoyl-ACP is used in the conversion of G3P to lysophosphatidic acid (LPA). Previous studies have shown that rickettsiae can either directly import G3P or import DHAP and subsequently generate G3P using GpsA (51, 52). Although our analysis failed to identify a dedicated acyltransferase for the actual generation of LPA (i.e., PlsB or PlsX/Y), enzymes are present for the conversion of LPA to most of the glycerophospholipids previously characterized from rickettsial membranes, including phosphatidylethanolamine (PE), phosphatidylglycerol, and phosphatidyl-l-serine (80, 81). The implications for this pathway hole are discussed in more detail below (“Whole pathways or pathway holes?”).
Despite prior reports of their presence in rickettsial membrane extracts, rickettsiae cannot synthesize cardiolipin or phosphatidylcholine (PC). This indicates that either these previous studies failed to completely eliminate host glycerophospholipids from isolated rickettsial membranes, or host cardiolipin and/or PC are incorporated into rickettsial membranes in trace amounts. Rickettsiae have been shown to hydrolyze PC during infection (82), and all rickettsial genomes do encode phospholipase D (Pld) (83), which was shown to release choline from PC in vitro (84). Rickettsia genomes also encode one or more choline transferases (LicD) that are known to ligate surface molecules with phosphorylcholine, a process often associated with modulation of host immunity (85). It is conceivable that Pld-mediated host membranolysis provides rickettsiae with choline. In theory, this process could also provide rickettsiae with a direct source of phosphatidic acid for the generation of glycerophospholipids, as would Pld-mediated recycling of PE (Fig. 2), intriguing concepts to consider in light of a missing G3P acyltransferase.
Rickettsia spp. must acquire host isoprenes and terpenoids to synthesize ubiquinone and the lipid carriers for PGN and LPS.
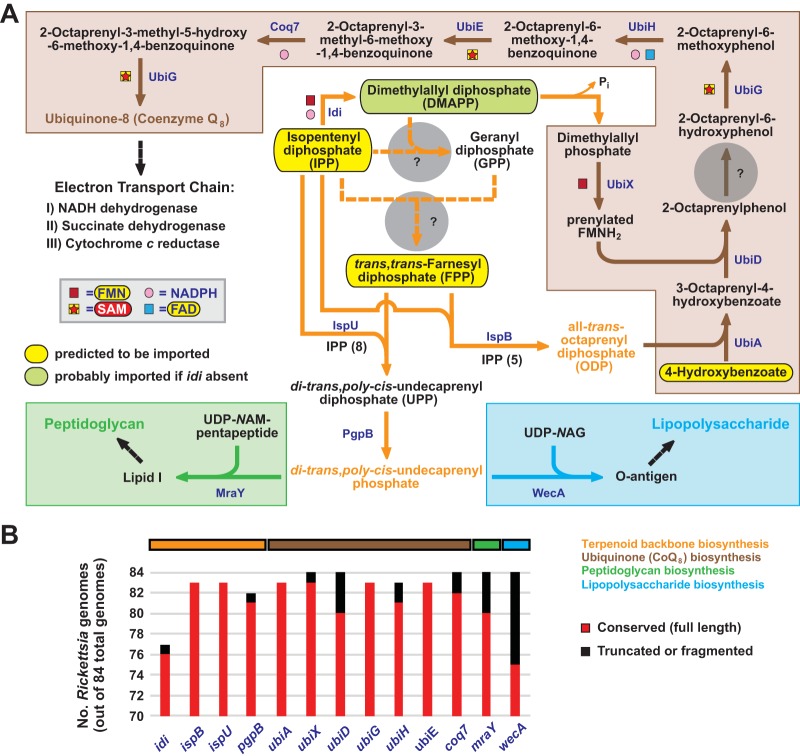
Isoprenes are the general precursors of all terpenoids, a diverse family of compounds involved in a variety of cell functions, including electron transport and membrane synthesis. Bacteria employ the mevalonate or nonmevalonate (MEP/DOXP) pathway to synthesize isoprenes, namely, isopentenyl diphosphate (IPP) and dimethylallyl diphosphate (DMAPP). DMAPP is the source of the dimethylallyl phosphate used in the first step of the ubiquinone (CoQ8) pathway. It is also combined with IPP to generate geranyl diphosphate (GPP), which in turn combines with IPP to yield trans,trans-farnesyl diphosphate (FPP), the precursor of terpenoid backbone synthesis. FPP combined with eight IPP molecules generates di-trans,poly-cis-undecaprenyl diphosphate (UPP), which is subsequently dephosphorylated to yield di-trans,poly-cis-undecaprenyl phosphate, the lipid carrier for lipid I of PGN (see Fig. S1E) and O-antigen of LPS (see Fig. S1F). Alternatively, FPP combined with five IPP molecules generates all-trans-octaprenyl diphosphate (ODP), a precursor of CoQ8 synthesis.
Rickettsiae lack enzymes of either the mevalonate or the MEP/DOXP pathway and thus must acquire isoprenes from the host (Fig. 3A). Most rickettsial species can generate DMAPP from IPP using IPP δ-isomerase (Idi); however, a relative lack of idi conservation (Fig. 3B) and the similar molecular weights of DMAPP and IPP isomers suggest the possibility, in some species, of a dual transport mechanism for these isoprenes, similar to that observed for DHAP and G3P (52). Our analysis revealed strong conservation of undecaprenyl-PP synthase (IspU) and octaprenyl-PP synthase (IspB), which synthesize UPP and ODP, respectively, but a significant pathway hole (absence of IspA; see Fig. S4A to C) that prevents the synthesis of either GPP or FPP. Consequently, rickettsiae need to import host FPP to synthesize these lipid carriers.
FIG 3 .
Rickettsia species must import host isoprenes and terpenoids for the synthesis of ubiquinone and the lipid carrier of lipid I and O-antigen. (A) In the absence of a mevalonate or MEP/DOXP pathway for terpenoid synthesis, Rickettsia species must import IPP from the host. If Idi is present, as it is in some species, DMAPP can be synthesized to provide dimethylallyl phosphate for the ubiquinone (CoQ8) pathway. Otherwise, DMAPP must also be imported from the host. Dashed orange lines indicate that no enzymes are present to use IPP and DMAPP for GPP generation, and thus, FPP must also be acquired from the host. Gray circles depict holes in the pathways for the generation of both GPP and FPP. Host-acquired IPP and FPP can then be used by undecaprenyl diphosphate synthase (IspU) and octaprenyl-diphosphate synthase (IspB) to generate terpenoid backbones UPP and ODP, respectively. Via phosphatidylglycerophosphatase B (PgpB), UPP is then converted to di-trans,poly-cis-undecaprenyl phosphate, the lipid carrier for lipid I (green) and O-antigen (light blue). OPP and PHBA are used by PHBA polyprenyltransferase (UbiA) to initiate CoQ8 synthesis (brown). The lack of enzymes to either synthesize chorismate or convert it to PHBA indicates that rickettsiae must import host PHBA, which is an essential host metabolite provided by diet and/or the microbiome. The gray circle indicates a hole (UbiC) in the CoQ8 synthesis pathway. Note that all rickettsial genomes encode UbiB, a putative kinase with an unknown role in CoQ8 biosynthesis (172). (B) Genes involved in terpenoid backbone and CoQ8 biosynthesis are largely conserved. The complete distributions of these genes in 84 Rickettsia genomes (see Fig. S4G) indicate that the most basal lineage of spotted fever group rickettsiae (R. tamurae, R. monacensis, REIP, and R. buchneri strains) lacks idi and thus must also acquire DMAPP from the host.
Pathways for Rickettsia terpenoid and ubiquinone biosynthesis contain holes that are not common in other bacterial genomes. Download FIG S4, PDF file, 1.4 MB (1.4MB, pdf) .
Copyright © 2017 Driscoll et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Synthesis of CoQ8 begins with the conversion of ODP and 4-hydroxybenzoate (PHBA) into 3-octaprenyl-PHBA, a reaction carried out by PHBA polyprenyltransferase (UbiA). Many bacteria synthesize PHBA from chorismate, a precursor of aromatic amino acid synthesis. Although they possess UbiA, rickettsiae lack the enzymes to either synthesize chorismate or convert it to PHBA, indicating that PHBA must be imported from the host (Fig. 3A). The remaining steps in CoQ8 synthesis are fairly well conserved, with the exception of a pathway hole at the hydroxylation of 2-octaprenylphenol (OPP) (see Fig. S4D to F). In E. coli, this reaction is catalyzed by the dedicated monooxygenase UbiI, while a ΔubiI mutant strain produces a low level of CoQ8 and a compound atypical of the CoQ8 pathway (86). A different enzyme is responsible for C-5 hydroxylation of OPP under anaerobic conditions (86), however, indicating that alternative ways to carry out this reaction exist. Taken together, the conservation of the CoQ8 pathway (Fig. 3B; see Fig. S4G), the import of PHBA, and the synthesis of ODP from host-acquired isoprenes and terpenoids indicate that rickettsiae can effectively supply the electron transport chain with canonical CoQ8.
Rickettsiae cannot synthesize B vitamins and most other cofactors.
In contrast to the conserved pathways for the synthesis of porphyrins, lipoate, and CoQ8 by rickettsiae, pathways for the synthesis of B vitamins and other cofactors are incomplete or entirely lacking. Our analysis indicates that at least 11 such molecules must be acquired directly from the host—and possibly more when derivatives are considered—on the basis of the pathways that do function in rickettsiae. Thiamine cannot be synthesized, and rickettsiae do not encode any enzymes that generate thiamine’s five phosphate derivatives; at a minimum, rickettsiae must import thiamine diphosphate (ThDP), as this cofactor is essential for the PDC and the oxoglutarate dehydrogenase complex of the TCA cycle (Fig. 1B). No enzymes are present for the generation of riboflavin from GTP and ribulose-5P or the conversion of riboflavin to flavin mononucleotide (FMN); consequently, rickettsiae must acquire FMN from the host. Flavin adenine dinucleotide (FAD) must also be imported, given that riboflavin kinase/FAD synthetase, which converts FMN to FAD, is absent. Pyridoxal phosphate (PLP) is another B vitamin that rickettsiae cannot synthesize, and given that animals are auxotrophic for PLP, rickettsiae must compete with the host for this substrate. This also applies to biotin and glutathione: rickettsiae contain the enzymes that ligate these substrates to their cellular targets—bifunctional ligase/repressor (BirA) and glutathione S-transferase (GstA), respectively—but not the capacity to synthesize the cofactors.
Rickettsiae can synthesize several other B vitamins and cofactors; however, even these pathways are dependent on precursors that they obtain from the host. The synthesis of pantothenate, the precursor of CoA, has likely been replaced by the ability to generate CoA from host-acquired dephospho-CoA as discussed above. Imported NAD+ (39) serves as a source for the generation of NADP+ via inorganic polyphosphate/ATP-NAD+ kinase, with NAD(P)+ transhydrogenase providing NADH and NADPH. Rickettsiae acquire SAM directly from the host via the highly conserved EamA transporter (see Fig. S10). Interestingly, R. prowazekii can import SAM (53) yet also generate it from host-acquired ATP and l-Met using the SAM synthase MetK (87). This appears to be the exception rather than the rule, however, as MetK-encoding genes are pseudogenized in most other Rickettsia species (data not shown). Our observations are in line with the proposition that the acquisition of metabolite transport systems facilitates the degradation of apposite biosynthetic pathways in rickettsiae (13). Remarkably, the EamA transporters were found piggybacking on integrative conjugative elements that appear to seed Rickettsia genomes with genes associated with intracellular invasion and survival (60); lateral transfer of such elements has occurred with obligate intracellular members of the phylum Bacteroidetes (88).
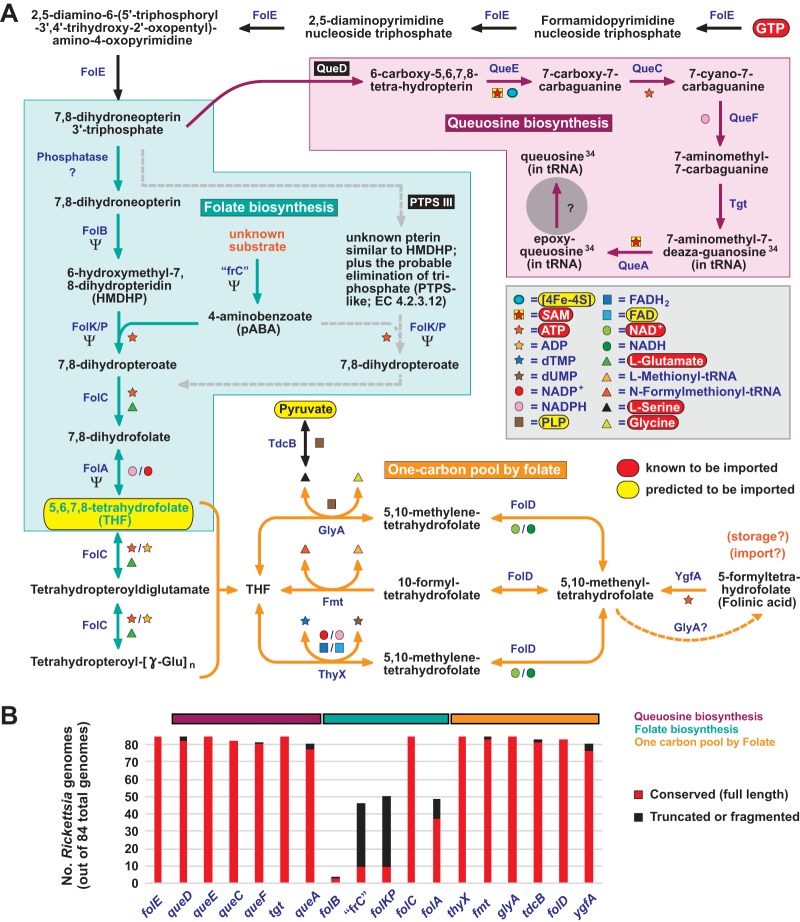
Regarding vitamin B9 (folate), comparative genomics originally indicated a degraded biosynthesis pathway in Rickettsia genomes (i.e., one or more genes encoding FolB, FolK/P, and FolA were missing) and suggested that folate derivatives must be imported from the host (7, 48). More recently, Hunter et al. (89) proposed that the Rickettsia endosymbiont of Ixodes pacificus (REIP) can synthesize the active form of the vitamin, tetrahydrofolate (THF), and provision it to its arthropod host. REIP is closely related to R. buchneri, a species found exclusively in Ixodes scapularis, the tick vector of Lyme disease (90). REIP and R. buchneri occur in high frequency in ticks and are not known to cause disease in vertebrates, collective factors reflecting a possible symbiosis between the host and microbe (90–94). The proposal for REIP provisioning THF to its tick host was based on the identification of a putative PTPS-III enzyme, a 6-pyruvoyltetrahydropterin synthase (PTPS) with an atypical active site, that can functionally replace FolB (95) (Fig. 4A). This “FolB bypass” is a strategy employed by some microbes wherein PTPS-III can directly convert 7,8-dihydroneopterin 3′-triphosphate (DHN-P3) to 6-hydroxymethyl-7,8-dihydropteridin (HMDHP), a function mediated by Glu replacing or accompanying the usual Cys in the PTPS active site (96). This subtle active-site modification alters the typical functions of PTPS enzymes, which include the synthesis of biopterin (97, 98) and queuosine (99, 100). Queuosine is a modified nucleoside that can occupy the first anticodon position of tRNAs for His, Asp, Asn, and Tyr (101), and its synthesis involves the conversion of DHN-P3 to 6-carboxy-5,6,7,8-tetra-hydropterin by the (canonical) PTPS enzyme QueD (Fig. 4A).
FIG 4 .
Rickettsia species lack the capability for de novo folate biosynthesis. (A) Rickettsiae use GTP cyclohydrolase I (FolE) to convert host-acquired GTP (red) to DHN-P3, a precursor of both queuosine (purple) and folate (aquamarine) biosynthesis. The classical THF synthesis pathway (aquamarine arrows), wherein DHN-P3 is dephosphorylated and subsequently converted to HMDHP by dihydropteridin aldolase (FolB), is disintegrating from Rickettsia genomes (Ψ denotes pseudogenization in over 50% of genomes). In the FolB bypass proposed by Hunter et al. (89) (gray dashed arrows), DHN-P3 is directly converted to HMDHP or a structurally similar molecule via PTPS-III (black box). Our analysis instead suggests that this enzyme is QueD (black box), which performs the first committed step in queuosine biosynthesis (see the text for further details). The one-carbon pool by folate (orange arrows) illustrates the role of host-acquired THF and several intermediates in the essential one-carbon transfer reactions that yield pyrimidine deoxynucleoside triphosphates, N-formylmethionyl-tRNA, and Ser/Gly. “frC,” fol_rel_CADD domain-containing protein (TIGR04305). The gray circle represents a hole in the pathway for queuosine synthesis (see Fig. S6). (B) Conservation of 18 genes involved in queuosine and THF biosynthesis and reactions within the one-carbon pool by folate. The complete distributions of these genes in 84 Rickettsia genomes reveals that no single Rickettsia species is capable of de novo folate biosynthesis, while the queuosine biosynthesis and one-carbon pool by folate pathways are highly conserved (see Fig. S5D).
In the present study, we present several lines of evidence that argue against de novo THF synthesis in rickettsiae. First, the REIP enzyme proposed by Hunter et al. to be PTPS-III (QueD in our results) actually contains the canonical PTPS active site (see Fig. S5A). Our analysis further reveals that rickettsial PTPS enzymes are highly conserved across Rickettsia genomes, and none of them contain the active-site modification associated with FolB bypass functionality (see Fig. S5B). Second, QueD catalyzes the first committed step in queuosine biosynthesis (Fig. 4A); surprisingly, this pathway is extremely conserved across the rickettsiae (including REIP), with the exception of a pathway hole at the terminal step converting epoxyqueuosine to queuosine (see Fig. S6). It seems unlikely that QueD would function in the synthesis of both 6-carboxy-5,6,7,8-tetrahydropterin (queuosine pathway) and HMDHP (folate pathway) from the same initial substrate (DHN-P3). Third, we show that it is unlikely that rickettsiae are unable to synthesize the 4-aminobenzoate (pABA) that is incorporated into the pterin ring of HMDHP, as the enzymes that would normally generate pABA from chorismate (namely, PabA/B and PabC) are not present, and chorismate itself is not synthesized and probably not needed by rickettsiae (discussed above for PHBA metabolism). Some genomes do encode “frC,” an atypical fol_rel_CADD domain-containing enzyme (TIGR04305) (102) that may provide a path to pABA generation from an unknown precursor (Fig. 4B; see Fig. S5D); however, the “frC” locus is degraded in almost all rickettsiae (including REIP) and is not likely to be functional. Fourth, phylogeny estimation indicates that rickettsiae once contained the canonical THF synthesis pathway that is complete in most other lineages of Rickettsiales (see Fig. S5C). Fifth, our analysis has found that every Rickettsia genome harbors one or more Fol-encoding pseudogenes (Fig. 4B; see Fig. S5D). While we did not explore the possibility that certain Rickettsia lineages have reacquired folate synthesis functionality via lateral transfer, collectively, our findings indicate that rickettsial genomes are characterized by an eroding folate biosynthesis pathway, with retention of only the enzymes necessary for the initiation of queuosine biosynthesis (FolE, QueD) and carrying out glutamylation of host-acquired THF (FolC). Thus, we predict that rickettsiae import host THF (and/or its derivatives, including folinic acid) to carry out the critical one-carbon transfer reactions involved in amino acid interconversion, pyrimidine synthesis, and translation.
Rickettsia species use DHN-P3 for queuosine biosynthesis and import host THF for one-carbon transfer reactions by folate. Download FIG S5, PDF file, 1.3 MB (1.4MB, pdf) .
Copyright © 2017 Driscoll et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
The Rickettsia queuosine biosynthesis pathway contains a hole for the reduction of epoxyqueuosine to queuosine. Download FIG S6, PDF file, 0.4 MB (414.6KB, pdf) .
Copyright © 2017 Driscoll et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Whole pathways or pathway holes?
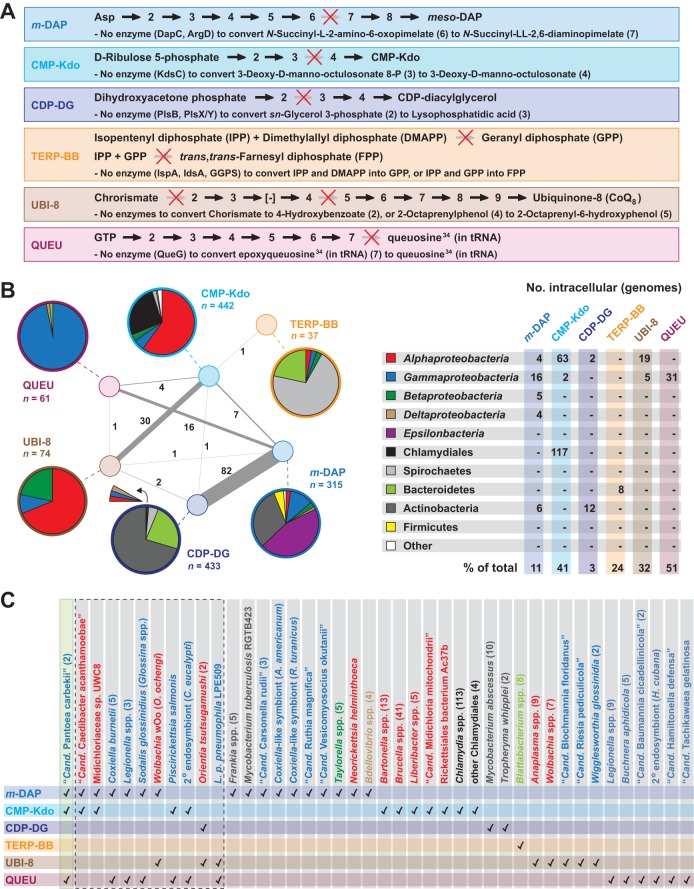
In total, our Rickettsia metabolic reconstruction identified six holes occurring in otherwise conserved pathways for the synthesis of DAP, CMP-Kdo, CDP-diacylglycerol (CDP-DG), terpenoid backbones, CoQ8, and queuosine (Fig. 5A). Evaluation of the occurrence of these pathway holes across other bacterial genomes revealed that rickettsiae are not unique in lacking homologs of the enzymes that typically carry out these reactions (Fig. 5B). Surprisingly, the “Rickettsia-like” pathways for DAP, CMP-Kdo, and CDP-DG biosynthesis were found in a large number of bacterial genomes. For DAP biosynthesis, several alternative routes for the conversion of 2,3,4,5-tetrahydrodipicolinate to DAP have been characterized (103–105), though rickettsiae do not encode any of the enzymes in these other pathways (see Fig. S2D). The large number (n = 360) and diversity of bacterial genomes simply lacking DapC within the predominant DAP pathway likely indicate the existence of an alternative aminotransferase that awaits experimental characterization. Similarly, within the CMP-Kdo biosynthesis pathway, the lack of KdsC homologs across divergent bacteria indicates that these species may use a different, possibly nonspecific, phosphatase to convert 3-deoxy-d-manno-octulosonate 8-P to 3-deoxy-d-manno-octulosonate. In support of this, KdsC is not essential in E. coli (106). Importantly, Rickettsia genomes encode multiple haloacid dehalogenase-like hydrolases (InterPro domain IPR023214) (60) that may provide functions similar to those of KdsC, which also possesses this domain.
FIG 5 .
Comparative analysis of six biosynthetic pathways containing holes. The reconstructed Rickettsia metabolic network revealed holes in six biosynthetic pathways, DAP (m-DAP), CMP-Kdo, CDP-diacylglycerol (CDP-DG), terpenoid backbones (TERP-BB), ubiquinone-8 (UBI-8), and queuosine (QUEU). The distribution of these pathways across other prokaryotic genomes was determined via comparative metabolic pathway analyses. (A) Illustration and description of each hole-containing biosynthetic pathway. A red X indicates the missing enzyme(s) within each pathway, compared to well-characterized biosynthetic pathways for other prokaryotes. (B) Distribution of Rickettsia-like biosynthetic pathways across other prokaryotic genomes. The pie charts at the left indicate the taxonomic breakdown of genomes per biosynthetic pathway, with connections between pathways illustrating the number of genomes containing multiple Rickettsia-like pathways. Note that Rickettsia genomes were excluded from these analyses. At the right, the taxonomic color scheme is shown, with the number of genomes from intracellular species provided. (C) Intracellular species containing one or more Rickettsia-like biosynthetic pathways. The green box depicts the only genome found to contain three Rickettsia-like pathways, that of “Ca. Pantoea carbekii”) (117), which is an extracellular primary symbiont of the brown marmorated stink bug, where it is found in the gastric cecal lumina (see the text for further details). The dashed box indicates genomes containing two Rickettsia-like pathways. Taxa are colored in accordance with the color scheme in panel B.
For CDP-DG synthesis, the lack of a G3P acyltransferase that converts G3P to LPA early in the synthesis of glycerophospholipids is puzzling. The ability to import G3P or import DHAP and convert it to G3P (51, 52), combined with the lack of other metabolic pathways that would use G3P directly, indicates that rickettsiae need to acquire G3P primarily for glycerophospholipid biosynthesis. For acylation of the G3P 1-position, bacteria typically use either PlsB, which primarily uses acyl-ACP as the fatty acyl donor, or PlsY, which uses acyl-PO4 produced from acyl-ACP by PlsX (107) (see Fig. S3A). In addition to rickettsiae, our analysis identified 433 other bacterial genomes that lack the genes encoding PlsB, PlsX, and PlsY (Fig. 5B), indicating that additional G3P acyltransferases remain to be characterized. The presence in 190 bacterial genomes of PlsX without either acyltransferase further supports this determination (see Fig. S3B) and may indicate that as-yet-unidentified G3P acyltransferases could use acyl-PO4 similar to PlsY. The 433 Rickettsia-like genomes that lack all three enzymes are found predominantly in species of Actinobacteria and Bacteroidetes (see Fig. S3C); comparative studies that include rickettsiae and these genomes may prove fruitful for identifying novel G3P acyltransferases.
The Rickettsia-like pathways for the synthesis of terpenoid backbones, CoQ8, and queuosine are far less common in other bacteria (Fig. 5B). For terpenoid backbone synthesis, 37 genomes were found that contained only genes encoding Idi, IspB, and IspU (see Fig. S4A and B), indicating that it is rare for bacteria to generate terpenoid backbones without the ability to synthesize isoprene (IPP, GPP) and terpenoid (FPP) precursors. Regarding CoQ8 synthesis, ubiI and ubiC occur far less frequently than other ubi genes in bacterial genomes (see Fig. S4D). Aside from the nonessentiality of UbiI in E. coli (discussed above), a very recent report proposed reannotation of the various proteobacterial CoQ8 flavin monooxygenases (UbiI, UbiH, UbiF, Coq7) into two novel groups named UbiM and UbiL (108). For UbiL (strictly Alphaproteobacteria proteins previously annotated as UbiH, such as all rickettsial UbiH proteins), the enzyme from Rhodospirillum rubrum was shown to complement ΔubiI and ΔubiH mutant E. coli strains, suggesting that the missing UbiI functionality may be carried out by UbiH in rickettsiae. Regarding UbiC (chorismate lyase), an alternative means to convert chorismate to PHBA was previously discovered for Xanthomonas species (109), indicating that other strategies may yet be uncovered to explain the lack of ubiC in so many bacterial genomes. Still, for rickettsiae and other intracellular bacteria that not only lack the shikimate pathway for the generation of chorismate but are devoid of the enzymes for the generation of aromatic amino acids from chorismate, transport systems must exist for PHBA acquisition from the host. Pathways that lack UbiC in other bacterial genomes may facilitate the identification of alternative mechanisms for the generation (or acquisition) of PHBA to fuel CoQ8 synthesis (see Fig. S4D and E).
The lack of the terminal enzyme (QueG) in the Rickettsia queuosine synthesis pathway implies either that the penultimate product, epoxyqueuosine, occupies position 34 (anticodon wobble position) in tRNAs with GUN anticodons or that another epoxyqueuosine reductase unrelated to QueG is found in rickettsial genomes. The latter scenario seems more plausible on the basis of two lines of evidence. First, QueG requires the cofactor cobalamin (vitamin B12) (110, 111), a large modified tetrapyrrole synthesized by dozens of enzymes (112). Rickettsiae cannot synthesize cobalamin, and no cobalamin-dependent enzymes are encoded within rickettsial genomes (data not shown), indicating that this cofactor is likely not imported. Thus, the absence of QueG correlates with the inability to provide its cofactor, cobalamin. In support of this, none of the 61 bacterial genomes that contain the Rickettsia-like queuosine pathway encode enzymes needed to synthesize cobalamin (data not shown). Second, 45 of these genomes that lack QueG, as well as all rickettsial genomes, encode DUF208-containing proteins (data not shown), which have very recently been characterized as cobalamin-independent epoxyqueuosine reductases unrelated to QueG (113). Genes encoding these proteins (renamed QueH) are often clustered with Que-encoding genes in bacterial genomes. In rickettsial genomes, queH is not physically linked to other Que-encoding genes, though it flanks a gene encoding another tRNA-associated protein, the glycine-tRNA ligase alpha subunit. Regardless, it seems reasonable to postulate that rickettsiae may use QueH to convert epoxyqueuosine to queuosine, although this awaits experimental confirmation.
We analyzed bacteria containing these six Rickettsia-like pathway holes on the basis of their lifestyle (i.e., extracellular versus intracellular) to explore whether reductive genome evolution as a consequence of an intracellular lifestyle might be a driving factor for gene decay within these pathways (Fig. 5B). In most cases, intracellular bacteria from only one or two phylogenetic groups contained Rickettsia-like pathways; for example, the Rickettsia-like queuosine pathway appeared solely in Gammaproteobacteria, while the terpenoid pathway was found only in Bacteroidetes. The exception is DAP, for which the Rickettsia-like pathway was distributed across intracellular species from five groups. The majority of intracellular bacteria that harbor Rickettsia-like pathways contain only one, with 10 taxa containing two (Fig. 5C). It is intriguing to consider that the first genes to decay within obsolete metabolic pathways might hinder but not entirely arrest metabolite biosynthesis, such that the microbe slowly becomes chemically addicted to a host-derived substrate. Suboptimal metabolic pathways may contribute to a low growth rate, a hallmark of rickettsiae (114, 115), which in turn may be advantageous for pilfering metabolites from the eukaryotic cytoplasm while preventing destruction of the host cell.
Nearly all extracellular bacteria that harbor Rickettsia-like pathway holes contain only one, with the remarkable exception of “Candidatus Pantoea carbekii,” which contains three (DAP, CMP-Kdo, and queuosine). Despite an extracellular lifestyle, this gammaproteobacterial species is a primary symbiont of the brown marmorated stink bug, where it resides primarily in the gastric cecal lumina (116). The “Ca. Pantoea carbekii” genome is minimal in size (1.2 Mb), and its gene repertoire contains characteristics of both highly specialized obligate mutualists and facultative species (117). Interestingly, we also identified a potential pseudogene for PlsB in the “Ca. Pantoea carbekii” glycerophospholipid pathway, suggesting a fourth possible Rickettsia-like pathway hole in this symbiont. This stunning degree of convergent evolution in metabolic pathways between rickettsieae and “Ca. Pantoea carbekii” may reflect common modes for pathway modification in the presence of host-acquired metabolites. Altogether, our work illuminates common and rare deviations from conserved metabolic pathways, with similar pathways in unrelated bacteria indicating convergence and possible novel enzymes awaiting characterization.
Black hole son: reductive evolution from mother Rickettsia.
To better understand the evolutionary trajectory of the six Rickettsia pathway holes, we analyzed their composition in the genomes of other taxa in the orders Rickettsiales and Holosporales (see Fig. S7A and B). Genes encoding KdsC, PlsB, UbiC, and UbiI were not detected in any other Rickettsiales/Holosporales genomes, indicating that rickettsial pathways for CMP-Kdo, CDP-DG, and CoQ8 likely never included these enzymes. In contrast, nearly all members of the orders Rickettsiales and Holosporales possess DapC, PlsX/Y, and IspA, with phylogeny estimation supporting their vertical inheritance from a proteobacterial ancestor (see Fig. S7C to F). Thus, it can be parsimoniously inferred that the pathway holes found in Rickettsia DAP, CDP-DG, and terpenoid backbone synthesis pathways reflect reductive evolution that has occurred since divergence from the Rickettsiales/Holosporales ancestor.
The evolutionary trajectory of six Rickettsia biosynthetic pathways that contain holes, or “missing enzymes.” Download FIG S7, PDF file, 2.2 MB (2.3MB, pdf) .
Copyright © 2017 Driscoll et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
The reduction in DAP, CDP-DG, and terpenoid backbone synthesis pathways is seen early within the family Rickettsiaceae, where “Candidatus Arcanobacter lacustris” (118) lacks PlsX/Y. DapC, PlsX/Y, and IspA are also missing from the Rickettsia sister lineage, which includes the scrub typhus agent (Orientia tsutsugamushi), Orientia chuto (119), and “Candidatus Occidentia massiliensis” (120). These results are not surprising, given that O. tsutsugamushi genomes are laden with proliferative/degradative mobile genetic elements (9, 121) and have more missing metabolic enzymes than Rickettsia species do (5, 8, 49). This Rickettsia sister clade lacks idi and must therefore acquire both IPP and DMAPP (in addition to FPP) from the host to initiate the CoQ8 and terpenoid backbone synthesis pathways. All other Rickettsiales/Holosporales genomes likewise lack idi, though they do encode enzymes of the MEP/DOXP pathway for the synthesis of isoprenes, indicating that IPP-DMAPP interconversion is unique to Rickettsia species. Remarkably, phylogeny estimation suggests that rickettsiae likely acquired idi from non-Alphaproteobacteria sources (Fig. S8A).
Phylogeny estimation of Idi and queuosine biosynthesis proteins. Download FIG S8, PDF file, 0.6 MB (651.5KB, pdf) .
Copyright © 2017 Driscoll et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Epoxyqueuosine reductases (QueG or QueH) were not identified in any other Rickettsiales/Holosporales genome besides that of Rickettsia (see Fig. S7A and B). Unexpectedly, only “Candidatus M. mitochondrii” was found to contain the other Que enzymes (QueD, QueE, QueC, QueF, and QueA but not QueH). The strict conservation of Tgt in all genomes suggests that species lacking Que-encoding genes incorporate host-acquired queuine directly into tRNAs with GUN anticodons, similar to the eukaryotic salvage mechanism (122). Phylogeny estimation indicates two origins of Tgt in Rickettsiales, with one clade consisting entirely of taxa that lack the Que-encoding genes (Anaplasmataceae and Holosporales) (Fig. S8B). The origins of the Que-encoding genes themselves are difficult to estimate with confidence (Fig. S8C to H); thus, it cannot be determined if they were present in the Rickettsiales ancestor and subsequently lost by all lineages except rickettsiae and “Ca. M. mitochondrii” or if these two lineages independently acquired them. Why rickettsiae and “Ca. M. mitochondrii” would generate queuosine from GTP rather than use less complex salvage mechanisms found in other members of the orders Rickettsiales and Holosporales is difficult to understand. Regardless, in conjunction with the estimated nonalphaproteobacterial origin of rickettsial idi, this analysis implies that rickettsiae can acquire metabolic enzymes throughout evolution to offset gene decay.
So many imported metabolites, so few characterized transporters.
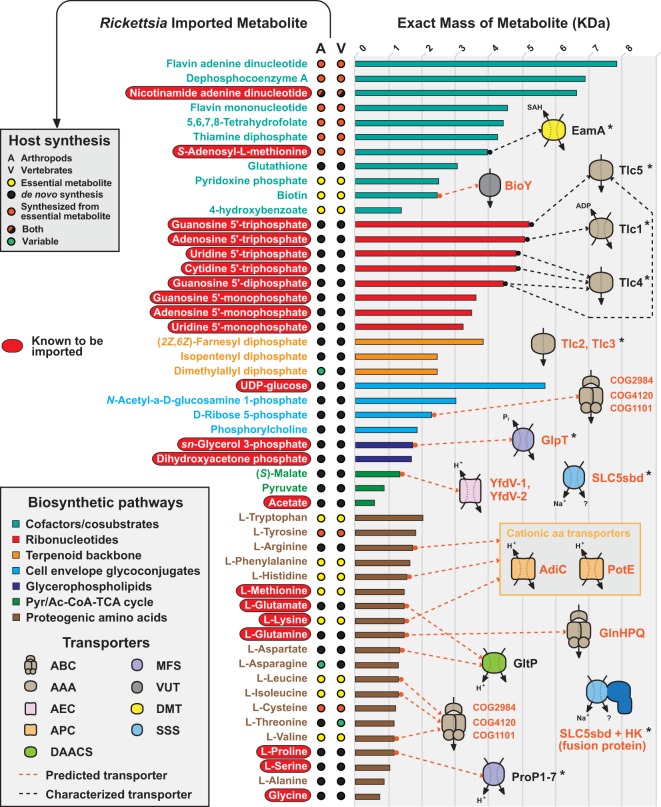
In total, our reconstructed metabolic network predicts that rickettsiae must import 51 host metabolites to supplement their patchwork metabolic pathways (Fig. 6). This adds 30 metabolites to the 21 previously shown to be imported by rickettsiae in either in vivo or in vitro assays. Roughly 80% of these metabolites are synthesized by both vertebrates and arthropods, with the other metabolites essential for both the host and the microbe (Fig. 6, circles at center). Importantly, the metabolites targeted by rickettsiae all belong to highly conserved eukaryotic pathways, which explains their ability to grow in a wide range of cell types. With such a high degree of metabolite thievery, rickettsiae must have in place an arsenal of transport systems to ensure that all of the essential metabolites are imported from the eukaryotic cytoplasm in sufficient quantities to support their growth. This also implies a remarkable level of regulatory control over these systems to avoid starving the host cell of its own essential metabolites too quickly.
FIG 6 .
Synopsis of known and predicted metabolites imported from the eukaryotic cytoplasm by rickettsiae. On the left, metabolites are grouped into biosynthetic pathways (colors are described in the inset at the bottom left), with red ellipses depicting 21 metabolites previously shown to be imported. The remaining 30 metabolites are predicted to be imported on the basis of the metabolic network reconstruction presented in this report. In the center are the biosynthesis capabilities of the metabolites in arthropod and vertebrate genomes (further described in the inset at the top left). Information was obtained from KEGG pathways for arthropods and vertebrates. On the right, within each group, metabolites are ranked by exact mass. Dashed lines connect metabolites with their known (black) or predicted (orange) transport systems (transporter families are listed in the inset at the bottom left). One ABC transporter (COG1101/COG4120/COG2984) is shown twice, as annotations indicate uptake of branched-chain amino acids, as well as monosaccharides (including ribose, galactose, and arabinose). SLC5sbd and SLC5sbd+HK (fusion protein with His kinase domain) transporters are not linked with specific metabolites because of their known broad range of substrates (e.g., sugars, amino acids, organo-cations such as choline, nucleosides, inositols, vitamins, urea, or anions). Asterisks indicate transporters previously shown to be associated with mobile genetic elements and/or predicted to be spread by lateral gene transfer across diverse intracellular bacteria (60, 88, 126, 127). Transporter names and family identifications (123) are as follows: ABC, ATP-binding cassette (3.A.1); AAA, ATP:ADP antiporter (2.A.12); DMT, drug/metabolite transporter (2.A.7); VUT, vitamin uptake transporter (2.A.88); AEC (2.A.69); MFS, major facilitator superfamily (2.A.1); APC, amino acid polyamine organo-cation (2.A.3); DAACS, dicarboxylate/amino acid:cation (Na+ or H+) symporter (2.A.23); SSS, solute:sodium symporter (2.A.21). Phylogenomics analysis indicates that these transporters are highly conserved in rickettsial genomes (see Fig. S10).
Based on a combination of previously characterized transporters and in silico transporter prediction (123), we compiled a modest set (n = 24) of transporters that are highly conserved across Rickettsia genomes (Fig. 6; see Fig. S10). Some transport systems (i.e., those involved in drug efflux, PGN recycling, phospholipid maintenance, etc.) were not considered. Alignment of this transporter set with our set of 51 imported metabolites illustrates that ribonucleotides have the most characterized transport pathways (38, 40–42, 54). Despite unknown transporters for the ribonucleotide monophosphates, our reconstruction shows that the eight ribonucleotides known to be imported can be used by rickettsiae to synthesize the remaining ribonucleotides and all of the deoxyribonucleotides required for replication, transcription, and regulation of the stringent response (see Fig. S9). The only other characterized transporter in our set is EamA, the SAM antiporter (53). Transporters for proteogenic amino acids have not been characterized, despite evidence for rickettsial import of Met, Glu, Lys, Gln, Pro, Ser, and Gly (22–24, 30–37). Predicted transporters of charged amino acids (AdiC, PotE, GltP), Gln (GlnHPQ), and branched-chain amino acids (COG2984/COG4120/COG1101) account for only half of the amino acids that rickettsiae need to import. The substrate ranges of seven pro/betaine symporters (ProP), which typically exchange proline and other osmolytes during osmoregulation (124), might account for the remaining amino acids, given that conserved ProP groups are highly divergent from one another (60). These and/or other transporters must be operational to counterbalance the absence of nearly all amino acid biosynthesis pathways.
Rickettsia transport and metabolism of host-acquired ribonucleotides. Download FIG S9, PDF file, 0.5 MB (490.3KB, pdf) .
Copyright © 2017 Driscoll et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Very few transporters could be predicted for other characterized (NAD, UDP-glucose, DHAP, G3P, and acetate) or putative (all others) imported metabolites. Both branched-chain amino acids and d-ribose 5-P are considered possible substrates of the ABC transporter COG2984/COG4120/COG1101. G3P is possibly transported by the known ATP-independent G3P transporter GlpT, which is highly conserved in rickettsiae; however, G3P import by R. prowazekii was shown to require ATP, indicating the likelihood of another transporter (52). As discussed above, duplicate YfdV transporters are predicted to import malate and possibly pyruvate. A single component of the BioMNY transporter (125), BioY, is conserved in rickettsial genomes and likely imports biotin. Finally, all rickettsial genomes encode at least one SLC5sbd transporter, which is a member of the solute:sodium symporter family. These transporters are widespread across prokaryotes and eukaryotes and work on a wide range of characterized substrates, including sugars, amino acids, organo-cations such as choline, nucleosides, inositols, vitamins, urea, and anions. Some rickettsial SLC5sbd transporters are fused with a BaeS-like, two-component signal transduction histidine kinase (HK) domain. Both SLC5sbd and SLC5sbd+HK proteins are highly proliferated in Orientia and Occidentia genomes (data not shown). While the substrates of these transporters are unknown, their importance in obligate intracellular living is evident, given their spread by lateral transfer across rickettsiae and unrelated obligate intracellular bacteria such as “Candidatus Amoebophilus asiaticus” and Cardinium endosymbionts of arthropods (60, 126, 127).
The requirement for several large metabolites (FAD, dephospho-CoA, NAD, FMN, THF, ThDP, FPP, UDP-glucose) implies that rickettsiae must have transport systems to import these molecules. We did not identify any of the energy-coupling factor transporters (128–130) typically associated with the uptake of large vitamins (or their derivatives) such as folate (131), riboflavin (132), and thiamine (133, 134). Despite demonstration that a TLC protein of environmental chlamydiae can transport NAD+ (135), there is little sequence similarity between this transporter and rickettsial TLC proteins (60). The Rickettsia SLC5sbd transporter shows mild similarity (~23% identical) to the E. coli sodium/pantothenate symporter PanF; however, as discussed above, rickettsiae most likely import dephospho-CoA and not pantothenate, since they cannot metabolize the latter. Finally, we did not identify any transporters for other carbohydrates, isporenes, or terpenoids.
The paucity of transporters relative to the high number of imported metabolites suggests that either the characterized and predicted transporters are not substrate specific or there are other transport systems that remain unidentified. It is worth considering as well that host transport systems may be hijacked by rickettsiae to ensure the delivery of metabolites that are not known to be transported by bacteria (e.g., FAD, UDP-glucose, dephospho-CoA, FPP). Given the possible shared ancestry of rickettsiae and the mitochondrial progenitor (136), a reasonable host transporter to be used by rickettsiae might be the mitochondrial porin, which consists of the voltage-dependent anion channel (VDAC). Involved in the regulation of metabolic and energetic flux across the mitochondrial outer membrane (137), VDAC is already known to transport most of the molecules that rickettsiae need to complement their patchwork metabolic network. Curiously, VDAC was previously found to associate with the rickettsial cell envelope (138, 139), though the functional significance remains unclear. Given that recombinant VDAC can assemble in the outer membrane of E. coli (140), it is possible that VDAC forms functional porins on rickettsial cells. Determining if VDAC or other host transporters are co-opted during rickettsial acquisition of host metabolites is a fascinating area for future research.
In light of metabolic parasitism, what, exactly, is a Rickettsia endosymbiont?
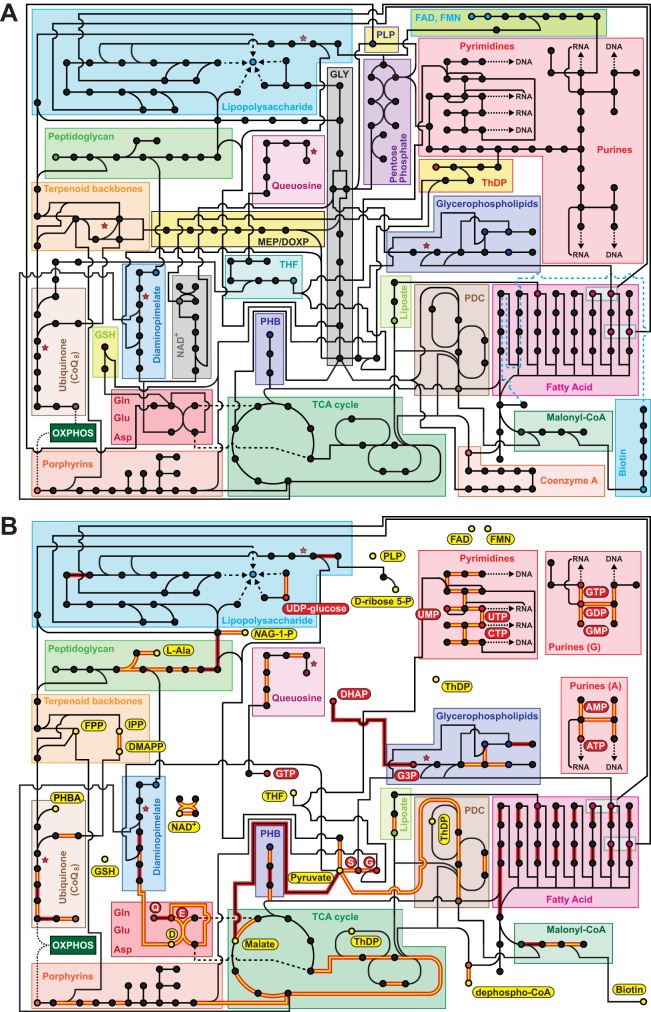
Our metabolic reconstruction illustrates the few biosynthesis pathways that remain in rickettsiae. In most free-living and facultative intracellular bacteria, these pathways are interconnected primarily by glycolysis and the pentose phosphate pathway, two of the most conserved metabolic processes across life’s three domains (Fig. 7A). Rickettsiae have replaced these core biosynthesis hubs by highly elaborate thievery of host metabolites to complement its patchwork metabolic network (Fig. 7B). Imported metabolites are used to initiate many other highly conserved biosynthetic pathways (e.g., PDC, TCA cycle, porphyrins, glycerophospholipids, nucleotides), as well as pathways unique to bacteria (e.g., LPS, PGN, DAP, PHB, CoQ8, queuosine, type II fatty acid synthesis). To this end, the Rickettsia metabolic network may approach the minimal metabolic unit for a bacterial parasite of the eukaryotic cytoplasm.
FIG 7 .
Rickettsia metabolic network reconstruction highlights reductive genome evolution and addiction to host cell metabolites. The network focuses on the biosynthesis pathways discussed in the text. For brevity, pathways for most amino acids are not shown. Red stars indicate six pathway holes (see Fig. 5). GLY, glycolysis; MEP/DOXP, nonmevalonate terpenoid biosynthesis; THF, 5,6,7,8-THF; GSH, glutathione. (A) Theoretical Rickettsia metabolic network in the absence of imported metabolites. Rickettsia metabolic pathways are supplemented with typical Gram-negative biosynthetic pathways to create a complete metabolic network. (B) Reconstructed Rickettsia metabolic network, including imported metabolites. Pathways removed from panel A have been purged from Rickettsia genomes throughout evolution, a consequence of pilfering of metabolites from the eukaryotic host. Red ellipses, metabolites known to be imported by Rickettsia species; yellow ellipses, metabolites predicted to be imported on the basis of metabolic network reconstruction. The import of S-adenosyl-l-methionine, phosphorylcholine, and the majority of amino acids is not included in the reconstruction. Pathway lines are highlighted in red to indicate cofactors that are synthesized directly from imported metabolites. Additionally, if the cofactor is directly imported from the host, the pathway line is yellow. The network is based on a phylogenomics analyses of 84 Rickettsia genomes (see Fig. S10).
Phylogenomics analysis of Rickettsia metabolic pathways and metabolite transporters. Download FIG S10, XLSX file, 0.4 MB (397.9KB, xlsx) .
Copyright © 2017 Driscoll et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Most reactions within the remnant pathways rely on vitamins and cofactors that are also pilfered from the host (Fig. 7B). While rickettsiae were previously considered facultative energy parasites because of their ability to not only steal host ATP but also generate their own via oxidative phosphorylation, their reliance on numerous host metabolites to drive ATP production accentuates the obligate parasitic nature of rickettsiae. Prior comparative genomics studies of diverse rickettsiae have identified differences between vertebrate pathogens and species/strains with little or no documented pathogenicity, with loss of virulence often attributed to the expansion of mobile genetic elements that have destroyed genes implicated in vertebrate cell colonization (60, 61). Furthermore, the genomes of attenuated pathogens, e.g., R. prowazekii strain Madrid E (141) and R. rickettsii strain Iowa (142, 143), contain defects in some genes implicated in vertebrate pathogenicity. Importantly, our metabolic network reconstruction included 84 Rickettsia genomes encompassing the full range from deadly pathogens to purported endosymbionts, including species from basal lineages associated with nonhematophagous arthropods (the ladybird beetle Adalia bipunctata [59, 144]) and a parasitic ciliate (Ichthyophthirius multifiliis [145, 146]), as well as a recently identified spotted fever group species discovered in an unlikely vector, the seal fur louse Proechinophthirus fluctus (147). Although complicated by some genome assemblies of poorer quality, our reconstructed metabolic network is highly conserved across all of these species and strains, indicating that, despite various impacts on host fitness, all sequenced rickettsiae fit the profile of a quintessential and comprehensive metabolic parasite of the eukaryotic cytoplasm.
Like rickettsiae, other obligate intracellular bacteria have reductive genomes, yet genuine endosymbionts tend to retain pathways for the synthesis of amino acids, vitamins, and cofactors (148)—even some endosymbionts of blood-feeding arthropods (149, 150). These metabolites are often provisioned to the host, with the host usually harboring the bacteria in a protective niche (e.g., a bacteriocyte). Rickettsia species associated with the silverleaf whitefly (Bemisia tabaci [151]) and the booklouse Liposcelis bostrychophila (152), both nonhematophagous arthropods, have been observed in bacteriocytes; however, only the L. bostrychophila system is considered a primary endosymbiosis, as its Rickettsia species is always present in parthenogenetic females (153). The booklouse Rickettsia was determined to be a strain of Rickettsia felis, a species that is usually associated with blood-feeding arthropods and may be an occasional human pathogen (154). Our previous phylogenomics study revealed that L. bostrychophila-associated R. felis differs from other species by the presence of a novel plasmid, pLbAR, that encodes several factors not seen in other sequenced rickettsiae (155). It remains to be determined whether pLbAR contributes to the maintenance of the L. bostrychophila-R. felis symbiosis.
To our knowledge, no Rickettsia species has been shown to provision its host(s) with metabolites; indeed, rickettsiae seem to be in competition with arthropod and vertebrate cells for at least 20 metabolites that are either essential or synthesized from essential nutrients (Fig. 6). For strains of R. buchneri, our earlier discovery of a plasmid harboring two complete biotin synthesis operons raised the possibility that biotin could be provisioned to I. scapularis as part of a potential mutualism (60, 90). This unique biotin operon has since been discovered in other diverse obligate intracellular bacteria, including the Cardinium endosymbiont of the parasitic wasp Encarsia pergandiella (156) and the Wolbachia endosymbiont of the bedbug Cimex lectularius (wCle) (157). Remarkably, wCle-cured bedbugs that were fed a biotin-supplemented blood meal showed no marked differences in fitness relative to wCle-infected bedbugs; together with more recent evidence that wCle provisions riboflavin to its host (158), this indicates that evolutionary transitions from facultative parasitism to obligate mutualism have occurred in Wolbachia species, and they have been mediated by metabolic interdependence across host and microbe. It remains to be determined whether or not R. buchneri provisions biotin to I. scapularis or if any of the numerous Rickettsia species identified in hosts from most eukaryotic lineages (1) have entangled metabolic networks with their hosts. Barring some other benefit afforded to their host outside metabolism (i.e., competitive exclusion of pathogens [159, 160]) and given their metabolic profile as quintessential parasites of the eukaryotic cytoplasm, we stress the use of “endoparasite” over “endosymbiont” for these nonpathogenic rickettsiae.
Conclusion.
Ancestral members of the order Rickettsiales were likely facultative intracellular species of protoeukaryotes, with a symbiosis established by one lineage that arguably facilitated eukaryogenesis (161). The remaining rickettsial lineages avoided spiraling into organelles or minimalist bacteria, instead relying on general features of the eukaryotic cell (including mitochondria) for growth. While many of the pioneering studies of Rickettsia metabolism analyzed bacteria purified away from host cells, it became clear that numerous essential host factors are required to adequately fuel the metabolic processes that support rickettsial growth. The advent of genome sequencing identified patchwork metabolic pathways and supported the notion that rickettsiae have become chemically addicted to the eukaryotic cell. To advance our knowledge beyond these observations, we reconstructed a surprisingly conserved metabolic core for rickettsiae and supplemented these patchwork metabolic pathways with metabolites predicted to be imported from the eukaryotic cell. As with any model, our reconstruction may have failed to predict some rickettsial metabolic potential or underestimated the number of metabolites acquired from the host; nevertheless, it provides an important framework upon which to structure future investigations. Some of the questions that arise from our model have broader application beyond rickettsiology as well, e.g., the potential discovery of novel transport systems or the identification of nonorthologous enzymes that function within canonical metabolic pathways.
Our study illustrates how little is known regarding rickettsial acquisition of host metabolites; in particular, it indicates that rickettsiae must use transport systems (possibly from the host) to import many large metabolites. Thus, future studies that characterize transporters in heterologous expression systems will continue to be invaluable tools in elucidating how rickettsiae pilfer host metabolites. We anticipate that careful evaluation of large-scale transcriptomic and proteomic data sets will reveal clues about how the host metabolism is hijacked throughout the course of rickettsial infection. Ultimately, establishing an axenic culture for rickettsiae will greatly facilitate genetic studies correlating transporters with imported substrates. Such an effort is conceptually appealing yet practically difficult. The present study represents an important contribution to that effort; it may be that differences between our estimated 51 essential metabolites and the complex media used in early metabolic assays (e.g., the recipe used by Bovarnick [71]) provide the “missing ingredients” necessary to support cell-free rickettsial growth.
Reductive genome evolution in rickettsiae has been interpreted by some as an ongoing process that, in its severest cases, correlates with an increase in vertebrate pathogenicity (162). We now understand that Rickettsia genomes continually acquire genes throughout their evolution and that genes encoding metabolic enzymes, metabolite transporters, and stringent response regulators are often components of mobile genetic elements (2, 60, 61, 155). This implies that there is strong selection on maintenance of the conserved rickettsial metabolome, which has evolved to allow rickettsiae to delicately parasitize the host cytoplasm, ensuring that large numbers of bacteria can persist before host cell destruction. This mode of parasitism allows for propagation throughout numerous arthropod tissues and, for some species, dissemination to vertebrates by blood feeding or fecal inoculation. Our work here provides novel insights into many aspects of rickettsial host-dependent metabolism. Metabolomics remains a poorly studied aspect of rickettsiology yet one where advancements would serve the fields of drug and vaccine design markedly well, particularly if certain host metabolites are scavenged using unique bacterial transport systems.
MATERIALS AND METHODS
Phylogenomics analysis. (i) Genus-level phylogeny estimation.
Protein sequences (n = 112,870) from 86 sequenced genomes were used to estimate a genus-level Rickettsia phylogeny. Rickettsia genomes were either retrieved from the NCBI Assembly database (n = 82) or obtained via personal communication (n = 2; Lucy Weinert, University of Cambridge) (see Fig. S10). Two non-Rickettsia genomes (O. tsutsugamushi strain Ikeda [accession no. GCF_000010205.1]; Rickettsiales bacterium Ac37b [GCF_000746585.1]) were included as outgroups. The RAST v 2.0 server (163) was used to annotate 16 Rickettsia assemblies that were not previously annotated. A total of 3,772 orthologous gene families were constructed from this data set using FastOrtho, a modified version of OrthoMCL (164), at an inflation of 1.5 and an identity threshold of 40%. A subset of 149 single-copy families conserved across all 86 taxa was independently aligned with MUSCLE v3.8.31 (165) using default parameters, and regions of poor alignment were masked using Gblocks (166). All modified alignments were concatenated into a single data set (41,975 positions) for phylogeny estimation using Phylobayes v4.1 (167), with the CAT model of substitution used as described previously (155). A phylogeny was also estimated under maximum likelihood with RAxML v8.2.4 (168) using a gamma model of rate heterogeneity and estimation of the proportion of invariable sites. Branch support was assessed with 1,000 pseudoreplications.
(ii) Order-level phylogeny estimation.
Protein sequences (n = 86,029) from 56 sequenced genomes were used to estimate an order-level phylogeny of Rickettsiales/Holosporales, sampling the groups Holosporales (n = 9), Anaplasmataceae (n = 22), Midichloriaceae (n = 4), and Rickettsiaceae (n = 18), as well as three outgroup genomes (Alphaproteobacteria strain BAL199, Azospirillum sp. strain B510, and Rhodospirillum rubrum ATCC 11170). Genomes were retrieved from NCBI (25) and PATRIC (28) (see Fig. S7). A total of 8,437 orthologous families were constructed using FastOrtho (inflation of 1.2), and a subset of 105 single-copy families conserved across at least 52 taxa (95%) was aligned, masked, and concatenated (23,990 positions) as described above. Phylogeny estimation was carried out with PhyloBayes and RAxML as described above.
Metabolic reconstruction.
For metabolic pathway analyses, the 84 Rickettsia genomes from our genus-level phylogeny estimation (outgroup genomes removed) were used to construct 3,576 orthologous gene families as described above. These families were subsequently assigned Kyoto Encyclopedia of Genes and Genomes (KEGG) K numbers by submitting all 109,191 individual protein sequences to the KEGG BlastKOALA service (169). K number assignments, as well as broader metabolic classifications, were extracted from the resulting HTML files (kcompile), transferred across orthologous families (ktransfer), and used for metabolic pathway reconstruction (kreconstruct). All of the software packages developed for this study are freely available in the keggerator repository on github.
Identification of pathway holes.
A pathway hole is simply defined here as a missing gene in an otherwise complete metabolic pathway. Pathways that are absent or highly fragmented are not considered. Candidate pathway holes were initially identified during inspection of the metabolic reconstruction results for Rickettsia species. To account for possible errors in our K number annotation, gene absences were confirmed by searching (blastp) our data set with orthologous sequences from E. coli. The taxonomic distribution of confirmed Rickettsia pathway holes was assessed by converting each pathway into a binary feature vector, where each position in the vector represents a pathway step containing either a 1 (gene present) or a 0 (gene absent). Pathway data for all KEGG genomes were subsequently analyzed (khole) for genomes that possess Rickettsia-like feature vectors.
Targeted phylogeny estimation.
To determine the evolutionary history of specific genes (e.g., those encoding Idi, enzymes of the folate and queuosine pathways, and Rickettsiales enzymes corresponding to Rickettsia pathway holes), individual protein phylogenies were estimated. To construct data sets for each protein, the rickettsial protein was used in blastp queries against several taxon-specific databases, i.e., (i) “Rickettsiales,” (ii) “Holosporales,” (iii) “Alphaproteobacteria (minus Rickettsiales and Holosporales),” (iv) “Proteobacteria (minus Alphaproteobacteria),” (v) “Bacteria (minus Proteobacteria),” and (vi) “minus Bacteria.” The top 5 to 10 (query-dependent) subjects from each search resulting in significant (>40 bits) alignments were all compiled and aligned with MUSCLE v3.8.31 using default parameters. Protein phylogenies were estimated under maximum likelihood with RAxML v8.2.4 using a gamma model of rate heterogeneity and estimation of the proportion of invariant sites. Both the Le and Gascuel (LG) and blocks of amino acid substitution matrix (BLOSUM62) models of amino acid substitution were used, and branch support was assessed using 1,000 pseudoreplications.
Transporter prediction.
Diverse transport systems were assembled and annotated in an iterative process involving bioinformatics predictions, manual evaluation, and comparative genomics analyses. For the R. typhi strain Wilmington genome, 96 transporters were predicted using TransportDB (170). The breakdown of these predictions was ATP-dependent systems (n = 38), ion channels (n = 3), secondary transporters (n = 48), and unclassified transporters (n = 7). Several Rickettsia genomes were also evaluated on the Transporter Classification Database (TCDB) (123). Transporter names and family identifications were assigned after comparison across TransportDB and TCDB and, in some cases, blastp searches against genomes with well-characterized transporters. Subsequent manual evaluation eliminated erroneous predictions, components of protein translocation pathways, and other transport systems that were deemed not likely to import the host metabolites under consideration. Additionally, for tripartite ABC transporters, missing components were manually added on the basis of genomic proximity to other components likely to be part of the same transporter. Putative substrates for each transport system were conservatively assigned on the basis of TCDB or close homology of the rickettsial transporter to transporters with well-characterized substrates.
ACKNOWLEDGMENTS
This work is dedicated to the late Herbert H. Winkler (1939–2016, former Professor of Microbiology and Immunology at the University of South Alabama College of Medicine), whose pioneering work on Rickettsia metabolism paved the way forward, and the memory of Chris Cornell, the author of “Black Hole Sun.” We are grateful to Lucy Weinert (University of Cambridge) for sharing the unpublished genomes of Rickettsia species isolated from the ladybird beetle A. bipunctata and the parasitic ciliate I. multifiliis.
This work was supported with funds from the National Institutes of Health National Institute of Allergy and Infectious Diseases grants (R01AI017828 and R01AI126853 to A.F.A., R21AI26108 to J.J.G. and M.S.R.). S.A.R., M.L.G., and S.S.L. were supported in part by the NIH NIAID grants T32AI095190 (Signaling Pathways in Innate Immunity) and T32AI007540 (Infection and Immunity). The content is solely the responsibility of the authors and does not necessarily represent the official views of the funding agencies. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Footnotes
Citation Driscoll TP, Verhoeve VI, Guillotte ML, Lehman SS, Rennoll SA, Beier-Sexton M, Rahman MS, Azad AF, Gillespie JJ. 2017. Wholly Rickettsia! Reconstructed metabolic profile of the quintessential bacterial parasite of eukaryotic cells. mBio 8:e00859-17. https://doi.org/10.1128/mBio.00859-17.
REFERENCES
- 1.Gillespie JJ, Nordberg EK, Azad AA, Sobral BW. 2012. Phylogeny and comparative genomics: the shifting landscape in the genomics era, p 84–141. In Azad AF, Palmer GH (ed), Intracellular pathogens II: rickettsiales. ASM Press, Washington, DC. [Google Scholar]
- 2.Driscoll T, Gillespie JJ, Nordberg EK, Azad AF, Sobral BW. 2013. Bacterial DNA sifted from the Trichoplax adhaerens (Animalia: Placozoa) genome project reveals a putative rickettsial endosymbiont. Genome Biol Evol 5:621–645. doi: 10.1093/gbe/evt036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Szokoli F, Castelli M, Sabaneyeva E, Schrallhammer M, Krenek S, Doak TG, Berendonk TU, Petroni G. 2016. Disentangling the taxonomy of Rickettsiales and description of two novel symbionts (“Candidatus Bealeia paramacronuclearis” and “Candidatus Fokinia cryptica”) sharing the cytoplasm of the ciliate protist Paramecium biaurelia. Appl Environ Microbiol 82:7236–7247. doi: 10.1128/AEM.02284-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Beier-Sexton M, Driscoll TP, Azad AF, Gillespie JJ. 2015. The family Rickettsiaceae, p 547–566. In Goldman E, Green LH (ed), Practical handbook of microbiology, 3rd edition. CRC Press, Boca Raton, FL. [Google Scholar]
- 5.Darby AC, Cho NH, Fuxelius HH, Westberg J, Andersson SGE. 2007. Intracellular pathogens go extreme: genome evolution in the Rickettsiales. Trends Genet 23:511–520. doi: 10.1016/j.tig.2007.08.002. [DOI] [PubMed] [Google Scholar]
- 6.Winkler HH. 1986. Early events in the interaction of the obligate intracytoplasmic parasite, Rickettsia prowazekii, with eukaryotic cells: entry and lysis. Ann Inst Pasteur Microbiol 137(Suppl A):333–336. doi: 10.1016/S0769-2609(86)80044-8. [DOI] [PubMed] [Google Scholar]
- 7.Dunning Hotopp JC, Lin M, Madupu R, Crabtree J, Angiuoli SV, Eisen JA, Eisen J, Seshadri R, Ren Q, Wu M, Utterback TR, Smith S, Lewis M, Khouri H, Zhang C, Niu H, Lin Q, Ohashi N, Zhi N, Nelson W, Brinkac LM, Dodson RJ, Rosovitz MJ, Sundaram J, Daugherty SC, Davidsen T, Durkin AS, Gwinn M, Haft DH, Selengut JD, Sullivan SA, Zafar N, Zhou L, Benahmed F, Forberger H, Halpin R, Mulligan S, Robinson J, White O, Rikihisa Y. 2006. Comparative genomics of emerging human ehrlichiosis agents. PLoS Genet 2:e21. doi: 10.1371/journal.pgen.0020021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fuxelius HH, Darby A, Min CK, Cho NH, Andersson SGE. 2007. The genomic and metabolic diversity of Rickettsia. Res Microbiol 158:745–753. doi: 10.1016/j.resmic.2007.09.008. [DOI] [PubMed] [Google Scholar]
- 9.Nakayama K, Yamashita A, Kurokawa K, Morimoto T, Ogawa M, Fukuhara M, Urakami H, Ohnishi M, Uchiyama I, Ogura Y, Ooka T, Oshima K, Tamura A, Hattori M, Hayashi T. 2008. The whole-genome sequencing of the obligate intracellular bacterium Orientia tsutsugamushi revealed massive gene amplification during reductive genome evolution. DNA Res 15:185–199. doi: 10.1093/dnares/dsn011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ray K, Marteyn B, Sansonetti PJ, Tang CM. 2009. Life on the inside: the intracellular lifestyle of cytosolic bacteria. Nat Rev Microbiol 7:333–340. doi: 10.1038/nrmicro2112. [DOI] [PubMed] [Google Scholar]
- 11.Walker DH, Ismail N. 2008. Emerging and re-emerging rickettsioses: endothelial cell infection and early disease events. Nat Rev Microbiol 6:375–386. doi: 10.1038/nrmicro1866. [DOI] [PubMed] [Google Scholar]
- 12.Winkler HH. 1990. Rickettsia species (as organisms). Annu Rev Microbiol 44:131–153. doi: 10.1146/annurev.mi.44.100190.001023. [DOI] [PubMed] [Google Scholar]
- 13.Audia JP. 2012. Rickettsial physiology and metabolism in the face of reductive evolution, p 221–242. In Palmer GH, Azad AF (ed), Intracellular pathogens II: Rickettsiales. ASM Press, Washington, DC. [Google Scholar]
- 14.Bovarnick MR, Snyder JC. 1949. Respiration of typhus rickettsiae. J Exp Med 89:561–565. doi: 10.1084/jem.89.6.561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Coolbaugh JC, Progar JJ, Weiss E. 1976. Enzymatic activities of cell-free extracts of Rickettsia typhi. Infect Immun 14:298–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Phibbs PV, Winkler HH. 1981. Regulatory properties of partially purified enzymes of the tricarboxylic acid cycle of Rickettsia prowazekii, p 421 In Burgdorfer W, Anacker RL (ed), Rickettsiae and rickettsial diseases. Academic Press, New York, NY. [Google Scholar]
- 17.Phibbs PV, Winkler HH. 1982. Regulatory properties of citrate synthase from Rickettsia prowazekii. J Bacteriol 149:718–725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wood DO, Atkinson WH, Sikorski RS, Winkler HH. 1983. Expression of the Rickettsia prowazekii citrate synthase gene in Escherichia coli. J Bacteriol 155:412–416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Smith DK, Winkler HH. 1979. Separation of inner and outer membranes of Rickettsia prowazeki and characterization of their polypeptide compositions. J Bacteriol 137:963–971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dasch GA, Samms JR, Weiss E. 1978. Biochemical characteristics of typhus group rickettsiae with special attention to the Rickettsia prowazekii strains isolated from flying squirrels. Infect Immun 19:676–685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Winkler HH, Daugherty RM. 1986. Acquisition of glucose by Rickettsia prowazekii through the nucleotide intermediate uridine 5′-diphosphoglucose. J Bacteriol 167:805–808. doi: 10.1128/jb.167.3.805-808.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Williams JC, Weiss E. 1978. Energy metabolism of Rickettsia typhi: pools of adenine nucleotides and energy charge in the presence and absence of glutamate. J Bacteriol 134:884–892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hopps HE, Hahn FE, Wisseman CL, Jackson EB, Smadel JE. 1956. Metabolic studies of rickettsiae. III. Studies of transamination, oxidative phosphorylation and glutamate-2-C14 incorporation by purified suspensions of Rickettsia mooseri. J Bacteriol 71:708–716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wisseman CL, Jackson EB, Hahn FE, Ley AC, Smadel JE. 1951. Metabolic studies of rickettsiae: I. The effects of antimicrobial substances and enzyme inhibitors on the oxidation of glutamate by purified rickettsiae. J Immunol 67:123–136. [PubMed] [Google Scholar]
- 25.Bovarnick MR, Miller JC. 1950. Oxidation and transamination of glutamate by typhus rickettsiae. J Biol Chem 184:661–676. [PubMed] [Google Scholar]
- 26.Hahn FE, Cohn ZA, Bozeman FM. 1960. Metabolic studies of rickettsiae. V. Metabolism of glutamine and asparagine in Rickettsia mooseri. J Bacteriol 80:400–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Williams J, Peterson J, Coolbaugh J. 1978. Relationship between glutamate metabolism and the incorporation of inorganic phosphate (Pi) into nucleotides by Rickettsia typhi, p 99 In Kazar J, Ormsbee R, Tarasevich I (ed), Rickettsiae and rickettsial diseases. VEDA, Bratislava, Slovakia. [Google Scholar]
- 28.Bovarnick MR. 1956. Phosphorylation accompanying the oxidation of glutamate by the Madrid E strain of typhus rickettsiae. J Biol Chem 220:353–361. [PubMed] [Google Scholar]
- 29.Zahorchak R, Winkler HH. 1981. Hydrolysis and synthesis of ATP by Rickettsia prowazekii, p 401 In Burgdorfi W, Anacker RL (ed), Rickettsiae and rickettsial diseases. Academic Press, New York, NY. [Google Scholar]
- 30.Austin FE, Winkler HH. 1988. Proline incorporation into protein by Rickettsia prowazekii during growth in Chinese hamster ovary (CHO-K1) cells. Infect Immun 56:3167–3172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Winkler HH, Daugherty RM. 1984. Proline transport and metabolism in Rickettsia prowazekii. J Bacteriol 158:460–463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zahorchak RJ, Winkler HH. 1983. Transmembrane electrical potential in Rickettsia prowazekii and its relationship to lysine transport. J Bacteriol 153:665–671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Smith DK, Winkler HH. 1977. Characterization of a lysine-specific active transport system in Rickettsia prowazeki. J Bacteriol 129:1349–1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bovarnick MR, Schneider L. 1960. The incorporation of glycine-1-C14 by typhus rickettsiae. J Biol Chem 235:1727–1731. [PubMed] [Google Scholar]
- 35.Austin FE, Turco J, Winkler HH. 1987. Rickettsia prowazekii requires host cell serine and glycine for growth. Infect Immun 55:240–244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bovarnick MR, Schneider L, Walter H. 1959. The incorporation of labeled methionine by typhus rickettsiae. Biochim Biophys Acta 33:414–422. doi: 10.1016/0006-3002(59)90131-3. [DOI] [PubMed] [Google Scholar]
- 37.Fujita K, Kohno S, Shishido A. 1959. The incorporation of methionine into purified rickettsiae. Jpn J Med Sci Biol 12:387–390. doi: 10.7883/yoken1952.12.387. [DOI] [PubMed] [Google Scholar]
- 38.Winkler HH. 1976. Rickettsial permeability. An ADP-ATP transport system. J Biol Chem 251:389–396. [PubMed] [Google Scholar]
- 39.Atkinson WH, Winkler HH. 1989. Permeability of Rickettsia prowazekii to NAD. J Bacteriol 171:761–766. doi: 10.1128/jb.171.2.761-766.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Winkler HH, Daugherty R, Hu F. 1999. Rickettsia prowazekii transports UMP and GMP, but not CMP, as building blocks for RNA synthesis. J Bacteriol 181:3238–3241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Atkinson WH, Winkler HH. 1985. Transport of AMP by Rickettsia prowazekii. J Bacteriol 161:32–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Speed RR, Winkler HH. 1991. Acquisition of thymidylate by the obligate intracytoplasmic bacterium Rickettsia prowazekii. J Bacteriol 173:1704–1710. doi: 10.1128/jb.173.5.1704-1710.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Austin FE, Winkler HH. 1988. Relationship of rickettsial physiology and composition to the Rickettsia-host cell interaction, p 29–50. In Walker DH. (ed), Biology of rickettsial diseases, vol. 2 CRC Press, Boca Raton, FL. [Google Scholar]
- 44.Andersson SG, Zomorodipour A, Andersson JO, Sicheritz-Pontén T, Alsmark UC, Podowski RM, Näslund AK, Eriksson AS, Winkler HH, Kurland CG. 1998. The genome sequence of Rickettsia prowazekii and the origin of mitochondria. Nature 396:133–140. doi: 10.1038/24094. [DOI] [PubMed] [Google Scholar]
- 45.Andersson JO, Andersson SG. 1999. Genome degradation is an ongoing process in Rickettsia. Mol Biol Evol 16:1178–1191. doi: 10.1093/oxfordjournals.molbev.a026208. [DOI] [PubMed] [Google Scholar]
- 46.Andersson JO, Andersson SG. 2001. Pseudogenes, junk DNA, and the dynamics of Rickettsia genomes. Mol Biol Evol 18:829–839. doi: 10.1093/oxfordjournals.molbev.a003864. [DOI] [PubMed] [Google Scholar]
- 47.Andersson JO, Andersson SG. 1999. Insights into the evolutionary process of genome degradation. Curr Opin Genet Dev 9:664–671. doi: 10.1016/S0959-437X(99)00024-6. [DOI] [PubMed] [Google Scholar]
- 48.Gillespie JJ, Williams K, Shukla M, Snyder EE, Nordberg EK, Ceraul SM, Dharmanolla C, Rainey D, Soneja J, Shallom JM, Vishnubhat ND, Wattam R, Purkayastha A, Czar M, Crasta O, Setubal JC, Azad AF, Sobral BS. 2008. Rickettsia phylogenomics: unwinding the intricacies of obligate intracellular life. PLoS One 3:e2018. doi: 10.1371/journal.pone.0002018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Min CK, Yang JS, Kim S, Choi MS, Kim IS, Cho NH. 2008. Genome-based construction of the metabolic pathways of Orientia tsutsugamushi and comparative analysis within the Rickettsiales Order. Comp Funct Genomics. 2008:623145. doi: 10.1155/2008/623145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Renesto P, Ogata H, Audic S, Claverie JM, Raoult D. 2005. Some lessons from Rickettsia genomics. FEMS Microbiol Rev 29:99–117. doi: 10.1016/j.femsre.2004.09.002. [DOI] [PubMed] [Google Scholar]
- 51.Frohlich KM, Roberts RAW, Housley NA, Audia JP. 2010. Rickettsia prowazekii uses an sn-glycerol-3-phosphate dehydrogenase and a novel dihydroxyacetone phosphate transport system to supply triose phosphate for phospholipid biosynthesis. J Bacteriol 192:4281–4288. doi: 10.1128/JB.00443-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Frohlich KM, Audia JP. 2013. Dual mechanisms of metabolite acquisition by the obligate intracytosolic pathogen Rickettsia prowazekii reveal novel aspects of triose phosphate transport. J Bacteriol 195:3752–3760. doi: 10.1128/JB.00404-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tucker AM, Winkler HH, Driskell LO, Wood DO. 2003. S-Adenosylmethionine transport in Rickettsia prowazekii. J Bacteriol 185:3031–3035. doi: 10.1128/JB.185.10.3031-3035.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Audia JP, Winkler HH. 2006. Study of the five Rickettsia prowazekii proteins annotated as ATP/ADP translocases (TLC): only Tlc1 transports ATP/ADP, while Tlc4 and Tlc5 transport other ribonucleotides. J Bacteriol 188:6261–6268. doi: 10.1128/JB.00371-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.McClure EE, Chávez ASO, Shaw DK, Carlyon JA, Ganta RR, Noh SM, Wood DO, Bavoil PM, Brayton KA, Martinez JJ, McBride JW, Valdivia RH, Munderloh UG, Pedra JHF. 2017. Engineering of obligate intracellular bacteria: progress, challenges and paradigms. Nat Rev Microbiol 15:544–558. doi: 10.1038/nrmicro.2017.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Omsland A, Cockrell DC, Howe D, Fischer ER, Virtaneva K, Sturdevant DE, Porcella SF, Heinzen RA. 2009. Host cell-free growth of the Q fever bacterium Coxiella burnetii. Proc Natl Acad Sci U S A 106:4430–4434. doi: 10.1073/pnas.0812074106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Omsland A, Heinzen RA. 2011. Life on the outside: the rescue of Coxiella burnetii from its host cell. Annu Rev Microbiol 65:111–128. doi: 10.1146/annurev-micro-090110-102927. [DOI] [PubMed] [Google Scholar]
- 58.Gillespie JJ, Beier MS, Rahman MS, Ammerman NC, Shallom JM, Purkayastha A, Sobral BS, Azad AF. 2007. Plasmids and rickettsial evolution: insight from Rickettsia felis. PLoS One 2:e266. doi: 10.1371/journal.pone.0000266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Murray GGR, Weinert LA, Rhule EL, Welch JJ. 2016. The phylogeny of rickettsia using different evolutionary signatures: how tree-like is bacterial evolution? Syst Biol 65:265–279. doi: 10.1093/sysbio/syv084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Gillespie JJ, Joardar V, Williams KP, Driscoll T, Hostetler JB, Nordberg E, Shukla M, Walenz B, Hill CA, Nene VM, Azad AF, Sobral BW, Caler E. 2012. A Rickettsia genome overrun by mobile genetic elements provides insight into the acquisition of genes characteristic of an obligate intracellular lifestyle. J Bacteriol 194:376–394. doi: 10.1128/JB.06244-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Felsheim RF, Kurtti TJ, Munderloh UG. 2009. Genome sequence of the endosymbiont Rickettsia peacockii and comparison with virulent Rickettsia rickettsii: identification of virulence factors. PLoS One 4:e8361. doi: 10.1371/journal.pone.0008361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Gehring AM, Lees WJ, Mindiola DJ, Walsh CT, Brown ED. 1996. Acetyltransfer precedes uridylyltransfer in the formation of UDP-N-acetylglucosamine in separable active sites of the bifunctional GlmU protein of Escherichia coli. Biochemistry 35:579–585. doi: 10.1021/bi952275a. [DOI] [PubMed] [Google Scholar]
- 63.Zhivkov V, Tosheva R, Zhivkova Y. 1975. Concentration of uridine diphosphate sugars in various tissues of vertebrates. Comp Biochem Physiol B 51:421–424. doi: 10.1016/0305-0491(75)90032-2. [DOI] [PubMed] [Google Scholar]
- 64.Whitfield C, Trent MS. 2014. Biosynthesis and export of bacterial lipopolysaccharides. Annu Rev Biochem 83:99–128. doi: 10.1146/annurev-biochem-060713-035600. [DOI] [PubMed] [Google Scholar]
- 65.Salema M, Poolman B, Lolkema JS, Dias MC, Konings WN. 1994. Uniport of monoanionic l-malate in membrane vesicles from Leuconostoc oenos. Eur J Biochem 225:289–295. doi: 10.1111/j.1432-1033.1994.00289.x. [DOI] [PubMed] [Google Scholar]
- 66.Young GB, Jack DL, Smith DW, Saier MH. 1999. The amino acid/auxin:proton symport permease family. Biochim Biophys Acta 1415:306–322. doi: 10.1016/S0005-2736(98)00196-5. [DOI] [PubMed] [Google Scholar]
- 67.Jendrossek D, Pfeiffer D. 2014. New insights in the formation of polyhydroxyalkanoate granules (carbonosomes) and novel functions of poly(3-hydroxybutyrate). Environ Microbiol 16:2357–2373. doi: 10.1111/1462-2920.12356. [DOI] [PubMed] [Google Scholar]
- 68.Silverman DJ, Wisseman CL, Waddell A. 1980. In vitro studies of rickettsia-host cell interactions: ultrastructural study of Rickettsia prowazekii-infected chicken embryo fibroblasts. Infect Immun 29:778–790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Barnard GN, Sanders JK. 1989. The poly-beta-hydroxybutyrate granule in vivo. A new insight based on NMR spectroscopy of whole cells. J Biol Chem 264:3286–3291. [PubMed] [Google Scholar]
- 70.Anderson AJ, Dawes EA. 1990. Occurrence, metabolism, metabolic role, and industrial uses of bacterial polyhydroxyalkanoates. Microbiol Rev 54:450–472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Bovarnick MR. 1960. Incorporation of acetate-1-C into lipid by typhus rickettsiae. J Bacteriol 80:508–512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kajimura J, Rahman A, Hsu J, Evans MR, Gardner KH, Rick PD. 2006. O acetylation of the enterobacterial common antigen polysaccharide is catalyzed by the product of the yiaH gene of Escherichia coli K-12. J Bacteriol 188:7542–7550. doi: 10.1128/JB.00783-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Brosnan JT. 2003. Interorgan amino acid transport and its regulation. J Nutr 133:2068S–2072S. [DOI] [PubMed] [Google Scholar]
- 74.Pang H, Winkler HH. 1994. Analysis of the peptidoglycan of Rickettsia prowazekii. J Bacteriol 176:923–926. doi: 10.1128/jb.176.3.923-926.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kato K, Tanaka R, Sano S, Tanaka A, Hosaka H. 2010. Identification of a gene essential for protoporphyrinogen IX oxidase activity in the cyanobacterium Synechocystis sp. PCC 6803 Proc Natl Acad Sci U S A 107:16649–16654. doi: 10.1073/pnas.1000771107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Lombard J, Moreira D. 2011. Early evolution of the biotin-dependent carboxylase family. BMC Evol Biol 11:232. doi: 10.1186/1471-2148-11-232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Hügler M, Krieger RS, Jahn M, Fuchs G. 2003. Characterization of acetyl-CoA/propionyl-CoA carboxylase in Metallosphaera sedula. Carboxylating enzyme in the 3-hydroxypropionate cycle for autotrophic carbon fixation. Eur J Biochem 270:736–744. doi: 10.1046/j.1432-1033.2003.03434.x. [DOI] [PubMed] [Google Scholar]
- 78.Chuakrut S, Arai H, Ishii M, Igarashi Y. 2003. Characterization of a bifunctional archaeal acyl coenzyme A carboxylase. J Bacteriol 185:938–947. doi: 10.1128/JB.185.3.938-947.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Lovullo E, Furlong R, Shirk P. 2015. The lipid biosynthesis hole in the rickettsiales. In 27th Meeting of the American Society for Rickettsiology, Lake Tahoe, CA. [Google Scholar]
- 80.Tzianabos T, Moss CW, McDade JE. 1981. Fatty acid composition of rickettsiae. J Clin Microbiol 13:603–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Winkler HH, Miller ET. 1978. Phospholipid composition of Rickettsia prowazeki grown in chicken embryo yolk sacs. J Bacteriol 136:175–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Winkler HH, Day L, Daugherty R. 1994. Analysis of hydrolytic products from choline-labeled host cell phospholipids during growth of Rickettsia prowazekii. Infect Immun 62:1457–1459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Gillespie JJ, Kaur SJ, Rahman MS, Rennoll-Bankert K, Sears KT, Beier-Sexton M, Azad AF. 2015. Secretome of obligate intracellular Rickettsia. FEMS Microbiol Rev 39:47–80. doi: 10.1111/1574-6976.12084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Renesto P, Dehoux P, Gouin E, Touqui L, Cossart P, Raoult D. 2003. Identification and characterization of a phospholipase D-superfamily gene in rickettsiae. J Infect Dis 188:1276–1283. doi: 10.1086/379080. [DOI] [PubMed] [Google Scholar]
- 85.Clark SE, Weiser JN. 2013. Microbial modulation of host immunity with the small molecule phosphorylcholine. Infect Immun 81:392–401. doi: 10.1128/IAI.01168-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Hajj Chehade M, Loiseau L, Lombard M, Pecqueur L, Ismail A, Smadja M, Golinelli-Pimpaneau B, Mellot-Draznieks C, Hamelin O, Aussel L, Kieffer-Jaquinod S, Labessan N, Barras F, Fontecave M, Pierrel F. 2013. ubiI, a new gene in Escherichia coli coenzyme Q biosynthesis, is involved in aerobic C5-hydroxylation. J Biol Chem 288:20085–20092. doi: 10.1074/jbc.M113.480368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Driskell LO, Tucker AM, Winkler HH, Wood DO. 2005. Rickettsial metK-encoded methionine adenosyltransferase expression in an Escherichia coli metK deletion strain. J Bacteriol 187:5719–5722. doi: 10.1128/JB.187.16.5719-5722.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Haferkamp I, Penz T, Geier M, Ast M, Mushak T, Horn M, Schmitz-Esser S. 2013. The endosymbiont Amoebophilus asiaticus encodes an S-adenosylmethionine carrier that compensates for its missing methylation cycle. J Bacteriol 195:3183–3192. doi: 10.1128/JB.00195-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Hunter DJ, Torkelson JL, Bodnar J, Mortazavi B, Laurent T, Deason J, Thephavongsa K, Zhong J. 2015. The rickettsia endosymbiont of ixodes pacificus contains all the genes of de novo folate biosynthesis. PLoS One 10:e0144552. doi: 10.1371/journal.pone.0144552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Kurtti TJ, Felsheim RF, Burkhardt NY, Oliver JD, Heu CC, Munderloh UG. 2015. Rickettsia buchneri sp. nov., a rickettsial endosymbiont of the blacklegged tick Ixodes scapularis. Int J Syst Evol Microbiol 65:965–970. doi: 10.1099/ijs.0.000047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Phan JN, Lu CR, Bender WG, Smoak RM, Zhong J. 2011. Molecular detection and identification of Rickettsia species in Ixodes pacificus in California. Vector Borne Zoonotic Dis 11:957–961. doi: 10.1089/vbz.2010.0077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Cheng D, Vigil K, Schanes P, Brown RN, Zhong J. 2013. Prevalence and burden of two rickettsial phylotypes (G021 and G022) in Ixodes pacificus from California by real-time quantitative PCR. Ticks Tick Borne Dis 4:280–287. doi: 10.1016/j.ttbdis.2012.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Cheng D, Lane RS, Moore BD, Zhong J. 2013. Host blood meal-dependent growth ensures transovarial transmission and transstadial passage of Rickettsia sp. phylotype G021 in the western black-legged tick (Ixodes pacificus). Ticks Tick Borne Dis 4:421–426. doi: 10.1016/j.ttbdis.2013.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Kurlovs AH, Li J, Cheng D, Zhong J. 2014. Ixodes pacificus ticks maintain embryogenesis and egg hatching after antibiotic treatment of rickettsia endosymbiont. PLoS One 9:e104815. doi: 10.1371/journal.pone.0104815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Dittrich S, Mitchell SL, Blagborough AM, Wang Q, Wang P, Sims PFG, Hyde JE. 2008. An atypical orthologue of 6-pyruvoyltetrahydropterin synthase can provide the missing link in the folate biosynthesis pathway of malaria parasites. Mol Microbiol 67:609–618. doi: 10.1111/j.1365-2958.2007.06073.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Pribat A, Jeanguenin L, Lara-Núñez A, Ziemak MJ, Hyde JE, de Crécy-Lagard V, Hanson AD. 2009. 6-Pyruvoyltetrahydropterin synthase paralogs replace the folate synthesis enzyme dihydroneopterin aldolase in diverse bacteria. J Bacteriol 191:4158–4165. doi: 10.1128/JB.00416-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Forrest HS, Baalen CV. 1970. Microbiology of unconjugated pteridines. Annu Rev Microbiol 24:91–108. doi: 10.1146/annurev.mi.24.100170.000515. [DOI] [PubMed] [Google Scholar]
- 98.Kong JS, Kang JY, Kim HL, Kwon OS, Lee KH, Park YS. 2006. 6-Pyruvoyltetrahydropterin synthase orthologs of either a single or dual domain structure are responsible for tetrahydrobiopterin synthesis in bacteria. FEBS Lett 580:4900–4904. doi: 10.1016/j.febslet.2006.08.006. [DOI] [PubMed] [Google Scholar]
- 99.McCarty RM, Somogyi A, Bandarian V. 2009. Escherichia coli QueD is a 6-carboxy-5,6,7,8-tetrahydropterin synthase. Biochemistry 48:2301–2303. doi: 10.1021/bi9001437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Reader JS, Metzgar D, Schimmel P, de Crécy-Lagard V. 2004. Identification of four genes necessary for biosynthesis of the modified nucleoside queuosine. J Biol Chem 279:6280–6285. doi: 10.1074/jbc.M310858200. [DOI] [PubMed] [Google Scholar]
- 101.Harada F, Nishimura S. 1972. Possible anticodon sequences of tRNA His, tRNA Asm, and tRNA Asp from Escherichia coli B. Universal presence of nucleoside Q in the first postion of the anticondons of these transfer ribonucleic acids. Biochemistry 11:301–308. doi: 10.1021/bi00752a024. [DOI] [PubMed] [Google Scholar]
- 102.Satoh Y, Kuratsu M, Kobayashi D, Dairi T. 2014. New gene responsible for para-aminobenzoate biosynthesis. J Biosci Bioeng 117:178–183. doi: 10.1016/j.jbiosc.2013.07.013. [DOI] [PubMed] [Google Scholar]
- 103.McCoy AJ, Adams NE, Hudson AO, Gilvarg C, Leustek T, Maurelli AT. 2006. l,l-Diaminopimelate aminotransferase, a trans-kingdom enzyme shared by Chlamydia and plants for synthesis of diaminopimelate/lysine. Proc Natl Acad Sci U S A 103:17909–17914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Misono H, Togawa H, Yamamoto T, Soda K. 1979. meso-αε-Diaminopimelate d-dehydrogenase: distribution and the reaction product. J Bacteriol 137:22–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Weinberger S, Gilvarg C. 1970. Bacterial distribution of the use of succinyl and acetyl blocking groups in diaminopimelic acid biosynthesis. J Bacteriol 101:323–324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Sperandeo P, Pozzi C, Dehò G, Polissi A. 2006. Non-essential KDO biosynthesis and new essential cell envelope biogenesis genes in the Escherichia coli yrbG-yhbG locus. Res Microbiol 157:547–558. doi: 10.1016/j.resmic.2005.11.014. [DOI] [PubMed] [Google Scholar]
- 107.Yao J, Rock CO. 2013. Phosphatidic acid synthesis in bacteria. Biochim Biophys Acta 1831:495–502. doi: 10.1016/j.bbalip.2012.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Pelosi L, Ducluzeau AL, Loiseau L, Barras F, Schneider D, Junier I, Pierrel F. 2016. Evolution of ubiquinone biosynthesis: multiple proteobacterial enzymes with various regioselectivities to catalyze three contiguous aromatic hydroxylation reactions. mSystems 1:e00091-16. doi: 10.1128/mSystems.00091-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Zhou L, Wang JY, Wu J, Wang J, Poplawsky A, Lin S, Zhu B, Chang C, Zhou T, Zhang LH, He YW. 2013. The diffusible factor synthase XanB2 is a bifunctional chorismatase that links the shikimate pathway to ubiquinone and xanthomonadins biosynthetic pathways. Mol Microbiol 87:80–93. doi: 10.1111/mmi.12084. [DOI] [PubMed] [Google Scholar]
- 110.Payne KAP, Fisher K, Sjuts H, Dunstan MS, Bellina B, Johannissen L, Barran P, Hay S, Rigby SEJ, Leys D. 2015. Epoxyqueuosine reductase structure suggests a mechanism for cobalamin-dependent tRNA modification. J Biol Chem 290:27572–27581. doi: 10.1074/jbc.M115.685693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Kittendorf JD, Sgraja T, Reuter K, Klebe G, Garcia GA. 2003. An essential role for aspartate 264 in catalysis by tRNA-guanine transglycosylase from Escherichia coli. J Biol Chem 278:42369–42376. doi: 10.1074/jbc.M304323200. [DOI] [PubMed] [Google Scholar]
- 112.Raux E, Schubert HL, Warren MJ. 2000. Biosynthesis of cobalamin (vitamin B12): a bacterial conundrum. Cell Mol Life Sci 57:1880–1893. doi: 10.1007/PL00000670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Zallot R, Ross R, Chen WH, Bruner SD, Limbach PA, de Crécy-Lagard V. 2017. Identification of a novel epoxyqueuosine reductase family by comparative genomics. ACS Chem Biol 12:844–851. doi: 10.1021/acschembio.6b01100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Wisseman CL, Edlinger EA, Waddell AD, Jones MR. 1976. Infection cycle of Rickettsia rickettsii in chicken embryo and L-929 cells in culture. Infect Immun 14:1052–1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Turco J, Winkler HH. 1989. Isolation of Rickettsia prowazekii with reduced sensitivity to gamma interferon. Infect Immun 57:1765–1772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Bansal R, Michel AP, Sabree ZL. 2014. The crypt-dwelling primary bacterial symbiont of the polyphagous pentatomid pest Halyomorpha halys (Hemiptera: Pentatomidae). Environ Entomol 43:617–625. doi: 10.1603/EN13341. [DOI] [PubMed] [Google Scholar]
- 117.Kenyon LJ, Meulia T, Sabree ZL. 2015. Habitat visualization and genomic analysis of “Candidatus Pantoea carbekii,”; the primary symbiont of the brown marmorated stink bug. Genome Biol Evol 7:620–635. doi: 10.1093/gbe/evv006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Martijn J, Schulz F, Zaremba-Niedzwiedzka K, Viklund J, Stepanauskas R, Andersson SGE, Horn M, Guy L, Ettema TJG. 2015. Single-cell genomics of a rare environmental alphaproteobacterium provides unique insights into Rickettsiaceae evolution. ISME J 9:2373–2385. doi: 10.1038/ismej.2015.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Izzard L, Fuller A, Blacksell SD, Paris DH, Richards AL, Aukkanit N, Nguyen C, Jiang J, Fenwick S, Day NPJ, Graves S, Stenos J. 2010. Isolation of a novel Orientia species (O. chuto sp. nov.) from a patient infected in Dubai. J Clin Microbiol 48:4404–4409. doi: 10.1128/JCM.01526-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Mediannikov O, Nguyen TT, Bell-Sakyi L, Padmanabhan R, Fournier PE, Raoult D. 2014. High quality draft genome sequence and description of Occidentia massiliensis gen. nov., sp. nov., a new member of the family Rickettsiaceae. Stand Genomic Sci 9:9. doi: 10.1186/1944-3277-9-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Cho NH, Kim HR, Lee JH, Kim SY, Kim J, Cha S, Kim SY, Darby AC, Fuxelius HH, Yin J, Kim JH, Kim J, Lee SJ, Koh YS, Jang WJ, Park KH, Andersson SGE, Choi MS, Kim IS. 2007. The Orientia tsutsugamushi genome reveals massive proliferation of conjugative type IV secretion system and host cell interaction genes. Proc Natl Acad Sci U S A 104:7981–7986. doi: 10.1073/pnas.0611553104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Zallot R, Yuan Y, de Crécy-Lagard V. 2017. The Escherichia coli COG1738 member YhhQ is involved in 7-cyanodeazaguanine (preQ0) transport. Biomolecules 7:E12. doi: 10.3390/biom7010012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Saier MH, Reddy VS, Tsu BV, Ahmed MS, Li C, Moreno-Hagelsieb G. 2016. The Transporter Classification Database (TCDB): recent advances. Nucleic Acids Res 44:D372–D379. doi: 10.1093/nar/gkv1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Culham DE, Lasby B, Marangoni AG, Milner JL, Steer BA, van Nues RW, Wood JM. 1993. Isolation and sequencing of Escherichia coli gene proP reveals unusual structural features of the osmoregulatory proline/betaine transporter, ProP. J Mol Biol 229:268–276. doi: 10.1006/jmbi.1993.1030. [DOI] [PubMed] [Google Scholar]
- 125.Hebbeln P, Rodionov DA, Alfandega A, Eitinger T. 2007. Biotin uptake in prokaryotes by solute transporters with an optional ATP-binding cassette-containing module. Proc Natl Acad Sci U S A 104:2909–2914. doi: 10.1073/pnas.0609905104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Schmitz-Esser S, Tischler P, Arnold R, Montanaro J, Wagner M, Rattei T, Horn M. 2010. The Genome of the amoeba symbiont “Candidatus Amoebophilus asiaticus” reveals common mechanisms for host cell interaction among amoeba-associated bacteria. J Bacteriol 192:1045–1057. doi: 10.1128/JB.01379-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Santos-Garcia D, Rollat-Farnier PA, Beitia F, Zchori-Fein E, Vavre F, Mouton L, Moya A, Latorre A, Silva FJ. 2014. The genome of Cardinium cBtQ1 provides insights into genome reduction, symbiont motility, and its settlement in Bemisia tabaci. Genome Biol Evol 6:1013–1030. doi: 10.1093/gbe/evu077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Erkens GB, Majsnerowska M, ter Beek J, Slotboom DJ. 2012. Energy coupling factor-type ABC transporters for vitamin uptake in prokaryotes. Biochemistry 51:4390–4396. doi: 10.1021/bi300504v. [DOI] [PubMed] [Google Scholar]
- 129.Rodionov DA, Hebbeln P, Eudes A, ter Beek J, Rodionova IA, Erkens GB, Slotboom DJ, Gelfand MS, Osterman AL, Hanson AD, Eitinger T. 2009. A novel class of modular transporters for vitamins in prokaryotes. J Bacteriol 191:42–51. doi: 10.1128/JB.01208-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Eitinger T, Rodionov DA, Grote M, Schneider E. 2011. Canonical and ECF-type ATP-binding cassette importers in prokaryotes: diversity in modular organization and cellular functions. FEMS Microbiol Rev 35:3–67. doi: 10.1111/j.1574-6976.2010.00230.x. [DOI] [PubMed] [Google Scholar]
- 131.Xu K, Zhang M, Zhao Q, Yu F, Guo H, Wang C, He F, Ding J, Zhang P. 2013. Crystal structure of a folate energy-coupling factor transporter from Lactobacillus brevis. Nature 497:268–271. doi: 10.1038/nature12046. [DOI] [PubMed] [Google Scholar]
- 132.Burgess CM, Slotboom DJ, Geertsma ER, Duurkens RH, Poolman B, van Sinderen D. 2006. The riboflavin transporter RibU in Lactococcus lactis: molecular characterization of gene expression and the transport mechanism. J Bacteriol 188:2752–2760. doi: 10.1128/JB.188.8.2752-2760.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Schauer K, Stolz J, Scherer S, Fuchs TM. 2009. Both thiamine uptake and biosynthesis of thiamine precursors are required for intracellular replication of Listeria monocytogenes. J Bacteriol 191:2218–2227. doi: 10.1128/JB.01636-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Schyns G, Potot S, Geng Y, Barbosa TM, Henriques A, Perkins JB. 2005. Isolation and characterization of new thiamine-deregulated mutants of Bacillus subtilis. J Bacteriol 187:8127–8136. doi: 10.1128/JB.187.23.8127-8136.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Haferkamp I, Schmitz-Esser S, Linka N, Urbany C, Collingro A, Wagner M, Horn M, Neuhaus HE. 2004. A candidate NAD+ transporter in an intracellular bacterial symbiont related to chlamydiae. Nature 432:622–625. doi: 10.1038/nature03131. [DOI] [PubMed] [Google Scholar]
- 136.Olsen GJ, Woese CR, Overbeek R. 1994. The winds of (evolutionary) change: breathing new life into microbiology. J Bacteriol 176:1–6. doi: 10.1128/jb.176.1.1-6.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Blachly-Dyson E, Forte M. 2001. VDAC channels. IUBMB Life 52:113–118. doi: 10.1080/15216540152845902. [DOI] [PubMed] [Google Scholar]
- 138.Emelyanov VV, Vyssokikh MY. 2006. On the nature of obligate intracellular symbiosis of rickettsiae—Rickettsia prowazekii cells import mitochondrial porin. Biochemistry (Mosc) 71:730–735. doi: 10.1134/S0006297906070054. [DOI] [PubMed] [Google Scholar]
- 139.Emelyanov VV. 2009. Mitochondrial porin VDAC 1 seems to be functional in rickettsial cells. Ann N Y Acad Sci 1166:38–48. doi: 10.1111/j.1749-6632.2009.04513.x. [DOI] [PubMed] [Google Scholar]
- 140.Walther DM, Bos MP, Rapaport D, Tommassen J. 2010. The mitochondrial porin, VDAC, has retained the ability to be assembled in the bacterial outer membrane. Mol Biol Evol 27:887–895. doi: 10.1093/molbev/msp294. [DOI] [PubMed] [Google Scholar]
- 141.Liu Y, Wu B, Weinstock G, Walker DH, Yu XJ. 2014. Inactivation of SAM-methyltransferase is the mechanism of attenuation of a historic louse borne typhus vaccine strain. PLoS One 9:e113285. doi: 10.1371/journal.pone.0113285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Clark TR, Noriea NF, Bublitz DC, Ellison DW, Martens C, Lutter EI, Hackstadt T. 2015. Comparative genome sequencing of Rickettsia rickettsii strains that differ in virulence. Infect Immun 83:1568–1576. doi: 10.1128/IAI.03140-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Ellison DW, Clark TR, Sturdevant DE, Virtaneva K, Porcella SF, Hackstadt T. 2008. Genomic comparison of virulent Rickettsia rickettsii Sheila Smith and avirulent Rickettsia rickettsii Iowa. Infect Immun 76:542–550. doi: 10.1128/IAI.00952-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Werren JH, Hurst GD, Zhang W, Breeuwer JA, Stouthamer R, Majerus ME. 1994. Rickettsial relative associated with male killing in the ladybird beetle (Adalia bipunctata). J Bacteriol 176:388–394. doi: 10.1128/jb.176.2.388-394.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Sun HY, Noe J, Barber J, Coyne RS, Cassidy-Hanley D, Clark TG, Findly RC, Dickerson HW. 2009. Endosymbiotic bacteria in the parasitic ciliate Ichthyophthirius multifiliis. Appl Environ Microbiol 75:7445–7452. doi: 10.1128/AEM.00850-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Zaila KE, Doak TG, Ellerbrock H, Tung CH, Martins ML, Kolbin D, Yao MC, Cassidy-Hanley DM, Clark TG, Chang WJ. 2017. Diversity and universality of endosymbiotic rickettsia in the fish parasite Ichthyophthirius multifiliis. Front Microbiol 8:189. doi: 10.3389/fmicb.2017.00189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Boyd BM, Allen JM, Koga R, Fukatsu T, Sweet AD, Johnson KP, Reed DL. 2016. Two bacterial genera, Sodalis and Rickettsia, associated with the seal louse Proechinophthirus fluctus (Phthiraptera: anoplura). Appl Environ Microbiol 82:3185–3197. doi: 10.1128/AEM.00282-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.McCutcheon JP, Moran NA. 2011. Extreme genome reduction in symbiotic bacteria. Nat Rev Microbiol 10:13–26. doi: 10.1038/nrmicro2670. [DOI] [PubMed] [Google Scholar]
- 149.Smith TA, Driscoll T, Gillespie JJ, Raghavan R. 2015. A Coxiella-like endosymbiont is a potential vitamin source for the lone star tick. Genome Biol Evol 7:831–838. doi: 10.1093/gbe/evv016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Rio RVM, Attardo GM, Weiss BL. 2016. Grandeur alliances: symbiont metabolic integration and obligate arthropod hematophagy. Trends Parasitol 32:739–749. doi: 10.1016/j.pt.2016.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Skaljac M, Zanic K, Ban SG, Kontsedalov S, Ghanim M. 2010. Co-infection and localization of secondary symbionts in two whitefly species. BMC Microbiol 10:142. doi: 10.1186/1471-2180-10-142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Perotti MA, Clarke HK, Turner BD, Braig HR. 2006. Rickettsia as obligate and mycetomic bacteria. FASEB J 20:2372–2374. doi: 10.1096/fj.06-5870fje. [DOI] [PubMed] [Google Scholar]
- 153.Yang Q, Kučerová Z, Perlman SJ, Opit GP, Mockford EL, Behar A, Robinson WE, Stejskal V, Li Z, Shao R. 2015. Morphological and molecular characterization of a sexually reproducing colony of the booklouse Liposcelis bostrychophila (Psocodea: Liposcelididae) found in Arizona. Sci Rep 5:10429. doi: 10.1038/srep10429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Labruna MB, Walker DH. 2014. Rickettsia felis and changing paradigms about pathogenic rickettsiae. Emerg Infect Dis 20:1768–1769. doi: 10.3201/eid2010.131797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Gillespie JJ, Driscoll TP, Verhoeve VI, Utsuki T, Husseneder C, Chouljenko VN, Azad AF, Macaluso KR. 2014. Genomic diversification in strains of Rickettsia felis isolated from different arthropods. Genome Biol Evol 7:35–56. doi: 10.1093/gbe/evu262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Penz T, Schmitz-Esser S, Kelly SE, Cass BN, Müller A, Woyke T, Malfatti SA, Hunter MS, Horn M. 2012. Comparative genomics suggests an independent origin of cytoplasmic incompatibility in Cardinium hertigii. PLoS Genet 8:e1003012. doi: 10.1371/journal.pgen.1003012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Nikoh N, Hosokawa T, Moriyama M, Oshima K, Hattori M, Fukatsu T. 2014. Evolutionary origin of insect-Wolbachia nutritional mutualism. Proc Natl Acad Sci U S A 111:10257–10262. doi: 10.1073/pnas.1409284111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Moriyama M, Nikoh N, Hosokawa T, Fukatsu T. 2015. Riboflavin provisioning underlies Wolbachia’s fitness contribution to its insect host. mBio 6:e01732-15. doi: 10.1128/mBio.01732-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Mattila JT, Munderloh UG, Kurtti TJ. 2007. Rickettsia peacockii, an endosymbiont of Dermacentor andersoni, does not elicit or inhibit humoral immune responses from immunocompetent D. andersoni or Ixodes scapularis cell lines. Dev Comp Immunol 31:1095–1106. doi: 10.1016/j.dci.2007.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Niebylski ML, Schrumpf ME, Burgdorfer W, Fischer ER, Gage KL, Schwan TG. 1997. Rickettsia peacockii sp. nov., a new species infecting wood ticks, Dermacentor andersoni, in western Montana. Int J Syst Bacteriol 47:446–452. doi: 10.1099/00207713-47-2-446. [DOI] [PubMed] [Google Scholar]
- 161.López-García P, Eme L, Moreira D. 2017. Symbiosis in eukaryotic evolution. J Theor Biol. doi: 10.1016/j.jtbi.2017.02.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Fournier PE, El Karkouri K, Leroy Q, Robert C, Giumelli B, Renesto P, Socolovschi C, Parola P, Audic S, Raoult D. 2009. Analysis of the Rickettsia africae genome reveals that virulence acquisition in Rickettsia species may be explained by genome reduction. BMC Genomics 10:166. doi: 10.1186/1471-2164-10-166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Aziz RK, Bartels D, Best AA, DeJongh M, Disz T, Edwards RA, Formsma K, Gerdes S, Glass EM, Kubal M, Meyer F, Olsen GJ, Olson R, Osterman AL, Overbeek RA, McNeil LK, Paarmann D, Paczian T, Parrello B, Pusch GD, Reich C, Stevens R, Vassieva O, Vonstein V, Wilke A, Zagnitko O. 2008. The RAST server: rapid annotations using subsystems technology. BMC Genomics 9:75. doi: 10.1186/1471-2164-9-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Feris KP, Ramsey PW, Frazar C, Rillig MC, Gannon JE, Holben WE. 2003. Structure and seasonal dynamics of hyporheic zone microbial communities in free-stone rivers of the Western United States. Microb Ecol 46:200–215. [DOI] [PubMed] [Google Scholar]
- 165.Edgar RC. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Talavera G, Castresana J. 2007. Improvement of phylogenies after removing divergent and ambiguously aligned blocks from protein sequence alignments. Syst Biol 56:564–577. doi: 10.1080/10635150701472164. [DOI] [PubMed] [Google Scholar]
- 167.Lartillot N, Lepage T, Blanquart S. 2009. PhyloBayes 3: a Bayesian software package for phylogenetic reconstruction and molecular dating. Bioinformatics 25:2286–2288. doi: 10.1093/bioinformatics/btp368. [DOI] [PubMed] [Google Scholar]
- 168.Stamatakis A, Ludwig T, Meier H. 2005. RAxML-III: a fast program for maximum likelihood-based inference of large phylogenetic trees. Bioinformatics 21:456–463. doi: 10.1093/bioinformatics/bti191. [DOI] [PubMed] [Google Scholar]
- 169.Kanehisa M, Sato Y, Morishima K. 2016. BlastKOALA and GhostKOALA: KEGG tools for functional characterization of genome and metagenome sequences. J Mol Biol 428:726–731. doi: 10.1016/j.jmb.2015.11.006. [DOI] [PubMed] [Google Scholar]
- 170.Ren Q, Chen K, Paulsen IT. 2007. TransportDB: a comprehensive database resource for cytoplasmic membrane transport systems and outer membrane channels. Nucleic Acids Res 35:D274–D279. doi: 10.1093/nar/gkl925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Fodorová M, Vadovič P, Toman R. 2011. Structural features of lipid A of Rickettsia typhi. Acta Virol 55:31–44. doi: 10.4149/av_2011_01_31. [DOI] [PubMed] [Google Scholar]
- 172.Aussel L, Pierrel F, Loiseau L, Lombard M, Fontecave M, Barras F. 2014. Biosynthesis and physiology of coenzyme Q in bacteria. Biochim Biophys Acta 1837:1004–1011. doi: 10.1016/j.bbabio.2014.01.015. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Rickettsia species import six host metabolites that are required for the synthesis of cell envelope glycoconjugates and glycerophospholipids. Download FIG S1, PDF file, 2 MB (2MB, pdf) .
Copyright © 2017 Driscoll et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
The Rickettsia DAP biosynthesis pathway contains a hole for the conversion of N-succinyl-l-2-amino-6-oxopimelate to N-succinyl-ll-2,6-DAP. Download FIG S2, PDF file, 0.4 MB (454.3KB, pdf) .
Copyright © 2017 Driscoll et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
The Rickettsia glycerophospholipid biosynthesis pathway contains a hole for the conversion of G3P to LPA. Download FIG S3, PDF file, 0.4 MB (422.4KB, pdf) .
Copyright © 2017 Driscoll et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Pathways for Rickettsia terpenoid and ubiquinone biosynthesis contain holes that are not common in other bacterial genomes. Download FIG S4, PDF file, 1.4 MB (1.4MB, pdf) .
Copyright © 2017 Driscoll et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Rickettsia species use DHN-P3 for queuosine biosynthesis and import host THF for one-carbon transfer reactions by folate. Download FIG S5, PDF file, 1.3 MB (1.4MB, pdf) .
Copyright © 2017 Driscoll et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
The Rickettsia queuosine biosynthesis pathway contains a hole for the reduction of epoxyqueuosine to queuosine. Download FIG S6, PDF file, 0.4 MB (414.6KB, pdf) .
Copyright © 2017 Driscoll et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
The evolutionary trajectory of six Rickettsia biosynthetic pathways that contain holes, or “missing enzymes.” Download FIG S7, PDF file, 2.2 MB (2.3MB, pdf) .
Copyright © 2017 Driscoll et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Phylogeny estimation of Idi and queuosine biosynthesis proteins. Download FIG S8, PDF file, 0.6 MB (651.5KB, pdf) .
Copyright © 2017 Driscoll et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Rickettsia transport and metabolism of host-acquired ribonucleotides. Download FIG S9, PDF file, 0.5 MB (490.3KB, pdf) .
Copyright © 2017 Driscoll et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Phylogenomics analysis of Rickettsia metabolic pathways and metabolite transporters. Download FIG S10, XLSX file, 0.4 MB (397.9KB, xlsx) .
Copyright © 2017 Driscoll et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.