ABSTRACT
The saw-toothed grain beetle, Oryzaephilus surinamensis (Silvanidae), is a cosmopolitan stored-product pest. Early studies on O. surinamensis in the 1930s described the presence of peculiar bacteriomes harboring endosymbiotic bacteria in the abdomen. Since then, however, the microbiological nature of the symbiont has been elusive. Here we investigated the endosymbiotic system of O. surinamensis in detail. In the abdomen of adults, pupae, and larvae, four oval bacteriomes were consistently identified, whose cytoplasm was full of extremely elongated tubular bacterial cells several micrometers wide and several hundred micrometers long. Molecular phylogenetic analysis identified the symbiont as a member of the Bacteroidetes, in which the symbiont was the most closely related to the endosymbiont of a grain pest beetle, Rhyzopertha dominica (Bostrichidae). The symbiont was detected in developing embryos, corroborating vertical symbiont transmission through host generations. The symbiont gene showed AT-biased nucleotide composition and accelerated molecular evolution, plausibly reflecting degenerative evolution of the symbiont genome. When the symbiont infection was experimentally removed, the aposymbiotic insects grew and reproduced normally, but exhibited a slightly but significantly more reddish cuticle and lighter body mass. These results indicate that the symbiont of O. surinamensis is not essential for the host’s growth and reproduction but contributes to the host’s cuticle formation. Symbiont genome sequencing and detailed comparison of fitness parameters between symbiotic and aposymbiotic insects under various environmental conditions will provide further insights into the symbiont’s biological roles for the stored-product pest.
KEYWORDS: bacterial symbiont, bacteriocyte, bacteriome, Bacteroidetes, cuticle formation, Oryzaephilus surinamensis, saw-toothed grain beetle, Silvanidae, stored-product pest
IMPORTANCE
Some beetles notorious as stored-product pests possess well-developed symbiotic organs called bacteriomes for harboring specific symbiotic bacteria, although their biological roles have been poorly understood. Here we report a peculiar endosymbiotic system of a grain pest beetle, Oryzaephilus surinamensis, in which four oval bacteriomes in the abdomen are full of extremely elongated tubular bacterial cells. Experimental symbiont elimination did not hinder the host’s growth and reproduction, but resulted in emergence of reddish beetles, uncovering the symbiont’s involvement in host’s cuticle formation. We speculate that the extremely elongated symbiont cell morphology might be due to the degenerative symbiont genome deficient in bacterial cell division and/or cell wall formation, which highlights an evolutionary consequence of intimate host-symbiont coevolution.
INTRODUCTION
Many insects host symbiotic bacteria in their gut, body cavity, or cells (1–3). Some bacterial symbionts are indispensable for growth, survival, and reproduction of their insect hosts via synthesis and supply of essential nutrients (4, 5), whereas other bacterial symbionts are dispensable, entailing either positive, neutral, or negative fitness consequences depending on environmental conditions, for their insect hosts as facultative microbial associates (6, 7). Among them, the most highly integrated host-symbiont associations are found in insects with specialized symbiotic organs, designated bacteriomes, consisting of host cells for harboring the symbiotic bacteria endocellularly, called bacteriocytes (1, 8, 9). Following early histological descriptions (reviewed in reference 1), bacteriome-associated endosymbiotic bacteria have been microbiologically characterized using modern molecular techniques from diverse insect orders, including the Hemiptera (aphids, coccids, whiteflies, leafhoppers, cicadas, stinkbugs, etc.) (10–16), the Diptera (tsetse flies, bat flies, louse flies, etc.) (17–19), the Blattodea (cockroaches, wood roaches, etc.) (20, 21), the Phthiraptera (sucking lice, chewing lice, etc.) (22, 23), and the Coleoptera (beetles). For a long time, the Curculionidae (weevils) and the Chrysomelidae (leaf beetles) have been the only beetle families whose bacteriome-associated endosymbiotic bacteria were microbiologically identified. As for weevils, an ancient gammaproteobacterial endosymbiont clade, Nardonella spp., found in diverse weevils (24, 25), a gammaproteobacterial endosymbiont, Sodalis pierantonius, that presumably replaced the original Nardonella endosymbiont in the lineage of grain weevils of the genus Sitophilus (26, 27), and another gammaproteobacterial endosymbiont, Curculioniphilus buchneri, that also replaced the original Nardonella endosymbiont in the lineage of acorn weevils of the genus Curculio and allied species (28–30), have been described. As for leaf beetles, a gammaproteobacterial endosymbiont clade, Macropleicola spp., associated with reed beetles of the subfamily Donaciinae (31–33), and several other gammaproteobacterial endosymbionts (34, 35) have been reported.
Beetles, characterized by their thick and hard exoskeleton, represent the majority of the insect species ever described (36). Some beetles are known to infest stored cereals and other durable commodities, being notorious as stored-product pests (37, 38). Possibly relevant to their peculiar ecology of continuously living on monotonous and non-fresh food sources, stored-product pest beetles are often associated with symbiotic microorganisms, including the Sitophilus grain weevils (Curculionidae) with bacteriome-harbored gammaproteobacterium Sodalis pierantonius (26, 27), the cigarette beetle Lasioderma serricorne and the drugstore beetle Stegobium paniceum (Anobiidae) with yeast-like fungal symbionts Symbiotaphrina spp. in the gut-associated symbiotic organs both endocellularly and extracellularly (39–41), and the lesser grain borer, Rhyzopertha dominica (Bostrichidae), whose bacteriome-associated endosymbiont was recently identified as belonging to the Bacteroidetes (42). Considering their easy rearing, experimental tractability, and economic importance, identification and understanding of biological roles of the bacteriome-associated endosymbionts in such stored-product pests are of general relevance to both basic and applied aspects of entomology and microbiology.
The saw-toothed grain beetle, Oryzaephilus surinamensis (Coleoptera: Silvanidae) (Fig. 1A), is a cosmopolitan stored-product pest (43, 44). The peculiar formation of the symbiotic system in O. surinamensis, consisting of four oval bacteriomes harboring a dense bacterial population and located in the abdomen, was first described histologically in the 1930s (45). Subsequent experimental works reported that symbiont elimination did not hinder growth and reproduction of O. surinamensis (46, 47), posing an enigma regarding the biological role of the symbiont for the insect host. Since then, however, the bacteriome-associated endosymbiosis in O. surinamensis has been totally untouched, leaving the understanding of the microbiological nature of the symbiont still elusive.
FIG 1 .
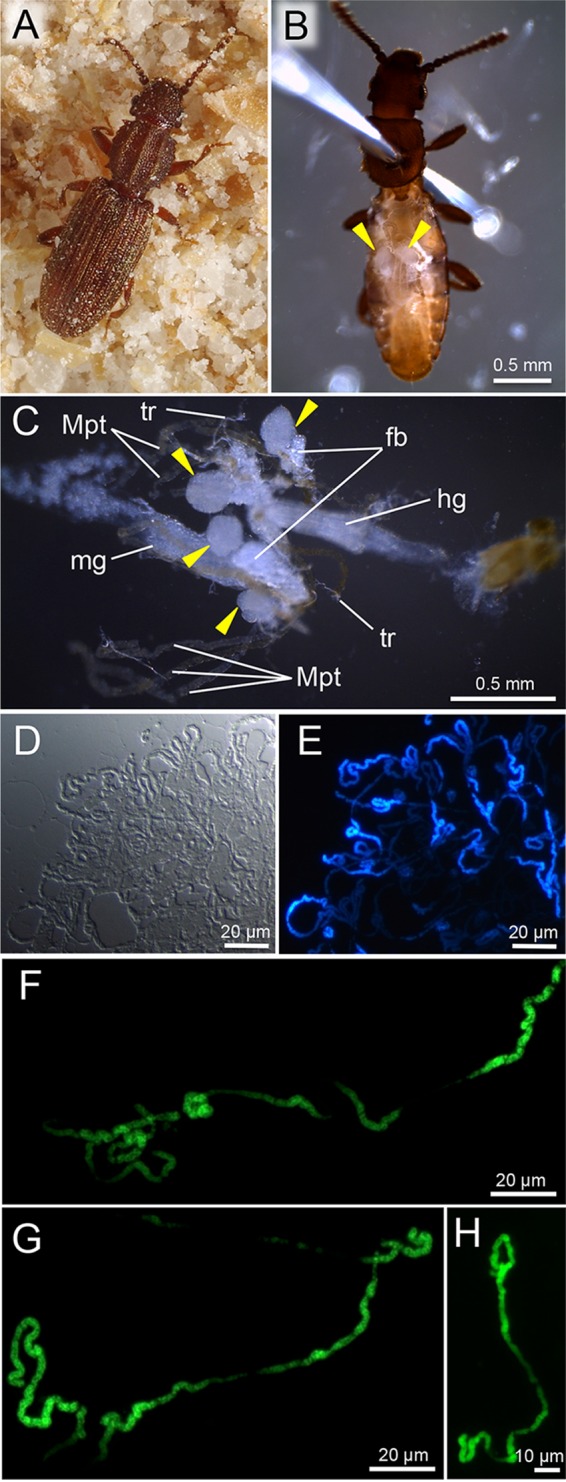
Bacteriomes and symbiotic bacteria of O. surinamensis. (A) Adult insect reared on whole-wheat flour. (B) Dissected adult insect, in which oval translucent bacteriomes are seen in the abdomen (arrowheads). (C) Internal organs dissected from an adult insect, in which four bacteriomes are highlighted (arrowheads). Note that reproductive tissues, fat body, and tracheae are mostly removed. Abbreviations: fb, fat body; hg, hindgut; mg, midgut; Mpt, Malpighian tubule; tr, trachea. (D and E) Differential interference microscopic image (D) and epifluorescence microscopic image (E) of symbiotic bacteria released from a dissected adult bacteriome. (F and G) Extremely elongated individual symbiont cells from dissected adult bacteriomes. (H) Elongated symbiont cell from a dissected pupal bacteriome. Bacterial DNA is stained with 4′,6-diamidino-2-phenylindole in panel E and SYTOX Green in panels F to H.
In this study, we investigated the endosymbiotic system of O. surinamensis to elucidate the microbiological nature of the symbiont, the fine structure of the symbiont, localization of the symbiont in the host tissues and cells, conditions for experimental symbiont elimination, and effects of symbiont removal on host phenotypes, by using modern molecular, histological, and morphometric techniques. We identified the symbiont as a novel bacterial lineage belonging to the Bacteroidetes, confirmed the histological descriptions in old literature, including endosymbiotic constitution with four oval bacteriomes and transovarial symbiont transmission to eggs, and newly discovered that the symbiont cells are extraordinarily elongated in shape and that the symbiont is certainly not essential for the host’s growth and reproduction but contributes to the host’s cuticle formation.
RESULTS
Extremely elongated symbiotic bacteria in bacteriomes.
When we dissected adult insects of the laboratory strain of O. surinamensis, translucent oval tissues were found in the abdomen (Fig. 1B), which were closely associated with Malpighian tubules, fat bodies, and tracheae (Fig. 1C). Smearing of the dissected tissues on a glass slide with DNA-binding fluorochromes visualized a dense population of tubular bacterial cells (Fig. 1D and E), verifying the tissues as constituting the bacteriomes of O. surinamensis. Strikingly, the symbiont cells were extremely elongated: in contrast to the cell width of a few micrometers, the cell length often reached some hundred micrometers, although the exact cell size was difficult to evaluate (Fig. 1F to H). The early histological study described that the symbiotic bacteria of O. surinamensis achieve their maximal length of 60 to 70 μm (45), but our observation unveiled much longer symbiont cell lengths than previously recognized.
Bacterial 16S rRNA gene of the symbiont.
The dissected adult abdomens were subjected to DNA extraction, PCR, and cloning and sequencing of the bacterial 16S rRNA gene, which yielded a 1,499-bp nucleotide sequence (sequence accession no. LC314798). BLASTN searches of the DNA databases using the sequence as query identified 16S rRNA gene sequences of Blattabacterium spp., the endosymbiont clade associated with cockroaches (e.g., GenBank accession no. CP005488, CP003605, and AP014609), as the top hits. From adult insects of different sources collected at Shizuoka and Niigata, Japan, the same 16S rRNA gene sequences were obtained (sequence accession no. LC314799 and LC314800), indicating that a specific bacterial symbiont is associated with O. surinamensis.
Phylogenetic placement of the symbiont.
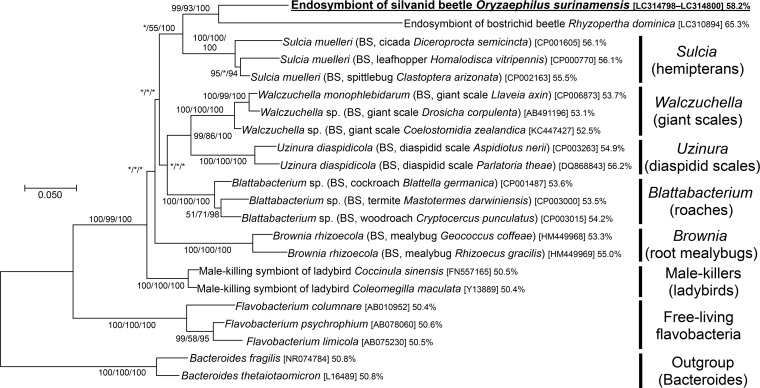
On the molecular phylogeny inferred from the 16S rRNA gene sequences, the symbiont of O. surinamensis was placed in the bacterial phylum Bacteroidetes, where the symbiont formed a distinct lineage clustering with endosymbiont clades associated with other insects, such as Sulcia of diverse hemipterans, Walczuchella of giant scale insects, Uzinura of diaspidid scale insects, Blattabacterium of cockroaches, Brownia of root mealybugs, etc. (Fig. 2). Notably, the symbiont of O. surinamensis (Silvanidae) was the most closely related to a recently reported, rosette-shaped bacterial endosymbiont of a stored-product pest, the lesser grain borer, R. dominica (Bostrichidae) (42) (Fig. 2).
FIG 2 .
Phylogenetic placement of the symbiont of O. surinamensis in the Bacteroidetes based on 16S rRNA gene sequences. A maximum likelihood phylogeny inferred from 1,185 aligned nucleotide sites is shown. Statistical support values for each clade are shown in the order of bootstrap probability of the neighbor-joining analysis, bootstrap probability of the maximum likelihood analysis, and posterior probability of the Bayesian analysis from left to right, where asterisks indicate values less than 50%. Host-related information in parentheses, accession number in brackets, and percentage of AT content follow each bacterial taxon. “BS” indicates bacteriocyte-associated endosymbiont. The explanation for each major clade is depicted on the right side.
In vivo localization and fine structure of the symbiont.
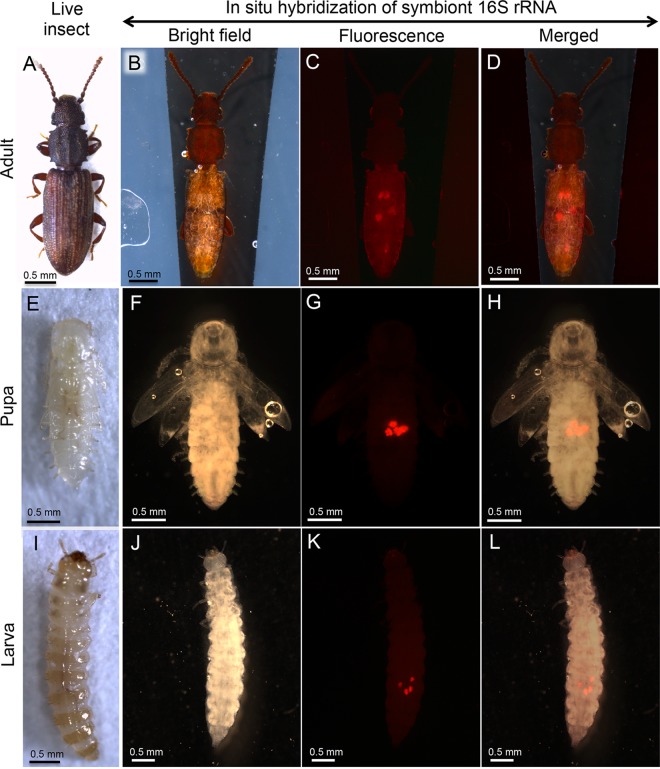
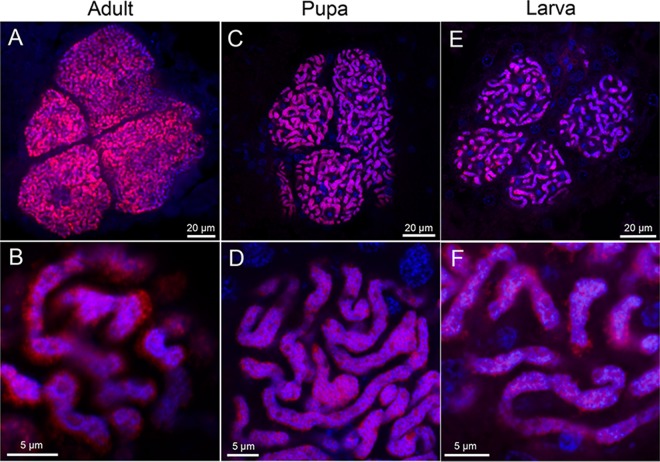
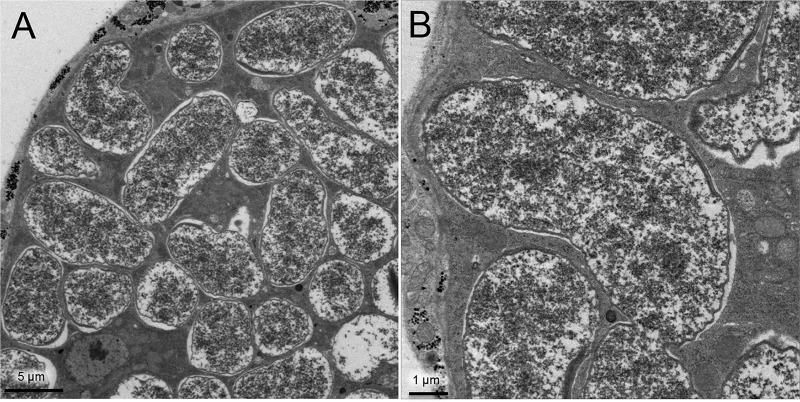
In order to visualize the symbiont localization in vivo, adults (Fig. 3A), pupae (Fig. 3E), and larvae (Fig. 3I) of O. surinamensis were subjected to whole-mount in situ hybridization targeting 16S rRNA of the symbiont. Four bacteriomes were consistently detected in the abdomen of adults (Fig. 3B to D), pupae (Fig. 3F to H), and larvae (Fig. 3J to L), of which two were situated dorsally (Fig. 4A and B) and the other two were located ventrally (Fig. 4C and D), in close association with fat body cells, tracheae, and Malpighian tubules (Fig. 1C; Fig. 4E). The peculiar formation of the symbiotic system is concordant with the previous histological descriptions of four oval bacteriomes in the abdomen of O. surinamensis (45, 46). Confocal fluorescence microscopy of the dissected bacteriomes of adults (Fig. 5A and B), pupae (Fig. 5C and D), and larvae (Fig. 5E and F) consistently visualized tubular bacterial cells filling the cytoplasm (Fig. 5). Transmission electron microscopy unveiled the fine structure of the symbiotic bacteria within the cytoplasm of the bacteriome (Fig. 6). In young embryos, a population of symbiotic bacteria was observed in a posterior region of the germ band (Fig. 7A and B). In mature embryos, four primordial bacteriomes were already formed in the abdomen (Fig. 7C and D). These observations favor the continuous host-symbiont association and the vertical symbiont transmission in O. surinamensis as described previously (45, 46).
FIG 3 .
Localization of bacteriomes in adults, pupae, and larvae of O. surinamensis. (A to D) Adults. (E to H) Pupae. (I to L) Larvae. (A, E, and I) Live insect images. (B, F, and J) Bright-field images of the insects subjected to whole-mount in situ hybridization targeting 16S rRNA of the symbiont. (C, G, and K) Epifluorescence dissection microscopic images of the same insects, in which four bacteriomes in the abdomen are visualized in red. (D, H, and L) Merged images. In panels B to D, F to H, and J to L, the insect bodies were gently pressed by a coverslip to locate the four bacteriomes to the same focal plane. In panels B to D, thin silicon rubber plates for stabilizing the position of the insect body are seen on both sides.
FIG 4 .
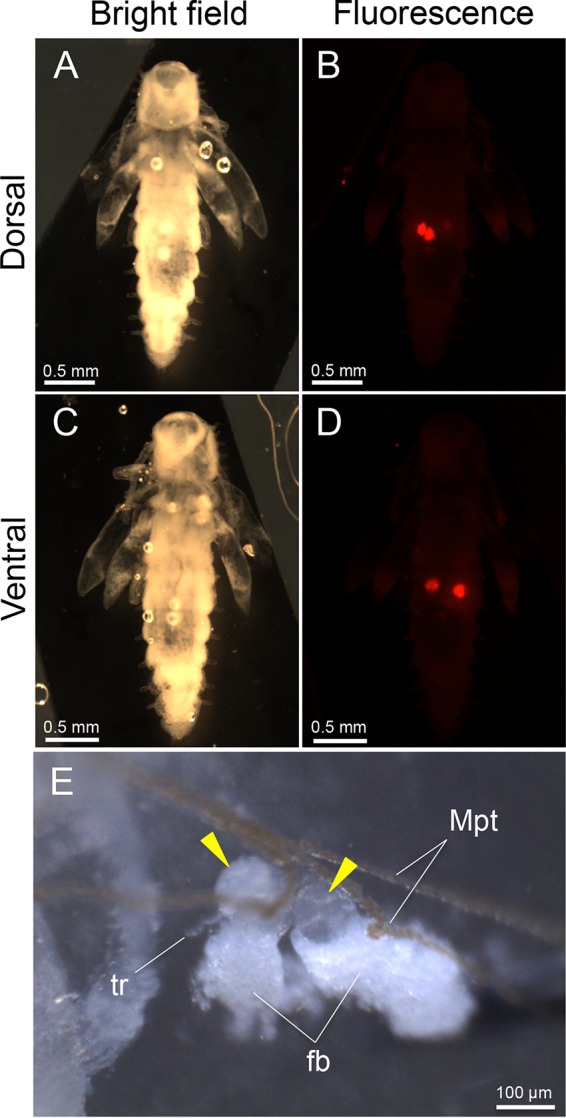
Localization and tissue construction of bacteriomes in pupae of O. surinamensis. (A) Bright-field image from the dorsal side. (B) Epifluorescence dissection microscopic image from the dorsal side, in which two dorsal bacteriomes are visualized in red by whole-mount in situ hybridization targeting 16S rRNA of the symbiont. (C) Bright-field image from the ventral side. (D) Epifluorescence dissection microscopic image from the ventral side, in which two ventral bacteriomes are seen in red. In panels A to D, different from Fig. 3F to H, the insect body was not pressed, thereby enabling the observation of the dorsal and ventral bacteriomes separately. (E) Magnified image of two dorsal bacteriomes (arrowheads) dissected from a pupa. Abbreviations: fb, fat body; Mpt, Malpighian tubule; tr, trachea.
FIG 5 .
Laser confocal microscopic images of bacteriomes and symbiotic bacteria of O. surinamensis. (A and B) Adult bacteriome. (C and D) Pupal bacteriome. (E and F) Larval bacteriome. (A, C, and E) Images of dissected bacteriomes. (B, D, and F) Enlarged images of symbiotic bacteria in the cytoplasm of the bacteriome. Red signals represent 16S rRNA of the symbiont visualized by fluorescence in situ hybridization, whereas blue signals are due to DNA staining.
FIG 6 .
Fine structure of the symbiont of O. surinamensis. (A) Transmission electron microscopic image of a pupal bacteriocyte. (B) Magnified image of the symbiotic bacteria.
FIG 7 .
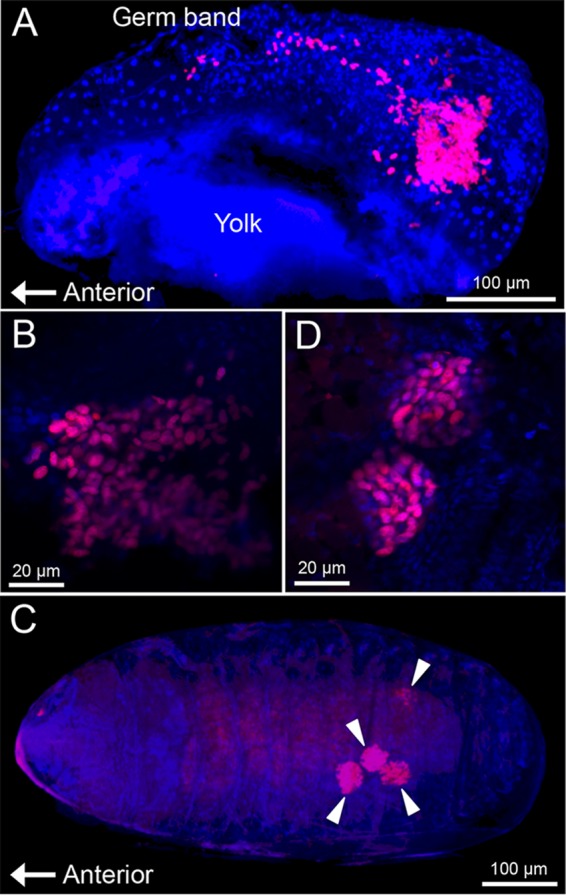
Localization of symbiotic bacteria in embryos of O. surinamensis. (A) Early embryo, in which a symbiont population in the germ band is seen in a posterior region. (B) Enlarged image of the bacterial population. (C) Late embryo, in which four primordial bacteriomes are seen in the abdomen (arrowheads). (D) Enlarged image of the primordial bacteriomes. Red signals represent 16S rRNA of the symbiont visualized by fluorescence in situ hybridization, whereas blue signals are due to DNA staining.
Molecular evolutionary aspects of the symbiont.
The AT content of 16S rRNA gene sequence of the symbiont of O. surinamensis, 58.2%, was higher than those of the other endosymbiont lineages in the Bacteroidetes, including Sulcia (55 to 56%), Brownia (53 to 55%), Blattabacterium (53 to 54%), and Uzinura (55 to 56%), and much higher than those of free-living Flavobacteria (50 to 51%) (Fig. 2). A relative rate test based on the 16S rRNA gene sequence showed that the evolutionary rate in the lineage of the symbiont of O. surinamensis was significantly higher than that in the lineage of free-living Flavobacteria, while being almost equivalent to those in the other endosymbiont lineages, like Sulcia and Blattabacterium (Table 1). These results suggest that the symbiont of O. surinamensis may have experienced reductive genome evolution like the other insect endosymbiont lineages in the Bacteroidetes. Notably, the symbiont of O. surinamensis exhibited remarkably lower AT content and a significantly slower molecular evolutionary rate than the symbiont of R. dominica (Table 1), suggesting the possibility that the symbiont genome reduction might have been less advanced in O. surinamensis than in R. dominica.
TABLE 1 .
Relative rate tests of 16S rRNA gene sequence of the symbiont of Oryzaephilus surinamensis in comparison with the symbiont of Rhyzopertha dominica, Sulcia symbionts, Blattabacterium symbionts, and allied free-living bacteria
| Lineage 1 | Lineage 2 | Outgroup | K1a | K2b | K1 − K2 | K1/K2 | P valuec |
|---|---|---|---|---|---|---|---|
| Symbiont of O. surinamensis
(LC314798) |
Symbiont of R. dominica
(LC310894) |
Bacteroides fragilis
(NR_074784) |
0.062 | 0.134 | −0.072 | 0.46 | 6.4 × 10−6 |
| Symbiont of O. surinamensis
(LC314798) |
Sulcia symbiontsd |
Bacteroides fragilis
(NR_074784) |
0.075 | 0.070 | 0.005 | 1.1 | 0.65 |
| Symbiont of O. surinamensis
(LC314798) |
Blattabacterium symbiontse |
Bacteroides fragilis
(NR_074784) |
0.068 | 0.051 | 0.017 | 1.3 | 0.12 |
| Symbiont of O. surinamensis
(LC314798) |
F. columnare and F. limicolaf |
Bacteroides fragilis
(NR_074784) |
0.126 | 0.092 | 0.034 | 1.4 | 0.024 |
Estimated mean distance between lineage 1 and the last common ancestor of lineages 1 and 2.
Estimated mean distance between lineage 2 and the last common ancestor of lineages 1 and 2.
P values were generated using the program RRTree (87). The analyses were performed using 1,425 aligned nucleotide sites of 16S rRNA gene sequences.
Based on 16S rRNA sequences of Sulcia symbionts from Homalodisca vitripennis (CP000770), Diceroprocta semicincta (CP001605), and Clastoptera arizonana (CP002163).
Generation of an aposymbiotic host line by antibiotic treatment.
In an attempt to investigate functional aspects of the endosymbiotic association, we tried to remove the symbiont infection from O. surinamensis by antibiotic treatment. Adult insects from the same laboratory colony were divided into two groups, and the control group was fed with unsupplemented whole-wheat flour, while the antibiotic-treated group was fed with whole-wheat flour supplemented with 0.05% tetracycline. After the feeding treatment for two generations, newly emerged adult insects of each group were transferred to and maintained on unsupplemented whole-wheat flour, thereby creating a control line and an antibiotic-treated line of O. surinamensis. Diagnostic PCR revealed that the insects of the antibiotic-treated line were all symbiont negative, whereas the insects of the control line were all symbiont positive (Fig. 8A), verifying removal of the symbiont in the antibiotic-treated line. Fluorescence in situ hybridization confirmed absence of the symbiont in the bacteriomes of the antibiotic-treated insects (Fig. 8B and C). Interestingly, the insects of the antibiotic-treated line grew and reproduced normally with no difficulty in rearing and maintenance. Thus far, we have been stably maintaining both the symbiotic control line and the aposymbiotic antibiotic-treated line for over 20 months (Fig. 8A), indicating that the symbiont is not essential for normal growth, survival, and reproduction of O. surinamensis.
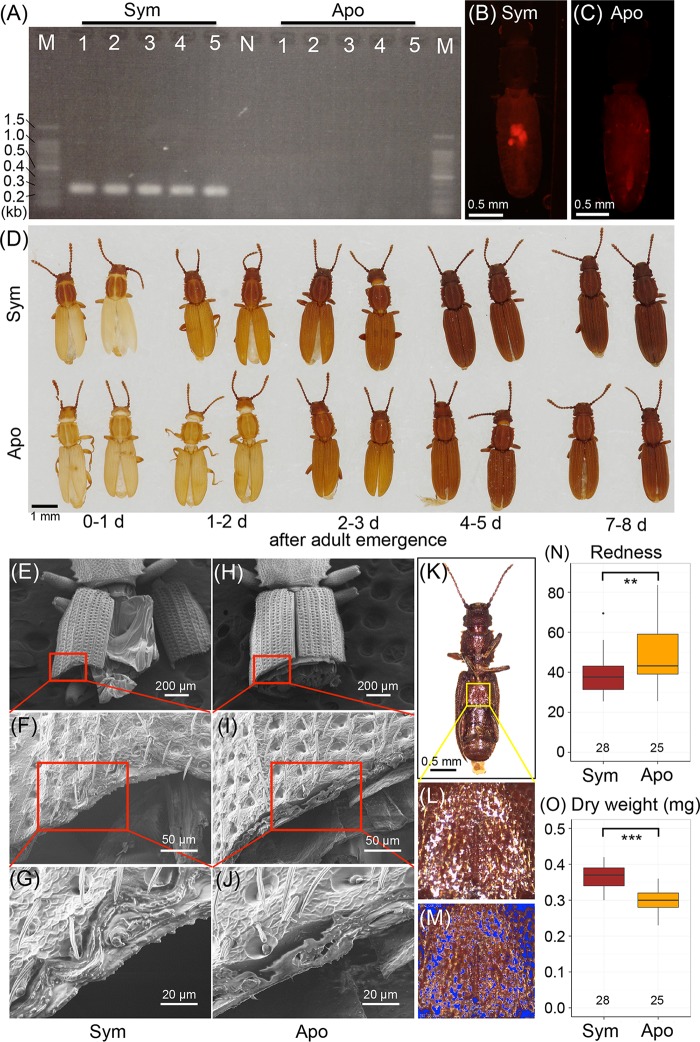
FIG 8 .
Effects of symbiont removal on adult phenotypes of O. surinamensis. (A) Diagnostic PCR detection of the symbiont from adult insects of the control symbiotic line (Sym) and the antibiotic-treated aposymbiotic line (Apo). Each lane in lanes 1 to 5 represents a DNA sample from a single insect. N and M represent the no-template control and DNA size markers, respectively. (B and C) Fluorescence in situ hybridization targeting 16S rRNA of the symbiont in adult females of the symbiotic strain (B) and the aposymbiotic strain (C). (D) Cuticle pigmentation after adult emergence of the symbiotic insects (upper row) and the aposymbiotic insects (lower row). (E to J) Scanning electron microscopic images of sectioned elytra of the symbiotic adult insects (E to G) and the aposymbiotic adult insects (H to J). Panels F and I are magnified images of panels E and H, as indicated by red rectangles; panels G and J are magnified images of panels F and I. (K to M) Process of quantifying redness of cuticle. From each of the ventral images of adult insects (K), a square area was extracted from the metasternum. On the square image (L), the pixels whose brightness was either above the top 10% or below the bottom 10% were masked in blue (M) and excluded from the analysis to minimize the effects of highlights and shadows. Then, RGB values for all (n) pixels were measured and averaged to obtain the redness index by Σ[R − mean (R, G, B)]/n. (N) Comparison of cuticle redness between the symbiotic insects and the aposymbiotic insects older than 2 weeks after adult emergence. (O) Comparison of dry body weight between the symbiotic insects and the aposymbiotic insects older than 2 weeks after adult emergence. In panels N and O, asterisks indicate statistically significant differences by t test: **, P < 0.01; ***, P < 0.001.
Effects of symbiont removal on host insect phenotypes.
However, our close inspection of the symbiotic insects and the aposymbiotic insects unveiled subtle but remarkable phenotypic differences between them. Upon eclosion, the pronotum of the symbiotic adult insects was pigmented in brown, in contrast to the pale pronotum of the aposymbiotic adult insects (Fig. 8D, 0 to 1 day). The differences in cuticle pigmentation between the symbiotic insects and the aposymbiotic insects were the most prominent on the next day (Fig. 8D, 1 to 2 days), subsequently became less conspicuous as the insects aged and the cuticular pigmentation proceeded (Fig. 8D, 2 to 3, 4 to 5, and 7 to 8 days), and finally became almost indiscernible in mature adult insects that were older than 2 weeks and dark in color. Even in the mature adult insects, however, detailed analyses uncovered notable differences between the symbiotic insects and the aposymbiotic insects. Scanning electron microscopy revealed that sectioned elytra of the symbiotic insects tended to be structurally denser than the sectioned elytra of the aposymbiotic insects, although the differences were rather qualitative and varied depending on individuals (Fig. 8E to J). Color imaging analysis of the adult metasternum (Fig. 8K to M) demonstrated that the cuticle of the aposymbiotic insects was significantly more reddish than the cuticle of the symbiotic insects (Fig. 8N). Weighing of acetone-preserved and dried insects revealed that the aposymbiotic insects were slightly but significantly lighter in body weight than the symbiotic insects (Fig. 8O).
DISCUSSION
Since the pioneering histological descriptions and some experimental studies dating back to the 1930s (45–47), we finally characterized the microbiological nature of the bacteriome-associated symbiont of the stored-product pest beetle, O. surinamensis, as a distinct bacterial lineage in the Bacteroidetes (Fig. 2). This study is the first to identify the bacteriome-associated endosymbiont from the beetle family Silvanidae.
Notably, the symbiont of O. surinamensis was the most closely related to the symbiont of R. dominica, forming a deeply diverged but statistically well-supported clade in the Bacteroidetes (Fig. 2). However, the phylogenetic affinity of the symbiotic bacteria is not concordant with the systematics of the host insects: the family Silvanidae is placed within the infraorder Cucujiformia, the family Bostrichidae belongs to the infraorder Bostrichiformia, and Cucujiformia is phylogenetically not allied to the Bostrichiformia in the Coleoptera (48). These phylogenetic patterns suggest the following possibilities: (i) a deep common ancestry of the symbiont originating from the common ancestor of the Cucujiformia and the Bostrichiformia, (ii) lateral symbiont transfer between the Silvanidae and the Bostrichidae, or (iii) independent symbiont acquisitions in the Silvanidae and the Bostrichidae from the common or closely related source bacteria belonging to the Bacteroidetes. Their common ecological aspect, living on cereal grains and other stored products, may have some relevance to these evolutionary patterns. A wider survey and molecular phylogenetic analyses of the bacteriome-associated endosymbionts in the Silvanidae and the Bostrichidae are needed to address which of these hypotheses is the most appropriate.
Judging from the AT-biased nucleotide composition (Fig. 2) and the accelerated molecular evolution (Table 1), it seems likely that the symbiont of O. surinamensis has experienced reductive genome evolution like the other endosymbiont lineages in the Bacteroidetes, like Sulcia (0.19 to 0.29 Mb) (49–51), Blattabacterium (0.59 to 0.64 Mb) (52–54), Uzinura (0.26 Mb) (55), and Walczuchella (0.31 Mb) (56). Although speculative, the extremely elongated cell shape of the symbiont of O. surinamensis may be relevant to the degenerative genome evolution and resultant deficiency in bacterial cell division and/or cell wall formation (57). Plausibly, the strange rosette-like cell shape of the phylogenetically related symbiont of R. dominica might be also accounted for in this context (42). Symbiont genome sequencing will provide pivotal clues to understanding these evolutionary issues.
A notable aspect of the endosymbiosis in O. surinamensis is that, although possessing the well-developed bacteriomes, the host insect is able to survive, grow, and reproduce even in the absence of the symbiont. This observation agrees with the following early experimental studies on O. surinamensis. Koch (46) established aposymbiotic colonies of O. surinamensis by high-temperature treatments at 36 to 38°C and reported that (i) the aposymbiotic insects were easily maintainable for as long as 5 years, (ii) during the period equivalent to some 25 insect generations, the aposymbiotic insects continued to form sterile bacteriomes in the abdomen, and (iii) no differences in growth and fecundity were detected between the aposymbiotic insects and the original symbiotic insects on a variety of foods, including oats, oatmeal, wheat, barley, buckwheat, hazelnut, and dried banana. Huger (47) reported that (i) antibiotic treatments also yielded aposymbiotic insects of O. surinamensis, (ii) a low-temperature treatment at 4°C for 24 days reduced the symbiont titer but did not eradicate it, and (iii) 45% of O. surinamensis strains collected from heavily infested grain warehouses turned out to be aposymbiotic, which might be attributable to overheating caused by massive proliferation of the insects in the closed grain stockrooms.
However, we strongly suspect that detailed fitness monitoring under a variety of environmental conditions, such as different diets, different nutritional conditions, different temperatures, different levels of humidity, with/without predators/parasites, etc., would lead to identification of ecological factors responsible for the persistence of the symbiont in O. surinamensis populations despite the dispensable nature of the symbiosis. In this context, our discovery of the slight but significant effects of aposymbiosis on the cuticle color and structure (Fig. 8) seems to be of importance. The sclerotized cuticle of beetles has been shown to confer mechanical strength, antipredator defense, tolerance to desiccation, and other beneficial consequences, which contribute to their survival and adaptation (58–61). Rearing experiments under these different conditions and strict fitness measurements are currently in progress to test these possibilities.
Previous studies on a variety of symbiont-dependent insects documented that experimental symbiont suppression often induced frail host insects with a pale and soft cuticle, as in, for example, aphids (62, 63), stinkbugs (15, 64–66), weevils (67–71), and others. These phenotypes may be due to general physiological or nutritional deficiency due to aposymbiosis (72, 73) or more specifically may be attributable to deficiency in symbiont-provisioned aromatic amino acids, namely, phenylalanine and tyrosine, which are the principal precursors needed for cuticle formation and hardening, as has been demonstrated in aphids (74, 75) and weevils (68, 70, 71). Genomic, transcriptomic, and metabolic analyses of the bacteriomes of O. surinamensis will provide pivotal clues to understanding what molecular and physiological mechanisms of the symbiont are involved in the cuticle formation of the host insect.
Finally, we point out that the easy rearing system for O. surinamensis, solely on whole-wheat flour in plastic containers almost free of laborious maintenance, in combination with the experimental tractability of both the symbiotic and aposymbiotic insects will facilitate experimental studies on nutritional, physiological, and metabolic aspects of host-symbiont interactions. In the flour beetle Tribolium castaneum, which is also maintainable on whole-wheat flour and renowned as a model insect in population biology, molecular genetics, and genomics (76–78), a nearly completely defined diet consisting of casein, glucose, cholesterol, vitamins, minerals, salts, and the insoluble fraction of yeast was developed for strict nutritional studies (79, 80). Development of a similar defined diet for O. surinamensis would enable detailed studies on functional aspects of the endosymbiosis by experimentally manipulating the diet components and by tracing the metabolites at the host-symbiont interface.
MATERIALS AND METHODS
Insects and rearing.
We mainly used a long-lasting laboratory strain of O. surinamensis, OsNFRI, which is of unknown origin, has been maintained for over 20 years at the National Food Research Institute, Tsukuba, Japan, and was provided by Akihiro Miyanoshita. The insects were reared on whole flour of domestically grown wheat (Ebetsu Flour Mill, Hokkaido, Japan) in plastic containers at 25°C under constant darkness. For symbiont elimination, the insects were reared on the whole-wheat flour supplemented with 0.05% tetracycline for two generations, and subsequently newly emerged adult insects were transferred to and maintained on the whole-wheat flour without the antibiotic. We also analyzed a strain of O. surinamensis provided by Kohjiro Tanaka, which was collected from sunflower seeds and maintained at Fuji Environment Service (Shizuoka, Japan), and a sample of O. surinamensis collected on 2 July 2017 at Ojiya, Niigata, Japan. Some insects were preserved in acetone as described previously (81) until DNA and phenotypic analyses.
DNA analysis.
The adult abdomens were dissected and subjected to DNA extraction using a QIAamp DNA minikit (Qiagen). A 1.5-kb segment of bacterial 16S rRNA gene was amplified by PCR with the primers 10FF (5′-AGT TTG ATC ATG GCT CAG GAT-3′) and 1515R (5′-GTA CGG CTA CCT TGT TAC GAC TTA G-3′) (14) under the temperature profile of 94°C for 5 min followed by 35 cycles of 94°C for 30 s, 55°C for 30 s, and 72°C for 1 min and a final incubation at 72°C for 5 min. The PCR products were cloned and sequenced as described previously (82). Diagnostic PCR detection of the symbiont was conducted with the primers OsFlav60F (5′-AAG TCG AGG GGC AAC ATG AA-3′) and OsFlav304R (5′-CTC AGG CCC CCT ACC GAT AA-3′) specifically targeting a 0.3-kb segment of 16S rRNA gene of the symbiont under the same temperature profile. The PCR products were electrophoresed in 1.5% agarose gels, stained with ethidium bromide, and photographed on a UV transilluminator.
Molecular phylogenetic and evolutionary analyses.
Multiple alignments of nucleotide sequences were constructed by ClustalW (83) implemented in MEGA v7.0.26 (84). The alignments were inspected and corrected manually and subjected to neighbor-joining analyses by MEGA v7.0.26 (84) with 1,000 bootstrap replicates, maximum likelihood analyses by MEGA v7.0.26 (84) with 1,000 bootstrap replicates, and Bayesian analyses by MrBayes v3.2.6 (85). The best-fit substitution model for the aligned sequences was evaluated by Kakusan v4 (86), which selected the generalized time-reversible (GTR) gamma model for both the maximum likelihood and Bayesian methods. Relative rate tests were performed by RRTree (87).
Histological procedures.
For observation of fresh cell images of the symbiont, the bacteriomes were dissected in phosphate-buffered saline (PBS: 0.8% NaCl, 0.02% KCl, 0.115% Na2HPO4, 0.02% KH2PO4 [wt/vol]), placed on a glass slide with a drop of SYTOX Green solution (1/1,000 dilution) or 4 μM 4′,6-diamidino-2-phenylindole solution, smeared with a coverslip, and observed under an epifluorescence microscope (Axiophot; Zeiss). For whole-mount fluorescence in situ hybridization, adults, pupae, larvae, and eggs were pretreated in 70% ethanol under a dissection microscope as follows. Adults were deprived of all wings by forceps, and a side of their abdomen was cut with a razor. Pupae and larvae were punctured by a needle at the anterior and posterior regions of the body. Eggs containing embryos were collected, treated with 50% bleach for 2 min, and washed thoroughly with distilled water. The pupal and larval samples were fixed in Carnoy’s solution (60% ethanol, 30% chloroform, and 10% acetic acid [vol/vol]) for 3 h at room temperature, washed thoroughly with 70% ethanol, treated with 6% H2O2 in 80% ethanol for 6 days to quench autofluorescence of the tissues (88), and washed thoroughly with 70% ethanol. The adult and egg samples were fixed with 4% formalin in PBS and washed thoroughly with PBS. For fluorescence in situ hybridization of dissected bacteriomes, adults, pupae, and larvae were dissected in 4% formalin in PBS, and the dissected tissues were fixed in fresh 4% formalin in PBS for 1 h at room temperature and washed thoroughly with PBS.
Fluorescence in situ hybridization.
Fluorescence in situ hybridization targeting 16S rRNA of the symbiont was conducted essentially as described previously (88). The samples were rehydrated with PBT (PBS containing 0.1% Tween 20 [vol/vol]) and incubated in hybridization buffer (20 mM Tris-HCl [pH 8.0], 0.9 M NaCl, 0.01% sodium dodecyl sulfate, 30% formamide [wt/vol]) containing 100 pmol/ml each probe at room temperature overnight. The following probes whose 5′ end was labeled with Alexa Fluor 555 were used either singly or in combination: EUB338 (5′-GCT GCC TCC CGT AGG AGT-3′), targeting diverse bacteria (89); EUB897 (5′-TTT GAG TTT YAV YCT TGC G-3′), targeting diverse bacteria designed in this study; Sul664R (5′-CCM CAC ATT CCA GYT ACT CC-3′) targeting Sulcia and allied bacteria of the Bacteroidetes (90); and OSFlav1256R (5′-ATC TAA TTA CTT AGT AGC AGC C-3′), specifically targeting the symbiont of O. surinamensis designed in this study. After being washed with PBT three times for 10 min each at room temperature, the samples were placed on glass slides, mounted in 80% glycerol or SlowFade antifade reagent (Thermo Fisher Scientific), and observed under a fluorescence dissection microscope (M165FC; Leica), an epifluorescence microscope (Axiophot; Zeiss), and/or a laser scanning confocal microscope (LSM710; Zeiss). Digital images were merged and adjusted manually using Adobe Photoshop CC 2017 (Adobe).
Electron microscopy.
For transmission electron microscopy, pupae were dissected in 2.5% glutaraldehyde in 0.1 M phosphate buffer (pH 7.4), and the dissected bacteriomes were prefixed with the fixative at 4°C overnight. Subsequently, the samples were postfixed with 2% osmium tetroxide in 0.1 M phosphate buffer (pH 7.4) at 4°C for 60 min, dehydrated through a water-ethanol series, and embedded in Epon 812 resin. Ultrathin sections (around 80 nm thick) were made by an ultramicrotome (EM UC7; Leica), mounted on copper meshes, stained with uranyl acetate and lead citrate, and observed under a transmission electron microscope (H-7600; Hitachi). For scanning electron microscopy, dry specimens of adult insects were cut with a razor blade. Subsequently, the samples were gold coated using Smart Coater (JEOL) and observed under a scanning electron microscope (JCM 6000; JEOL) operated at high vacuum and at 5 kV with use of the secondary electron signal.
Qualitative and quantitative analyses of adult phenotypes.
From mass-cultured symbiotic and aposymbiotic colonies of O. surinamensis, all adult insects were removed and transferred to new rearing containers. Then, newly emerged adult insects were collected from the old rearing containers every day, by which we obtained age-defined symbiotic and aposymbiotic adult insects. The age-defined adult insects were preserved in acetone, dried in air, placed on a piece of double-sided adhesive tape, and photographed by a digital camera. Symbiotic and aposymbiotic mature adult insects older than 2 weeks after emergence were dehydrated in acetone for several days, dried in air for a day, and weighed on a microbalance (MT5; Mettler Toledo, Columbus, OH). Subsequently, the adult insects were photographed from the ventral side by a digital camera (EC3; Leica) connected to a dissection microscope (S8APO; Leica) under a constant-lighting condition. Image processing operations and measurements were carried out with the software Natsumushi v0.99 (https://sites.google.com/site/mtahashilucanid/program/natsumushi). From each image of the metasternum (Fig. 8K), a square area of maximal size was extracted (Fig. 8L). On the square image, the pixels whose brightness was either over the top 10% or below the bottom 10% were masked in blue and excluded from the analysis to minimize the effects of highlights and shadows (Fig. 8M). Then, 8-bit-integer RGB (red, green, blue) values (0 ≦ R, G, B ≦ 255) for all (n) pixels were measured and averaged to obtain the redness index by Σ[R − mean (R, G, B)]/n.
Accession number(s).
The 16S rRNA gene sequences of the symbiont of O. surinamensis determined in this study have been deposited in the DNA Data Bank of Japan database under accession no. LC314798 to LC314800.
ACKNOWLEDGMENTS
We thank Akihiro Miyanoshita and Kohjiro Tanaka for providing O. surinamensis samples.
This study was supported by JSPS Kakenhi grant no. JP25221107 to T.F.
Footnotes
Citation Hirota B, Okude G, Anbutsu H, Futahashi R, Moriyama M, Meng X-Y, Nikoh N, Koga R, Fukatsu T. 2017. A novel, extremely elongated, and endocellular bacterial symbiont supports cuticle formation of a grain pest beetle. mBio 8:e01482-17. https://doi.org/10.1128/mBio.01482-17.
REFERENCES
- 1.Buchner P. 1965. Endosymbiosis of animals with plant microorganisms. Interscience, New York, NY. [Google Scholar]
- 2.Boutzis K, Miller TA. 2003. Insect symbiosis. CRC Press, Boca Raton, FL. [Google Scholar]
- 3.Zchori-Fein E, Bourtzis K. 2012. Manipulative tenants: bacteria associated with arthropods. CRC Press, Boca Raton, FL. [Google Scholar]
- 4.Moran NA, McCutcheon JP, Nakabachi A. 2008. Genomics and evolution of heritable bacterial symbionts. Annu Rev Genet 42:165–190. doi: 10.1146/annurev.genet.41.110306.130119. [DOI] [PubMed] [Google Scholar]
- 5.Douglas AE. 2009. The microbial dimension in insect nutritional ecology. Funct Ecol 23:38–47. doi: 10.1111/j.1365-2435.2008.01442.x. [DOI] [Google Scholar]
- 6.Werren JH, Baldo L, Clark ME. 2008. Wolbachia: master manipulators of invertebrate biology. Nat Rev Microbiol 6:741–751. doi: 10.1038/nrmicro1969. [DOI] [PubMed] [Google Scholar]
- 7.Oliver KM, Degnan PH, Burke GR, Moran NA. 2010. Facultative symbionts in aphids and the horizontal transfer of ecologically important traits. Annu Rev Entomol 55:247–266. doi: 10.1146/annurev-ento-112408-085305. [DOI] [PubMed] [Google Scholar]
- 8.Koga R, Meng XY, Tsuchida T, Fukatsu T. 2012. Cellular mechanism for selective vertical transmission of an obligate insect symbiont at the bacteriocyte-embryo interface. Proc Natl Acad Sci U S A 109:E1230–E1237. doi: 10.1073/pnas.1119212109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Matsuura Y, Kikuchi Y, Miura T, Fukatsu T. 2015. Ultrabithorax is essential for bacteriocyte development. Proc Natl Acad Sci U S A 112:9376–9381. doi: 10.1073/pnas.1503371112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Munson MA, Baumann P, Kinsey MG. 1991. Buchnera gen. nov. and Buchnera aphidicola sp. nov., a taxon consisting of the mycetocyte-associated, primary endosymbionts of aphids. Int J Syst Bacteriol 41:566–568. doi: 10.1099/00207713-41-4-566. [DOI] [Google Scholar]
- 11.Thao ML, Moran NA, Abbot P, Brennan EB, Burckhardt DH, Baumann P. 2000. Cospeciation of psyllids and their primary prokaryotic endosymbionts. Appl Environ Microbiol 66:2898–2905. doi: 10.1128/AEM.66.7.2898-2905.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Moran NA, Dale C, Dunbar H, Smith WA, Ochman H. 2003. Intracellular symbionts of sharpshooters (Insecta: Hemiptera: Cicadellinae) form a distinct clade with a small genome. Environ Microbiol 5:116–126. doi: 10.1046/j.1462-2920.2003.00391.x. [DOI] [PubMed] [Google Scholar]
- 13.Thao ML, Baumann P. 2004. Evolutionary relationships of primary prokaryotic endosymbionts of whiteflies and their hosts. Appl Environ Microbiol 70:3401–3406. doi: 10.1128/AEM.70.6.3401-3406.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Moran NA, Tran P, Gerardo NM. 2005. Symbiosis and insect diversification: an ancient symbiont of sap-feeding insects from the bacterial phylum Bacteroidetes. Appl Environ Microbiol 71:8802–8810. doi: 10.1128/AEM.71.12.8802-8810.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hosokawa T, Kikuchi Y, Nikoh N, Shimada M, Fukatsu T. 2006. Strict host-symbiont cospeciation and reductive genome evolution in insect gut bacteria. PLoS Biol 4:e337. doi: 10.1371/journal.pbio.0040337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kikuchi Y, Hosokawa T, Nikoh N, Meng XY, Kamagata Y, Fukatsu T. 2009. Host-symbiont co-speciation and reductive genome evolution in gut symbiotic bacteria of acanthosomatid stinkbugs. BMC Biol 7:2. doi: 10.1186/1741-7007-7-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Aksoy S. 1995. Wigglesworthia gen. nov. and Wigglesworthia glossinidia sp. nov., taxa consisting of the mycetocyte-associated, primary endosymbionts of tsetse flies. Int J Syst Bacteriol 45:848–851. doi: 10.1099/00207713-45-4-848. [DOI] [PubMed] [Google Scholar]
- 18.Hosokawa T, Nikoh N, Koga R, Satô M, Tanahashi M, Meng XY, Fukatsu T. 2012. Reductive genome evolution, host-symbiont co-speciation, and uterine transmission of endosymbiotic bacteria in bat flies. ISME J 6:577–587. doi: 10.1038/ismej.2011.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chrudimský T, Husník F, Nováková E, Hypša V. 2012. Candidatus sodalis melophagi sp. nov.: phylogenetically independent comparative model to the tsetse fly symbiont Sodalis glossinidius. PLoS One 7:e40354. doi: 10.1371/journal.pone.0040354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bandi C, Damiani G, Magrassi L, Grigolo A, Fani R, Sacchi L. 1994. Flavobacteria as intracellular symbionts in cockroaches. Proc Biol Sci 257:43–48. doi: 10.1098/rspb.1994.0092. [DOI] [PubMed] [Google Scholar]
- 21.Clark JW, Kambhampati S. 2003. Phylogenetic analysis of Blattabacterium, endosymbiotic bacteria from the wood roach, Cryptocercus (Blattodea: Cryptocercidae), including a description of three new species. Mol Phylogenet Evol 26:82–88. doi: 10.1016/S1055-7903(02)00330-5. [DOI] [PubMed] [Google Scholar]
- 22.Sasaki-Fukatsu K, Koga R, Nikoh N, Yoshizawa K, Kasai S, Mihara M, Kobayashi M, Tomita T, Fukatsu T. 2006. Symbiotic bacteria associated with stomach discs of human lice. Appl Environ Microbiol 72:7349–7352. doi: 10.1128/AEM.01429-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fukatsu T, Hosokawa T, Koga R, Nikoh N, Kato T, Hayama S, Takefushi H, Tanaka I. 2009. Intestinal endocellular symbiotic bacterium of the macaque louse Pedicinus obtusus: distinct endosymbiont origins in anthropoid primate lice and the Old World monkey louse. Appl Environ Microbiol 75:3796–3799. doi: 10.1128/AEM.00226-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lefèvre C, Charles H, Vallier A, Delobel B, Farrell B, Heddi A. 2004. Endosymbiont phylogenesis in the Dryophthoridae weevils: evidence for bacterial replacement. Mol Biol Evol 21:965–973. doi: 10.1093/molbev/msh063. [DOI] [PubMed] [Google Scholar]
- 25.Conord C, Despres L, Vallier A, Balmand S, Miquel C, Zundel S, Lemperiere G, Heddi A. 2008. Long-term evolutionary stability of bacterial endosymbiosis in Curculionoidea: additional evidence of symbiont replacement in the Dryophthoridae family. Mol Biol Evol 25:859–868. doi: 10.1093/molbev/msn027. [DOI] [PubMed] [Google Scholar]
- 26.Heddi A, Grenier AM, Khatchadourian C, Charles H, Nardon P. 1999. Four intracellular genomes direct weevil biology: nuclear, mitochondrial, principal endosymbiont, and Wolbachia. Proc Natl Acad Sci U S A 96:6814–6819. doi: 10.1073/pnas.96.12.6814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Oakeson KF, Gil R, Clayton AL, Dunn DM, von Niederhausern AC, Hamil C, Aoyagi A, Duval B, Baca A, Silva FJ, Vallier A, Jackson DG, Latorre A, Weiss RB, Heddi A, Moya A, Dale C. 2014. Genome degeneration and adaptation in a nascent stage of symbiosis. Genome Biol Evol 6:76–93. doi: 10.1093/gbe/evt210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Toju H, Hosokawa T, Koga R, Nikoh N, Meng XY, Kimura N, Fukatsu T. 2010. ‘Candidatus Curculioniphilus buchneri’, a novel clade of bacterial endocellular symbionts from weevils of the genus Curculio. Appl Environ Microbiol 76:275–282. doi: 10.1128/AEM.02154-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Toju H, Fukatsu T. 2011. Diversity and infection prevalence of endosymbionts in natural populations of the chestnut weevil: relevance of local climate and host plants. Mol Ecol 20:853–868. doi: 10.1111/j.1365-294X.2010.04980.x. [DOI] [PubMed] [Google Scholar]
- 30.Toju H, Tanabe AS, Notsu Y, Sota T, Fukatsu T. 2013. Diversification of endosymbiosis: replacements, co-speciation and promiscuity of bacteriocyte symbionts in weevils. ISME J 7:1378–1390. doi: 10.1038/ismej.2013.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kölsch G, Matz-Grund C, Pedersen BV. 2009. Ultrastructural and molecular characterization of endosymbionts of the reed beetle genus Macroplea (Chrysomelidae, Donaciinae), and proposal of “Candidatus Macropleicola appendiculatae” and “Candidatus Macropleicola muticae.” Can J Microbiol 55:1250–1260. doi: 10.1139/w09-085. [DOI] [PubMed] [Google Scholar]
- 32.Kölsch G, Pedersen BV. 2010. Can the tight co-speciation between reed beetles (Col., Chrysomelidae, Donaciinae) and their bacterial endosymbionts, which provide cocoon material, clarify the deeper phylogeny of the hosts? Mol Phylogenet Evol 54:810–821. doi: 10.1016/j.ympev.2009.10.038. [DOI] [PubMed] [Google Scholar]
- 33.Kleinschmidt B, Kölsch G. 2011. Adopting bacteria in order to adapt to water—how reed beetles colonized the wetlands (Coleoptera, Chrysomelidae, Donaciinae). Insects 2:540–554. doi: 10.3390/insects2040540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kölsch G, Synefiaridou D. 2012. Shared ancestry of symbionts? Sagrinae and Donaciinae (Coleoptera, Chrysomelidae) harbor similar bacteria. Insects 3:473–491. doi: 10.3390/insects3020473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fukumori K, Koga R, Nikoh N, Fukatsu T. 2017. Symbiotic bacteria associated with gut symbiotic organs and female genital accessory organs of the leaf beetle Bromius obscurus (Coleoptera: Chrysomelidae). Appl Entomol Zool doi: 10.1007/s13355-017-0513-0. [DOI] [Google Scholar]
- 36.Grimaldi D, Engel MS. 2005, Evolution of the insects. Cambridge University Press, New York, NY. [Google Scholar]
- 37.Hagstrum DW, Subramanyam B. 2009. A review of stored-product entomology information sources. Am Entomol 55:174–183. doi: 10.1093/ae/55.3.174. [DOI] [Google Scholar]
- 38.Hagstrum D. 2017. Atlas of stored-product insects and mites. AACC International, St.-Paul, MN. [Google Scholar]
- 39.Breitsprecher E. 1928. Beiträge zur Kenntnis der Anobiidensymbiose. Z Morphol Oekol Tiere 11:495–538. doi: 10.1007/BF02433460. [DOI] [Google Scholar]
- 40.Kühlwein H, Jurzitza G. 1961. Studien an der Symbiose der Anobiiden Archiv Mikrobiol 40:247–260. doi: 10.1007/BF00540577. [DOI] [PubMed] [Google Scholar]
- 41.Noda H, Kodama K. 1996. Phylogenetic position of yeastlike endosymbionts of Anobiid beetles. Appl Environ Microbiol 62:162–167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Okude G, Koga R, Hayashi T, Nishide Y, Meng XY, Nikoh N, Miyanoshita A, Fukatsu T. 2017. Novel bacteriocyte-associated pleomorphic symbiont of the grain pest beetle Rhyzopertha dominica (Coleoptera: Bostrychidae). Zool Lett 3:13. doi: 10.1186/s40851-017-0073-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Back EA, Cotton RT. 1926. Biology of the saw-toothed grain beetle, Oryzaephilus surinamensis Linne. J Agric Res 33:435–452. [Google Scholar]
- 44.Howe RW. 1956. The biology of the two common storage species of Oryzaephilus (Coleoptera, Cucujidae). Ann Appl Biol 44:341–355. doi: 10.1111/j.1744-7348.1956.tb02127.x. [DOI] [Google Scholar]
- 45.Koch A. 1931. Die Symbiose von Oryzaephilus surinamensis L. (Cucujidae, Coleoptera). Z Morphol Oekol Tiere 23:389–424. doi: 10.1007/BF00446355. [DOI] [Google Scholar]
- 46.Koch A. 1936. Symbiosestudien. II. Experimentelle Untersuchungen an Oryzaephilus surinamensis L. [Cucujidae, Coleopt]. Z Morphol Oekol Tiere 32:137–180. [Google Scholar]
- 47.Huger A. 1956. Experimentelle Untersuchungen über die künstliche Symbiontenelimination bei Vorratsschädlingen: Rhizopertha dominica F. (Bostrychidae) und Oryzaephilus surinamensis L. (Cucujidae). Z Morphol Oekol Tiere 44:626–701. doi: 10.1007/BF00390698. [DOI] [Google Scholar]
- 48.Hunt T, Bergsten J, Levkanicova Z, Papadopoulou A, John OS, Wild R, Hammond PM, Ahrens D, Balke M, Caterino MS, Gómez-Zurita J, Ribera I, Barraclough TG, Bocakova M, Bocak L, Vogler AP. 2007. A comprehensive phylogeny of beetles reveals the evolutionary origins of a superradiation. Science 318:1913–1916. doi: 10.1126/science.1146954. [DOI] [PubMed] [Google Scholar]
- 49.McCutcheon JP, Moran NA. 2007. Parallel genomic evolution and metabolic interdependence in an ancient symbiosis. Proc Natl Acad Sci U S A 104:19392–19397. doi: 10.1073/pnas.0708855104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bennett GM, Moran NA. 2013. Small, smaller, smallest: the origins and evolution of ancient dual symbioses in a phloem-feeding insect. Genome Biol Evol 5:1675–1688. doi: 10.1093/gbe/evt118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Koga R, Moran NA. 2014. Swapping symbionts in spittlebugs: evolutionary replacement of a reduced genome symbiont. ISME J 8:1237–1246. doi: 10.1038/ismej.2013.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sabree ZL, Kambhampati S, Moran NA. 2009. Nitrogen recycling and nutritional provisioning by Blattabacterium, the cockroach endosymbiont. Proc Natl Acad Sci U S A 106:19521–19526. doi: 10.1073/pnas.0907504106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.López-Sánchez MJ, Neef A, Peretó J, Patiño-Navarrete R, Pignatelli M, Latorre A, Moya A. 2009. Evolutionary convergence and nitrogen metabolism in Blattabacterium strain Bge, primary endosymbiont of the cockroach Blattella germanica. PLoS Genet 5:e1000721. doi: 10.1371/journal.pgen.1000721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sabree ZL, Huang CY, Arakawa G, Tokuda G, Lo N, Watanabe H, Moran NA. 2012. Genome shrinkage and loss of nutrient-providing potential in the obligate symbiont of the primitive termite Mastotermes darwiniensis. Appl Environ Microbiol 78:204–210. doi: 10.1128/AEM.06540-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sabree ZL, Huang CY, Okusu A, Moran NA, Normark BB. 2013. The nutrient supplying capabilities of Uzinura, an endosymbiont of armoured scale insects. Environ Microbiol 15:1988–1999. doi: 10.1111/1462-2920.12058. [DOI] [PubMed] [Google Scholar]
- 56.Rosas-Pérez T, Rosenblueth M, Rincón-Rosales R, Mora J, Martínez-Romero E. 2014. Genome sequence of “Candidatus Walczuchella monophlebidarum” the flavobacterial endosymbiont of Llaveia axin axin (Hemiptera: Coccoidea: Monophlebidae). Genome Biol Evol 6:714–726. doi: 10.1093/gbe/evu049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Errington J. 2013. L-form bacteria, cell walls and the origins of life. Open Biol 3:120143. doi: 10.1098/rsob.120143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Crowson RA. 1981, The biology of the Coleoptera. Academic Press, London, United Kingdom. [Google Scholar]
- 59.Weissling TJ, Giblin-Davis RM. 1993. Water loss dynamics and humidity preference of Rhynchophorus cruentatus (Coleoptera: Curculionidae) adults. Environ Entomol 22:93–98. doi: 10.1093/ee/22.1.93. [DOI] [Google Scholar]
- 60.Tseng HY, Lin CP, Hsu JY, Pike DA, Huang WS. 2014. The functional significance of aposematic signals: geographic variation in the responses of widespread lizard predators to colourful invertebrate prey. PLoS One 9:e91777. doi: 10.1371/journal.pone.0091777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.van de Kamp T, Riedel A, Greven H. 2016. Micromorphology of the elytral cuticle of beetles, with an emphasis on weevils (Coleoptera: Curculionoidea). Arthropod Struct Dev 45:14–22. doi: 10.1016/j.asd.2015.10.002. [DOI] [PubMed] [Google Scholar]
- 62.Ishikawa H, Yamaji M. 1985. Symbionin, an aphid endosymbiont-specific protein. I. Production of insects deficient in sybiont. Insect Biochem 15:155–163. doi: 10.1016/0020-1790(85)90003-4. [DOI] [Google Scholar]
- 63.Ohtaka C, Ishikawa H. 1991. Effects of heat treatment on the symbiotic system of an aphid mycetocyte. Symbiosis 11:19–30. [Google Scholar]
- 64.Hosokawa T, Hironaka M, Inadomi K, Mukai H, Nikoh N, Fukatsu T. 2013. Diverse strategies for vertical symbiont transmission among subsocial stinkbugs. PLoS One 8:e65081. doi: 10.1371/journal.pone.0065081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hosokawa T, Ishii Y, Nikoh N, Fujie M, Satoh N, Fukatsu T. 2016. Obligate bacterial mutualists evolving from environmental bacteria in natural insect populations. Nat Microbiol 1:15011. doi: 10.1038/nmicrobiol.2015.11. [DOI] [PubMed] [Google Scholar]
- 66.Kikuchi Y, Tada A, Musolin DL, Hari N, Hosokawa T, Fujisaki K, Fukatsu T. 2016. Collapse of insect-microbe symbiosis under simulated climate change. mBio 7:e01578-16. doi: 10.1128/mBio.01578-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Nardon P. 1973. Obtention d’une souche asymbiotique chez le charançon Sitophilus sasakii: différentes méthodes et comparaison avec la souche symbiotique d’origine. C R Acad Sci Paris D 3:65–67. [Google Scholar]
- 68.Wicker C, Nardon P. 1982. Development responses of symbiotic and aposymbiotic weevils Sitophilus oryzae L. (Coleoptera, Curculionidae) to a diet supplemented with aromatic amino acids. J Insect Physiol 28:1021–1024. doi: 10.1016/0022-1910(82)90008-7. [DOI] [Google Scholar]
- 69.Kuriwada T, Hosokawa T, Kumano N, Shiromoto K, Haraguchi D, Fukatsu T. 2010. Biological role of Nardonella endosymbiont in its weevil host. PLoS One 5:e13101. doi: 10.1371/journal.pone.0013101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Vigneron A, Masson F, Vallier A, Balmand S, Rey M, Vincent-Monégat C, Aksoy E, Aubailly-Giraud E, Zaidman-Rémy A, Heddi A. 2014. Insects recycle endosymbionts when the benefit is over. Curr Biol 24:2267–2273. doi: 10.1016/j.cub.2014.07.065. [DOI] [PubMed] [Google Scholar]
- 71.Anbutsu H, Moriyama M, Nikoh N, Hosokawa T, Futahashi R, Tanahashi M, Meng XY, Kuriwada T, Mori N, Oshima K, Hattori M, Fujie M, Satoh N, Maeda T, Shigenobu S, Koga R, Fukatsu T. Symbiosis for beetle’s hardness: symbiont’s tiny genome for supplying a single amino acid, and host’s control of the synthesis pathway. Proc Natl Acad Sci U S A, in press. [Google Scholar]
- 72.Douglas AE. 1989. Mycetocyte symbiosis in insects. Biol Rev 64:409–434. doi: 10.1111/j.1469-185X.1989.tb00682.x. [DOI] [PubMed] [Google Scholar]
- 73.Douglas AE. 1998. Nutritional interactions in insect-microbial symbioses: aphids and their symbiotic bacteria Buchnera. Annu Rev Entomol 43:17–37. doi: 10.1146/annurev.ento.43.1.17. [DOI] [PubMed] [Google Scholar]
- 74.Rabatel A, Febvay G, Gaget K, Duport G, Baa-Puyoulet P, Sapountzis P, Bendridi N, Rey M, Rahbé Y, Charles H, Calevro F, Colella S. 2013. Tyrosine pathway regulation is host-mediated in the pea aphid symbiosis during late embryonic and early larval development. BMC Genomics 14:235. doi: 10.1186/1471-2164-14-235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Simonet P, Gaget K, Parisot N, Duport G, Rey M, Febvay G, Charles H, Callaerts P, Colella S, Calevro F. 2016. Disruption of phenylalanine hydroxylase reduces adult lifespan and fecundity, and impairs embryonic development in parthenogenetic pea aphids. Sci Rep 6:34321. doi: 10.1038/srep34321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Tribolium Genome Sequencing Consortium, Richards S, Gibbs RA, Weinstock GM, Brown SJ, Denell R, Beeman RW, Gibbs R, Beeman RW, Brown SJ, Bucher G, Friedrich M, Grimmelikhuijzen CJ, Klingler M, Lorenzen M, Richards S, Roth S, Schröder R, Tautz D, Zdobnov EM, Muzny D, Gibbs RA, Weinstock GM, Attaway T, Bell S, Buhay CJ, Chandrabose MN, Chavez D, Clerk-Blankenburg KP, Cree A, Dao M, Davis C, Chacko J, Dinh H, Dugan-Rocha S, Fowler G, Garner TT, Garnes J, Gnirke A, Hawes A, Hernandez J, Hines S, Holder M, Hume J, Jhangiani SN, Joshi V, Khan ZM, Jackson L, Kovar C, Kowis A, Lee S, et al. . 2008. The genome of the model beetle and pest Tribolium castaneum. Nature 452:949–955. doi: 10.1038/nature06784. [DOI] [PubMed] [Google Scholar]
- 77.Brown SJ, Shippy TD, Miller S, Bolognesi R, Beeman RW, Lorenzen MD, Bucher G, Wimmer EA, Klingler M. 2009. The red flour beetle, Tribolium castaneum (Coleoptera): a model for studies of development and pest biology. Cold Spring Harb Protoc 2009:pdb.emo126. doi: 10.1101/pdb.emo126. [DOI] [PubMed] [Google Scholar]
- 78.Costantino R, Desharnais R. 2012. Population dynamics and the Tribolium model: genetics and demography. Springer-Verlag, New York, NY. [Google Scholar]
- 79.Fraenkel G, Blewett M. 1943. The basic food requirements of several insects. J Exp Biol 20:28–34. [Google Scholar]
- 80.Fraenkel G, Blewett M. 1943. The vitamin B-complex requirements of several insects. Biochem J 37:686–692. doi: 10.1042/bj0370686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Fukatsu T. 1999. Acetone preservation: a practical technique for molecular analysis. Mol Ecol 8:1935–1945. doi: 10.1046/j.1365-294x.1999.00795.x. [DOI] [PubMed] [Google Scholar]
- 82.Fukatsu T, Nikoh N. 1998. Two intracellular symbiotic bacteria of the mulberry psyllid Anomoneura mori (Insecta, Homoptera). Appl Environ Microbiol 64:3599–3606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Thompson JD, Higgins DG, Gibson TJ. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res 22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Kumar S, Stecher G, Tamura K. 2016. MEGA7: Molecular Evolutionary Genetics Analysis version 7.0 for bigger datasets. Mol Biol Evol 33:1870–1874. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Ronquist F, Teslenko M, van der Mark P, Ayres DL, Darling A, Höhna S, Larget B, Liu L, Suchard MA, Huelsenbeck JP. 2012. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol 61:539–542. doi: 10.1093/sysbio/sys029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Tanabe AS. 2011. Kakusan4 and Aminosan: two programs for comparing nonpartitioned, proportional, and separate models for combined molecular phylogenetic analyses of multilocus sequence data. Mol Ecol Resour 11:914–921. doi: 10.1111/j.1755-0998.2011.03021.x. [DOI] [PubMed] [Google Scholar]
- 87.Robinson-Rechavi M, Huchon D. 2000. RRTree: relative-rate tests between groups of sequences on a phylogenetic tree. Bioinformatics 16:296–297. doi: 10.1093/bioinformatics/16.3.296. [DOI] [PubMed] [Google Scholar]
- 88.Koga R, Tsuchida T, Fukatsu T. 2009. Quenching autofluorescence of insect tissues for in situ detection of endosymbionts. Appl Entomol Zool 44:281–291. doi: 10.1303/aez.2009.281. [DOI] [Google Scholar]
- 89.Amann RI, Krumholz L, Stahl DA. 1990. Fluorescent-oligonucleotide probing of whole cells for determinative, phylogenetic, and environmental studies in microbiology. J Bacteriol 172:762–770. doi: 10.1128/jb.172.2.762-770.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Koga R, Bennett GM, Cryan JR, Moran NA. 2013. Evolutionary replacement of obligate symbionts in an ancient and diverse insect lineage. Environ Microbiol 15:2073–2081. doi: 10.1111/1462-2920.12121. [DOI] [PubMed] [Google Scholar]