Abstract
Gibberellins (GAs), a class of diterpenoid phytohormones, play a key role in regulating diverse processes throughout the life cycle of plants. Bioactive GA levels are rapidly regulated by Gibberellin-dioxygenases (GAox), which are involved in the biosynthesis and deactivation of gibberellin. In this manuscript, a comprehensive genome-wide analysis was carried out to find all GAox in Camellia sinensis. For the first time in a tea plant, 14 CsGAox genes, containing two domains, DIOX_N (PF14226) and 2OG-FeII_Oxy, were identified (PF03171). These genes all belong to 2-oxoglutarate-dependent dioxygenases (2-ODD), including four CsGA20ox (EC: 1.14.11.12), three CsGA3ox (EC: 1.14.11.15), and seven CsGA2ox (EC: 1.14.11.13). According to the phylogenetic classification as in Arabidopsis, the CsGAox genes spanned five subgroups. Each CsGAox shows tissue-specific expression patterns, although these vary greatly. Some candidate genes, which may play an important role in response to external abiotic stresses, have been identified with regards to patterns, such as CsGA20ox2, CsGA3ox2, CsGA3ox3, CsGA2ox1, CsGA2ox2, and CsGA2ox4. The bioactive GA levels may be closely related to the GA20ox, GA3ox and GA2ox genes. In addition, the candidate genes could be used as marker genes for abiotic stress resistance breeding in tea plants.
Keywords: genome-wide analysis, gibberellin, Gibberellins-dioxygenases, abiotic stress, Camellia sinensis
1. Introduction
Gibberellins (GAs), a class of diterpenoid phytohormones, play a key role in regulating diverse processes throughout the life cycle of plants, such as stem elongation, anther development, and flower induction [1,2,3,4]. Although up to 136 different gibberellin molecules have hitherto been discovered, most of these molecules have been identified as biosynthetic intermediates or catabolites of bioactive GAs, with GA1, GA3, GA4, and GA7 being the only bioactive GAs [1,5]. Recent studies on GA biosynthesis and deactivation, as well as crosstalk of GA with other plant hormones and environmental cues have achieved great progress, along with advancements in molecular genetics and functional genomics [1,6].
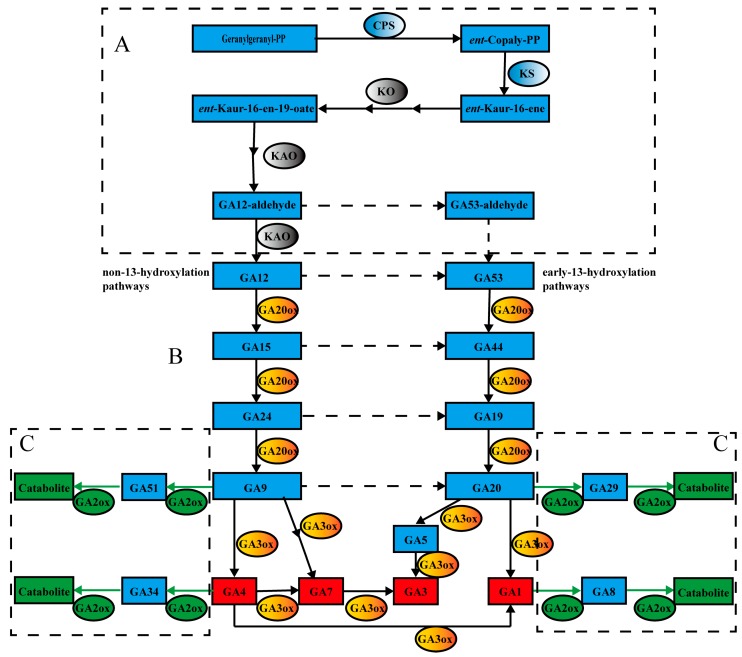
In higher plants, the biosynthesis and deactivation of GA mainly involves three stages of reactions (Figure 1) [1,3]. In the first stage, the synthesis of GAs begins from geranylgeranyl diphosphate (Geranylgeranyl-PP), which creates the metabolite ent-kaur-16-ene through two synthetases, ent-copalyl diphosphate synthase (CPS) and ent-kaurene synthase (KS). Following this, GA12 and GA53 are synthesized from ent-kaur-16-ene by cytochrome P450-dependent monoxygenases—these are ent-kaurene oxidase (KO) and ent-kaurenoic acid oxidase (KAO) respectively—through three consecutive steps (Figure 1A) [1,7]. In the final stage of biosynthesis, the synthesis of various GAs occurs via two pathways: the early-13-hydroxylation and the non-13-hydroxylation pathways (Figure 1) [6,8]. The 2-oxoglutarate dependent dioxygenases (2-ODDs), including GA 20-oxidases (GA20ox) and GA 3-oxidases (GA3ox), are the key enzymes in a series of oxidation steps, which convert GA12 and GA53 to various GA intermediates and bioactive GAs (GA1 and GA4) (Figure 1B). In some species, bioactives (GA3 and GA7) probably derive from GA9 and GA20 under the oxidation catalyzed by GA3ox enzymes, respectively, via 2,3-didehydro-GA9 and GA5 [1]. In the pathways and regulation of GA degradation, GA 2-oxidases (GA2ox) are the unique enzymes, which also belong to 2-ODDs. These enzymes inactivate the bioactives (GA1 and GA4) and their immediate precursors (GA20 and GA9) (Figure 1C) [1].
Figure 1.
The principal pathways of gibberellin (GA) biosynthesis and deactivation GA-dioxygenases in higher plants. In this figure, Ggeranylgeranyl-PP = geranylgeranyl diphosphate; ent-Copaly-PP = ent-copalyl diphosphate; CPS = ent-copalyl diphosphate synthase; KS = ent-kaurene synthase; KAO = ent-kaurenoic acid oxidase; KO = ent-kaurene oxidase; GA20ox = GA 20-oxidase; GA3ox = GA 3-oxidase; GA2ox = GA 2-oxidase.
Interestingly, the enzymes involved in the first step in the degradation of bioactive GAs and the final step of GA biosynthesis were found to be similar (Figure 1B,C). Furthermore, previous analyses found that all the GA20ox, GA3ox and GA2ox sequences belonged to the 2-ODDs superfamily, which share high homology with the functional domains [9,10,11,12]. Most of the GA20ox genes of the biosynthesis and catabolism of gibberellin have been now cloned and identified [2,8]. GA20ox, identified as a multi-catalytic enzyme, may play a regulatory role in the biosynthetic flux of the pathway [12]. The putative sequence of GA20ox contains the proposed consensus sequence NXYPXCXXP and three histidine residues, which are involved in binding the common co-substrate 2-oxoglutarate and Fe2+, respectively [13]. The sequence LPWKET, which may be related to the binding of the GA substrate, is also conserved in the GA20oxes of pumpkin [14] and Arabidopsis [13]. In Arabidopsis, the first three GA2ox genes cloned were annotated as AtGA2ox1, AtGA2ox2 and AtGA2ox3 [15]. After this, the genes identified in Hedden and Phillips [6] were described as AtGA2ox4 and AtGA2ox6. Furthermore, AtGA2ox7 and AtGA2ox8, distinct from AtGA2ox1–AtGA2ox6 [6], were noted by Schomburg et al. [11]. In summary, GA20ox, GA3ox, and GA2ox each encoded by a small gene family, with all CsGAox gene expression patterns showing tissue-specific and different responses to abiotic stresses [6]. Expression patterns of GA20ox and GA3ox could be involved in the negative feedback regulation of bioactive GAs [1,2], while GA2ox may be regulated by positive feedback regulation [1,16]. Many environmental responses are regulated through GA abundance, while GA metabolism is regulated by environmental signals as well as by abiotic and biotic stresses [1]. GA metabolism and signaling seem to be closely regulated through feedback mechanisms to maintain GA homeostasis [1,2].
Endogenous GA and abscisic acid (ABA) strongly influence the growth of tea plants [17,18,19]. However, many studies focus on physiology, with few having examined gene expression. Results of the Camellia sinensis var. assamica tea tree genome sequence, which provided the opportunity to perform a genome-wide scan of gene families, were published in 2017, while the genome database was opened publicly at the same time [20]. In this study, a genome-wide survey of the Gibberellin-dioxygenase (GAox) gene family was conducted using the Tea Tree (C. sinensis var. assamica) Genome Database [20]. Meanwhile, sequences were supplied from the tea plant (C. sinensis) transcriptome databases [21]. Predicted sequences of all tea GAox genes were identified with the tool SMART, Pfam 31.0, and the NCBI database. Markedly, 14 full-length GAox genes were confirmed for the first time in tea plants (C. sinensis), using cloning and sequencing. Some characteristics and putative protein sequences were predicted and identified. Moreover, to gain insights into the evolutionary diversity of GAox genes, a comparative phylogenetic analysis of the tea plant with the Arabidopsis and the other ten species plant (Citrus sinensis, Cucumls sativus, Glycine max, Medicago truncatula, Physcomitrella patens, Populus trichocarpa, Selaginella moellendorffii, Sorghum bicolor, Vitis vinifera and Zea mays) GAox gene family was performed. Tissue-specific expression and expression patterns of CsGAox genes in response to abiotic stresses, including high- or low-temperature stress, exogenous GA or ABA, polyethylene glycol (PEG), and high salinity treatments were analyzed. In tea plants, CsGAoxes has different patterns of response in tissues and abiotic stress. For future research, this study will serve as a foundation into the functional roles of CsGAox genes.
2. Materials and Methods
2.1. Plant Materials and Abiotic Treatments
The plant material we picked was the tea cultivar ‘Wuniuzao’ (C. sinensis cv. Wuniuzao). For tissue and abiotic stress treatments, two-year-old cutting seedlings were planted in a pot and grown with a natural photoperiod under greenhouse conditions at the State Key Laboratory of Tea Plant Biology and Utilization, Tea Science Research Institute, College of Tea and Food technology, Anhui Agricultural University (Hefei, China). We also provided thorough pest and fertilizer management [22].
For the GA or ABA treatment, a freshly prepared working solution of 100 μM GA or ABA was sprayed on the leaves. For high- or low-temperature treatment, tea plants grown in plant growth chambers (temperature 25 °C ± 2 °C) were transferred to chambers maintained at 38 °C or 4 °C for 24 h [23]. The second and/or third mature leaves were obtained at 0, 4, 12 and 24 h under the above treatment for gene expression analysis. To initiate drought stress, whole plants were removed from the pots, washed with tap water to remove the soil from the roots, and rinsed with purified water for 15 min. The plants were transferred to a 10% (w/v) PEG-6000 solution for 24 h [24,25]. Finally, stress was alleviated by washing the plants with tap water to remove the PEG-6000, with the plants recovered without PEG for another 48 h. The second and/or third mature leaves were obtained at 0, 4, 8, 12, 24 h as well as and recovery at 48 h under drought stress treatment for gene expression analysis. To initiate salt stress, two-year-old plants were irrigated with 100 mM NaCl for 24 h [24,25]. The second and/or third mature leaves were obtained at 0, 4, 8, 12, 24, 48, 72 and 120 h under salt stress treatment for gene expression analysis. Controls (non stress-treated plants) were set up at each time point, and for all the above assays, plants of approximately equal size were selected for all treatments. The three independent biological replicates of each varieties were harvested. The samples were harvested, quickly frozen in liquid nitrogen, and stored at −80 °C for RNA extraction.
2.2. RNA Isolation and cDNA Synthesis
The sample was ground into powder with liquid nitrogen, before the total RNA was isolated in accordance with the method of a commercial RNAprep pure plant kit (QIAGEN Co., Dusseldorf, Germany). A Nanodrop 2000 spectrophotometer (Thermo Scientific, Wilmington, DE, USA) was used to measure the concentration of isolated RNA, and the quality were assessed using 1.2% formaldehyde–agarose gel electrophoresis. For quantitative reverse transcriptase polymerase chain reaction (qRT-PCR), the first-strand copy DNA (cDNA) of the sample was synthesized with the PrimeScript™ II 1st Strand cDNA Synthesis Kit (TaKaRa, Tokyo, Japan). The cDNA was diluted 10-fold for PCR amplification.
2.3. Data Mining for GAox Protein Genes
A multiple database search was performed to find all of the members of the GAox family in Sorghum bicolor, Physcomitrella patens, Populus trichocarpa, Cucumls sativus, Medicago truncatula, G.max, Citrus sinensis, Vitis vinifera, Zea mays, Selaginella moellendorffii and Arabidopsis [26]. The strategy to obtain every gene of the GAox family in an Arabidopsis genome involved the following steps.
The A. thaliana GAox proteins were retrieved from the The Arabidopsis Information Resource (TAIR) [27] database. For GA-dioxygenase protein genes in Arabidopsis (AtGAox), the key word “gibberellin” was first used as a query to search in the TAIR database [28,29]. Following this, the sequences obtained were used as queries to search in the TAIR10 Protein databases using BioEdit software [30], with the BLASTP program at the e-value of 10−3 to search all similar sequences [31]. All sequences obtained were then used as queries to search against the TAIR Protein databases using TAIR BLAST 2.2.8 [27] to avoid false positives [32]. After this, we verified these sequences using the tool SMART [33], the Pfam 31.0 database [34], and the NCBI database [35].
For GA-dioxygenase protein genes in tea plant (CsGAox), all predicted GAox protein sequences of multiple databases were used as query sequences to search against the Tea Tree Genome Database (Available online: http://www.plantkingdomgdb.com/tea_tree/) [20] using the BLASTP program. The e-value used in the BLASTP was 10−3. The tool SMART, Pfam 31.0, and NCBI database were finally used to confirm each predicted CsGAox protein sequence as the GAox protein family.
2.4. Cloning the Full-Length of cDNA of GAox Protein Genes
All predicted CsGAox protein sequences were used as query sequences to search in transcriptome databases (previously generated from tea plant cultivar ‘Shuchazao’) [21] using TBLASTN program. All the target unigenes were identified by BLASTx (NCBI), and sequences with more than 60% matching identity were used for assembly using SeqMan software (DNAStar, Inc. Madison, WI, USA) in the DNAStar package. Finally, all predicted protein of assembled sequences were compared with known GAox sequences by applying ClustalX [36] to verify that the sequences were candidates. PCR primer (Supplementary Table S1) pairs were designed, using the software Primer Premier 5 (Premier Biosoft International, Palo Alto, CA, USA), to clone the full-length cDNA sequences of the CsGAox genes by RT-PCR. The PCR product was purified with an Axygen Gel Extraction Kit (Axygen, Union City, CA, USA), and was ligated into a pMD-19T vector (TaKaRa, Tokyo, Japan), from which single clones were sequenced by Sangon Biotech Co. (Shanghai, China). The sequence accuracy was confirmed by sanger dideoxy sequencing.
2.5. Analysis of Sequences
Information about the length of coding sequences and amino acid sequenced was determined by DNAMAN and DNAstar software. The molecular weights and theoretical pIs were predicted using the ProtParam [37] tool. WoLF PSORT [38] and TargetP [39] were combined to predict the subcellular localization of the proteins. Predicted amino acid sequence alignment and phylogenetic tree of the CsGAoxes with the GAoxes from other plant species were aligned using the ClustalW method with default parameters, in Bioedit and MEGA6.0 [30,36,40]. To determine the subgroup classification of CsGAox genes, the phylogenetic tree and conserved motifs were assessed for the CsGAox-encoded proteins. GAox proteins from Arabidopsis and ten plant species (Citrus sinensis, Cucumls sativus, G. max, M. truncatula, P. patens, P. trichocarpa, S. moellendorffii, S. bicolor, V. vinifera, and Z. mays) GAox proteins were used to classify the tea plant proteins into different groups using the neighbor-joining method with 1500 bootstrap replicates in MEGA6.0. The full-length amino acid sequences of CsGAox genes were entered into the MEME analysis tool to find their conserved motifs with the maximum number of motifs to identify set to 15 [41]. Each de novo detected motif was further subjected to a search in the Interpro database [42] to find any resemblance with known domains. Consensus sequences were also separately scanned in the Interpro database to find the domains present in pre-identified GAoxes.
2.6. Gene Expression Analysis by qRT-PCR
Specific primers (Supplementary Table S2) for qRT-PCR were designed using the software Primer Premier 5, and synthesized by Sangon Biotech Co. (Shanghai, China). The primer sequences with the cDNA template were checked by PCR. To ensure the efficiency of optimal polymerization, the amplification length for each gene was restricted to 150–300 bp. The reaction program of qRT-PCR was performed under the following conditions: 95 °C for 30 s, followed by 40 cycles at 95 °C for 5 s, and 60 °C for 30 s. The reaction volume was 25 μL, which contained 4 μL of diluted cDNA strand, 6.5 μL of deionized water, 12.5 μL of SYBR®Premix Ex Taq™II (Tli RNaseH Plus; TaKaRa), 1 μL of forward primer, and 1 μL of reverse primer. The mean values and standard deviation were calculated based on three independent biological replicates. CsActin [43,44] was used as a reference gene to normalize the expression of related genes involved in GAs biosynthesis and recycling of tea plants.
The relative gene expression levels were calculated using the comparative 2−ΔΔCt method [45]. Regarding the heatmaps, the 2−ΔΔCt values of the transcripts of the CsGAox genes from various tea plant tissues samples were normalized to bud tissues and expressed as a log2-fold change. In the stress-treated plant samples, the values were normalized to plant samples of the 0 h treatment and expressed as a log2-fold change. Concerning the histogram of qRT-PCR analysis, the relative gene expression levels were calculated and expressed as the fold change relative to expression of the 0 h treatment (expression = 1). Differences in gene expression levels were detected by Tukey’s multiple-range test at a 0.05 probability level. The qPCR experiments were conducted with three independent total RNA samples.
3. Results
3.1. Isolation, Identification and Annotation Information of the GAox Family Genes in Tea Plants
Based on genome and transcriptome databases, more than 60 candidate GAox genes were obtained in the tea plant. Several sequences, which shared approximately 99% open reading frame (ORF) identity with other candidate CsGAox genes (data not shown), were excluded, and 14 genes were further analyzed. Afterwards, we named genes based on the nomenclature and the name of Arabidopsis. The gene names, accession numbers, Gene ID, number of deduced amino acid, molecular weights, predicted subcellular localizations and groups classifications are summarized in Table 1. CsGA20ox1, CsGA3ox1 and CsGA3ox2 isoforms have been submitted to NCBI (Direct submission). The table shows that the identified GAox family genes encode proteins ranging from 332 (GA2ox2) to 383 (GA20ox1) amino acids (aa) in length, with an average of 353 aa. There was a predicted molecular mass range of 37.18–43.03 kDa, and a pI ranging from 5.30 (CsGA20ox4) to 8.22 (CsGA3ox2). Most CsGAox family genes were predicted to be located in the nucleus and cytoplasm, which is related to their functional location [1,2,3,6,46].
Table 1.
The 14 CsGAox family genes in the tea plant.
| Gene names | Accession Numbers | Gene ID a | Number of Deduced Amino Acid | Molecular Weight (kDa) | Subcellular Location (WoLF PSORT/TargetP) | Groups | Theoretical pI |
|---|---|---|---|---|---|---|---|
| CsGA20ox1 | KC193604 | CSA007392 | 383 | 42.94 | nucl | B | 6.06 |
| CsGA20ox2 | KY296366 | NF | 376 | 43.02 | nucl/cyto | B | 6.98 |
| CsGA20ox3 | MF370231 | CSA000105 | 378 | 43.03 | nucl/cyto | B | 6.94 |
| CsGA20ox4 | MF370232 | CSA002490 | 356 | 40.31 | cyto | B | 5.30 |
| CsGA2ox1 | KY296367 | CSA032124 | 333 | 37.18 | nucl/cyto | A1 | 5.93 |
| CsGA2ox2 | KY296368 | CSA026961 | 332 | 37.31 | cyto/nucl | A1 | 7.61 |
| CsGA2ox3 | KY296369 | CSA004444 | 334 | 37.56 | nucl/cyto/chlo | A1 | 5.47 |
| CsGA2ox4 | MF370234 | NF | 344 | 38.67 | nucl | A2 | 5.73 |
| CsGA2ox6 | MF370235 | CSA014596 | 340 | 38.02 | nucl/chol/mito | A2 | 7.94 |
| CsGA2ox7 | MF370236 | CSA013052 | 346 | 39.24 | cyto/nucl | A3 | 6.79 |
| CsGA2ox8 | MF370237 | CSA034015 | 335 | 38.53 | cyto/nucl | A3 | 5.33 |
| CsGA3ox1 | KF703743 | CSA002905 | 361 | 40.29 | chlo/mito/nucl | C | 6.67 |
| CsGA3ox2 | KF703744 | CSA020111 | 373 | 41.08 | nucl/cyto | C | 8.22 |
| CsGA3ox3 | MF370233 | CSA034282 | 345 | 38.72 | cyto/nucl | C | 5.66 |
WoLF PSORT [38] and TargetP [39] were used to predict the subcellular localization of the 14 CsGAox family genes; the most likely locations are listed. nucl: nucleus; chlo: chloroplast; cyto: cytoplasmic; mito: mitochondrion. Groups were classified based on phylogenetic trees with their corresponding numbers in Arabidopsis. a Gene identifier of tea tree genome database [20] is available or not. NF: not found.
3.2. Similarity and Phylogenetic Gene Structure Analysis of the GAox Genes
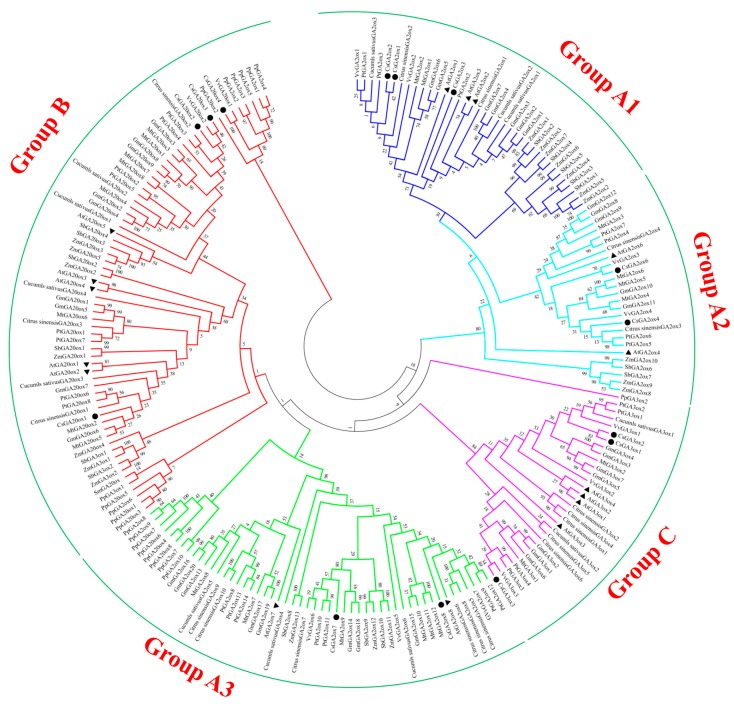
In order to investigate evolutionary relationships, a total of 213 GAox protein sequences from C. sinensis, Arabidopsis and other plant species were subjected to phylogenetic analysis. All sequences derived from dicotyledonous plants are well divided, but for monocotyledon plants, some sequences (SbGA3ox1, SbGA3ox2, ZmGA3ox1 and ZmGA3ox2) were not accurately classified. However, most sequences of P. patens were not well divided following the same distinction. The results indicated that all the CsGAox sequences belonged to the 2-ODDs superfamily. From a functional point of view, the CsGAox genes were classified into three subfamilies: the biosynthesis of gibberellin gene family containing the GA20ox and GA3ox genes encoding GA20ox protein (EC: 1.14.11.12) and GA3ox protein (EC: 1.14.11.15). Furthermore, the most studied deactivation pathway is 2β-hydroxylation, which is catalyzed by a class of GA2ox enzymes (EC: 1.14.11.13). These are soluble 2-ODDs [1,2,3,6,7,15,46,47,48,49]. Particularly, GA2ox genes were classified into three subgroups. Therefore, CsGAox genes were classified into three final subfamilies and five different subgroups (Figure 2 and Table 1).
Figure 2.
Phylogenetic analysis of putative GAox family proteins in C. sinensis with other plant GAoxes. The following species were analyzed: Arabidopsis, C. sinensis, Citrus sinensis, Cucumls sativus, G.max, M. truncatula, P. patens, P. trichocarpa, S. moellendorffii, S. bicolor, V. vinifera, and Z. mays. The predicted amino acid sequences of the 14 CsGAoxs and their corresponding sequences from Arabidopsis and from 10 other plant species were aligned using the ClustalW [36] sequence alignment program. The phylogenetic tree was constructed using MEGA6.0 [40] software with the neighbor-joining tree method with 1500 bootstrap replicates. Five subgroups are shown in various colors. The sequences of C. sinensis and Arabidopsis are highlighted with black dots and triangles, respectively.
Predicted amino acid sequence alignment and phylogenetic tree of the CsGAoxes with the GAoxes from other plant species were aligned using the ClustalW method. Results of their similarities and differences were listed in Supplementary Figure S1 (group B), Figure S2 (group C), Figure S3 (group A1), Figure S4 (group A2) and Figure S5 (group A3). Except CsGA2ox7, most of the CsGA3ox (group C) and CsGA2ox (group A) genes showed that the phylogenetic relationships of CsGAoxes were closer to edicots compared to monocots, indicating that the evolutionary rate of CsGAox gene family was faster than previously thought. With regards to GA20ox genes (group B), CsGA20ox1, CsGA20ox2, and CsGA20ox3 showed a closer relationship with edicots. CsGA20ox4 and VvGA20ox1 however, which were clustered together in the group of Physcomitrella patens, showed notable differences from the GA20ox genes of edicot and monocot. The variant phylogenetic relationships of CsGAoxes indicated that the GAox genes had different evolutionary rates during tea plant evolution. Using a multiple sequence alignments program to analyze the 14 ORFs and their encoded amino acid sequences, the results of their similarities were listed in Supplementary Table S3. The nucleotide sequence similarities varied from 38.9% (CsGA3ox2 and CsGA2ox8) to 87.0% (CsGA2ox2 and CsGA2ox1), while proteins exhibited similarities from 22.3% (CsGA2ox3 and CsGA2ox7) to 86.1% (CsGA2ox1 and CsGA2ox2).
The alignment in Figure 3 shows that these sequences contained the two domains DIOX_N (PF14226) and 2OG-FeII_Oxy (PF03171). GA20ox, GA3ox and GA2ox are members of the 2-ODDs. The proposed consensus sequence NXYPXCXXP of 2-ODDs, which may be involved in the binding of a common cosubstrate (2-oxoglutaric acid) [13,50], was highly conserved in all CsGA20ox and CsGA3ox (Supplementary Figures S1 and S2), but only a partial (NXYPXC) sequence was highly conserved in CsGA2ox (Supplementary Figures S3–S5). Meanwhile, the three histidine residues for binding Fe2+ were all conserved in all putative CsGAoxes [13,50]. The sequence LPWKET, which is highly conserved in all GA20ox so far (Supplementary Figure S1) has never been certified to exist other 2-ODDs [1,9,11], and may be involved in the binding of the GA substrate [13]. The related enzymes, GA3ox and GA2ox, did not contain this proposed consensus sequence (Supplementary Figures S2–S5) [13]. The proposed consensus sequence of CsGA3ox may be the sequence MWSEGXT (Figure 3 and Supplementary Figure S2). However, the sequence of CsGA2ox may be classified into three subgroups (XGWVEYLL, XGEXEYLL and XSWSEAXH). Lee and Zeevaart [51] suggest that GA2ox family can be divided into three classes on the basis of the phylogenetic relationships. The CsGAox genes were also classified into three subfamilies and five different subgroups based on this partial sequence. These individual genes play specific roles in different developmental processes that are regulated by GA. The diversified sequence may be related to the binding of the GA substrate in all GAoxes and thus, more studies are needed to clarify this relation.
Figure 3.
Alignment of the predicted amino acid sequences for the CsGAoxs. Putative tea plant GAox amino acid sequences were aligned using ClustalW [36] method and were edited in Bioedit [30]. Identical and similar residues are shaded in black and gray, respectively. The DIOX_N (PF14226) and 2OG-FeII_Oxy (PF03171) domains are highlighted with a bold line. The three histidine residues for binding Fe2+ are all conserved, which are marked under the alignment. The proposed consensus sequence NXYPXCXXP and LPWKET are highlighted by boxes.
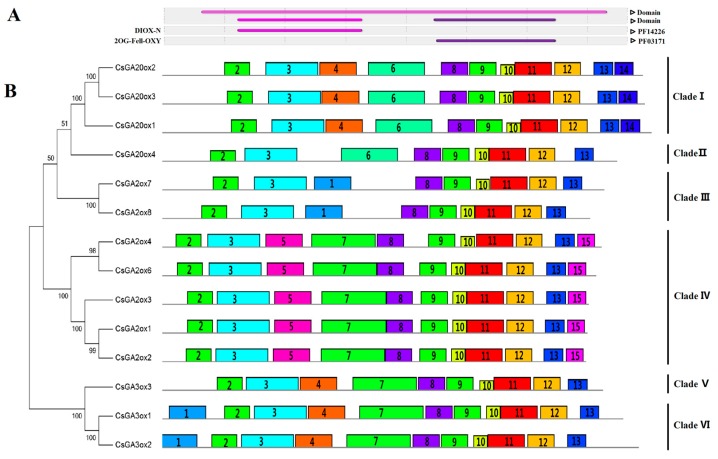
A phylogenetic tree was constructed and 15 discrete motifs were found, with 14 CsGAox proteins used to further assess and classify the CsGAox proteins (Figure 4). The CsGAox proteins could be classified into six clades when analyzed together with the motifs. Due to the lack of Motifs 4 and 14, CsGA20ox4 was separated from other members of GA20ox into Clade II. GA3ox3 was classified to Clade V, due to the lack of Motif 1. Motifs 2 and 3 may be contained in the DIOX_N (PF14226) domain, while the 2OG-FeII_Oxy (PF03171) domain may consist of the motifs 8–12. Motifs 4–7 were highly diversified in all CsGAox proteins. These motifs may be involved in the binding of the GA substrate in all GAoxes and thus, more studies are needed to clarify this relation. The results of the MEME prediction may suggest evolutionary conservation in the basal architecture of GAox family members.
Figure 4.
Phylogenetic analysis and conserved motif identification in CsGAox proteins from tea plants. (A) Interpro analysis revealed two types of domains as a part of the 2-oxoglutarate-dependent dioxygenase superfamily in previously defined GAox proteins; (B) De novo motif identification of GAox proteins. Motifs 2 and 3 show resemblance to the DIOX_N (PF14226) domain, while motifs 8–12 show resemblance to the 2OG-FeII_Oxy (PF03171) domain. Finally, motifs 4–7 may be involved in the binding of the GA substrate.
3.3. Tissue-Specific Expression of CsGAoxes
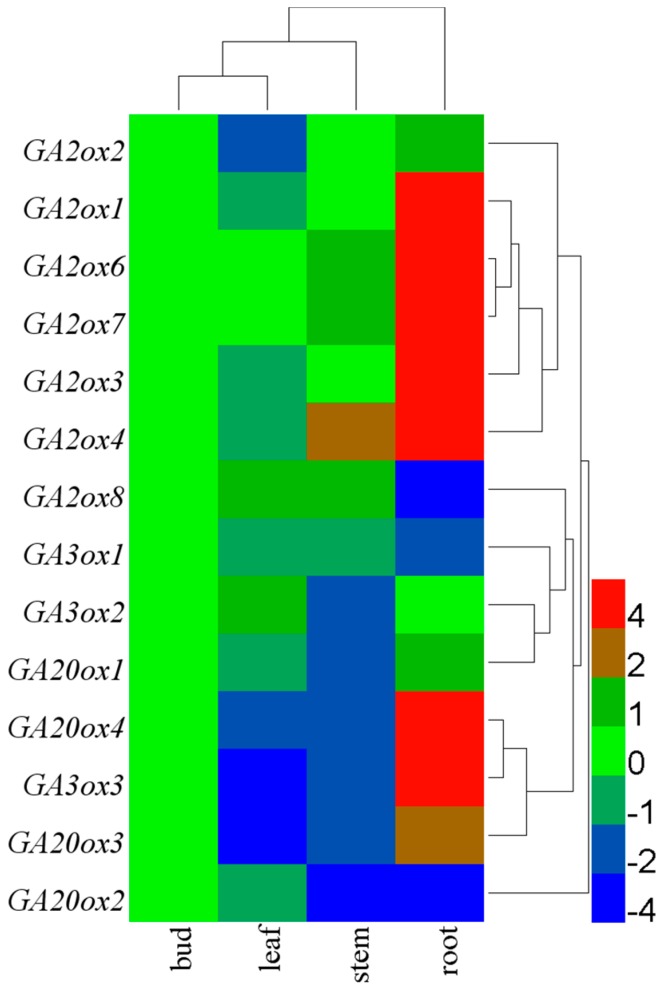
Gibberellins play a key role in regulating diverse processes throughout the life cycle of plants [1,2,3,4]. The tissue specificity of GAox gene expression may be related to the physiological and biochemical functions of each tissue. Hence, it is important that the CsGAox gene expression in specialized tissue should be documented and analyzed. The transcript abundance of 14 CsGAoxes in four different tissues, including roots, stems, leaves and bud, was obtained. Variable transcription levels showed that these genes were ubiquitously and specially expressed in all tissues (Figure 5 and Supplementary Figure S6). CsGAox genes, such as CsGA2ox1, CsGA2ox3, CsGA2ox4, CsGA2ox6, CsGA2ox7, CsGA3ox3, CsGA20ox3, and CsGA20ox4 showed maximum expression levels in the root tissues, while some other CsGAox genes, such as CsGA2ox8, CsGA3ox1, and CsGA20ox2 showed several-fold lower expression in root tissues compared to the other three tissues. The similar expression levels of all CsGAox genes have been showed in leaf and stem tissues with few exceptions. Overall, each CsGAox shows tissue-specific expression patterns [6], although these patterns varied greatly.
Figure 5.
Tissue-specific expression of CsGAoxs. The relative gene expression levels were calculated using the 2−ΔΔCt method and expressed as the fold change relative to expression of the bud tissue. Actin was used as housekeeping gene. The mean expression values were again normalized with logarithm with the base of 2 using the HemI [52] software. The color bar in all heat maps represents the expression values: blue represents low expression, green represents no significant difference in expression, and red denotes high expression.
3.4. Differential Expression of CsGAox Genes under Abiotic Stresses
In tea plants, CsGAoxes take part in numerous abiotic stress responses. qRT-PCR was used to analyze the expression patterns of CsGAox genes in tea plants subjected to various abiotic stresses, such as cold (4 °C), heat (38 °C), ABA (100 μM), GA (100 μM), drought, and salinity (100 mM NaCl). Differential expression levels of CsGAox genes were observed under various stress conditions (Figure 6, Figure 7 and Figure 8).
Figure 6.
Effect of cold, heat, exogenous GA and ABA stresses on CsGAox gene expression in leaves of tea plants. The relative gene expression levels were calculated using the 2−ΔΔCt method and expressed as the fold change relative to expression of the 0-h treatment. Actin was used as housekeeping gene. The mean expression values were again normalized using logarithm with the base of 2 using the HemI software [52]. The color bar in all heat maps represents the expression values: blue represents low expression, green represents no significant difference in expression, and red denotes high expression.
Figure 7.
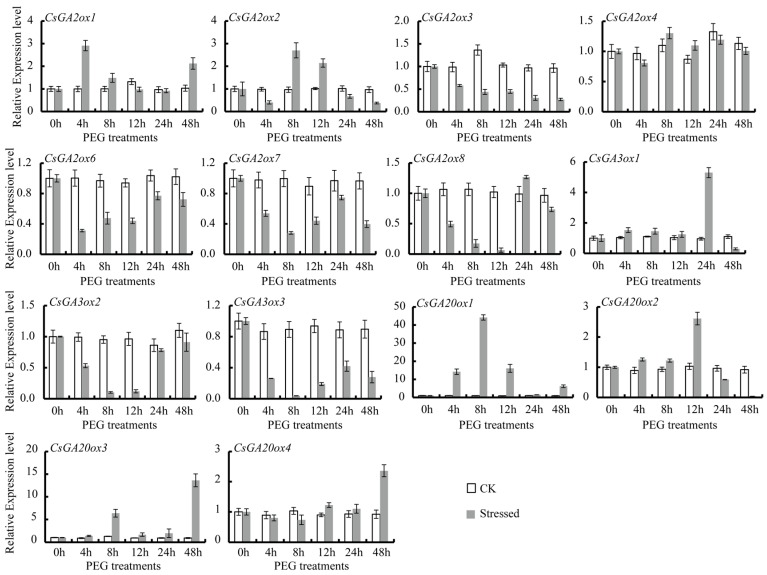
Effect of drought stresses on the CsGAox gene expression in leaves of tea plants. CK represents controls (non stress-treated plants) and stressed represents polyethylene glycol (PEG) stress-treated plants. The relative gene expression levels were calculated using the 2−ΔΔCt method and expressed as the fold change relative to expression of the 0-h treatment. Actin was used as a housekeeping gene.
Figure 8.
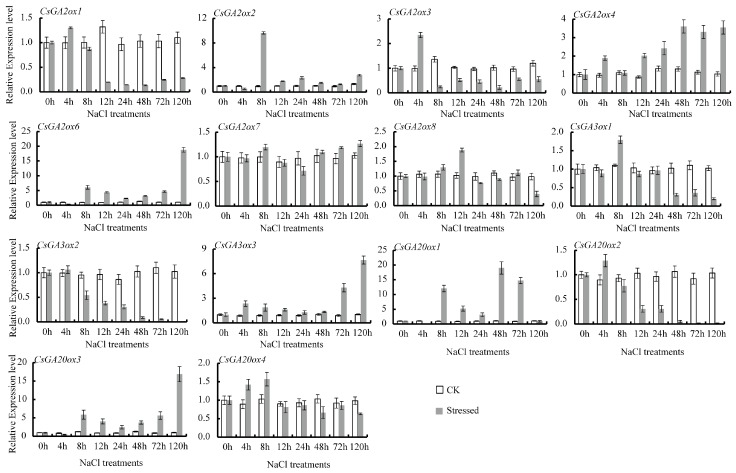
Effect of high salinity stresses on CsGAox gene expression in leaves of tea plants. CK represents controls (non stress-treated plants) and stressed represents NaCl stress-treated plants. The relative gene expression levels were calculated using the 2−ΔΔCt method and expressed as the fold change relative to expression of the 0-h treatment. Actin was used as a housekeeping gene.
In the present study, the expression profile of CsGAoxes showed that the tea plants were markedly affected by conditions of short-term stress (0, 4, 12, and 24 h). As shown in Figure 6, several CsGAox genes were downregulated in leaves after cold and heat treatments. Notably, CsGA2ox3, CsGA2ox8, CsGA3ox2, CsGA3ox3, and CsGA20ox2 showed several-fold lower expression after temperature stress. In contrast, another CsGAox gene (CsGA2ox1) was induced after 12 h of cold stress, while CsGA2ox2 and CsGA2ox4 were induced after 12 h of heat stress. Overall, CsGAox exhibited highly sensitive upregulation or downregulation of gene expression in response to temperature stress compared to controls (Figure 6, Supplementary Figures S7 and S8).
The susceptibility and resistance of plants to diseases can be increased with exogenous application of GA3 and an inhibitor of GA synthesis (uniconazole), respectively [53]. Endogenous GA and ABA strongly influence tea plant growth. The antagonistic roles played by GA and ABA regulate numerous developmental processes [54,55]. Under exogenous GA or ABA, CsGAoxes expression profiles are complicated. The expression pattern in response to GA or ABA of many genes were remarkably upregulated during 24 h of stress. The highest levels of CsGA2ox3 and CsGA20ox4 were observed after 12 h of GA treatment, while CsGA20ox1 was dramatically induced after 4 h of GA or ABA stress. Meanwhile, GA or ABA stress suppressed the transcription of certain CsGAoxes, such as CsGA2ox2, CsGA2ox8, CsGA3ox2 and CsGA20ox2. In general, seven genes containing CsGA2ox3, CsGA2ox8, CsGA3ox2 and CsGA20ox1-4 were upregulated or downregulated by at least four-fold after GA or ABA stress (Figure 6, Supplementary Figures S9 and S10). Similar conclusions have been obtained from previous studies. Most of GA2oxes are upregulated by GA treatment [15,16]. While, most of GA20ox and GA3ox genes are downregulated by applying GA [6,49]. The GA metabolism may be influenced by a feedback mechanism regulating bioactive GAs [6,56,57].
Under PEG treatment to leaves, almost all CsGAoxes had diminished expression levels during a 24-h treatment period. Notably, CsGA2ox3, CsGA2ox6, CsGA2ox7, CsGA2ox8, CsGA3ox2, and CsGA3ox3 showed several fold lower expression after PEG stress (Figure 7). However, the expression patterns of certain genes varied greatly. For example, the expression of CsGA2ox1, CsGA3ox1, CsGA20ox1 and CsGA20ox3 was significantly induced at different points in time. In particular, CsGA20ox1 was upregulated by at least 10-fold after PEG stress, and maintained relatively high levels after 48-h rehydration (Figure 7). In comparison, the other twelve genes, excluding the constitutively expressed CsGA2ox3 and CsGA3ox1 genes, displayed no significant changes in response to PEG after 24 h of treatment.
Upon NaCl stress to leaves, there were different responses from the CsGAoxes (Figure 8). The transcription level of four genes (CsGA2ox2, CsGA2ox6, CsGA20ox1, CsGA20ox3) was downregulated by high salinity stresses after 4 h, but increased significantly (at least five-fold) after this time point and maintained relatively high levels until 120 h (Figure 8). However, CsGA2ox1, CsGA2ox3, CsGA2ox8, CsGA3ox1, CsGA3ox2, CsGA20ox2 and CsGA20ox4 had diminished expression levels. In particular, four genes (CsGA2ox1, CsGA2ox3, CsGA3ox2 and CsGA20ox2) were downregulated by at least four-fold after 8 h high salinity treatment (Figure 8). CsGA3ox3 expression rapidly increased by at least four-fold at 72 and 120 h (Figure 8).
4. Discussion
GAs play a key role in regulating diverse processes throughout the life cycle of plants, such as stem elongation, anther development, and flower induction [1,2,3,4]. GA metabolism may be influenced by a feedback mechanism that regulates bioactive GAs [6,56,57]. Recently, several plant genomes have been sequenced with the development of next-generation sequencing. As a result, the tea tree genome sequencing was published in 2017 [20], and the genome database fortunately opened publicly at the same time, which created opportunities for us to identify 14 full-length CsGAox protein-coding genes with genome-wide analysis in this study. We observed that these CsGAoxes can be classified into three final subfamilies and five different subgroups, including the groups A1, A2, A3, B and C. Most CsGAox family proteins were predicted to be located in the nucleus and cytoplasmic. Huang et al. [46] found that the OsGA2ox6-GFP protein had the same localization as the control GFP construct, giving fluorescent signals in both the nucleus and cytoplasm. Perhaps, there was no correlation between function and location. However, further research is also warranted for CsGAoxes in order to improve and perfect the tea plant genome database, such as chromosomal localization and the exon–intron organization of the corresponding CsGAox genes [58,59].
The GAox protein sequences from C. sinensis, Arabidopsis and G. max were subjected to phylogenetic analysis. The results indicated that these sequences contained the two domains of DIOX_N (PF14226) and 2OG-FeII_Oxy (PF03171), which belong to the 2-ODDs superfamily (Figure 3). Meanwhile, all CsGAox enzymes have been involved in the principal pathways of GA biosynthesis (group B and C) and deactivation (group A1, A2 and A3) in higher plants [1].
For group B and C, GA20ox and GA3ox converted GA12 and GA53 into various GA intermediates and bioactive GAs through two parallel pathways in the third stage of GA biosynthesis. Both GA20ox and GA3ox are soluble 2-ODDs that are present in the cytosol [13,60,61,62,63]. The two parallel pathways are: the non-13-hydroxylation branch, which leads to the production of 13-H GAs; including GA4, and the 13-hydroxylation branch, which leads to the biosynthesis of 13-OH GAs, including GA1 (Figure 1B) [1]. In group B, the GA20ox enzymes (EC: 1.14.11.12) are encoded by multigene families that are responsible for the production of GA9 and GA20, which are precursors of bioactive GAs (Figure 1B) [1,6,14,64,65]. To date, Arabidopsis contains five paralogous GA20ox genes (AtGA20ox1–AtGA20ox5), while rice contains four paralogous GA20ox genes (OsGA20ox1–OsGA20ox4). In the study, similar numbers of CsGA20oxes were isolated and identified, including CsGA20ox1–CsGA20ox4. In the next reaction via the GA3ox enzymes (EC: 1.14.11.15), GA9 and GA20 are hydroxylated to form the biologically active hormones GA4 and GA1 (Figure 1B) [1,65]. GA3ox enzymes are also encoded by multiple genes in all plant species. There are four GA3ox enzymes in Arabidopsis and two GA3ox enzymes in rice [6,60,66,67,68]. Three CsGA3ox isoforms have been confirmed and sequenced by RT-PCR amplification with genome-wide analysis in C. sinensis. The production of bioactive GAs has been shown to be limited by the GA20ox step [61,64,69,70]. There is controversy as to whether GA3ox may be a rate-limiting step in GA biosynthesis.
For group A, the most studied deactivation pathway is catalyzed by a class of GA2ox enzymes. [1,2,3,6,7,15,46,47,48,49]. In this study, seven CsGA2ox, isolated and identified in tea plants, were classified into three groups for the analysis of Arabidopsis genes (Figure 2). The present findings were consistent with findings by Lee and Zeevaart [51], who found that the GA2ox family can be divided into three classes on the basis of phylogenetic relationships. Members of class I and II catabolize C19-GAs, while class III members can only hydroxylate C20-GAs [6,11,15,38,71]. Arabidopsis has five C19GA2ox genes, including the group A1 (AtGA2ox1, AtGA2ox2 and AtGA2ox3) and group A2 (class II) (AtGA2ox4 and AtGA2ox6) [3,15,49]. The group A3 (class III) (AtGA2ox7 and AtGA2ox8) belong to C20GA2ox genes [11]. The C20GA2ox subgroup only acts on C20-GA precursors, such as GA12 and GA53, to form GA110 and GA97, but not on C19-GAs. However, SoGA2ox1 (Spinacia oleracea) can act on both C19-GA and C20-GA [72]. The C20GA2ox enzymes contain three unique conserved motifs that are absent in the class of C19GA2ox [51]. Motifs 5, 7 and 15 may be unique conserved motifs, which were found to be important for the activity of this class of GA2ox [37]. Deactivation mechanisms and pathways for effectively regulating bioactive hormone levels are critical for proper plant growth and development. Several different GA inactivation pathways have been revealed, which are critical for controlling endogenous GA levels.
In the present study, the expression levels of 14 CsGAox genes were observed in tea plant tissues, although these levels varied greatly depending on the tissue type. In root tissues, CsGAox genes, such as CsGA2ox1, CsGA2ox3, CsGA2ox4, CsGA2ox6, CsGA2ox7, CsGA3ox3, CsGA20ox3, and CsGA20ox4 showed maximum expression levels, while some other CsGAox genes, such as CsGA2ox8, CsGA3ox1 and CsGA20ox2, showed several-fold lower expression compared to the other three tissues. The similar expression levels of all CsGAox genes have been showed in leaf and stem tissues with a few exceptions. From these results, the different expression levels in all the tea plant tissues is indicated that some CsGAox genes might play important roles in plant development and have unique functions in specific developmental stages.
Some candidate genes, which may play important response to low- and high-temperature, have been identified with regards to expression patterns. Heat maps show that CsGAox genes were downregulated in leaves after cold and heat treatments. Notably, CsGA2ox3, CsGA2ox8, CsGA3ox2, CsGA3ox3, and CsGA20ox2 showed several-fold lower expression after temperature stress. In contrast, another CsGAox gene (CsGA2ox1) was induced after 12 h of cold stress, while CsGA2ox2 and CsGA2ox4 were induced after 12 h of heat stress. Similar to Arabidopsis, cold treatment also induced a transient increase in AtGA2ox1 transcription levels [73]. However, AtGA3ox1 gene is induced by cold temperatures in seeds soaked in the dark [74]. At high temperatures, GA20ox (AtGA20ox1, AtGA20ox2, and AtGA20ox3) and GA3ox (AtGA3ox1 and AtGA3ox2) are suppressed, which leads to low levels of bioactive GAs [75]. High temperatures repress GA synthesis in Arabidopsis [1]. Meanwhile, cold treatments modulate GA metabolism via upregulation of AtGA2ox3 and AtGA2ox6 gene transcription levels, which decreases bioactive GAs [73].
Endogenous GA and ABA strongly influence tea plant growth [17,18,19]. The antagonistic roles played by GA and ABA regulate numerous developmental processes [54,55]. In Arabidopsis seeds, higher amounts of ABA were found to accumulate in GA-deficient GA1-3 mutant seeds [76], whereas activation of GA biosynthesis genes, such as AtGA3ox1 and AtGA3ox2, was observed in the ABA2-2 mutant during seed development [77]. AtGA3ox1 and AtGA3ox2 were severely repressed at high temperatures, but their expression was completely de-repressed in ABA-deficient mutants [75]. In this study, seven genes containing CsGA2ox3, CsGA2ox8, CsGA3ox2 and CsGA20ox1-4 were upregulated or downregulated by at least four-fold after GA or ABA stress (Figure 6, Supplementary Figures S9 and S10). Most GA2oxes were upregulated by GA treatment [15,16], while most GA20ox and GA3ox genes were downregulated by applied GA [6,49]. ABA reduces bioactive GA levels by decreasing AtGA20ox1 and increasing AtGA2ox6 transcription levels [78]. Therefore, ABA plays a key role in the suppression of GA biosynthesis. Meanwhile, endogenous and exogenous ABA could induce GA deactivation through regulating the expression of some critical genes [79]. This evidence suggests that GAs play an important role in the abiotic stress response, which probably occurs through modulating the ABA signaling pathway [1].
We also examined CsGAox gene expression patterns in leaves under drought and high salinity stress. GAs influence drought tolerance in plants [80]. Almost all CsGAoxes had diminished expression levels during the 24-h treatment period. Notably, CsGA2ox3, CsGA2ox6, CsGA2ox7, CsGA2ox8, CsGA3ox2, and CsGA3ox3 showed several-fold lower expression after PEG stress. However, the expression of CsGA2ox1, CsGA3ox1, CsGA20ox1, and CsGA20ox3 was significantly induced at different points in time. Arabidopsis mutants over-expressing AtGAox1 and AtGAox2 exhibited decreased drought tolerance, while AtGA20ox1 AtGA20ox2 and AtGA3ox1 AtGA3ox2 double mutants, or the AtGA20ox1 AtGA20ox2 AtGA20ox3 triple mutant presented dramatically increased drought tolerance compared to the wild-type Arabidopsis [80]. In Arabidopsis, six AtGA2ox genes were upregulated under high-salinity stress, including AtGA2ox1, AtGA2ox2, AtGA2ox4, AtGA2ox6, AtGA2ox7 and AtGA2ox8 [81]. In comparison, the transcription level of four genes (CsGA2ox2, CsGA2ox6, CsGA20ox1 and CsGA20ox3) was downregulated by high salinity stresses after 4 h, but increased significantly (by at least five-fold) after that time point and maintained relatively high levels until 120 h. However, four genes (CsGA2ox1, CsGA2ox3, CsGA3ox2 and CsGA20ox2) were downregulated by at least four-fold after 8 h of high salinity treatment (Figure 8). Additionally, CsGA3ox3 rapidly increased by at least four-fold of expression after 72 and 120 h.
5. Conclusions
For the first time in tea plants, 14 CsGAox genes, containing the two domains of DIOX_N (PF14226) and 2OG-FeII_Oxy, were identified (PF03171), all of which were 2-oxoglutarate dependent dioxygenases (2-ODD). These included four CsGA20ox (EC: 1.14.11.12), three CsGA3ox (EC: 1.14.11.15) and seven CsGA2ox (EC: 1.14.11.13). These 14 CsGAox genes were phylogenetically clustered into three subfamilies and five subgroups (Figure 2). The genes in these subfamilies displayed differential expression profiles in the studied tissues. Each CsGAox shows tissue-specific expression patterns, although the level of expression varied greatly. Moreover, the qRT-PCR experiment was used to analyze the expression patterns of CsGAox genes in tea plants subjected to various abiotic stresses. Some candidate genes, which may play important roles in responding to cold, heat, PEG, high salinity, or exogenous GA or ABA stresses, have been identified with regards to expression patterns, such as CsGA20ox2, CsGA3ox2, CsGA3ox3, CsGA2ox1, CsGA2ox2, and CsGA2ox4. However, CsGA2ox3, CsGA2ox6, and CsGA2ox8 may have some different expression patterns in response to external abiotic stresses. The production of bioactive GAs may be limited by the GA20ox step. It is controversial whether GA3ox may be a rate-limiting step in GA biosynthesis. Different GA2ox enzymes convert bioactive GAs and their precursors to limit bioactive GA levels and regulate many stages of plant development. In addition, the candidate genes could be used as marker genes for abiotic stress resistance breeding in tea plants.
Acknowledgments
This work was supported by the National Natural Science Foundation of China [grant number: 31270729]. The authors are very grateful to the editors and reviewers for their constructive comments on this paper.
Supplementary Materials
The following are available online at www.mdpi.com/2073-4425/8/9/235/s1. Table S1: Primers of CsGAox genes in RT-PCR. Table S2: Primers of CsGAox genes in qRT-PCR. Table S3: Comparison of the 14 CsGAox ORFs and putative amino acid sequences. Figure S1: Predicted amino acid sequence alignment and phylogenetic tree of the CsGA20oxes with the GA20oxes from other plant species. Figure S2: Predicted amino acid sequence alignment and phylogenetic tree of the CsGA3oxes with the GA3oxes from other plant species. Figure S3: Predicted amino acid sequence alignment and phylogenetic tree of the CsGA2ox1, 2, 3 with the GA2oxes from other plant species. Figure S4: Predicted amino acid sequence alignment and phylogenetic tree of the CsGA2ox4, 6 with the GA2oxes from other plant species. Figure S5: Predicted amino acid sequence alignment and phylogenetic tree of the CsGA2ox7, 8 with the GA2oxes from other plant species. Figure S6: Tissue-specific expression profiles of CsGAoxes. Figure S7: Expression profiles of CsGAox genes in tea plants under cold stresses in leaves. Figure S8: Expression profiles of CsGAox genes in tea plants under heat stresses in leaves. Figure S9: Expression profiles of CsGAox genes in tea plants under exogenous GA stresses in leaves. Figure S10: Expression profiles of CsGAox genes in tea plants under exogenous ABA stresses in leaves. List of protein sequences used in this study: List of putative GAox genes in Sorghum bicolor, Physcomitrella patens, Populus trichocarpa, Cucumis sativus, Medicago truncatula, G.max, Citrus sinensis, Vitis vinifera, Zea mays, Selaginella moellendorffii, and Arabidopsis.
Author Contributions
C.P. performed majority of the experiments and conducted the data analysis, C.P., K.T. and Q.B. prepared and wrote the manuscript; C.P., K.T., L.W. and Y.H. participated in sample collection; K.T., Q.S., Y.Y. and Y.P. involved in the preparation of RNA samples for transcriptome data analysis. Y.L., J.J. and C.J. designed this study.
Conflicts of Interest
The authors declare no conflict of interests.
References
- 1.Gao X., Zhang Y., He Z., Fu X. In: Hormone Metabolism & Signaling in Plants. Li J.Y., Li C.Y., Smith S.M., editors. Academic Press; New York, NY, USA: 2017. pp. 107–160. [DOI] [Google Scholar]
- 2.Hedden P., Kamiya Y. Gibberellin biosynthesis: enzymes, genes and their regulation. Annu. Rev. Plant Physiol. Plant Mol. Biol. 1997;48:431–460. doi: 10.1146/annurev.arplant.48.1.431. [DOI] [PubMed] [Google Scholar]
- 3.Olszewski N., Gubler F. Gibberellin signaling: biosynthesis, catabolism, and response pathways. Plant Cell. 2002;14:S61–S80. doi: 10.1105/tpc.010476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vera-Sirera F., Gomez M.D., Perez-Amador M.A. DELLA proteins, a group of GRAS transcription regulators that mediate gibberellin signaling. Plant Transcr. Factors. 2016:313–328. [Google Scholar]
- 5.Hedden P., Thomas S.G. Gibberellin biosynthesis and its regulation. Biochem. J. 2012;444:11–25. doi: 10.1042/BJ20120245. [DOI] [PubMed] [Google Scholar]
- 6.Hedden P., Phillips A.L. Gibberellin metabolism: new insights revealed by the genes. Trends Plant Sci. 2000;5:523–530. doi: 10.1016/S1360-1385(00)01790-8. [DOI] [PubMed] [Google Scholar]
- 7.Huerta L., Garcialor A., Garciamartinez J.L. Characterization of gibberellin 20-oxidases in the citrus hybrid carrizo citrange. Tree Physiol. 2009;29:569–577. doi: 10.1093/treephys/tpn049. [DOI] [PubMed] [Google Scholar]
- 8.Sponsel V.M., Hedden P. In: Plant Hormones. Biosynthesis, Signal Transduction, Action. Davies P.J., editor. Kluwer Academic Publishers; Dordrecht, The Netherlands: 2004. pp. 63–94. [Google Scholar]
- 9.De C.E., De L.V. 2-oxoglutarate-dependent dioxygenase and related enzymes: biochemical characterization. Phytochemistry. 1994;36:1093–1107. doi: 10.1016/s0031-9422(00)89621-1. [DOI] [PubMed] [Google Scholar]
- 10.Phillips A.L., Ward D.A., Uknes S., Appleford N.E., Lange T., Huttly A.K., Gaskin P., Graebe J.E., Hedden P. Isolation and expression of three gibberellin 20-oxidase cDNA clones from Arabidopsis. Plant Physiol. 1995;108:1049–1057. doi: 10.1104/pp.108.3.1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schomburg F.M., Bizzell C.M., Lee D.J., Zeevaart J.A., Amasino R.M. Overexpression of a novel class of gibberellin 2-oxidases decreases gibberellin levels and creates dwarf plants. Plant Cell. 2003;15:151–163. doi: 10.1105/tpc.005975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vidal A.M., Gisbert C., Talón M., Primomillo E., Lópezdíaz I., Garcíamartínez J.L. The ectopic overexpression of a citrus gibberellin 20-oxidase enhances the non-13-hydroxylation pathway of gibberellin biosynthesis and induces an extremely elongated phenotype in tobacco. Physiol. Plant. 2001;112:251–260. doi: 10.1034/j.1399-3054.2001.1120214.x. [DOI] [PubMed] [Google Scholar]
- 13.Xu Y.L., Li L., Wu K., Peeters A.J., Gage D.A., Zeevaart J.A. The GA5 locus of Arabidopsis thaliana encodes a multifunctional gibberellin 20-oxidase: molecular cloning and functional expression. Proc. Natl. Acad. Sci. USA. 1995;92:6640–6644. doi: 10.1073/pnas.92.14.6640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lange T., Hedden P., Graebe J.E. Expression cloning of a gibberellin 20-oxidase, a multifunctional enzyme involved in gibberellin biosynthesis. Proc. Natl. Acad. Sci. USA. 1994;91:8552–8556. doi: 10.1073/pnas.91.18.8552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Thomas S., Phillips A., Hedden P. Molecular cloning and functional expression of gibberellin 2-oxidases, multifunctional enzymes involved in gibberellin deactivation. Proc. Natl. Acad. Sci. USA. 1999;96:4698–4703. doi: 10.1073/pnas.96.8.4698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Elliott R.C., Ross J.J., Smith J.J., Lester D.R., Reid J.B. Feed-forward regulation of gibberellin deactivation in pea. J. Plant Growth Regul. 2001;20:87–94. doi: 10.1007/s003440010004. [DOI] [Google Scholar]
- 17.Barua D.N. Seasonal Dormancy in Tea (Camellia sinensis L.) Nature. 1969;224:514. doi: 10.1038/224514a0. [DOI] [Google Scholar]
- 18.Nagar P.K., Kumar A. Changes in endogenous gibberellin activity during winter dormancy in tea (Camellia sinensis (L.) O. Kuntze) Acta Physiol. Plant. 2000;22:439–443. doi: 10.1007/s11738-000-0086-2. [DOI] [Google Scholar]
- 19.Yue C., Zeng J.M., Zhang Z.F., Wang X.C., Cao H.L. Research Progress in the Phytohormone of Tea Plant (Camellia sinensis) J. Tea Sci. 2012;32:382–392. [Google Scholar]
- 20.Xia E.H., Zhang H.B., Sheng J., Li K., Zhang Q.J., Kim C., Zhang Y., Liu Y., Zhu T., Li W., et al. The tea tree genome provides insights into tea flavor and independent evolution of caffeine biosynthesis. Mol. Plant. 2017;10:866–877. doi: 10.1016/j.molp.2017.04.002. [DOI] [PubMed] [Google Scholar]
- 21.Wang Y.N., Tang L., Hou Y., Wang P., Yang H., Wei C.L. Differential transcriptome analysis of leaves of tea plant (Camellia sinensis) provides comprehensive insights into the defense responses to Ectropis oblique attack using RNA-Seq. Funct. Integr. Genom. 2016;16:383–398. doi: 10.1007/s10142-016-0491-2. [DOI] [PubMed] [Google Scholar]
- 22.Luo Y.P. Textbook of Tea Cultivation. China Agriculture Press; Beijing, China: 2008. pp. 176–177. [Google Scholar]
- 23.Li H., Huang W., Wang G.L., Wu Z.J., Zhuang J. Expression profile analysis of ascorbic acid-related genes in response to temperature stress in the tea plant, Camellia sinensis (L.) O. Kuntze. Genet. Mol. Res. 2016;15 doi: 10.4238/gmr.15048756. [DOI] [PubMed] [Google Scholar]
- 24.Li X.W., Feng Z.G., Yang H.M., Zhu X.P., Liu J., Yuan H.Y. A novel cold-regulated gene from Camellia sinensis, CsCOR1, enhances salt- and dehydration-tolerance in tobacco. Biochem. Biophys. Res. Commun. 2010;394:354–359. doi: 10.1016/j.bbrc.2010.03.011. [DOI] [PubMed] [Google Scholar]
- 25.Yue C., Cao H., Wang L., Zhou Y., Hao X., Zeng J., Wang X., Yang Y. Molecular cloning and expression analysis of tea plant aquaporin (AQP) gene family. Plant Physiol. Biochem. 2014;83:65–76. doi: 10.1016/j.plaphy.2014.07.011. [DOI] [PubMed] [Google Scholar]
- 26.The University of California Phytozome. [(accessed on 9 May 2016)]; Available online: https://phytozome.jgi.doe.gov/pz/portal.html.
- 27.The Arabidopsis Biological Resource Center TAIR. [(accessed on 9 July 2017)]; Available online: http://www.arabidopsis.org/
- 28.Wang D., Pei K., Fu Y., Sun Z., Li S., Liu H., Tang K., Han B., Tao Y. Genome-wide analysis of the auxin response factors (ARF) gene family in rice (Oryza sativa) Gene. 2007;394:13–24. doi: 10.1016/j.gene.2007.01.006. [DOI] [PubMed] [Google Scholar]
- 29.Zhu X., Wang M., Li X., Jiu S., Wang C., Fang J. Genome-wide analysis of the sucrose synthase gene family in grape (Vitis vinifera): structure, evolution, and expression profiles. Genes. 2017;8:111. doi: 10.3390/genes8040111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Altschul S.F., Madden T.L., Schafferi A.A., Zhang J., Zhang Z., Miller W., Lipman D.J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hall T.A. BioEdit: A User-Friendly Biological Sequence Alignment Editor and Analysis Program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 1999;41:95–98. [Google Scholar]
- 32.Altschul S.F., Wootton J.C., Gertz E.M., Agarwala R., Morgulis A., Schäffer A.A., Yu Y.K. Protein database searches using compositionally adjusted substitution matrices. FEBS J. 2005;272:5101–5109. doi: 10.1111/j.1742-4658.2005.04945.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Letunic I., Doerks T., Bork P. SMART 7: recent updates to the protein domain annotation resource. Nucleic Acids Res. 2012;40:302–305. doi: 10.1093/nar/gkr931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Finn R.D., Coggill P., Eberhard R.Y., Eddy S.R., Mistry J., Mitchell A.L., Potter S.C., Punta M., Qureshi M., Sangrador-Vegas A., et al. The Pfam protein families database: towards a more sustainable future. Nucleic Acids Res. 2016;44:279–285. doi: 10.1093/nar/gkv1344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Johnson M., Zaretskaya I., Raytselis Y., Merezhuk Y., McGinnis S., Madden T.L. NCBI BLAST: a better web interface. Nucleic Acids Res. 2008;36:5–9. doi: 10.1093/nar/gkn201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Thompson J.D., Gibson T.J., Plewniak F., Jeanmougin F., Higgins D.G. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;25:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wilkins M.R., Gasteiger E., Bairoch A., Sanchez J.C., William K.L., Appel R.D., Hochstrasser D.F. Protein identification and analysis tools in the ExPASy server. Methods Mol. Biol. 1999;112:531–532. doi: 10.1385/1-59259-584-7:531. [DOI] [PubMed] [Google Scholar]
- 38.Horton P., Park K.J., Obayashi T., Fujita N., Harada H., Adams-Collier C.J., Nakai k. WoLF PSORT: protein localization predictor. Nucleic Acids Res. 2007;35:585–587. doi: 10.1093/nar/gkm259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Emanuelsson O., Brunak S., von Heijne G., Nielsen H. Locating proteins in the cell using TargetP, SignalP and related tools. Nat. Protoc. 2007;2:953–971. doi: 10.1038/nprot.2007.131. [DOI] [PubMed] [Google Scholar]
- 40.Tamura K., Stecher G., Peterson D., Filipski A., Kumar S. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol. Biol. Evol. 2013;30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bailey T.L., Boden M., Buske F.A., Frith M., Grant C.E., Clementi L., Ren J., Li W.W., Noble W.S. MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res. 2009;37:202–208. doi: 10.1093/nar/gkp335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hunter S., Apweiler R., Attwood T.K., Bairoch A., Bataman A., Binns D., Bork P., Das U., Daugherty L., Duquenne L., et al. InterPro: the integrative protein signature database. Nucleic Acids Res. 2009;37:211–215. doi: 10.1093/nar/gkn785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Erik V.B., Asad M., Marc D.L. Validation of reference genes for gene expression analysis in chicory (Cichorium intybus) using quantitative real-time PCR. BMC Mol. Biol. 2010;11:15. doi: 10.1186/1471-2199-11-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gohain B., Bandyopadhyay T., Borchetia S., Bharalee R., Gupta S., Bhorali P., Agarwala N., Das S. Identification and validation of stable reference genes in Camellia Species. J. Bioteachnol. Pharm. Res. 2011;2:9–18. [Google Scholar]
- 45.Livak K.J., Schmittgen T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 46.Huang J., Tang D., Shen Y., Qin B., Hong L., You A., Li M., Wang X., Yu H., Gu M., et al. Activation of gibberellin 2-oxidase 6 decreases active gibberellin levels and creates a dominant semi-dwarf phenotype in rice (Oryza sativa L.) J. Genet. Genom. 2010;37:23–36. doi: 10.1016/S1673-8527(09)60022-9. [DOI] [PubMed] [Google Scholar]
- 47.Lo S.F., Yang S.Y., Chen K.T., Hsing Y.I., Zeevaart J.A., Chen L.J., Yu S.M. A novel class of gibberellin 2-oxidases control semidwarfism, tillering, and root development in rice. Plant Cell. 2008;20:2603–2618. doi: 10.1105/tpc.108.060913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shan C., Mei Z., Duan J., Chen H., Feng H., Cai W. OsGA2ox5, a gibberellin metabolism enzyme, is involved in plant growth, the root gravity response and salt stress. PLoS One. 2014;9:e87110. doi: 10.1371/journal.pone.0087110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yamaguchi S., Kamiya Y. Gibberellin biosynthesis: its regulation by endogenous and environmental signals. Plant Cell Physiol. 2000;41:251–257. doi: 10.1093/pcp/41.3.251. [DOI] [PubMed] [Google Scholar]
- 50.Xu J., Lange T., Altpeter F. Cloning and characterization of a cDNA encoding a multifunctional gibberellin 20-oxidase from perennial ryegrass (Lolium perenne L.) Plant Sci. 2002;163:147–155. doi: 10.1016/S0168-9452(02)00088-2. [DOI] [Google Scholar]
- 51.Lee D.J., Zeevaart J.A. Molecular cloning of GA 2-oxidase3 from spinach and its ectopic expression in Nicotiana sylvestris. Plant Physiol. 2005;138:243–254. doi: 10.1104/pp.104.056499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Deng W., Wang Y., Liu Z., Cheng H., Xue Y. HemI: a toolkit for illustrating heatmaps. PLoS ONE. 2014;9:e111988. doi: 10.1371/journal.pone.0111988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yang D.L., Li Q., Deng Y.W., Lou Y.G., Wang M.Y., Zhou G.X., Zhang Y.Y., He Z.H. Altered disease development in the eui mutants and Eui overexpressors indicates that gibberellins negatively regulate rice basal disease resistance. Mol. Plant. 2008;1:528–537. doi: 10.1093/mp/ssn021. [DOI] [PubMed] [Google Scholar]
- 54.Razem F.A., Baron K., Hill R.D. Turning on gibberellin and abscisic acid signaling. Curr. Opin. Plant Biol. 2006;9:454–459. doi: 10.1016/j.pbi.2006.07.007. [DOI] [PubMed] [Google Scholar]
- 55.Weiss D., Ori N. Mechanisms of cross talk between gibberellin and other hormones. Plant Physiol. 2007;144:1240–1246. doi: 10.1104/pp.107.100370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bethke P.C., Jones R.L. Gibberellin signaling. Curr. Opin. Plant Biol. 1998;1:440–446. doi: 10.1016/S1369-5266(98)80270-7. [DOI] [PubMed] [Google Scholar]
- 57.Phillips A. Gibberellins in Arabidopsis. Plant Physiol. Biochem. 1998;36:115–124. doi: 10.1016/S0981-9428(98)80096-X. [DOI] [Google Scholar]
- 58.Venkatesh J., Park S.W. Genome-wide analysis and expression profiling of DNA-binding with one zinc finger (Dof) transcription factor family in potato. Plant Physiol. Biochem. 2015;94:73–85. doi: 10.1016/j.plaphy.2015.05.010. [DOI] [PubMed] [Google Scholar]
- 59.Xi Y., Liu J., Dong C., Cheng Z.M. The CBL and CIPK gene family in grapevine (Vitis vinifera): genome-wide analysis and expression profiles in response to various abiotic stresses. Front. Plant Sci. 2017;8:978. doi: 10.3389/fpls.2017.00978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Chiang H., Hwang l., Goodma H. Isolation of the Arabidopsis GA4 locus. Plant Cell. 1995;7:195–201. doi: 10.1105/tpc.7.2.195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Plackett A., Powers S., Fernandez-Garcia N., Urbanova T., Takebayashi Y., Seo M., Jikumaru Y., Benlloch R., Nilsson O., Ruiz-Rivero O., et al. Analysis of the developmental roles of the Arabidopsis gibberellin 20-oxidases demonstrates that GA20ox1, -2, and -3 are the dominant paralogs. Plant Cell. 2012;24:941–960. doi: 10.1105/tpc.111.095109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Reinecke D.M., Wickramarathna A.D., Ozga J.A., Kurepin L.V., Jin A.L., Good A.G., Pharis R.P. Gibberellin 3-oxidase gene expression patterns influence gibberellin biosynthesis, growth, and development in pea. Plant Physiol. 2013;163:929–945. doi: 10.1104/pp.113.225987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sponsel V., Schmidt F., Porter S., Nakayama M., Kohlstruk S., Estelle M. Characterization of new gibberellin-responsive semidwarf mutants of Arabidopsis. Plant Physiol. 1997;115:1009–1020. doi: 10.1104/pp.115.3.1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Coles J.P., Phillips A.L., Croker S.J., Garcia-Lepe R., Lewis M.J., Hedden P. Modification of gibberellin production and plant development in Arabidopsis by sense and antisense expression of gibberellin 20-oxidase genes. Plant J. 1999;17:547–556. doi: 10.1046/j.1365-313x.1999.00410.x. [DOI] [PubMed] [Google Scholar]
- 65.Yamaguchi S. Gibberellin metabolism and its regulation. Annu. Rev. Plant Biol. 2008;59:225–251. doi: 10.1146/annurev.arplant.59.032607.092804. [DOI] [PubMed] [Google Scholar]
- 66.Itoh H., Ueguchi-Tanaka M., Sentoku N., Kitano H., Matsuoka M., Kobayashi M. Cloning and functional analysis of two gibberellin 3β-hydroxylase genes that are differently expressed during the growth of rice. Proc. Natl. Acad. Sci. USA. 2001;98:8909–8914. doi: 10.1073/pnas.141239398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Plackett A., Thomas S., Wilson Z., Hedden P. Gibberellin control of stamen development: a fertile field. Trends Plant Sci. 2011;16:568–578. doi: 10.1016/j.tplants.2011.06.007. [DOI] [PubMed] [Google Scholar]
- 68.Yamaguchi S., Smith M.W., Brown R.G.S., Kamiya Y., Sun T. Phytochrome regulation and differential expression of gibberellin 3β-hydroxylase genes in germinating Arabidopsis seeds. Plant Cell. 1998;10:2115–2126. doi: 10.1105/tpc.10.12.2115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Huang S., Raman A.S., Ream J.E., Fujiwara H., Cerny R.E., Brown S.M. Overexpression of 20-oxidase confers a gibberellin overproduction phenotype in Arabidopsis. Plant Physiol. 1998;118:773–781. doi: 10.1104/pp.118.3.773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Israelsson M., Mellerowicz E., Chono M., Gullberg J., Mortz T. Cloning and overproduction of gibberellin 3-oxidase in hybrid aspen trees. Effects on gibberellin homeostasis and development. Plant Physiol. 2004;135:221–230. doi: 10.1104/pp.104.038935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Sakamoto T., Kobayashi M., Itoh H., Tagiri A., Kayaka H., Lwahori S., Matsuoka M. Expression of a gibberellin 2-oxidase gene around the shoot apex is related to phase transition in rice. Plant Physiol. 2001;125:1508–1516. doi: 10.1104/pp.125.3.1508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Lee D.J., Zeevaart J.A. Differential regulation of RNA levels of gibberellin dioxygenases by photoperiod in spinach. Plant Physiol. 2002;130:2085–2094. doi: 10.1104/pp.008581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Achard P., Gong F., Cheminant S., Alioua M., Hedden P., Genschik P. The cold-inducible CBF1 factor-dependent signaling pathway modulates the accumulation of the growth-repressing DELLA proteins via its effect on gibberellin metabolism. Plant Cell. 2008;20:2117–2129. doi: 10.1105/tpc.108.058941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Yamauchi Y., Ogawa M., Kuwahara A., Hanada A., Kamiya Y., Yamaguchi S. Activation of gibberellin biosynthesis and response pathways by low temperature during imbibition of Arabidopsis thaliana seeds. Plant Cell. 2004;16:367–378. doi: 10.1105/tpc.018143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Toh S., Imamura A., Watanabe A., Nakabayashi K., Okamoto M., Jikumaru Y., Hanada A., Aso Y., Ishiyama K., Tamura N., et al. High temperature-induced abscisic acid biosynthesis and its role in the inhibition of gibberellin action in Arabidopsis seeds. Plant Physiol. 2008;146:1368–1385. doi: 10.1104/pp.107.113738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Oh E., Yamaguchi S., Hu J., Yusuke J., Jung B., Paik L., Lee H.S., Sun T.P., Kamiya Y., Choi G. PIL5, a phytochrome-interacting bHLH protein, regulates gibberellin responsiveness by binding directly to the GAI and RGA promoters in Arabidopsis seeds. Plant Cell. 2007;19:1192–1208. doi: 10.1105/tpc.107.050153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Seo M., Hanada A., Kuwahara A., Endo A., Okamoto M., Yamauchi Y., North H., Marion-Poll A., Sun T.P., Koshiba T., et al. Regulation of hormone metabolism in Arabidopsis seeds: phytochrome regulation of abscisic acid metabolism and abscisic acid regulation of gibberellin metabolism. Plant J. 2006;48:354–366. doi: 10.1111/j.1365-313X.2006.02881.x. [DOI] [PubMed] [Google Scholar]
- 78.Zentella R., Zhang Z.L., Park M., Thomas S.G., Endo A., Murase K., Fleet C.M., Jikumary Y., Nambara E., Kamiya Y., Sun T.P. Global analysis of DELLA direct targets in early gibberellin signaling in Arabidopsis. Plant Cell. 2007;19:3037–3057. doi: 10.1105/tpc.107.054999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Yaish M.W., El-Kereamy A., Zhu T., Beatty P.H., Good A.G., Bi Y.M., Rothstein S.J. The APETALA-2-like transcription factor OsAP2-39 controls key interactions between abscisic acid and gibberellin in rice. PLoS Genet. 2010;6:e1001098. doi: 10.1371/journal.pgen.1001098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Colebrook E.H., Thomas S.G., Phillips A.L., Hedden P. The role of gibberellin signalling in plant responses to abiotic stress. J. Exp. Biol. 2014;217:67–75. doi: 10.1242/jeb.089938. [DOI] [PubMed] [Google Scholar]
- 81.Magome H., Yamaguchi S., Hanada A., Kamiya Y., Oda K. The DDF1 transcriptional activator upregulates expression of a gibberellin-deactivating gene, GA2ox7, under high-salinity stress in Arabidopsis. Plant J. 2008;56:613–626. doi: 10.1111/j.1365-313X.2008.03627.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.