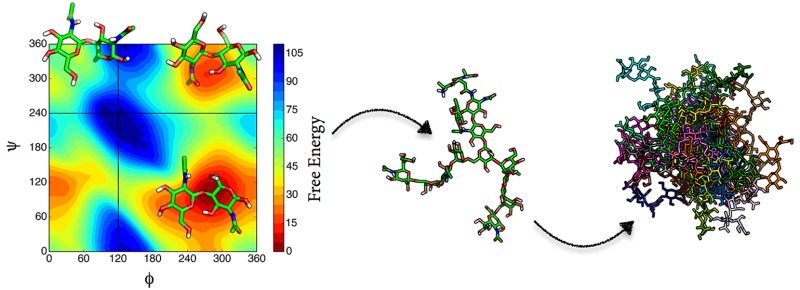
Abstract
In spite of the abundance of glycoproteins in biological processes, relatively little three-dimensional structural data is available for glycan structures. Here, we study the structure and flexibility of the vast majority of mammalian oligosaccharides appearing in N- and O-glycosylated proteins using a bottom up approach. We report the conformational free-energy landscapes of all relevant glycosidic linkages as obtained from local elevation simulations and subsequent umbrella sampling. To the best of our knowledge, this represents the first complete conformational library for the construction of N- and O-glycan structures. Next, we systematically study the effect of neighboring residues, by extensively simulating all relevant trisaccharides and one tetrasaccharide. This allows for an unprecedented comparison of disaccharide linkages in large oligosaccharides. With a small number of exceptions, the conformational preferences in the larger structures are very similar as in the disaccharides. This, finally, allows us to suggest several efficient approaches to construct complete N- and O-glycans on glycoproteins, as exemplified on two relevant examples.
Introduction
Most of the processes in living cells take place via some form of carbohydrate interaction.1 Therefore, understanding the effect of glycosylation, which occurs on more than 50% of the eukaryotic proteins, is crucial to reveal biological mysteries. Glycans, the carbohydrate moieties of glycoproteins, serve as biological markers at various stages of cellular differentiation and proliferation and also play significant roles in cell–cell recognition, cell adhesion, and signal transduction. This broad range of functionality is attributed to their structural diversity in terms of size, sequence, branching, and linkage types that can be formed from a variety of possible monomeric units. However, there is a lack of reliable information on the structure of glycoproteins for several reasons. Most of the experimentally solved biomolecular structures have undergone extensive manipulation of oligosaccharides before X-ray crystallography or NMR spectroscopy due to their inherent flexibility and high degree of coordination with water. The available X-ray structures either have different types of glycoforms than the physiologically relevant forms or show only short sequences of glycan units. Also, structures from X-ray crystallography offer the crystalline form of glycoproteins, lacking a more diverse solution representation. NMR can offer structures in solution; however, these represent averages of simultaneously occurring conformers, limiting the amount of structural information. Furthermore, glycan structures in the databases often contain wrong or missing information about stereochemistry and nomenclature.2 Due to its detailed spatial and temporal resolution, molecular dynamics simulation emerges as a powerful tool for the modeling of glycoproteins. Some progress has been made in search of the conformational preferences of glycan units with different carbohydrate force-fields; including AMBER,3 GLYCAM,4 CHARMM,5 and GROMOS.6,7 In the current work, we will focus on the conformational preferences of carbohydrate structures that are relevant for glycoproteins.
It has become increasingly accepted that the tertiary structure of a protein around a glycosylation site plays a more crucial role than the primary structure in determining the occurrence and level of glycosylation.8 Even though glycosylation is a cotranslational process, processing of glycan units and their variation also depends on the accessibility of the glycosylation site and the glycans to the related processing enzymes.9 Therefore, modeling of the glycoproteins not only contributes a three-dimensional visualization of the glycosylated proteins but also offers explanations for variations in the glycan units as characterized by experimental techniques.10
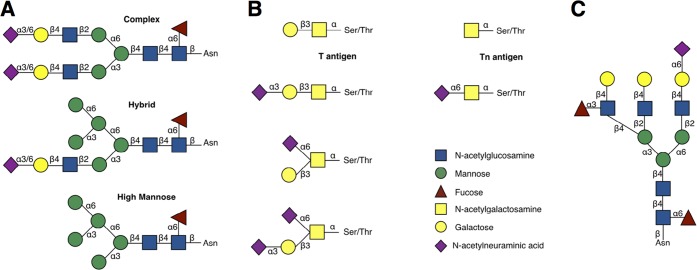
Depending on the variations of the 1,3- or 1,6-branches, N-linked oligosaccharides are classified into three groups: high mannose, hybrid, and complex types (see Figure 1). All types start with two N-acetylglucosamine (β-d-GlcpNAc) units that are linked to an asparagine side chain. The high mannose type subsequently consists of only α-d-Manp residues. In the complex type there are different types of monosaccharides present (for example β-d-GlcpNAc, β-d-Galp, α-d-Neup5Ac, β-d-GalpNAc, and α-l-Fucp) occurring in multiple levels of branching or antennas. In hybrid types, the 1,6-branch may be typically high mannose while the 1,3-branch is complex.
Figure 1.
(A) Major classes of N-glycan. (B) Some core types of O-glycan. (C) Typical complex type of N-glycan in mature glycoproteins.
O-Glycosidic linkages are formed between different monosaccharides and amino acidresidues. The most common glycoproteins carrying O-glycans are mucins and in some cases they carry only a GalNAc residue linked to serine or threonine residues, while in others the O-glycan may be a disaccharide (α-d-Neup5Ac-(2 → 6)-β-d-GalpNAc; β-d-Galp-(1 → 3)-α-d-GalpNAc) or an oligosaccharide consisting of β-d-Galp or β-d-GlcpNAc, elongated from the core units.
We will use a bottom up approach, investigating the conformational preferences of all relevant disaccharides, then moving on to trisaccharides and larger structures. In particular, we will focus on the conformational preferences of the glycosidic linkages and the influence of additional substituents on the central units.
We start by analyzing 17 disaccharide or amino acid-monosaccharide units that are fragments of the vast majority of mammalian glycan trees belonging to mature glycoproteins, involving N- and O-glycans. Furthermore, for the first time, we systematically study the effect of neighboring residues on the conformational preferences of the disaccharides by extensively sampling all relevant trisaccharides. The observed conformational preferences of the disaccharides can be subsequently used to build the common major classes of N- and O-glycans, including typical modifications (like core fucosylation). With the selected disaccharide units, typical N-glycans found in mature glycoproteins can be created. Additionally, for O-glycans, the Tn and T antigen, blood group related antigens Lewis x, and their Sialyl versions can be build. The full list of the studied fragments is presented in Table 1.
Table 1. Overview of the Studied Dimers, along with the Relative Free Energies of the Most Relevant Conformationsa.
| GA | GB | GC | GD | |
|---|---|---|---|---|
| [kJ/mol] | [kJ/mol] | [kJ/mol] | [kJ/mol] | |
| α-d-Neup5Ac-(2 → 6)-β-d-Galp | 0.0 | 4.4 | 3.2 | |
| α-d-Neup5Ac-(2 → 6)-β-d-GalpNAc | 0.0 | 14.9 | 4.0 | |
| α-d-Neup5Ac-(2 → 3)-β-d-Galp | 14.6 | 0.0 | 36.2 | 8.6 |
| α-d-Neup5Ac-(2 → 8)-α-d-Neup5Ac | 0.0 | 24.0 | 9.6 | |
| β-d-Galp-(1 → 4)-β-d-GlcpNAc | 18.9 | 42.2 | 0.0 | 12.6 |
| β-d-GlcpNAc-(1 → 2)-α-d-Manp | 61.7 | 21.9 | 39.2 | 0.0 |
| α-d-Manp-(1 → 6)-β-d-Manp | 0.0 | 39.0 | ||
| α-d-Manp-(1 → 3)-β-d-Manp | 23.9 | 0.0 | 70.4 | 42.7 |
| β-d-Manp-(1 → 4)-β-d-GlcpNAc | 13.4 | 39.8 | 0.0 | 12.2 |
| β-d-GlcpNAc-(1 → 4)-β-d-GlcpNAc | 21.3 | 42.0 | 0.0 | 14.0 |
| α-l-Fucp-(1 → 6)-β-d-GlcpNAc | 45.3 | 0.0 | ||
| α-l-Fucp-(1 → 3)-β-d-GlcpNAc | 72.8 | 39.3 | 12.9 | 0.0 |
| α-d-Manp-(1 → 2)-α-d-Manp | 31.4 | 0.0 | 81.6 | 44.4 |
| β-d-Galp-(1 → 3)-α-d-GalpNAc | 43.5 | 14.4 | 25.6 | 0.0 |
| β-d-GlcpNAc → ASN | 16.6 | 0.0 | ||
| α-d-GalpNAc → SER | 0.0 | 44.2 | ||
| α-d-GalpNAc → THR | 0.0 | 19.5 | 45.4 | 86.0 |
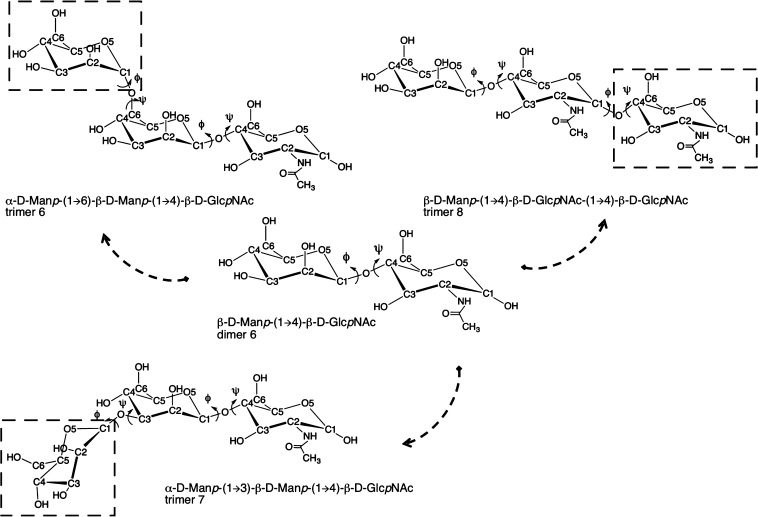
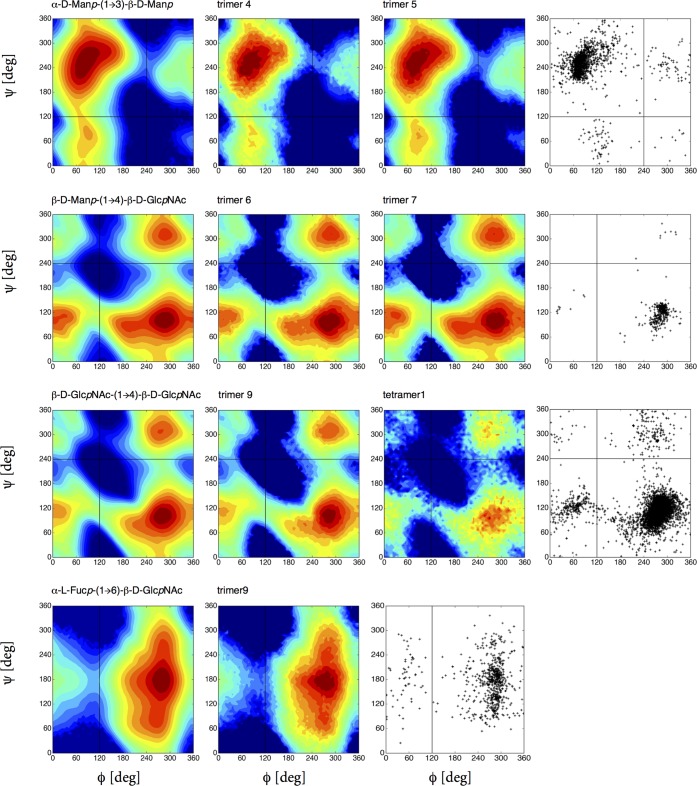
The most relevant conformations of the dimers are characterized through the glycosidic ϕ and ψ angles. For a (1 → X) linkage, where X is 2, 3, 4, or 6 for a reducing disaccharide, the glycosidic dihedral angles, ϕ and ψ, are defined as O5–C1–OX–CX′ and C1–OX–CX′–C(X – 1)′, respectively. For α-d-Neup5Ac-(2 → X) linkage, ϕ and ψ, are defined as O6–C2–OX–CX′ and C2–OX–CX′–C(X – 1)′, respectively. See Figure 2 for the numbering in the β-d-Manp-(1 → 4)-β-d-GlcpNAc dimer as an example. This figure also shows the subsequent steps of our analysis. To investigate the effect of neighboring residues on the conformational preferences of the dimer, all relevant trimers are studied as well, in the case of the dimer in Figure 2; three different dimers are studied.
Figure 2.
Representation of the β-d-Manp-(1 → 4)-β-d-GlcpNAc dimer and demonstration of the nomenclature (center). All the disaccharide fragments from complex type of glycan are simulated with their connecting third monosaccharide; trimer6 represents an additional α-d-Manp-(1 → 6) link, trimer7 a α-d-Manp-(1 → 3) link, and trimer8 a β-d-GlcpNAc link. See Table 2 for the rest of the set.
Previously, similar simulations investigating the conformational preferences of disaccharides as well as N-glycans have been described. Fernandes et al.11 simulated some of the related linkages using the GROMOS 43a1 force field with plain MD. Initial structures were taken from a systematic search in vacuum and used in 100 ns explicit solvent simulation. Perić-Hassler et al.12 investigated glucose-based disaccharides using the same enhanced sampling method as the current work. Yang et al.13 applied a protocol based on replica exchange to sample the conformations of N-glycans. The same technique was used to extensively sample complex glycans14 and high mannose oligosaccharides.15 Even though these studies provide understanding about conformational preferences of several disaccharides, either the studied systems and techniques are different or the sampling of all relevant conformations is insufficient. To the best of our knowledge, the current work represents a first description of a consistent conformational library for the vast majority of relevant linkages in mammalian glycans.
The organization of this paper is as follows. In the methods section, the theory of the enhanced sampling method used is presented, followed by the computational details for its application. Then, the procedure for the analysis of the simulations is introduced. In the Results and Discussion section, a comparison of the conformational preferences as obtained by the simulations with the available experimental data and literature are reported. Furthermore, the effect of neighboring residues on these preferences are systematically studied. We subsequently propose protocols for the construction of the higher oligosaccharides and describe how our approach may be used in future work. Finally, the main findings are summarized in the Conclusion.
Methods
This work aims to suggest protocols to build carbohydrate moieties of glycoproteins in the absence of previous structural information on carbohydrates. As outlined above, we start with disaccharides as they carry all the rotational degrees of freedom of the glycosidic linkage. The conformational characteristics of a disaccharide can be represented by a plot of ϕ against ψ, in analogy to the Ramachandran plot for amino acids. As the relative orientations of the successive monosaccharides differ in each linkage type, the ϕ, ψ map of a disaccharide is distinct. Therefore, an analysis of all possible conformations of a disaccharide is a key for creating higher oligosaccharides. Molecular dynamics (MD) simulations are capable of sampling broad ranges of conformations and are therefore the method of choice. However, it turns out that relevant, distinct conformations, represented by different regions on the ϕ, ψ map are often separated by high barriers, hampering the computational efficiency of the method. Similarly, some conformations may be thermodynamically accessible, but only observed very rarely in plain MD simulations, even if they are experimentally observed. Therefore, we apply an enhanced sampling method to ensure a near-to-complete sampling of the ϕ, ψ maps, local elevation umbrella sampling (LEUS).16,17 Application of this method offers enhanced coverage of those regions in the ϕ, ψ maps which are sterically allowed. It relies on first building up a memory-based biasing potential in the desired subspace and a subsequent umbrella sampling phase to sample the conformational space using the “frozen” biased potentials. We used the adaptable nature of the LEUS method by creating disaccharide biasing potential libraries in the glycosidic torsional angle subspace (ϕ and ψ) and construct the larger carbohydrate moiety of glycoproteins using the relevant biasing potentials from the glycosidic linkages of all disaccharide fragments. A similar kind of approach has been proposed for proteins with peptide ϕ and ψ dihedral angles.18,19
MD Simulation Settings
All MD simulations were performed using the GROMOS11 biomolecular simulation package (http://www.gromos.net)20 with the 53A6glyc parameter set7,21 of the GROMOS force field for carbohydrates. Minor modifications of this parameter set were applied to ensure compatibility with the protein force field and to ensure the stereochemistry of sialic acid. These modifications are listed in Table S2 of the Supporting Information. In particular, we have updated the N-Acetyl linkage in GlcNAc and Neu5Ac to the current protein force field and adjusted the GlcNAc-Asn linkage accordingly.
Initial structures of the disaccharide units of complex glycans were modeled in the molecular operating environment (MOE; Chemical Computing Group, Inc. 2014) from the crystal structure of IgG Fc carrying a sialylated complex type of glycan (PDB ID 4BYH(22)). The rest of the initial structures of units were modeled using the glycosidic dihedral angle preferences according to the Glycosciences portal.23 Hydrogen atoms were added according to geometric criteria followed by a short energy minimization using the steepest-descent algorithm. The compounds were placed in a periodic cubic water box with simple point charge (SPC) water molecules24 and initialized with a 1.4 nm minimum distance of the solute to the box walls. The system was further relaxed by a steepest descent minimization with position restraints on the solute atoms. Prior to the production simulations, the systems were equilibrated with initial random velocities generated from a Maxwell–Boltzmann distribution at 60 K. The systems were heated up to 300 K in five discrete steps with a simulation length of 20 ps, while simultaneously, position restrains on the solute atoms were reduced from 2.5 × 104 to 0.0 kJ mol–1 nm–2.
The simulations were performed at a constant temperature of 300 K and a constant pressure of 1 atm using a weak coupling scheme for both temperature and pressure with coupling times τT = 0.1 ps and τP = 0.5 ps and an isothermal compressibility of 4.575 × 10–4 kJ–1 mol nm3. Newton’s equations of motion were integrated using the leapfrog scheme with a time step of 2 fs. The SHAKE algorithm25 was used to maintain the bond distances at their desired values. Long-range electrostatic interactions beyond a cutoff of 1.4 nm were truncated and approximated by a generalized reaction field with a dielectric permittivity of 61.26 Nonbonded interactions up to a distance of 0.8 nm were computed at every time step using a pairlist that was updated every 5 steps. Interactions up to 1.4 nm were computed at pairlist updates and kept constant in between.
Creating Biased Potential Libraries with LE and Sampling with US
The local elevation (LE) biasing potential is defined as the sum of grid-based functions along the LE coordinates Q, that is (eq 1)
| 1 |
In the original implementation g is defined as a one-dimensional truncated Gaussian function;17 in this study, a polynomial type is used instead (eq 2), following its advantages discussed in ref (16).
| 2 |
Ng is the number of grid points along the Nl dimensional subspace and nk is the number of visits to the corresponding grid cell. MI is the minimum image function ensuring the choice of Qi when it is periodic. H is the Heaviside step function. The force constant, c, and width, σ, are the free parameters which need to be tuned for the building up procedure.
After the LE phase comes an equilibrium umbrella sampling (US) phase during which the biased Hamiltonian is time-independent. That is the values of nk are no longer increased
| 3 |
The real (unbiased) probability to find the system at a value Q can be recovered from the LEUS (biased) simulations by reweighting:
| 4 |
where δ is the Dirac delta function, and β is 1/kBT with kB as the Boltzmann constant and T, the temperature. The angular brackets indicate an ensemble average over the biased US simulation. The corresponding free-energy profile can be obtained as
| 5 |
In this study, the LEUS subspace is defined by the glycosidic dihedral angles ϕ and ψ. After testing a number of different possible combinations, parameters for the LEUS method are chosen as Ng = 36, σ = 360°/Ng, c = 0.005 kJ mol–1, Nl = 2 for the ϕ and ψ angle of a glycosidic linkage. The LE phase is run for tLE = 100 ns and the US phase for tUS = 100 ns. During the US phase, the trajectory is saved for every 0.1 ps resulting in 106 frames for 100 ns to meet the statistical efficiency as discussed in ref (12).
For each of the 17 disaccharides, plain MD simulations were carried out for 50 ns after equilibration. Glycosidic linkage libraries were created by LE and free-energy maps were constructed after US. To assess the effect of neighboring linkages on consecutive units, we used disaccharide frozen potentials and applied umbrella sampling on trisaccharides. Thereby, proving our concept that disaccharide motion libraries based on glycosidic linkages can be taken as a model for oligosaccharides. For each of the 10 trisaccharides simulated, the initial structures were modeled based on the free-energy maps of the disaccharides by setting their relevant glycosidic linkage angles to the global minimum value in the disaccharide. After equilibration, US sampling was applied on two glycosidic linkages with four ϕ and ψ dihedral angles of the trimer unit by using the respective frozen LE potentials evaluated from the corresponding disaccharides.
Analysis
Free-energy maps G(ϕ, ψ) were created from the LEUS simulations after reweighting of the biased energy following eqs 4 and 5. The global minimum of each map was set to G = 0 kJ mol–1 and the value for grid points that were never visited set to Gmax.
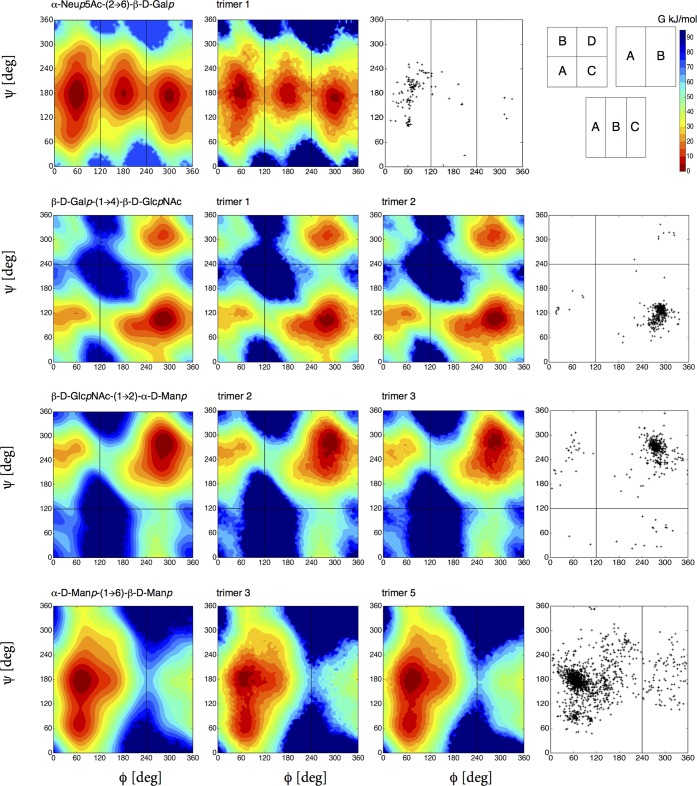
The free-energy maps were divided into distinct conformational regions (A, B, C, up to D) with different ϕ, ψ cutoff values for different disaccharides as illustrated in Figure 3. This scheme has previously been used in refs (6) and12 for glucose-based disaccharides. After the unbiasing procedure, the free energies of these states (GA, GB, GC, and GD) are calculated by integration over regions enclosed by the cutoff values. Subsequently, the values were rescaled by setting the state having the minimum free energy to 0.
Figure 3.
Free-energy maps G(ϕ, ψ) in the glycosidic dihedral angle subspace from unbiased LEUS simulations compared with available crystal and NMR data of the related linkages taken from glycanstructure.org.27 The first column represents the disaccharide simulation, and the second and/or third column corresponds to the disaccharide within a trisaccharide simulation (for trimer codes see Table 2). The division into distinct conformational states (A, B, C, and D) is illustrated at right top panel with colorbar used for free-energy maps with 5 kJ mol–1 spacing starting from global minimum. Regions that were never visited are shown in dark blue. For the rest of the free-energy maps, see Figures 4, 6, and 7 as well as Figure S1 in the Supporting Information.
Results and Discussion
The conformational landscapes of 17 dissaccharides (see Table 1), 10 trisaccharides, and 1 tetramer were studied (see Table 2). The analyzed disaccharides constitute the most typical mammalian N- and O-glycans (see Figure 1). Glycoproteins carrying those units have crucial biological functions, often performed through their glycans. However, how different types of glycans change the function of the glycoproteins often remains unsolved because of incomplete information on their structure. To shed light on the structure–function relation we explored how addition of a subsequent monosaccharide affects the conformational preferences. Our approach is to examine this effect through comparing the free-energy landscape of trisaccharides with their relevant disaccharide units.
Table 2. Relative Free Energy Values of Conformational States from Trisaccharide and Disaccharide LEUS Simulations after Unbiasinga.
| GA | GB | GC | GD | ||
|---|---|---|---|---|---|
| [kJ/mol] | [kJ/mol] | [kJ/mol] | [kJ/mol] | ||
| α-d-Neup5Ac-(2 → 6)-β-d-Galp -(1 → 4)-β-d-GlcpNAc | trimer1 | 0.0 | 3.7 | 2.1 | |
| α-d-Neup5Ac-(2 → 6)-β-d-Galp | dimer1 | 0.0 | 4.4 | 3.2 | |
| α-d-Neup5Ac-(2 → 6)-β-d-Galp-(1 → 4)-β-d-GlcpNAc | trimer1 | 20.6 | 41.0 | 0.0 | 11.7 |
| β-d-Galp-(1 → 4)-β-d-GlcpNAc(1 → 2)-α-d-Manp | trimer2 | 17.7 | 42.0 | 0.0 | 11.6 |
| β-d-Galp-(1 → 4)-β-d-GlcpNAc | dimer2 | 18.9 | 42.2 | 0.0 | 12.6 |
| β-d-Galp-(1 → 4)-β-d-GlcpNAc-(1 → 2)-α-d-Manp | trimer2 | 60.1 | 20.9 | 38.9 | 0.0 |
| β-d-GlcpNAc-(1 → 2)-α-d-Manp -(1 → 6)-β-d-Manp | trimer3 | 58.7 | 20.7 | 36.7 | 0.0 |
| β-d-GlcpNAc-(1 → 2)-α-d-Manp-(1 → 3)-β-d-Manp | trimer4 | 60.5 | 20.8 | 37.9 | 0.0 |
| β-d-GlcpNAc-(1 → 2)-α-d-Manp | dimer3 | 61.7 | 21.9 | 39.2 | 0.0 |
| β-d-GlcpNAc-(1 → 2)-α-d-Manp-(1 → 6)-β-d-Manp | trimer3 | 0.0 | 38.8 | ||
| α-d-Manp-(1 → 3)-[α-d-Manp-(1 → 6)]-β-d-Manp | trimer5 | 0.0 | 37.7 | ||
| α-d-Manp-(1 → 6)-β-d-Manp-(1 → 4)-β-d-GlcpNAc | trimer6 | 0.0 | 42.5 | ||
| α-d-Manp-(1 → 6)-β-d-Manp | dimer4 | 0.0 | 39.0 | ||
| β-d-GlcpNAc-(1 → 2)-α-d-Manp-(1 → 3)-β-d-Manp | trimer4 | 27.9 | 0.0 | 72.4 | 48.2 |
| α-d-Manp-(1 → 3)- [α-d-Manp-(1 → 6)]-β-d-Manp | trimer5 | 24.0 | 0.0 | 71.2 | 43.5 |
| α-d-Manp-(1 → 3)-β-d-Manp-(1 → 4)-β-d-GlcpNAc | trimer7 | 23.2 | 0.0 | 70.0 | 44.6 |
| α-d-Manp-(1 → 3)-β-d-Manp | dimer5 | 23.9 | 0.0 | 70.4 | 42.7 |
| α-d-Manp-(1 → 3)-β-d-Manp-(1 → 4)-β-d-GlcpNAc | trimer7 | 13.2 | 41.1 | 0.0 | 10.2 |
| α-d-Manp-(1 → 6)-β-d-Manp-(1 → 4)-β-d-GlcpNAc | trimer6 | 15.4 | 42.1 | 0.0 | 10.8 |
| β-d-Manp-(1 → 4)-β-d-GlcpNAc -(1 → 4)-β-d-GlcpNAc | trimer8 | 14.5 | 40.2 | 0.0 | 11.8 |
| β-d-Manp-(1 → 4)-β-d-GlcpNAc -(1 → 4)-β-d-GlcpNAc-ASN | tetramer | 16.5 | 39.5 | 0.0 | 12.4 |
| β-d-Manp-(1 → 4)-β-d-GlcpNAc | dimer6 | 13.4 | 39.8 | 0.0 | 12.2 |
| β-d-Manp-(1 → 4)-β-d-GlcpNAc-(1 → 4)-β-d-GlcpNAc | trimer8 | 21.8 | 39.6 | 0.0 | 13.4 |
| β-d-GlcpNAc-(1 → 4)- [α-l-Fucp-(1 → 6)]-β-d-GlcpNAc | trimer9 | 21.8 | 42.0 | 0.0 | 14.2 |
| β-d-GlcpNAc-(1 → 4)-[α-l-Fucp-(1 → 3)]-β-d-GlcpNAc | trimer10 | 27.1 | 38.9 | 0.0 | 18.8 |
| β-d-Manp-(1 → 4)-β-d-GlcpNAc-(1 → 4)-β-d-GlcpNAc-ASN | tetramer | 23.2 | 44.8 | 0.0 | 15.0 |
| β-d-GlcpNAc-(1 → 4)-β-d-GlcpNAc | dimer7 | 21.3 | 42.0 | 0.0 | 14.0 |
| β-d-GlcpNAc-(1 → 4)-[α-l-Fucp-(1 → 6)]-β-d-GlcpNAc | trimer9 | 45.4 | 0.0 | ||
| α-l-Fucp-(1 → 6)-β-d-GlcpNAc | dimer8 | 45.3 | 0.0 | ||
| β-d-GlcpNAc-(1 → 4)-[α-l-Fucp-(1 → 3)]-β-d-GlcpNAc | trimer10 | 69.8 | 38.4 | 20.1 | 0.0 |
| α-l-Fucp-(1 → 3)-β-d-GlcpNAc | dimer9 | 72.8 | 39.3 | 12.9 | 0.0 |
Comparison of Disaccharide Free-Energy Maps with Experimental Data
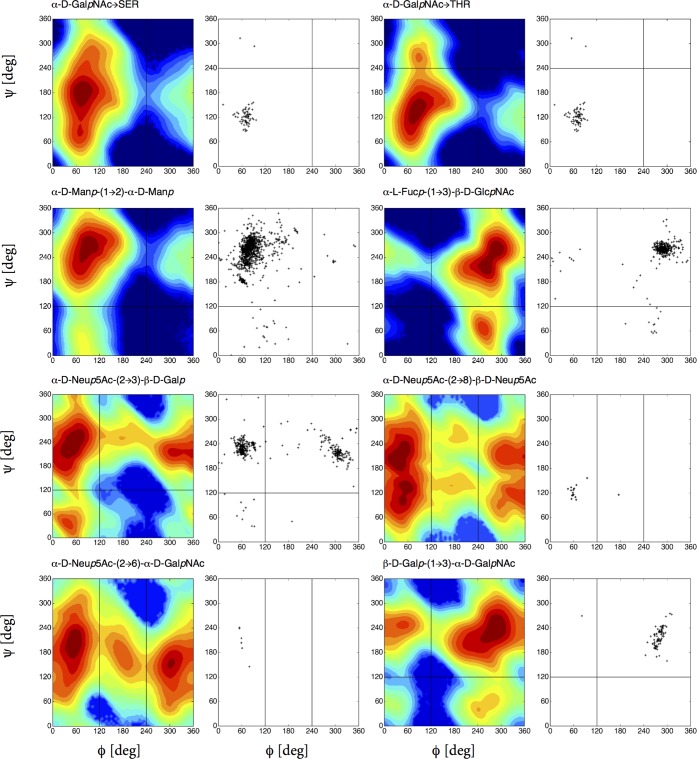
The glycosidic linkage conformations of 17 disaccharides and 10 trisaccharides evaluated from LEUS simulations are compared with available experimental data in Figures 3–7. In Figures 3 and 4, the first column represents the free-energy map of the disaccharide from LEUS simulation, while the second and third column corresponds to the conformation of the same linkage as observed in the trisaccharide LEUS simulations. In the last column, available crystal and NMR structure data of the corresponding linkage gathered from glycanstructure.org are plotted.27 In the free-energy maps, conformational landscapes are divided into states (A, B, C, up to D) which is illustrated in the right top panel of Figure 3. Contour maps were drawn with 5 kJ mol–1 spacing starting from the global minimum which is set to 0 kJ mol–1. The regions that were never visited are shown in dark blue in all free-energy maps. In Figure 5, the remaining disaccharide free-energy maps are reported and compared to ϕ, ψ maps from experimental data. Comparison of the free-energy maps shows that the simulations of disaccharides are in good agreement with the available crystal and NMR structures. Not only the most populated regions but also the shape of the conformational maps and the density of observations was captured when there were enough available structures. Besides the most stable states, there are other thermally accessible states present. Some of the disaccharide free-energy landscapes which are previously reported from plain MD simulations are also in agreement considering the most visited energy basin.11 We applied two-dimensional local elevation along ϕ and ψ dihedral angle; however, in the 1 → 6, 2 → 6, and 2 → 8-linked disaccharides, sampling along the extra dihedral angles (ω for 1 → 6, 2 → 6; χ1 and χ2 for 2 → 8) was free (see Figures S1 and S2) as was also seen in the work of Perić-Hassler et al.12 For the β-N-glycosidic linkage, a one-dimensional local elevation potential was built along O5–C1–Nδ2–Cγ torsional angle.
Figure 7.
Comparison of free-energy maps G(ϕ, ψ) of dimer9 and dimer7 to the values in trimer10 in disaccharide and trisaccharide simulations along with the comparison of available X-ray and NMR structures with exact matches of the β-d-GlcpNAc-(1 → 4)-[α-l-Fucp-(1 → 3)]-β-d-GlcpNAc trimer. A hypothetical molecule was constructure using nonobserved conformations of ϕ, ψ, leading to an obvious steric clash. The color coding is as in Figure 3.
Figure 4.
Free-energy maps G(ϕ, ψ) in the glycosidic dihedral angle subspace from unbiased LEUS simulations compared with available crystal and NMR data of the related linkages taken from glycanstructure.org (last column).27 The first column represents disaccharide simulation, and the second and/or third column corresponds to the disaccharide within a trisaccharide simulation and one for tetramer (for trimer/tetramer codes, see Table 2). Color coding as in Figure 3.
Figure 5.
Free-energy maps G(ϕ, ψ) of disaccharide fragments of O-glycan cores and sialic acid (Neup5Ac) linkage variations in the glycosidic dihedral angle subspace from LEUS simulations compared with available crystal and NMR data of the related linkages taken from glycanstructure.org.27
Analysis of Disaccharides
The free energies of the individual conformational states is given in Table 1, and the respective minimum energy conformations in terms of the glycosidic angles is presented in Table S1. We consider states within 25 kJ mol–1 (∼10kBT) to be thermally accessible. Except from disaccharides containing sialic acid, αNeup5Ac, for all the α-linked disaccharides and O-linkages with a nonreducing residue involved in α linkage (α-d-Manp-(1 → 6)-β-d-Manp, α-d-Manp-(1 → 3)-β-d-Manp, α-d-Manp-(1 → 2)-α-d-Manp, and linkages α-d-GalpNAc → SER and α-d-GalpNAc → THR) we observe only one significant minimum for the ϕ dihedral angle, in the [60°; 100°] range. While for β-linked (also for α-l) disaccharides and N-linkage with the nonreducing residue involved in β linkage (β-d-Galp-(1 → 4)-β-d-GlcpNAc, β-d-GlcpNAc-(1 → 4)-β-d-GlcpNAc, β-d-GlcpNAc-(1 → 4)-β-d-Manp, β-d-Manp-(1 → 4)-β-d-Manp, β-d-Galp-(1 → 3)-β-d-GalpNAc, α-l-Fucp-(1 → 3)-β-d-GlcpNAc, α-l-Fucp-(1 → 6)-β-d-GlcpNAc, and β-d-GlcpNAc-ASN) the ϕ dihedral angle is observed in the [0°; 100°] and [270°; 300°] ranges, with the latter one being the most populated. These observations of the ϕ dihedral angle are in agreement with conformations predicted by the exoanomeric effect. The minima of the ϕ dihedral angle in g+ and g– conformations are preferred, whereby the latter one is disfavored for the nonreducing residue involved in α-linkage due to steric considerations.8
When we compare X-(1 → 4)-β-d-GlcpNAc where X is β-d-GlcpNAc, β-d-Manp, and β-d-Galp, we observe that for all disaccharides conformation C is preferred, with conformation D representing the second energetically favored state at 12–14 kJ mol–1 above the minimum. However, when β-d-Manp is the nonreducing residue (X), the difference between state D (second energetically favored) and state A (third energetically favored) is only 1.2 kJ mol–1, significantly smaller than the rest, for which state A is around 7 kJ mol–1 higher.
Turning our attention to the ψ dihedral angle, we observe for disaccharides with a R-configuration at CX′ (1 → 4 linkages having d-GlcpNAc at the reducing end) preferred values in the [60°; 120°] range and a secondary visited state with a ψ dihedral angle of about 300°. The ones with a S-configuration at CX′ (1 → 3 or 2 → 3 linkages having D-Manp, d-Galp, d-GalpNAc, and d-GlcpNAc at the reducing end; 1 → 2 linkage having d-Manp at the reducing end), however, prefer conformations in the [220°; 300°] range. For (1 → 6)-linked disaccharides, preferred values of the ψ dihedral angle lie within the [160°; 180°] range. These results are also compatible with the previous studies from the glucose-based disaccharide simulations12 and also with MM3 maps.28
Disaccharides containing neuraminic (sialic) acid at the nonreducing end (α-d-Neup5Ac-(2 → 6)-β-d-Galp and its (2 → 6)-α-d-Galp isomer) are commonly found in the carbohydrate moiety of glycoproteins and glycolipids, typically found at outermost ends of glycan units. It is a key component of receptors in molecular recognition for many viruses and bacterial toxins29 and the receptor specificity of influenza viruses is defined in terms of their recognition of the type of sialic acid glycosidic linkage.30 Ligabue-Braun et al. evaluated the conformational ring puckering properties of sialic acid along with 53A6glyc compatible parameters using metadynamics indicating 2C5 as preferred and confirmed this behavior by 1000 ns of unbiased MD simulations.31 Free-energy maps of the disaccharides show three distinct allowed regions for the ϕ dihedral angle (60°, 180°, and 300°) unlike other α linked disaccharides. The related states (A, B, C) have very close free-energy values of 0.0, 4.4, and 3.2 kJ mol–1, respectively. The additional degree of freedom, ω, in these linkages is almost freely rotating as can be seen in Figure S1.
The α-d-Neup5Ac-(2 → 8)-α-d-Neup5Ac disaccharide commonly occurs in gangliosides. They are mostly found at the end of the oligosaccharides of glycoproteins and they also form polysialic acid chains which have important function in cell adhesion and differentiation events. There are only few structures in the pdb carrying this linkage (see Figure 5); therefore, the conformational preferences of this linkage could not be compared with X-ray structural data. However, our results having the lowest energy for ϕ in g+ and for ψ taking a broader range is compatible with existing NMR data.32,33 Our simulations showed extensive sampling along the χ1, while χ2 remains at a value of 120° throughout (see Figure S2). The predominant state for χ1 was at g+ conformation which is in agreement with NMR studies.32,33 Since the sampling is rather complex because of the additional two dihedral angles (χ1, χ2) in the glycosidic linkage, we also built a local elevation potential along the ϕ, χ2 space, thereby including χ2 in the LE biasing stage. Again the predominant χ2 region was 120°; however, we observed a preference for χ2 at 300° when ϕ is in g+. Forcing χ2 angle to take 300° with a ϕ value of 60° required 20 kJ mol–1. Our results suggest that sialic acid containing 2 → 8 and 2 → 6 linkages are more flexible and span a larger area than the 2 → 3 linkage, because of the additional degrees of freedom.
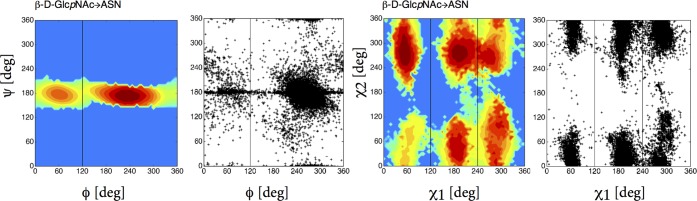
The α-d-Manp-(1 → 2)-α-d-Manp linkage is highly abundant in high-mannosidic glycans. Säwén et al.34 studied this disaccharide extensively with MD and NMR, derived its most probable conformation with glycosidic angles ϕ = 80° and ψ = 273°. Our results for this disaccharide shows the most energetically favored state as B and minimum conformation at this state has ϕ = 96° and ψ = 276° (see Table S1) showing strong agreement with their results. α-d-GalpNAc → Thr and α-d-GalpNAc → Ser have biologically important functions and also are of interest for therapeutics, especially in the development of vaccines for cancer treatment.35 Also, this glycosylation has been known to force the peptide backbone to an extended conformation.35 According to the work of Corzana et al., a change in the sequence of the glycopeptide from Thr to Ser can disturb the function of it. The only difference between these two linkages is the additional methyl group in the Thr. One can see the impact of this methyl group on the change in ψ angle preference at the top of Figure 5. In the presence of a methyl group (Thr linkage) the free-energy landscape in the ψ dihedral angle splits into ranges [110°; 170°] and [250°; 270°] compared to one larger range [90°; 200°] observed in the Ser linkage. In the Ser linkage, the most populated state has a preference for the ψ angle around 180° resulting in a trans conformation while in Thr, it is at 120°. Also, N-linkage which occurs in most of the cases between Asn and N-acetylglucosamine (GlcNAc) has a global minimum at 240° and secondary preference of 60° along the O5–C1–Nδ2–Cγ and C1–Nδ2–Cγ–Cβ torsional angle. The C1–Nδ2–Cγ–Cβ torsional angle has a strong double bonded character (in analogy to a peptide bond). It remains around 180° and was not included in LE biasing. Coverage of preferences of the Asn side chain torsional angles χ1:Nδ2–Cγ–Cβ–Cα and χ2:Cγ–Cβ–Cα–N was also captured as can be seen from Figure 6.
Figure 6.
Free-energy map G(ϕ, ψ) of ASN linkage in sampled ϕ, ψ subspace. Comparison along glycosidic (ϕ, ψ) and torsional (χ1, χ2) angles that are available in pdb structures.
Although pyranose rings can adapt a number of ring conformations (chair, half-chair, envelope, boat, and skew-boat) for saturated rings, a chair is preferred since it relieves much of the torsional strain. Since there are high energy barriers between ring conformational states, they often adopt their minimum energy puckerings; for d-sugars, it is 4C1, and for l-sugars, 1C4. According to metadynamic studies of Pol-Fachin et al.7 with the 53A6glyc force field, the free energies of transition from 4C1 to 1C4 is −77, −54, and −59 kJ/mol for β-d-Glcp, β-d-Galp, and β-d-Manp, respectively. During our LEUS simulations, each monosaccharide accordingly remained in their expected ring puckering conformation, and no transitions were observed toward minor states. All the simulations started from their expected ring puckering conformations which were 1C4 for l-Fucp, 2C5 for d-Neup5Ac and 4C1 for d-sugars. While the transition free energies7 between differently puckered states is probably too high for this force field, restricting the simulations to the most relevant conformations avoids significant complications when analyzing the preferences along the glycosidic linkages.
Comparison of the Conformational Preferences of Trisaccharides
The trisaccharides are compared to their respective dimer units in terms of their free-energy landscapes in Figures 3 and 4. While the unbiased free-energy landscapes are slightly more noisy in the trisaccharides the sampling using the disaccharide LE potentials is clearly sufficient to reconstruct the conformational preferences. The relative free-energies of the states are compared in Table 2. As can be seen from this table, the most populated state in the trisaccharides always remains the same as the ones observed for the corresponding disaccharide. Not only the global minimum state, but also the preferred order of the states did not change. The preference for second, third, and fourth energetically favored states remained the same in the presence of an additional linkage. Overall, the relative free-energy values of the linkages as observed in the trisaccharides deviated by around 2 kJ mol–1 up to a maximum of 7.2 kJ mol–1 from their disaccharide simulations. This maximum deviation was seen in the trisaccharides where the two flanking monosaccharides were on adjacent carbon atoms of the central one (trimer4 and trimer10). Besides from these two cases, the maximum deviation in the thermally accessible states was around 3 kJ mol–1. Subsequently, we studied the occurrence of intramolecular hydrogen bonds in the di- and trisaccharides. The presence of a H-bond was determined using a geometric criterion, requiring a hydrogen–acceptor distance shorter than 0.25 nm and a donor hydrogen–acceptor angle larger than 135°. The occurrences as observed in the LEUS simulations were unbiased using umbrella sampling, to obtain average occurrences for the unbiased ensembles. In Table S3 in the Supporting Information, we report the occurrences of the most prominent (P1) and the second most relevant (P2) hydrogen bonds, using a cutoff of 2%.
Strong H-bonds are observed in the β(1 → 4) linkage between the hydroxyl group vicinal to the linkage to the ring oxygen atom of the nonreducing residue (HO3′–O5), with occurrences between 80 and 90%. The highest probability was observed in the β-d-GlcpNAc-(1 → 4)-β-d-GlcpNAc linkages, with 93%, which may be attributed to its rigidity. Strikingly, for the β-d-Galp-(1 → 4)-β-d-GlcpNAc linkage, this hydrogen bond was observed in the dimer for only 12%, which was increased to 70% in the trimer. Note that, according to Table 2, this was not resulting in a significant shift in the conformational preferences. For the α(1 → 3) linkage, the corresponding H-bonds were much weaker, with occurrences of 2–9% (HO4′–O5 and HO2′–O5). Similarly, the HO3′–O5 hydrogen bond was populated for about 5% in the β(1 → 2) linkage, for which an additional HO4′–O6 H-bond was seen in the trimers. No H-bonds with a probability higher than 2% were observed for the (1 → 6) linkages, as also reported by Perić-Hassler et al.12 and Patel et al.36 in their studies. Finally, sialic acid showed intramonomer H-bonding between its HO8 hydroxyl group and either one of the carboxyl oxygens O1A/O1B. This H-bond also seems to be more pronounced in the trisaccharide than in the disaccharide.
From the similarity between conformational preferences of the linkages in disaccharides and trisaccharides, one can conclude that the effect of consecutive linkages on the previous linkage is limited and taking the disaccharides as a model for oligosaccharides is applicable. However, in the exceptional case β-d-GlcpNAc-(1 → 4)-[α-l-Fucp-(1 → 3)]-β-d-GlcpNAc there were regions that were not sampled in the trisaccharide simulations which are sampled in the disaccharide. We further analyzed those regions which were sterically not allowed in the trisaccharide case, as illustrated in Figure 7. This was expectable since the linkages are introduced right next to each other. However, when α-l-Fucp was 1 → 6 linked to the β-d-GlcpNAc-(1 → 4)-β-d-GlcpNAc disaccharide (trimer9), all the states that were sampled in the disaccharide LEUS simulations are covered (see Figure 4). This comparison also confirms the additional conformational freedom in 1 → 6 linkages. In the other trimer simulation where the linkage is connected to carbon atoms right next to each other, β-d-GlcpNAc-(1 → 2)-α-d-Manp-(1 → 3)-β-d-Manp (trimer4) the conformational preferences compared to their disaccharide behavior were not effected as much as for trimer10. See Figure S3 for a detailed comparison of the conformational preferences of the dimer linkages in trimer4. The reduced effect for this trimer may be explained in terms of the stereochemical orientation in this trimer as shown in Figure 8. Trimer10 was the case where the maximum deviation of 7.2 kJ/mol in free energy from its dimers were seen; at the α-l-Fucp-(1 → 3)-β-d-GlcpNAc linkage, state C. This result emphasizes the role of the N-acetyl group in the monosaccharides, restricting the conformational space in the trisaccharides where the linkages are on the adjacent carbon atoms (see Figure 8). Besides from the presence of the extra N-acetyl group in trimer10, additional strain comes from the position of the linkages where the β(1 → 4) linkage stays between the α(1 → 3) linkage at C3 and the methyl group at C5 (illustrated in Figure 8), thereby, affecting the free-energy landscape of β(1 → 4) linkage as can be seen from Figure 7. An additional trimer unit α-d-Manp-(1 → 2)-α-d-Manp-(1 → 2)-α-d-Manp commonly seen in high-mannose glycans was simulated to further study the effect of close-by linkages. The free-energy landscape of its dimer units showed similar conformational preferences compared to its dimer simulation (see Figure S3). The relative free energy values of both linkages showed ∼3 kJ/mol difference in its conformational states compared to the disaccharide simulation (data not shown). For the relative free energy values of conformational states of α-d-Manp-(1 → 2)-α-d-Manp disaccharide refer to Table 1.
Figure 8.
Representation of trimer4, β-d-GlcpNAc-(1 → 2)-α-d-Manp-(1 → 3)-β-d-Manp (A) and trimer10, β-d-GlcpNAc-(1 → 4)-[α-l-Fucp-(1 → 3)]-β-d-GlcpNAc (B) where the glycosidic linkages are connected to carbon atoms next to each other in their global minimum conformations. In these two special cases, the free energy difference deviated most from their corresponding disaccharide values. For trimer4, maximum deviation compared to its dimers occurred in the α-d-Manp-(1 → 3)-β-d-Manp linkage with 5.5 kJ/mol at state D. For trimer10, the maximum deviation compared to its dimers occurred in α-l-Fucp-(1 → 3)-β-d-GlcpNac linkage with 7.2 kJ/mol at state C.
Construction of Oligosaccharides
While the analysis in the previous paragraph implies that for most linkages, the local effect of neighboring monosaccharides will be limited, longer range influences in larger structure are still to be expected.33,37,38 We developed several approaches to construct glycoproteins without any prior structural knowledge of its carbohydrate moiety. First, the glycan unit of a desired glycoprotein may be built according to the minimum free-energy conformations of its individual glycosidic linkages. This offers an initial model that can be used as it is, or refined further (Figure 9A). Alternatively, optimal trees of a glycan can subsequently be constructed either by randomly picking states for each glycosidic linkage according to their probabilities (computed from the relative free energies) or by applying US for each linkage with a frozen disaccharide LE potential energy library. This generates a broad bundle of glycan structures, which can be filtered in a protein environment after alignment with the glycosylation site of the protein (either Asn or Ser/Thr). Longer range effects of the conformational preferences, most explicitly steric clashes, will be reflected in this bundle of feasible glycan structures. In a filtering step all structures from the bundle are discarded that have a nonbonded energy (intraglycan and glycan–protein) above a given cutoff value. The remaining glycoprotein structures are minimized in vacuum and optimal trees may be selected according to the minimum energy values.
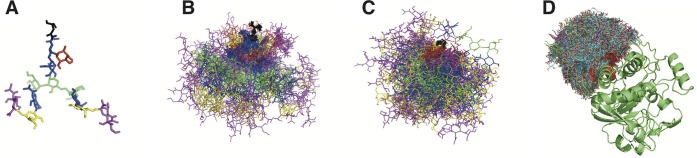
Figure 9.
(A) Initial structure of a complex type of glycan created from related global minima of each glycosidic linkage after unbiasing LEUS simulations. (B) 50 frames aligned on ASN (shown in black) from plain MD and (C) from LEUS simulations. (D) 2100 Filtered structures of all the trajectory from LEUS which are fitted to protein environment (here it is ST6GAL-I, an important enzyme catalyzing the α2,6 sialylation on N-glycans, having a glycosylation site at ASN149, PDB ID 4JS1).
As an example we demonstrate this procedure by constructing a sialylated complex-type N-glycan tree on the human β-galactoside α2,6-sialyltransferase-I (ST6GAL-I, PDB code 4JS1) see Figure 9. Human ST6GAL-I has two glycosylation sites on ASN149 and ASN161 which were suggested to be critical for its folding39 and have an impact on in its enzymatic function.40 Here the glycan tree is built only on ASN149 for representation. The initial structure of the complex type of a glycan tree was created from the global minimum of each glycosidic linkages (after unbiasing of disaccharide LEUS simulations). For the construction of the initial glycan structure, Fernandes et al. used the most populated conformations of the disaccharides obtained from 100 ns plain MD simulations. In their work, they are building the glycoprotein only with these units and subsequently apply plain MD to refine the carbohydrate environment. However, since plain MD simulations may get caught in one energy minimum, we rather apply the US stage on all linkages simultaneously to generate a diverse trajectory. Such a fragment-based approach has been described before.19 Construction of the glycan tree through enhanced methods is the crucial step, since there are evidence of experimentally obtained N-glycans taking highly flexible dynamic conformations. A fold-back conformer of high-mannose-type oligosaccharides explored by combination of NMR and REMD is one of the examples.15 Use of LEUS enables us to capture such conformers as well (see Figure S4). The difference between plain MD simulation and the LEUS simulation can be seen from Figure 9B and C where 50 frames of the trajectories from plain MD and LEUS were randomly selected and aligned on the Asn residue. In the plain MD simulation, the oligosaccharide was only fluctuating around its initial structure, while the LEUS trajectories carried more diversity. After LEUS simulation of the oligosaccharide, these diverse structures were fitted on a protein environment, by alignment of the Asn residue. Using an energy cutoff of +104 kJ mol–1, these trajectories were filtered. Next the protein (PDB ID 4JS1,41 ST6GAL-1) carrying different possible conformations of its oligosaccharide unit are minimized in vacuum. In Figure 9D, one can see 2100 minimized glycan structures of the protein with its glycan. For clarity, only a single protein structure is shown.
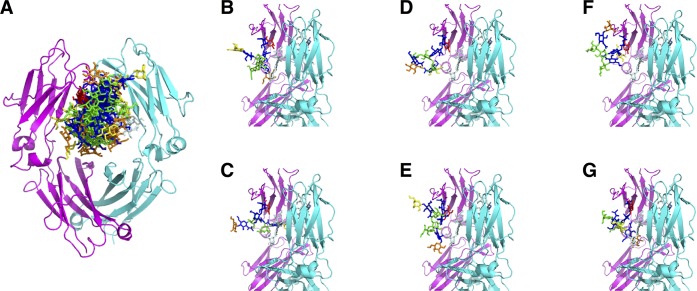
In most of the glycoproteins, glycans are exposed to the surface and they are relatively free to move, as seen in Figure 9D. However, there are therapeutically important glycoproteins where glycans are not on the surface but rather embedded within the glycoprotein environment. In a second application, such a complicated system was chosen; the IgG Fc fragment carrying two complex-type biantennary N-glycans attached to ASN297 of the dimeric Fc fragment, buried in the cavity between the two chains. The Fc glycans are heterogeneous leading to diverse glycoforms which correlates with certain diseases.42 In this application, we have studied the IgG Fc fragment containing a fucosylated complex-type N-glycan, terminated with galactose, due to its high occurrence in healthy humans.43 Since not only the two glycan units are interacting with each other but they are also embedded within the core of glycoprotein, the number of optimal trees that was obtained was significantly reduced compared to fitting of glycans on the exposed protein surface, but some diversity remained among the structures after filtering (see Figure 10A). In addition to the conformation observed in the crystal structure (PDB ID 4BYH(22)), we have found other energetically possible conformers for the glycan units that are more exposed to the surface and more accessible, as has also been suggested by a NMR study.44 The conformations obtained after minimization of the fitted structures showed that glycan units can take fully exposed conformations, either having the 1,6-branch (Figure 10B) or the 1,3-branch exposed (Figure 10C), and a more dynamic glycan conformation with both ends exposed (Figure 10D and E). Furthermore, conformations of glycan units having contacts with the protein and with the other glycan chain were seen and illustrated in Figure 10F and G, respectively. From previous NMR analysis of the Fc glycan it was concluded that the 1,6-branch terminus of the glycan is anchored to the protein surface while the 1,3-branch is exposed and readily accessible for enzymatic addition of sialic acid.45,46 However, in the recent NMR spin relaxation study of Barb and Prestegard,44 it was proposed that not only the 1,3-branch but also the 1,6-branch is highly dynamic and can take exposed conformations as well.
Figure 10.
(A) Representation of a fitting of the galactosylated glycoform on IgG Fc protein carrying glycans on both chains at its core. Protein and glycans units are represented in cartoon and stick form, respectively. N-Acetylglucosamine, mannose, and fucose units are shown in blue, green, and red, respectively. Galactose in the 1,6-branch is shown in yellow and in the 1,3-branch in orange. (B–G) Related views with different levels of glycan exposure: (B) the 1,6-branch is exposed; (C) the 1,3-branch is exposed; (D, E) both ends of the glycan are exposed; (F, G) examples of conformations where the glycan interacts with the protein or with the other glycan tree.
Note that the presampled bundle of glycan trees for a given chemical composition be reused time and again for different protein systems. By generating a library of typically occurring glycan tree bundles, the model building can be done rather efficiently. These approaches are readily applicable using the GROMOS package for molecular simulations. The analysis programs to filter unfavorable glycan trees from the larger conformational bundle will also be made available in the GROMOS++ library of analysis programs. Finally, all local elevation biasing potentials of the disaccharides in Table 1 are made available in the Supporting Information.
Conclusion
The conformations of glycans in conjugation with glycoproteins form a challenge both for experimental and theoretical methods. Their complexity is the result of the variety of possible monomeric units which are linked in a branched way and have differently populated conformational states. There is a pronounced lack of spatial information about them causing an obstacle to grasp the full picture about their biological functions. Especially toward the end of the glycan chains, the amount of available structural data is scarce; see for instance the rightmost panels for the α-d-Neup5NAc-(2 → 6)-β-d-Galp linkage in Figure 3 and for various O-glycan core units in Figure 5.
In this article, we studied the glycosidic linkages that occur in virtually all of the biologically relevant N- and O-linked glycan units. The individual free-energy landscapes were derived from extensive local elevation and umbrella sampling molecular dynamics simulations. For the first time, we reported a complete conformational library in the form of the free-energy landscapes of all disaccharide linkages needed to construct biologically relevant glycans. Since the glycan structures have a great flexibility, not only the lowest-energy conformation is relevant, but also energetically less favorable conformations can be expected to be accessible. For linkages for which structural data is available, our simulations are in excellent agreement with observed conformational preferences, while linkages for which experimental data is sparse, such as the α-d-Neup5NAc-(2 → 6)-α-d-Galp, α-d-Neup5NAc-(2 → 8)-α-d-Neup5NAc linkage and various O-glycan linkages, we provide conformational preferences that complement the experimental data. Many of these linkages have not been studied thoroughly before, other than using vacuum conformational searches or very short MD simulations. All relevant initial structures, topologies, and local-elevation biasing potentials of the disaccharides are made available in the Supporting Information.
In a next step, we systematically studied the effect of an additional linkage on the free-energy landscape of the disaccharide. All relevant trisaccharides and one tetrasaccharide were extensively simulated and the free-energy landscapes were compared. We show that the conformational preferences are largely independent of the consecutive linkages. However, we identified branching linkages, for which neighboring molecules do limit the accessible conformational space (see Figure 7).
The conformational library for the disaccharides, combined with the LEUS method, enables us to build models of higher oligosaccharides without extensive additional sampling of the conformational space. At the hand of two realistic examples, we demonstrate possible modeling methods for glycans that are conjugated to glycoproteins. Overall, our work offers important handles to realistically model the three-dimensional structures and flexibility of glycans in the absence of experimental data. As such, our work opens the way to more realistically study the effect of protein glycosylation on its structure, dynamics, and function.
Acknowledgments
This work was supported by the Austrian Science Fund in the context of the Doctoral Program BioToP (Grant W 1224). The authors thank Maria Pechlaner for help with programming the gromos++ program fit_ener used to filter glycan structures based on their energies.
Supporting Information Available
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acs.jcim.7b00351.
Glycosidic linkage conformations at free-energy minima of the corresponding states (Table S1). Overview of changes in the force field that was used in the current work as compared to the 53A6glyc parameters (Table S2). Average occurrence of intramolecular hydrogen bonds (Table S3). Free-energy maps along the additional degrees of freedom in (1–6) and (2–8) linkages (Figures S1 and S2). Detailed comparison of dimer conformational preferences to those in trimer4 (Figure S3). Examples of fold-back conformations observed in the conformational bundle of a complex glycan tree (Figure S4) (PDF)
Topologies, input structures, and local elevation biasing potentials for all dimers listed in Table 1 (ZIP)
The authors declare no competing financial interest.
Supplementary Material
References
- Mechref Y.; Novotny V. M. In Handbook of glycomics; Cummings R., Pierce J. M., Eds.; Academic Press: San Diego, 2010; pp 1–43. [Google Scholar]
- Lütteke T.; von der Lieth C.-W. PDB-Care (PDB Carbohydrate Residue Check): a Program to Support Annotation of Complex Carbohydrate Structures in PDB Files. BMC Bioinf. 2004, 5, 69. 10.1186/1471-2105-5-69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Momany F. A.; Willett J. L.; Schnupf U. Molecular Dynamics Dimulations of a Dyclic-DP-240 Amylose Fragment in a Periodic Cell: Glass Transition Temperature and Water Diffusion. Carbohydr. Polym. 2009, 78, 978–986. 10.1016/j.carbpol.2009.07.034. [DOI] [Google Scholar]
- Kirschner K. N.; Yongye A. B.; Tschampel S. M.; González-Outeiriño J.; Daniels C. R.; Foley B. L.; Woods R. J. GLYCAM06: A Generalizable Biomolecular Force Field. Carbohydrates. J. Comput. Chem. 2008, 29, 622–655. 10.1002/jcc.20820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guvench O.; Hatcher E.; Venable R. M.; Pastor R. W.; MacKerell A. D. CHARMM Additive All-Atom Force Field for Glycosidic Linkages between Hexopyranoses. J. Chem. Theory Comput. 2009, 5, 2353–2370. 10.1021/ct900242e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen H. S.; Hünenberger P. H. A Reoptimized GROMOS Force Field for Hexopyranose-Based Carbohydrates Accounting for the Relative Free Energies of Ring Conformers, Anomers, Epimers, Hydroxymethyl Rotamers, and Glycosidic Linkage Conformers. J. Comput. Chem. 2011, 32, 998–1032. 10.1002/jcc.21675. [DOI] [PubMed] [Google Scholar]
- Pol-Fachin L.; Rusu V. H.; Verli H.; Lins R. D. GROMOS 53A6 GLYC, an Improved GROMOS Force Field for Hexopyranose-based Carbohydrates. J. Chem. Theory Comput. 2012, 8, 4681–4690. 10.1021/ct300479h. [DOI] [PubMed] [Google Scholar]
- Rao V. S. R.; Qasba P. K.; Balaji P. V.; Chandrasekaran R.. Conformation of Carbohydrates; Harwood Academic Publishers: Amsterdam, The Netherlands, 1998. [Google Scholar]
- Arnold J. N.; Radcliffe C. M.; Wormald M. R.; Royle L.; Harvey D. J.; Crispin M.; Dwek R. A.; Sim R. B.; Rudd P. M. The Glycosylation of Human Serum IgD and IgE and the Accessibility of Identified Oligomannose Structures for Interaction with Mannan-Binding Lectin. J. Immunol. 2004, 173, 6831–6840. 10.4049/jimmunol.173.11.6831. [DOI] [PubMed] [Google Scholar]
- Montero-Morales L.; Maresch D.; Castilho A.; Turupcu A.; Ilieva K. M.; Crescioli S.; Karagiannis S. N.; Lupinek C.; Oostenbrink C.; Altmann F.; Steinkellner H. Recombinant Plant-Derived Human IgE Glycoproteomics. J. Proteomics 2017, 161, 81–87. 10.1016/j.jprot.2017.04.002. [DOI] [PubMed] [Google Scholar]
- Fernandes C. L.; Sachett L. G.; Pol-Fachin L.; Verli H. GROMOS96 43a1 Performance in Predicting Oligosaccharide Conformational Ensembles within Glycoproteins. Carbohydr. Res. 2010, 345, 663–671. 10.1016/j.carres.2009.12.018. [DOI] [PubMed] [Google Scholar]
- Perić-Hassler L.; Hansen H. S.; Baron R.; Hünenberger P. H. Conformational Properties of Glucose-based Disaccharides Investigated Using Molecular Dynamics Simulations with Local Elevation Umbrella Sampling. Carbohydr. Res. 2010, 345, 1781–1801. 10.1016/j.carres.2010.05.026. [DOI] [PubMed] [Google Scholar]
- Yang M.; Huang J.; MacKerell A. D. Enhanced Conformational Sampling Using Replica Exchange with Concurrent Solute Scaling and Hamiltonian Biasing Realized in One Dimension. J. Chem. Theory Comput. 2015, 11, 2855–2867. 10.1021/acs.jctc.5b00243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Re S.; Miyashita N.; Yamaguchi Y.; Sugita Y. Structural Diversity and Changes in Conformational Equilibria of Biantennary Complex-Type N-Glycans in Water Revealed by Replica-Exchange Molecular Dynamics Simulation. Biophys. J. 2011, 101, L44–L46. 10.1016/j.bpj.2011.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi T.; Sakae Y.; Zhang Y.; Yamamoto S.; Okamoto Y.; Kato K. Exploration of Conformational Spaces of High-Mannose-Type Oligosaccharides by an NMR-Validated Simulation. Angew. Chem., Int. Ed. 2014, 53, 10941–10944. 10.1002/anie.201406145. [DOI] [PubMed] [Google Scholar]
- Hansen H. S.; Hünenberger P. H. Using the Local Elevation Method to Construct Optimized Umbrella Sampling Potentials: Calculation of the Relative Free Energies and Interconversion Barriers of Glucopyranose Ring Conformers in Water. J. Comput. Chem. 2010, 31, 1–23. 10.1002/jcc.21253. [DOI] [PubMed] [Google Scholar]
- Huber T.; Torda A. E.; van Gunsteren W. F. Local Elevation: A Method for Improving the Searching Properties of Molecular Dynamics Simulation. J. Comput.-Aided Mol. Des. 1994, 8, 695–708. 10.1007/BF00124016. [DOI] [PubMed] [Google Scholar]
- Kannan S.; Zacharias M. Enhanced Sampling of Peptide and Protein Conformations Using Replica Exchange Simulations with a Peptide Backbone Biasing-Potential. Proteins: Struct., Funct., Genet. 2007, 66, 697–706. 10.1002/prot.21258. [DOI] [PubMed] [Google Scholar]
- Hansen H. S.; Daura X.; Hünenberger P. H. Enhanced Conformational Sampling in Molecular Dynamics Simulations of Solvated Peptides: Fragment-Based Local Elevation Umbrella Sampling. J. Chem. Theory Comput. 2010, 6, 2598–2621. 10.1021/ct1003059. [DOI] [PubMed] [Google Scholar]
- Schmid N.; Christ C. D.; Christen M.; Eichenberger A. P.; Van Gunsteren W. F. Architecture, Implementation and Parallelisation of the GROMOS Software for Biomolecular Simulation. Comput. Phys. Commun. 2012, 183, 890–903. 10.1016/j.cpc.2011.12.014. [DOI] [Google Scholar]
- Pol-Fachin L.; Verli H.; Lins R. D. Extension and Validation of the GROMOS 53A6(GLYC) Parameter Set for Glycoproteins. J. Comput. Chem. 2014, 35, 2087–2095. 10.1002/jcc.23721. [DOI] [PubMed] [Google Scholar]
- Crispin M.; Yu X.; Bowden T. A. Crystal Structure of Sialylated IgG Fc: Implications for the Mechanism of Intravenous Immunoglobulin Therapy. Proc. Natl. Acad. Sci. U. S. A. 2013, 110, E3544–E3546. 10.1073/pnas.1310657110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lütteke T.; Bohne-Lang A.; Loss A.; Goetz T.; Frank M.; von der Lieth C. W. GLYCOSCIENCES.de: An Internet Portal to Support Glycomics and Glycobiology Research. Glycobiology 2006, 16, 71R–81R. 10.1093/glycob/cwj049. [DOI] [PubMed] [Google Scholar]
- Berendsen H. J. C.; Postma J. P. M.; van Gunsteren W. F. J. H. In Intermolecular Forces; Pullman B., Ed.; Reidel: Dordrecht, 1981; p 331–342. [Google Scholar]
- Ryckaert J. P.; Ciccotti G.; Berendsen H. J. Numerical Integration of the Cartesian Equations of Motion of a System with Constraints: Molecular Dynamics of N-alkanes. J. Comput. Phys. 1977, 23, 327–341. 10.1016/0021-9991(77)90098-5. [DOI] [Google Scholar]
- Heinz T. N.; Van Gunsteren W. F.; Hünenberger P. H. Comparison of Four Methods to Compute the Dielectric Permittivity of Liquids from Molecular Dynamics Simulations. J. Chem. Phys. 2001, 115, 1125–1136. 10.1063/1.1379764. [DOI] [Google Scholar]
- Jo S.; Im W. Glycan Fragment Database: A Database of PDB-Based Glycan 3D Structures. Nucleic Acids Res. 2013, 41, D470–D474. 10.1093/nar/gks987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank M.; Lütteke T.; von der Lieth C. W. GlycoMapsDB: A Database of the Accessible Conformational Space of Glycosidic Linkages. Nucleic Acids Res. 2007, 35, 287–290. 10.1093/nar/gkl907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stencel-Baerenwald J. E.; Reiss K.; Reiter D. M.; Stehle T.; Dermody T. S. The Sweet Spot: Defining Virus-Sialic Acid Interactions. Nat. Rev. Microbiol. 2014, 12, 739–749. 10.1038/nrmicro3346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Priyadarzini T. R. K.; Selvin J. F. A.; Gromiha M. M.; Fukui K.; Veluraja K. Theoretical Investigation on the Binding Specificity of Sialyldisaccharides with Hemagglutinins of influenza A Virus by Molecular Dynamics Simulations. J. Biol. Chem. 2012, 287, 34547–34557. 10.1074/jbc.M112.357061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ligabue-Braun R.; Sachett L. G.; Pol-Fachin L.; Verli H. V. The Calcium Goes Meow: Effects of Ions and Glycosylation on Fel d 1, the Major Cat Allergen. PLoS One 2015, 10, 1–19. 10.1371/journal.pone.0132311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michon F.; Brisson J. R.; Jennings H. J. Conformational Differences Between Linear α(2→8)-linked Homosialooligosaccharides and the Epitope of the Group B Meningococcal Polysaccharide. Biochemistry 1987, 26, 8399–405. 10.1021/bi00399a055. [DOI] [PubMed] [Google Scholar]
- Brisson J. R.; Baumann H.; Imberty A.; Perez S.; Jennings H. J. Helical Epitope of the Group B Meningococcal α(2–8)-linked Sialic acid Polysaccharide. Biochemistry 1992, 31, 4996–5004. 10.1021/bi00136a012. [DOI] [PubMed] [Google Scholar]
- Säwén E.; Massad T.; Landersjö C.; Damberg P.; Widmalm G. Population Distribution of Flexible Molecules from Maximum Entropy Analysis Using Different Priors as Background Information: Application to the ϕ, ψ-Conformational Space of the α-(1→2)-linked Mannose Disaccharide Present in N- and O-linked Glycoproteins. Org. Biomol. Chem. 2010, 8, 3684. 10.1039/c003958f. [DOI] [PubMed] [Google Scholar]
- Corzana F.; Busto J. H.; Jiménez-Osés G.; De Luis M. G.; Asensio J. L.; Jiménez-Barbero J.; Peregrina J. M.; Avenoza A. Serine versus Threonine Glycosylation: The Methyl Group Causes a Drastic Alteration on the Carbohydrate Orientation and on the Surrounding Water Shell. J. Am. Chem. Soc. 2007, 129, 9458–9467. 10.1021/ja072181b. [DOI] [PubMed] [Google Scholar]
- Patel D. S.; Pendrill R.; Mallajosyula S. S.; Widmalm G.; MacKerell A. D. Conformational Properties of α- or β-(1→6)-Linked Oligosaccharides: Hamiltonian Replica Exchange MD Simulations and NMR Experiments. J. Phys. Chem. B 2014, 118, 2851–2871. 10.1021/jp412051v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franca E. F.; Lins R. D.; Freitas L. C. G.; Straatsma T. P. Characterization of Chitin and Chitosan Molecular Structure in Aqueous Solution. J. Chem. Theory Comput. 2008, 4, 2141–2149. 10.1021/ct8002964. [DOI] [PubMed] [Google Scholar]
- Franca E. F.; Freitas L. C. G.; Lins R. D. Chitosan Molecular Structure as a Function of N-Acetylation. Biopolymers 2011, 95, 448–460. 10.1002/bip.21602. [DOI] [PubMed] [Google Scholar]
- Chen C.; Colley K. J. Minimal Structural and Glycosylation Requirements for ST6Gal I Activity and Trafficking. Glycobiology 2000, 10, 531–583. 10.1093/glycob/10.5.531. [DOI] [PubMed] [Google Scholar]
- Fast D. G.; Jamieson J. C.; McCaffrey G. The Role of the Carbohydrate Chains of Gal beta-1,4-GlcNAc alpha 2,6-Sialyltransferase for Enzyme Activity. Biochim. Biophys. Acta, Protein Struct. Mol. Enzymol. 1993, 1202, 325–330. 10.1016/0167-4838(93)90023-K. [DOI] [PubMed] [Google Scholar]
- Kuhn B.; Benz J.; Greif M.; Engel A. M.; Sobek H.; Rudolph M. G. The Structure of Human α-2,6-Sialyltransferase Reveals the Binding Mode of Complex Glycans. Acta Crystallogr., Sect. D: Biol. Crystallogr. 2013, 69, 1826–1838. 10.1107/S0907444913015412. [DOI] [PubMed] [Google Scholar]
- Shade K.-T.; Anthony R. Antibody Glycosylation and Inflammation. Antibodies 2013, 2, 392–414. 10.3390/antib2030392. [DOI] [Google Scholar]
- Arnold J. N.; Wormald M. R.; Sim R. B.; Rudd P. M.; Dwek R. A. The Impact of Glycosylation on the Biological Function and Structure of Human Immunoglobulins. Annu. Rev. Immunol. 2007, 25, 21–50. 10.1146/annurev.immunol.25.022106.141702. [DOI] [PubMed] [Google Scholar]
- Barb A. W.; Prestegard J. H. NMR Analysis Demonstrates Immunoglobulin G N-glycans are Accessible and Dynamic. Nat. Chem. Biol. 2011, 7, 147–153. 10.1038/nchembio.511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wormald M. R.; Rudd P. M.; Harvey D. J.; Chang S. C.; Scragg I. G.; Dwek R. A. Variations in Oligosaccharide-Protein Interactions in Immunoglobulin G Determine the Site-Specific Glycosylation Profiles and Modulate the Dynamic Motion of the Fc Oligosaccharides. Biochemistry 1997, 36, 1370–1380. 10.1021/bi9621472. [DOI] [PubMed] [Google Scholar]
- Yamaguchi Y.; Kato K.; Shindo M.; Aoki S.; Furusho K.; Koga K.; Takahashi N.; Arata Y.; Shimada I. Dynamics of the Carbohydrate Chains Attached to the Fc Portion of Immunoglobulin G as Studied by NMR Spectroscopy Assisted by Selective 13C Labeling of the Glycans. J. Biomol. NMR 1998, 12, 385–394. 10.1023/A:1008392229694. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.