Abstract
While RNA interference (RNAi) therapy has demonstrated significant potential for cancer treatment, effective and safe systemic delivery of RNAi agents such as small interfering RNA (siRNA) into tumor cells in vivo remains challenging. We herein reported a unique multistaged siRNA delivery nanoparticle (NP) platform, which is comprised of (i) a polyethylene glycol (PEG) surface shell, (ii) a sharp tumor microenvironment (TME) pH-responsive polymer that forms the NP core, and (iii) charge-mediated complexes of siRNA and tumor cell-targeting- and penetrating-peptide-amphiphile (TCPA) that are encapsulated in the NP core. When the rationally designed, long circulating polymeric NPs accumulate in tumor tissues after intravenous administration, the targeted siRNA-TCPA complexes can be rapidly released via TME pH-mediated NP disassembly for subsequent specific targeting of tumor cells and cytosolic transport, thus achieving efficient gene silencing. In vivo results further demonstrate that the multistaged NP delivery of siRNA against bromodomain 4 (BRD4), a recently discovered target protein that regulates the development and progression of prostate cancer (PCa), can significantly inhibit PCa tumor growth.
Keywords: Nanoparticle, Multistaged delivery, TME-responsive, siRNA, Cancer therapy
INTRODUCTION
Nanomedicine has shown great promise for more effective and safer cancer therapy.1–6 However, the successful clinical translation of cancer nanotherapeutics still faces considerable challenges due to the complexities and heterogeneity of tumors, therefore requiring the rational design of nanoparticle (NP) delivery systems and patient selection.6 To address the barriers involved in systemic NP delivery to solid tumors (such as blood circulation, tumor accumulation and penetration, cellular uptake, and intracellular release), stimuli-responsive NP-based multistaged delivery techniques have recently emerged for effective cancer treatment.6–12 These multistaged NP delivery systems can respond to tumor microenvironment (TME) (e.g., acidic pH, over-expressed enzymes and hypoxia)13–17 to change their physicochemical properties including size, zeta potential and hydrophilic-hydrophobic balance, thereby leading to enhanced diffusion, cellular uptake, and/or intracellular cargo release.18–24
At present, multistaged NP technologies have been successfully applied to the systemic delivery of small molecule drugs, and have shown the capability of efficient intracellular drug delivery for better anticancer effect.20–23, 25 Nevertheless, moderate effort has been made to develop NPs for multistaged in vivo delivery of therapeutic nucleic acids such as small interfering RNA (siRNA), which has shown great potential on cancer treatment by silencing the expression of target gene(s), especially those that encode “undruggable” proteins.26–30 Currently, the de-PEGylation technique is a commonly used strategy to promote siRNA delivery efficacy, in which polyethylene glycol (PEG) chains can be cleaved by the acidic pH31–33 or over-expressed matrix metalloprotease (MMP)34, 35 in tumor tissues to simultaneously achieve long blood circulation and enhanced cellular uptake. However, the complicated TME-responsive chemistry involved in these de-PEGylation techniques may introduce additional complexities in the synthesis and scale-up of therapeutic NPs.
Herein, we developed a unique and robust TME pH-responsive multistaged NP platform for systemic targeted siRNA delivery and effective cancer therapy (Figure 1). This NP platform is composed of a sharp TME pH-responsive PEGylated polymer and a tumor cell-targeting and -penetrating peptide-amphiphile (TCPA). After encapsulating the siRNA/TCPA complexes, the resulting NP platform shows the following features for multistaged siRNA delivery (Figure 1A): i) PEG outer shell prolongs blood circulation and thus enhances tumor accumulation; ii) sensitive response of the hydrophobic poly(2-(hexamethyleneimino) ethyl methacrylate) (PHMEMA) to TME pH36, 37 induces the rapid disassembly of NPs and exposure of siRNA-TCPA complexes at tumor site; iii) tumor cell-targeting ability of TCPA attributable to its RGD ligand segment38, 39 improves the cellular uptake of the siRNA-TCPA complexes; iv) cell-penetrating ability of TCPA attributable to its cationic polyarginine segment40 enhances the cytosolic siRNA transport to achieve efficient gene silencing; and v) facile synthesis of the PHMEMA polymer and TCPA as well as robust NP formulation enable the scale-up of this NP platform. In this work, we further chose bromodomain 4 (BRD4) as a proof-of-concept target and systematically evaluated the multistaged NPs for BRD4 siRNA (siBRD4) delivery and its anticancer efficacy. BRD4 is a conserved member of the bromodomain and extra-terminal (BET) family of chromatin readers, which plays a critical role in transcription by RNA polymerase II by facilitating recruitment of the positive transcription elongation factor b (P-TEFb).41, 42 For metastatic castration-resistant prostate cancer (mCRPC), BRD4 physically interacts with the N-terminal domain of androgen receptor (AR), a key factor involved in the progression of primary prostate cancer (PCa) to mCRPC after androgen-deprivation therapy.43–45 Recent studies demonstrated that BRD4 inhibition can disrupt AR recruitment to target gene loci46–48 and exhibits much more effective mCRPC treatment than direct AR antagonism (e.g., enzalutamide).49 Our in vivo results show that the systemic delivery of siBRD4 with the multistaged polymeric NP platform can also efficiently inhibit BRD4 expression in the tumor tissue and significantly inhibit PCa tumor growth.
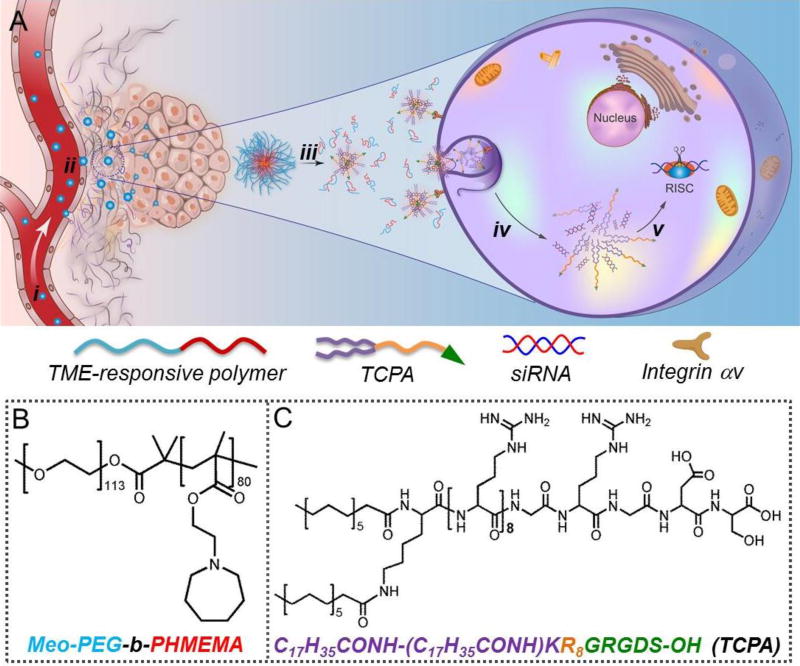
Figure 1.
(A) Schematic illustration of the TME pH-responsive multistaged NP platform for systemic targeted siRNA delivery and effective cancer therapy. After intravenous injection (i), the siRNA-loaded NPs can extravasate from leaky tumor vasculature and accumulate in the tumor tissue (ii). In response to TME pH, the NPs rapidly disassemble to release siRNA-TCPA complexes (iii), which then target and penetrate tumor cells (iv) to achieve efficient cytosolic siRNA transport and gene silencing (v). (B) Molecular structure of TME pH-responsive polymer Meo-PEG-b-PHMEMA. (C) Molecular structure of TCPA with tumor cell-targeting and -penetrating functions attributable to the R8 and RGDS segments, respectively.
RESULTS AND DISCUSSION
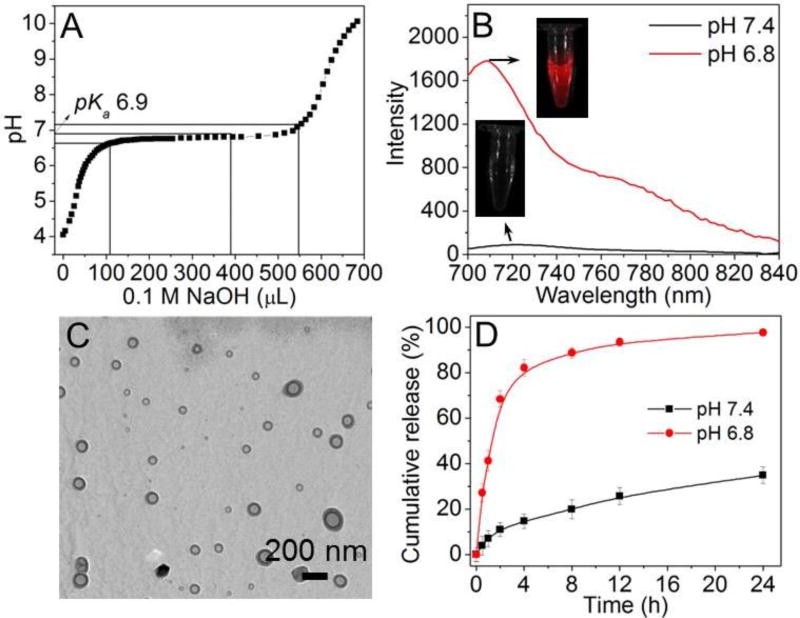
The TME pH-responsive polymer, methoxyl-poly(ethylene glycol)-conjugated PHMEMA (Meo-PEG-b-PHMEMA) was synthesized via atom-transfer radical polymerization (ATRP), using PEG-Br as a macroinitiator (Figure 1B, Scheme S1–S3). The molecular weight of the polymer was examined by gel permeation chromatography (GPC) and the polymer structure was analyzed by nuclear magnetic resonance (Figures S1–S3) to confirm successful synthesis. Classic acid-base titration was used to examine the pKa of this polymer, and the pKa value is determined as ~6.9 (Figure 2A), which is close to the pH of tumor extracellular fluid (6.5~6.8).50 This result suggests that a TME pH-responsive cargo release can be achieved when using a carrier formulated with the Meo-PEG-b-PHMEMA polymer.36 To further support this, a near-infrared dye Cy5.5 was incorporated into the hydrophobic PHMEMA moiety (Meo-PEG-b-P(HMEMA-AMA-Cy5.5), Scheme S4 and Figure S4). When mixing the Cy5.5-labelled polymer with Meo-PEG-b-PHMEMA (1:1 in molar ratio), due to the amphiphilic nature of these two polymers, they can self-assemble into well-dispersed NPs (Figure S5), with an average size of ~40 nm as determined by dynamic light scattering (DLS, Figure S5). Owing to the self-quenching of the aggregated fluorophores inside the hydrophobic cores of these NPs, there is very weak fluorescence signal at a pH (e.g., pH 7.4) above pKa of Meo-PEG-b-PHMEMA (Figure 2B). In contrast, when dispersing these NPs into PBS buffer at a pH (e.g., pH 6.8) below pKa, the electrostatic repulsion among the protonated PHMEMA moieties leads to the disassembly of the NPs (Figure S5) and a dramatic increase in the fluorescence signal within 3 min (Figure 2B). Measurement of the fluorescence intensity upon pH change shows that the pH difference from 10 to 90% fluorescence activation (ΔpH10–90%) is 0.24 (Figure S5),36, 37 which is similar to the previous report36 and much smaller than that of small molecule dyes (about 2 pH units),51 suggesting the sharp TME pH response of the Meo-PEG-b-PHMEMA polymer.
Figure 2.
(A) Acid-base titration profile of the TME pH-responsive polymer Meo-PEG-b-PHMEMA. (B) Emission fluorescence spectrum of Cy5.5-labelled TME pH-responsive NPs incubated in PBS buffer at different pHs for 3 min. (C) TEM image of the DY677-siRNA-loaded TCPA2-NPs at pH 7.4. (D) Cumulative siRNA release from the DY677-siRNA-loaded TCPA2-NPs at different pHs.
We next examined the siRNA loading ability and TME pH-responsive behavior of the siRNA NPs. A modified nanoprecipitation method52 was employed to prepare the NPs by mixing siRNA aqueous solution with dimethylformamide (DMF) solution containing both Meo-PEG-b-PHMEMA and TCPA. In this mixture, the negatively charged siRNA molecules spontaneously assemble with the positively charged TCPA into complexes via electrostatic interaction. By adding the above mixture to rapidly stirring deionized water, the amphiphilic Meo-PEG-b-PHMEMA polymer and siRNA-TCPA complexes were co-nanoprecipitated to form NP with a hydrophilic PEG shell and a siRNA-TCPA complex-containing PHMEMA core. Two TCPAs (TCPA1: C17H35CONH-GR8GRGDS-OH shown in Figure S6; and TCPA2: C17H35CONH-(C17H35CONH)-KR8GRGDS-OH shown in Figure 1C) were used to adjust the siRNA loading efficiency and physiochemical properties of the NPs (denoted TCPA1-NPs and TCPA2-NPs). Under the same conditions, the siRNA encapsulation efficiency (EE%, Table S1) of the TCPA1-NPs (~39%) is lower than that of TCPA2-NPs (~52%). In contrast, the size of the TCPA1-NPs (~90.1 nm, Figure S7) is larger than that of the TCPA2-NPs (~72.8 nm). One possible reason may be due to the two hydrophobic tails of TCPA2 that could facilitate the formation of more compact siRNA-TCPA2 complexes to improve the siRNA loading ability and decrease the size of the NPs.53, 54 In addition, the TCPA2-NPs show a strong protection of siRNA stability. When encapsulating fluorescein (FL) and its quencher (Dabcyl)-labelled siRNA into the NPs, there is nearly no change in the FL fluorescence after 6 h incubation with RNase (Figure S8). As a comparison, naked siRNA can be rapidly degraded by RNase within 15 min, which induces the dissociation between FL and Dabcyl, and thereby significant increase in the fluorescence intensity of the FL. We then chose the TCPA2-NPs to evaluate their TME pH-responsive behavior. As shown in the transmission electron microscopy (TEM) image of Figure 2C, the NPs exhibit a spherical morphology at pH 7.4. After incubating these NPs in pH 6.8 PBS buffer for 3 min, there are some large amorphous aggregates and small size particles (Figure S9A), which may possibly correspond to the ionized polymer and exposed siRNA-TCPA2 complexes. This result is confirmed by DLS analysis, in which particles ranging from several nanometers to thousand nanometers can be detected (Figure S9B). The fluorescence spectra of the NPs loaded with FL and Dabcyl-labelled siRNA also demonstrate the presence of exposed siRNA-TCPA2 complexes after TME pH-triggered NP disassembly (Figure S9C). Similar as the fluorescence change at pH 7.4 (Figure S8), significant increase in the fluorescence intensity can be observed after incubating the naked siRNA with RNase at pH 6.8 for 15 min. In contrast, with the TCPA2 to condense siRNA and form complexes that can protect siRNA from degradation by RNase, there is no obvious change in the fluorescence intensity after incubating the TCPA2-NPs with RNase at pH 6.8 for 1 h. This TME pH-triggered NP disassembly also offers a fast release of loaded siRNA (Figure 2D). More than 80% of DY677-labelled siRNA (DY677-siRNA) is released after 4 h at pH 6.8. Within the same time frame, less than 20% of the loaded siRNA is released at pH 7.4.
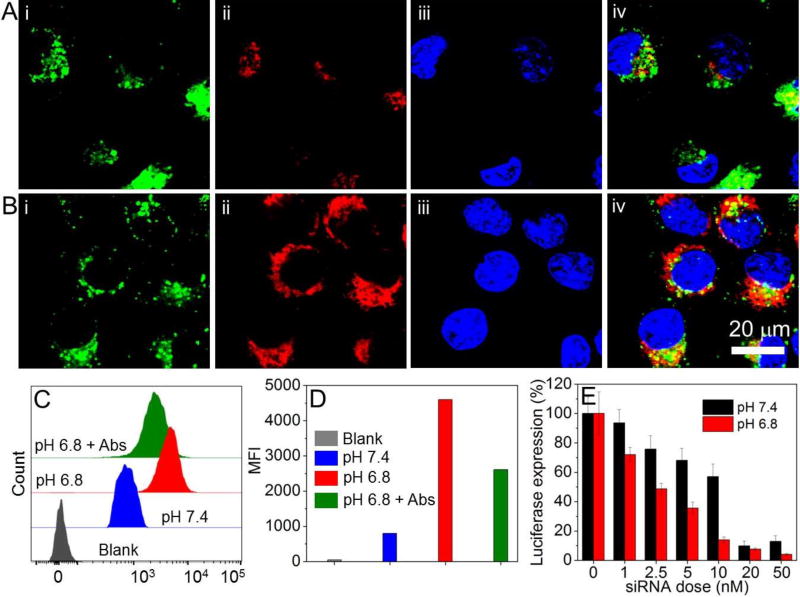
To investigate whether this TME pH-triggered NP disassembly can improve cellular uptake of loaded siRNA and enhance gene silencing, luciferase-expressing HeLa (Luc-HeLa) cells were incubated with the DY677-siRNA-loaded TCPA2-NPs at pH 6.8 or 7.4 for 2 h. The cellular uptake was observed by confocal laser scanning microscopy (CLSM). Compared to the cells incubated at pH 7.4 (Figure 3A), the brighter red fluorescence indicates a higher siRNA uptake at pH 6.8 (Figure 3B). More importantly, the internalized siRNA molecules at pH 6.8 show a much higher distribution in the cytoplasm compared to that of the internalized siRNA molecules at pH 7.4. We further employed flow cytometry to quantitatively examine the uptake at different pHs. As shown in Figures 3C and 3D, the siRNA uptake at pH 6.8 is > 5-fold more than that of the cells incubated at pH 7.4. When pre-treating the cells with the integrin αvβ3 and αvβ5 antibodies for 15 min and then incubating with the siRNA-loaded NPs at pH 6.8, there is around 2-fold decrease in the siRNA uptake. Notably, due to the positive charge of the polyarginine segments in the exposed siRNA-TCPA2 complexes at pH 6.8, the antibody-treated cells still show stronger siRNA uptake than the cells incubated at pH 7.4. These results indicate that the exposed siRNA-TCPA2 complexes resulting from the pH-triggered disassembly of NPs can increase the siRNA uptake by RGD-mediated targeting of the over-expressed integrins (αvβ3 and αvβ5) on Luc-HeLa cells.55 In addition, the positively charged polyarginine segments also contribute to the increased siRNA uptake and can simultaneously penetrate the endosomal membranes to improve cytosolic siRNA delivery.32, 56, 57
Figure 3.
(A, B) CLSM images of Luc-HeLa cells incubated with DY677-siRNA-loaded TCPA2-NPs at pH of 7.4 (A) or 6.8 (B) for 2 h. Endosomes are stained by lysotracker green, and nuclei were stained by Hoechst 33342. (i) Endosomes with green fluorescence; (ii) DY677-siRNA with red fluorescence; (iii) Nuclei with blue fluorescence; and (iv) Overlap of (i), (ii) and (iii). (C) Flow cytometry profile and (D) mean fluorescence intensity (MFI) of Luc-HeLa cells incubated with DY677-siRNA-loaded TCPA2-NPs at different pHs for 2 h, and the cells incubated with integrin αvβ3 and αvβ5 antibodies for 15 min followed by incubation with the DY677-siRNA-loaded TCPA2-NPs at pH 6.8 (pH 6.8 + Abs) for 2h. (E) Luc expression in Luc-HeLa cells treated with siLuc-loaded TCPA2-NPs at different pHs.
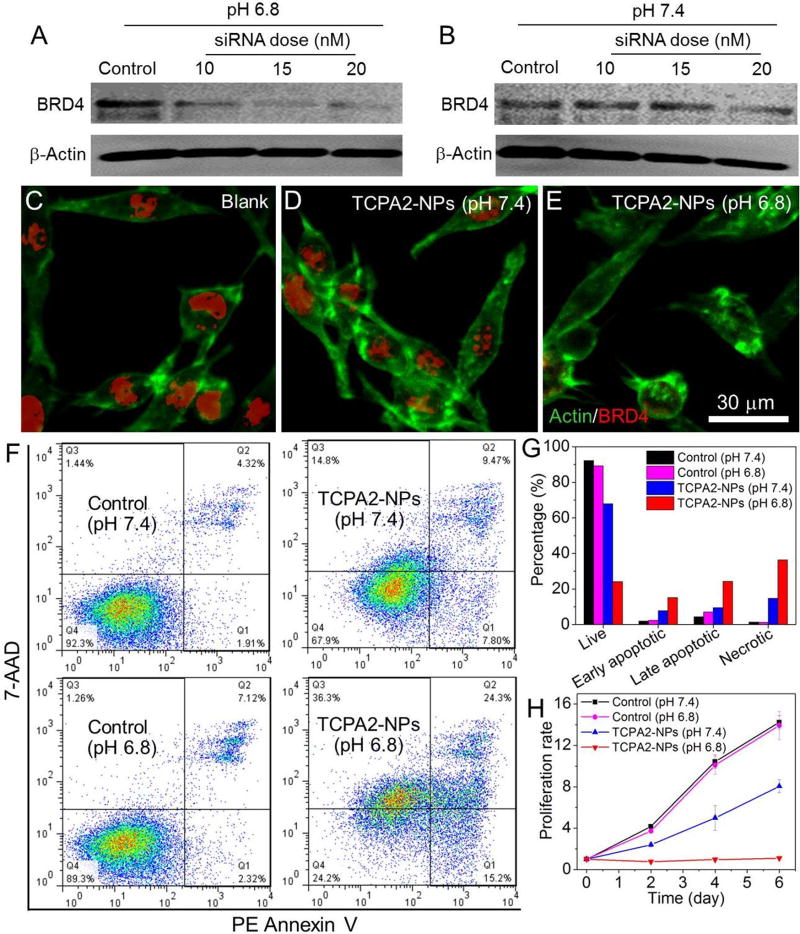
We then encapsulated Luc siRNA (siLuc) into the TCPA2-NPs and evaluated their gene silencing efficacy in Luc-HeLa cells. As shown in Figure 3E, the siLuc-loaded NPs reduced Luc expression at both pH 7.4 and 6.8. Notably, due to rapid disassembly of the NPs at pH 6.8 to increase the cytosolic siRNA delivery (Figures 3B and 3C), a much better gene silencing efficacy was shown vs. that at pH 7.4, and the NPs can knock down ~90% Luc expression with low cytotoxicity at a 10 nM siRNA dose (Figure S11). We further examined the TCPA2-NPs for silencing the expression of BRD4, a conserved member of the BET family of chromatin readers that interacts with the N-terminal domain of AR to regulate AR signaling network in mCRPC.49 LNCaP cells, an AR positive PCa cell line with high level of BRD4 (Figure S12)49 and integrins (αvβ3 and αvβ5) expression,58 were used as a model cell line. Figures 4A and 4B showed that the siBRD4-loaded NPs had a higher efficacy in BRD4 silencing at pH 6.8, with very low BRD4 expression at the 15 nM siRNA dose (Figure 4A). In comparison, for the cells treated with the siBRD4-loaded NPs at pH 7.4, there is still a high level of BRD4 expression with 20 nM siRNA (Figure 4B). Similar results can be also found in the immunofluorescence staining analysis (Figures 4C–4E). Red fluorescence corresponding to the residual BRD4 can still be observed in the cells treated with the siRNA-loaded NPs at pH 7.4 (Figure 4D), but very weak fluorescence is observable in the cells treated with the same NPs at pH 6.8 (Figure 4E). With this efficient BRD4 silencing at pH 6.8, the percentage of apoptotic (Annexin-V positive) or necrotic (Annexin V-negative and 7-ADD-postivie) cells increases markedly to 39.5% or 36.3%, respectively (Figures 4F and 4G), which is around 2.5-flod higher than that of the cells treated with the siBRD4 NPs at pH 7.4. In addition, the BRD4 silencing also induces significant inhibition of cell proliferation. Only 20% of the LNCaP cells treated with NPs for 24 h at pH 6.8 are alive after 6 days (Figure 4H), while there is about 8-fold increase in the number of cells for the pH 7.4 group.
Figure 4.
(A, B) Western blot analysis of BRD4 expression in LNCaP cells treated with siBRD4-loaded TCPA2-NPs at pH 6.8 (A) and 7.4 (B). (C-E) Immunofluorescence analysis of BRD4 expression (red fluorescence) in LNCaP cells treated with siBRD4-loaded TCPA2-NPs (20 nM siRNA dose) at pH 6.8 vs. 7.4. Blank: cells incubated in free culture medium. (F) Flow cytometry analysis and (G) quantification of apoptosis of LNCaP cells treated with siBRD4-loaded TCPA2-NPs at a 20 nM siRNA dose. (H) Proliferation profile of LNCaP cells treated with siBRD4-loaded TCPA2-NPs at a 20 nM siRNA dose. The siLuc-loaded TCPA2-NPs were used as the control in these experiments.
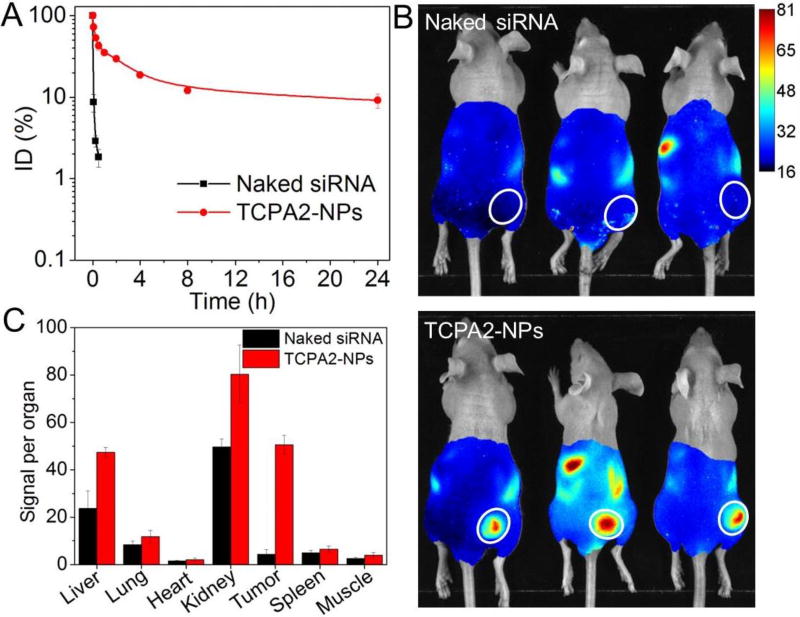
After validating the efficient gene silencing of the TCPA2-NPs in vitro, we next assessed their pharmacokinetics (PK) and biodistribution (BioD). PK was examined by intravenous injection of DY677-siRNA-loaded NPs to health mice (1 nmol siRNA dose per mouse, n = 3). As shown in Figure 5A, with the protection of PEG outer layer,52, 55, 59 the TCPA2-NPs show long blood circulation with a half-life (t1/2) of around 4.38 h. In contrast, the naked siRNA or siRNA-TCPA2 complexes (Figure S13) are rapidly cleared from the blood and their blood t1/2 is less than 10 min. The BioD was evaluated by intravenously injecting DY677-siRNA-loaded NPs into LNCaP xenograft tumor-bearing mice. Figure 5B shows the fluorescent image of the mice at 24 h post injection. Given the long blood circulation characteristic of the TCPA2-NPs, they showed a much higher tumor accumulation than naked siRNA or the siRNA-TCPA2 complexes. The tumors and major organs were harvested at 24 h post injection (Figure S14) and the quantification of BioD is shown in Figure 5C. The TCPA2-NPs demonstrated an approximately 10-fold higher siRNA accumulation in tumors than the naked siRNA.
Figure 5.
(A) Blood circulation profile of naked DY677-siRNA vs. DY677-siRNA-loaded TCPA2-NPs. (B) Overlaid fluorescent image of LNCaP xenograft tumor-bearing nude mice at 24 h post injection of naked DY677-siRNA vs. DY677-siRNA-loaded TCPA2-NPs. Tumors are indicated by ellipses. (C) Biodistribution of the NPs in the tumors and major organs of LNCaP xenograft tumor-bearing nude mice sacrificed at 24 h post injection of the DY677-siRNA-loaded TCPA2-NPs.
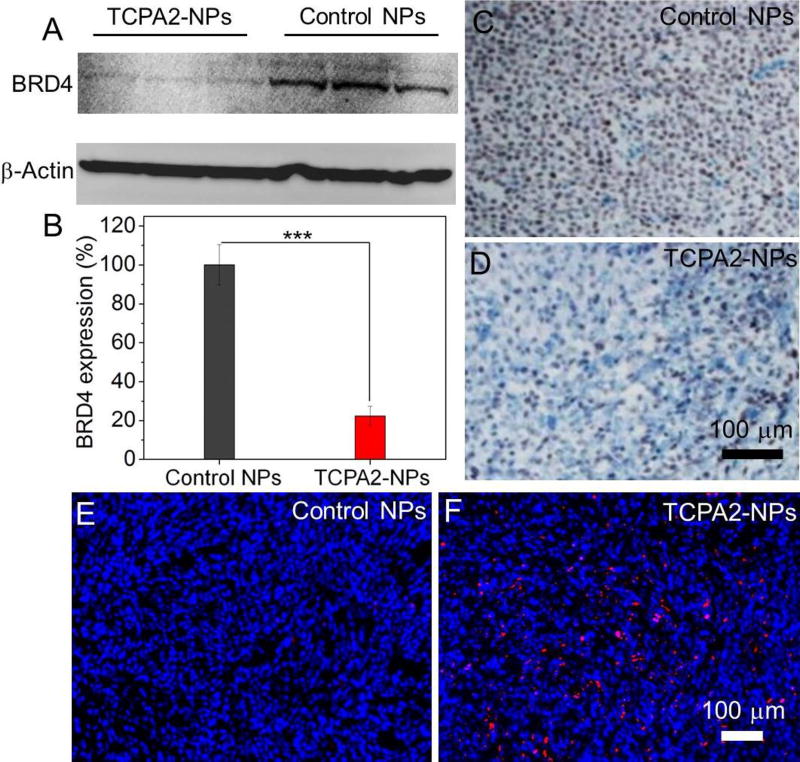
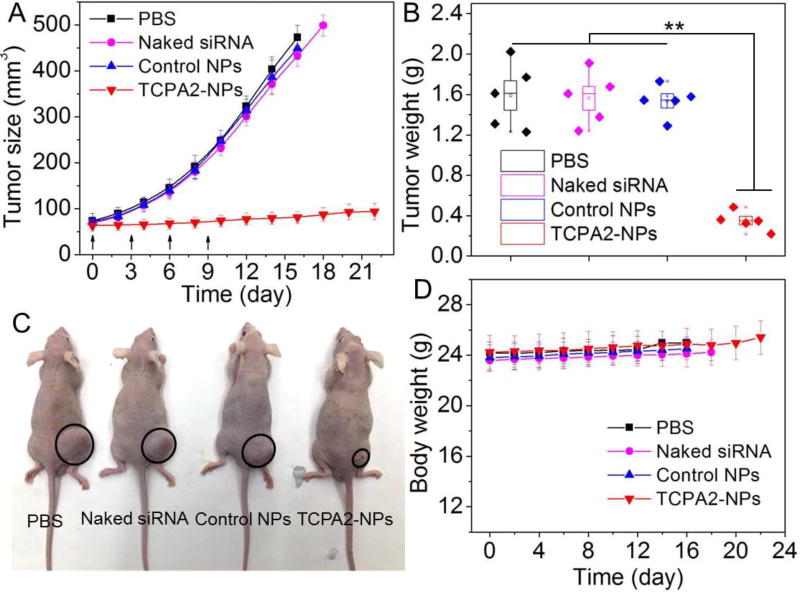
With these promising in vitro and PK/BioD results, we further evaluated whether our multistaged siRNA delivery platform can silence BRD4 expression in vivo and show anticancer effect. To do this, the siBRD4-loaded NPs were intravenously injected into the LNCaP xenograft tumor-bearing mice (1 nmol siRNA dose per mouse, n = 3) for three consecutive days. Western blot analysis of the tumor tissue shows that the siBRD4-loaded NPs led to ~78% knockdown in BRD4 expression compared to the control NPs loaded with siLuc (Figures 6A and 6B). A similar tendency can be also found in the immunohistochemistry staining analysis (Figures 6C and 6D). Along with the suppressed BRD4 expression, there is a significant increase in tumor cell apoptosis confirmed by TUNEL staining (Figures 6E and 6F). To confirm whether the NP-mediated BRD4 silencing has an anticancer effect, the siBRD4-loaded NPs were intravenously injected into the LNCaP xenograft tumor-bearing mice once every three days at a 1 nmol siRNA dose per mouse (n = 5). After four consecutive injections, the tumor growth was significantly inhibited compared to the mice treated with PBS, naked siBRD4, or siLuc-loaded NPs (Figure 7). There is less than 1.5-fold increase (from ~63 to ~93 mm3) in tumor size of the mice treated with the siBRD4-loaded NPs at day 21 (Figure 7A). For the mice in the three control groups, their tumor size (Figure 7A) and tumor weight (Figure 7B) are more than 4-fold larger than those in the siBRD4-loaded NP group. Noteworthy, the siBRD4-loaded NPs showed no obvious influence on mouse body weight (Figure 7D). Moreover, to further evaluate the potential in vivo side effects of TCPA2-NPs, healthy mice received the injection of siBRD4-loaded NPs (1 nmol siRNA dose per mouse, n = 3). Blood serum analysis showed that TNF-α, IFN-γ, IL-6, and IL-12 levels were in the normal range at 24 h post injection (Figure S15). After three daily injections, no noticeable histological changes were noticed in the tissues from heart, liver, spleen, lung or kidney (Figure S16). All of these results indicate the good biocompatibility of this NP platform.
Figure 6.
(A, B) Western blot and (C, D) immunohistochemistry analysis of BRD4 expression in the LNCaP tumor tissue after systemic treatment by control NPs and siBRD4-loaded TCPA2-NPs. *** P < 0.001 (E, F) TUNEL staining of the LNCaP tumor tissue after systemic treatment by control NPs and siBRD4 TCPA2-NPs. TUNEL-positive apoptotic cells were stained with red fluorescence. The siLuc-loaded TCPA2-NPs were used as control NPs.
Figure 7.
(A) Tumor size and (B) tumor weight of the LNCaP xenograft tumor-bearing nude mice (n = 5) after systemic treatment by PBS, naked siBRD4, control NPs, and siBRD4-loaded TCPA2-NPs. Intravenous injections are indicated by the arrows. ** P < 0.01 (C) Representative photograph of the LNCaP xenograft tumor-bearing nude mice in each group at day 16. Tumors are indicated by ellipses. (D) Body weight of the LNCaP xenograft tumor-bearing nude mice in each group. The siLuc-loaded TCPA2-NPs were used as control NPs.
CONCLUSION
We have successfully developed a unique and robust TME pH-responsive multistaged NP platform for systemic and targeted siRNA delivery to solid tumors. In vivo results show that this multistaged NP platform with long blood circulation can first highly accumulate at tumor site and then rapidly respond to TME pH to expose the siRNA-TCPA complexes, which subsequently target and penetrate tumor cells to achieve efficient cytosolic siRNA transport and gene silencing. Along the multistaged delivery concept reported herein, other TME-responsive (e.g., enzyme- and hypoxia-sensitive) NPs formulated with various biomacromolecules (e.g., siRNA, miRNA, mRNA, and protein) could be further pursued for the treatment of a wide range of solid tumors.
Supplementary Material
Acknowledgments
This work was supported by the NIH grants CA200900 (J.S.), R00CA160350 (J.S.), and HL127464 (O.C.F.); the David H. Koch-Prostate Cancer Foundation (PCF) Program in Cancer Nanotherapeutics (O.C.F.); the Movember-PCF Challenge Award (O.C.F. and J.S.); the PCF Young Investigator Award (J.S.); and the National Research Foundation of Korea grant K1A1A2048701 (O.C.F.). We also thank Dr. Yingjie Xu from Boston Children’s Hospital for help with the apoptosis experiment.
Footnotes
ASSOCIATED CONTENT
Supporting Information. Synthesis and characterization of the polymers, characterization of the Cy5.5-labelled NPs, cell viability, BRD4 expression in various PCa cell lines, PK and BioD of the siRNA-TCPA2 complexes, histology, and immune response are presented in the Supporting Information. This material is available free of charge via the Internet at http://pubs.acs.org.
O.C.F. has financial interest in Selecta Biosciences, Tarveda Therapeutics, and Placon Therapeutics.
References
- 1.Farokhzad OC, Langer R. ACS Nano. 2009;3:16–20. doi: 10.1021/nn900002m. [DOI] [PubMed] [Google Scholar]
- 2.Petros RA, DeSimone JM. Nat. Rev. Drug Discov. 2010;9:615–627. doi: 10.1038/nrd2591. [DOI] [PubMed] [Google Scholar]
- 3.Yu MK, Park J, Jon S. Theranostics. 2012;2:3–44. doi: 10.7150/thno.3463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fang RH, Hu C-MJ, Luk BT, Gao W, Copp JA, Tai Y, O’Connor DE, Zhang L. Nano Lett. 2014;14:2181–2188. doi: 10.1021/nl500618u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mo R, Jiang T, Di J, Tai W, Gu Z. Chem. Soc. Rev. 2014;43:3595–3629. doi: 10.1039/c3cs60436e. [DOI] [PubMed] [Google Scholar]
- 6.Shi J, Kantoff PW, Wooster R, Farokhzad OC. Nat. Rev. Cancer. 2017;17:20–37. doi: 10.1038/nrc.2016.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pacardo DB, Ligler FS, Gu Z. Nanoscale. 2015;7:3381–3391. doi: 10.1039/c4nr07677j. [DOI] [PubMed] [Google Scholar]
- 8.Wang S, Huang P, Chen X. ACS Nano. 2016;10:2991–2994. doi: 10.1021/acsnano.6b00870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang S, Huang P, Chen X. Adv. Mater. 2016;28:7340–7364. doi: 10.1002/adma.201601498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen B, Dai W, He B, Zhang H, Wang X, Wang Y, Zhang Q. Theranostics. 2017;7:538–558. doi: 10.7150/thno.16684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tong R, Chiang HH, Kohane DS. Proc. Natl. Acad. Sci. U S A. 2013;110:19048–19053. doi: 10.1073/pnas.1315336110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ragelle H, Danhier F, Préat V, Langer R, Anderson DG. Expert Opin. Drug Deliv. 2016:1–14. doi: 10.1080/17425247.2016.1244187. [DOI] [PubMed] [Google Scholar]
- 13.Polyak K, Haviv I, Campbell IG. Trends Genet. 2009;25:30–38. doi: 10.1016/j.tig.2008.10.012. [DOI] [PubMed] [Google Scholar]
- 14.Albini A, Sporn MB. Nat. Rev. Cancer. 2007;7:139–147. doi: 10.1038/nrc2067. [DOI] [PubMed] [Google Scholar]
- 15.Danhier F, Feron O, Preat V. J. Control. Release. 2010;148:135–146. doi: 10.1016/j.jconrel.2010.08.027. [DOI] [PubMed] [Google Scholar]
- 16.Quail DF, Joyce JA. Nat. Med. 2013;19:1423–1437. doi: 10.1038/nm.3394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Miao L, Wang Y, Lin CM, Xiong Y, Chen N, Zhang L, Kim WY, Huang L. J. Control. Release. 2015;217:27–41. doi: 10.1016/j.jconrel.2015.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wong C, Stylianopoulos T, Cui J, Martin J, Chauhan VP, Jiang W, Popović Z, Jain RK, Bawendi MG, Fukumura D. Proc. Natl. Acad. Sci. U S A. 2011;108:2426–2431. doi: 10.1073/pnas.1018382108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Du J-Z, Du X-J, Mao C-Q, Wang J. J. Am. Chem. Soc. 2011;133:17560–17563. doi: 10.1021/ja207150n. [DOI] [PubMed] [Google Scholar]
- 20.Ju C, Mo R, Xue J, Zhang L, Zhao Z, Xue L, Ping Q, Zhang C. Angew. Chem. Int. Ed. 2014;53:6253–6258. doi: 10.1002/anie.201311227. [DOI] [PubMed] [Google Scholar]
- 21.Li L, Sun W, Zhong J, Yang Q, Zhu X, Zhou Z, Zhang Z, Huang Y. Adv. Funct. Mater. 2015;25:4101–4113. [Google Scholar]
- 22.Li H-J, Du J-Z, Du X-J, Xu C-F, Sun C-Y, Wang H-X, Cao Z-T, Yang X-Z, Zhu Y-H, Nie S, Wang J. Proc. Natl. Acad. Sci. U S A. 2016;113:4164–4169. doi: 10.1073/pnas.1522080113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li H-J, Du J-Z, Liu J, Du X-J, Shen S, Zhu Y-H, Wang X, Ye X, Nie S, Wang J. ACS Nano. 2016;10:6753–6761. doi: 10.1021/acsnano.6b02326. [DOI] [PubMed] [Google Scholar]
- 24.Haynes MT, Huang L. Mol. Ther. 2016;24:849–851. doi: 10.1038/mt.2016.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xu R, Zhang G, Mai J, Deng X, Segura-Ibarra V, Wu S, Shen J, Liu H, Hu Z, Chen L, Huang Y, Koay E, Huang Y, Liu J, Ensor JE, Blanco E, Liu X, Ferrari M, Shen H. Nat. Biotech. 2016;34:414–418. doi: 10.1038/nbt.3506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yin H, Kanasty RL, Eltoukhy AA, Vegas AJ, Dorkin JR, Anderson DG. Nat. Rev. Genet. 2014;15:541–555. doi: 10.1038/nrg3763. [DOI] [PubMed] [Google Scholar]
- 27.Zuckerman JE, Davis ME. Nat. Rev. Drug Discov. 2015;14:843–856. doi: 10.1038/nrd4685. [DOI] [PubMed] [Google Scholar]
- 28.Tseng Y-C, Mozumdar S, Huang L. Adv. Drug Deliv. Rev. 2009;61:721–731. doi: 10.1016/j.addr.2009.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang Y, Satterlee A, Huang L. Mol. Ther. 2012;20:1298–1304. doi: 10.1038/mt.2012.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kanasty R, Dorkin JR, Vegas A, Anderson D. Nat. Mater. 2013;12:967–977. doi: 10.1038/nmat3765. [DOI] [PubMed] [Google Scholar]
- 31.Yang X-Z, Du J-Z, Dou S, Mao C-Q, Long H-Y, Wang J. ACS Nano. 2012;6:771–781. doi: 10.1021/nn204240b. [DOI] [PubMed] [Google Scholar]
- 32.Sun C-Y, Shen S, Xu C-F, Li H-J, Liu Y, Cao Z-T, Yang X-Z, Xia J-X, Wang J. J. Am. Chem. Soc. 2015;137:15217–15224. doi: 10.1021/jacs.5b09602. [DOI] [PubMed] [Google Scholar]
- 33.Gujrati M, Vaidya AM, Mack M, Snyder D, Malamas A, Lu ZR. Adv. Healthc. Mater. 2016;5:2882–2895. doi: 10.1002/adhm.201600677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hatakeyama H, Akita H, Ito E, Hayashi Y, Oishi M, Nagasaki Y, Danev R, Nagayama K, Kaji N, Kikuchi H, Baba Y, Harashima H. Biomaterials. 2011;32:4306–4316. doi: 10.1016/j.biomaterials.2011.02.045. [DOI] [PubMed] [Google Scholar]
- 35.Wang H-X, Yang X-Z, Sun C-Y, Mao C-Q, Zhu Y-H, Wang J. Biomaterials. 2014;35:7622–7634. doi: 10.1016/j.biomaterials.2014.05.050. [DOI] [PubMed] [Google Scholar]
- 36.Wang Y, Zhou K, Huang G, Hensley C, Huang X, Ma X, Zhao T, Sumer BD, DeBerardinis RJ, Gao J. Nat. Mater. 2014;13:204–212. doi: 10.1038/nmat3819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhou K, Wang Y, Huang X, Luby-Phelps K, Sumer BD, Gao J. Angew. Chem. Int. Ed. 2011;50:6109–6114. doi: 10.1002/anie.201100884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chen K, Chen X. Theranostics. 2011;1:189–200. doi: 10.7150/thno/v01p0189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Danhier F, Le Breton A, Préat V. Mol. Pharm. 2012;9:2961–2973. doi: 10.1021/mp3002733. [DOI] [PubMed] [Google Scholar]
- 40.Fuchs SM, Raines RT. Cell. Mol. Life Sci. 2006;63:1819–1822. doi: 10.1007/s00018-006-6170-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jang MK, Mochizuki K, Zhou M, Jeong H-S, Brady JN, Ozato K. Mol. Cell. 2005;19:523–534. doi: 10.1016/j.molcel.2005.06.027. [DOI] [PubMed] [Google Scholar]
- 42.Yang Z, Yik JHN, Chen R, He N, Jang MK, Ozato K, Zhou Q. Mol. Cell. 2005;19:535–545. doi: 10.1016/j.molcel.2005.06.029. [DOI] [PubMed] [Google Scholar]
- 43.Taylor BS, Schultz N, Hieronymus H, Gopalan A, Xiao Y, Carver BS, Arora VK, Kaushik P, Cerami E, Reva B, Antipin Y, Mitsiades N, Landers T, Dolgalev I, Major JE, Wilson M, Socci ND, Lash AE, Heguy A, Eastham JA, Scher HI, Reuter VE, Scardino PT, Sander C, Sawyers CL, Gerald WL. Cancer Cell. 2010;18:11–22. doi: 10.1016/j.ccr.2010.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chen CD, Welsbie DS, Tran C, Baek SH, Chen R, Vessella R, Rosenfeld MG, Sawyers CL. Nat. Med. 2004;10:33–39. doi: 10.1038/nm972. [DOI] [PubMed] [Google Scholar]
- 45.Visakorpi T, Hyytinen E, Koivisto P, Tanner M, Keinanen R, Palmberg C, Palotie A, Tammela T, Isola J, Kallioniemi O-P. Nat. Genet. 1995;9:401–406. doi: 10.1038/ng0495-401. [DOI] [PubMed] [Google Scholar]
- 46.Delmore Jake E, Issa Ghayas C, Lemieux Madeleine E, Rahl Peter B, Shi J, Jacobs Hannah M, Kastritis E, Gilpatrick T, Paranal Ronald M, Qi J, Chesi M, Schinzel Anna C, McKeown Michael R, Heffernan Timothy P, Vakoc Christopher R, Bergsagel PL, Ghobrial Irene M, Richardson Paul G, Young Richard A, Hahn William C, Anderson Kenneth C, Kung Andrew L, Bradner James E, Mitsiades Constantine S. Cell. 2011;146:904–917. doi: 10.1016/j.cell.2011.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Filippakopoulos P, Qi J, Picaud S, Shen Y, Smith WB, Fedorov O, Morse EM, Keates T, Hickman TT, Felletar I, Philpott M, Munro S, McKeown MR, Wang Y, Christie AL, West N, Cameron MJ, Schwartz B, Heightman TD, La Thangue N, French CA, Wiest O, Kung AL, Knapp S, Bradner JE. Nature. 2010;468:1067–1073. doi: 10.1038/nature09504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wu S-Y, Lee AY, Lai H-T, Zhang H, Chiang C-M. Mol. Cell. 2013;49:843–857. doi: 10.1016/j.molcel.2012.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Asangani IA, Dommeti VL, Wang X, Malik R, Cieslik M, Yang R, Escara-Wilke J, Wilder-Romans K, Dhanireddy S, Engelke C, Iyer MK, Jing X, Wu Y-M, Cao X, Qin ZS, Wang S, Feng FY, Chinnaiyan AM. Nature. 2014;510:278–282. doi: 10.1038/nature13229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Griffiths JR, McIntyre DJ, Howe FA, Stubbs M. Novartis Found. Symp. 2001;240:46–62. doi: 10.1002/0470868716.ch4. discussion 62, 7, 152–153. [DOI] [PubMed] [Google Scholar]
- 51.Urano Y, Asanuma D, Hama Y, Koyama Y, Barrett T, Kamiya M, Nagano T, Watanabe T, Hasegawa A, Choyke PL, Kobayashi H. Nat. Med. 2009;15:104–109. doi: 10.1038/nm.1854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhu X, Xu Y, Solis LM, Tao W, Wang L, Behrens C, Xu X, Zhao L, Liu D, Wu J, Zhang N, Wistuba II, Farokhzad OC, Zetter BR, Shi J. Proc. Natl. Acad. Sci. U S A. 2015;112:7779–7784. doi: 10.1073/pnas.1505629112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lim Y-b, Lee E, Lee M. ngew. Chem. Int. Ed. 2007;46:9011–9014. doi: 10.1002/anie.200702732. [DOI] [PubMed] [Google Scholar]
- 54.Xu X-D, Chen J-X, Cheng H, Zhang X-Z, Zhuo R-X. Polym. Chem. 2012;3:2479–2486. [Google Scholar]
- 55.Xu X, Wu J, Liu Y, Yu M, Zhao L, Zhu X, Bhasin S, Li Q, Ha E, Shi J, Farokhzad OC. Angew. Chem. Int. Ed. 2016;55:7091–7094. doi: 10.1002/anie.201601273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ren Y, Cheung HW, von Maltzhan G, Agrawal A, Cowley GS, Weir BA, Boehm JS, Tamayo P, Karst AM, Liu JF, Hirsch MS, Mesirov JP, Drapkin R, Root DE, Lo J, Fogal V, Ruoslahti E, Hahn WC, Bhatia SN. Sci. Transl. Med. 2012;4:147ra112. doi: 10.1126/scitranslmed.3003778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Xu X, Wu J, Liu Y, Saw PE, Tao W, Yu M, Zope H, Si M, Victorious A, Rasmussen J, Ayyash D, Farokhzad OC, Shi J. ACS Nano. 2017;11:2618–2627. doi: 10.1021/acsnano.6b07195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sutherland M, Gordon A, Shnyder SD, Patterson LH, Sheldrake HM. Cancers. 2012;4:1106–1145. doi: 10.3390/cancers4041106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Knop K, Hoogenboom R, Fischer D, Schubert US. Angew. Chem. Int. Ed. 2010;49:6288–6308. doi: 10.1002/anie.200902672. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.