Abstract
Background
Keratoconus normally presents as a sporadic disease. Although different studies have found sequence variants of the visual system homeobox 1 (VSX1) gene associated with keratoconus in humans, no research has detected such variants in sporadic keratoconus patients from China. To investigate the possibility of VSX1 being a candidate susceptibility gene for Chinese patients with sporadic keratoconus, we performed sequence screening of this gene in such patients.
Methods
Whole DNA was obtained from the leukocytes in the peripheral venous blood of 50 patients with sporadic keratoconus and 50 control subjects without this ocular disorder. Polymerase chain reaction single-strand conformation polymorphism analysis and direct DNA sequencing technology were used to detect sequence variation in the five exons and splicing regions of the introns of the VSX1 gene. The sequencing results were analyzed using DNAstar software.
Results
One novel missense heterozygous sequence variant (p.Arg131Pro) was found in the first exon of the VSX1 gene in one keratoconus patient. Another heterozygous sequence variant (p.Gly160Val) in the second exon was found in two keratoconus patients. These variants were not detected in the control subjects. In the third intron of the VSX1 gene, c.8326G > A nucleotide substitution (including heterozygous and homozygous change) was also discovered. The frequency of this variation did not differ significantly between patients and controls, it should belong to single-nucleotide polymorphism of the VSX1 gene. Bioinformatic analysis also predicted that one missense sequence variation (p.Arg131Pro) may not cause a pathogenic change.
Conclusions
In this study, we added one novel missense sequence variation (p.Arg131Pro) in the coding region of the VSX1 gene to the range of VSX1 coding region variations observed in patients with sporadic keratoconus from China. Our work suggests that VSX1 sequence variants might be involved in the pathogenesis of sporadic keratoconus, but their precise role in disease causation requires further investigation.
Keywords: Sporadic keratoconus, VSX1 gene, Sequence variant, Single-nucleotide polymorphism
Background
Keratoconus is a congenital disease involving noninflammatory corneal ectasia. Its prevalence is estimated to be 1/2000 in the general population, and the incidence ratio between males and females is 1.7:1 [1, 2]. Although most keratoconus cases are sporadic, about 6–10% of cases have been reported to have a positive family history. Keratoconus can exhibit dominant or recessive inheritance; with autosomal dominant inheritance, the disease exhibits variable phenotypes with incomplete penetrance [3, 4]. The pathological features of keratoconus include decreased corneal collagen or the abnormal distribution of collagen fiber, which reduce the mechanical resistance of the cornea and therefore cause the center of the cornea to protrude, leading to a thin conical shape. Slit lamp microscope examinations of keratoconus patients have demonstrated Vogt’s line, Fleischer’s ring, and corneal scarring [5–7]. The onset of the disease occurs during puberty, involving both eyes, and with highly irregular astigmatism. In the late phase, patients show acute corneal edema and scar formation, along with significant vision loss. Although several treatment modalities are available, severe keratoconus remains an indication for corneal transplantation [8–10].
Genome-wide linkage analyses have identified several chromosomal loci and genes that may be related to keratoconus; some of these were eventually ruled out, while for others, an association with this disease could not be confirmed. One of the candidate genes strongly implicated in keratoconus is visual system homeobox gene 1 (VSX1) localized to chromosome 20 p11–q11 [11–14]. The VSX1 gene encodes a paired-like homeodomain transcription factor. It is associated with eye development and was first isolated and cloned from a cDNA library of the retina of adult goldfish. In the human body, the VSX1 gene is expressed in the embryonic craniofacial region, the granular layer of adult retina, and corneal tissue. The human VSX1 gene has five exons and encodes a 365-amino-acid protein with a homeobox DNA binding domain and a CVC domain, being highly conserved among vertebrates. Barbaco et al. [15] reported that the VSX1 gene was activated in corneal stroma during the process of corneal wound repair in vitro and in vivo, accompanied by the transformation of corneal stromal cells into myofibroblasts. These results implied that VSX1 gene variants may play an important role in the development of keratoconus. Several variants of the VSX1 gene [16–19] have been reported from various parts of the world, but a definitive pathogenic role of these variants in keratoconus has not yet been established because segregation of these variants was also seen in some unaffected individuals.
To our knowledge, this is the first study to perform screening for variants of the VSX1 gene in the Chinese population and to explore their role in keratoconus. Nucleotide variations in the five exons and the splicing regions of introns of the VSX1 gene were examined by polymerase chain reaction single-strand conformation polymorphism (PCR-SSCP) analysis and direct DNA sequencing in 50 Chinese patients with keratoconus, which were compared with the sequences in 50 controls.
Methods
Subjects and clinical examination
All sporadic keratoconus patients and control subjects were identified at the Department of Ophthalmology, Taizhou Municipal Hospital. A total of 50 patients were diagnosed with sporadic keratoconus during the period of March 2013 to July 2015, including 29 males and 21 females, aged from 15 to 35 years old. The diagnosis was based on symptoms and the results of slit lamp microscope examinations; the diagnostic features included progressive curvature of the anterior corneal surface steepening, corneal thinning in the same region, Fleischer’s ring, and Vogt’s stripes, combined with the corneal surface topography and a caudal surface of the cornea thicker than 20 μm. Patients were considered as sporadic cases after examining the immediate family members and identifying the patient as an isolated case of keratoconus. Another 50 healthy people who underwent detailed ocular and ophthalmological examinations that ruled out any ophthalmic diseases were selected as a control group to determine whether the detected variation was SNP of VSX1 gene. The control subjects included 31 males and 19 females, aged from 20 to 36 years old. The age and gender generally matched between the two groups and systemic physical examinations were performed in both groups to rule out other concurrent diseases. This study followed the Declaration of Helsinki and was reviewed by the ethics committee of Taizhou Municipal Hospital. Written informed consent was obtained from all of the subjects in this study.
Genetic study
Three milliliters of peripheral venous blood was extracted from the subjects and stored in EDTA anticoagulation tubes. Genomic DNA was isolated from blood leukocytes using the Dzup (Blood) Genomic DNA Isolation Reagent (B518205; Sangon Biotech Co., Ltd., Shanghai, China), in accordance with the manufacturer’s standard methods.
Based on previous studies [20, 21] and the DNA sequence of the human VSX1 gene from the NCBI database, primers were designed for the five exons of this gene. These primers were synthesized by Sangon Biotech Co., Ltd. (Shanghai, China). Sequences of the primers are listed in Table 1.
Table 1.
List of primers and PCR amplification conditions used for VSX1 gene amplification
| Exon | Sequence of primers (5′-3′) | Annealing temperature (°C) | Product length (bp) |
|---|---|---|---|
| 1 | CAGCTGATTGGAGCCCTTC(sense) | 62 | 599 |
| CTCAGAGCCTAGGGGACAGG(antisense) | |||
| 2 | CTTAAGTACCCAAGAGGTTCATAACT(sense) | 62 | 269 |
| GAAACCACTGGGCCTGCTATCAT(antisense) | |||
| 3 | AAGCAGGCACGGTGGTCCTTA(sense) | 63 | 293 |
| AAGGGACTGCTGATTGGCTCACT(antisense) | |||
| 4 | GCTCGGGAGAGAAGATCC(sense) | 62 | 399 |
| TCAGTAAACTGACGTTGCTTG(antisense) | |||
| 5 | GGAAATTTACTTCATTGCTGAATTT(sense) | 60 | 495 |
| TGGCATTGCATTTTATCTTGACA(antisense) |
The HBP 220 PCR cycler (HYBAID, England) was used for a 50-μL PCR reaction system containing 200 ng of DNA template, 5 μL of 10× buffer solution, 1.5 mmol/L MgCl2, 0.25 μmol/L primers, 100 μmol/L of each dNTP, and 2 U of Taq DNA polymerase (Sangon Biotech Co., Ltd., Shanghai, China). The PCR procedure was as follows: 94 °C for 5 min; 35 amplification cycles of denaturation at 94 °C for 45 s, annealing for 45 s, and extension at 72 °C for 50 s; and finally extension at 72 °C for 10 min. PCR products were examined by 2% agarose gel electrophoresis and EB (Ethidium Bromide) staining. The annealing temperatures of each exon and the fragment sizes of PCR products are listed in Table 1.
An 8% (49:1) nondenaturing polyacrylamide gel with 5% glycerol was used for SSCP analysis and silver staining. Here, 5-μL PCR products mixed with sample buffer (90% formamide, 0.05% bromophenol blue, and 0.05% xylene blue) were processed for denaturation at 95 °C for 10 min, placed in an ice bath, and then subjected to electrophoresis (100 V) at room temperature for 5 to 6 h (when xylene blue reached the bottom of the gel). Then, the gel was fixed in 10% ethanol for 5 min and soaked in 1% silver nitrate for 5 min, followed by staining in 0.012 mol/L silver nitrate and 0.05% formaldehyde for 10 min. Next, the gel was processed in 0.28 mol/L sodium carbonate, 0.1% formaldehyde, and 0.1 g/L thiosulfate sodium for colorization, which was stopped by immersion in 10% acetic acid.
DNA with an abnormal banding pattern in electrophoresis was detected by SSCP and silver staining analysis. Its PCR product was then purified using isopropyl alcohol and bidirectionally screened to identify the sequence variant using the ABI3730XL DNA Analyzer (Sangon Biotech Co., Ltd., Shanghai, China). Nucleotide sequences were then compared with the published VSX1 cDNA sequence (GenBank NM_014588).
Bioinformatic analysis
The in silico prediction programs Polyphen-2 (http://genetics.bwh.harvard.edu/pph2/) and PROVEAN (http://provean.jcvi.org/index.php) were used to predict the effect of the VSX1 gene sequence variants on protein structure and function [22–24].
Statistics
To determine statistically significant differences between the groups, data analysis was performed using SPSS version 18.0. Continuous data are displayed here as mean ± standard deviation. Differences between two groups were examined by t-test. Repeated measure ANOVA was used to analyze the differences among multiple groups. Values of P < 0.05 were considered statistically significant.
Results
PCR-SSCP results
All exons of the VSX1 gene in sporadic keratoconus patients were amplified by PCR. The PCR products showed high specificity and efficiency in amplification by 2% agarose gel electrophoresis, with a single band corresponding to the correct fragment length (Fig. 1). This suggested that there were no sequence variants with a large insertion or deletion in the five exons of the VSX1 gene in the sporadic keratoconus patients. By SSCP analysis and silver staining, four types of abnormal banding pattern were clearly shown in the electrophoresis gels, suggesting the presence of four sequence variants in the five exons of the VSX1 gene.
Fig. 1.
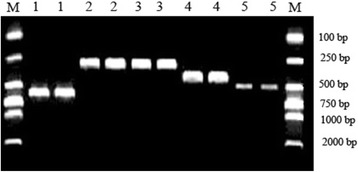
Electrophoresis pattern of PCR products of the VSX1 gene. M: DNA markers, 1: first exon (599 bp), 2: second exon (269 bp), 3: third exon (293 bp), 4: fourth exon (399 bp), 5: fifth exon (495 bp)
DNA sequencing results
The 50 sporadic keratoconus patients and 50 control subjects recruited for this study underwent screening of the entire coding region and exon–intron junctions of the VSX1 gene by direct sequencing of DNA. The DNA sequencing results confirmed the abnormal banding patterns found in the SSCP analysis and silver staining as described above. The results revealed four nucleotide sequence variants of the VSX1 gene in Chinese sporadic keratoconus patients, including two missense sequence variants and one SNP change (heterozygous and homozygous) (Table 2).
Table 2.
List of sequence variants of the VSX1 gene in Chinese sporadic keratoconus patients
| Nucleotide change | Amino acid change | Change site | Change type | Keratoconus (n = 50) | Controls (n = 50) |
|---|---|---|---|---|---|
| c.5427G > C | p.Arg131Pro | First exon | Heterozygous missense variant | 1 (2.0%) | 0 (0.0%) |
| c.7672G > T | p.Gly160Val | Second exon | Heterozygous missense variant | 2 (4.0%) | 0 (0.0%) |
| c.8326G > A | Noncoding region | Third intron | Heterozygous SNP variant | 3 (6.0%) | 4 (8.0%) |
| c.8326G > A | Noncoding region | Third intron | Homozygous SNP variant | 2 (4.0%) | 1 (2.0%) |
The sequence variant c.5427G > C was found in the first exon of the VSX1 gene in one case of sporadic keratoconus. This involved a change in the codon from CGC to CCC, leading to a missense substitution of arginine to proline at codon 131 (p.Arg131Pro sequence variant). p.Arg131Pro was not detected in any control subjects (Fig. 2) and had not previously been identified. We registered it in a public database at https://www.ncbi.nlm.nih.gov/projects/SNP/ (ID: rs267597889). Using the PolyPhen-2 tool, p.Arg131Pro was assessed as being benign, in terms of its phenotypic effects, with a score of 0.344; using the PROVEAN tool, it was assessed as being neutral, with a score of −0.048. Both tools predicted that the replaced amino acid would have no adverse effects and would be tolerated.
Fig. 2.
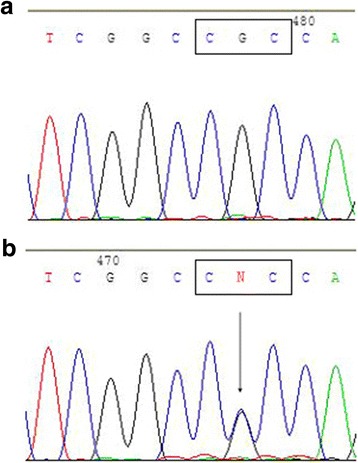
The p.Arg131Pro variant of the VSX1 gene. The sequence of a normal control (a) included the nucleotides CGC at codon 131 in exon 1 of VSX1. The missense variant sequence (b) showed a G to C (c. 5427 G > C) heterozygous transition at the second nucleotide position in this codon (arrow), which results in a change from arginine (CGC) to proline (CCC) (p.Arg131Pro)
The other missense sequence variant c.7672G > T was found in the second exon of the VSX1 gene in two cases of sporadic keratoconus. This involves a change in codon 160 from GGC to GTC, leading to a missense substitution of glycine to valine (p.Gly160Val sequence variant) (Fig. 3). No similar change was found in any of the 50 normal controls.
Fig. 3.
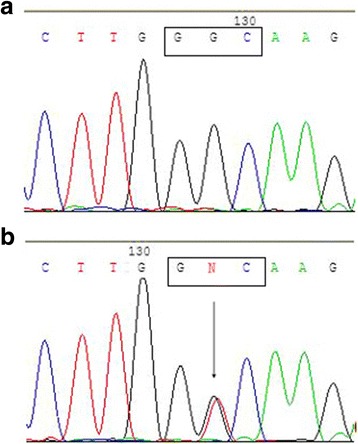
The p.Gly160Val variant of the VSX1 gene. The sequence of a normal control (a) included the nucleotides GGC at codon 160 in the second exon of VSX1. The missense variant sequence (b) showed a G to T (c. 7672 G > T) heterozygous transition at the second nucleotide position in this codon (arrow), which results in a change from glycine (GGC) to valine (GTC) (p.Gly160Val)
In the third intron of the VSX1 gene, the substitution of G > A at nucleotide position 8326 (c.8326G > A) was identified, which was present in heterozygous form in three cases of sporadic keratoconus and in four controls (Fig. 4). It was also present in homozygous form in two sporadic keratoconus patients and one control (Fig. 5). There was no significant difference in the incidence of c.8326G > A between the two groups.
Fig. 4.
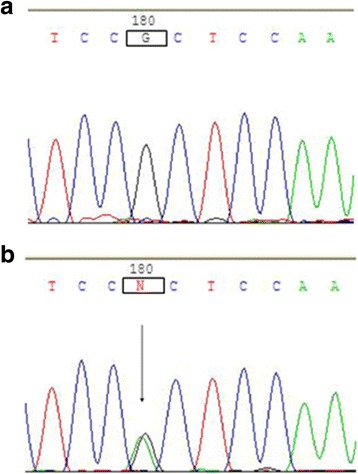
The c.8326G > A heterozygous change of the VSX1 gene. The sequence of a normal control (a) included the nucleotide G at position 8326 in the third intron of VSX1. The variant sequence (b) showed a G to A heterozygous transition (arrow), which results in a c.8326G > A SNP change
Fig. 5.
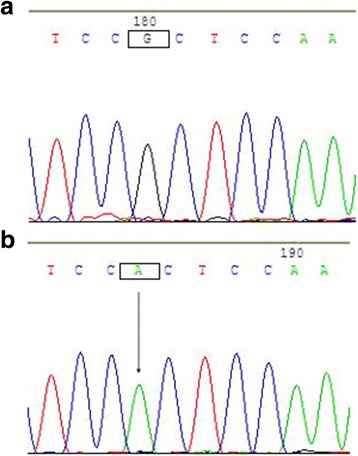
The c.8326G > A homozygous change of the VSX1 gene. The sequence of a normal control (a) included the nucleotide G at position 8326 in the third intron of VSX1. The variant sequence (b) showed a G to A homozygous transition (arrow), which results in a c.8326G > A SNP change
Discussion
Keratoconus is a complex condition with a multi-factorial etiology that normally occurs as a sporadic disorder [25]. Recent studies have revealed the involvement of various genes in its development, such as VSX1, SOD1, COL6A1, TGFBI, DOCK9, STK24, and IPO5 [26–28]. Despite elucidate the etiology and the disease progression were conducted by numerous research, VSX1 is the only gene indicated as a vital genetic factor in determining the keratoconus [29]. Verma et al. ruled out the involvement of the VSX1 gene in South Indian patients with sporadic keratoconus [30]. However, Shetty et al. found that p.Leu268His, a novel missense variation in the coding region of VSX1, might be involved in the pathogenesis of sporadic keratoconus in Indians [22]. These earlier studies indicated the need for further study on the relationship between VSX1 pathogenic variations and sporadic keratoconus in different subsets of the keratoconus-affected population. Shetty et al. also summarized the VSX1 coding variants that are associated with keratoconus in different ethnic groups, but the Chinese were not included in this work (Table 3) [16–19, 22, 29, 31–38]. In the present study, PCR-SSCP and direct DNA sequencing techniques were used to examine nucleotide variations in the five exons and splicing regions of introns of the VSX1 gene in 50 unrelated Chinese patients with keratoconus, which were compared with the sequences in 50 controls. The results showed the presence of four nucleotide variations in sporadic keratoconus patients, consisting of a missense p.Arg131Pro sequence variant (heterozygous), a missense p.Gly160Val sequence variant (heterozygous), and nucleotide c.8326G > A sequence variation (in both heterozygous and homozygous forms). p.Gly160Val was located in the second exon of the VSX1 gene. In 2008, Mok et al. [33] first reported that p.Gly160Val variation was found in 13 out of 249 cases (5.3%) of sporadic keratoconus in South Korea, being located in the homeobox DNA binding domain of VSX1 gene coding regions and involving a polar amino acid being replaced by a neutral one. This may affect the binding of the VSX1 protein to DNA and modify the transcription rate. Therefore, it was believed that p.Gly160Val of the VSX1 gene increases the risk of the onset of keratoconus. In this study, we identified p.Gly160Val in two cases of 50 sporadic keratoconus patients, the sequence variant rate was 4.0%, but was absent in the control group. The findings suggested that p.Gly160Val may be the cause of two cases of sporadic keratoconus among these Chinese subjects. However, further biophysical studies are necessary to evaluate the precise molecular mechanism behind the effects of this variant form of VSX1.
Table 3.
Summary of VSX1 coding variants identified in patients with keratoconus [22]
| Coding variants | Clinical significance | Phenotype | Unrelated Controls | Ethnic groups | References |
|---|---|---|---|---|---|
| p.Leu 17 Pro | Pathogenica | Keratoconus | – | Italian | [17] |
| p.Leu 17 Val | Nonpathogenic | Keratoconus | + | Korean | [31] |
| p. Pro 58 Leu | Pathogenica, b | Keratoconus | – | Caucasian | [32] |
| p. Leu 159 Met | Unknown | Keratoconus | – | Caucasian | [16, 18] |
| p. Asn 151 Ser | Pathogenica | Keratoconus | – | Korean | [33] |
| p. Gly 160 Val | Nonpathogenic | Keratoconus | + | Korean | [31, 33] |
| p. Val 199 Leu | Nonpathogenic | Keratoconus | + | Korean | [31] |
| p. Arg 166 Trp | Unknown | Keratoconus | + | Caucasian, Iranian | [16, 34] |
| p. Gln 175 His | Unknown | Keratoconus | – | Indian | [35] |
| p. Arg 217 His | Nonpathogenic | Keratoconus | + | Indian, Pakistan, European | [19, 36] |
| p. Gly 239 Arg | Pathogenica, b | Keratoconus | – | Italian | [29] |
| p. His 244 Arg | Unknown | Keratoconus | + | Caucasian, Iranian | [16, 18, 34, 37] |
| p. Ser 251 Thr | Unknown | Keratoconus | – | Indian | [22] |
| p. Pro 247 Arg | Nonpathogenic | Keratoconus | + | Italian | [16, 17, 38] |
| p. Leu 268 His | Pathogenica, b | Keratoconus | – | Indian | [22] |
Coding variants of the VSX1 gene have been reported in studies based on an original reporta and bioinformatic predictionsb. The symbols “+” and “−” represent present and absent, respectively
The c.8326G > A sequence variation occurred in the third intron of the VSX1 gene. Abu-Amero et al. [25] detected c.8326G > A heterozygous variation in the Saudi Arabian population. They identified this variation in keratoconus patients as well as in controls, with no difference in its incidence between the two groups. This variation is located within an intron and does not cause a change in the amino acid sequence encoded by the VXS1 gene. Therefore, they considered that c.8326G > A variation belonged to SNP change of VSX1 gene. In our study, the c.8326G > A variant of the VSX1 gene was identified in both heterozygous and homozygous form, and it did not differ significantly in its incidence between the two groups. Overall, our results demonstrate the existence of SNPs in the VSX1 gene in the Chinese population, and suggest that sporadic keratoconus patients from different ethnic groups may have the same pathogenesis.
In the present study, we identified one novel heterozygous variant c.5427G > C (p.Arg131Pro; rs267597889) of the VSX1 gene in a 20-year-old affected male, which was absent from all 50 controls. This patient had a history of keratoconus lasting more than 5 years. The slit lamp microscope examination showed conical protrusion (opacity at the top) of the cornea in both eyes, with corneal thinning and a vertical linear scar, as well as complete Fleischer’s ring. p.Arg131Pro (replacing the amino acid arginine with proline) has not been reported previously, so this work adds to the range of variants in the VSX1 gene and suggests the role of this novel variant in keratoconus. However, predictions made using the PolyPhen-2 and PROVEAN tools suggested that this variant would not have detrimental phenotypic effects. This is explained by the fact that the 131st amino acid arginine is located in a noncritical open reading frame region of the VSX1 gene, so its character is not particularly important and it has not been conserved among the gene’s orthologs. The presence of p.Arg131Pro in just 2% of the patients in this study reinforces the assertion that this is a minor gene, but its exact role during development needs to be clarified.
Conclusions
In summary, the p.Arg131Pro, p.Gly160Val, and c.8326G > A variations in the VSX1 gene were identified for the first time in Chinese patients with sporadic keratoconus. We added one novel missense sequence variation (p.Arg131Pro) in the coding region of the VSX1 gene to the existing range of identified VSX1 variations. However, it was predicted that this novel variant may not cause a pathogenic change. Large-scale studies recruiting more patients are required to obtain a deeper understanding of the genetic mechanisms underlying the pathogenesis of keratoconus.
Acknowledgments
We are grateful to the patients and their families for participating in this study. We also thank staff at the Second Hospital of Ningbo Yingzhou for their help and support.
Funding
The study was supported by the Medical Scientific Research Foundation of Zhejiang Province, China (Grant No. 2013KYA233).
Availability of data and materials
The datasets supporting the conclusions of this article are included within the article. The datasets used and analyses performed during the current study are available from the corresponding author or the first author on reasonable request, but confidential patient data will not be shared.
Abbreviations
- PCR-SSCP
Polymerase chain reaction single-strand conformation polymorphism
- SNP
Single-nucleotide polymorphism
- VSX1
Visual system homeobox gene 1
Authors’ contributions
YFY conceived and designed the study. TG analyzed and interpreted the patient data, and wrote the manuscript. XW performed a critical review and helped to revise the manuscript. LBZ used bioinformatic tools to predict the effects of the mutations. HJW performed statistical analysis and helped to collect the data. All authors read and approved the final manuscript.
Ethics approval and consent to participate
This study followed the Declaration of Helsinki and was reviewed by the ethics committee of Taizhou Municipal Hospital. Written informed consent was obtained from all of the subjects in this study.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Tao Guan, Email: 120641612@qq.com.
Xue Wang, Email: wangxue19860206@163.com.
Li-Bin Zheng, Email: 360976910@qq.com.
Hai-Jian Wu, Email: 2336401319@qq.com.
Yu-Feng Yao, Phone: +86-574-13605780483, Email: 1054484763@qq.com.
References
- 1.Rabinowitz YS. Keratoconus. Surv Ophthalmol. 1998;42:297–319. doi: 10.1016/S0039-6257(97)00119-7. [DOI] [PubMed] [Google Scholar]
- 2.Fung SS, Aiello F, Maurino V. Outcomes of femtosecond laser-assisted mushroom-configuration keratoplasty in advanced keratoconus. Eye (Lond) 2016;30:553–561. doi: 10.1038/eye.2015.273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nowak DM, Gajecka M. The genetics of keratoconus. Middle East Afr J Ophthalmol. 2011;18:2–6. doi: 10.4103/0974-9233.75876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rabinowitz YS. The genetics of keratoconus. Ophthalmol Clin N Am. 2003;16:607–620. doi: 10.1016/S0896-1549(03)00099-3. [DOI] [PubMed] [Google Scholar]
- 5.Szczotka-Flynn L, Slaughter M, McMahon T, et al. Disease severity and family history in keratoconus. Br J Ophthalmol. 2008;92:1108–1111. doi: 10.1136/bjo.2007.130294. [DOI] [PubMed] [Google Scholar]
- 6.Mocan MC, Yilmaz PT, Irkec M, et al. The significance of Vogt's striae in keratoconus as evaluated by in vivo confocal microscopy. Clin Exp Ophthalmol. 2008;36:329–334. doi: 10.1111/j.1442-9071.2008.01737.x. [DOI] [PubMed] [Google Scholar]
- 7.Abu-Amero KK, Al-Muammar AM, Kondkar AA. Genetics of keratoconus: where do we stand? J Ophthalmol. 2014;2014:641708. doi: 10.1155/2014/641708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Stabuc-Silih M, Strazisar M, Ravnik-Glavac M, et al. Genetics and clinical characteristics of keratoconus. Acta Dermatovenerol Alp Pannonica Adriat. 2010;19:3–10. [PubMed] [Google Scholar]
- 9.Patel S. Characterisation of keratoconus. Br J Ophthalmol. 2011;95:759–760. doi: 10.1136/bjo.2010.200618. [DOI] [PubMed] [Google Scholar]
- 10.Maharana PK, Agarwal K, Jhanji V, et al. Deep anterior lamellar keratoplasty for keratoconus: a review. Eye Contact Lens. 2014;40:382–389. doi: 10.1097/ICL.0000000000000076. [DOI] [PubMed] [Google Scholar]
- 11.Gajecka M, Radhakrishna U, Winters D, et al. Localization of a gene for keratoconus to a 5.6-Mb interval on 13q32. Invest Ophthalmol Vis Sci. 2009;50:1531–1539. doi: 10.1167/iovs.08-2173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bisceglia L, De Bonis P, Pizzicoli C, et al. Linkage analysis in keratoconus: replication of locus 5q21.2 and identification of other suggestive Loci. Invest Ophthalmol Vis Sci. 2009;50:1081–1086. doi: 10.1167/iovs.08-2382. [DOI] [PubMed] [Google Scholar]
- 13.Burdon KP, Coster DJ, Charlesworth JC, et al. Apparent autosomal dominant keratoconus in a large Australian pedigree accounted for by digenic inheritance of two novel loci. Hum Genet. 2008;124:379–386. doi: 10.1007/s00439-008-0555-z. [DOI] [PubMed] [Google Scholar]
- 14.Abu-Amero KK, Kondkar AA, Azad TA, et al. Keratoconus is associated with increased copy number of mitochondrial DNA. Mol Vis. 2014;20:1203–1208. [PMC free article] [PubMed] [Google Scholar]
- 15.Barbaro V, Di Iorio E, Ferrari S, et al. Expression of VSX1 in human corneal keratocytes during differentiation into myofibroblasts in response to wound healing. Invest Ophthalmol Vis Sci. 2006;47:5243–5250. doi: 10.1167/iovs.06-0185. [DOI] [PubMed] [Google Scholar]
- 16.Heon E, Greenberg A, Kopp KK, et al. VSX1: a gene for posterior polymorphous dystrophy and keratoconus. Hum Mol Genet. 2002;11:1029–1036. doi: 10.1093/hmg/11.9.1029. [DOI] [PubMed] [Google Scholar]
- 17.Bisceglia L, Ciaschetti M, De Bonis P, et al. VSX1 mutational analysis in a series of Italian patients affected by keratoconus: detection of a novel mutation. Invest Ophthalmol Vis Sci. 2005;46:39–45. doi: 10.1167/iovs.04-0533. [DOI] [PubMed] [Google Scholar]
- 18.Tang YG, Picornell Y, Su X, et al. Three VSX1 gene mutations, L159M, R166W, and H244R, are not associated with keratoconus. Cornea. 2008;27:189–192. doi: 10.1097/ICO.0b013e31815a50e7. [DOI] [PubMed] [Google Scholar]
- 19.Tanwar M, Kumar M, Nayak B, et al. VSX1 gene analysis in keratoconus. Mol Vis. 2010;16:2395–2401. [PMC free article] [PubMed] [Google Scholar]
- 20.Paliwal P, Singh A, Tandon R, et al. A novel VSX1 mutation identified in an individual with keratoconus in India. Mol Vis. 2009;15:2475–2479. [PMC free article] [PubMed] [Google Scholar]
- 21.Stabuc-Silih M, Strazisar M, Hawlina M, et al. Absence of pathogenic mutations in VSX1 and SOD1 genes in patients with keratoconus. Cornea. 2010;29:172–176. doi: 10.1097/ICO.0b013e3181aebf7a. [DOI] [PubMed] [Google Scholar]
- 22.Shetty R, Nuijts RM, Nanaiah SG, et al. Two novel missense substitutions in the VSX1 gene: clinical and genetic analysis of families with Keratoconus from India. BMC Med Genet. 2015;16:33–42. doi: 10.1186/s12881-015-0178-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Delaveri A, Rapti A, Poulou M, et al. BTNL2 gene SNPs as a contributing factor to sarcoidosis pathogenesis in a cohort of Greek patients. Meta Gene. 2014;2:619–630. doi: 10.1016/j.mgene.2014.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hamasaki-Katagiri N, Salari R, Wu A, et al. A gene-specific method for predicting hemophilia-causing point mutations. J Mol Biol. 2013;425:4023–4033. doi: 10.1016/j.jmb.2013.07.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Abu-Amero KK, Kalantan H, Al-Muammar AM. Analysis of the VSX1 gene in keratoconus patients from Saudi Arabia. Mol Vis. 2011;17:667–672. [PMC free article] [PubMed] [Google Scholar]
- 26.Karolak JA, Polakowski P, Szaflik J, et al. Molecular Screening of Keratoconus Susceptibility Sequence Variants in VSX1, TGFBI, DOCK9, STK24, and IPO5 Genes in Polish Patients and Novel TGFBI Variant Identification. Ophthalmic Genet. 2016;37:37–43. doi: 10.3109/13816810.2014.926375. [DOI] [PubMed] [Google Scholar]
- 27.Galvis V, Tello A, Prada AM, et al. Changing Trends in Keratoconus Management. Cornea. 2016;35:e6–e7. doi: 10.1097/ICO.0000000000000748. [DOI] [PubMed] [Google Scholar]
- 28.Pena-Garcia P, Peris-Martinez C, Abbouda A, et al. Detection of subclinical keratoconus through non-contact tonometry and the use of discriminant biomechanical functions. J Biomech. 2016;49:353–363. doi: 10.1016/j.jbiomech.2015.12.031. [DOI] [PubMed] [Google Scholar]
- 29.De Bonis P, Laborante A, Pizzicoli C, et al. Mutational screening of VSX1, SPARC, SOD1, LOX, and TIMP3 in keratoconus. Mol Vis. 2011;17:2482–2494. [PMC free article] [PubMed] [Google Scholar]
- 30.Verma A, Das M, Srinivasan M, et al. Investigation of VSX1 sequence variants in South Indian patients with sporadic cases of keratoconus. BMC Res Notes. 2013;6:103. doi: 10.1186/1756-0500-6-103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jeoung JW, Kim MK, Park SS, et al. VSX1 gene and keratoconus: genetic analysis in Korean patients. Cornea. 2012;31:746–750. doi: 10.1097/ICO.0b013e3181e16dd0. [DOI] [PubMed] [Google Scholar]
- 32.Vincent AL, Jordan C, Sheck L, et al. Screening the visual system homeobox 1 gene in keratoconus and posterior polymorphous dystrophy cohorts identifies a novel variant. Mol Vis. 2013;19:852–860. [PMC free article] [PubMed] [Google Scholar]
- 33.Mok JW, Baek SJ, Joo CK. VSX1 gene variants are associated with keratoconus in unrelated Korean patients. J Hum Genet. 2008;53:842–849. doi: 10.1007/s10038-008-0319-6. [DOI] [PubMed] [Google Scholar]
- 34.Saee-Rad S, Hashemi H, Miraftab M, et al. Mutation analysis of VSX1 and SOD1 in Iranian patients with keratoconus. Mol Vis. 2011;17:3128–3136. [PMC free article] [PubMed] [Google Scholar]
- 35.Paliwal P, Tandon R, Dube D, et al. Familial segregation of a VSX1 mutation adds a new dimension to its role in the causation of keratoconus. Mol Vis. 2011;17:481–485. [PMC free article] [PubMed] [Google Scholar]
- 36.Dash DP, George S, O'Prey D, et al. Mutational screening of VSX1 in keratoconus patients from the European population. Eye (Lond) 2010;24:1085–1092. doi: 10.1038/eye.2009.217. [DOI] [PubMed] [Google Scholar]
- 37.Dehkordi FA, Rashki A, Bagheri N, et al. Study of VSX1 mutations in patients with keratoconus in southwest Iran using PCR-single-strand conformation polymorphism/heteroduplex analysis and sequencing method. Acta Cytol. 2013;57:646–651. doi: 10.1159/000353297. [DOI] [PubMed] [Google Scholar]
- 38.Eran P, Almogit A, David Z, et al. The D144E substitution in the VSX1 gene: a non-pathogenic variant or a disease causing mutation? Ophthalmic Genet. 2008;29:53–59. doi: 10.1080/13816810802008242. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets supporting the conclusions of this article are included within the article. The datasets used and analyses performed during the current study are available from the corresponding author or the first author on reasonable request, but confidential patient data will not be shared.


