Abstract
Background
Kaempferol is a flavonol with broad bioactivity of anti-oxidant, anti-cancer, anti-diabetic, anti-microbial, cardio-protective and anti-asthma. Microbial synthesis of kaempferol is a promising strategy because of the low content in primary plant source.
Methods
In this study, the biosynthesis pathway of kaempferol was constructed in the budding yeast Saccharomyces cerevisiae to produce kaempferol de novo, and several biological measures were taken for high production.
Results
Firstly, a high efficient flavonol synthases (FLS) from Populus deltoides was introduced into the biosynthetic pathway of kaempferol. Secondly, a S. cerevisiae recombinant was constructed for de novo synthesis of kaempferol, which generated about 6.97 mg/L kaempferol from glucose. To further promote kaempferol production, the acetyl-CoA biosynthetic pathway was overexpressed and p-coumarate was supplied as substrate, which improved kaempferol titer by about 23 and 120%, respectively. Finally, a fed-batch process was developed for better kaempferol fermentation performance, and the production reached 66.29 mg/L in 40 h.
Conclusions
The titer of kaempferol in our engineered yeast is 2.5 times of the highest reported titer. Our study provides a possible strategy to produce kaempferol using microbial cell factory.
Electronic supplementary material
The online version of this article (doi:10.1186/s12934-017-0774-x) contains supplementary material, which is available to authorized users.
Keywords: Kaempferol, Flavonol synthase, Acetyl-CoA, Saccharomyces cerevisiae
Background
Kaempferol is a polyphenol anti-oxidant found in many edible plants, which have been commonly used in traditional medicine (e.g. Ginkgo biloba, Tilia spp., Equisetum spp., Moringa oleifera, Sophora japonica and propolis) [1, 2]. Dietary kaempferol has attracted extensive attention because of the beneficial effects on human health, including anti-oxidant, anti-inflammatory, anti-microbial, anti-cancer, cardio-protective, neuro-protective, anti-diabetic, anti-osteoporotic, estrogenic, anti-estrogenic, anxiolytic, analgesic and anti-allergic activities [1–4]. Interestingly, although kaempferol inhibits cancer cell growth and induces cancer cell apoptosis, it appears to preserve or protect normal cell viability [2]. However, the content of kaempferol in plants is very low, which results in high cost for kaempferol production from traditional plant extraction [5, 6]. Thus, it is a promising alternative strategy to produce kaempferol using microbial cell factory.
Biosynthetic pathway of kaempferol has been identified in plant [5]. At the initial stage, phenylalanine is converted into p-coumaryl-CoA by phenylalanine ammonia lyase (PAL), cinnamic acid 4-hydroxylase (C4H) and 4-coumaric acid ligase (4CL). Then, naringenin is generated by condensation reaction between one molecular p-coumaryl-CoA and three molecules of malonyl-CoA by chalcone synthesis (CHS) and chalcone isomerase (CHI). At last, the naringenin is converted into kaempferol via dihydrokaempferol by flavanone 3β-hydroxylase (F3H) and flavonol synthase (FLS). Although kaempferol has been successfully synthesized by engineered microbes, the titer of kaempferol is still lower than many other microbial produced flavonoids, such as naringenin and eriodictyol [7–11]. One of the possible causes is that the carbon flux toward precursors is usually not enough [12]. Engineering metabolic pathway toward acetyl-CoA or malonyl-CoA would partially solve this problem [13–15]. In addition, the low efficiency of key enzymes in kaempferol biosynthetic pathway could also cause this issue. It has been reported that although the precursor naringenin was sufficiently supplied, the recombinant cell factory still produced kaempferol at a very low level [10].
In this study, we proposed to construct a yeast cell factory to improve kaempferol production. Firstly, we would screen higher efficient FLS from different plants to improve kaempferol conversion rate from the precursor naringenin. Secondly, the selected FLS would be combined with other pathway genes to build a microbial cell factory for de novo kaempferol synthesis. Moreover the acetyl-CoA and malonyl-CoA pathways would be engineered to further improve kaempferol biosynthesis. p-Coumarate would be also supplemented as substrate to improve the precursors supply. Finally, the fermentation condition would be optimized for better kaempferol production.
Results and discussion
Biosynthesis of kaempferol from naringenin
Despite the significant pharmaceutical effect of kaempferol, the low content in plant or engineered microbes has restricted its application. We firstly attempted to construct the kaempferol pathway in S. cerevisiae from naringenin, a common intermediate of flavonoid biosynthesis. Naringenin is converted into kaempferol via dihydrokaempferol, catalyzed with flavanone 3β-hydroxylase (F3H) and flavonol synthase (FLS) (Fig. 1a). In this research, F3H from Arabidopsis thaliana [16] and FLS from Malus domestica [17] (encoded by AtF3H and MdFLS respectively) were expressed in S. cerevisiae to build a kaempferol-producing recombinant. Indeed, using (2S)-naringenin as substrate, the recombinant yeast expressing AtF3H produced (+)-dihydrokaempferol (Fig. 1b, c). On the other hand, both (+)-dihydrokaempferol and kaempferol were detected in the medium fermented by the recombinant expressing AtF3H and MdFLS (Fig. 1b, d). Moreover, the molecular weights of (+)-dihydrokaempferol and kaempferol were confirmed by liquid chromatograph–mass spectrometer (LC–MS) (Fig. 1c, d). However, when 100 mg/L initial (2S)-naringenin was supplemented, 23.62 mg/L (+)-dihydrokaempferol and only 6.03 mg/L kaempferol were produced through 24 h conversion. The relatively low titer of kaempferol indicated that the step from (+)-dihydrokaempferol to kaempferol is the bottleneck of kaempferol production from naringenin.
Fig. 1.
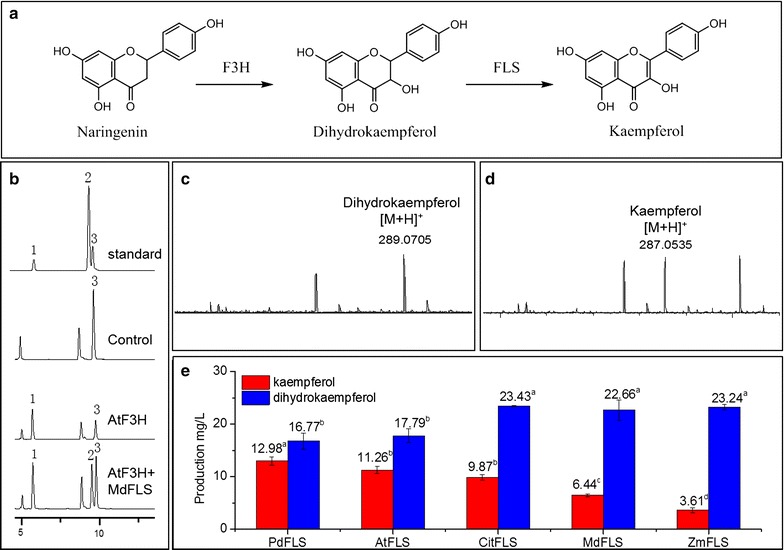
Functional identification of two key enzymes of F3H and FLS in kaempferol biosynthetic pathway. a The biosynthetic pathway from naringenin to kaempferol. Naringenin is converted into dihydrokaempferol with flavanone 3β-hydroxylase (F3H), and dihydrokaempferol is converted into kaempferol via flavonol synthase (FLS). b HPLC analysis of the product in whole cell catalysis with overexpression of F3H and FLS in yeast. Chromatogram data were collected at 335 nm. Peak 1, (+)-dihydrokaempferol; peak 2, kaempferol; peak 3, (2S)-naringenin. (2S)-naringenin was the substrate and was detected in the wild type W3-Y22. c LC–MS analysis of dihydrokaempferol produced by W3-AtF3H in whole cell catalysis. d LC–MS analysis of kaempferol produced by W3-MdFLS in whole cell catalysis. e Production of kaempferol and dihydrokaempferol of recombinant yeast strain expressing FLS from five plant species. Recombinant yeast strain expressing AtF3H and FLS from different species were fermented. Data are shown as average in triplicate assays. Error bar shows standard division. Different superscript letter indicates significant difference in Tukey analysis (α = 0.05)
To solve this problem, we proposed to find a more efficient FLS from other plant sources. FLS genes from five plant species, including A. thaliana, Citrus unshiu, M. domestica, P. deltoides and Zea Mays [10, 18–21] (encoded by AtFLS, CitFLS, MdFLS, PdFLS and ZmFLS, respectively), were overexpressed with AtF3H in yeast, respectively. The viability of these FLS in kaempferol production were compared according to the final kaempferol productions in whole cell catalysis. The strain expressing PdFLS resulted in the highest kaempferol production (12.98 mg/L, Fig. 1e) and the lowest dihydrokaempferol accumulation (16.77 mg/L, Fig. 1e), indicating that PdFLS has the highest efficiency to produce kaempferol from dihydrokaempferol (Additional file 1: Figure S1A, S1B). Besides, the recombinants expressing AtFLS and CitFLS produced lower kaempferol than that expressing PdFLS, but significant higher than that expressing MdFLS and ZmFLS (Fig. 1e). According to the previous studies, FLS from P. deltoides, A. thaliana and C. unshiu performed both FLS and F3H function, which are able to use naringenin as a substrate to produce kaempferol as well as dihydrokaempferol [20–23]. However, this F3H function was not observed in FLS from M. domestica and Z. Mays [17, 18]. This could partially explain that the recombinants expressing PdFLS, AtFLS and CitFLS produced significant higher kaempferol than those expressing MdFLS and ZmFLS. Although the sole expression of bifunctional FLS would enable the recombinant to convert naringenin into kaempferol, the co-expression with F3H would promote flavonol titer and has been widely adopted in the previous studies [9, 24]. Considering the co-expression of PdFLS and AtF3H resulted in the highest kaempferol production from naringenin, this gene cluster was retained for further kaempferol biosynthesis.
Construction of de novo synthetic pathway of kaempferol
Based on the high efficient kaempferol conversion with AtF3H and PdFLS, we further attempted to build a pathway for de novo biosynthesis of kaempferol. A naringenin synthesis strain, W3NP, was constructed as parent strain through expressing phenylalanine ammonia lyase, cinnamic acid 4-hydroxylase, 4-coumaric acid ligase, chalcone synthase, and chalcone isomerase from Erigeron breviscapus (encoded by PAL, C4H, 4CL, CHS, CHI, respectively). Indeed, W3NP was able to synthesize naringenin from glucose, which was confirmed by LC–MS and ultraviolet (UV) absorption (Fig. 2b and Additional file 1: Figure S2). Thus, by introducing AtF3H and PdFLS to W3NP, a yeast recombinant W3NP-FF was generated, which carries a pathway for de novo kaempferol biosynthesis (Fig. 2c). In batch fermentation, W3NP produced 2.29 mg/L naringenin, while W3NP-FF produced 6.97 mg/L kaempferol, which was accompanied with 3.55 mg/L dihydrokaempferol and 3.32 mg/L naringenin (Additional file 1: Table S1). The total flavonoids production was largely improved by introducing kaempferol biosynthetic pathway (Additional file 1: Table S1). This is probably because the expression of PdFLS and AtF3H strengthened the driving force toward flavonoid synthesis from glucose. Besides, the higher kaempferol production compared to dihydrokaempferol in W3NP-FF also indicated that PdFLS is efficient in kaempferol production.
Fig. 2.
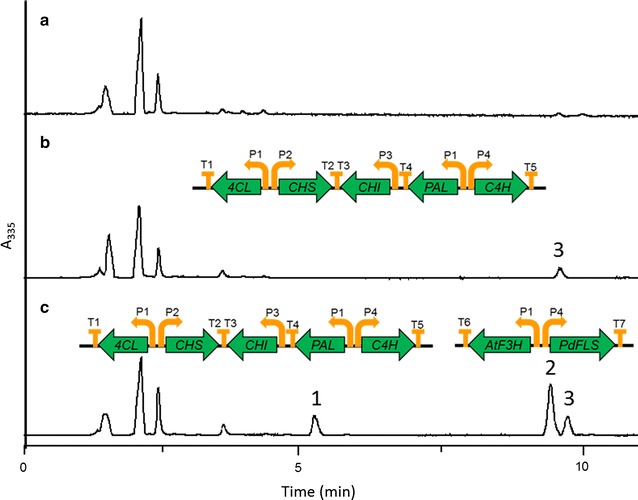
Pathway construction for de novo biosynthesis of kaempferol in yeast. HPLC analysis of the broth fermented by W303-1A (a), W3NP (b) and W3NP-FF (c) from glucose. W303-1A, wild type; W3NP, a recombinant harboring the pathway of naringenin biosynthesis; W3NP-FF, a recombinant harboring the pathway of kaempferol biosynthesis. Peak 1, dihydrokaempferol; peak 2, kaempferol; peak 3, naringenin. Organization of expressed gene clusters are shown at the top of the chromatogram. P1, ADH1 promoter; P2, HXT7 promoter; P3, PGI1 promoter; P4, TDH3 promoter; T1, TPI1 terminator; T2, TPG1 terminator; T3, ADH1 terminator; T4, FBA1 terminator; T5 PDC1 terminator; T6, RPS2 terminator; T7, TDH1 terminator
Metabolic engineering for kaempferol production
In order to further improve the production of kaempferol, we proposed to enhance the metabolic pathway of precursors. Increasing the intracellular acetyl-CoA pool and (or) malonyl-CoA pool was beneficial for high-level flavonoids production [14, 25, 26]. The biosynthetic pathway of malonyl-CoA from ethanol consists of four steps, catalyzed by alcohol dehydrogenase, aldehyde dehydrogenase, acetyl-coA synthetase and acetyl-CoA carboxylase (encoded by ADH2, ALD6, ACS SE and ACC1, respectively Fig. 3a). To increase endogenous supply of acetyl-CoA or malonyl-CoA, biosynthetic genes were overexpressed in the kaempferol producing strain W3NP-FF. The gene cluster harboring ADH2, ALD6, and ACS SE expression cassettes (Additional file 1: Figure S3B), and another harboring ADH2, ALD6, ACS SE and ACC1 (Additional file 1: Figure S3C) expression cassettes were separately transformed to W3NP-FF, generating W3NP-FF-A3 and W3NP-FF-A4, respectively. In batch fermentation, W3NP-FF-A3 produced 8.6 mg/L kaempferol from glucose, which increased 23% compared to W3NP-FF (Fig. 3b and Additional file 1: Table S1). However, W3NP-FF-A4 significantly decreased the production of kaempferol by 48%, and also decreased the production of naringenin and dihydrokaempferol by 75 and 63% respectively, compared to W3NP-FF (Fig. 3b and Additional file 1: Table S1). Besides, the ACC1 overexpression caused a 33% reduction in final OD600, and reduce by 37% in specific kaempferol production compared to these of W3NP-FF-A3 in fermentation from glucose (Fig. 3b and Additional file 1: Table S1).
Fig. 3.
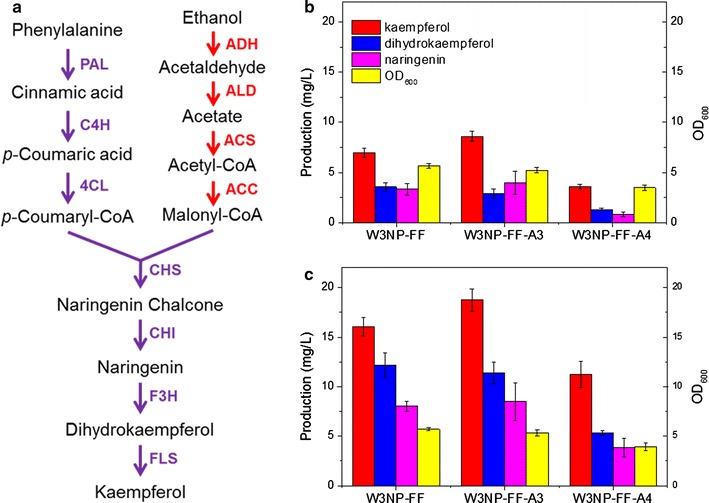
Metabolic engineering for high kaempferol production in S. cerevisiae. a Metabolic pathway and engineering scheme for kaempferol production. Purple arrows represent reactions catalyzed by exogenous enzymes from plant in kaempferol synthesis pathway. Red arrows represent reactions catalyzed by endogenous enzymes in yeast, which are involved in the precursor synthesis, and overexpressed for kaempferol production. PAL, phenylalanine ammonia lyase; C4H, cinnamic acid 4-hydroxylase; 4CL, 4-coumaric acid ligase; CHS, chalcone synthase; CHI, chalcone isomerase; F3H, flavanone 3β-hydroxylase; FLS, flavonol synthase; ADH, alcohol dehydrogenase; ALD, aldehyde dehydrogenase; ACS, acetyl-coA synthetase; ACC, acetyl-CoA carboxylase. b OD600 and production of naringenin, dihydrokaempferol and kaempferol in batch fermentation at 60 h. Glucose (20 g/L) was used as the sole carbon source. c OD600 and production of naringenin, dihydrokaempferol and kaempferol in batch fermentation at 60 h. The media were supplemented with 20 g/L glucose and 1 mM p-coumarate. Data are shown as average ± standard division in triplicate tests
ACC1 encodes cytosolic acetyl-CoA carboxylase (ACCase), catalyzing the malonyl-CoA formation from acetyl-CoA in S. cerevisiae, which is regarded as the rate limiting enzyme in fatty acid synthesis, and is thus tightly regulated in transcription and post-translation levels [27, 28]. In most case, overexpressing a native or a mutant ACC1 increased the malonyl-CoA pool or the derived products [29–32]. By contrast, some studied suggested that the ACCase overexpression impaired cell growth [29, 32, 33], or did not promote the production of malonyl-CoA derived products significantly [34, 35]. The reasons for this phenomenon have generally attributed to an imbalanced synthesis of long-chain fatty acids, depletion of intermediates or high metabolic burden [27, 29, 32]. In this study, an ACC1 mutant with Ser659Ala and Ser1157Ala, which improves ACCase activity by abolishing Snf1-dependent regulation, was driven by a PGK1 promoter. Thus, expression of ACCase had been enhanced in both transcription and post-translation levels. The excessively increased ACCase activity may cause imbalanced synthesis of long-chain fatty acids and/or abnormal intracellular levels of acetyl-CoA and AMP, and interfere cell growth and metabolism, including flavonoids synthesis (Fig. 3b and Additional file 1: Table S1). Our results suggest that a simple and straightforward ACCase overexpression could not increase malonyl-CoA derived products as expected, and introducing a dynamic ACCase regulation or heterologous malonyl-CoA synthesis pathway would be a more promising strategy [36, 37].
Another important precursor of flavonoids biosynthesis is phenylalanine. The intracellular l-phenylalanine synthesis is tightly regulated in S. cerevisiae [10, 38]. Here, we supplemented p-coumarate in the fermentation medium in order to partially alleviate the limitation of precursor supply [10, 38]. The p-coumarate supplement significantly increased total flavonoids production by about 2–3.5 times for W3NP-FF, W3NP-FF-A3 and W3NP-FF-A4 (Fig. 3b, c and Additional file 1: Table S1). Particularly, the kaempferol production of W3NP-FF-A3 reached 18.76 mg/L, which is close to the highest reported value (22.57 mg/L) [39].
Fermentation optimization for kaempferol production
Fermentation conditions, such as pH, media components, temperature, stirring speed and ventilation rate, also largely affect the titer of products in microbial cell factory. In this research, a Quasi exponent feed strategy [40] was adopted in fed-batch fermentation for high kaempferol production with W3NP-FF and W3NP-FF-A3. The OD600 and the production of flavonoids were measured and shown in Fig. 4a, b. Both of W3NP-FF and W3NP-FF-A3 produced much higher kaempferol in fed-batch fermentation than that in batch fermentation (Figs. 3b, c, and 4a, b). To increase substrate supply, 1 mM p-coumarate was supplemented after 24 h fermentation. Then the production of kaempferol, dihydrokaempferol and naringenin were consequently increased for both W3NP-FF-A3 and W3NP-FF (Fig. 4a, b). For W3NP-FF-A3, the kaempferol production continuously increased in the early 40 h (Fig. 4b); while for W3NP-FF, the kaempferol production reached the top at 32 h, but with a stationary phase from 12 to 24 h (Fig. 4a). This result showed that overexpression of acetyl-CoA synthetic pathway (ADH2, ALD6, and ACS SE) not only increased the titer of kaempferol, but also improved the persistence of kaempferol production. The highest kaempferol titer in fed-batch fermentation achieved 66.29 mg/L by W3NP-FF-A3, which is 3.5-fold over that in batch fermentation (Fig. 4b, Additional file 1: Table S1 and S2). To our knowledge, it is about 2.5 times of the reported highest titer (26.57 mg/L kaempferol produced by an engineered S. cerevisiae [39]).
Fig. 4.
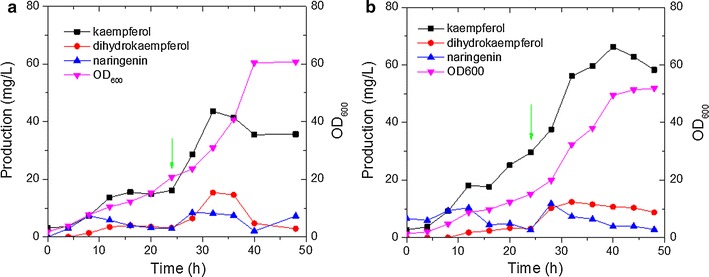
Fed-batch fermentation of kaempferol in well controlled fermenters. OD600 and production of naringenin, dihydrokaempferol and kaempferol are shown in fed-batch fermentation with W3NP-FF (a) and W3NP-FF-A3 (b). The fermentations were performed at 30 °C, and pH was maintained at 5.0. The feed was started when residual ethanol and glucose were completely depleted. A Quasi exponent feed strategy was adopted. Green arrow indicates 1 mM p-coumarate was supplemented at 24 h
In the flavonoid synthetic pathway, malonyl-CoA and p-coumaroyl-CoA are two direct precursors in chalcone forming. In this research, the intracellular supply of malonyl-CoA has been enhanced by over-expression the acetyl-CoA synthetic pathway; on the other hand, the supply of p-coumaroyl-CoA has been satisfied by supplementing p-coumarate in the medium, which trends to add extra cost in fermentation. Alternatively, it has been reported that engineering the aromatic amino acid biosynthesis pathway and overexpressing the tyrosine ammonia-lyase (TAL) increased the formation and accumulation of p-coumaric acid in yeast cells [41]. This is an attractive strategy for promoting the production of p-coumaric acid derived products from glucose, and saving p-coumaric acid supplement in fermentation. Recently, Rodriguez et al. expressed 4CL, CHS, CHI, CHR (encoding chalcone reductase), F3H (from Astragalus mongholicus), FLS (from Arabidopsis thaliana) and CPR (encoding cytochrome P450 reductase) and FMO (encoding a cytochrome P450 flavonoid monooxygenases) genes in p-coumaric acid over-accumulated strains, to produce several flavonoids including kaempferol [39]. Taking advantage of the enhanced p-coumaric acid synthesis, the engineered strain produced 26.57 mg/L kaempferol [39], which is the highest reported titer to our best knowledge. In this research, we comprehensively applied pathway construction, enzyme selection, metabolic engineering, intermediate supplement and fermentation optimization to realize and upgrade kaempferol production gradually, and achieved the highest kaempferol production at 66.29 mg/L. Here, we emphasize the combination of multi biological solutions for efficient product synthesis in a microbial cell factory.
Conclusions
In this study, we successfully synthesized kaempferol through S. cerevisiae recombinants expressing FLS from P. deltoides, which resulted in higher kaempferol production than those from A. thaliana, C. unshiu, M. domestica, and Z. mays. Through expressing PAL, C4H, 4CL, CHS, CHI, AtF3H and PdFLS, we constructed a yeast recombinant that synthesizes kaempferol de novo. Furthermore, we demonstrated that overexpressing the acetyl-CoA synthetic pathway (consisted of ADH2, ALD6 and ACS SE) in cytoplasm and p-coumarate supplement in media would significantly increase kaempferol titer of the recombinant. Fermentation conditions are also closely related to the kaempferol biosynthesis that fed-batch fermentation in a bioreactor with a Quasi exponent feed strategy largely improved kaempferol production compared to batch fermentation in flask. The titer of kaempferol reached up to 66.29 mg/L after 40 h fermentation. To our knowledge, it is the highest kaempferol titer in microbial cell factories currently.
Methods
Genes and strains
Flavonol synthases from A. thaliana (accessing number NP_196481.1, encoded by AtFLS), C. unshiu (accessing number: Q9ZWQ9.1, encoded by CitFLS), M. domestica (accessing number: NP_001306179.1, encoded by MdFLS),Z. Mays (accessing number: XP_008646309.1, encoded by ZmFLS) and P. deltoides (TIGR accession number: TC74233 [23], encoded by PdFLS) were investigated in this work. Flavanone 3β-hydroxylase (F3H) was from A. thaliana (encoded by AtF3H, accessing number: NP_190692.1). PAL, C4H, 4CL, CHS and CHI were from Erigeron breviscapus (Vant.) Hand-Mazz. (kindly provided by Guang-Hui Zhang [42]). The codon usage of CitFLS, MdFLS, ZmFLS, PdFLS, PAL, C4H, 4CL, CHS and CHI were optimized for S. cerevisiae (Additional file 1: Table S3) and synthesized by a local company.
Escherichia coli DH5α was used for gene cloning. S. cerevisiae W303-1A was used for genetic engineering and kaempferol production. All strains and plasmids used in this study are shown in Table 1.
Table 1.
Plasmids and strains used in this study
| Plasmid or strain | Genotype | Source or reference |
|---|---|---|
| Plasmids | ||
| YCplac22 | Amp Trp1 | Gietz [50] |
| YCplac33 | AmpURA3 | Gietz [50] |
| Y22-T4 | TADH1/TTDH3 | This work |
| Y22-AtF3H | PPGK1-AtF3H-TADH1 | This work |
| Y22-T4-AtF3H-AtFLS | PPGK1-AtF3H-TADH1/PTDH3-AtFLS-TTDH1 | This work |
| Y22-T4-AtF3H-CitFLS | PPGK1-AtF3H-TADH1/PTDH3-CitFLS-TTDH1 | This work |
| Y22-T4-AtF3H-MdFLS | PPGK1-AtF3H-TADH1/PTDH3-MdFLS-TTDH1 | This work |
| Y22-T4-AtF3H-PdFLS | PPGK1-AtF3H-TADH1/PTDH3-PdFLS-TTDH1 | This work |
| Y22-T4-AtF3H-ZmFLS | PPGK1-AtF3H-TADH1/PTDH3-ZmFLS-TTDH1 | This work |
| Y33-ALAC-ADH2 | PTEF2-ACS SE-TRPS2/PPGK1-ALD6-TTDH1/PHXT7-ADH2-TRPL9A | This work |
| Y33-ALAC-ACAD | PTEF2-ACS SE-TRPS2/PPGK1-ALD6-TTDH1/PHXT7-ADH2-TRPL9A/PPGK1-ACC1-TCCW12 | This work |
| Strains | ||
| W303-1A | MATa leu2-3112 ura3-1 trp1-92 his3-11,15 ade2-1 can1-100 | Thomas [51] |
| W3-AtF3H | W303-1A carrying Y22-AtF3H | This work |
| W3-AtFLS | W303-1A carrying Y22-T4-AtF3H-AtFLS | This work |
| W3-CitFLS | W303-1A carrying Y22-T4-AtF3H-CitFLS | This work |
| W3-MdFLS | W303-1A carrying Y22-T4-AtF3H-MdFLS | This work |
| W3-PdFLS | W303-1A carrying Y22-T4-AtF3H-PdFLS | This work |
| W3-ZmFLS | W303-1A carrying Y22-T4-AtF3H-ZmFLS | This work |
| W3NP | YORWdelta17::His3/PADH1-4CL-TTPI1/PHXT7-CHS-TTPG1/PPGI1-CHI-TADH1/PADH1-PAL-TFBA1/PTDH3-C4H-TPDC1 | This work |
| W3NP-FF | W3NP carrying Y22-T4-AtF3H-PdFLS and YCplac33 | This work |
| W3NP-FF-A3 | W3NP carrying Y22-T4-AtF3H-PdFLS and Y33-ALAC-ADH2 | This work |
| W3NP-FF-A4 | W3NP carrying Y22-T4-AtF3H-PdFLS and Y33-ALAC-ACAD | This work |
Gene cloning and plasmid construction
All DNA manipulations were performed according to standard procedures [43]. Phusion High-Fidelity DNA Polymerase (Thermo Fisher Scientific) was used for PCR amplification. A DNA fragment, named Ter-4, was designed for Golden gate cloning, which contained two terminators (T RSP2 and T TDH1), being separated by two BsaI digest sites (the scheme was shown in Additional file 1: Figure S3A). The Ter-4 fragment was synthesized by a local company (Synbio Technologies), and cloned to pUC57. Then the Ter-4 fragment and YCplac22 were amplified (primers: T4-YCP-F/T4-YCP-R, Y22-T4-F/Y22-T4-R, Additional file 1: Table S4) and assembled together using CPEC (Circular Polymerase Extension Cloning [44]), generating Y22-T4 (Additional file 1: Figure S4), the backbone for FLS and F3H expression. In CPEC, the vector and the insert share overlapping regions at the ends, and the hybridized insert and vector extend using each other as a template until they complete a full circle in a PCR system, and finally the PCR product is transformed directly to DH5α. For assembly of gene expression cassettes, 3 DNA fragments, including a head-to-head promoter fragment, AtF3H and each of the FLS genes (AtFLS, CitFLS, MdFLS, PdFLS, ZmFLS), were amplified (primers: P1-F/P1-R for promoter fragment, AtF3H-GG2-F/AtF3H-GG2-R for AtF3H, FLS-GG1-F/FLS-GG1-R for each FLS gene, Additional file 1: Table S4) respectively, and ligated to Y22-T4 through golden gate cloning [45], generating FLS and F3H expression cassettes (Additional file 1: Figures S3A, S4). Thus, Y22-T4-F3H-AtFLS, Y22-T4-AtF3H-CitFLS, Y22-T4-AtF3H-MdFLS, Y22-T4-AtF3H-PdFLS and Y22-T4-AtF3H-ZmFLS was constructed. Then, expression cassette of AtF3H was amplified (primer: F3H-CPF/F3H-CPR, Additional file 1: Table S4) and ligated to YCplac22 through CPEC [44], resulting in Y22-AtF3H. Similarly, expression cassettes of ALD6, ACS SE [46], ADH2 and ACC1 (a mutant with Ser659Ala and Ser1157Ala, [29]) were constructed through golden gate cloning. Then, the cassettes of ALD6, ACS SE and ADH2 were inserted into YCplac33 through Gibson assembling [47] (primers: Y33-F/Y33-R, ALAC-GibF/ALAC-GibR, ADH2-GibF/ADH2-GibR, Additional file 1: Table S4), resulting in the plasmid Y33-ALAC-ADH2 (Additional file 1: Figure S3B). Cassettes of ALD6, ACS SE, ADH2 and ACC1 were inserted into YCplac33 through Gibson assembling [47] (primers: Y33-F/Y33-R, ALAC-GibF/ALAC-GibR, ACAD-GibF/ADH2-GibR, Additional file 1: Table S4), generating the plasmid Y33-ALAC-ACAD (Additional file 1: Figure S3C).
Strain construction
To generate a naringenin-producing S. cerevisiae recombinant, a modularized two-step (M2S) chromosome integration technique [45] was applied for PAL, C4H, 4CL, CHS and CHI expression. In brief, PAL and C4H; 4CL and CHS; and CHI was ligated with promoters and terminators respectively through golden gate cloning, forming the expression cassettes. Then, DNA fragments harboring these cassettes were transformed together with His marker and homologous arms into wild strain (W303-1A), and integrated into genome through DNA assembler [48] (Additional file 1: Figure S3D), resulting in the naringenin-producing recombinant, W3NP. Then, Y22-T4-AtF3H-PdFLS was co-transformed into W3NP with YCplac33, Y33-ALAC-ADH2 and Y33-ALAC-ACAD respectively, resulting in W3NP-FF, W3NP-FF-A3 and W3NP-FF-A4.
Media and culture condition
Escherichia coli was grown in Luria–Bertani (LB) medium at 37 °C. Ampicillin (50 μg/mL) was added to the medium when required. Yeast strains were grown at 30 °C in YPD medium (10 g/L yeast extract, 20 g/L Bacto peptone, and 20 g/L glucose) or defined mineral medium (YSCD), containing 6.7 g/L yeast nitrogen base (YNB) without amino acids (Difco, Detroit, Michigan), supplemented with the appropriate auxotrophic requirements and 20 g/L glucose. YSCD supplemented with 5 mM sodium ascorbate and 0.1 mM Fe2SO4 was used for batch fermentation in flask, and 1 mM p-coumarate was added when required.
The medium for fed-batch fermentation contained: 20 g/L glucose, 15 g/L (NH4)2SO4, 8 g/L KH2PO4, 6.2 g/L MgSO4·7H2O, 12 mL/L vitamin solution and 10 mL/L trace metal solution, where the vitamin solution contained 0.05 g/L biotin, 1 g/L calcium pantothenate, 1 g/L nicotinic acid, 25 g/L inositol, 1 g/L thiamine HCl, 1 g/L pyridoxal HCl, 0.2 g/L p-aminobenzoic acid and 2.5 g/L adenine; the trace metal solution contained: 5.75 g/L ZnSO4·7H2O, 0.32 g/L MnCl2·4H2O, 0.47 g/L CoCl2·6H2O, 0.48 g/L Na2MoO4·2H2O, 2.9 g/L CaCl2·2H2O, 2.8 g/L FeSO4·7H2O and 80 mL/L 0.5 M EDTA, pH 8.0. The same medium was used for seed culture in fed-batch fermentation [49].
Fermentation
Whole cell catalysis
Inoculum was cultured in YSCD medium at 30 °C for 12 h. The pre-culture was refreshed in 20 mL YSCD to OD600 = 1. Cells were then collected and re-suspended in 3 mL YSCD supplemented with 5 mM sodium ascorbate, 0.1 mM Fe2SO4 and 100 mg/L (2S)-naringenin (Solarbio Life Sciences), to a final cell concentration of OD600 = 5. The reaction was incubated at 30 °C in an orbital shaker (220 rpm) for 24 h.
Batch fermentation
Inoculum was cultured in YSCD medium at 30 °C for 12 h. The pre-culture was then used to inoculate 20 mL batch fermentation medium (YSCD supplemented with 5 mM sodium ascorbate and 0.1 mM Fe2SO4) in 250 mL shaker flasks to a final cell concentration of OD600 = 1. One mM p-coumarate was added as precursor when required. The fermentation lasted for 60 h.
Fed-batch fermentation
Glycerol-stocked cells were inoculated into 40 mL YSCD and cultured at 30 °C, 220 rpm for 24 h. The culture was transferred to 360 mL fed-batch fermentation medium, and cultured for another 24 h. 300 mL seeds were inoculated to 1.5 L fed-batch fermentation medium in Baoxin bioreactor with a maximal working volume of 3 L. The fermentations were performed at 30 °C, and pH was maintained at 5.0 with automatic addition of ammonium hydroxide or 1 M H2SO4. The agitation rate was kept between 300 and 800 rpm, and the air flow was set as 1.5 vvm. The dissolved oxygen concentration was controlled above 40% throughout regulation the agitation rate [40]. The feed was started after residual ethanol and glucose were completely depleted. A Quasi exponent feed strategy was adopted as described [40]. Feed reagent contained: 386 g/L glucose, 9 g/L KH2PO4, 5.12 g/L MgSO4·7H2O, 3.5 g/L K2SO4, 0.28 g/L Na2SO4, 5 g/L adenine, 12 mL/L vitamin solution and 10 mL/L trace metal solution. 1 mM p-coumarate was supplemented at 24 h.
Detection and quantification of the products
Flavonoids were extracted directly from the culture with an equal volume of methanol. The extraction were analyzed by High Performance Liquid Chromatography (HPLC, Agilent), using Phenomenex Kinetex Biphenyl Column (5 μm, 250 × 4.6 mm) equipped with a photodiode array detector. The mobile phase consisted of acetonitrile and water (0.1% phosphoric acid) using a gradient elution of 30–40% acetonitrile for 10 min, 40–95% acetonitrile for 2.5 min,95–30% acetonitrile for 2.5 min and 30% acetonitrile for 5 min, at flow rate of 1 mL/min. Samples were analyzed by LC–MS using a Thermo U3000-LTQ XL (Thermo Scientific) system coupled to the ion trap mass spectrometer with an ESI source operating in the positive mode. LC–MS analysis was operated with the same LC operation method, except 0.1% formic acid was substitute to phosphoric acid. Quantification of the kaempferol was based on the peak areas of absorbance at 335 nm. Quantification of dihydrokaempferol and naringenin was based on the peak areas of absorbance at 290 nm.
Authors’ contributions
LD and WD contributes equally in this work. WD, JC and HJ conceived and designed the manuscript. LD and WD performed the experimental research and drafted the manuscript. XL constructed the recombinant W3NP. XC carried out the fed-batch fermentation. HJ, EH and JC outlined the structure and reviewed the manuscript. All authors read and approved the final manuscript.
Acknowledgements
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Availability of data and materials
All data generated or analyzed during this study are included in this published article.
Consent for publication
Not applicable.
Ethics approval and consent to participate
Not applicable.
Funding
This work was supported by a 973 Program (2015CB755704), the Hundred Talent Program of the Chinese Academy of Sciences and a National Natural Science Foundation of China (31670100) to HJ, and a Tianjin Research Program of Application Foundation and Advanced Technology (15JCYBJC24200), and a National Natural Science Foundation of China (31501041) to WD, and a start-up research grant from University of Macau (SRG2015-00062-ICMS-QRCM) to JC.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Additional file
Additional file 1: Table S1. The production of flavonoids from engineered yeasts in batch fermentation. Table S2. Maximum OD600 and production of kaempferol, dihydrokaempferol and naringenin of W3NP-FF and W3NP-FF-A3 in fed-batch fermentation. Table S3. Optimized gene sequence for S. cerevisiae. Table S4. Primers used in this work. Figure S1. Mole ratio of kaempferol to dihydrokaempferol (A) and mole ratio of kaempferol to total naringenin (B) in whole cell catalysis with the recombinants expressing FLS orthologs. Figure S2. Identification of naringenin produced by W3NP from glucose. A, LC-MS analysis of naringenin produced by W3NP. B, UV absorption of naringenin produced by W3NP (solid line) and standard (dash line). Figure S3. Scheme of plasmids construction and gene manipulation for kaempferol production. A, Sketch drawing for F3H and FLS expression. B, Y33-ALAC-ADH2, a plasmid expressing ALD6, ACS SE and ADH2. C, Y33-ALAC-ACAD, a plasmid expressing ALD6, ACS SE, ADH2 and ACC1. D, Genome manipulation in W3NP for naringenin production. Figure S4. Scheme of cassette assembly through golden gate cloning. A. The design of Ter-4 fragment and the cloning vector Y22-T4. The vector Y22-T4, DNA fragments of promoter, F3H and FLS were digested by BsaI and ligated through sticky ends.
Footnotes
Electronic supplementary material
The online version of this article (doi:10.1186/s12934-017-0774-x) contains supplementary material, which is available to authorized users.
Lijin Duan and Wentao Ding contributed equally to this work
Contributor Information
Lijin Duan, Email: duan_lj@tib.cas.cn.
Wentao Ding, Email: ding_wt@tib.cas.cn.
Xiaonan Liu, Email: liu_xn@tib.cas.cn.
Xiaozhi Cheng, Email: cheng_xzh@tib.cas.cn.
Jing Cai, Email: jingcai@umac.mo.
Erbing Hua, Email: huarb@tust.edu.cn.
Huifeng Jiang, Email: jiang_hf@tib.cas.cn.
References
- 1.Calderón-Montaño JM, Burgos-Morón E, Pérez-Guerrero C, López-Lázaro M. A review on the dietary flavonoid kaempferol. Mini Rev Med Chem. 2011;11:298. doi: 10.2174/138955711795305335. [DOI] [PubMed] [Google Scholar]
- 2.Chen AY, Chen YC. A review of the dietary flavonoid, kaempferol on human health and cancer chemoprevention. Food Chem. 2013;138:2099–2107. doi: 10.1016/j.foodchem.2012.11.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Granado-Serrano AB, Martin MA, Bravo L, Goya L, Ramos S. Quercetin induces apoptosis via caspase activation, regulation of Bcl-2, and inhibition of PI-3-kinase/Akt and ERK pathways in a human hepatoma cell line (HepG2) J Nutr. 2006;136:2715–2721. doi: 10.1093/jn/136.11.2715. [DOI] [PubMed] [Google Scholar]
- 4.Chandramohan G, Al-Numair KS, Alsaif MA, Veeramani C. Antidiabetic effect of kaempferol a flavonoid compound, on streptozotocin-induced diabetic rats with special reference to glycoprotein components. Prog Nutr. 2015;17:50–57. [Google Scholar]
- 5.Muthukrishnan SD, Kaliyaperumal A, Subramaniyan A. Identification and determination of flavonoids, carotenoids and chlorophyll concentration in Cynodon dactylon (L.) by HPLC analysis. Nat Prod Res. 2015;29:785–790. doi: 10.1080/14786419.2014.986125. [DOI] [PubMed] [Google Scholar]
- 6.Agar OT, Dikmen M, Ozturk N, Yilmaz MA, Temel H, Turkmenoglu FP. Comparative studies on phenolic composition, antioxidant, wound healing and cytotoxic activities of selected Achillea L. Species Growing in Turkey. Molecules. 2015;20:17976–18000. doi: 10.3390/molecules201017976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Koopman F, Beekwilder J, Crimi B, Van HA, Hall RD, Bosch D, van Maris AJ, Pronk JT, Daran JM. De novo production of the flavonoid naringenin in engineered Saccharomyces cerevisiae. Microb Cell Fact. 2012;11:155. doi: 10.1186/1475-2859-11-155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhu S, Wu J, Du G, Zhou J, Chen J. Efficient synthesis of eriodictyol from l-tyrosine in Escherichia coli. Appl Environ Microbiol. 2014;80:3072. doi: 10.1128/AEM.03986-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Trantas E, Panopoulos N, Ververidis F. Metabolic engineering of the complete pathway leading to heterologous biosynthesis of various flavonoids and stilbenoids in Saccharomyces cerevisiae. Metab Eng. 2009;11:355–366. doi: 10.1016/j.ymben.2009.07.004. [DOI] [PubMed] [Google Scholar]
- 10.Leonard E, Yan Y, Koffas MA. Functional expression of a P450 flavonoid hydroxylase for the biosynthesis of plant-specific hydroxylated flavonols in Escherichia coli. Metab Eng. 2006;8:172–181. doi: 10.1016/j.ymben.2005.11.001. [DOI] [PubMed] [Google Scholar]
- 11.Miyahisa I, Funa N, Ohnishi Y, Martens S, Moriguchi T, Horinouchi S. Combinatorial biosynthesis of flavones and flavonols in Escherichia coli. Appl Microbiol Biotechnol. 2006;71:53–58. doi: 10.1007/s00253-005-0116-5. [DOI] [PubMed] [Google Scholar]
- 12.Janssen HJ. Fatty acid synthesis in Escherichia coli and its applications towards the production of fatty acid based biofuels. Biotechnol Biofuels. 2014;7:7. doi: 10.1186/1754-6834-7-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cronan JE, Thomas J. Bacterial fatty acid synthesis and its relationships with polyketide synthetic pathways. Complex Enzym Microbial Nat Prod Biosynth Part B Polyketides Aminocoumarins Carbohydr. 2009;459:395–433. doi: 10.1016/S0076-6879(09)04617-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Leonard E, Lim KH, Saw PN, Koffas MA. Engineering central metabolic pathways for high-level flavonoid production in Escherichia coli. Appl Environ Microbiol. 2007;73:3877–3886. doi: 10.1128/AEM.00200-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu W, Zhang B, Jiang R. Improving acetyl-CoA biosynthesis in Saccharomyces cerevisiae via the overexpression of pantothenate kinase and PDH bypass. Biotechnol Biofuels. 2017;10:41. doi: 10.1186/s13068-017-0726-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Owens DK, Crosby KC, Runac J, Howard BA, Winkel BSJ. Biochemical and genetic characterization of Arabidopsis flavanone 3β-hydroxylase. Plant Physiol Biochem Ppb. 2008;46:833–843. doi: 10.1016/j.plaphy.2008.06.004. [DOI] [PubMed] [Google Scholar]
- 17.Halbwirth H, Fischer TC, Schlangen K, Rademacher W, Schleifer K-J, Forkmann G, Stich K. Screening for inhibitors of 2-oxoglutarate-dependent dioxygenases: flavanone 3β-hydroxylase and flavonol synthase. Plant Sci. 2006;171:194–205. doi: 10.1016/j.plantsci.2006.03.014. [DOI] [Google Scholar]
- 18.Falcone Ferreyra ML, Rius S, Emiliani J, Pourcel L, Feller A, Morohashi K, Casati P, Grotewold E. Cloning and characterization of a UV-B-inducible maize flavonol synthase. Plant J. 2010;62:77–91. doi: 10.1111/j.1365-313X.2010.04133.x. [DOI] [PubMed] [Google Scholar]
- 19.Pelletier MK, Murrell JR, Shirley BW. Characterization of flavonol synthase and leucoanthocyanidin dioxygenase genes in Arabidopsis. Further evidence for differential regulation of “early” and “late” genes. Plant Physiol. 1997;113:1437–1445. doi: 10.1104/pp.113.4.1437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lukačin R, Wellmann F, Britsch L, Martens S, Matern U. Flavonol synthase from Citrus unshiu is a bifunctional dioxygenase. Phytochemistry. 2003;62:287–292. doi: 10.1016/S0031-9422(02)00567-8. [DOI] [PubMed] [Google Scholar]
- 21.Yang SM, Han SH, Kim BG, Ahn JH. Production of kaempferol 3-O-rhamnoside from glucose using engineered Escherichia coli. J Ind Microbiol Biotechnol. 2014;41:1311–1318. doi: 10.1007/s10295-014-1465-9. [DOI] [PubMed] [Google Scholar]
- 22.Prescott AG, Stamford NP, Wheeler G, Firmin JL. In vitro properties of a recombinant flavonol synthase from Arabidopsis thaliana. Phytochemistry. 2002;60:589–593. doi: 10.1016/S0031-9422(02)00155-3. [DOI] [PubMed] [Google Scholar]
- 23.Kim BG, Joe EJ, Ahn JH. Molecular characterization of flavonol synthase from poplar and its application to the synthesis of 3-O-methylkaempferol. Biotechnol Lett. 2010;32:579–584. doi: 10.1007/s10529-009-0188-x. [DOI] [PubMed] [Google Scholar]
- 24.Malla S, Pandey RP, Kim BG, Sohng JK. Regiospecific modifications of naringenin for astragalin production in Escherichia coli. Biotechnol Bioeng. 2013;110:2525–2535. doi: 10.1002/bit.24919. [DOI] [PubMed] [Google Scholar]
- 25.Leonard E, Yan Y, Fowler ZL, Li Z, Lim CG, Lim KH, Koffas MAG. Strain improvement of recombinant Escherichia coli for efficient production of plant flavonoids. Mol Pharm. 2007;5:257–265. doi: 10.1021/mp7001472. [DOI] [PubMed] [Google Scholar]
- 26.Xu P, Ranganathan S, Fowler ZL, Maranas CD, Koffas MAG. Genome-scale metabolic network modeling results in minimal interventions that cooperatively force carbon flux towards malonyl-CoA. Metab Eng. 2011;13:578–587. doi: 10.1016/j.ymben.2011.06.008. [DOI] [PubMed] [Google Scholar]
- 27.Tehlivets O, Scheuringer K, Kohlwein SD. Fatty acid synthesis and elongation in yeast. Biochim Biophys Acta. 2007;1771:255–270. doi: 10.1016/j.bbalip.2006.07.004. [DOI] [PubMed] [Google Scholar]
- 28.Woods A, Munday MR, Scott J, Yang X, Carlson M, Carling D. Yeast SNF1 is functionally related to mammalian AMP-activated protein kinase and regulates acetyl-CoA carboxylase in vivo. J Biol Chem. 1994;269:19509–19515. [PubMed] [Google Scholar]
- 29.Shi S, Chen Y, Siewers V, Nielsen J. Improving production of malonyl coenzyme A-derived metabolites by abolishing Snf1-dependent regulation of Acc1. MBio. 2014;5:e01130-14. doi: 10.1128/mBio.01130-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shi S, Valle-Rodriguez JO, Khoomrung S, Siewers V, Nielsen J. Functional expression and characterization of five wax ester synthases in Saccharomyces cerevisiae and their utility for biodiesel production. Biotechnol Biofuels. 2012;5:7. doi: 10.1186/PREACCEPT-1932279820621895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Choi JW, Da Silva NA. Improving polyketide and fatty acid synthesis by engineering of the yeast acetyl-CoA carboxylase. J Biotechnol. 2014;187:56–59. doi: 10.1016/j.jbiotec.2014.07.430. [DOI] [PubMed] [Google Scholar]
- 32.Kildegaard KR, Jensen NB, Schneider K, Czarnotta E, Ozdemir E, Klein T, Maury J, Ebert BE, Christensen HB, Chen Y, et al. Engineering and systems-level analysis of Saccharomyces cerevisiae for production of 3-hydroxypropionic acid via malonyl-CoA reductase-dependent pathway. Microb Cell Fact. 2016;15:53. doi: 10.1186/s12934-016-0451-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Davis MS, Solbiati J, Cronan JE., Jr Overproduction of acetyl-CoA carboxylase activity increases the rate of fatty acid biosynthesis in Escherichia coli. J Biol Chem. 2000;275:28593–28598. doi: 10.1074/jbc.M004756200. [DOI] [PubMed] [Google Scholar]
- 34.Bhan N, Xu P, Khalidi O, Koffas MAG. Redirecting carbon flux into malonyl-CoA to improve resveratrol titers: proof of concept for genetic interventions predicted by OptForce computational framework. Chem Eng Sci. 2013;103:109–114. doi: 10.1016/j.ces.2012.10.009. [DOI] [Google Scholar]
- 35.Li X, Guo D, Cheng Y, Zhu F, Deng Z, Liu T. Overproduction of fatty acids in engineered Saccharomyces cerevisiae. Biotechnol Bioeng. 2014;111:1841–1852. doi: 10.1002/bit.25239. [DOI] [PubMed] [Google Scholar]
- 36.Xu P, Li L, Zhang F, Stephanopoulos G, Koffas M. Improving fatty acids production by engineering dynamic pathway regulation and metabolic control. Proc Natl Acad Sci USA. 2014;111:11299–11304. doi: 10.1073/pnas.1406401111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chen WN, Tan KY. Malonate uptake and metabolism in Saccharomyces cerevisiae. Appl Biochem Biotechnol. 2013;171:44–62. doi: 10.1007/s12010-013-0334-8. [DOI] [PubMed] [Google Scholar]
- 38.Malla S, Koffas MA, Kazlauskas RJ, Kim BG. Production of 7-O-Methyl aromadendrin, a medicinally valuable flavonoid, in Escherichia coli. Appl Environ Microbiol. 2012;78:684–694. doi: 10.1128/AEM.06274-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rodriguez A, Strucko T, Stahlhut SG, Kristensen M, Svenssen DK, Forster J, Nielsen J, Borodina I. Metabolic engineering of yeast for fermentative production of flavonoids. Bioresour Technol. 2017 doi: 10.1016/j.biortech.2017.06.043. [DOI] [PubMed] [Google Scholar]
- 40.Tippmann S, Scalcinati G, Siewers V, Nielsen J. Production of farnesene and santalene by Saccharomyces cerevisiae using fed-batch cultivations with RQ-controlled feed. Biotechnol Bioeng. 2016;113:72–81. doi: 10.1002/bit.25683. [DOI] [PubMed] [Google Scholar]
- 41.Rodriguez A, Kildegaard KR, Li M, Borodina I, Nielsen J. Establishment of a yeast platform strain for production of p-coumaric acid through metabolic engineering of aromatic amino acid biosynthesis. Metab Eng. 2015;31:181–188. doi: 10.1016/j.ymben.2015.08.003. [DOI] [PubMed] [Google Scholar]
- 42.Jiang NH, Zhang GH, Zhang JJ, Shu LP, Zhang W, Long GQ, Liu T, Meng ZG, Chen JW, Yang SC. Analysis of the transcriptome of Erigeron breviscapus uncovers putative scutellarin and chlorogenic acids biosynthetic genes and genetic markers. PLoS ONE. 2014;9:e100357. doi: 10.1371/journal.pone.0100357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shubeita HE, Sambrook JF, Mccormick AM. Molecular-cloning and analysis of functional cdna and genomic clones encoding bovine cellular retinoic acid-binding protein. Proc Natl Acad Sci USA. 1987;84:5645–5649. doi: 10.1073/pnas.84.16.5645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Quan JY, Tian JD. Circular polymerase extension cloning for high-throughput cloning of complex and combinatorial DNA libraries. Nat Protoc. 2011;6:242–251. doi: 10.1038/nprot.2010.181. [DOI] [PubMed] [Google Scholar]
- 45.Li S, Ding W, Zhang X, Jiang H, Bi C. Development of a modularized two-step (M2S) chromosome integration technique for integration of multiple transcription units in Saccharomyces cerevisiae. Biotechnol Biofuels. 2016;9:232. doi: 10.1186/s13068-016-0645-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shiba Y, Paradise EM, Kirby J, Ro DK, Keasing JD. Engineering of the pyruvate dehydrogenase bypass in Saccharomyces cerevisiae for high-level production of isoprenoids. Metab Eng. 2007;9:160–168. doi: 10.1016/j.ymben.2006.10.005. [DOI] [PubMed] [Google Scholar]
- 47.Gibson DG, Young L, Chuang RY, Venter JC, Rd HC, Smith HO. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat Methods. 2009;6:343–345. doi: 10.1038/nmeth.1318. [DOI] [PubMed] [Google Scholar]
- 48.Shao Z, Zhao H, Zhao H. DNA assembler, an in vivo genetic method for rapid construction of biochemical pathways. Nucleic Acids Res. 2009;37:e16. doi: 10.1093/nar/gkn991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Paddon CJ, Westfall PJ, Pitera DJ, Benjamin K, Fisher K, McPhee D, Leavell MD, Tai A, Main A, Eng D, et al. High-level semi-synthetic production of the potent antimalarial artemisinin. Nature. 2013;496:528–532. doi: 10.1038/nature12051. [DOI] [PubMed] [Google Scholar]
- 50.Gietz RD, Akio S. New yeast-Escherichia coli shuttle vectors constructed with in vitro mutagenized yeast genes lacking six-base pair restriction sites. Gene. 1989;74:527–534. doi: 10.1016/0378-1119(88)90185-0. [DOI] [PubMed] [Google Scholar]
- 51.Thomas BJ, Rothstein R. Elevated recombination rates in transcriptionally active DNA. Cell. 1989;56:619. doi: 10.1016/0092-8674(89)90584-9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data generated or analyzed during this study are included in this published article.


