Abstract
There remains an urgent need for rapid diagnostic methods that can evaluate antibiotic resistance for pathogenic bacteria in order to deliver targeted antibiotic treatments. Toward this end, we present a rapid and integrated single-cell biosensing platform, termed dropFAST, for bacterial growth detection and antimicrobial susceptibility assessment. DropFAST utilizes a rapid resazurin-based fluorescent growth assay coupled with stochastic confinement of bacteria in 20 pL droplets to detect signal from growing bacteria after 1 h incubation, equivalent to 2–3 bacterial replications. Full integration of droplet generation, incubation, and detection into a single, uninterrupted stream also renders this platform uniquely suitable for in-line bacterial phenotypic growth assessment. To illustrate the concept of rapid digital antimicrobial susceptibility assessment, we employ the dropFAST platform to evaluate the antibacterial effect of gentamicin on E. coli growth.
Keywords: Droplet, microfluidics, antimicrobial, susceptibility, test, picoliter
1. Introduction
The emergence of multi-drug resistant bacteria due to indiscriminate use of broad-spectrum antibiotics has grown into a global healthcare crisis (Boucher et al., 2009; Carlet et al., 2011; Finley et al., 2013; Kunin et al., 1973; Roca et al., 2015). To combat this threat, there is an urgent need for diagnostic methods that can rapidly evaluate antibiotic resistance of these dangerous bacteria to ensure timely and targeted antibiotic treatments (Exec. Order No. 13676, 3 C.F.R., 2014; Laxminarayan et al., 2013; Spellberg et al., 2011). To this end, various genetic techniques have been proposed based on amplification and detection of known resistance-conferring genes in bacteria (Tenover, 2010). However, due to constantly evolving genetic mechanisms of resistance, such approaches are less reliable than direct phenotypic detection of bacterial growth (Mach et al., 2011). Phenotypic methods for evaluating antibiotic resistance, in which bacteria are directly grown in the presence of various antibiotics to determine their sensitivity or resistance, are limited by a lengthy assay procedure. Indeed, current standard phenotypic antimicrobial susceptibility tests (AST) in clinical laboratories (e.g., broth dilution and disk diffusion) require a 16 to 20 h incubation period (Davenport et al., 2017; Performance Standards for Antimicrobial Susceptibility Testing; Twenty-Fourth Informational Supplement, 2014). Although many strains of bacteria grow rapidly and some (e.g., E. coli) can even replicate in as little as every 20 minutes (Fossum et al., 2007; Parija, 2016; Powell, 1956), conventional AST still requires a lengthy incubation period. This is in part attributed to the large volumes in which bacteria are cultured and measured (100 μL to 20 mL), which necessitate growing a high number of bacteria to ensure reliable interpretation of bacterial growth. This observation suggests that a reduction of analysis volume can reduce the incubation time necessary for assessing antibiotic resistance. Moreover, this incubation time may be further reduced to the timescale of individual bacterial replication if such an event can be reliably observed at the single-cell level. In short, a technology capable of handling small sample volumes and detecting the replication of individual bacteria can drastically accelerate phenotypic assessment of antimicrobial susceptibility.
Microfluidics is well-suited for handling reaction volumes at microliters and below. Measuring reactions confined in small volumes benefits from high local signal-to-background ratio, which can accelerate single-cell detection and measurements. Indeed, researchers have reported various microchambers (Balaban et al., 2004; Choi et al., 2013; Mohan et al., 2013; Sun et al., 2011) and microchannels (Chen et al., 2010; Li et al., 2014; Lu et al., 2013; Mai et al., 2014; Wakamoto et al., 2013) that can couple with microscopy to detect the growth and replication of single bacterial cells. Unfortunately, with throughputs limited to the order of tens of channels and chambers per device, these devices lack the statistical capacity to analyse many cells and account for the inherent heterogeneity of the bacterial population (Delvigne et al., 2014; Nichols et al., 2011). Furthermore, these devices frequently rely on additional materials (e.g., cellulose membranes (Balaban et al., 2004; Wakamoto et al., 2013) and agarose gels (Choi et al., 2013; Eun et al., 2011; Li et al., 2014)) or external forces (e.g., electric force (Lu et al., 2013)) to trap bacteria, which increase the complexity of device fabrication and operation. In part to address such challenges, droplet microfluidics recently emerged as an approach for single-cell isolation, detection, and analysis (Kaminski et al., 2016; Yan et al., 2016). Similar to microchambers and microchannels, microfluidic droplets enable volume reduction, high local signal-to-background ratio, and accelerated time to detection (Boedicker et al., 2008; Ng et al., 2016; Tushar D. Rane et al., 2012). Critically, droplet-based platforms employ passive “flow-focusing” structures to generate droplets that can stochastically encapsulate single bacteria at around 100 to 1000 Hz and detection modules that can exclude empty droplets from analysis. Droplet-based platforms therefore offer a compelling alternative for accelerating single-cell detection and measurements.
Despite these promising attributes, droplet microfluidics has not been able to reduce the analysis time of antimicrobial susceptibility assessment to the timescale of bacterial replication. In an early example, Boedicker et al. tested antimicrobial susceptibility of S. aureus after incubating the bacteria for 7 h in the presence of antibiotics via a single-step fluorescence assay (Boedicker et al., 2008). The relatively large droplet volume of 4 nL in this work necessitated 7 h incubation for reliable growth detection. In a later work, Liu et al. used smaller droplets of 330 pL for detecting a mutant resistant to fusidic acid, but required 8 h incubation due to a less-sensitive non-fluorescent detection approach based on light scattering (Liu et al., 2016). Notably, these techniques were demonstrated on experimental workflows requiring a separate device for droplet generation, an incubator for off-chip droplet incubation, and an additional device for droplet detection (Boedicker et al., 2008; Liu et al., 2009, 2016). To accelerate the analysis time for assessing antimicrobial susceptibility to the timescale of bacterial replication, an integrated workflow that ensures precise temporal control from single-bacteria confinement to detection must also be implemented.
Herein, we present a rapid and integrated method for single-bacteria growth analysis and antimicrobial susceptibility assessment, dropFAST (droplet-based Fluorescent Antimicrobial Susceptibility Test). In dropFAST, we encapsulate and incubate single bacterial cells in picoliter-sized droplets that are more than ten-fold smaller than those reported in earlier works, and we use a fluorescence assay for detection of bacterial growth. This combination allows us to substantially reduce the required time for detecting signal from growing bacteria to approach the fundamental limit of growth determination – the bacterial replication time. Importantly, the shortened incubation time minimizes the device footprint, which facilitates the integration of single bacterial cell encapsulation, bacteria culturing, and fluorescence-based bacterial growth detection into a streamlined workflow within a monolithic device. As an additional benefit, dropFAST routinely analyzes ~104–105 individual bacterial droplets in each experiment; such throughput confers the statistical capacity to account for potential inaccuracies due to bacterial population heterogeneity. As a demonstration, we assessed the antibacterial effect of the antibiotic gentamicin on the growth of E. coli with single-cell resolution after only 1 h of on-chip incubation.
2. Materials and methods
2.1. Design and fabrication of microfluidic device
The microfluidic device design consists of a 20 μm flow-focusing nozzle for droplet generation, followed by a 500-μm-wide serpentine incubation channel that spans roughly 1.75 m and then a 10 μm droplet detection window. A casting mold was fabricated by spinning a 20 μm layer of SU8-3050 photoresist (MicroChem) onto a 4 in. silicon wafer and patterning using standard photolithography. The microfluidic devices were made of polydimethylsiloxane (PDMS) by pouring 30 g of 10:1 ratio of Sylgard 184 (Dow Corning) base to curing agent onto the SU8 mold. After curing the PDMS replica, it was permanently bonded to cover glass (130 μm thickness, Ted Pella) through oxygen plasma treatment in order to seal the channels. Prior to operation, the microfluidic chips were treated with Aquapel and baked at 80 °C for ≥ 20 minutes to render microfluidic channel surfaces hydrophobic.
2.2. Operation of microfluidic device
Frozen stocks of E. coli (ATCC 25922, ATCC BAA 2471) were thawed, washed twice, and diluted to 107 CFU/mL in Muller-Hinton II cation adjusted broth (Sigma-Aldrich). Separately, 400 μM resazurin (Sigma-Aldrich) was mixed with either 0 μg/mL or 8 μg/mL gentamicin (Sigma-Aldrich) in Mueller-Hinton broth. Both bacterial sample and resazurin/antibiotic solution were then drawn into separate 1-m-long sections of Tygon tubing (Cole-Parmer) with an inner diameter of around 500 μm. Both Tygon tubing sections were individually connected to Hamilton 1000 glass syringes (Sigma-Aldrich) containing FC-40 oil (Sigma-Aldrich), which served as the displacement fluid for injecting both aqueous samples from Tygon tubings into the device using a syringe pump at 15 μL/h (Harvard Apparatus). An oil phase consisting of FC-40 oil and 5% poly(ethylene glycol) di-(krytox-FSH amide) surfactant by weight was pumped into the device through the oil inlet of the device at 60 μL/h by a separate syringe pump. To confirm stable droplet generation, the device was imaged using a 4× objective lens and a CCD camera during droplet generation and after droplet incubation. Droplet incubation was conducted on chip at 37 °C using a controllable peltier heater on which the microfluidic device rested.
2.3. Continuous-flow droplet detection
Continuous-flow droplet detection was conducted using a custom optical stage consisting of a 552 nm laser excitation source (OBIS, Coherent, Inc.) and a silicon avalanche photodiode detector (APD) (SPCM-AQRH13, ThorLabs). The laser was operated at 1 mW power and was focused into the detection zone of the device using a 40× objective (Thorlabs RMS40X-PF, NA 0.75, focal depth ~ 0.6 μm). As droplets flowed through the detection zone, fluorescence data was continuously acquired by the APD with 0.1 ms sampling time. A custom LabVIEW program was used to control fluorescence data acquisition.
3. Results and Discussion
3.1. Rapid and integrated dropFAST platform
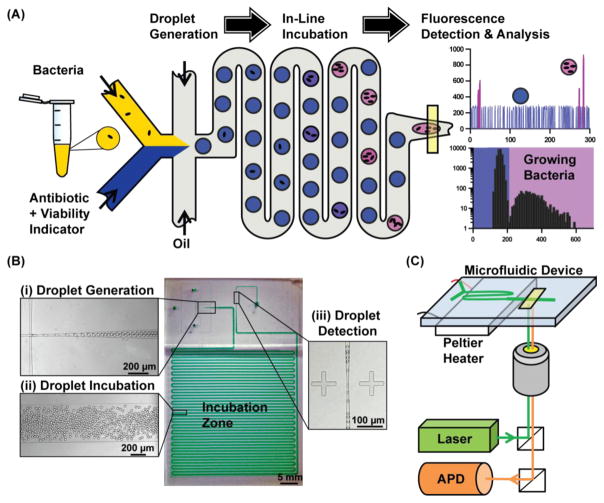
Our dropFAST platform achieves single-cell isolation, culture, and detection within picoliter droplets in a streamlined, single-step workflow (Fig. 1(A)). In this platform, a droplet microfluidic device is used to stochastically confine single bacteria in picoliter droplets that contain a culture broth (Mueller-Hinton broth), a fluorescent bacterial growth indicator dye, and/or an antibiotic. After droplet encapsulation, bacteria can grow within these droplets as they flow through a 37 °C incubation region. The platform utilizes a commonly-used growth indicator dye, resazurin, to directly detect bacterial growth within droplets in a single step via fluorescence. In the presence of cellular metabolic functions of any species or strains of bacteria, resazurin is reduced by the intracellular electron receptors NADH and FADH to form the brightly fluorescent resorufin (Bueno et al., 2002). This indicator allows us to clearly differentiate droplets that have encapsulated actively growing bacteria (represented as pink in Fig. 1(A)), from droplets which contain no bacteria or where antibiotics may have prevented growth (represented as blue in Fig. 1(A)). By leveraging picoliter volume droplets and the resazurin-based fluorescence assay, dropFAST is able to detect bacterial growth after only a few bacterial replication events. Fluorescence signals from all droplets are analyzed via a histogram to assess and quantify the growth of the bacterial population after incubation. Importantly, our platform operates in a continuous flow, whereby droplet generation, incubation, and detection take place in a single, uninterrupted stream. This feature makes our platform suitable for high-throughput analysis, and in principle, continuous in-line operation for handling large sample volumes.
Fig. 1.
dropFAST integrated platform. (A) For a bacterial sample, dropFAST enables encapsulation of single bacterial cells along with a fluorescent bacterial growth indicator dye and antibiotics into picoliter-sized droplets. This is immediately followed by short on-chip incubation that allow for a few bacterial replications and then in-line fluorescence detection. Empty droplets emit weak fluorescence, while droplets with growing bacteria emit strong fluorescence. Growing bacteria in a sample is then quantified via histogram analysis of droplet fluorescence. (B) Integrated dropFAST device parallelizes droplet generation, incubation, and detection in a continuous flow. The droplet generator consists of a 20 μm flow-focusing region that facilitates stable droplet generation (inset i), followed by a 500-μm-wide incubation zone (inset ii), and finally a 10-μm-wide detection zone that allows for sequential detection of droplets (inset iii). (C) Integrated dropFAST platform includes instrumentation to facilitate heated incubation using a Peltier heater and streamlined fluorescence detection using a laser excitation source and an avalanche photodiode (APD) detector.
We have developed a PDMS-based, monolithic microfluidic device that is coupled with a heating and optical detection instrument to realize the integrated dropFAST platform. Our device is fabricated via PDMS soft-lithography (Tang and Whitesides, 2010; Xia and Whitesides, 1998) (Fig. 1(B)) and features a 20-μm-wide flow-focusing junction where equal parts of the sample and the reagent (injected via independent inlets) combine and are sheared by oil into ~20 pL droplets with high uniformity and stability (Fig. 1(B), inset (i)). A single bacterium confined in a 20-pL droplet drastically elevates the local bacterial concentration (equivalent to 5×107 cells/mL in bulk analysis), which enables early detection of bacterial growth after only a few replication events. After generation, the droplets flow through a serpentine incubation channel for on-chip incubation (Fig. 1(B), inset (ii)). Finally, each droplet sequentially funnels through a 10-μm-wide detection zone where the fluorescence emitted from each droplet can be individually measured. The narrow detection zone stretches each passing droplet, which increases the transit time of each droplet through the observation volume and enables collection of additional data points for a reliable fluorescence-based quantification of each droplet (Fig. 1(B), inset (iii)). We have also developed an instrument that operates in tandem with the dropFAST device and facilitates in-line droplet incubation and detection. The instrument is composed of a Peltier heater and a custom-built fluorescence detector with a laser illumination source and a silicon avalanche photodiode (APD) detector (Fig. 1(C)). The dropFAST device is mounted on the Peltier heater such that the incubation channel can be uniformly heated at 37 °C to facilitate bacterial growth within droplets as they flow through the incubation channel. A laser is focused within the detection zone of the device to continuously illuminate traversing droplets, and an APD fluorescence detector is used to monitor fluorescent events as the droplets are passed from the incubation channel. The combination of a microfluidic device and instrumentation enables full integration of the dropFAST assay under a simple, streamlined workflow, obviating the need for complex robotic fluid handling systems or labor-intensive protocols to integrate processes in separate modules.
3.2. Single-cell encapsulation, bacterial growth, and fluorescence in picoliter droplets
Prior to realizing the dropFAST workflow, we validated single bacterial cell encapsulation, bacterial growth, and fluorescence emission in picoliter droplets. In these experiments, we aimed to encapsulate single bacterial cells in ~10% of droplets (i.e., mean occupancy λ = 0.1) by loading 107 CFU/mL of E.coli (ATCC 25922) diluted in Mueller-Hinton broth into one of the two inlets of our device. We loaded 400 μM resazurin in Mueller-Hinton broth into the other inlet of our device and commenced droplet generation. E. coli encapsulated in droplets were directly incubated at 37 °C as they flowed through the incubation channel in ~1 h. We then stopped all flows and examined the stationary droplets in the device via microscopy. Approximately 1 in every 10 droplets located immediately downstream to the flow-focusing junction contains one E. coli cell and the rest of the droplets contain no cells (Fig. S1(A)), which is on par with the expected mean occupancy of 0.1. Near the end of the incubation zone, about every 1 in 10 droplets contains as many as four E. coli cells and the rest of the droplets are empty (Fig. S1(B)), indicating that the encapsulated cells had replicated during incubation. Furthermore, the droplets containing growing bacteria (Fig. S2(A)) emit strong fluorescence under fluorescence microscopy (Fig. S2(B)). These results indicate that as few as 2 bacterial replications can produce detectable fluorescence signal in droplets.
We also showed that resazurin and the surfactant used to stabilize droplets pose negligible inhibition to E. coli growth. We exposed E. coli samples to either 200 μM resazurin or no resazurin (i.e., control), as well as to either a 4× solution of oil/surfactant or no oil/surfactant (i.e., control) for 1 h at 37 °C and plated these samples overnight. We counted comparable number of colonies between the controls and the test samples (Fig. S3A, S3B), thus confirming that both resazurin and the surfactant are benign to E. coli growth.
3.3 Rapid bacterial growth detection and quantification via fluorescence
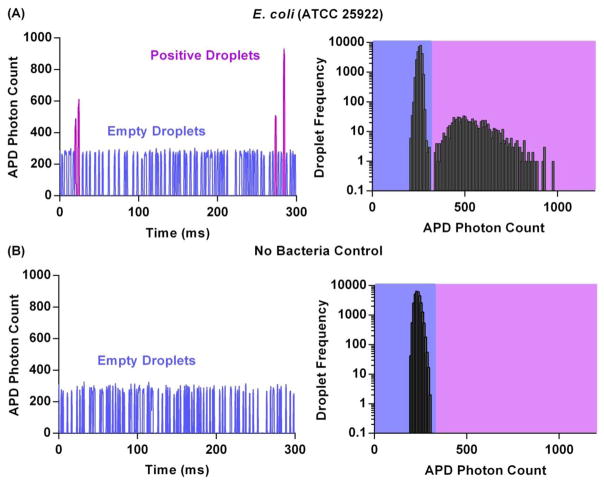
The dropFAST platform is capable of detecting bacterial growth in a sample after 1 h incubation by measuring and recording fluorescence emitted from individual picoliter droplets generated from the original sample. To demonstrate rapid detection of growth, we introduced E. coli spiked in Mueller-Hinton broth and resazurin (without any antibiotics) into our device and performed the dropFAST operating routine, which consists of droplet generation followed by 1 h incubation at 37 °C and then in-line fluorescence detection. While we maintained a relatively high mean occupancy of λ = 0.1 and E. coli concentration of 107 CFU/mL in this demonstration, it should be noted that our platform can readily detect rare positive droplets from ~106 droplets, and therefore has the capacity to operate with much lower mean occupancies and input bacterial concentration. As the droplets complete incubation and sequentially flow into the detection zone of the device, the APD registers and records a fluorescence time trace where individual peaks represent each droplet passing through the detection zone. When E. coli cells are encapsulated and grown in droplets, they emit a strong fluorescence signal due to local accumulation of fluorescent resorufin, resulting in tall “positive” peaks in the fluorescence time trace. In contrast, for droplets that contain no bacteria, a shorter, weakly fluorescent baseline signal is detected (Fig. 2(A)).
Fig. 2.
Rapid bacterial growth quantification with E. coli (A) The fluorescence intensity of each droplet is quantified. Droplets containing growing bacteria (positive droplets) emit greater fluorescence than empty droplets. The data is further plotted in a histogram with bin size of ~10 for population distribution analysis. For an input concentration of 107 CFU/mL of E.coli, an average of 903 ± 137 droplets out of 30000 total droplets comprise of the positive droplet population. Standard deviation is from triplicate experiments. (B) When no bacteria are input into the platform, no positive peaks are detected and only the negative population of background/empty droplets is present.
Histogram analysis of droplet fluorescence offers an effective means to quantify the population of growing bacteria in a sample. From the recorded fluorescence time trace of the E. coli containing droplets, a histogram of the average droplet fluorescence intensity was plotted for 30000 of the detected droplets. This results in a bimodal distribution, consisting of two distinct subpopulations, which represent the weakly fluorescent empty droplets (relative standard deviation (RSD) = 4.3%) and strongly fluorescent “positive” droplets (RSD = 21.8%). The wider spread in the positive droplet population can be explained by the heterogeneity inherent to the bacterial population. Because the histogram of droplet fluorescence is distinctly bimodal, consisting of a mixture of subpopulations, a Gaussian mixture model (GMM) is employed to fit the data and extract the individual subpopulation means and their relative weights (Chan et al., 2008; Lin et al., 2016) (Fig. S4). Using this approach, we calculate that from the 30000 detected droplets, on average 903 (± 137) are positive droplets, containing growing colonies of E. coli. Histogram analysis thus enables quantification of growing bacteria in each sample.
As a negative control, we verified that in the absence of bacteria, no “positive” peaks are detected. We loaded our device without any bacteria and only Mueller-Hinton broth along with resazurin and performed the dropFAST operating routine. By studying the time trace of fluorescence emitted by these droplets, we notice that all detected droplets exhibit approximately the same weak fluorescence intensity – that of empty droplets (Fig. 2(B)). Histogram analysis here results in a unimodal distribution consisting of only a single empty droplet population.
3.4 Rapid assessment of antibiotic effect on bacterial growth
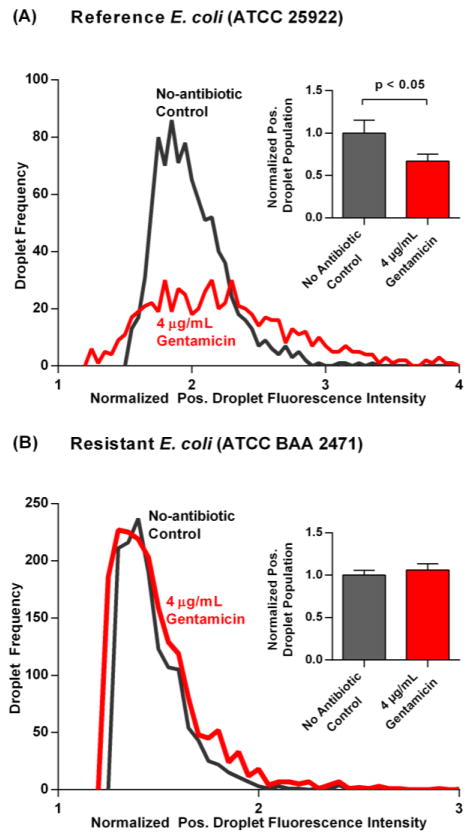
Inhibition of bacterial growth due to antibiotics can be detected after 1 h incubation using the dropFAST platform. To demonstrate this, we tested our reference E. coli strain against an inhibitory concentration of gentamicin (Performance Standards for Antimicrobial Susceptibility Testing; Twenty-Fourth Informational Supplement, 2014), a commonly used intravenous antibiotic for infections caused by gram-negative bacteria such as E. coli. We first confirmed that gentamicin inhibited the growth of our reference strain of E. coli using a bulk broth dilution assay. After 16 h incubation of E.coli with 4 μg/mL gentamicin, there is no discernable growth of bacteria, as determined by the sample’s lack of turbidity by visual inspection (Fig. S5). Next, the antibacterial effect of gentamicin on the reference strain of E. coli was rapidly determined through the dropFAST platform. We spiked E. coli in Mueller-Hinton broth and resazurin along with gentamicin into our device and performed the dropFAST assay. After measuring fluorescence emitted from 30000 droplets, we performed histogram analysis to determine the negative and positive subpopulations of our droplet data. For both no-antibiotic control and the gentamicin-containing sample, we normalized the fluorescence intensities of the positive droplets to the average fluorescence intensities of the negative droplets and then plotted histograms of the normalized fluorescence intensities of the positive droplets. Comparing the resulting histograms, it is clear that the addition of gentamicin reduces the frequency of positive droplets (Fig. 3(A)). Indeed, when E. coli was exposed to gentamicin, only 606 ± 71 droplets out of 30000 droplets emit strong fluorescence (from triplicated experiments). To facilitate direct comparison, this positive droplet population was then normalized to the positive droplet population of the no-antibiotic control. Compared to the control (1.00 ± 0.15), there is a significant reduction (p < 0.05) of the positive droplet population in the presence of gentamicin (0.67 ± 0.08) (Fig. 3(A), inset). Of note, we observe a residual population of positive droplets even in this high dose of gentamicin, which presumably arise from E. coli metabolizing and converting resazurin to fluorescent resorufin before their growth is fully inhibited by gentamicin. Considering that the biochemical assay employed in the dropFAST platform is identical to bulk resazurin-based AST, it is likely that the drastic enhancement in sensitivity afforded by the dropFAST platform reveals a subpopulation that is slow-acting to the effects of gentamicin at higher doses. We anticipate that these observations may help to encourage further investigation of phenotypic heterogeneity in drug-susceptible bacteria. It should be noted that in addition to the enhanced sensitivity, the dropFAST platform is able to assess the antibacterial effect of gentamicin with a much higher throughput and shorter incubation time than any other phenotypic AST methods found in literature.
Fig. 3.
(A) Histograms of the fluorescence intensity detected from positive droplets normalized to their respective average negative droplet fluorescence are plotted for a reference strain of E. coli without and with the antibiotic gentamicin. In the presence of 4 μg/mL of gentamicin, a smaller positive droplet population is observed. The positive droplet population is then normalized to the average positive droplet population from the no-antibiotic control and plotted side-by-side for comparison. There is a significant reduction in percentage of positive droplets (p < 0.05) for the reference strain of E. coli when subject to gentamicin; (B) For a multi-drug resistant strain of E. coli subject to the same antibiotic conditions, histograms of positive droplet fluorescence looks very similar without and with gentamicin. Moreover, from triplicate experiments for each condition, the positive droplet population without and with gentamicin stayed approximately equal. 30000 droplets are interrogated in each experiment, and error bars correspond to triplicate measurements.
Exposing a multi-drug resistant strain of E. coli (ATCC BAA 2471) to gentamicin revealed negligible difference in positive droplet population when compared to the no-antibiotic control. Growth of the multi-drug resistant strain of E. coli in the presence of gentamicin was first confirmed by visual inspection of sample turbidity in benchtop broth dilution (Fig. S5). Next, as with the reference E. coli, the multi-drug resistant strain of E. coli was evaluated without and with gentamicin in the dropFAST platform. Thirty thousand droplets were analyzed for each case, and histograms of the normalized positive droplet fluorescence were plotted. Here, no apparent difference in frequency is noticed for the positive droplets in the presence of gentamicin (Fig. 3(B)). Indeed, the normalized positive droplet population for the no-antibiotic control case (1.00 ± 0.06) is not significantly different from the gentamicin-exposed positive droplet population (1.06 ± 0.07) (Fig. 3(B), inset). Thus, the platform accurately distinguishes the effect of gentamicin on two different strains of E. coli after only 1 h growth.
4. Conclusions
In this work, we demonstrate a novel droplet microfluidic platform for rapid bacterial growth detection and assessment of antibiotic susceptibility. By leveraging the drastically reduced background enabled by picoliter volume droplets, as well as the rapidity and high sensitivity of the resazurin based fluorescent growth assay, we accelerate the incubation time of bacterial growth for reliable detection to approximately 1 h, equivalent to only 2–3 replications of E. coli. Our results represent the fastest phenotype-based approach to assess drug resistant pathogens at a speed that is only confined by the fundamental limit of growth determination. Moreover, our dropFAST platform seamlessly integrates single bacteria isolation during picoliter droplet generation, bacterial incubation and growth in droplets, and droplet detection into a single-step workflow and standalone device. As a demonstration, we detected the antibacterial effect of gentamicin on E. coli at the single-cell resolution. Taken as a whole, dropFAST outpaces previous implementations of droplet microfluidic technology for the purpose of antimicrobial sensitivity assessment in speed, throughput, and integration.
While the dropFAST technology represents a substantial improvement upon the state of the art, several new considerations arise as a result of the enhanced ability to detect bacterial metabolism. For instance, for a small subpopulation of bacterial cells in the presence of gentamicin, metabolism prior to cell death may generate an apparently positive fluorescent signal that cannot be distinguished from drug-resistant cells under the current workflow. To address this challenge, we may first further optimize assay parameters (e.g., resazurin concentration and droplet volume) to suppress metabolic signal generation from dying bacteria, and in essence, threshold a subpopulation with low metabolic activity as negatives. Alternatively, placing additional detection zones throughout the length of the incubation channel may enable the detection of metabolic activity at various time points to help differentiate between resistant and slow-dying populations. The linear architecture of the dropFAST platform is readily amenable to such expansion. As dropFAST is designed to be a universal platform capable of testing any species of bacteria against any type of antibiotic, additional validations using different strains of bacteria against several different antibiotics are also warranted. Considering practical needs in clinical microbiology, several additions to the current dropFAST device may be necessary, including the ability to test multiple combinations of bacteria and antibiotics in the same device while maintaining droplet stability and uniformity (Kaminski et al., 2012), as well as the ability to automate a sample-to-answer workflow by integrating a sample loading system (T. D. Rane et al., 2012; Zec et al., 2012). With a potential to improve functionality and scale, we envision the dropFAST platform becoming a useful clinical tool for accelerating phenotypic assessment of antimicrobial susceptibility.
Supplementary Material
Highlights.
Confinement of single-bacteria in picoliter droplets accelerates growth detection
Integrated biosensing platform consists of monolithic device and single-step workflow
High-throughput platform can be operated in continuous flow
Platform used to determine E. coli susceptibility to gentamicin after 1 h. incubation
Acknowledgments
This work has been supported by National Institutes of Health (R01AI117032) and National Science Foundation (1159771 and 1033744).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Balaban NQ, Merrin J, Chait R, Kowalik L, Leibler S. Bacterial Persistence as a Phenotypic Switch. Science (80-) 2004;305:1622 L–P-1625. doi: 10.1126/science.1099390. [DOI] [PubMed] [Google Scholar]
- Boedicker JQ, Li L, Kline TR, Ismagilov RF. Detecting bacteria and determining their susceptibility to antibiotics by stochastic confinement in nanoliter droplets using plug-based microfluidics. Lab Chip. 2008;8:1265–1272. doi: 10.1039/b804911d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boucher HW, Talbot GH, Bradley JS, Edwards JE, Gilbert D, Rice LB, Scheld M, Spellberg B, Bartlett J. Bad bugs, no drugs: no ESKAPE! An update from the Infectious Diseases Society of America. Clin Infect Dis. 2009;48:1–12. doi: 10.1086/595011. [DOI] [PubMed] [Google Scholar]
- Bueno C, Villegas ML, Bertolotti SG, Previtali CM, Neumann MG, Encinas MV. The excited-state interaction of resazurin and resorufin with amines in aqueous solutions. Photophysics and photochemical reactions. Photochem Photobiol. 2002;76:385–390. doi: 10.1562/0031-8655(2002)076<0385:tesior>2.0.co;2. [DOI] [PubMed] [Google Scholar]
- Carlet J, Collignon P, Goldmann D, Goossens H, Gyssens IC, Harbarth S, Jarlier V, Levy SB, N’Doye B, Pittet D, Richtmann R, Seto WH, Van Der Meer JW, Voss A. Society’s failure to protect a precious resource: Antibiotics. Lancet. 2011;378:369–371. doi: 10.1016/S0140-6736(11)60401-7. [DOI] [PubMed] [Google Scholar]
- Chan C, Feng F, Ottinger J, Foster D, West M, Kepler TB. Statistical mixture modeling for cell subtype identification in flow cytometry. Cytometry A. 2008;73:693–701. doi: 10.1002/cyto.a.20583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen CH, Lu Y, Sin MLY, Mach KE, Zhang DD, Gau V, Liao JC, Wong PK. Antimicrobial susceptibility testing using high surface-to-volume ratio microchannels. Anal Chem. 2010;82:1012–1019. doi: 10.1021/ac9022764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi J, Jung YG, Kim J, Kim S, Jung Y, Na H, Kwon S. Rapid antibiotic susceptibility testing by tracking single cell growth in a microfluidic agarose channel system. Lab Chip. 2013;13:280–7. doi: 10.1039/c2lc41055a. [DOI] [PubMed] [Google Scholar]
- Collins DJ, Neild A, deMello A, Liu A-Q, Ai Y. The Poisson distribution and beyond: methods for microfluidic droplet production and single cell encapsulation. Lab Chip. 2015:3439–3459. doi: 10.1039/C5LC00614G. [DOI] [PubMed] [Google Scholar]
- Davenport M, Mach KE, Shortliffe LMD, Banaei N, Wang T-H, Liao JC. New and developing diagnostic technologies for urinary tract infections. Nat Rev Urol. 2017 doi: 10.1038/nrurol.2017.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delvigne F, Zune Q, Lara AR, Al-Soud W, Sørensen SJ. Metabolic variability in bioprocessing: Implications of microbial phenotypic heterogeneity. Trends Biotechnol. 2014 doi: 10.1016/j.tibtech.2014.10.002. [DOI] [PubMed] [Google Scholar]
- Eun YJ, Utada AS, Copeland MF, Takeuchi S, Weibel DB. Encapsulating bacteria in agarose microparticles using microfluidics for high-throughput cell analysis and isolation. ACS Chem Biol. 2011;6:260–266. doi: 10.1021/cb100336p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Exec. Order No. 13676, 3 C.F.R., 2014.
- Finley RL, Collignon P, Larsson DGJ, Mcewen SA, Li XZ, Gaze WH, Reid-Smith R, Timinouni M, Graham DW, Topp E. The scourge of antibiotic resistance: The important role of the environment. Clin Infect Dis. 2013 doi: 10.1093/cid/cit355. [DOI] [PubMed] [Google Scholar]
- Fossum S, Crooke E, Skarstad K. Organization of sister origins and replisomes during multifork DNA replication in Escherichia coli. EMBO J. 2007;26:4514–4522. doi: 10.1038/sj.emboj.7601871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan W, Chen L, Rane TD, Wang TH. Droplet Digital Enzyme-Linked Oligonucleotide Hybridization Assay for Absolute RNA Quantification. Sci Rep. 2015;5:13795. doi: 10.1038/srep13795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaminski TS, Jakiela S, Czekalska MA, Postek W, Garstecki P. Automated generation of libraries of nL droplets. Lab Chip. 2012;12:3995–4002. doi: 10.1039/c2lc40540g. [DOI] [PubMed] [Google Scholar]
- Kaminski TS, Scheler O, Garstecki P. Droplet microfluidics for microbiology: techniques, applications and challenges. Lab Chip. 2016;16:2168–2187. doi: 10.1039/C6LC00367B. [DOI] [PubMed] [Google Scholar]
- Köster S, Angilè FE, Duan H, Agresti JJ, Wintner A, Schmitz C, Rowat AC, Merten Ca, Pisignano D, Griffiths AD, Weitz Da. Drop-based microfluidic devices for encapsulation of single cells. Lab Chip. 2008;8:1110–1115. doi: 10.1039/b802941e. [DOI] [PubMed] [Google Scholar]
- Kunin CM, Tupasi T, Craig WA. A brief exposition of the problem and some tentative solutions. Ann Intern Med. 1973 doi: 10.7326/0003-4819-79-4-555. [DOI] [PubMed] [Google Scholar]
- Laxminarayan R, Duse A, Wattal C, Zaidi AKM, Wertheim HFL, Sumpradit N, Vlieghe E, Hara GL, Gould IM, Goossens H, Greko C, So AD, Bigdeli M, Tomson G, Woodhouse W, Ombaka E, Peralta AQ, Qamar FN, Mir F, Kariuki S, Bhutta ZA, Coates A, Bergstrom R, Wright GD, Brown ED, Cars O. Antibiotic resistance-the need for global solutions. Lancet Infect Dis. 2013 doi: 10.1016/S1473-3099(13)70318-9. [DOI] [PubMed] [Google Scholar]
- Li B, Qiu Y, Glidle A, McIlvenna D, Luo Q, Cooper J, Shi HC, Yin H. Gradient microfluidics enables rapid bacterial growth inhibition testing. Anal Chem. 2014;86:3131–3137. doi: 10.1021/ac5001306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin L, Chan C, West M. Discriminative variable subsets in Bayesian classification with mixture models, with application in flow cytometry studies. Biostatistics. 2016;17:40–53. doi: 10.1093/biostatistics/kxv021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu W, Kim HJ, Lucchetta EM, Du W, Ismagilov RF. Isolation, incubation, and parallel functional testing and identification by FISH of rare microbial single-copy cells from multi-species mixtures using the combination of chemistrode and stochastic confinement. Lab Chip. 2009;9:2153–62. doi: 10.1039/b904958d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Painter RE, Enesa K, Holmes D, Whyte G, Garlisi CG, Monsma FJ, Rehak M, Craig FF, Smith CA. High-throughput screening of antibiotic-resistant bacteria in picodroplets. Lab Chip. 2016:1636–1643. doi: 10.1039/C6LC00180G. [DOI] [PubMed] [Google Scholar]
- Lu Y, Gao J, Zhang DD, Gau V, Liao JC, Wong PK. Single cell antimicrobial susceptibility testing by confined microchannels and electrokinetic loading. Anal Chem. 2013;85:3971–3976. doi: 10.1021/ac4004248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mach KE, Wong PK, Liao JC. Biosensor diagnosis of urinary tract infections: A path to better treatment? Trends Pharmacol Sci. 2011 doi: 10.1016/j.tips.2011.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mai J, Abhyankar VV, Piccini ME, Olano JP, Willson R, Hatch AV. Rapid detection of trace bacteria in biofluids using porous monoliths in microchannels. Biosens Bioelectron. 2014;54:435–441. doi: 10.1016/j.bios.2013.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohan R, Mukherjee A, Sevgen SE, Sanpitakseree C, Lee J, Schroeder CM, Kenis PJA. A multiplexed microfluidic platform for rapid antibiotic susceptibility testing. Biosens Bioelectron. 2013;49:118–125. doi: 10.1016/j.bios.2013.04.046. [DOI] [PubMed] [Google Scholar]
- Ng EX, Miller MA, Jing T, Chen CH. Single cell multiplexed assay for proteolytic activity using droplet microfluidics. Biosens Bioelectron. 2016;81:408–414. doi: 10.1016/j.bios.2016.03.002. [DOI] [PubMed] [Google Scholar]
- Nichols RJ, Sen S, Choo YJ, Beltrao P, Zietek M, Chaba R, Lee S, Kazmierczak KM, Lee KJ, Wong A, Shales M, Lovett S, Winkler ME, Krogan NJ, Typas A, Gross CA. Phenotypic landscape of a bacterial cell. Cell. 2011;144:143–156. doi: 10.1016/j.cell.2010.11.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parija S. Textbook of Microbiology and Immunology. 3. Elseiver; 2016. [Google Scholar]
- Performance Standards for Antimicrobial Susceptibility Testing; Twenty-Fourth Informational Supplement. Clinical Laboratory Standards Institute. CLSI; Wayne, PA, USA: 2014. [Google Scholar]
- Powell EO. Growth rate and generation time of bacteria, with special reference to continuous culture. J Gen Microbiol. 1956;15:492–511. doi: 10.1099/00221287-15-3-492. [DOI] [PubMed] [Google Scholar]
- Rane TD, Zec HC, Puleo C, Lee AP, Wang TH. Droplet microfluidics for amplification-free genetic detection of single cells. Lab Chip. 2012;12:3341. doi: 10.1039/c2lc40537g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rane TD, Zec HC, Wang TH. A Serial Sample Loading System: Interfacing Multiwell Plates with Microfluidic Devices. J Lab Autom. 2012;17:370–377. doi: 10.1177/2211068212455169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roca I, Akova M, Baquero F, Carlet J, Cavaleri M, Coenen S, Cohen J, Findlay D, Gyssens I, Heure OE, Kahlmeter G, Kruse H, Laxminarayan R, Liébana E, López-Cerero L, MacGowan A, Martins M, Rodríguez-Baño J, Rolain JM, Segovia C, Sigauque B, Taconelli E, Wellington E, Vila J. The global threat of antimicrobial resistance: Science for intervention. New Microbes New Infect. 2015 doi: 10.1016/j.nmni.2015.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spellberg B, Blaser M, Guidos RJ, Boucher HW, Bradley JS, Eisenstein BI, Gerding D, Lynfield R, Reller LB, Rex J, Schwartz D, Septimus E, Tenover FC, Gilbert DN. Combating antimicrobial resistance: Policy recommendations to save lives. Clin Infect Dis. 2011:52. doi: 10.1093/cid/cir153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun P, Liu Y, Sha J, Zhang Z, Tu Q, Chen P, Wang J. High-throughput microfluidic system for long-term bacterial colony monitoring and antibiotic testing in zero-flow environments. Biosens Bioelectron. 2011;26:1993–1999. doi: 10.1016/j.bios.2010.08.062. [DOI] [PubMed] [Google Scholar]
- Tang S, Whitesides G. Basic microfluidic and soft lithographic techniques. Optofluidics Fundam Devices, Appl. 2010:7–32. [Google Scholar]
- Tenover FC. Potential impact of rapid diagnostic tests on improving antimicrobial use. Ann N Y Acad Sci. 2010;1213:70–80. doi: 10.1111/j.1749-6632.2010.05827.x. [DOI] [PubMed] [Google Scholar]
- Wakamoto Y, Dhar N, Chait R, Schneider K, Signorino-Gelo F, Leibler S, McKinney JD. Dynamic Persistence of Antibiotic-Stressed Mycobacteria. Science (80-) 2013;339:91–96. doi: 10.1126/science.1229858. [DOI] [PubMed] [Google Scholar]
- Xia Y, Whitesides GM. Soft Lithography. Annu Rev Mater Sci. 1998;28:153–184. doi: 10.1146/annurev.matsci.28.1.153. [DOI] [Google Scholar]
- Yan Y, Boey D, Ng LT, Gruber J, Bettiol A, Thakor NV, Chen CH. Continuous-flow C. elegans fluorescence expression analysis with real-time image processing through microfluidics. Biosens Bioelectron. 2016;77:428–434. doi: 10.1016/j.bios.2015.09.045. [DOI] [PubMed] [Google Scholar]
- Zec H, Rane TD, Wang TH. Microfluidic platform for on-demand generation of spatially indexed combinatorial droplets. Lab Chip. 2012;12:3055–3062. doi: 10.1039/c2lc40399d. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.