Abstract
Cu(0)-mediated polymerization was employed to synthesize a library of structurally varied cationic polymers and their application as antibacterial peptide mimics was assessed. Eight platform polymers were first synthesized with low degrees of polymerization (DP) using (2-Boc-amino)ethyl acrylate as the monomer and either ethyl α-bromoisobutyrate or dodecyl 2-bromoisobutyrate as the initiator (thus providing hydrocarbon chain termini of C2 or C12, respectively). A two-step modification strategy was then employed to generate the final sixteen-member polymer library. Specifically, an initial deprotection was employed to reveal the primary amine cationic polymers, followed by guanylation. The biocidal activity of these cationic polymers was assessed against various strains of Escherichia coli, Klebsiella pneumoniae, Acinetobacter baumannii, Pseudomonas aeruginosa, Staphylococcus aureus, and Streptococcus pneumoniae. Polymers having a short segment of guanidine units and a C12 hydrophobic terminus were shown to provide the broadest antimicrobial activity against the panel of isolates studied, with MIC values approaching those for Gram-positive targeting antibacterial peptides: daptomycin and vancomycin. The C12-terminated guanidine functional polymers were assayed against human red blood cells, and a concomitant increase in haemolysis was observed with decreasing DP. Cytotoxicity was tested against HEK293 and HepG2 cells, with the lowest DP C12-terminated polymer exhibiting minimal toxicity over the concentrations examined, except at the highest concentration. Membrane disruption was identified as the most probable mechanism of bacteria cell killing, as elucidated by membrane permeability testing against E. coli.
Graphical abstract
A new class of oligomeric cationic polymers with lipophilic tails were developed as antibacterial lipopeptide mimics, and revealed structurally dependent bacterial killing.
Introduction
The emergence of antibiotic resistant bacteria and a lack of new antibiotics in the drug discovery pipeline has led to an antibiotic crisis.1 Bacteria commonly gain resistance to antibiotics by either picking up genetic material from other bacteria or through mutations that impart new mechanisms of resistance; (e.g. changing vulnerable cellular targets to reduce their susceptibility to antibiotics, up-regulating enzymes which degrade the antibiotic or reducing the intracellular concentration to below toxic levels via efflux pumps).2,3 As a result of the decreasing efficacy of available antibiotics, there is a critical need for new effective agents and bacteria-resistant medical implant/device surfaces.4–6 Cationic antimicrobial peptides (AMPs) have gained research attention due to their broad spectrum of antibacterial activity, and lower susceptibility to resistance.7 Antibacterial peptides owe their activity to the combination of ionizable nitrogenous moieties, which allow for the binding to the outer surface of the bacteria, and the lipophilic segment, which is able to insert into the cell membrane causing cell death through osmotic swelling and lysis. However, such peptides come with significant downsides: they are costly to manufacture in large quantities and possess short half-lives due to their degradation by proteases in the body.8–10 To overcome these limitations a number of natural antimicrobial peptide mimics have been developed that have better or similar activities to AMPs, such as synthetic AMPs11, β-peptides12,13, peptoids14 and AApeptides15. However these mimics are still hampered by costly synthetic approaches, fast proteolytic degradation, low bioavailability and undesirable toxicity profiles. As such their clinical utility is limited.7,16 One appealing alternative is the use of wholly synthetic cationic polymers to mimic the structure and function of AMPs. In comparison to peptides, these materials can be produced cheaply and on a large scale, and can be easily modified. Moreover, recent work has demonstrated they can exhibit potent biological activity and stability compared with other analogs of AMPs.17–19
Within the literature, there is increasing focus on synthetic materials incorporating lysine and arginine mimicking groups, as these moieties are commonly found in AMPs. In particular, increased activity has been observed with lysine mimicking groups (i.e., primary amines) compared to tertiary amine containing polymers.20,21 Further, substitution of the lysine mimicking group with an arginine mimic (i.e., guanidine) has been shown to increase antimicrobial activity and selectivity by both Tew and colleagues22, and Locock et al.8 In contrast, Morgan and co-workers recently observed that polymers incorporating only amines or amines with small amounts of guanidine had greater antimicrobial activity and lower mammalian cell toxicity than polymers that contained a higher proportion of guanidine functionalized groups.23
Overall, guanidine functional polymers offer numerous benefits such as: high water solubility, a wide spectrum antimicrobial activity, excellent activity, and low toxicity.24–27 As such further exploration of guanidine functional materials is warranted.
Recently, we demonstrated the facile synthesis of a new class of antibacterial peptide mimics using Cu(0)-mediated polymerization. These materials comprised a short block of cationic monomer and a lipophilic tail of tunable chain length afforded by the initiator. Monomers with differing pKa were employed, and we identified that a short 2-(dimethylamino)ethyl acrylate (DMAEA) block and a C12 aliphatic tail yielded greater antibacterial activity than polymers with either a C2 tail and/or cationic units of lower pKa against Staphylococcus aureus, Klebsiella pneumoniae, Acinetobacter baumannii, and Pseudomonas aeruginosa. Importantly, these bacteria make up four of the six so-called “ESKAPE” pathogens that currently cause the majority of hospital-acquired multidrug-resistant infections.28,29
In this study, we report the synthesis of low degree of polymerization (DP) guanidine functional polymers with two different hydrophobic tail lengths (C2 or C12). We focused our attention on utilizing a C12 tail as work by Mowery et al. had previously shown that increasing or decreasing the length of the tail from 12 carbons negatively affected the antibacterial activity.30 Specifically herein we describe the synthesis of a series of 2-(Boc-amino)ethyl acrylate (2-BocAEA) polymers employing Cu(0)-mediated polymerization with low DP (DP < 27). These polymers were then deprotected to afford a primary amine, which was subsequently converted to a guanidine group. The minimum inhibitory concentration (MIC) for each polymer was determined against S. aureus, Escherichia coli, K. pneumoniae, A. baumannii, P. aeruginosa, and Streptococcus pneumoniae (details of bacteria used are shown in Tables SI 2 and SI 4), and the results compared with both the primary amine polymer analogues, and the best performing PDMAEA antibacterial peptide analogue we had previously identified.28 The lead compounds were assayed against human red blood cells, and Human Embryonic Kidney 293 (HEK293) cells and Human hepatocellular carcinoma (HepG2) cells, to elucidate levels of haemolysis and cytotoxicity, respectively. The mechanism of cell death was tested by the inner membrane depolarization assay and outer membrane permeabilization assay using E. coli.
Results and Discussion
Polymer Synthesis and Characterization
A library of low DP (low molecular weight) well-defined cationic polymers with hydrophobic tails of either C12 or C2 length was synthesized via Cu(0)-mediated polymerization followed by post-synthetic modification. Initially eight 2-BocAEA polymers with DPs ranging from 27 to 5 were synthesized as summarized in Table SI 1. Deprotection of the P(2-BocAEA) with an excess of trifluoroacetic acid (TFA), revealed the primary amine functional polymer (P(2-AEA)) (Scheme 1) in quantitative yield. Deprotection was confirmed by 1H NMR spectroscopy (see Figures SI 1 and SI 2), with successful deprotection revealed by the absence of the tert-butyl hydrogen peak at 1.45 ppm (‘h’ in Figure SI 1). Guanylation was performed by reacting the revealed primary amine groups with 1.5 equiv. of 1H-pyrazole-1-carboxamidine hydrochloride and 3 equiv. of N,N-diisopropylethylamine in anhydrous ethanol to yield guanidine groups (P(2-GEA)) (Scheme 1). 1H NMR spectroscopy confirmed successful guanylation via the downfield shift of peak ‘g’ from ~3.30 ppm to ~3.60 ppm (Figures SI 2 and SI 3). Further, ATR FT-IR analysis confirmed that the guanidine group had been successfully incorporated into the polymer by the appearance of a new peak characteristic of the C=N stretch (1624 cm−1)8 (as shown in Figure SI 4).
Scheme 1.
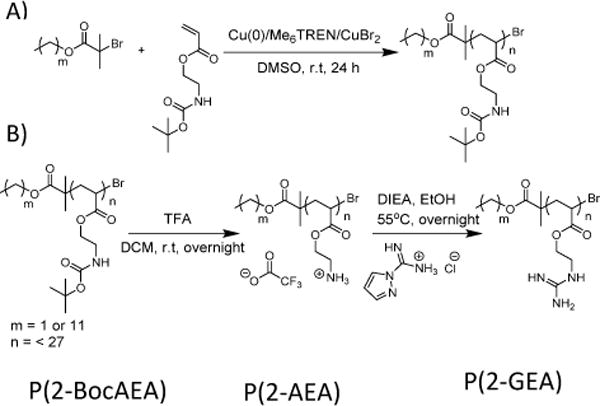
A) Synthesis of P(2-BocAEA) using Cu(0)-mediated polymerization; B)Deprotection of boc-groups of P(2-BocAEA) by TFA and subsequent guanylation of the deprotected polymer using 1H-pyrazole-1-carboxamidine hydrochloride.
Single point bacterial inhibition assay
To assess the antibacterial killing efficacy of the synthesized materials, a single point bacterial inhibition assay was conducted against E. coli, K. pneumoniae, A. baumannii, P. aeruginosa, and S. aureus with a standard concentration for the polymers of 32 μg/mL (see Table SI 4). The active compounds were classified as polymers that completely inhibited growth, while those identified as partially active allowed only partial growth. Compounds classified as “inactive” allowed full growth of the bacteria. From the data presented in Table SI 4, at 32 μg/mL the leading active compounds were the C12-terminated guanidine functional polymers, with the lowest DPs (DP 7 and 13) and C12-terminated polymers being active against every bacterial strain in the panel. The primary amine containing polymers exhibited minimal activity against the bacteria when compared to the guanidine containing polymers. Further, the C2-terminated guanidine functionalized polymers displayed no activity against any of the bacteria strains studied, indicating that a longer hydrophobic tail length was required in order to exert activity over the bacteria. When compared to the best-performing DMAEA polymer from our previous study, the C12-terminated guanidine functionalized polymers synthesized herein displayed activity against more of the bacteria tested, with the PDMAEA only found to be active against E. coli and A. baumannii.28 This is consistent with the results of our previous paper demonstrating that PDMAEA was more active against Gram-negative bacteria rather than Gram-positive bacteria. Of the 17 polymers that were tested, 7 polymers were identified to be either active or partially active. These were then used in an MIC assay against the standard panel of bacteria strains: E. coli, K. pneumoniae, A. baumannii, P. aeruginosa, and S. aureus.
MIC (Minimum Inhibitory Concentration) Assay
The MIC values were determined against the standard panel of bacteria (Table SI 5). The MIC results reveal that, in general, the polymers exhibited higher activity against the Gram-positive bacteria S. aureus, which is likely due to the lack of the lipopolysaccharide (LPS) layer found only in Gram-negative bacteria.31 Of the Gram-negative bacteria studied, the polymers exhibited greater activity against E. coli and A. baumannii, compared to K. pneumoniae and P. aeruginosa.
The increased effect of the guanidine functional polymer against the Gram-positive bacteria is potentially attributable to the guanidine group undergoing multidentate binding to the phosphate head groups (exposed teichoic acids) on the outside of the Gram-positive bacteria.8 However, for Gram-negative bacteria it is postulated that the guanidine forms multiple bonds with the LPS layer, but lacks the hydrophobic character necessary to enter the cell and induce destruction.21 An interesting alternative explanation or contribution to the Gram-positive bacteria selectivity seen has been proposed by Lienkamp et al., that the double membrane in the Gram-negative bacteria causes a polymer concentration gradient at this interface, preventing the polymer reaching the plasma membrane at a necessary concentration for killing.32
The literature generally reports that primary amine containing polymers exhibit greater activity than secondary or tertiary amines, although this effect does appear to be dependent on other monomers found in the polymer.20,33 Polymers (7G, 8G and 9G) exhibiting an MIC of ≤ 16 μg/mL when tested against the standard panel were then tested against an extended panel of drug-resistant bacteria, including P. aeruginosa (Polymyxin-resistant), K. pneumoniae (NDM-1 positive), S. aureus (GISA, MRSA, VRSA), and St. pneumoniae (MDR). The results from this MIC assay are shown in Table 1.
Table 1.
Antibacterial activity of cationic acrylate polymers against the extended panel of bacteria
| Polymer Nº | Compound | Initiator chain length | DP |
P. aeruginosa (Polymyxin resistant) |
K. pneumonia (NDM-1 positive) |
S. aureus (GISA) |
S. aureus (GISA, MRSA) |
S. aureus (VRSA) |
St. pneumoniae (MDR) |
|---|---|---|---|---|---|---|---|---|---|
| MIC [μg/mL] | |||||||||
| – | Colistin | – | – | >32 | 0.125–0.5 | – | – | – | – |
| – | Polymyxin B | – | – | 32 | 0.125–0.5 | – | – | – | – |
| – | Vancomycin | – | – | – | – | 4 | 4–8 | >32 | 1–2 |
| – | Daptomycin | – | – | – | – | 8 | 8–16 | 2 | 2 |
| 7G | P(2-GEA) | 12 | 18 | >32 | >32 | 16 | 16 | 16 | 16 |
| 8G | P(2-GEA) | 12 | 13 | >32 | >32 | 8 | 16 | 8 | 8–16 |
| 9G | P(2-GEA) | 12 | 7 | >32 | >32 | 4–8 | 8 | 4–8 | 8 |
GISA; Glycopeptide Intermediate (resistant) S. aureus, VRSA; Vancomycin resistant S. aureus, MRSA; Methicillin resistant S. aureus. Colistin and polymyxin B were tested against Gram-negative bacteria, and vancomycin and daptomycin were tested against Gram-positive bacteria due to their spectrum of activity/specificity.
The polymers were also compared to four clinically relevant antibacterial peptides. Colistin and polymyxin B are effective towards Gram-negative bacteria as they bind to the LPS and disrupt membrane integrity, but ineffective against Gram-positive bacteria.34 While daptomycin and vancomycin target Gram-positive bacteria, they do this through different mechanisms of action to each other.35,36 From Table 1 the polymers generally show greater activity towards Gram-positive bacteria. For the Gram-negative isolates, the MIC values were higher than the maximum concentration tested, indicating that the guanidine functionalized polymers are inactive against these Gram-negative isolates. These polymers though not being as effective against Gram-negative bacteria as the polymyxins, are approaching the MIC values of daptomycin and vancomycin against these drug-resistant Gram-positive bacteria.
Haemolytic Activity and Cytotoxicity
As the dodecyl-terminated guanidine functionalized polymers exhibited higher activity against the standard panel of bacteria compared to the other polymers, these were selected for analysis of haemolytic activity and mammalian cell toxicity to determine whether they possess specific activity for bacterial membranes or are generally toxic to cells. The haemolytic activity of the polymer was investigated against human red blood cells over a concentration range of 15 – 1500 μg/mL. The results of this assay (Figure SI 5) revealed that the materials tested caused only low levels of haemolysis over the MIC range. However, as the DP of the guanidine functionalized segment decreased, there was a concomitant increase in haemolysis induced at the higher concentration values tested. This suggests that the guanidine group lowers the haemolysis induced. Moreover, as shown in Figure SI 6, for an equivalent DP the haemolytic activity of the amine polymers is lower than for the guanidine functionalized polymers. Nevertheless, within the series of amine-functionalized polymers higher DP polymers exhibit lower haemolytic activity (as was observed for the guanidine functionalized materials). Importantly, the MICs observed for these polymers are either around this concentration of 15 μg/mL (or lower for S. aureus) indicating that these polymers will not induce substantial levels of haemolysis if administered at the MIC concentrations.
To further examine the mammalian cell toxicity, a cell viability study was conducted to examine HEK293 and HepG2 cells against the C12-terminated DP 7 guanidine functionalized polymer (Polymer 9G). The serum concentration in the media was kept as low as possible (1% w/v) to maximize free drug concentration. The cytotoxic concentration 50 (CC50) value for HepG2 cells was determined to be 56 μg/mL. For HEK293 cells there was no significant toxicity (< 80% growth) observed at the highest concentration tested (100 μg/mL), and as such the tested concentration range did not allow determination of a CC50 value (as shown in Figure SI 7A). The C12-terminated DP 7 amine- and guanidine-functionalized polymers were tested against HEK293 cells, with similar trends being observed as for the haemolysis assay. Specifically, the amine polymer showed lower toxicity than the guanidine functionalized polymer at an equivalent DP (as shown in Figure SI 7B). The CC50 value for Polymer 9G was determined to be 129 μg/mL, while Polymer 9D showed limited toxicity over the concentration range studied.
These results demonstrate the low mammalian cell toxicity of these materials over the MIC concentration range.
Selectivity: Antimicrobial Activity versus Haemolysis
Ideally, an antimicrobial compound that is intended for human medical applications should combine both high antimicrobial activity (low MIC) with low haemolytic activity. By comparing the haemolytic activity against the MIC for a given bacterium, we can gain an understanding of the selectivity of the lead compounds for the bacteria tested compared to human red blood cells. The selectivity index is used routinely to describe the selectivity of the compound towards a particular bacterium.37,38 This is obtained by dividing the haemolysis concentration 50 (HC50) value by the MIC value for a given bacterium: these values are shown in Table SI 7.
From the data presented in Table SI 7, it can be observed that Polymer 8G shows the greatest selectivity index (between 172 and 343) for methicillin-resistant S. aureus (MRSA).
Mechanism of action
To determine the polymers mechanism of action, a representative polymer was tested against E. coli. E. coli was used due to it being optimized and routinely used for 1-N-phenylnaphthylamine (NPN) and 3,3′-dipropylthiadicarbocyanine iodide (diSC3-5) assays, as well as the representative polymer (Polymer 9G) exhibiting high antibacterial activity against E. coli compared to other bacteria in the standard panel.
Inner membrane Depolarization Assay
In order to explore the mechanism of action of a representative member of the library synthesized (Polymer 9G), a cytoplasmic membrane depolarization assay was performed using diSC3-5. This assay is widely used to measure changes in bacterial cell walls as an indicator for cell wall disruption. In this assay diSC3-5 accumulates in cells on hyperpolarized membranes where it exhibits self-quenched fluorescence.39 Fluorescence is generated if the membrane loses its potential or integrity, with the dye being released from the cell. Membrane potential disruption was observed at around 1 μg/mL of polymer 9G, and increased dramatically as the concentration increased, reaching saturation at around 33 μg/mL (Figure 1). This indicates that the polymer does indeed interact with the bacterial cytoplasmic membrane, and causes inner membrane depolarization/disruption which likely contributes to bacterial killing.40,41
Figure 1.
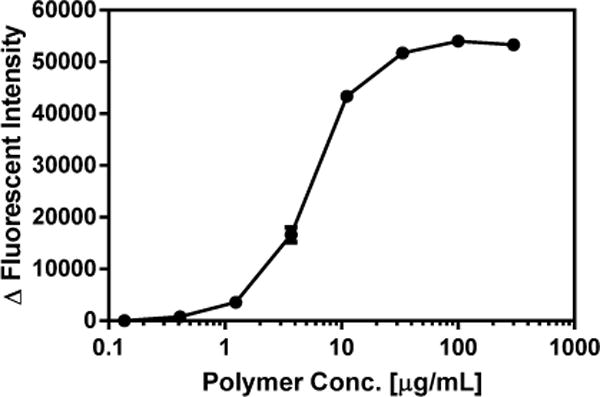
DiSC3-5 assay of Polymer 9G. Data are presented as Mean ± Standard Deviation
Outer membrane Permeabilization Assay
NPN was employed as a molecular probe to investigate the interaction of polymer 9G with the outer bacterial membrane of E. coli. NPN is a small hydrophobic molecule that fluoresces in the hydrophobic environment of lipid membranes, but only weakly fluoresces in aqueous environments. The observed increase in fluorescence on the addition of the polymer confirms that the polymer has permeabilized the outer membrane of the bacteria, as NPN is excluded from intact bacterial outer membranes.40,42,43 Similar to the membrane potential assay, outer membrane disruption was observed between 0.3 – 1 μg/mL of Polymer 9G, and further leakage was induced as the concentration increased, with saturation reached at approximately 100 μg/mL (Figure 2). These results indicate that the polymer does interact with the outer membrane of E. coli, and causes the membrane to become permeabilized. In combination with the diSC3-5 assay results, this reveals that the likely mechanism of killing involves the disruption of the bacterial cell wall.
Figure 2.
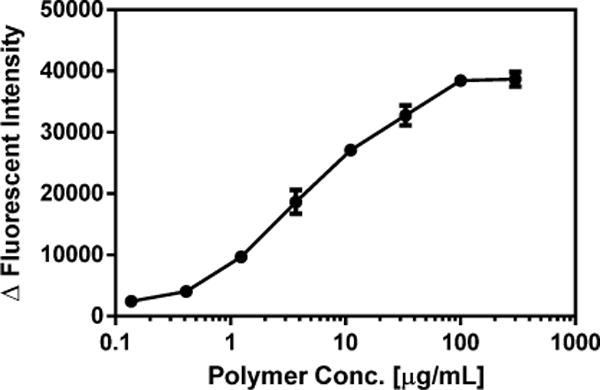
NPN uptake of Polymer 9G. Data are presented as Mean ± Standard Deviation
Conclusions
In summary, a library of guanidine and primary amine functionalized polymers was synthesized and their effectiveness against a series of bacteria examined. Polymers with a C12 terminus and short segment of guanidine pendants exhibited superior activity compared to polymers incorporating a C2 terminus and/or pendant primary amines. In particular, C12-terminated guanidine-functional polymers exhibited greater activity against Gram-positive bacteria, while the polymers tested exhibited high activity against the E. coli and A. baumannii, but were relatively inactive against the other Gram-negative bacteria. It was found that a low DP of guanidine caused higher amounts of haemolysis compared to polymers containing a higher DP of guanidine units. A representative polymer showed no significant toxicity towards HEK293 cells till the highest concentration of 100 μg/mL, and produced a CC50 value of 56 μg/mL against HepG2 cells. Membrane perturbation assays using polymer 9G suggested that these materials likely cause disruption of the bacterial cell wall. Furthermore, comparison with a tertiary amine-functionalized analogue (PDMAEA) demonstrated similar activity to the primary amine polymers, while the polymers incorporating guanidine pendants showed substantially better antibacterial activity.
Supplementary Material
Acknowledgments
This collaborative research was conducted by the Australian Research Council Centre of Excellence in Convergent Bio-Nano Science and Technology (project number CE140100036). JLG wishes to acknowledge receiving an APA scholarship. TPD wishes to acknowledge the award of an Australian Laureate Fellowship. Jian Li and Tony Velkov are supported by a research grant from the National Institute of Allergy and Infectious Diseases of the National Institutes of Health (RO1 AI111965). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institute of Allergy and Infectious Diseases or the National Institutes of Health. Jian Li is an Australian NHMRC Senior Research Fellow. Tony Velkov is an Australian NHMRC Industry Career Development Research Fellow. Matthew Cooper would like to acknowledge a grant from the NHMRC PRF APP 1059354 (2014–2018). JLG would like to acknowledge Dr. Jeroen Goos (ARC CoE CBNS, Monash University) for the kind gift of 2-(Boc-amino)ethyl acrylate.
Footnotes
Electronic Supplementary Information (ESI) available: Materials, experimental methods and other additional data. See DOI: 10.1039/x0xx00000x
Notes and references
- 1.CDC-Get Smart: Know When Antibiotics Work 2013. 2014 Apr 17; Available from: http://www.cdc.gov/getsmart/antibiotic-use/antibiotic-resistance-faqs.html#how-bacteria-resist.
- 2.Nolte O. Protein Pept Lett. 2014;21:330. doi: 10.2174/09298665113206660106. [DOI] [PubMed] [Google Scholar]
- 3.Rice LB. Clin Infect Dis. 2006;43:S100. doi: 10.1086/504487. [DOI] [PubMed] [Google Scholar]
- 4.Venkateswaran S, Wu M, Gwynne PJ, Hardman A, Lilienkampf A, Pernagallo S, et al. Journal of Materials Chemistry B. 2014;2:6723. doi: 10.1039/c4tb01129e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang BL, Jin TW, Han YM, Shen CH, Li Q, Lin QK, et al. Journal of Materials Chemistry B. 2015;3:5501. doi: 10.1039/c5tb00597c. [DOI] [PubMed] [Google Scholar]
- 6.Hadjesfandiari N, Yu K, Mei Y, Kizhakkedathu JN. Journal of Materials Chemistry B. 2014;2:4968. doi: 10.1039/c4tb00550c. [DOI] [PubMed] [Google Scholar]
- 7.Yeung ATY, Gellatly SL, Hancock REW. Cell Mol Life Sci. 2011;68:2161. doi: 10.1007/s00018-011-0710-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Locock KES, Michl TD, Valentin JDP, Vasilev K, Hayball JD, Qu Y, et al. Biomacromolecules. 2013;14:4021. doi: 10.1021/bm401128r. [DOI] [PubMed] [Google Scholar]
- 9.Nederberg F, Zhang Y, Tan JP, Xu K, Wang H, Yang C, et al. Nature chemistry. 2011;3:409. doi: 10.1038/nchem.1012. [DOI] [PubMed] [Google Scholar]
- 10.Giuliani A, Pirri G, Nicoletto SF. Central European Journal of Biology. 2007;2:1. [Google Scholar]
- 11.Fjell CD, Hiss JA, Hancock REW, Schneider G. Nat Rev Drug Discov. 2012;11:37. doi: 10.1038/nrd3591. [DOI] [PubMed] [Google Scholar]
- 12.Porter EA, Wang XF, Lee HS, Weisblum B, Gellman SH. Nature. 2000;404:565. doi: 10.1038/35007145. [DOI] [PubMed] [Google Scholar]
- 13.Godballe T, Nilsson LL, Petersen PD, Jenssen H. Chemical Biology & Drug Design. 2011;77:107. doi: 10.1111/j.1747-0285.2010.01067.x. [DOI] [PubMed] [Google Scholar]
- 14.Chongsiriwatana NP, Patch JA, Czyzewski AM, Dohm MT, Ivankin A, Gidalevitz D, et al. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:2794. doi: 10.1073/pnas.0708254105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Niu YH, Wu HF, Li YQ, Hu YG, Padhee S, Li Q, et al. Org Biomol Chem. 2013;11:4283. doi: 10.1039/c3ob40444g. [DOI] [PubMed] [Google Scholar]
- 16.Ong ZY, Wiradharma N, Yang YY. Adv Drug Deliv Rev. 2014;78:28. doi: 10.1016/j.addr.2014.10.013. [DOI] [PubMed] [Google Scholar]
- 17.Ganewatta MS, Tang CB. Polymer. 2015;63:A1. [Google Scholar]
- 18.Yang C, Krishnamurthy S, Liu J, Liu SQ, Lu XH, Coady DJ, et al. Advanced Healthcare Materials. 2016;5:1272. doi: 10.1002/adhm.201600070. [DOI] [PubMed] [Google Scholar]
- 19.Engler AC, Shukla A, Puranam S, Buss HG, Jreige N, Hammond PT. Biomacromolecules. 2011;12:1666. doi: 10.1021/bm2000583. [DOI] [PubMed] [Google Scholar]
- 20.Palermo EF, Kuroda K. Biomacromolecules. 2009;10:1416. doi: 10.1021/bm900044x. [DOI] [PubMed] [Google Scholar]
- 21.Palermo EF, Kuroda K. Applied Microbiology and Biotechnology. 2010;87:1605. doi: 10.1007/s00253-010-2687-z. [DOI] [PubMed] [Google Scholar]
- 22.Gabriel GJ, Madkour AE, Dabkowski JM, Nelson CF, Nusslein K, Tew GN. Biomacromolecules. 2008;9:2980. doi: 10.1021/bm800855t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Exley SE, Paslay LC, Sahukhal GS, Abel BA, Brown TD, McCormick CL, et al. Biomacromolecules. 2015;16:3845. doi: 10.1021/acs.biomac.5b01162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Qian LY, Guan Y, He BH, Xiao HN. Polymer. 2008;49:2471. [Google Scholar]
- 25.Zhang YM, Jiang JM, Chen YM. Polymer. 1999;40:6189. [Google Scholar]
- 26.Wei DF, Zhou RH, Zhang YW, Guan Y, Zheng AN. Journal of Applied Polymer Science. 2013;130:419. [Google Scholar]
- 27.Oule MK, Azinwi R, Bernier AM, Kablan T, Maupertuis AM, Mauler S, et al. Journal of Medical Microbiology. 2008;57:1523. doi: 10.1099/jmm.0.2008/003350-0. [DOI] [PubMed] [Google Scholar]
- 28.Grace JL, Huang JX, Cheah S, Truong NP, Cooper MC, Li J, et al. RSC Adv. 2016;6:15469. doi: 10.1039/C5RA24361K. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Boucher HW, Talbot GH, Bradley JS, Edwards JE, Gilbert D, Rice LB, et al. Clin Infect Dis. 2009;48:1. doi: 10.1086/595011. [DOI] [PubMed] [Google Scholar]
- 30.Mowery BP, Lindner AH, Weisblum B, Stahl SS, Gellman SH. Journal of the American Chemical Society. 2009;131:9735. doi: 10.1021/ja901613g. [DOI] [PubMed] [Google Scholar]
- 31.Engler AC, Wiradharma N, Ong ZY, Coady DJ, Hedrick JL, Yang YY. Nano Today. 2012;7:201. [Google Scholar]
- 32.Lienkamp K, Kumar KN, Som A, Nusslein K, Tew GN. Chemistry-a European Journal. 2009;15:11710. doi: 10.1002/chem.200802558. [DOI] [PubMed] [Google Scholar]
- 33.Rubén Tejero DL, López-Fabal Fátima, Gómez-Garcés José L, Fernández-García Marta. Polymer Chemistry. 2015 [Google Scholar]
- 34.Landman D, Georgescu C, Martin DA, Quale J. Clin Microbiol Rev. 2008;21:449. doi: 10.1128/CMR.00006-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Silverman JA, Perlmutter NG, Shapiro HM. Antimicrobial Agents and Chemotherapy. 2003;47:2538. doi: 10.1128/AAC.47.8.2538-2544.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Watanakunakorn C. Reviews of Infectious Diseases. 1981;3:S210. [PubMed] [Google Scholar]
- 37.Ilic N, Novkovic M, Guida F, Xhindoli D, Benincasa M, Tossi A, et al. Biochimica Et Biophysica Acta-Biomembranes. 2013;1828:1004. doi: 10.1016/j.bbamem.2012.11.017. [DOI] [PubMed] [Google Scholar]
- 38.Li YM, Bionda N, Yongye A, Geer P, Stawikowski M, Cudic P, et al. ChemMedChem. 2013;8:1865. doi: 10.1002/cmdc.201300232. [DOI] [PubMed] [Google Scholar]
- 39.Anderson RC, Hancock REW, Yu PL. Antimicrobial Agents and Chemotherapy. 2004;48:673. doi: 10.1128/AAC.48.2.673-676.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang LJ, Dhillon P, Yan H, Farmer S, Hancock REW. Antimicrobial Agents and Chemotherapy. 2000;44:3317. doi: 10.1128/aac.44.12.3317-3321.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang Y, Zhou Z, Zhu J, Tang Y, Canady TD, Chi EY, et al. Polymers. 2011;3:1199. [Google Scholar]
- 42.Johnson L, Mulcahy H, Kanevets U, Shi Y, Lewenza S. Journal of Bacteriology. 2012;194:813. doi: 10.1128/JB.05230-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Helander IM, Nurmiaho-Lassila EL, Ahvenainen R, Rhoades J, Roller S. International Journal of Food Microbiology. 2001;71:235. doi: 10.1016/s0168-1605(01)00609-2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


